Abstract
The TGFβ signaling pathway is a crucial regulator of developmental processes and disease. The activity of TGFβ ligands is modulated by various families of soluble inhibitors that interfere with the interactions between ligands and receptors. In an unbiased, genome-wide RNAi screen to identify genes involved in ligand-dependent signaling, we unexpectedly identified the BMP/Activin/Nodal inhibitor Coco as an enhancer of TGFβ1 signaling. Coco synergizes with TGFβ1 in both cell culture and Xenopus explants. Molecularly, Coco binds to TGFβ1 and enhances TGFβ1 binding to its receptor Alk5. Thus, Coco acts as both an inhibitor and an enhancer of signaling depending on the ligand it binds. This finding raises the need for a global reconsideration of the molecular mechanisms regulating TGFβ signaling.
KEY WORDS: Coco, TGFβ signaling modulators, Dual-activity pathway modulators
Summary: A genome-wide RNAi screen identifies the BMP/Activin/Nodal inhibitor Coco as an enhancer of the binding of TGFβ1 to its receptor Alk5, thus potentiating TGFβ1 signaling in murine cells and Xenopus explants.
INTRODUCTION
The TGFβ pathway plays important roles in development and disease. Ligand binding by TGFβ superfamily members to type II receptors leads to recruitment and activation of a type I receptor, which in turn phosphorylates and activates receptor-regulated Smads (R-Smads). The activated R-Smads form complexes with Smad4, translocate to the nucleus and directly regulate the transcription of target genes (Schmierer and Hill, 2007; Massagué, 2012). The canonical TGFβ pathway consists of two branches. BMP/GDF ligands signal through the type I receptors Alk1/2/3/6 to activate R-Smads1/5/8, whereas TGFβ/Activin/Nodal ligands signal through the type I receptors Alk4/5/7 to activate R-Smad2/3. Notably, TGFβ1, TGFβ2 and TGFβ3 ligands signal through Alk5, whereas Nodal and Activin signal through Alk4 and Alk7, thus raising the possibility of differential regulation of these signals.
A large number of secreted inhibitors of the TGFβ signaling pathway have been identified. The correct timing and spatial localization of these inhibitors is crucial for proper embryonic development (Wu and Hill, 2009). Coco (also known as Dante, Cer2, Dand5, Cerl2, Grem3), which belongs to the DAN family of secreted proteins (Hsu et al., 1998; Pearce et al., 1999), is expressed maternally in the animal pole of the oocyte and fertilized egg, and its expression declines by mid-gastrulation (Bell et al., 2003). By contrast, other inhibitors, such as Noggin, Chordin and Follistatin, are expressed zygotically, in the embryonic organizer (node in mammals) and delineate the future neural territory (De Robertis and Kuroda, 2004). We originally discovered Coco in a gain-of-function screen for injected mRNAs with the ability to induce secondary head structures in Xenopus (Bell et al., 2003). Maternal Coco controls germ layer specification by inhibition of Activin, Nodal, and BMP signaling (Bates et al., 2013). Zygotic Coco expression, by contrast, is involved in proper specification of the right-left axis by inhibiting Nodal and Derriere (Hashimoto et al., 2004; Marques et al., 2004; Vonica and Brivanlou, 2007). Inhibition of BMP signaling by Coco also plays an important role in metastatic breast cancer (Gao et al., 2012). Thus, in all contexts examined to date, Coco acts as an inhibitor of TGFβ ligands.
We recently developed a system to measure TGFβ signaling dynamics by monitoring the localization of Smad4 (Warmflash et al., 2012; Sorre et al., 2014). We showed that Smad4 is a faithful reporter for TGFβ1 signaling dynamics and that TGFβ1 signaling responds to changes in ligand concentration before returning to baseline levels within 4 h (Warmflash et al., 2012). We took advantage of this system to investigate the molecular mechanism involved in the regulation of TGFβ1 signaling dynamics in a genome-wide RNAi screen. Among the factors that affect nuclear residency of Smad4, we rediscovered Coco. Unexpectedly, the results from this screen suggested that Coco is an enhancer of signaling through the TGFβ1 ligand. Here, we demonstrate that Coco is a dual-activity modulator of TGFβ signaling, enhancing activity through TGFβ1 while inhibiting Nodal and BMP ligands.
RESULTS
An siRNA screen identifies Coco as an enhancer of TGFβ1 signaling
To identify genes involved in ligand-dependent Smad4 nuclear translocation, we performed an unbiased, genome-wide RNAi screen using the C2C12 GFP-Smad4 reporter cell line (Warmflash et al., 2012) as a platform to monitor TGFβ1-induced changes in Smad4 localization (Fig. 1A). Application of 2 ng/ml TGFβ1 resulted in the rapid nuclear translocation of GFP-Smad4 that peaked within 1 h and its cytoplasmic relocalization within 6 h (Warmflash et al., 2012). Thus, we searched for genes that could either prevent GFP-Smad4 nuclear localization at 1 h or prevent its return to the cytoplasm by 6 h. To perform the screen, C2C12 GFP-Smad4 reporter cells were transfected with pools of three small interfering RNAs (siRNAs) targeting individual genes. Cells were stimulated with 2 ng/ml TGFβ1 48 h after transfection and fixed and analyzed for GFP-Smad4 localization at 1 and 6 h after TGFβ1 application. As controls for impairing and for enhancing GFP-Smad4 localization, we used the TGFβ1 type I receptor TGFβR1 (Alk5) and Exportin1 (Xpo1), a protein essential for nuclear export of Smad4 (Schmierer and Hill, 2007), respectively. Knockdown of TGFβR1 blocked translocation of GFP-Smad4 in the nucleus (Fig. 1B). Conversely, knockdown of Xpo1 led to the maintenance of GFP-Smad4 in the nucleus (Fig. 1B).
Fig. 1.
