Abstract
Background
The reproductive mechanisms of mollusk species have been interesting targets in biological research because of the diverse reproductive strategies observed in this phylum. These species have also been studied for the development of fishery technologies in molluscan aquaculture. Although the molecular mechanisms underlying the reproductive process have been well studied in animal models, the relevant information from mollusks remains limited, particularly in species of great commercial interest. Crassostrea hongkongensis is the dominant oyster species that is distributed along the coast of the South China Sea and little genomic information on this species is available. Currently, high-throughput sequencing techniques have been widely used for investigating the basis of physiological processes and facilitating the establishment of adequate genetic selection programs.
Results
The C.hongkongensis transcriptome included a total of 1,595,855 reads, which were generated by 454 sequencing and were assembled into 41,472 contigs using de novo methods. Contigs were clustered into 33,920 isotigs and further grouped into 22,829 isogroups. Approximately 77.6% of the isogroups were successfully annotated by the Nr database. More than 1,910 genes were identified as being related to reproduction. Some key genes involved in germline development, sex determination and differentiation were identified for the first time in C.hongkongensis (nanos, piwi, ATRX, FoxL2, β-catenin, etc.). Gene expression analysis indicated that vasa, nanos, piwi, ATRX, FoxL2, β-catenin and SRD5A1 were highly or specifically expressed in C.hongkongensis gonads. Additionally, 94,056 single nucleotide polymorphisms (SNPs) and 1,699 simple sequence repeats (SSRs) were compiled.
Conclusions
Our study significantly increased C.hongkongensis genomic information based on transcriptomics analysis. The group of reproduction-related genes identified in the present study constitutes a new tool for research on bivalve reproduction processes. The large group of molecular markers discovered in this study will be useful for population screening and marker assisted selection programs in C.hongkongensis aquaculture.
Introduction
Mollusks represent a major branch of lophotrochozoan organisms and have been an interesting target for the study of reproductive biology because diverse reproductive strategies have evolved in this phylum. Mollusks include dioecious and hermaphroditic species, and species that are capable of sex changes [1,2]. Studies on the reproduction of mollusk species may provide insights into the reproduction system and its evolution.
Crassostrea hongkongensis, a member of the phylum Mollusca, is a dominant oyster species on the coast of the South China Sea, with a cultivation history of ~700 years [3]. It is considered a popular seafood with a market demand that’s been growing over time, and it is one of the three major oyster species produced by aquaculture in China. In 2011, the production of C.hongkongensis reached 1,100,000 tons [4]. However, in recent years, the C.hongkongensis cultivation industry has been affected by the emerging shortage of seeds due to region planning, industrialization and urbanization in some seed-producing areas as well as population degradation. Therefore, there is urgent demand for better and higher seed production. The identification of reproduction-related genes will advance our understanding of C.hongkongensis reproduction and possibly contribute to the production of high-quality seeds. It will also help us understand the general underlying molecular mechanisms of bivalve reproduction, which will provide a basis for the development of aquaculture technology for bivalves.
To date, our knowledge of the molecular mechanisms of oyster reproduction is still limited. Several genes involved in germ cell development (vasa and nanos) have been identified in the Pacific oyster C. gigas and pearl oyster P. fucata [5,6]. Genes related to the sex determining pathways (Dmrt-like, SoxH, FoxL2and fem-1like) have been reported in C.gigas and P.margaritifera [7,8,9], and it has been shown that oysters share some key sex determination genes with vertebrates [7]. Hormones related to gonad development, oocyte maturation and vitellogenesis, including insulin [10], estradiol [11], 5-hydroxytryptamine (5-HT) [12] and gonadotropin-releasing hormone (GnRH) [13], and hormone receptors (estrogen receptor, 5-HT receptor, GnRH receptor) [14,15,16] have also been studied in oysters. Neuropeptides, which include egg-laying hormones (ELH) that control egg-laying behavior, were reported recently in P.fucata and C.gigas [17].
However, in-depth studies on the oyster reproduction mechanisms are hindered by the lack of genetic information. Although several studies have produced EST sequences in C.hongkongensis [18,19,20], gene sequences for this species are still scarce in GenBank. Recently, high-throughput next-generation sequencing (NGS) technologies, which include Solexa/Illumina, SOLiD/Applied technologies and 454/Roche, have created unprecedented opportunities for generating genetic information in non-model species and have been utilized to survey the genes related to reproduction. Roche’s 454 GS-FLX platform, which now provides a sequence read size approaching the length of conventional Sanger sequencing and sequencing coverage that is much deeper than that available with Sanger sequencing, has been widely used for de novo transcriptome sequencing in species including fish (turbot) [21]; shrimp (Litopenaeus vannamei) [22]; and mollusks (Bathymodiolus azoricus [23], Ruditapes philippinarum [24], Patinopecten yessoensis [25], and Mytilus edulis [26]). In particular, reproduction-related genes have been discovered in turbot and P. yessoensis by 454 pyrosequencing and have been useful for understanding the molecular process of reproduction in these species [21,25].
In the present study, we sequenced and assembled the C.hongkongensis transcriptome obtained by 454 sequencing, and studied the genes related to reproduction. The group of reproduction-related genes identified through this analysis constitutes a new tool for research on the bivalve reproduction process and provided insights into the origin and ancient character of the reproduction-related genes. We also characterized SNPs and SSRs to be employed for genetic improvement purposes. Hopefully, our study would be of great importance by providing resources for functional gene discovery, genetic and genomic studies on this species.
Materials and Methods
Sample Preparation
Adult C.hongkongensis were purchased from an aquaculture farm in Zhanjiang, Guangdong Province, China. Tissues including gill, adductor muscle, digestive gland, hemocytes, mantle, heart, and male and female gonads were dissected from sexually mature adult oysters. Embryos at the4-cell stage, blastula, trochophore and D-shaped larval stages were collected by filtration on a 30 μm mesh. All samples were homogenized in TRIzol (Invitrogen, Carlsbad, USA) and stored at -80°C (total RNA isolation), or fixed in 4% paraformaldehyde buffer (in situ hybridization). Isolated RNA was quality and quantity checked using electrophoresis and a Nanodrop 2000c Spectrophotometer (Thermo Scientific, USA).
454 cDNA Library Construction and Transcriptome Sequencing
To maximize the discovery of C.hongkongensis genes, a normalized cDNA library was constructed. RNA from each sample was combined into a single pool and mixed well (see detail in S1 Table).The SMART (Switching Mechanism At 5’ end of RNA Template) kit (BD Clonetech, Mountain View, CA) was used to retrotranscribe total poly-adenylated RNA. First-strand cDNA was synthesized with SMART Oligo Ⅱoligonucleotide (5’-AAGCAGTGGTATCAACGCAGAGTACGGGGG-3’) and a modified oligo-dT primer (5’-AAGCAGTGGTATCAACGCAGAGTACTTTTGTTTTTTTTTCTTTTTTTTTTVN- 3’). Double-strand cDNA was obtained from 1μl of the first-strand PCR reaction with SMART PCR primer (5’-AAGCAGTGGTATCAACGCAGAGT-3’). Then, the amplified dsDNA products were purified using the QIAquick PCR Purification Kit (Qiagen, USA) and normalized using the Trimmer-Direct Kit (Evrogen, Moscow). The quality of the normalized cDNA library was determined on a 1.4% agarose gel. Approximately 2.5 μg of the normalized cDNA pool was used for a titration run using one quarter of a plate on the Roche 454 GS FLX sequencer (Roche, Basel, Switzerland), and then 15 μg of cDNA was sequenced with a whole-plate run on the same equipment at the University of Illinois, USA. The reads from this sequencing were deposited in the NIH Short Read Archive database with Run accession number SRR949615.
De Novo Assembly of the C.hongkongensis Transcriptome
GS FLX data were processed using the Roche GS FLX software (v2.6). De novo assembly of the transcriptome was performed using the GS De Novo Assembler v2.6 (Roche).The overlap settings used for this assembly were 40bp and 90% identity, with all other parameters set at the default values by the-cdna and-urt options. Prior to assembly, the primers and adaptors were trimmed accordingly.
Sequence Annotation
Sequence annotation was performed against the Swiss-Prot and Non-redundant (Nr) protein databases, using BlastX (version2.2.28+) with a significance threshold cut off of E-value ≤ 1e-06. A maximum of 20 hits were taken into account for each blast query. To avoid redundant annotations, only the longest isotig from each isogroup was selected. Gene names were assigned to each selected isotig based on the best BLAST hit (highest score).
The set of longest isotigs was also used to identify Gene Ontology (GO) designations using the program Blast2GO [27].The BLAST result (xml file) from the Swiss-Prot annotation was imported into the Blast2GO program, and the GO terms associated with blast hits were retrieved. GO annotations were conducted only for isotigs with significant BLAST hits below an E-value of 1e-06, with 55 as the annotation cut-off and 5 as the GO weight. No HSP-hit coverage cut-off was used. InterproScan annotation was also conducted using Blast2GO.The obtained information on protein domains and motifs were included to improve global annotations.
Sequences of the longest isotig from each isogroup were uploaded to the online KEGG Automatic Annotation Server (KAAS; http://www.genome.jp/tools/kaas/) for ortholog assignment and pathway mapping. The bi-directional best hit (BBH) method was used to compare the C.hongkongensis transcriptome against each genome in the reference set of the KEGG GENES database.
RT-qPCR
Total RNA was isolated from mantle, gill, heart, hemocytes, adductor muscle, digestive gland, male and female gonads (each sample was pooled from five individuals). After the digestion of genomic DNA with DNase I to prevent genomic DNA contamination, 1μg of total RNA was reverse transcribed using PrimeScript RT reagent Kit with gDNA eraser (Perfect Real Time) (Takara, Japan).The resulting cDNAs were diluted, and amount equivalent to 5 ng of starting RNA was assayed for reproduction-related gene expression using EF1α as the reference gene. SYBR-based qRT-PCR reactions (Light Cycler 480 Master Mix I, Roche, Basel, Switzerland) were performed on a Light Cycler 480 system (Roche, Basel, Switzerland) using the following reaction conditions: 95°C for 1 m in followed by 45 cycles of 95°C for 10 s, 58°C for 10 s and 72°C for 10 s. The primers used are listed in S2 Table. A melting curve was generated at the end of the reaction to check for the accurate amplification of the target amplicon. The PCR efficiencies of the target gene and reference gene were verified and were approximately equal. The relative gene expression was calculated according to the formula: 2 -(Ct target gene–Ct reference gene). Water was used instead of cDNA as a negative control for amplification, and DNAase-untreated cDNA was used to check for the absence of genomic DNA contamination. All the results are expressed as the mean ± s.e.m (standard error to the mean). The results were analyzed for statistical significance using one-way ANOVA (significance was considered when p < 0.01). Data were analyzed using the GraphPad Prism software version 5.0. The relative expression data of sex determination/differentiation genes were imported into the MultiExperiment Viewer software (http://www.tm4.org/mev.html) for heatmap production.
mRNA in situ hybridization
A713-bp fragment for Ch-nanos and a1146-bp fragment for Chpiwil1were amplified by RT-PCR with the primers listed in S2 Table. These fragments were subcloned into the pGEM-easy T vector (Promega, USA) and sequenced. The resulting plasmids were linearized using SacII and NdeI in separate reactions. The linearized products were purified and used as a template to generate sense and antisense DIG-labeled RNA probes using a DIG RNA labeling kit (SP6/T7; Roche Diagnostics, Germany).
