Abstract
Protein misfolding neurodegenerative diseases arise through neurotoxicity induced by aggregation of host proteins. These conditions include Alzheimer’s disease, Huntington’s disease, Parkinson’s disease, motor neuron disease, tauopathies and prion diseases. Collectively, these conditions are a challenge to society because of the increasing aged population and through the real threat to human food security by animal prion diseases. It is therefore important to understand the cellular and molecular mechanisms that underlie protein misfolding-induced neurotoxicity as this will form the basis for designing strategies to alleviate their burden. Prion diseases are an important paradigm for neurodegenerative conditions in general since several of these maladies have now been shown to display prion-like phenomena. Increasingly, cell cycle activity and the DNA damage response are recognised as cellular events that participate in the neurotoxic process of various neurodegenerative diseases, and their associated animal models, which suggests they are truly involved in the pathogenic process and are not merely epiphenomena. Here we review the role of cell cycle activity and the DNA damage response in neurodegeneration associated with protein misfolding diseases, and suggest that these events contribute towards prion-induced neurotoxicity. In doing so, we highlight PrP transgenic Drosophila as a tractable model for the genetic analysis of transmissible mammalian prion disease.
Keywords: Neurodegenerative disease, Protein misfolding, Prion, Transmissible, Cell cycle, DNA repair, Chromatin, PrP transgenic Drosophila
Core tip: It is important to understand the cellular and molecular mechanisms of protein misfolding-induced neurotoxicity in order to combat conditions such as Alzheimer’s, Huntington’s, Parkinson’s, and motor neuron disease, tauopathies and prion diseases. Here, we review the role of cell cycle activity and the DNA damage response in neurodegeneration associated with protein misfolding diseases, including prion diseases. In doing so, we highlight PrP transgenic Drosophila as a tractable model of transmissible mammalian prion disease. Our review provides a new impetus to the study of prion diseases, which are increasingly seen as an important paradigm for neurodegenerative conditions in general.
INTRODUCTION
While many diseases can cause degeneration of nervous system tissue, including human immunodeficiency virus infection and acquired immune deficiency syndrome, multiple sclerosis or rabies, the designation of protein misfolding neurodegenerative disease is typically assigned to those induced by aberrant folding and aggregation of disease-specific host proteins. These conditions, which include Alzheimer’s disease, Huntington’s disease, Parkinson’s disease, motor neuron disease, tauopathies and prion diseases, are invariably fatal as there are no known treatments[1,2]. Each of these conditions is characterised by the misfolding of a disease-specific protein[3] and accumulation of misfolded protein in the brain is central to the pathological process that typically manifests as synaptic loss, neuronal dysfunction, with resultant clinical symptoms. Prion diseases include scrapie of sheep, bovine spongiform encephalopathy (BSE) of cattle, together with Creutzfeldt-Jakob disease (CJD) and fatal familial insomnia (FFI) in humans[4]. Prion diseases are an important paradigm for protein misfolding neurodegenerative conditions in general since Alzheimer’s, Huntington’s, Parkinson’s and motor neuron disease, as well as tauopathies all possess features of prion-like transmission in experimental settings, evidenced by transcellular spread of misfolded disease-specific protein[5]. However, prion diseases are unique since they are transmissible between individuals of the same and different species, that sometimes occurs unintentionally. Protein misfolding neurodegenerative diseases typically cause clinical disease late in life and are therefore a major concern to society because of the increasing size of the ageing population. In addition, prion diseases are a significant concern to food security since they occur in animals destined for human consumption. Understanding the mechanism of neurotoxicity induced by protein misfolding will allow the design of strategies to alleviate the burden of these conditions.
Many aspects of prion-induced neurotoxicity remain incompletely understood. During prion diseases the normal host protein PrPC is converted into the abnormal form, PrPSc, the transmissible prion agent[4,6] (Figure 1). This conversion event appears to be an essential requirement for prion disease neurotoxicity, evidenced by the failure of exogenous PrPSc to cause pathology in brain tissue devoid of PrPC[7,8] and the reversal of neurodegeneration when PrPC expression is ablated during prion infection[9-11]. The essential requirement for PrP expression in prion-induced neurotoxicity may suggest that an intermediate in the conversion of PrPC to PrPSc is the neurotoxic agent[12,13]. Alternatively, neurotoxicity may result from an interference with the normal biosynthesis and metabolism of PrPC mediated by the presence of PrPSc[14]. For example, PrP can accumulate in the cytosol in a misfolded form when proteasomal activity is compromised[15,16] and cytosolic PrP has been reported to be neurotoxic in some neurons[17-20]. A feature of prion-induced neurotoxicity is its effect on protein synthesis. For example, it has been shown that accumulation of PrPSc in cells[21] and mice[22] with an ongoing prion infection triggers over-activation of the PERK/eIF2a branch of the unfolded protein response. This in turn leads to persistently high levels of phosphorylated eIF2a and consequently a block of protein translation. Pharmacological inhibition of PERK can reverse the prion disease-induced block in protein synthesis and alleviate this toxic phenotype despite the continued accumulation of PrPSc[23].