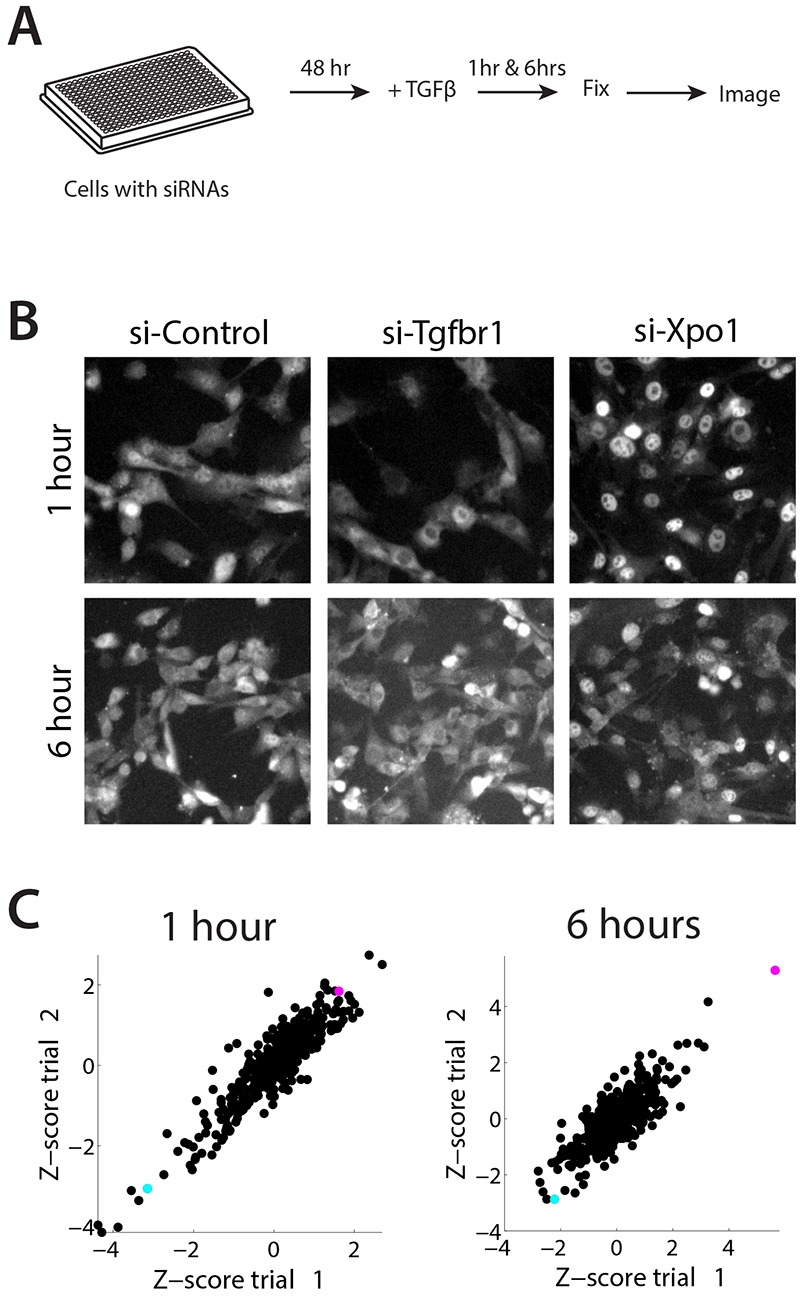
A genome-wide siRNA screen identifies modulators of TGFβ1 signaling. See also supplementary material Fig. S1 and Table S1. (A) Schematic of the siRNA screen. C2C12 cells expressing GFP-Smad4 were incubated with pools of three siRNAs for each gene for 48 h, at which point cells were treated with 2 ng/ml TGFβ1 for 1 or 6 h. Cells were then fixed and imaged. (B) Validation of the siRNA screen. Images show the localization of GFP-Smad4 in C2C12 cells after 1 or 6 h of treatment with 2 ng/ml TGFβ1 incubated for 2 days with a non-targeting siRNA (si-Control), or siRNAs targeting the TGFβ receptor 1 (si-Tgfbr1) or Exportin1 (si-Xpo1). Knockdown of TGFβR1 leads to complete inhibition of GFP-Smad4 nuclear translocation at 1 h, while knockdown of Xpo1 prevents the cytoplasmic relocalization of GFP-Smad4 at 6 h. (C) Scatter plots showing the reproducibility of the screen results between the two replicates at 1 h and 6 h. The Z-score indicates a normalized measure of the nuclear/cytoplasmic ratio of GFP-Smad4 fluorescence. Data for Xpo1 is indicated in blue, data for TGFβR1 is indicated in purple.
Screening of 17,582 genes, in two independent experiments, identified 321 genes (1.8%) that significantly affected the translocation of GFP-Smad4 when knocked down (supplementary material Fig. S1A and Table S1). Statistical analysis demonstrated a robust reproducibility between trials (r=0.93) (Fig. 1C). Of the genes impacting signaling, 166 (51.7%) blocked GFP-Smad4 translocation, and 155 (48.3%) enhanced the accumulation of GFP-Smad4 in the nucleus. Gene Ontology analysis and categorization of the candidates demonstrated that the genes identified in the screen belonged to a variety of classes (supplementary material Fig. S1B). These included transcriptional and translational regulators; channels, receptors, and other membrane proteins; secreted proteins; kinases and signaling proteins; and several proteins with unknown functions (supplementary material Table S1).
Modulation of Smad4 translocation can be due to an upstream effect on the pathway, for example modulation of R-Smads, or it can be due to an effect on Smad4 itself. To distinguish between these two possibilities, we examined the ability of R-Smad2 to translocate in response to TGFβ1 ligand in RFP-Smad2 reporter cells with knockdown of each of the 321 candidates. Nearly all genes for which knockdown prevented Smad4 translocation also prevented Smad2 translocation, whereas those enhancing Smad4 translocation were varied in their effect on Smad2 (supplementary material Fig. S1C and Table S1).
Among the genes that we discovered as modifiers of TGFβ1 signaling was a gene known as Coco (Dand5/Dte/Dante), which we originally identified as a BMP4 and Nodal inhibitor (Bell et al., 2003). Therefore, we expected Coco to behave in a similar manner in this context and also inhibit TGFβ1 signaling. However, knockdown of Coco strongly impaired, rather than enhanced, GFP-Smad4 translocation and thus TGFβ1 signaling. This finding led us to investigate Coco's mechanism of action further.