Ovaries of early developing stage, late developing stage and mature stage were fixed in 4% paraformaldehyde buffered with 1×phosphate saline (pH7.4) at 4°C overnight. Paraffin-embedded ovary samples were sectioned (7 μm thick) and mounted on glass slides coated with 1% Poly-L-Lysine solution. Slices were deparaffinized through two washes of xylene, hydrated through an ethanol gradient, washed with 1×PBS and then digested with proteinase K (10 μg/ml) for 10 min at 37°C. Following wash and prehybridization, the serial sections were hybridized overnight at 60°C with either antisense or sense probes in hybridization solution. After an extensive wash, the sections were incubated in 1% blocking reagent in PBS for 1 hr and then transferred into 1/5000 dilution of anti-DIG alkaline phosphatase conjugated fab fragment at room temperature for 2 hrs. A mixture of BCIP/NBT was then used for color development. All slices were mounted with Mowiol and imaged under a Leica DMR HC (Leica, Germany) microscope.
Sequence and Phylogenetic Analysis
Sequences from various organisms (see S3 Table for accession numbers) were obtained from the NCBI genome server (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and aligned using the DNAMAN 8 program (http://www.lynnon.com/). The optimal phylogenetic tree of each gene was constructed using the neighbor-joining method, implemented in the MEGA5.0 program [28]. One thousand boot strap trials were run for each node. The protein motifs were analyzed with SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1).
SNP and SSR Discovery
Potential SNPs were detected using GS Reference Mapper v2.6 with default parameters (cDNA mode). The set of the longest isotig was used as a reference sequence. Primers and adaptors were trimmed before mapping. SNP identification was limited to isotigs or contigs: (i) the total number of reads that fully span the difference location (Total Depth) ≥8; (ii) the percentage of different reads versus total reads that fully span the difference location (Var Freq) ≥ 25%.For SSR detection, the set of longest isotig was fed into the MicroSAtellite (MISA) program [29]. All types of SSRs, from dinucleotides to hexanucleotides, were searched using default settings (the minimum repeat number was six for dinucleotide and five for tri-, tetra-, penta- and hexanucleotide).
Results and Discussion
454 Transcriptome Sequencing and Assembly
The normalized pooling library was prepared from larvae of four different developmental stages (4-cell stage, blastula, trochophore and D-shaped larvae) and various tissues (gill, adductor muscle, digestive gland, hemocytes, mantle, heart, ovary and testis) and sequenced using the Roche 454 GS FLX sequencer. A single run of the 454 sequencing generated 1,595,855 raw sequencing reads with an average length of 382 bp and total output of 609,282,210 bp (Table 1).
Table 1. Summary statistics of the transcriptome assembly for Crassostrea hongkongensis.
| Category | Value |
|---|---|
| Total number of raw reads | 1,595,855 |
| Total length of raw sequences | 609,282,210 bp |
| Total number of clean reads | 1,405,240 |
| Total length of clean reads(bp) | 522,591,765 |
| reads assembled | 1,266,466 |
| Total contigs | 41,472 |
| Average length of contigs | 958bp |
| Contig size N50 | 1,571bp |
| Total isotigs | 33,920 |
| Average length of isotigs | 1,924bp |
| Isotig size N50 | 2,752bp |
| Total Isogroups | 22,829 |
| Total singletons | 138,631 |
| Total unigenes | 161,460 |
The 41,472 contigs were assembled by GS De Novo Assembler v2.6, with an average length of 958 bp (Table 1).These contigs were then joined into 33,920 isotigs with an average length of 1,492 bp (N50 = 2,752 bp), and the length of 21,830 isotigs (64.4% of total isotigs) were longer than 1,000 bp (S1 Fig). The isotigs were further grouped into 22,829 isogroups. The remaining 138,631 reads that were not assembled by the GS De Novo Assembler v2.6 were considered singletons. In total, 161,460 unigenes (#isogroups + #singletons) were produced. Because previous reports indicated that most singletons represent lowly expressed transcripts, artifacts derived from cDNA synthesis, sequencing and contamination [30], they were excluded from the following analysis in this study.
Our 454 sequencing data and GS De Novo assembly of C.hongkongensis transcriptome were compared favorably with transcriptomes from other mollusks (produced with NGS). The average length of the raw reads and contigs in C.hongkongensis transcriptome was 382 bp and 958 bp, respectively, which are better than or similar to those of molluscan transcriptomes obtained from 454 sequencing: 283 bp and 509 bp in B.azoricus [31], 369 bp and 535 bp in L.elliptica [32], 319 bp and 723 bp in C.angulata [33], 313 bp and 618 bp in P.yessoensis [25]. They were better than those obtained from Illumina: 61 bp and 536 bp in Radix balthica [34], 90 bp and 874 bp in C.virginica [35]. Overall, the C.hongkongensis transcriptome obtained from 454 sequencing here is a high-quality one in either sequencing or assembly.
Gene Annotation and Functional Classification
To identify the putative functions of the C.hongkongensis genes, the longest isotig from each isogroup was chosen as a representative and compared with those in Swiss-prot protein database and NCBI Non-redundant (Nr) database. A total of 12,273 isotigs (53.8% of total isogroups) showed significant matches to known proteins in the Swiss-prot database, corresponding to 10,079 different well-annotated proteins (S4 Table). A higher percentage of isogroups (17,721 isotigs, 77.6% of total isogroups) had significant matches to known proteins in the Nr database (S4 Table), corresponding to 13,631 unique proteins. The remaining isogroups that were not annotated appeared to be either C.hongkongensis-specific genes or homologous genes with unknown functions in other species.
Gene Ontology (GO), which is an international standardized gene functional classification system, was used to classify the predicted C.hongkongensis genes in terms of their associated biological processes, cellular components and molecular functions. GO terms were retrieved from the association to best-hit for 9,909 (43.4%) of the overall 22,829 isogroups. Protein domain and motif information were retrieved by InterProScan via Blast2GO, and corresponding annotations were merged with already existing GO terms. A total of 15,050 isogroups provided significant InterProScan information, with 7,498 of them resulting in GO annotation. After merging, a total of 11,633 isogroups (51.0% of total isogroups) were assigned at least one GO term. In the biological process category, genes involved in the cellular process (GO:0009987) and metabolic process (GO:0008152) were prominently represented. Cells (GO:0005623) and organelles (GO:0043226) represented the majority of terms in the cellular component category, whereas in the molecular function category, the vast majority is related to binding (GO:0005488) and catalytic activity (GO:0003824; Fig 1). The GO classification results in each category here were similar to the previously sequenced P.yessoensis and Chlamys farreri via 454 sequencing [25,36].
Fig 1. Gene Ontology(GO) analysis of the Crassostrea hongkongensis transcriptome on level 2.
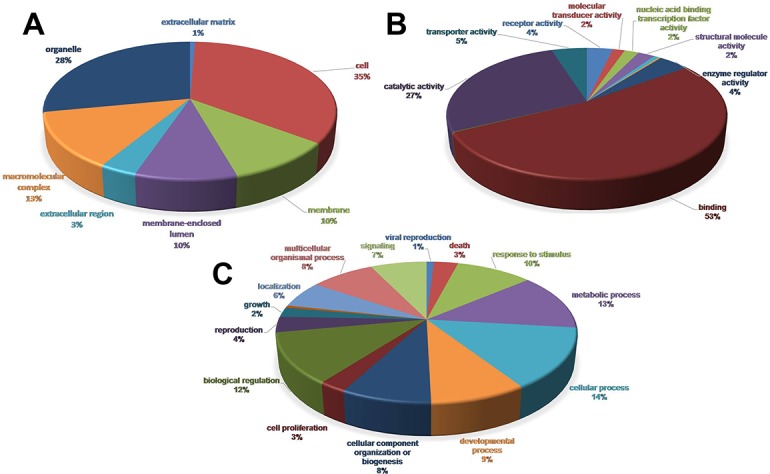
The percentage and distribution of top-level GO-terms were portrayed in the three categories: (A) Cellular component; (B) Molecular function and (C) Biological process.
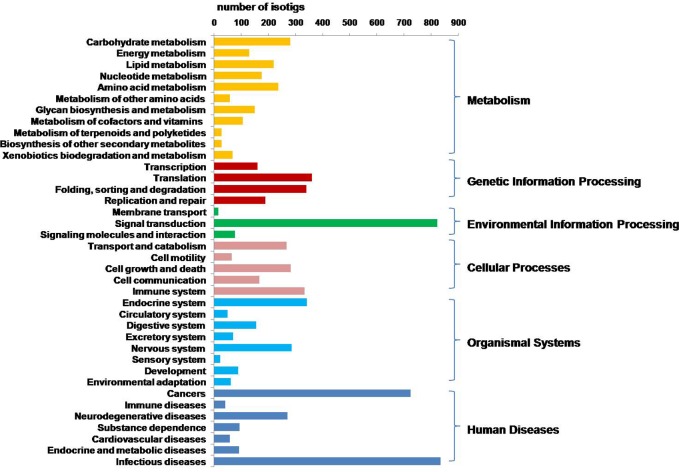
The KEGG orthology (KO) is a classification system that provides an alternative functional annotation of genes based on their associated biological pathways. The KO annotations for C.hongkongensis were based on sequence similarity searches for reference sequences in the NCBI database. Overall, 5,197 isogroups were assigned to KOs, which are involved in 257 different pathways (S5 Table). The most populated pathways were “infectious diseases” with 835 isogroups involved, “signal transduction” with 823 isogroups and “cancers” with 725isogroups (Fig 2). The KEGG pathways distribution in the C.hongkongensis transcriptome was identical to those in the Mizuhopecten yessoensis and C. virginica transcriptomes [35,37].
Fig 2. KEGG biochemical mappings for C. hongkongensis transcriptome.