Figure 1.
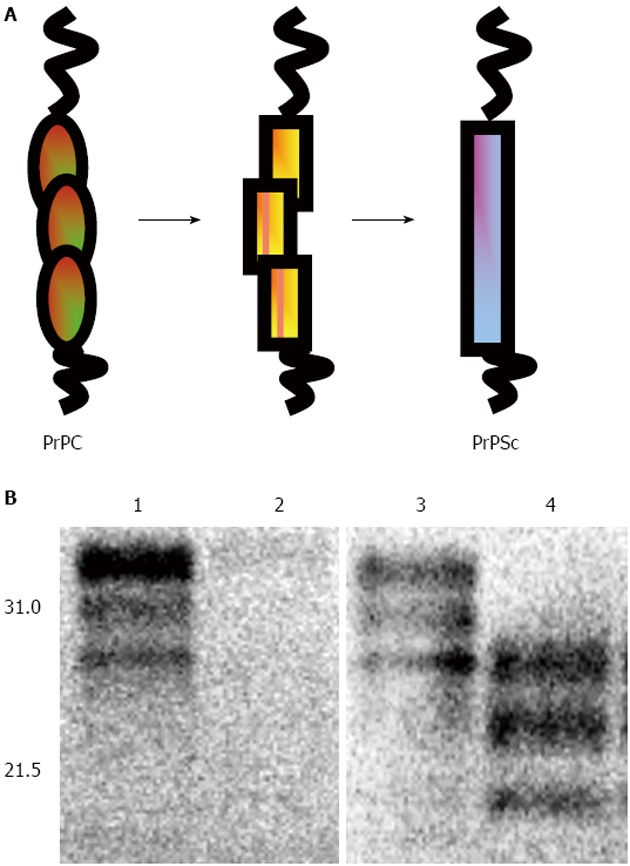
Conversion of PrPC into PrPSc. A: Schematic diagram of the conversion of PrPC into PrPSc. A major structural event occurs in the C-terminal domain of PrPC as it converts from a predominantly α-helical form into one enriched for β-sheet. This conformational change may involve the formation of intermediate structures of the protein; B: Western blot detection of ovine PrP. VRQ/VRQ sheep brain homogenate from animals that were scrapie-free (tracks 1 and 2) or scrapie-infected (tracks 3 and 4) were pre-treated with (tracks 2 and 4) or without (tracks 1 and 3) PK at 32 μg/mL at 37 °C for 30 min and the products analysed by SDS/PAGE, and Western blot probed with anti-PrP monoclonal antibody 683. Molecular weight markers (in kDa) are shown on the left (Reproduced by kind permission of CAB Reviews).
The value of these discoveries would be amplified by a more complete understanding of the sequence of cellular events that occur during the early stages of prion disease. This applies particularly to those acting prior to the onset of, and which may lead to, inhibition of protein synthesis. Knowledge in this area will be of fundamental importance to the understanding of prion biology per se and facilitate the search for early acting genetic modifiers of the neurotoxic process associated with these conditions. Interestingly, a number of reports have documented cell cycle activity and the DNA damage response (DDR) in post mitotic, terminally differentiated neurons during various neurodegenerative diseases[24-27], which represent potential candidates for such early acting pathways. This appears paradoxical since these are events traditionally associated with dividing cells. Here we discuss a potential role of cell cycle activity and DDR in prion-induced neurotoxicity. In support of this viewpoint, we present a novel Drosophila model of transmissible mammalian prion disease that provides a new animal system to study protein misfolding disease, one that combines the robust tools of experimental prion disease and fly genetics.
DDR
During the cell cycle, proliferating cells replicate their DNA and undergo division. This process is a highly organised series of cellular events that are tightly coordinated through the phase-specific expression of positive and negative regulatory proteins. Various quality control checkpoints operate to ensure faithful progression through the cell cycle. In addition to DNA replication errors, all cells whether proliferating or not, are constantly exposed to stimuli that can induce damage to DNA. These genotoxic stimuli may arise from exogenous events such as exposure to irradiation or carcinogens, or alternatively from endogenous events such as intracellular metabolism and associated reactive oxygen species (ROS)[28-30].