Coco knockdown impairs TGFβ1 signaling
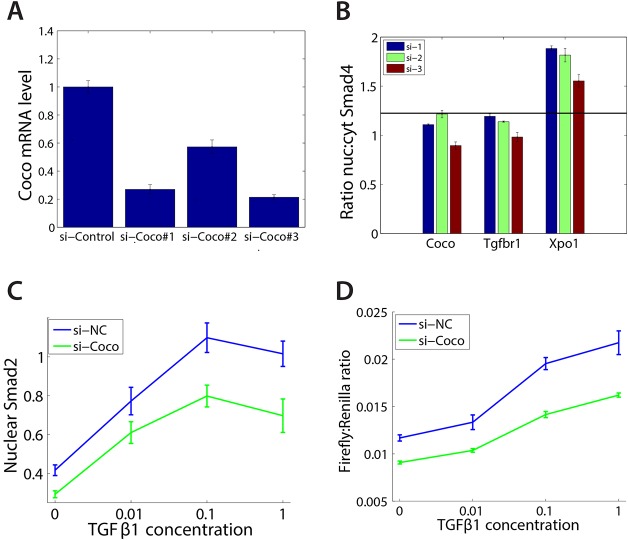
To exclude nonspecific effects from the siRNAs, we first tested each individual siRNA from the pool of three in C2C12 cells. Transfection of two independent Coco siRNAs led to a significant reduction in Coco expression levels and also caused a reduction in Smad4 nuclear translocation (21-24% of the original expression level and 20-40% of the original Smad4 translocation; Fig. 2A,B). By contrast, the third siRNA as well as a non-targeting control had no significant effect on either Coco expression or Smad4 localization. siRNA-mediated Coco knockdown also significantly reduced the level of nuclear Smad2 as measured by immunofluorescence (Fig. 2C), further supporting the idea that Coco is required for TGFβ1 signaling and demonstrating that Coco exerts its function upstream of Smad4. Knockdown of Coco also impaired TGFβ1-mediated transcription, as measured by the CAGA-luciferase reporter (Fig. 2D). Taken together, our data suggest that Coco does not inhibit, but rather is necessary for, signaling by the TGFβ1 ligand. This is in contrast to the inhibitory activity of Coco on Nodal and BMP4 (Bell et al., 2003).
Fig. 2.
Coco is required for TGFβ1 signaling. (A) Validation of the Coco siRNAs used in the screen. Transfection of C2C12 cells with two of the three siRNAs from the screen significantly reduces the levels of Coco mRNA by >75%. (B) Knockdown of Coco impairs Smad4 nuclear translocation. Transfection of C2C12 cells with the two siRNAs that reduce mRNA levels by >75% leads to a significant reduction in the Smad4 translocation in response to TGFβ1, similar to the effect caused by siRNAs targeting TGFβR1. Conversely, siRNAs targeting Xpo1 cause an accumulation of GFP-Smad4 in the nucleus. (C) Knockdown of Coco impairs Smad2 nuclear translocation, as measured by immunofluorescence. siRNA-mediated knockdown of Coco significantly decreases the nuclear accumulation of Smad2 after 1 h of treatment with various doses of TGFβ1. (D) Knockdown of Coco impairs TGFβ1-induced transcription. C2C12 cells expressing the CAGA12-firefly luciferase reporter were exposed to increasing doses of TGFβ1. siRNA-mediated knockdown of Coco decreases the transcriptional induction of the TGFβ1 reporter. Firefly luciferase signal was normalized to the co-expressed Renilla luciferase. Error bars represent s.d. from at least five images per condition or three independent replicates.
Coco overexpression enhances TGFβ1 signaling
We next examined whether overexpression of Coco would enhance TGFβ1 signaling. To address this, we used the ePiggyBac transposable element system (Lacoste et al., 2009) to generate a C2C12 cell line that expresses Coco under the control of the Tet-responsive-element (TRE) (Fig. 3A). This allows inducible Coco expression by addition of doxycycline (Dox). We then monitored the effects of Coco overexpression on GFP-Smad4 nuclear translocation in cells, in the presence or absence of TGFβ1 ligand. We found that Coco overexpression led to a significant increase in Smad4 nuclear localization after TGFβ1 treatment (Fig. 3B). These effects were dependent on the Dox dose, and thus dependent on the amount of Coco induction (Fig. 3B). Notably, Coco addition did not prolong Smad4 nuclear localization, but rather enhanced the magnitude of TGFβ1 signaling (Fig. 3B). Coco overexpression also enhanced activation of both the CAGA12-luciferase reporter construct as well as the direct transcriptional targets Ctgf and Pai1 (also known as Serpine1) (Fig. 3C,D). Consistent with Coco exerting its effects extracellularly, in a non-cell-autonomous manner, conditioned media from Coco-expressing cells increased TGFβ1-induced reporter activity (Fig. 3E). Together, these results suggest that Coco overexpression enhances TGFβ1 signaling in C2C12 cells.
Fig. 3.