Overall, the annotation and functional classification of C.hongkongensis transcripts provided a valuable dataset for the identification of functional genes, investigating specific bioprocesses and pathways as well as for further genome-wide research and analyses in this species.
Genes and Pathways Related to Reproduction
Reproduction is a biological process by which new individual orgamisms–“offspring”–are produced from “parents”. The reproduction process can be divided into the following categories: germline development, sex determination/differentiation, oocyte maturation and spawning, fertilization and others. Both GO and KEGG analyses identified transcripts that are potentially involved in reproduction. GO classification identified 1910 genes related to reproduction (GO:0000003, S6 Table), which was much more than those (318 reproduction-related genes) identified in P. yessoensis by 454 sequencing [25]. KEGG annotation identified 205 genes related to reproduction, distributed in 8 pathways (Table 2). These genes covered all major processes of reproduction, including germline development, sex determination/differentiation, oocyte maturation, fertilization and others. Because germline development, sex determination/differentiation and oocyte maturation are basic processes of reproduction for C.hongkongensis, genes functioning in these processes were analyzed in priority below.
Table 2. Reproduction-related pathways identified in the C. hongkongensis transcriptome.
| Description | KEGG code | No. of genes in the pathway |
|---|---|---|
| Insulin signaling pathway | ko04910 | 56 |
| Oocyte meiosis | ko04114 | 50 |
| Progesterone-mediated oocyte maturation | ko04914 | 41 |
| Estrogen signaling pathway | ko04915 | 35 |
| GnRH signaling pathway | ko04912 | 32 |
| Prolactin signaling pathway | ko04917 | 26 |
| Ovarian Steroidogenesis | ko04913 | 15 |
| Steroid hormone biosynthesis | ko00140 | 12 |
Germline development genes
The 84 genes related to germline development were identified by GO analysis (GO:0007281, S6 Table) and literature supported searching (S7 Table). Table 3 shows some relevant genes with well-known functions in the process, and most of them were identified for the first time in C.hongkongensis, including some core germline genes such as vasa, nanos and piwi.
Table 3. Selection of some novel germline development genes identified in the C.hongkongensis transcriptome.
| Gene name | Isotig name | Isotig length | E-value | Species | Gene ontology |
|---|---|---|---|---|---|
| Bmp4 | isotig19635 | 1998 | 3.36E-78 | Gallus gallus | GO:0008083 GO:0007281 |
| Bru | isotig17186 | 3483 | 1.85E-107 | Danio rerio | GO:0007283 GO:0007281 |
| isot ig22487 | 1373 | 2.37E-98 | Danio rerio | GO:0007286 GO:0008016 | |
| gcl | isotig19380 | 2076 | 2.06E-145 | Mus musculus | GO:0007275 GO:0007277 |
| lin28 | isotig22508 | 1370 | 8.70E-44 | Drosophila melanogaster | GO:0007281 GO:0007549 |
| mago | isotig30591 | 618 | 6.46E-90 | Homo sapiens | GO:0008103 GO:0007267 |
| mex-3 | isotig16497 | 5168 | 3.69E-79 | Xenopus laevis | GO:0008270 GO:0005509 |
| nanos | isotig23837 | 3166 | 2.13E-175 | Mus musculus | GO:0007444GO:0007314 |
| par-1 | isotig31179 | 583 | 2.47E-09 | Caenorhabditis elegans | |
| piwi | isotig17193 | 3468 | 0 | Mus musculus | GO:0003729 GO:0005654 |
| pum | isotig11287 | 3581 | 1.84E-114 | Rattus norvegicus | GO:0007291 GO:0007475 |
| isotig16484 | 5232 | 0 | Mus musculus | GO:0003730 GO:0005829 | |
| stau | isotig16834 | 4044 | 6.56E-108 | Rattus norvegicus | GO:0003723 GO:0005875 |
| tud | isotig09805 | 3781 | 5.31E-18 | Homo sapiens | GO:0005737GO:0000003 |
| isotig16579 | 4788 | 1.23E-12 | Homo sapiens | GO:0009987GO:0000003 | |
| isotig16345 | 6854 | 4.22E-47 | Oryzias latipes | GO:0005515GO:0007275 | |
| isotig17301 | 3335 | 2.93E-30 | Homo sapiens | GO:0005739GO:0005515 | |
| isotig18032 | 2692 | 4.05E-28 | Oryzias latipes | GO:0009987GO:0005737 | |
| isotig22822 | 1318 | 3.43E-11 | Homo sapiens | ||
| vasa | isotig11445 | 3166 | 2.13E-175 | Mus musculus | GO:0007281GO:0007283 |
Vasa is a key determinant in germline formation in eukaryotes. It encodes an RNA helicase that is characterized by the presence of zinc knuckle motifs, a DEAD box helicase domain and a helicase conserved C-terminal domain. In the C.hongkongensis transcriptome, a transcript of 3,166 bp (isotig11445) encoding a Vasa-related protein was identified and showed an ORF of 2,118 bp. The deduced amino acid sequence is 705 aa long and contains three zinc knuckle motifs, a DAED box helicase domain and a helicase C domain (Fig 3A). Phylogenetic analysis performed with related proteins indicated that the Vasa-related protein encoded by isotig11445 clustered to molluscan Vasa, particularly to bivalve Vasa (Fig 3B). Therefore, we named the C.hongkongensis Vasa-related protein “Chvasa”. Chvasa was highly expressed in female and male gonads with weak expression in other tissues (Fig 3C). In bivalves, a vasa ortholog has been characterized in C. gigas (oyster vasa-like gene, Oyvlg), and its expression was restricted to germline cells both in males and females, including germinal stem cells and auxiliary cells [38]. Knockdown of Oyvlg by RNAi resulted in germ cell underproliferation and prematurely arrested meiosis, indicating a key role of vasa in bivalve germ cell development [38,39]. Our experiment showed that a high expression level of Chvasa was detected in both female and male gonads, indicating a role for Chvasa in reproduction and possibly in the development and maintenance of germ cells. Further investigations are needed to understand the role of Chvasa in C.hongkongensis germline development.
Fig 3. Vasa gene identified in the Crassostrea hongkongensis transcriptome and its expression profile.
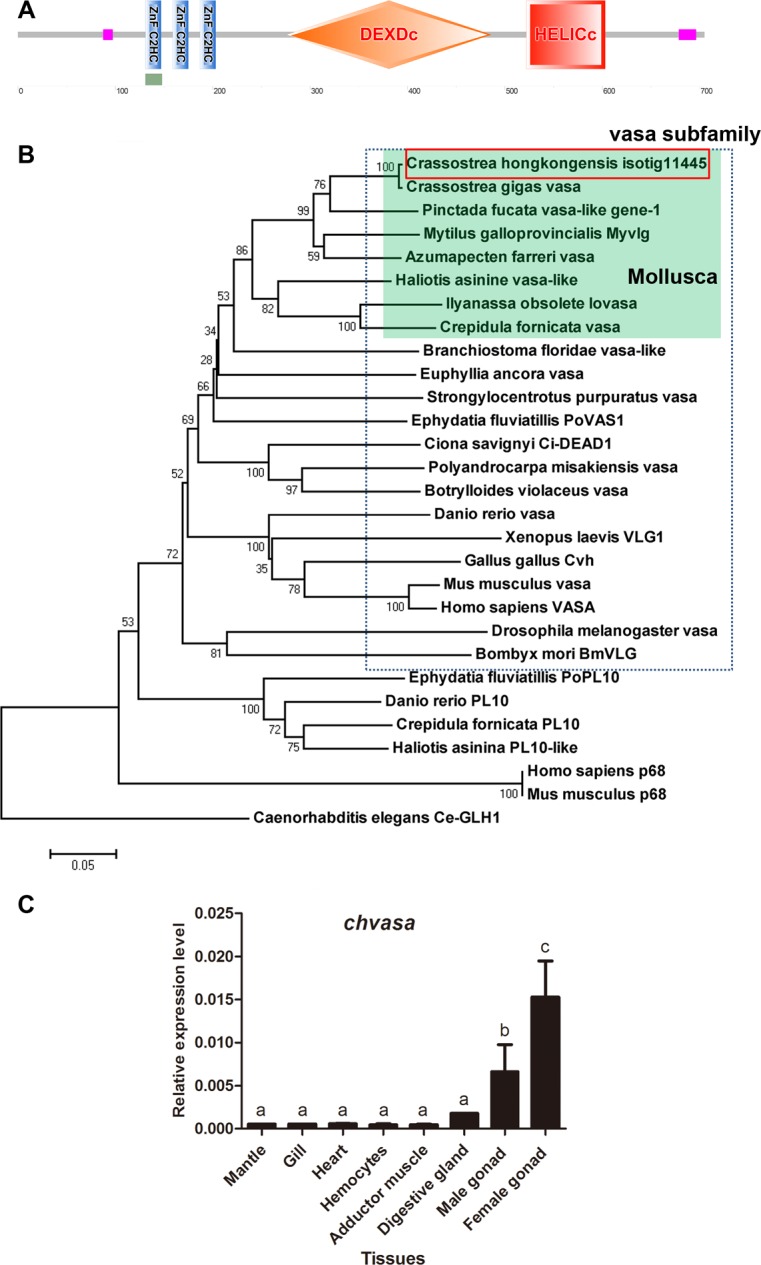
(A) Domain structure of the C.hongkongensis vasa ortholog. (B) Molecular phylogenetic tree of Vasa subfamily and related proteins. Bootstrap values from 1000 trials are indicated at each branch node. The scale bar indicates 0.05 amino acid replacements per site. C elegans Ce-GLH-1 was used as an outgroup. The transcript encoding Chvasa, indicated by a red open box, is properly aligned to the clade of molluscan Vasa colored by green. The Vasa subfamily proteins are enclosed by the dotted lines. For the GenBank accession numbers of the reference sequences, see S3 Table. (C) Expression profile of C.hongkongensis vasa ortholog (Chvasa) in adult tissues with mean ± s.e.m. as error bars (n = 5).