DNA damage in metazoan cells is deleterious: it may initiate mutagenesis or chromosomal re-arrangements that result in de-regulated cell cycle activity and neoplasia, or aberrant gene expression concomitant with cellular dysfunction and senescence or cell death[31-33]. In order to avoid these hazardous effects, cells have evolved a variety of molecular mechanisms for the repair of DNA damage[34]. For example, base excision repair (BER) is used to correct oxidative lesions[35] while nucleotide excision repair (NER) can excise UV light-induced thymidine dimers[36]. Single strand breaks (SSBs) in DNA, which may occur through ROS-mediated lesions or intermediates in BER, are repaired by polB and various ligases[37]. Double strand breaks (DSBs), that can arise through failures in DNA transcription or replication, are repaired by two different mechanisms: non-homologous end joining (NHEJ), which is error prone, or homologous recombination (HR), which is error-free but is restricted to the S/G2 phase of the cell cycle in dividing cells[38]. DSBs in DNA arise relatively infrequently, though are particularly hazardous as they can induce a significant loss of genomic integrity[39].
The maintenance of genome integrity is critical to organismal function and survival. As a consequence, cells co-ordinate an elaborate set of mechanisms that function in the surveillance and repair of DNA lesions with cell cycle progression. These integrated pathways are collectively referred to as the DNA damage response (DDR). In proliferating cells, checkpoint control mechanisms mediate cell cycle arrest to allow DNA repair when damage is detected, although senescence or apoptosis may ensue in the case of extensive lesions[40-42]. In contrast, post mitotic terminally differentiated neurons appear to display a lower capacity for DNA repair than proliferating cells, and they are thought to accumulate and tolerate comparatively high levels of DNA damage, since they are unable to replace damaged cells by division[43,44]. However, increasing evidence suggests that cell cycle activity and DDR are features of post mitotic neurons in neurodegenerative conditions[25-27,45,46]. For example, post mitotic neurons, when exposed to genotoxic stimuli, can replicate DNA and initiate apoptosis associated with cell cycle activation[47]. In addition, evidence of cell cycle activity and DNA damage can be found in natural and experimental hosts undergoing protein misfolding diseases, such as Alzheimer’s disease[48-51]; amyotrophic lateral sclerosis[52,53]; Huntington’s disease[54,55] and Parkinson’s disease[56-58].
THE CONTRIBUTION OF DNA DAMAGE AND DDR TO NEUROTOXICITY
Neurons like all other cell types are subject to a variety of stimuli that can potentially induce deleterious DNA damage. In dividing cells DNA damage activates cell cycle arrest concomitant with DDR so that the integrity of the cellular genome is maintained between successive generations. A major cell cycle checkpoint control operates at the G2/M interface to allow for DNA damaged during replication to be repaired prior to mitosis. Since post mitotic neurons are unable to divide, the expression of cell cycle associated genes in these cells may promote the DDR and facilitate access to DNA for repair in order to maintain genome integrity and appropriate regulation of gene expression. An emerging view is that structural modulation of chromatin associated with these processes, together with genome integrity, have a major influence on the neurotoxic process in post mitotic neurons during neurodegenerative disease[27,59]. In this context, important unanswered questions include: Do the same processes and events also occur in protein misfolding neurodegenerative diseases? And if so, what precisely are the molecular mechanisms that confer neurotoxicity and that culminate in neuronal dysfunction and neurodegeneration?
Chromatin is a repeat structure of nuclear DNA and histone proteins with nucleosomes representing the fundamental core unit[60,61]. The structure of chromatin is strongly influenced by post translational modifications of the histone proteins through the addition of various chemical groupings including phosphate, acetyl or methyl moieties[62]. In addition, sequence variants of core histone proteins (e.g., H2A.X) exist that further enhance chromatin structural diversity[63]. Chemical modification of histones, or the inclusion of their sequence variants, influence nucleosome-DNA or inter-nucleosome interactions and thereby regulate the degree of chromatin compaction and consequentially DNA transcriptional activity. Heterochromatin is relatively compacted and transcriptionally silent, whereas euchromatin is a more relaxed and open structure that is permissive for gene activation[64-67]. Chromatin structure and its modulation are therefore fundamental features in the maintenance of DNA integrity and regulation of gene expression.
DNA contained in compacted chromatin is relatively well protected from genotoxic stimuli and is typically inaccessible to transcription and DDR machinery. During DDR, chromatin undergoes transient dis-aggregation at the sites of DNA lesion to facilitate access of repair and cell cycle checkpoint proteins[68-70]. In some cases of DNA repair, chromatin modulation may be quite extensive and extend over several kilobases[71]. Since open chromatin is evident in regions of actively transcribed DNA, heterochromatin relaxation in response to DDR can trigger aberrant gene expression of normally silenced regions of the genome. Indeed, it has been shown that wide spread loss of heterochromatin occurs in Drosophila and mouse tauopathy models (tau transgenics), and human Alzheimer’s disease, and that this is associated with aberrant gene expression in CNS neurons[72]. Conversely, genetic rescue of tau-induced heterochromatin loss substantially reduced tau-induced neurodegeneration in Drosophila. It has been postulated that post mitotic neurons undergoing DDR and associated changes in chromatin organisation, may have the potential to revert to a de-differentiated state, and that this might be linked to activation of apoptotic pathways[73,74]. Mechanistically, oxidative stress and subsequent DNA damage were identified as causes of heterochromatin loss in tau neurotoxicity[72]. These studies suggest an etiological progression from neurotoxic stimuli to chromatin-mediated gene regulation and subsequent neurodegeneration.