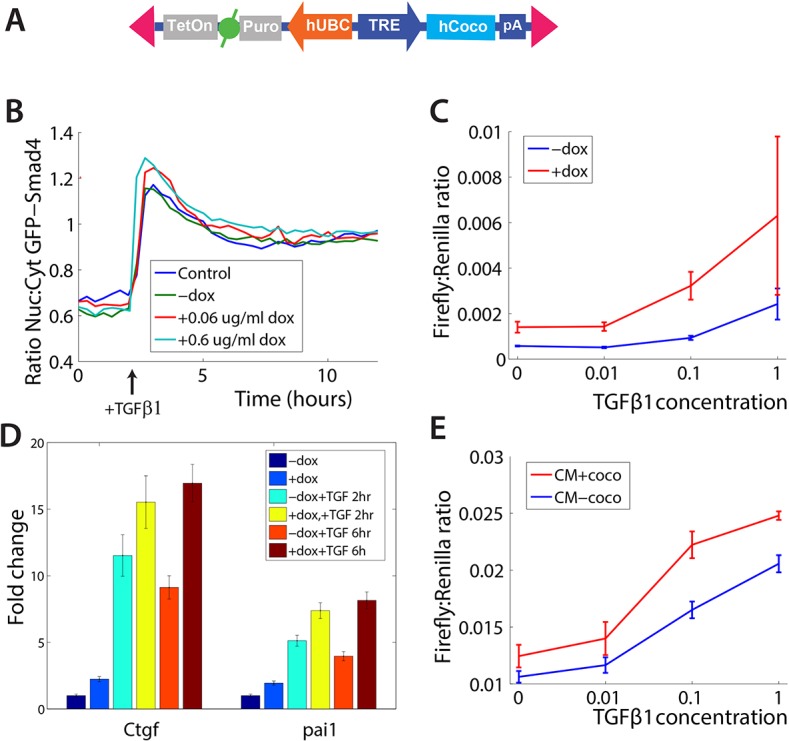
Coco enhances TGFβ1 signaling. (A) Schematic of the cassette for the inducible expression of human COCO in ePiggyBac. (B) Coco enhances nuclear translocation of Smad4 in a dose-dependent manner. Live-cell imaging of C2C12 expressing GFP-Smad4 shows that expression of COCO increases nuclear accumulation of Smad4 in a dose-dependent manner. Cells were stimulated with TGFβ1 at the time indicated by the arrow. (C) COCO enhances TGFβ1-induced transcription. Expression of Coco leads to increased transcriptional output from the CAGA12-firefly luciferase reporter in response to TGFβ1 treatment. (D) Coco enhances TGFβ1 activity. COCO expression leads to higher levels of the endogenous TGFβ1 targets Ctgf and Pai1. (E) Coco functions extracellularly. Conditioned medium from COCO-expressing cells (CM+coco) leads to an increase in the transcription of the CAGA12-luciferase reporter compared with conditioned medium from cells not expressing Coco (CM–coco). Error bars represent s.d. from three independent replicates.
Coco inhibits BMP signaling in C2C12
Coco was originally classified as an inhibitor of TGFβ signaling based on its function blocking Nodal and BMP signaling in Xenopus embryos (Bell et al., 2003). However, this is in contrast with the effect of Coco on the TGFβ1 ligand in mammalian cells. To rule out the possibility that the differential activity is due to species-specific differences between the amphibian and the mammalian system, we assessed the effects of mammalian Coco on BMP4 signaling in C2C12 cells. We monitored activation of the BMP4 signaling cascade by measuring the levels of BMP4-induced phosphorylated Smad1 (P-Smad1). Treatment of C2C12 cells with increasing doses of BMP4 led to the accumulation of P-Smad1 as determined by immunostaining (Fig. 4A). However, expression of Coco dramatically reduced P-Smad1 levels, confirming the Xenopus finding that Coco is indeed an inhibitor of BMP4 signaling (Bell et al., 2003). Additionally, microinjection of synthetic mRNA encoding human COCO into early Xenopus embryos led to the emergence of two-headed tadpoles, similar to the phenotype observed after injection of the Xenopus Coco (Bell et al., 2003) (Fig. 4B). This excludes differences in Coco activity between the two species. Therefore, our data indicate that Coco activity is ligand specific, with Coco inhibiting some TGFβ ligands, such as Nodal and BMP4, while enhancing others, such as TGFβ1.
Fig. 4.

Coco inhibits BMP signaling. (A) Coco impairs BMP4 signaling. Expression of Coco significantly reduces the levels of phospho-Smad1 (P-Smad1) in response to BMP4 in C2C12 cells. (B) Human COCO induces the formation of a secondary head in Xenopus. Injection of 2-cell-stage Xenopus embryos with mRNA encoding human or Xenopus Coco leads to the same phenotype of induction of a secondary head, consistent with Coco being an inhibitor of BMP signaling in vivo. Error bars represent s.d. from at least five images per condition.
Interaction between Coco and TGFβ1 modulates cell fate decisions
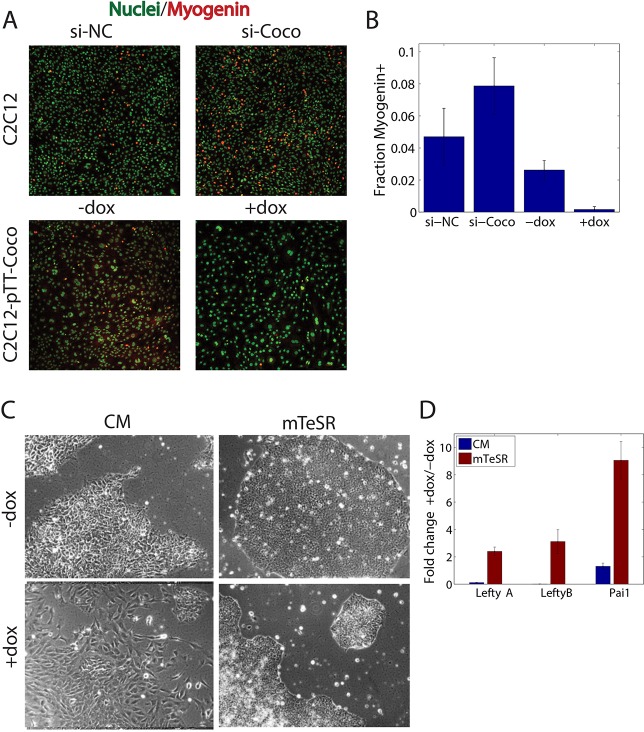
To understand whether Coco enhancement of TGFβ1 signaling has biological relevance, we investigated how Coco levels affect TGFβ1 activity in two different contexts. We first determined the effects of Coco on the differentiation of C2C12 cells to myoblasts, a process that is inhibited by TGFβ1 (Kollias et al., 2006). We found that C2C12 cells transfected with a Coco siRNA have a twofold higher rate of myoblast differentiation, as determined by myogenin immunostaining, compared with cells transfected with a control siRNA (Fig. 5A,B). Consistently, overexpression of Coco completely inhibited myoblast differentiation (Fig. 5A,B). This further supports the idea that Coco enhancement of TGFβ1 signaling has biological relevance.
Fig. 5.