Nanos, another conserved core germline gene, is a translational repressor characterized by a nanos RNA-binding domain with two conserved Cys-Cys-His-Cys zinc finger motifs. It plays important roles in early development and, more specifically, in primordial germ cell (PGC) development [40]. A transcript encoding a nanos ortholog (isotig23837) was identified in the C.hongkongensis transcriptome, with an ORF of 708 bp. The deduced amino acid sequence is 235 aa and contains a nanos RNA binding domain. Alignment of the amino acid sequence of the predicted C.hongkongensis nanos RNA binding domain with other orthologs indicated high conservation of two characteristic CCHC zinc finger motifs (Fig 4A and 4B). This C.hongkongensis nanos mRNA was named “Ch-nanos”.Ch-nanos was specifically expressed in both male and female gonads when examined by RT-qPCR, suggesting an important role of Ch-nanos in C.hongkongensis reproduction. The expression level of Ch-nanos in female gonads was 2.3-fold higher than that in male gonads (Fig 4C). mRNA in situ hybridization showed that Ch-nanos was specifically expressed in developing germ cells including oogonia and early vitellogenic oocytes, but not in somatic cells (Fig 4D–4F), indicating a possible role of Ch-nanos in the formation of germ cells; it might be used as a molecular marker for germ cells in early developing stage female gonads. In mollusks, nanos orthologs have been isolated in Ilyanassa obsolete [41], Haliotis asinine [42] and P.fucata [6]. In these studies, the expression patterns of molluscan nanos orthologs during embryonic and larval development have been reported. However, the expression patterns and functions of molluscan nanos orthologs in germline development are unclear. Here, we identified a nanos homologue in C.hongkongensis and studied its expression pattern in different tissues and during germ cell development. It will be interesting to further follow the expression pattern of Ch-nanos during germline development and study its specific role in germ cells.
Fig 4. Analysis of Nanos ortholog identified in C.hongkongensis transcriptome.
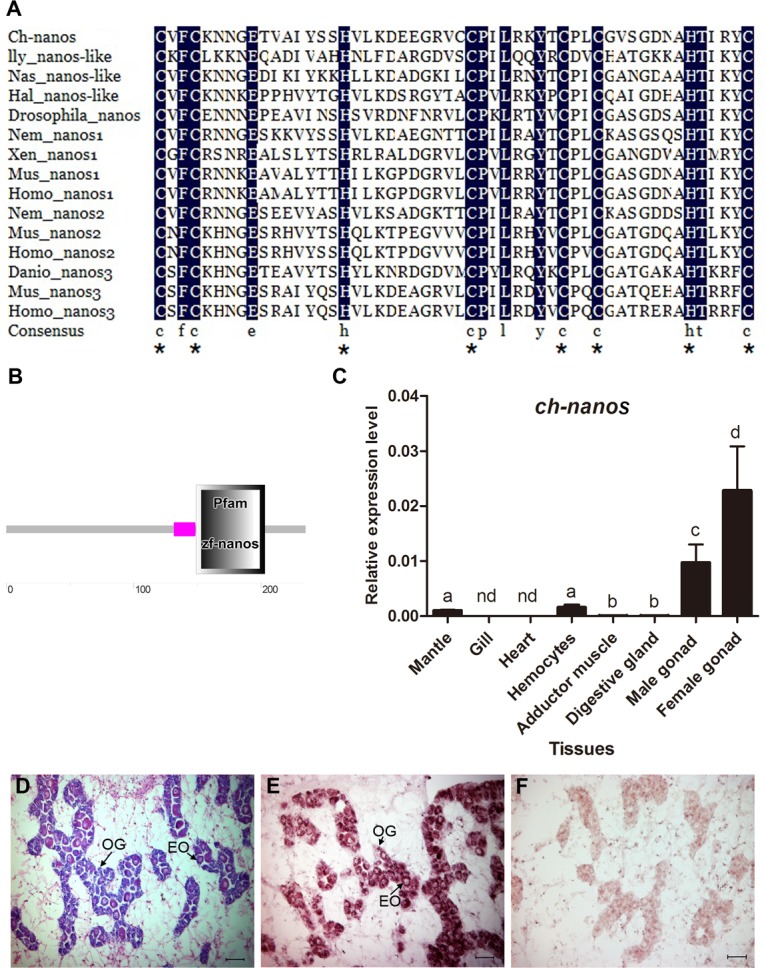
(A) Amino acid sequence alignment of the nanos RNA-binding domain predicted in the Ch-nanos with that of other Nanos orthologs. Two highly conserved CCHC zinc finger motifs are indicated by asterisks. For the GenBank accession numbers of the reference sequences, see S3 Table. (B) Domain structure of Ch-nanos. (C) Expression profile of in Ch-nanos adult tissues by qRT-PCR with mean±s.e.m. as error bars (n = 5). nd: not detected. (D) Histological analysis of C.hongkongensis female gonad at early developing stage. (E) Expression profile of Ch-nanos in early developing stage female gonad detected by ISH with antisense and sense probe. (F) Positive cells are stained in purple.OG: oogonia; EO: early vitellogenic oocyte.
Piwi (P-element induced wimpy testis) is a member of the argonaute family of small RNA-binding proteins that possesses a PAZ domain in the middle and a PIWI-domain at the C-terminal end. In Drosophila and C.elegans, the lack of Piwi results in complete depletion of germline stem cells and sterility in both males and females. In contrast, overexpression of piwi increases both the number and mitosis rate of germline stem cellsin Drosophila [43,44]. A PAZ domain and PIWI-domain containing transcript (isotig17193) was identified in the C.hongkongensis transcriptome (Fig 5A). Phylogenetic analysis revealed that isotig17193 was a close relative of vertebrate Piwi-like 1 protein (Piwil1) rather than other members of the Piwi-like protein family and Ago1 (Fig 5B). Isotig17193 was named “ChPiwil1”. ChPiwil1was expressed in gonads but at a low level in somatic organs (Fig 5C). Its expression in the ovary was 2.8-fold higher than that in the testis. We then studied the expression patterns of ChPiwil1 during oogenesis using mRNA in situ analysis. We found that ChPiwil1was only expressed in developing germ cells such as oogonia and early vitellogenic oocytes, but not in mature oocytes (Fig 5D–5L). These results support a possible role of ChPiwil1 in female germline stem cell development. In mollusks, information on piwi homologues is still scarce. To our knowledge, this is the first time that a piwi homologue and its expression pattern during germline development were reported in mollusk. Further investigations are needed to demonstrate the precise role of piwi genes in molluscan germline development.
Fig 5. Analysis of Piwi genes identified in C.hongkongensis transcriptome.
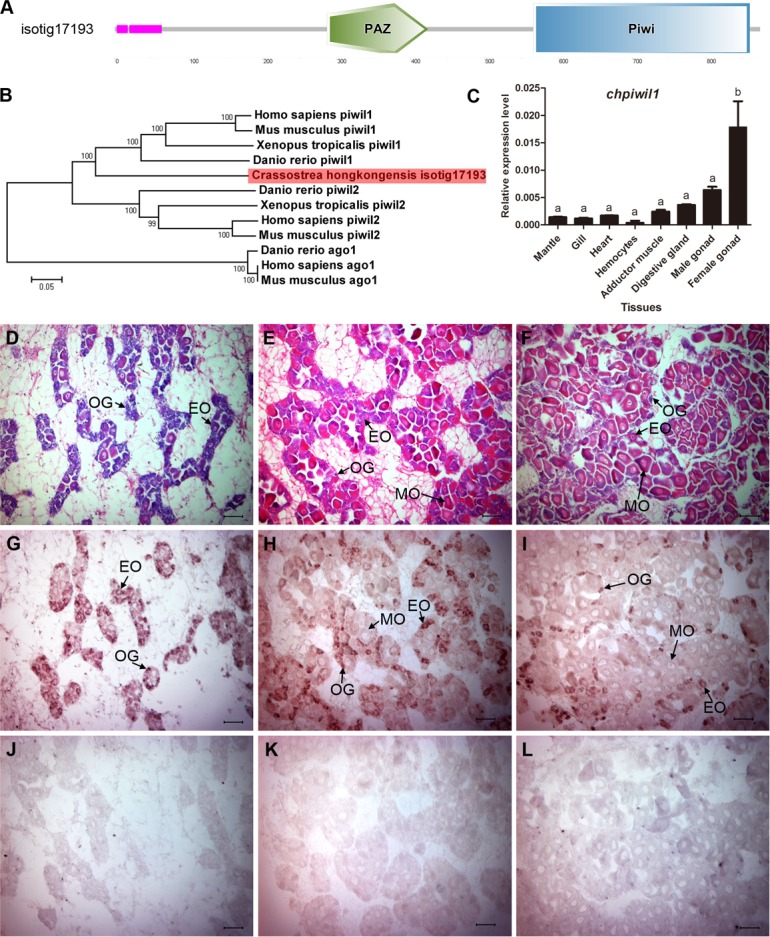
(A) Domain structure of the C.hongkongensis Piwi orthologs. (B) Phylogenetic tree constructed using the neighbor-joining method on the basis of the amino acid sequences alignment of C.hongkongensis Piwi ortholog with Piwi-like proteins of vertebrates. A total of 1000 bootstrap trials were run. Bootstrap values were indicated at each branch node. The scale bar represents an evolutionary distance of 0.1 amino acid substitutions per position. The transcript encoding C.hongkongensis Piwi ortholog, colored by red, was aligned to the clade of vertebrate Piwil1proteins. For the GenBank accession numbers of the reference sequences, see S3 Table. (C) Expression profile of ChPiwil1 in adult tissues by qRT-PCR with mean±s.e.m. as error bars (n = 5). (D-L) Expression profile of ChPiwil1during C.hongkongensis oogenesis by ISH. Histological analysis of C.hongkongensis female gonad at early developing stage (D), late developing stage (E) and mature stage (F) stained with HE. Expression profile of ChPiwil1during C.hongkongensis oogenesis by ISH with antisense (G-I) and sense probe (J-L). Positive cells are stained with brown or purple. OG: oogonia; EO: early vitellogenic oocyte; MO: mature ova.
Overall, the analysis of the C.hongkongensis transcriptome identified 84 germline development related genes. Among these, vasa, nanos and piwi are the three conserved core germline genes. The presence of vasa, nanos and piwi in C.hongkongensis indicates that it may share a conserved set of core genes with other animals for the generation of new germ cells in development.
Sex-determination/differentiation genes
Sex determination and sex differentiation are two major processes that occur during sexual development. While the sex determination process determines whether the bipotential primordium will develop into a testis or an ovary, the sex differentiation process occurs immediately after the sex determination process and involves the actual development of the testes or ovaries from the undifferentiated gonad [45].GO analyses of ‘Sex determination’ and ‘sex differentiation’ term annotation identified only 4 isotigs as orthologues of the GO0007530 (sex determination, S6 Table) annotation and 11 isotigs as orthologues of the GO0007544 (sex differentiation, S6 Table) annotation.