General instability of the cellular genome, as a consequence of damage to mitochondrial or nuclear DNA, or to chromatin, is also a potential cause of neurotoxicity[75]. Since post mitotic terminally differentiated neurons are unable to divide, these cells are forced to endure genotoxic insults. However, if the level of DNA damage exceeds the capacity of the DDR, or if DDR function is compromised, mutations and incorrect repair may lead to inappropriate DNA metabolism and, de-regulated gene expression or harmful mutations[32]. This view is supported by the correlation between neurodegeneration and sensitivity to DNA damage and/or DDR deficiencies[76-81]. DNA damage that compromises mitochondrial function could lead to disturbances in the cellular energy balance and have a detrimental effect on neuronal function including synaptic defects, as occurs in various inherited neurological disorders[82]. Since the brain has a high metabolic activity neurons are thought to be particularly prone to oxidative stress, a recognised cause for DNA damage. Oxidative stress and mitochondrial dysfunction are increasingly implicated in protein misfolding-induced neurodegeneration although the molecular events of this association have not yet been defined[83]. Mitochondria are the principal source of cellular ROS and mitochondrial DNA is particularly sensitive to ROS-mediated damage[84]. The mutation rate of mitochondrial DNA, which lacks histone proteins, is > 15 fold higher than that of nuclear DNA[85]. Mutations in mitochondrial DNA can perturb the expression and function of oxidative phosphorylation complexes and thereby precipitate mitochondrial dysfunction, which in turn may lead to accelerated ROS generation[86,87].
Many studies have shown that ageing, a major risk factor for neurodegenerative disease, is associated with an accumulation of DNA lesions in the mature brain. DNA lesions may additionally arise from an age-dependent reduction in DNA repair capacity[88] and contribute to a reduction in genome integrity[43,89]. These DNA lesions, which are envisaged to occur in individual neurons, may result in the expression of mutant proteins that either fold or traffic incorrectly. This will result in an increasing demand on the cellular protein quality control machinery that functions to detect and triage these molecules, a situation already exacerbated in the case of protein misfolding diseases. In this situation, activation of the unfolded protein response may occur in order to attempt to maintain protein homeostasis[21,22]. The effects of aberrant misfolded protein accumulation that arise in protein misfolding diseases presumably enhance DNA damage and accelerate the loss of genome integrity and thereby promote the onset of neurodegenerative disease.
CELL CYCLE-ASSOCIATED PROTEINS WITH A ROLE AT THE SYNAPSE
Mature nerve cells are derived from neural progenitors that undergo proliferation, exit the cell cycle and mature into terminally differentiated neurons. Under normal circumstances, post mitotic neurons do not participate in any further cell cycle activity. Any attempt by post mitotic neurons to undergo cell cycle re-entry is considered to be detrimental to these cells. However, it has become evident that terminally differentiated neurons express a variety of proteins with important roles in cell cycle regulation that have a normal function in diverse post mitotic neuronal events under physiological conditions[90]. Significantly, some of these cell cycle-associated proteins localise to synapses in post mitotic neurons. For example, the Orc2-5 core subunits of the origin of recognition complex (Orc), which is key to initiating DNA replication, are highly expressed in differentiated mammalian neurons. Orc3 and Orc5 are enriched in the postsynaptic dendritic compartment, and regulate the dendritic filopodia and spine formation[91]. The anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase, locates to both pre- and postsynaptic sites in post mitotic neurons, regulating synaptic terminal growth and differentiation as well as synapse formation and function (reviewed in[92]). Other cell cycle associated gene products implicated in regulating synaptic function include the PI3Kinase family member ataxia telangiectasia mutated (ATM), which in post mitotic neurons associates with synaptic vesicle proteins[93], and Cyclin E that acts as a repressor of the synaptic regulator Cdk5[94].
While it is accepted that these various proteins, initially discovered as central to cell division, can have additional roles in post mitotic cells, it remains unclear whether dysregulation of their expression or function is linked to neurotoxicity and cell death in protein misfolding neurodegenerative diseases[24]. One suggestion has been that synaptic loss early in neurodegenerative conditions, results in upregulation of cell cycle-associated gene expression in a bid to maintain synaptic function and plasticity, but that this might lead to inappropriate action of these proteins in the nucleus, promoting neuronal dedifferentiation and apoptosis[24]. For example, shuttling of Cdk5 from the nucleus to cytoplasm has been postulated as critical for the breakdown of the post mitotic state in neurons[95]. Alternatively, it is conceivable that dysregulation of cell cycle-associated proteins at the synapse and concomitant sub-optimal synaptic communication may lead to increased metabolism as neurons struggle to remain within their homeostatic activity range. This in turn could lead to increased production of ROS, with an ensuing cycle of genotoxicity and associated dysregulation of gene expression.