Coco modulates TGFβ1-dependent cell fate decisions. See also supplementary material Fig. S2. (A) Coco levels affect myoblast differentiation. C2C12 transfected with an siRNA against Coco (si-Coco) differentiate more readily into myotube precursors compared with cells treated with a non-targeting siRNA (si-NC) as determined by myogenin immunostaining. Overexpression of Coco (+dox) completely abolishes myoblast differentiation. These results support a role for Coco as an enhancer of TGFβ1. Green, nuclei; red, myogenin. (B) Quantification of results shown in A. (C) Coco expression affects maintenance of pluripotency in hESCs. Expression of Coco (+dox) leads to the rapid differentiation of stem cells grown in conditioned media (CM) but not in mTeSR, further suggesting that Coco inhibits Activin/Nodal while enhancing TGFβ1 activity. (D) Coco induces the expression of TGFβ1-responsive genes in hESCs. Expression of the TGFβ1-responsive genes LeftyA, LeftyB and Pai1 is increased in Coco-expressing hESCs grown in mTeSR. Data are shown as a ratio of the mRNA levels in Coco-expressing (+dox) versus not expressing (−dox) cells. Error bars represent s.d. from at least five images per condition or three independent replicates.
Next, we investigated the effects of Coco overexpression in the context of maintenance of pluripotency in human embryonic stem cells (hESCs) using the RUES2 cell line (James et al., 2006). Maintenance of pluripotency requires signaling through Smad2/3, which can either be provided by Activin/Nodal ligands for hESCs cultured in conditioned medium (CM+FGF2) or TGFβ1 for hESCs cultured in the defined medium mTeSR1. Consistent with Coco inhibiting Activin/Nodal signaling, COCO overexpression in cells cultured in CM+FGF2 led to rapid loss of pluripotent morphology (Fig. 5C). Conversely, cells cultured in mTeSR1 did not change morphology, consistent with the idea that Coco does not inhibit TGFβ1 signaling (Fig. 5C). Furthermore, COCO expression in RUES2 cells grown in mTeSR strongly upregulated the TGFβ1-responsive genes LEFTYA, LEFTYB and PAI1 (LEFTY 2, LEFTY 1 and SERPINE1, respectively – Human Gene Nomenclature Database) (Fig. 5D), while downregulating the BMP4-responsive genes ID1, ID2 and ID3 (supplementary material Fig. S2A). Taken together, our data demonstrate that selective activity of Coco on different TGFβ ligands has a significant consequence on cell fate determination.
Using a variety of assays, we previously demonstrated that Coco is a potent inhibitor of the BMP and Smad1 branch of signaling of the TGFβ pathway in Xenopus (Bell et al., 2003). To address the role of Coco on TGFβ1 in the same system, we investigated whether Coco expression is able to synergize with TGFβ1 in inducing the dorsal-specific gene Goosecoid (gsc). Increasing concentrations of synthetic coco mRNA were injected together with RNA encoding TGFβ1 and TGFβRII, which are necessary for TGFβ1 signaling at this stage, and gsc expression was measured by qRT-PCR. We found that gsc expression is enhanced by Coco in a dose-dependent manner (supplementary material Fig. S2B). Consistent with its inhibitory role in BMP signaling, Coco expression significantly decreased the levels of the BMP-responsive gene vent2.2 (also known as ventx2.2) (supplementary material Fig. S2C). In addition, we determined the effects of Coco on cement gland formation. The cement gland is an organ located at the extreme anterior of the frog embryo that can be induced by both inhibitory and stimulatory signals (Bradley et al., 1996). We found that Coco induces expression of the cement gland marker Xag (also known as ag1; supplementary material Fig. S2D). This induction is increased by co-expression of TGFβ1, suggesting that Coco and TGFβ1 have a synergistic role in Xag induction. Similarly, Coco and TGFβ1 synergize in inducing ectopic cement glands in the developing embryo (supplementary material Fig. S2E,F). These results confirm that Coco functions as a dual activity modulator of TGFβ ligands in Xenopus embryonic explants.
Coco binds to TGFβ1 and enhances TGFβ1-ALK5 receptor interactions
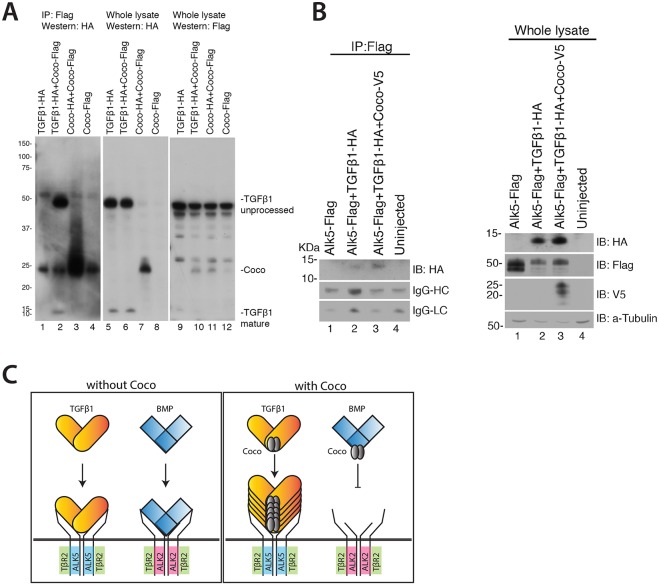
The synergistic effect of Coco on TGFβ1 prompted us to investigate the mechanism by which Coco functions. We had previously demonstrated that Coco is able to bind BMP4 and Nodal (Bell et al., 2003; Vonica and Brivanlou, 2007), and therefore asked first if Coco can also bind TGFβ1. Extracts of Xenopus embryos injected with HA-tagged TGFβ1 and Flag-tagged coco mRNA were immunoprecipitated. Precipitation with anti-Flag antibody resulted in co-precipitation of HA-tagged TGFβ1, indicating that Coco does indeed bind to TGFβ1 (Fig. 6A). This finding made us wonder whether Coco enhances TGFβ1 signaling by affecting ligand-receptor binding. To test this possibility, we microinjected Xenopus embryos with mRNAs encoding TGFβ1, Coco and ALK5 and performed immunoprecipitation experiments. We found that Coco enhances the binding of TGFβ1 to ALK5 (Fig. 6B). Taken together, the mechanistic evidence presented above strongly suggests that Coco-mediated enhancement of TGFβ1 signaling is due to an increase in the level of Smad2 phosphorylation and nuclear translocation of Smad2-Smad4 complexes (Fig. 6C).