We then searched the C.hongkongensis transcriptome for such genes that were previously identified in model organisms (Table 4, S8 Table). Of the 49 genes examined, homologues were found for 20 genes, indicating that C.hongkongensis share some common sex determination/differentiation genes with other animals on the sequence level. To our surprise, we did not find homologues of vertebrate Dmrt1 (Doublesex and MAB-3 related transcription factor 1) and Sry (sex-determining region on the Y-chromosome), although we found homologues for other Dmrt and Sox family members (e.g., Dmrt2a, Sox4, Sox6 and Sox8). Dmrt1 is a transcription factor that contains a zinc finger DNA-binding motif (DM domain) and plays conserved roles in male sex determination and differentiation [46]. Members of this family also include the doublesex (dsx) gene in Drosophila and MAB-3 in C.elegans. Recently, an oyster Dmrt1 homologue was identified in C.gigas (CgDsx) and was found to be exclusively expressed in gonads [7]. Sry is a member of the Sox (Sry-related HMG box) protein family and is the single genetic trigger that regulates male sex determination and testicular differentiation [47]. A gene with a close relative to Sox30 and Sry in vertebrates has been reported in the C.gigas genome (CgSoxH) and was exclusively expressed in testis [7]. The absence of Dmrt1 and Sry homologues in our analysis may be a result of the low expression level of these genes in our samples or an artifact due to the incompleteness of the C.hongkongensis transcriptome dataset.
Table 4. Presence of sex determination/differentiation genes from Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, Mus musculus and Crassostrea hongkongensis.
| Gene common name | Species with homologues (homologue names) | ||||
|---|---|---|---|---|---|
| Fly (D) | Worm (C) | Fish (Dr) | Mouse (M) | Oyster(Ch) | |
| WT1 | yes | yes | |||
| Sf1 | yes | yes | |||
| CBX2 | yes | yes | yes | ||
| LHX9 | yes | yes | yes | ||
| EMX2 | yes | yes | |||
| GATA4 | yes | yes | yes | ||
| SRY | yes | ||||
| Sox9 | sox100B | yes | yes | ||
| FOG2 | yes | yes | |||
| AMH | yes | yes | |||
| DMRT1 | dsx | MAB-3 | yes | yes | |
| DMRT3 | yes | yes | |||
| DMRT6 | yes | ||||
| DHH | yes | yes | yes | ||
| MAP3K1 | yes | yes | yes | ||
| Map3k4 | yes | yes | yes | ||
| ATRX | dATRX | xnp-1 | yes | yes | |
| Fgf9 | yes | ||||
| Gadd45g | yes | yes | yes | ||
| Hhat | yes | yes | yes | yes | yes |
| Kdm3a | yes | yes | |||
| Dax1 | yes | yes | yes | ||
| Six1–Six4 | yes | yes | yes | yes | yes |
| Sox3 | yes | yes | |||
| Sox8 | yes | yes | |||
| Sox10 | yes | ||||
| GSDF | yes | ||||
| PDGF α and β | yes | ||||
| AMHR2 | yes | yes | |||
| AR | yes | yes | |||
| SRD5A1, SRD5A2, SRD5A3 | yes | yes | yes | ||
| CYP11B | yes | yes | |||
| WNT4 | yes | yes | yes | yes | |
| FOXL2 | yes | yes | yes | ||
| RSPO1 | yes | ||||
| β-catenin | armadillo | yes | yes | yes | |
| FST | yes | yes | yes | ||
| Cyp19A1 | yes | yes | |||
| ERa | yes | yes | yes | ||
| Xol-1 | yes | ||||
| Sdc | yes | yes | yes | yes | |
| Her | yes | yes | |||
| Tra | yes | yes | yes | yes | yes |
| Fem | yes | yes | yes | yes | |
| Fru | yes | ||||
| Sis | yes | ||||
| Run | yes | yes | yes | yes | yes |
| Sxl | yes | ||||
| Doa | yes | CLK | CLK | yes | |
C, Caenorhabditis elegans (nematode); D, Drosophila melanogaster (fruit fly); Dr, Danio rerio (zebrafish); M, Mus musculus (mouse);Ch, Crassostrea hongkongensis (Oyster)
To identify whether these 20 genes function in the C.hongkongensis reproduction process, we studied the expression profiles of these genes in different tissues by qRT-PCR. We found that 4 genes (ATRX, Foxl2, β-catenin and SRD5A1) were expressed at a higher level in female gonads than in other tissues, with Foxl2 specifically expressed in female gonads, suggesting that they may play important roles in female sex determination/differentiation or maintenance of female mature gonads in C.hongkongensis. The other 16 genes (Doa, ER a, Fem, FST, GATA4, Gadd45g, Hhat, LHX9, MAP3k1, Map3k4, Dax1, Run, Six1, Sox8, Tra and WNT4) did not show high expression levels in gonads (Fig 6), suggesting that they may not function in the C.hongkongensis reproduction process. However, we cannot rule out the possibility that they may exhibit high expression levels at earlier stages.
Fig 6. Heat map of sex determination/differentiation genes expressed at different adult tissues of C.hongkongensis.
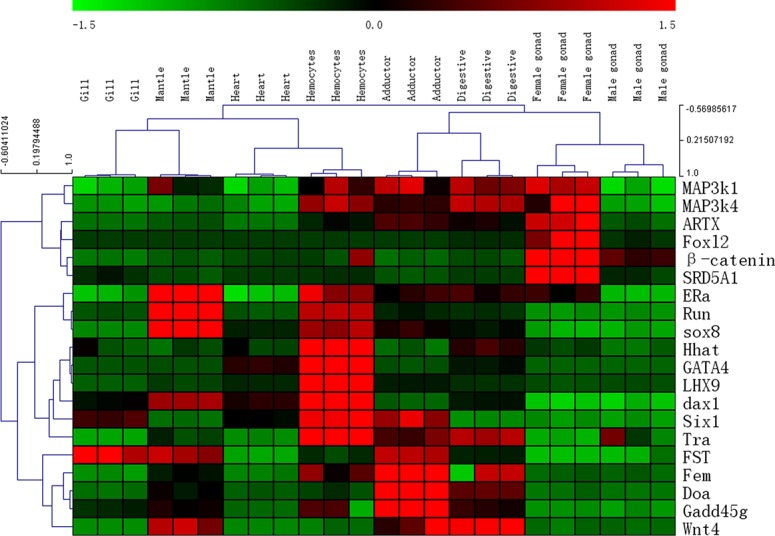
Genes showing similar expression profiles on all samples (columns) were clustered together. Color represents the normalized expression after variance-stabilizing transformation. Expression levels are depicted with a color scale, in which shades of red represent higher expression and shades of green represent lower expression.
α-thalassemia/mental retardation syndrome X-linked(ATRX) gene ATRX is a chromatin remodeling protein that plays a critical and conserved role in mammalian and nematode sexual differentiation [48,49]. Mutations in human ATRX cause mental retardation, craniofacial deformities, psychomotor failure, alpha-thalassemia as well as urogenital abnormalities ranging from undescended testes to testicular dysgenesis and female or ambiguous external genitalia [50]. In C. elegans, the ATRX ortholog (xnp-1) acts in concert with lin35 (the ortholog to mammalian retinoblastoma protein) to regulate target gene expression. Knockout of either xnp-1 or lin35 alone does not produce a phenotype. However, the double mutants were sterile with severe defects in gonadal development and a decrease in germ cell numbers in both male and female individuals [49]. The human ATRX contains an ADD (ATRX, DNMT3b and DNMT3L) zinc finger domain and a Sucrose Non Fermenting 2 (SNF2)-like DNA-dependent ATPase domain [51,52]. The ADD domain is composed of two zinc fingers (PHD-like and GATA-like zinc fingers) that bind to histone tails and mediate ATRX binding to chromatin. However, in C.elegans and D.melanogaster, the putative ATRX homolog only has a conserved SNF2-like DNA-dependent ATPase domain and lack the ADD domain [53,54]. However, little is known about ATRX in mollusks.
A sequence of 6,767 bp (isotig16330) encoding an ATRX ortholog was identified, showing an ORF of 5,607 bp. The deduced amino acid sequence is 1,868 aa long and contains an ADD domain and a SNF2-like DNA-dependent ATPase domain, suggesting that ATRX ortholog in C.hongkongensis has the same structural feature as mammalian ATRX rather than C.elegans and D.melanogaster ATRX homologs. This C.hongkongensis ATRX ortholog mRNA was named “Ch-ATRX”. Alignment of the amino acid sequence of the ADD domain from oyster ATRX ortholog with vertebrate ATRX indicated high conservation of a C2-C2 zinc finger and a C4-C4 zinc finger (Fig 7). Ch-ATRX is highly expressed in female gonads with relatively low expression levels in other organs. In mammals, ATRX was strongly expressed in both female and male gonads [48], but in our investigation, we observed that ChATRX exhibits higher mRNA levels in the ovary than in the testis, suggesting that ChATRX may play a more important role in ovary development in C.hongkongensis. The role of ATRX in the molluscan ovary remains unknown and awaits further investigation.
Fig 7. Amino acid sequence alignment of the ADD domain from C.hongkongensis ATRX ortholog with vertebrate ATRX.
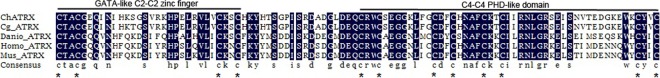
The highly conserved C2-C2 zinc finger and C4-C4 zinc finger are indicated by asterisks.
β-catenin is a subunit of the cadherin protein complex and acts as an intracellular signal transducer in the Wnt signaling pathway. Wnt4 and Rspo1, two components of the Wnt signaling pathway, activate β-Catenin, which in turn regulates the transcription of a variety of genes, among them important ovarian components, such as Wnt4 and Fst [55,56]. The expression of the stabilized form of β-Catenin in the developing mouse XY gonad leads to male-to-female sex-reversal [55]. In mollusks, a β-catenin ortholog has been identified in the Pacific oyster C. gigas (Cg-β-catenin). Cg-β-catenin is strongly expressed in mature female gonads with low expression levels in male gonads [57].
A 3,888 bp transcript encoding a β-cateninortholog (isotig11203) was identified in C.hongkongensis transcriptome, having an ORF of 2,499 bp in length. The deduced amino acid sequence is 832 aa long and contains an armadillo repeat region, which is a common feature of β-catenin in other species. Phylogenetic analysis based on the amino acid sequences of β-catenin from various species indicate that the C.hongkongensis β-catenin ortholog was closely related to Cg-β-catenin and was clustered into the molluscan β-catenin group (Figure A in S2 Fig). Therefore, we named this C.hongkongensis β-catenin ortholog (isotig11203) “Ch-β-catenin”. Ch-β-catenin mRNA expression, measured by real-time quantitative RT-PCR, was detected in all tested adult tissues, but was maximal in female gonads. The expression pattern of Ch-β-catenin was similar to that of Cg-β-catenin [57], suggesting a more important role of β-catenin in oyster female sex differentiation rather than male sex differentiation.