CELL CYCLE ACTIVITY AND DDR IN PRION-INDUCED NEUROTOXICITY?
It is not yet established whether cell cycle activity and DDR are features of prion-mediated neurotoxicity. Evidence this might be the case derives from observations of mammalian models of prion disease. For example, nuclear accumulation of proliferating cell nuclear antigen (PCNA) and phosphorylated histone H2A.X proteins, which in other cell types are indicative of DNA replication and/or repair, have been detected in CNS neurons of mice that model familial CJD and FFI prion diseases[96]. In addition, the brains of scrapie-affected hamsters show evidence of cell cycle activity with an increase in the proteins polo-like kinase (PLK) 1 and cyclin B1, and a decrease of PLK3 and Cdc25C[97]. PLKs, which function as key regulators of the cell cycle and its checkpoint response to genotoxic stress, are regulated by synaptic activity in post mitotic neurons[98]. Prion infectivity experiments in vivo have shown that mice deficient in BER activity displayed an accelerated clinical course of prion disease as compared to wild type animals[99]. These various animal models of prion disease are supportive of the view that DNA damage plays a pivotal role in prion-induced neurotoxicity. It will be important to verify this is the case, in order to determine the extent of commonality in the mechanism(s) of neurotoxicity between different neurodegenerative conditions and prion diseases. This is underlined by the fact that bona fide prion diseases are seen as important paradigms for other protein misfolding diseases, and common underlying mechanisms would suggest the possibility of common therapeutic strategies for these presently invariably fatal diseases. However, prion diseases are difficult to study in their natural hosts, such as ruminants and humans, because these diseases can take many years to develop, resulting in progress being slow and cumbersome[4]. In addition, the natural forms of prion diseases tend to occur in outbred populations that render genetic analysis of complex biochemical pathways difficult. Even in the more tractable experimental system of mouse models, the significant expenses of time and husbandry restrict the scope of genetic experimentation for dissection of prion disease mechanisms.
A DROSOPHILA MODEL OF TRANSMISSIBLE MAMMALIAN PRION DISEASE
In order to circumvent the difficulties associated with the genetic analysis of prion diseases in their natural hosts, we have established Drosophila as a new tractable animal model of transmissible mammalian prion disease. Importantly, because of the high evolutionary conservation of most cellular signaling pathways and processes, our Drosophila model system allows exploitation of the power of fly genetics to probe the mechanisms of prion-induced neurotoxicity.
We have used pUAST/PhiC31-mediated site-directed germ line transformation to generate Drosophila transgenic for topological and polymorphic variants of ovine PrP under expression control of the bipartite UAS-GAL4 system[100-102]. The topological variants of ovine PrP were targeted to the plasma membrane, to the cytosol, or for secretion. Site-specific PCR using genomic DNA from ovine PrP transgenic flies as substrate, together with DNA sequence analysis, was used to confirm that a single copy of each PrP transgene had been inserted at a single site in the genome of each appropriate fly line. Expression control of ovine PrP in Drosophila via the UAS-GAL4 system allowed the prion protein to be targeted to defined cell populations during a specific period of development and ageing. For example, UAS-ovine PrP flies crossed with the elav-GAL4 driver fly line achieves efficient expression of cell-surface anchored ovine prion protein in all neurons of Drosophila[100,102].
Our Drosophila model allowed us to test the hypothesis that exogenous ovine prions can induce toxicity in flies transgenic for ovine PrP. Remarkably, adult Drosophila, which express ovine PrP pan neuronally and that are exposed to ovine prions at the larval stage, show a neurotoxic phenotype as compared to control non-transgenic flies that have been similarly exposed to prion inocula. The prion-induced neurotoxicity in PrP transgenic Drosophila is evidenced by an accelerated decline in locomotor activity[100,101,103] (Figure 2). In addition, we have used protein misfolding cyclic amplification (PMCA) to show that this prion-induced phenotype is accompanied by accumulation of proteinase K (PK)-resistant PrPSc in fly brains[100]. The presence of PrPSc is a pathogonomic feature of prion diseases. However, the most sensitive hallmark of transmissible prion diseases, is the transmission of these conditions to new hosts, since in some prion-infected hosts, neuropathology can develop in the apparent absence of PrPSc and conversely, PrPSc can accumulate in the absence of neuropathology[4,104]. Importantly therefore, we have demonstrated that the prion-induced fly phenotype is transmissible to PrP transgenic Drosophila[100,101,103]. In mammalian hosts, prion-mediated toxicity has been shown to be inextricably linked to prion replication[4,12,105] and these two events only occur in PrP expressing hosts. In our experiments, scrapie-infected sheep brain material did not induce toxicity in control non-PrP transgenic flies, and head homogenate from these prion-exposed control flies did not transmit any toxicity to fresh PrP transgenic recipient flies. Collectively, these data are consistent with the formation of transmissible prions in Drosophila transgenic for PrP expression. Furthermore, while the conversion of PrPC to PrPSc has been reported to occur either at the cell surface or within the endocytic pathway[106-108], our novel studies in Drosophila show that PrP targeted to the plasma membrane, to the cytosol, or for secretion, can participate in the generation of prion-induced toxicity.