Fig. 6.
Coco enhances the interactions of TGFβ1 with its receptor Alk5. See also supplementary material Fig. S3. (A) Coco binds to TGFβ1. Immunoprecipitation of proteins extracted from animal caps injected with HA-tagged TGFβ1 and Flag-tagged coco mRNA shows that Coco forms complexes with TGFβ1. (B) Coco expression leads to enhanced binding of TGFβ1 to Alk5. Flag-tagged Alk5, HA-tagged TGFβ1 and V5-tagged Coco were co-expressed in Xenopus embryos. Alk5 immunoprecipitation leads to pull-down of TGFβ1, with more TGFβ1 being pulled down in the presence of Coco. (C) Model for the mechanism of action of Coco as an enhancer of TGFβ1 signaling and inhibition of BMP signaling.
DISCUSSION
We took advantage of our previously described fluorescent assay for TGFβ signaling dynamics to perform an RNAi-based screen for factors that modulate Smad4 nuclear translocation in response to the TGFβ1 ligand in C2C12 cells. Among the 321 genes that we identified, we discovered Coco as an enhancer of TGFβ1 signaling that increases the magnitude of Smad4 translocation into the nucleus. Coco is required for signaling by the TGFβ1 ligand and synergizes with TGFβ1 in modulating cell-fate decisions. At the molecular level, Coco binds to TGFβ1 ligand and enhances the interaction of TGFβ1 with its receptor Alk5, thus leading to increased signaling.
We had originally discovered Coco based on a gain-of-function screen for factors that induce secondary dorsal and head structures in Xenopus embryos (Bell et al., 2003). Coco was shown to be an inhibitor of Activin, Nodal and BMP4, providing a molecular explanation for the embryonic phenotype. However, here we demonstrate that Coco has the opposite activity with regard to the TGFβ1 ligand. We therefore propose that Coco functions as both an inhibitor and an enhancer, depending on the TGFβ ligand it is exposed to. To our knowledge, Coco is the first secreted protein that has been shown to exert opposing activities on signaling by different TGFβ ligands.
We had previously proposed two embryonic roles for Coco. Maternal Coco functions by blocking incoming mesendoderm-inducing signals in the early blastula to delineate the boundaries of the marginal zone. During this time window, TGFβ1 ligand is not expressed and therefore Coco acts solely as an inhibitor. Zygotic Coco is involved in the specification of the right-left axis at the neurula stage (Vonica and Brivanlou, 2007). Coco restricts Nodal signaling to the left side of the embryo by inhibiting Xnr1 (Nodal1 – Xenbase) and derrière (Gdf3 – Xenbase) signaling on the right side. Given that TGFβ1 is also expressed in this region and that Coco enhances TGFβ1 signaling, the interaction of TGFβ1 and Coco might play an important role in the specification of the right-left axis. Consistent with these results is the fact that ectopic expression of TGFβ1 randomizes the right-left axis of the embryo. Therefore, during early embryogenesis, the activating and inhibitory functions of Coco are functionally and temporally separated. The function performed by Coco depends on which TGFβ ligands are concurrently expressed (supplementary material Fig. S3A,B).
Alternatively, it is possible to speculate that TGFβ1 could compete with BMPs and Nodal for binding to Coco in the embryo. In the absence of TGFβ1-specific receptors, TGFβ1 might saturate Coco and prevent the inhibitory effects of Coco on BMPs and Nodal.
We find that knockdown of Coco leads to increased myogenin expression in a myoblast differentiation paradigm in C2C12 cells. Conversely, Coco overexpression completely abolishes myogenin expression. As the process of myoblast differentiation is negatively regulated by TGFβ1 signaling, our results support the idea that Coco enhances the activity of TGFβ1. Indeed, these results cannot be explained by only considering the inhibitory effects of Coco on BMP signaling, as BMP signaling has been shown to inhibit myoblast formation in favor of osteoblast formation (Katagiri et al., 1994).
Coco expression has also been shown to be sufficient for dormant breast cancer cells to undergo reactivation and metastasis in the lung of adult mice (Gao et al., 2012). This effect was ascribed to the function of Coco in blocking lung-derived BMP ligands. Our data, however, raise the possibility that the role of Coco in metastasis might also include an effect mediated via enhancement of TGFβ1 signaling, in addition to BMP inhibition.
Our data show that Coco binds TGFβ1, and that the presence of Coco enhances the interactions between TGFβ1 and its receptor Alk5. This strongly supports a model in which Coco functions as a direct enhancer of TGFβ1. However, we cannot exclude the possibility that, in addition to directly enhancing TGFβ1 signal transduction, Coco might also inhibit an inhibitor of TGFβ1, thus making more TGFβ1 available for interactions with the Alk5 receptor.
It is tempting to speculate that the dual function of Coco might also be shared by other ‘classic’ inhibitors of TGFβ ligands, such as cerberus, noggin, chordin and follistatin. If this were the case, it would require reconsideration of our current understanding of the extracellular regulation of the TGFβ signaling pathway.
Coco was only one of the 321 hits that we identified in the loss-of-function screen for modulators of Smad4 nuclear translocation. Dissection of the function of the remaining hits might lead to further understanding of the molecular mechanisms controlling TGFβ1 signaling and will most likely highlight new players and regulators of the pathway.