Forkhead box L2-related gene The forkhead box L2 gene (FoxL2), which encodes a winged helix/forkhead transcription factor, is a key gene in ovarian determination in vertebrates [58]. In adult mammals, FoxL2 is mainly expressed in the ovary where it functions to suppress genes involved in testis differentiation from early embryonic gonad differentiation throughout adult life [59,60,61]. Homologues of FoxL2 have also been reported in invertebrates [62,63,64]. In mollusks, a FoxL2 ortholog has been identified in C.gigas (CgFoxL2). CgFoxL2 is expressed in labial palps, female and male gonads, with significantly higher expression levels in female gonads [7,63].
In C.hongkongensis transcriptome, a 662 bp transcript (isotig29919) related to FoxL2 were identified by BlastX against the Swiss-prot and Nr database. The ORF is 641 bp encoding a 215 aa product that harbors a forkhead box domain. In phylogenetic analysis using amino acid sequences of FoxL1 and FoxL2 from various species, the product of isotig29919 was aligned to the clade of FoxL2 proteins and was closely related to CgFoxL2 (Figure B in S2 Fig). Therefore, we named isotig29919 “ChFoxL2”. ChFoxL2 was specifically expressed in ovary, suggesting a key role in ovary development. It will be very interesting to study the regulatory role of ChFoxL2 in female sex determination/differentiation of C.hongkongensis in the future.
5a-reductase1 gene (SRD5A1) 5a-reductase 1 enzyme, encoded by the SRD5A1 gene, has two important physiological functions in model animals: (i) catalyze the conversion of testosterone into a more potent androgen, dihydrotestosterone (DHT), which participates in the sexual differentiation processes [65]; and (ii) convert progesterone and deoxycorticosterone (DOC) to the irrespective 5-reduced derivatives, precursors of allopregnanolone and tetrahydroDOC, potent allosteric modulators of the γ-aminobutyricacid receptor(GABAA-R) [66], which participates in the regulation of various psychophysiological phenomena [67,68]. Homologues of SRD5A1 have been reported in human [69], rat [70], bird [71], fish [72] and frog [73]. The expression pattern of SRD5A1 was different among the different species. In the adult rat, SRD5A1was expressed at high levels in non androgen target tissues (e.g., liver, brain, ovary and skin) and at lower levels in theprostate, epididymis, seminal vesicles, testis [65,70]. In Xenopus laevis, SRD5A1 mRNA was expressed in all examined tissues with the highest expression in brain, gonads and kidney and lower expression in liver, heart and spleen [74].To date, there have been few reports on the expression pattern of SRD5A1 in mollusks and its role in molluscan reproduction.
In the transcriptome, we identified a transcript (isotig26744) that encodes 5a-reductase 1 enzyme (SRD5A1) with strong similarity to mouse SRD5A1 (E-value = 1.34E-82). The transcript was 897 bp long with an ORF of 804 bp, which encoded a protein of 267 amino acids. In phylogenetic analysis based on amino acid sequences of SRD5A1 orthologs, the product encoded by isotig26744 was clustered into the SRD5A1 group and was most closely related to C.gigasSRD5A (Figure C in S2 Fig). Therefore, the isotig26744 was identified for Ch-SRD5A1. Ch-SRD5A1 mRNA was expressed highly in female gonad. The high expression in the ovary supports a possible role of Ch-SRD5A1 in determining or promoting female-specific development.
Oocyte maturation pathway genes
The mechanism underlying oocyte maturation in oysters is poorly understood. In oyster C.gigas and C.hongkongensis, oocytes are naturally arrested at the prophase of meiosis I, undergo meiosis re-initiation and germinal vesicle breakdown (GVBD) upon environmental stimuli or hormonal stimulation, are secondarily arrested in metaphase I and become mature, fertilizable eggs [75]. Here, 41 genes of the oocyte maturation pathway, which include MAPK pathway genes, CDC2, cyclin B, polo-like kinase and CDC25 etc., were found in the C.hongkongensis transcriptome (S3 Fig). Stephano and Gould found that oocytes immediately after removal from the C.gigas ovary exhibit little MAPK activity, but this increases as the oocytes mature to metaphase I. When MAPK activation is inhibited, meiosis is abnormal, suggesting that MAPK plays an important role in C.gigas oocyte maturation [76]. In model organisms such as Xenopus, the entry into meiosis I depends on the activation of maturation promoting factor (MPF or Cdc2/cyclin B), which triggers GVBD. The MAPK pathway and the polo-like kinase/CDC25 pathway are responsible for the activation of MPF in meiosis [77]. However, in C.hongkongensis, further investigations are needed to determine the functional roles of genes identified here in the oocyte maturation process.
Overall, our results show that 454 sequencing of the C.hongkongensis transcriptome was useful in identifying reproduction related genes because a large number of reproduction-related genes and pathways have been identified in C.hongkongensis.
Characterization of SNPs and SSRs
Molecular markers such as SNPs or SSRs are the basis for genetic mapping and comparative genomic analysis, which are in turn used for the detection of quantitative trait loci (QTL) and for marker assisted selection (MAS) programs [78]. Although some microsatellite markers have been developed in C.hongkongensis [79,80,81], few studies have been conducted to investigate cDNA associated SNPs and SSRs in this species, despite the potential for targeting candidate genes.
A total of 94,056 SNPs and 3,357 indels were detected in 11,427 isotigs of the C.hongkongensis transcriptome. The overall frequency of all SNP types was 1 per 392bp.The proportions of transition substitutions were 30.3% for A/G and 30.2% for C/T, compared with smaller proportions of transversions for A/C (8.6%), G/T (8.3%),A/T (14.0%) and C/G (5.1%; Fig 8).
Fig 8. Classification of single nucleotide polymorphisms (SNPs) identified from the C.hongkongensis transcriptome.
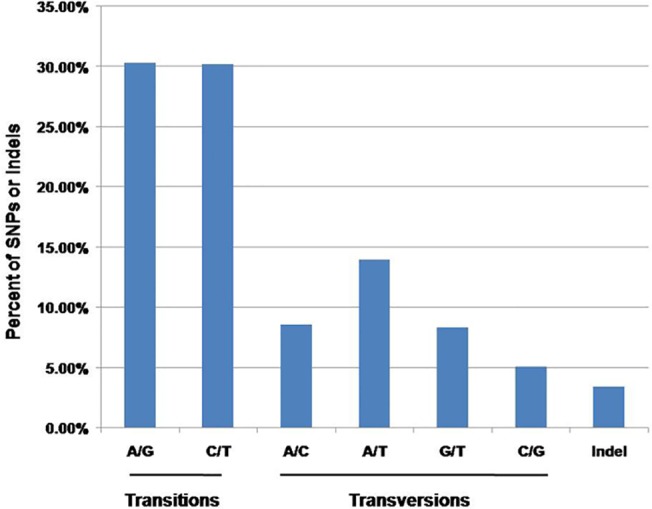
Transitions occurred more frequently than transversions. The overall frequency of all types of SNPs including indels was one per 392 bp.
In addition, our search revealed 1,452 isotigs containing 1,699 SSRs in the 22,829 isotigs, with 208 isotigs containing at least two SSRs. Of these, 789 showed significant hits in BlastX against the Swiss-prot database with an E-value cutoff of ≤ 1e-6 and were thus annotated. The frequency of EST-SSRs observed in the transcriptome was 6.4%, and the distribution density was one SSR per 21.7 kb of expressed sequences. The most abundant repeat type was AG (30.2%, 513) followed by AT (20.9%, 356), AC (11.4%, 194) and ATC (0.09%, 157). Regarding the length of the motif, dinucleotide microsatellites were the most common ones and hexanucleotides were the least abundant (Table 5). In addition, SSRs with a lower number of repeats were more common than those with a higher number of repeats, with the most common class being n = 6 (579 loci: 34.1%). Furthermore, 14.2% of loci contained more than 10 repeat units.
Table 5. Frequency distribution of SSRs by motif length in the C.hongkongensis transcriptome.
| SSR motif length | Repeat unit number | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 8 | 9 | 10 | >10 | Total | % | |
| Di | - | 439 | 187 | 107 | 62 | 43 | 225 | 1,063 | 62.5% |
| Tri | 331 | 119 | 39 | 28 | 16 | 10 | 10 | 553 | 32.5% |
| Tetra | 32 | 17 | 10 | 3 | 0 | 0 | 7 | 69 | 4.1% |
| Penta | 7 | 4 | 1 | 1 | 0 | 0 | 0 | 13 | 0.07% |
| Hexa | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.005% |
| Total | 371 | 579 | 237 | 139 | 78 | 53 | 242 | 1699 | 100% |
We next searched the seven genes (Chvasa, Ch-nanos, ChPiwil1, Ch-ATRX, Ch-β-catenin, ChFoxL2, Ch-SRD5A1) which were expressed highly in C.hongkongensis gonad for possible SNPs and SSRs. 17 SNPs were identified in Ch-ATRX, followed by 9 in Chvasa, 7 in Ch-nanos, 2 in Ch-SRD5A1 and 1 in Ch-β-catenin (S9 Table). No SNPs was detected in ChPiwil1 and ChFoxL2. One TCC type SSR was identified in Chvasa and no SSRs was identified in other genes.
The large number of potential molecular markers found in this study will be useful for gene mapping in this species and for comparative mapping and oyster evolutionary studies.
Conclusions
In conclusion, our study provided the first assembled transcriptome for C. hongkongensis, an economically important shellfish in South China. Based on GO, KEGG classification and literature-based searches for known reproduction-related genes, we conclude that our study captured a significant number of genes that may be involved in reproduction. The group of reproduction-related genes identified here constitutes a new tool for research on the bivalve reproduction process and provided insights into the origin and ancient characteristics of the reproduction-related genes. The large set of molecular markers discovered here will be useful for population studies and marker-assisted selection programs in C.hongkongensis aquaculture.
Supporting Information
(PPTX)
(PPTX)
(PPTX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(XLSX)
Acknowledgments
We thank Prof. Chaoqun Hu (South China Sea Institute of Oceanology, Chinese Academy of Sciences) for providing the facilities for sectioning and imaging.
Data Availability
Sequencing data are deposited in the NIH Short Read Archive database with Run accession number SRR949615.