Figure 2.
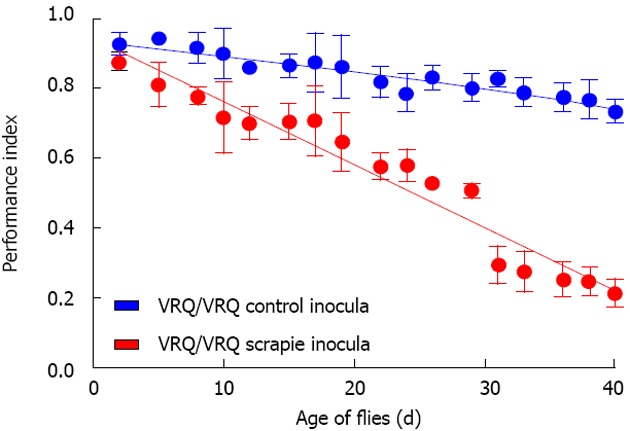
Prion-exposed ovine PrP transgenic Drosophila show enhanced locomotor defect. Drosophila with pan neuronal expression of ovine VRQ(cyt) were fed VRQ/VRQ scrapie-free (blue circles, blue line) or scrapie-infected (red circles, red line) sheep brain homogenate at the larval stage of development. The locomotor activity of adult flies was assessed by a negative geotaxis climbing assay. The performance index is shown for each genotype of fly per time point (Reproduced with permission, from Thackray et al[100] 2014. © the Biochemical Society).
Our observations validate PrP transgenic Drosophila as a new animal model to study the mechanisms of prion-induced neurotoxicity. One of the key benefits of this model system is its rapid and highly reproducible progression to symptomatic stages. This opens the door to a detailed cellular and molecular analysis of the sequence of changes that occur from immediately after infection until symptoms of neurotoxicity become overt. To this end we have performed a functional genomic analysis of prion-infected Drosophila transgenic for ovine PrP, membrane bound by a glycosylphosphatidyl-inositol (GPI) anchor in order to search for biochemical pathways and genetic modifiers of prion-induced neurotoxicity[109]. Our preliminary RNA-Seq-based analysis has revealed that during the early phase of prion infection in PrP transgenic Drosophila, the expression of genes associated with cell cycle re-entry and DNA damage repair were up-regulated in the fly brain. This observation is indicative of cell cycle activity and DDR in the early phase of prion-induced neurotoxicity. Significantly, during the early phase of prion infection in our fly model, cell cycle activation genes (e.g., PCNA) and double-stranded DNA repair genes (e.g., H2Av) are up-regulated, as also seen in brains of prion-diseased mice[96]. Importantly, we found that this response precedes a dramatic down-regulation of genes associated with protein synthesis, including those involved with eIF2a and mTOR pathways. These are interesting observations in light of the reports of translational defects in prion-infected mice[22]. Our novel observations show that prion infection in Drosophila has the potential to re-capitulate prion-induced events in mammalian hosts. Our data further suggest that cell cycle re-entry and inhibition of protein synthesis are temporally linked events in prion-induced neurotoxicity. In this context our hypothesis (Figure 3) is that neurotoxicity in post-mitotic neurons, stressed by prion replication, arises through aberrant cell cycle re-entry that contributes to the effect of sustained inhibition of protein synthesis and eventual neuronal dysfunction.
Figure 3.
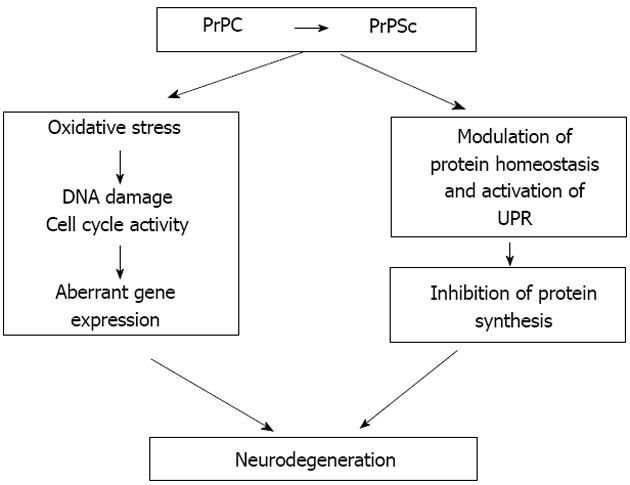
Hypothetical model for prion-induced neurodegeneration. The conversion of PrPC into PrPSc is an essential requirement for the neurotoxicity that occurs during prion disease. Neurodegeneration in post mitotic neurons, stressed by prion replication, may arise through various cellular events including aberrant cell cycle re-entry and sustained inhibition of protein synthesis. These two processes may operate in parallel or may potentially represent temporally linked events. In both cases, aberrant cell cycle re-entry may contribute to the effect of sustained inhibition of protein synthesis evident in prion-induced neurotoxicity.