MATERIALS AND METHODS
Screening
The siRNA screen was performed with the GFP-Smad4 C2C12 line described previously (Warmflash et al., 2012) using a library of Ambion silencer siRNAs. Each well of a 384-well plate contained three siRNAs targeted to the same gene for a total of 1.5 pmol of siRNA in 5 µl volume. On each plate, we manually added controls to two wells – siRNA against TGFβR1 and siRNA against Xpo1 in the same volume and concentration as the remainder of the siRNAs. To each well, we added 5 µl Lipofectamine2000 solution (Invitrogen; 1:100 in Optimem), incubated for 20 min, added 40 µl of cell suspension (30,000 cells/ml in growth medium) and incubated the plates for 48 h. TGFβ1 solution was prepared at a concentration of 20 ng/ml in growth medium, and 5 µl of this solution was dispensed into each well (final concentration of 2 ng/ml). After either 1 or 6 h the plates were washed once with PBS, fixed for 20 min with 4% paraformaldehyde and then washed twice with PBS. Plates were sealed and imaged with an ArrayScan instrument. Images were analyzed using custom software written in MATLAB (Mathworks) with an algorithm that we have described previously (Warmflash et al., 2012). Gene Ontology analysis was performed using DAVID Functional Annotation Tool (NCBI). Annotation terms were derived from the Swiss-Prot and Protein Information Resource database (SR-PIR-keywords). The RNAi dataset has been submitted to the GEO database under the accession number GSE70924.
Constructs
The ePiggyBac (Lacoste et al., 2009) construct for conditional expression of hCoco was cloned into the BamHI-NotI sites of the ePB-TT vector described previously (Arduini and Brivanlou, 2012). Xenopus tgfb1 (formerly tgfb5) cDNA was obtained from GE Healthcare Dharmacon (Clone Id:8548338) and subcloned into pCS105. This tgfb1 was HA-tagged at three amino acids downstream of the cleavage site (RKKR) by standard PCR methods. Human TGFBR2 was obtained from Addgene (#15012). Xenopus coco and coco-Flag were previously described (Bell et al., 2003). Xenopus coco was HA-tagged and V5-tagged at the C-terminus by standard PCR methods. Human Coco was obtained from GE (8069306) and subcloned into pCS105.
Cell culture, transfections, and selection of stable lines
C2C12 cells were maintained in DMEM medium containing 10% fetal bovine serum. The GFP-Smad4 and RFP-Smad2 reporter cell lines were described previously. The C2C12-hCoco-expressing line was created by transfecting cells with the ePB-P-TT-hCoco plasmid together with the ePB-transposase plasmid followed by selection with 4 µg/ml puromycin for 3 days. The RUES2 line of human embryonic stem cells was maintained on Matrigel (BD biosciences; 1:40 in DMEM-F12)-coated dishes either in mouse embryonic fibroblast conditioned medium (CM) supplemented with 20 ng/ml bFGF or in mTeSR1 as indicated. The hCoco-expressing RUES2 line was created by nucleofecting RUES2 cells with ePB-P-TT-hCoco and the ePB-transposase plasmids using an Amaxa nucleofector and nucleofection solution L followed by 3 days of selection with 4 µg/ml puromycin.
Xenopus explant dissection, cell culture and RNA preparation
Xenopus laevis embryos were obtained by in vitro fertilization and staged according to Nieuwkoop and Faber (1967). Microinjection, explant dissection and cell culture were performed as described (Hemmati-Brivanlou and Melton, 1994; Wilson and Hemmati-Brivanlou, 1995). RNA for injections was prepared using the mMessage mMachine in vitro SP6 Transcription Kit (Life Technologies).
Immunofluorescence
Cells were plated in glass-bottomed dishes (MatTek) at least one day before the experiment. After stimulation, cells were rinsed once with PBS, fixed with 4% paraformaldehyde for 20 min, rinsed twice with PBS, blocked for 30 min with 3% donkey serum, 0.1% Triton X-100 in PBS and then incubated overnight at 4°C with primary antibodies diluted in blocking buffer. Antibodies used were against Smad2/3 (BD Transduction Labs 610842; 1:100), pSmad1 (Cell Signaling; 1:100) and Myogenin (clone F5D; abcam 1835; 1:100). Cells were then washed three times with PBS+0.1% Tween20 for 30 min each wash, incubated for 30 min with secondary antibodies (Alexa 488 or Alexa 647; 1:500) and Sytox (1:50,000) diluted in blocking buffer, and washed twice more with PBS+0.1% Tween20 for 30 min each wash. When performing immunofluorescence against pSmad1, the protocol was modified to add a 30 min incubation with 1% SDS in PBS at 37°C before blocking.
Live cell imaging
C2C12 cells were plated in glass-bottomed dishes (MatTek) at least one day before imaging. Immediately before imaging, we switched the cell medium to custom culture medium (DMEM without Phenol Red or riboflavin) containing 10% fetal bovine serum. We collected images every 15 min using an Olympus Vivaview microscope with a 20×, 0.80NA lens.
Gene expression analysis
C2C12 cells were grown in 60-mm dishes and RUES2 cells were grown in 35-mm dishes. Total RNA was isolated with the TRIzol reagent (Life Technologies), treated with DNase (Ambion), and cDNA was synthesized with the Transcriptor First Strand cDNA Synthesis Kit (Roche). Real-time qRT-PCR analysis was performed with the LightCycler 480 SYBR Green I kit (Roche). A complete list of primers is provided in the supplementary materials and methods.
For cell culture, gene expression was normalized to the expression of mouse or human ATP50 as appropriate. For Xenopus experiments, ornithine decarboxylase 1 (odc1) was used to normalize gene expression.
Luciferase assays
C2C12 cells were transfected with 3.6 µg of CAGA12-luc plasmid and 0.4 µg of pRL-Tk for normalization. Cells were treated as described in the text and lysed and analyzed for luciferase activity using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions.
Co-immunoprecipitation assay
Immunoprecipitation assays were performed as described (Hama et al., 2002) with minor modifications. Two nanograms of Alk5-Flag and/or 2 ng TGFβ1-HA and/or 250 pg of Coco-V5 were injected into the animal hemisphere of two-cell-stage embryos. Embryos were lysed when sibling embryos had reached stage 23. Lysates were incubated overnight at 4°C with anti-Flag polyclonal antibody (1:1000) followed by incubation with Dynabeads Protein G (Life Technologies) for 1 h at 4°C. After four washes with lysis buffer, proteins were boiled and eluted in NuPAGE LDS Sample Buffer (Life Technologies) with 0.1 M DTT. Antibodies used were anti-HA (Abcam, ab18181), anti-Alk2 (LifeSpan Biosciences, LS-B2010), anti-Alk5 (R&D Systems, AF587), anti-Flag (Sigma-Aldrich, F3165, F7425) and anti-V5 (Abcam, ab9116).