Funding Statement
Support was provided by High Tech Research and Development Program of China (863 program) [http://www.863.gov.cn/] no.2012AA10A405-3 to YZ; National Natural Science Foundation of China [http://www.nsfc.gov.cn/] no.41306145 to YT; National Natural Science Foundation of China [http://www.nsfc.gov.cn/] no. 31101927 to JC; Pearl River S&T Nova Program of Guangzhou [http://www.gzsi.gov.cn/] no. 2013J2200095 to YZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Guo X, Allen SK Jr. Sex determination and polyploid gigantism in the dwarf surfclam (Mulinia lateralis Say). Genetics. 1994. December;138(4):1199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chavez-Villalba J, Soyez C, Huvet A, Gueguen Y, Lo C, Le Moullac G. Determination of Gender in the Pearl Oyster Pinctada Margaritifera . J Shellfish Res. 2011. August;30(2):231–40. [Google Scholar]
- 3. Lam K, Morton B. Mitochondrial DNA and morphological identification of a new species of Crassostrea (Bivalvia: Ostreidae) cultured for centuries in the Pearl River Delta, Hong Kong, China. Aquaculture. 2003. December 1;228(1–4):1–13. [Google Scholar]
- 4.Chinese Fishery Yearbook. 2012. Available: http://wenku.baidu.com/link?url=PueA5qaqZZv1zx-CwBdVEWig06g-7j2lt3v3_S_8pFLw0c-ZiOt-k2t37amy-qmDuahlD6LxWnKdReO9jSIcDEwvB79froobLj17N-c8MvO
- 5. Fabioux C, Huvet A, Lelong C, Robert R, Pouvreau S, Daniel JY, et al. Oyster vasa-like gene as a marker of the germline cell development in Crassostrea gigas . Biochem Biophys Res Commun. 2004. July 23;320(2):592–8. [DOI] [PubMed] [Google Scholar]
- 6. Matsumoto T, Masaoka T, Fujiwara A, Nakamura Y, Satoh N, Awaji M. Reproduction-related genes in the pearl oyster genome. Zoolog Sci. 2013. October;30(10):826–50. 10.2108/zsj.30.826 [DOI] [PubMed] [Google Scholar]
- 7. Zhang N, Xu F, Guo X. Genomic Analysis of the Pacific Oyster (Crassostrea gigas) Reveals Possible Conservation of Vertebrate Sex Determination in a Mollusc. G3 (Bethesda). 2014. September 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Teaniniuraitemoana V, Huvet A, Levy P, Klopp C, Lhuillier E, Gaertner-Mazouni N, et al. Gonad transcriptome analysis of pearl oyster Pinctada margaritifera: identification of potential sex differentiation and sex determining genes. BMC Genomics. 2014;15:491 10.1186/1471-2164-15-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Naimi A, Martinez AS, Specq ML, Mrac A, Diss B, Mathieu M, et al. Identification and expression of a factor of the DM family in the oyster Crassostrea gigas . Comp Biochem Physiol A Mol Integr Physiol. 2009. February;152(2):189–96. 10.1016/j.cbpa.2008.09.019 [DOI] [PubMed] [Google Scholar]
- 10. Hamano K, Awaji M, Usuki H. cDNA structure of an insulin-related peptide in the Pacific oyster and seasonal changes in the gene expression. J Endocrinol. 2005. October;187(1):55–67. [DOI] [PubMed] [Google Scholar]
- 11. Andrew MN, Dunstan RH, O'Connor WA, Van Zwieten L, Nixon B, MacFarlane GR. Effects of 4-nonylphenol and 17alpha-ethynylestradiol exposure in the Sydney rock oyster, Saccostrea glomerata: Vitellogenin induction and gonadal development. Aquat Toxicol. 2008. June 2;88(1):39–47. 10.1016/j.aquatox.2008.03.003 [DOI] [PubMed] [Google Scholar]
- 12. Alavi SM, Matsumura N, Shiba K, Itoh N, Takahashi KG, Inaba K, et al. Roles of extracellular ions and pH in 5-HT-induced sperm motility in marine bivalve. Reproduction. 2014. March;147(3):331–45. 10.1530/REP-13-0418 [DOI] [PubMed] [Google Scholar]
- 13. Bigot L, Zatylny-Gaudin C, Rodet F, Bernay B, Boudry P, Favrel P. Characterization of GnRH-related peptides from the Pacific oyster Crassostrea gigas . Peptides. 2012. April;34(2):303–10. 10.1016/j.peptides.2012.01.017 [DOI] [PubMed] [Google Scholar]
- 14. Matsumoto T, Nakamura AM, Mori K, Akiyama I, Hirose H, Takahashi Y. Oyster estrogen receptor: cDNA cloning and immunolocalization. Gen Comp Endocrinol. 2007. April;151(2):195–201. [DOI] [PubMed] [Google Scholar]
- 15. Wang Q, He M. Molecular characterization and analysis of a putative 5-HT receptor involved in reproduction process of the pearl oyster Pinctada fucata . Gen Comp Endocrinol. 2014. August 1;204:71–9. 10.1016/j.ygcen.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 16. Rodet F, Lelong C, Dubos MP, Costil K, Favrel P. Molecular cloning of a molluscan gonadotropin-releasing hormone receptor orthologue specifically expressed in the gonad. Biochim Biophys Acta. 2005. September 25;1730(3):187–95. [DOI] [PubMed] [Google Scholar]
- 17. Stewart MJ, Favrel P, Rotgans BA, Wang T, Zhao M, Sohail M, et al. Neuropeptides encoded by the genomes of the Akoya pearl oyster Pinctata fucata and Pacific oyster Crassostrea gigas: a bioinformatic and peptidomic survey. BMC Genomics. 2014;15:840 10.1186/1471-2164-15-840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Z, Zhang Q. Molecular cloning, characterization and expression of heat shock protein 70 gene from the oyster Crassostrea hongkongensis responding to thermal stress and exposure of Cu(2+) and malachite green. Gene. 2012. April 15;497(2):172–80. 10.1016/j.gene.2012.01.058 [DOI] [PubMed] [Google Scholar]
- 19. Zha G, Chen VP, Luk WK, Zou X, Choi RC, Tsim KW. Characterization of acetylcholinesterase in Hong Kong oyster (Crassostrea hongkongensis) from South China Sea. Chem Biol Interact. 2013. March 25;203(1):277–81. 10.1016/j.cbi.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y, Fu D, Yu F, Liu Q, Yu Z. Two catalase homologs are involved in host protection against bacterial infection and oxidative stress in Crassostrea hongkongensis . Fish Shellfish Immunol. 2011. December;31(6):894–903. 10.1016/j.fsi.2011.08.005 [DOI] [PubMed] [Google Scholar]
- 21. Ribas L, Pardo BG, Fernandez C, Alvarez-Dios JA, Gomez-Tato A, Quiroga MI, et al. A combined strategy involving Sanger and 454 pyrosequencing increases genomic resources to aid in the management of reproduction, disease control and genetic selection in the turbot (Scophthalmus maximus). BMC Genomics. 2013;14:180 10.1186/1471-2164-14-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen X, Zeng D, Xie D, Zhao Y, Yang C, Li Y, et al. Transcriptome analysis of Litopenaeus vannamei in response to white spot syndrome virus infection. Plos One. 2013;8(8):e73218 10.1371/journal.pone.0073218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bettencourt R, Pinheiro M, Egas C, Gomes P, Afonso M, Shank T, et al. High-throughput sequencing and analysis of the gill tissue transcriptome from the deep-sea hydrothermal vent mussel Bathymodiolus azoricus . BMC Genomics. 2010;11:559 10.1186/1471-2164-11-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Milan M, Coppe A, Reinhardt R, Cancela LM, Leite RB, Saavedra C, et al. Transcriptome sequencing and microarray development for the Manila clam, Ruditapes philippinarum: genomic tools for environmental monitoring. BMC Genomics. 2011;12:234 10.1186/1471-2164-12-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hou R, Bao Z, Wang S, Su H, Li Y, Du H, et al. Transcriptome sequencing and de novo analysis for Yesso scallop (Patinopecten yessoensis) using 454 GS FLX. Plos One. 2011;6(6):e21560 10.1371/journal.pone.0021560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Philipp EER, Kraemer L, Melzner F, Poustka AJ, Thieme S, Findeisen U, et al. Massively Parallel RNA Sequencing Identifies a Complex Immune Gene Repertoire in the lophotrochozoan Mytilus edulis . Plos One. 2012. March 20;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005. September 15;21(18):3674–6. [DOI] [PubMed] [Google Scholar]
- 28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. 2011. October;28(10):2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999. January 15;27(2):573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meyer E, Aglyamova GV, Wang S, Buchanan-Carter J, Abrego D, Colbourne JK, et al. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics. 2009;10:219 10.1186/1471-2164-10-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bettencourt R, Pinheiro M, Egas C, Gomes P, Afonso M, Shank T, et al. High-throughput sequencing and analysis of the gill tissue transcriptome from the deep-sea hydrothermal vent mussel Bathymodiolus azoricus . BMC Genomics. 2010. October 11;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clark MS, Thorne MAS, Vieira FA, Cardoso JCR, Power DM, Peck LS. Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genomics. 2010. June 7;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qin J, Huang Z, Chen J, Zou Q, You W, Ke C. Sequencing and de novo analysis of Crassostrea angulata (Fujian oyster) from 8 different developing phases using 454 GSFlx. Plos One. 2012;7(8):e43653 10.1371/journal.pone.0043653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feldmeyer B, Wheat CW, Krezdorn N, Rotter B, Pfenninger M. Short read Illumina data for the de novo assembly of a non-model snail species transcriptome (Radix balthica, Basommatophora, Pulmonata), and a comparison of assembler performance. BMC Genomics. 2011;12:317 10.1186/1471-2164-12-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang L, Li L, Zhu Y, Zhang G, Guo X. Transcriptome analysis reveals a rich gene set related to innate immunity in the Eastern oyster (Crassostrea virginica). Mar Biotechnol (NY). 2014. February;16(1):17–33. [DOI] [PubMed] [Google Scholar]
- 36. Wang S, Hou R, Bao Z, Du H, He Y, Su H, et al. Transcriptome sequencing of Zhikong scallop (Chlamys farreri) and comparative transcriptomic analysis with Yesso scallop (Patinopecten yessoensis). Plos One. 2013;8(5):e63927 10.1371/journal.pone.0063927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meng XL, Liu M, Jiang KY, Wang BJ, Tian X, Sun SJ, et al. De novo characterization of Japanese scallop Mizuhopecten yessoensis transcriptome and analysis of its gene expression following cadmium exposure. Plos One. 2013;8(5):e64485 10.1371/journal.pone.0064485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fabioux C, Pouvreau S, Le Roux F, Huvet A. The oyster vasa-like gene: a specific marker of the germline in Crassostrea gigas . Biochem Biophys Res Commun. 2004. March 19;315(4):897–904. [DOI] [PubMed] [Google Scholar]