CONCLUSION
Prion diseases are an important paradigm for protein misfolding neurodegenerative diseases. It is important to establish the sequence and causal links of cellular events that underlie prion-induced neurotoxicity. This will help determine how protein misfolding and aggregation causes neurotoxicity and how this devastating process may be alleviated. Emerging evidence suggests that cell cycle activity and the DNA damage response are cellular processes that may be involved in prion-induced neurodegeneration, as appears to be the case in other neurodegenerative diseases. With the power of Drosophila genetics now in play, many important questions can be systematically addressed. Important questions to be answered include what is the temporal order of the cellular events that are responsible for the progression of prion-induced neurotoxicity. In addition, what is the relationship between the accumulation of cell-cycle related proteins in prion-infected post mitotic neurons, the suppression of translation and resultant neurotoxicity? Future research in this area will be enhanced by the use of a Drosophila model of transmissible mammalian prion disease.
Footnotes
P- Reviewer: Jeong BH, Musci G S- Editor: Ji FF L- Editor: A E- Editor: Yan JL
Supported by The NC3Rs, No. NC/K000462/1 (in part).
Conflict-of-interest statement: The authors declare that there are no conflicts of interest.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: December 26, 2014
First decision: February 7, 2015
Article in press: April 30, 2015
References
- 1.Forman MS, Trojanowski JQ, Lee VM. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 2.Guo JL, Lee VM. Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nat Med. 2014;20:130–138. doi: 10.1038/nm.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner SB. Prion biology and diseases. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 5.Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64:783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 6.Aguzzi A, Baumann F, Bremer J. The prion’s elusive reason for being. Annu Rev Neurosci. 2008;31:439–477. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- 7.Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, Marino S, Weissmann C, Aguzzi A. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- 8.Brandner S, Raeber A, Sailer A, Blättler T, Fischer M, Weissmann C, Aguzzi A. Normal host prion protein (PrPC) is required for scrapie spread within the central nervous system. Proc Natl Acad Sci USA. 1996;93:13148–13151. doi: 10.1073/pnas.93.23.13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallucci G, Dickinson A, Linehan J, Klöhn PC, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 10.Mallucci GR, White MD, Farmer M, Dickinson A, Khatun H, Powell AD, Brandner S, Jefferys JG, Collinge J. Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron. 2007;53:325–335. doi: 10.1016/j.neuron.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 11.White MD, Farmer M, Mirabile I, Brandner S, Collinge J, Mallucci GR. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci USA. 2008;105:10238–10243. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature. 2011;470:540–542. doi: 10.1038/nature09768. [DOI] [PubMed] [Google Scholar]
- 13.Zhou M, Ottenberg G, Sferrazza GF, Lasmézas CI. Highly neurotoxic monomeric α-helical prion protein. Proc Natl Acad Sci USA. 2012;109:3113–3118. doi: 10.1073/pnas.1118090109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakrabarti O, Ashok A, Hegde RS. Prion protein biosynthesis and its emerging role in neurodegeneration. Trends Biochem Sci. 2009;34:287–295. doi: 10.1016/j.tibs.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma J, Lindquist S. Conversion of PrP to a self-perpetuating PrPSc-like conformation in the cytosol. Science. 2002;298:1785–1788. doi: 10.1126/science.1073619. [DOI] [PubMed] [Google Scholar]
- 16.Ma J, Lindquist S. Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc Natl Acad Sci USA. 2001;98:14955–14960. doi: 10.1073/pnas.011578098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma J, Wollmann R, Lindquist S. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science. 2002;298:1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Bowers SL, Wang F, Pu XA, Nelson RJ, Ma J. Cytoplasmic prion protein induces forebrain neurotoxicity. Biochim Biophys Acta. 2009;1792:555–563. doi: 10.1016/j.bbadis.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fioriti L, Dossena S, Stewart LR, Stewart RS, Harris DA, Forloni G, Chiesa R. Cytosolic prion protein (PrP) is not toxic in N2a cells and primary neurons expressing pathogenic PrP mutations. J Biol Chem. 2005;280:11320–11328. doi: 10.1074/jbc.M412441200. [DOI] [PubMed] [Google Scholar]
- 20.Roucou X, Guo Q, Zhang Y, Goodyer CG, LeBlanc AC. Cytosolic prion protein is not toxic and protects against Bax-mediated cell death in human primary neurons. J Biol Chem. 2003;278:40877–40881. doi: 10.1074/jbc.M306177200. [DOI] [PubMed] [Google Scholar]