Supplementary Material
Acknowledgements
We would like to thank Qixiang Zhang for technical support, all members of the Brivanlou and Siggia laboratories at The Rockefeller University for helpful discussions, and the RNAi core facility at New York University for assistance with screening.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
A.D., A.W., T.H. and A.H.B. planned experiments; A.D., A.W., T.H. and B.S. performed and analyzed experiments; A.D., A.W. and A.H.B. wrote the manuscript.
Funding
This work was supported by the National Institutes of Health (NIH) [R01-GM101653]; and funds from the Tri-I Starr Foundation for the Human Pluripotent Core Facility. The NYU RNAi Core is supported by the Laura and Isaac Perlmutter Cancer Center Support Grant [NIH/NCI P30CA16087] and NYSTEM Contract C026719. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.122358/-/DC1
References
- Arduini B. L. and Brivanlou A. H. (2012). Modulation of FOXD3 activity in human embryonic stem cells directs pluripotency and paraxial mesoderm fates. Stem Cells 30, 2188-2198. 10.1002/stem.1200 [DOI] [PubMed] [Google Scholar]
- Bates T. J. D., Vonica A., Heasman J., Brivanlou A. H. and Bell E. (2013). Coco regulates dorsoventral specification of germ layers via inhibition of TGFbeta signalling. Development 140, 4177-4181. 10.1242/dev.095521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E., Muñoz-Sanjuán I., Altmann C. R., Vonica A. and Brivanlou A. H. (2003). Cell fate specification and competence by Coco, a maternal BMP, TGFbeta and Wnt inhibitor. Development 130, 1381-1389. 10.1242/dev.00344 [DOI] [PubMed] [Google Scholar]
- Bradley L., Wainstock D. and Sive H. (1996). Positive and negative signals modulate formation of the Xenopus cement gland. Development 122, 2739-2750. [DOI] [PubMed] [Google Scholar]
- De Robertis E. M. and Kuroda H. (2004). Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell Dev. Biol. 20, 285-308. 10.1146/annurev.cellbio.20.011403.154124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Chakraborty G., Lee-Lim A. P., Mo Q., Decker M., Vonica A., Shen R., Brogi E., Brivanlou A. H. and Giancotti F. G. (2012). The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell 150, 764-779. 10.1016/j.cell.2012.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama J., Suri C., Haremaki T. and Weinstein D.C. (2002). The molecular basis of Src kinase specificity during vertebrate mesoderm formation. J. Biol. Chem. 277, 19806-19810. 10.1074/jbc.M110637200 [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Rebagliati M., Ahmad N., Muraoka O., Kurokawa T., Hibi M. and Suzuki T. (2004). The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development 131, 1741-1753. 10.1242/dev.01070 [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A. and Melton D. A. (1994). Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell 77, 273-281. 10.1016/0092-8674(94)90319-0 [DOI] [PubMed] [Google Scholar]
- Hsu D. R., Economides A. N., Wang X., Eimon P. M. and Harland R. M. (1998). The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol. Cell 1, 673-683. 10.1016/S1097-2765(00)80067-2 [DOI] [PubMed] [Google Scholar]
- James D., Noggle S. A., Swigut T. and Brivanlou A. H. (2006). Contribution of human embryonic stem cells to mouse blastocysts. Dev. Biol. 295, 90-102. 10.1016/j.ydbio.2006.03.026 [DOI] [PubMed] [Google Scholar]
- Katagiri T., Yamaguchi A., Komaki M., Abe E., Takahashi N., Ikeda T., Rosen V., Wozney J. M., Fujisawa-Sehara A. and Suda T. (1994). Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 127, 1755-1766. 10.1083/jcb.127.6.1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollias H. D., Perry R. L. S., Miyake T., Aziz A. and McDermott J. C. (2006). Smad7 promotes and enhances skeletal muscle differentiation. Mol. Cell. Biol. 26, 6248-6260. 10.1128/MCB.00384-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste A., Berenshteyn F. and Brivanlou A. H. (2009). An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell 5, 332-342. 10.1016/j.stem.2009.07.011 [DOI] [PubMed] [Google Scholar]
- Marques S., Borges A. C., Silva A. C., Freitas S., Cordenonsi M. and Belo J. A. (2004). The activity of the Nodal antagonist Cerl-2 in the mouse node is required for correct L/R body axis. Genes Dev. 18, 2342-2347. 10.1101/gad.306504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. (2012). TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616-630. 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P. D. and Faber J. (1967). Normal table of Xenopus laevis. Amsterdam, The Netherlands: North Holland Publishing. [Google Scholar]
- Pearce J. J. H., Penny G. and Rossant J. (1999). A mouse cerberus/Dan-related gene family. Dev. Biol. 209, 98-110. 10.1006/dbio.1999.9240 [DOI] [PubMed] [Google Scholar]
- Schmierer B. and Hill C. S. (2007). TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8, 970-982. 10.1038/nrm2297 [DOI] [PubMed] [Google Scholar]
- Sorre B., Warmflash A., Brivanlou A. H. and Siggia E. D. (2014). Encoding of temporal signals by the TGF-β pathway and implications for embryonic patterning. Dev. Cell. 30, 334-342. 10.1016/j.devcel.2014.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonica A. and Brivanlou A. H. (2007). The left–right axis is regulated by the interplay of Coco, Xnr1 and derrière in Xenopus embryos. Dev. Biol. 303, 281-294. 10.1016/j.ydbio.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Warmflash A., Zhang Q., Sorre B., Vonica A., Siggia E. D. and Brivanlou A. H. (2012). Dynamics of TGF-beta signaling reveal adaptive and pulsatile behaviors reflected in the nuclear localization of transcription factor Smad4. Proc. Natl. Acad. Sci. USA 109, E1947-E1956. 10.1073/pnas.1207607109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson P. A. and Hemmati-Brivanlou A. (1995). Induction of epidermis and inhibition of neural fate by Bmp-4. Nature 376, 331-333. 10.1038/376331a0 [DOI] [PubMed] [Google Scholar]
- Wu M. Y. and Hill C. S. (2009). Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev. Cell 16, 329-343. 10.1016/j.devcel.2009.02.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.