- 39. Fabioux C, Corporeau C, Quillien V, Favrel P, Huvet A. In vivo RNA interference in oyster—vasa silencing inhibits germ cell development. FEBS J. 2009. May;276(9):2566–73. 10.1111/j.1742-4658.2009.06982.x [DOI] [PubMed] [Google Scholar]
- 40. Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, et al. Conserved role of nanos proteins in germ cell development. Science. 2003. August 29;301(5637):1239–41. [DOI] [PubMed] [Google Scholar]
- 41. Rabinowitz JS, Chan XY, Kingsley EP, Duan Y, Lambert JD. Nanos is required in somatic blast cell lineages in the posterior of a mollusk embryo. Current Biology. 2008. March 11;18(5):331–6. 10.1016/j.cub.2008.01.055 [DOI] [PubMed] [Google Scholar]
- 42. Kranz AM, Tollenaere A, Norris BJ, Degnan BM, Degnan SM. Identifying the germline in an equally cleaving mollusc: Vasa and Nanos expression during embryonic and larval development of the vetigastropod Haliotis asinina . J Exp Zool B Mol Dev Evol. 2010. June 15;314(4):267–79. 10.1002/jez.b.21336 [DOI] [PubMed] [Google Scholar]
- 43. Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997. June;124(12):2463–76. [DOI] [PubMed] [Google Scholar]
- 44. Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000. February;127(3):503–14. [DOI] [PubMed] [Google Scholar]
- 45. Hayes TB. Sex determination and primary sex differentiation in amphibians: genetic and developmental mechanisms. J Exp Zool. 1998. August 1;281(5):373–99. [PubMed] [Google Scholar]
- 46. Kopp A. Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 2012. April;28(4):175–84. 10.1016/j.tig.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tanaka SS, Nishinakamura R. Regulation of male sex determination: genital ridge formation and Sry activation in mice. Cell Mol Life Sci. 2014. December;71(24):4781–802. 10.1007/s00018-014-1703-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huyhn K, Renfree MB, Graves JA, Pask AJ. ATRX has a critical and conserved role in mammalian sexual differentiation. BMC Dev Biol. 2011;11:39 10.1186/1471-213X-11-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bender AM, Wells O, Fay DS. lin-35/Rb and xnp-1/ATR-X function redundantly to control somatic gonad development in C. elegans . Dev Biol. 2004. September 15;273(2):335–49. [DOI] [PubMed] [Google Scholar]
- 50. Badens C, Lacoste C, Philip N, Martini N, Courrier S, Giuliano F, et al. Mutations in PHD-like domain of the ATRX gene correlate with severe psychomotor impairment and severe urogenital abnormalities in patients with ATRX syndrome. Clin Genet. 2006. July;70(1):57–62. [DOI] [PubMed] [Google Scholar]
- 51. Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996. December;5(12):1899–907. [DOI] [PubMed] [Google Scholar]
- 52. Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S, et al. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum Mol Genet. 2011. June 1;20(11):2195–203. 10.1093/hmg/ddr107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lopez-Falcon B, Meyer-Nava S, Hernandez-Rodriguez B, Campos A, Montero D, Rudino E, et al. Characterization of the Drosophila Group Ortholog to the Amino-Terminus of the Alpha-Thalassemia and Mental Retardation X-Linked (ATRX) Vertebrate Protein. PLoS One. 2014;9(12):e113182 10.1371/journal.pone.0113182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Villard L, Fontes M, Ewbank JJ. Characterization of xnp-1, a Caenorhabditis elegans gene similar to the human XNP/ATR-X gene. Gene. 1999. August 5;236(1):13–9. [DOI] [PubMed] [Google Scholar]
- 55. Maatouk DM, DiNapoli L, Alvers A, Parker KL, Taketo MM, Capel B. Stabilization of beta-catenin in XY gonads causes male-to-female sex-reversal. Hum Mol Genet. 2008. October 1;17(19):2949–55. 10.1093/hmg/ddn193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chassot AA, Ranc F, Gregoire EP, Roepers-Gajadien HL, Taketo MM, Camerino G, et al. Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum Mol Genet. 2008. May 1;17(9):1264–77. 10.1093/hmg/ddn016 [DOI] [PubMed] [Google Scholar]
- 57. Santerre C, Sourdaine P, Adeline B, Martinez AS. Cg-SoxE and Cg-beta-catenin, two new potential actors of the sex-determining pathway in a hermaphrodite lophotrochozoan, the Pacific oyster Crassostrea gigas. Comp Biochem Physiol A Mol Integr Physiol. 2014. January;167:68–76. 10.1016/j.cbpa.2013.09.018 [DOI] [PubMed] [Google Scholar]
- 58. Georges A, Auguste A, Bessiere L, Vanet A, Todeschini AL, Veitia RA. FOXL2: a central transcription factor of the ovary. J Mol Endocrinol. 2014. February;52(1):R17–33. 10.1530/JME-13-0159 [DOI] [PubMed] [Google Scholar]
- 59. Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nature Genetics. 2001. February;27(2):159–66. [DOI] [PubMed] [Google Scholar]
- 60. Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011. August 4;476(7358):101–4. 10.1038/nature10239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009. December 11;139(6):1130–42. 10.1016/j.cell.2009.11.021 [DOI] [PubMed] [Google Scholar]
- 62. Adell T, Muller WE. Isolation and characterization of five Fox (Forkhead) genes from the sponge Suberites domuncula . Gene. 2004. June 9;334:35–46. [DOI] [PubMed] [Google Scholar]
- 63. Naimi A, Martinez AS, Specq ML, Diss B, Mathieu M, Sourdaine P. Molecular cloning and gene expression of Cg-Foxl2 during the development and the adult gametogenetic cycle in the oyster Crassostrea gigas . Comp Biochem Physiol B Biochem Mol Biol. 2009. September;154(1):134–42. 10.1016/j.cbpb.2009.05.011 [DOI] [PubMed] [Google Scholar]
- 64. Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: Phylogeny and embryonic expression. Developmental Biology. 2006. December 1;300(1):49–62. [DOI] [PubMed] [Google Scholar]
- 65. Russell DW, Wilson JD. Steroid 5 alpha-reductase: two genes/two enzymes. Annu Rev Biochem. 1994;63:25–61. [DOI] [PubMed] [Google Scholar]
- 66. Mellon SH, Griffin LD. Neurosteroids: biochemistry and clinical significance. Trends Endocrinol Metab. 2002. Jan-Feb;13(1):35–43. [DOI] [PubMed] [Google Scholar]
- 67. Segovia S, del Cerro MC, Ortega E, Perez-Laso C, Rodriguez-Zafra C, Izquierdo MA, et al. Role of GABAA receptors in the organization of brain and behavioural sex differences. Neuroreport. 1996. November 4;7(15–17):2553–7. [DOI] [PubMed] [Google Scholar]
- 68. Patte-Mensah C, Kibaly C, Mensah-Nyagan AG. Substance P inhibits progesterone conversion to neuroactive metabolites in spinal sensory circuit: a potential component of nociception. Proc Natl Acad Sci U S A. 2005. June 21;102(25):9044–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thiele S, Hoppe U, Holterhus PM, Hiort O. Isoenzyme type 1 of 5alpha-reductase is abundantly transcribed in normal human genital skin fibroblasts and may play an important role in masculinization of 5alpha-reductase type 2 deficient males. European Journal of Endocrinology. 2005. June;152(6):875–80. [DOI] [PubMed] [Google Scholar]
- 70. Normington K, Russell DW. Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for distinct physiological functions. J Biol Chem. 1992. September 25;267(27):19548–54. [PubMed] [Google Scholar]
- 71. Soma KK, Bindra RK, Gee J, Wingfield JC, Schlinger BA. Androgen-metabolizing enzymes show region-specific changes across the breeding season in the brain of a wild songbird. Journal of Neurobiology. 1999. November 5;41(2):176–88. [PubMed] [Google Scholar]
- 72. Pasmanik M, Callard GV. Changes in brain aromatase and 5 alpha-reductase activities correlate significantly with seasonal reproductive cycles in goldfish (Carassius auratus). Endocrinology. 1988. April;122(4):1349–56. [DOI] [PubMed] [Google Scholar]
- 73. Andersson S, Bishop RW, Russell DW. Expression cloning and regulation of steroid 5 alpha-reductase, an enzyme essential for male sexual differentiation. J Biol Chem. 1989. September 25;264(27):16249–55. [PMC free article] [PubMed] [Google Scholar]
- 74. Urbatzka R, Lutz I, Kloas W. Aromatase, steroid-5-alpha-reductase type 1 and type 2 mRNA expression in gonads and in brain of Xenopus laevis during ontogeny. Gen Comp Endocrinol. 2007. Aug-Sep;153(1–3):280–8. [DOI] [PubMed] [Google Scholar]
- 75. Colas P, Dube F. Meiotic maturation in mollusc oocytes. Semin Cell Dev Biol. 1998. October;9(5):539–48. [DOI] [PubMed] [Google Scholar]
- 76. Stephano JL, Gould MC. MAP kinase, a universal suppressor of sperm centrosomes during meiosis? Dev Biol. 2000. June 15;222(2):420–8. [DOI] [PubMed] [Google Scholar]
- 77. Frank-Vaillant M, Haccard O, Ozon R, Jessus C. Interplay between Cdc2 kinase and the c-Mos/MAPK pathway between metaphase I and metaphase II in Xenopus oocytes. Dev Biol. 2001. March 1;231(1):279–88. [DOI] [PubMed] [Google Scholar]
- 78. Liu ZJ, Cordes JF. DNA marker technologies and their applications in aquaculture genetics (vol 238, pg 1, 2004). Aquaculture. 2004. December 20;242(1–4):735–6. [Google Scholar]
- 79. Li L, Xiao S, Yu Z. Development of twenty-six microsatellite loci from Crassostrea hongkongensis and cross-species amplification in two closely related species. J Genet. 2011. August;90(2):e58–61. [PubMed] [Google Scholar]
- 80. Xia JJ, He XC, Yu ZN. Isolation and characterization of fourteen novel microsatellite loci in the Hong Kong oyster, Crassostrea hongkongensis . Conserv Genet. 2009. December;10(6):1829–32. [Google Scholar]
- 81. Li L, Yu ZN. Isolation and characterization of 24 microsatellite loci in oyster Crassostrea hongkongensis . Conserv Genet Resour. 2010. September;2:93–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PPTX)
(PPTX)
(PPTX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
Sequencing data are deposited in the NIH Short Read Archive database with Run accession number SRR949615.