- 21.Roffé M, Beraldo FH, Bester R, Nunziante M, Bach C, Mancini G, Gilch S, Vorberg I, Castilho BA, Martins VR, et al. Prion protein interaction with stress-inducible protein 1 enhances neuronal protein synthesis via mTOR. Proc Natl Acad Sci USA. 2010;107:13147–13152. doi: 10.1073/pnas.1000784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, Halliday M, Morgan J, Dinsdale D, Ortori CA, et al. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature. 2012;485:507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno JA, Halliday M, Molloy C, Radford H, Verity N, Axten JM, Ortori CA, Willis AE, Fischer PM, Barrett DA, et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci Transl Med. 2013;5:206ra138. doi: 10.1126/scitranslmed.3006767. [DOI] [PubMed] [Google Scholar]
- 24.Arendt T. Cell cycle activation and aneuploid neurons in Alzheimer’s disease. Mol Neurobiol. 2012;46:125–135. doi: 10.1007/s12035-012-8262-0. [DOI] [PubMed] [Google Scholar]
- 25.Madabhushi R, Pan L, Tsai LH. DNA damage and its links to neurodegeneration. Neuron. 2014;83:266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- 27.Brochier C, Langley B. Chromatin modifications associated with DNA double-strand breaks repair as potential targets for neurological diseases. Neurotherapeutics. 2013;10:817–830. doi: 10.1007/s13311-013-0210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iliakis G, Wang Y, Guan J, Wang H. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22:5834–5847. doi: 10.1038/sj.onc.1206682. [DOI] [PubMed] [Google Scholar]
- 29.Mazouzi A, Velimezi G, Loizou JI. DNA replication stress: causes, resolution and disease. Exp Cell Res. 2014;329:85–93. doi: 10.1016/j.yexcr.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 30.Yan S, Sorrell M, Berman Z. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell Mol Life Sci. 2014;71:3951–3967. doi: 10.1007/s00018-014-1666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 32.McKinnon PJ. Maintaining genome stability in the nervous system. Nat Neurosci. 2013;16:1523–1529. doi: 10.1038/nn.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vermeij WP, Hoeijmakers JH, Pothof J. Aging: not all DNA damage is equal. Curr Opin Genet Dev. 2014;26:124–130. doi: 10.1016/j.gde.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 35.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 36.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 37.Caldecott KW. DNA damage responses and neurological disease. Preface. DNA Repair (Amst) 2008;7:1009. doi: 10.1016/j.dnarep.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 40.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 41.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 42.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 43.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 44.Rao KS. DNA repair in aging rat neurons. Neuroscience. 2007;145:1330–1340. doi: 10.1016/j.neuroscience.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Herrup K. Cell division in the CNS: protective response or lethal event in post-mitotic neurons? Biochim Biophys Acta. 2007;1772:457–466. doi: 10.1016/j.bbadis.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrup K, Neve R, Ackerman SL, Copani A. Divide and die: cell cycle events as triggers of nerve cell death. J Neurosci. 2004;24:9232–9239. doi: 10.1523/JNEUROSCI.3347-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kruman II, Wersto RP, Cardozo-Pelaez F, Smilenov L, Chan SL, Chrest FJ, Emokpae R, Gorospe M, Mattson MP. Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron. 2004;41:549–561. doi: 10.1016/s0896-6273(04)00017-0. [DOI] [PubMed] [Google Scholar]
- 48.Vincent I, Rosado M, Davies P. Mitotic mechanisms in Alzheimer’s disease? J Cell Biol. 1996;132:413–425. doi: 10.1083/jcb.132.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y, Geldmacher DS, Herrup K. DNA replication precedes neuronal cell death in Alzheimer’s disease. J Neurosci. 2001;21:2661–2668. doi: 10.1523/JNEUROSCI.21-08-02661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullaart E, Boerrigter ME, Ravid R, Swaab DF, Vijg J. Increased levels of DNA breaks in cerebral cortex of Alzheimer’s disease patients. Neurobiol Aging. 1990;11:169–173. doi: 10.1016/0197-4580(90)90542-8. [DOI] [PubMed] [Google Scholar]
- 51.Lyras L, Cairns NJ, Jenner A, Jenner P, Halliwell B. An assessment of oxidative damage to proteins, lipids, and DNA in brain from patients with Alzheimer’s disease. J Neurochem. 1997;68:2061–2069. doi: 10.1046/j.1471-4159.1997.68052061.x. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen MD, Boudreau M, Kriz J, Couillard-Després S, Kaplan DR, Julien JP. Cell cycle regulators in the neuronal death pathway of amyotrophic lateral sclerosis caused by mutant superoxide dismutase 1. J Neurosci. 2003;23:2131–2140. doi: 10.1523/JNEUROSCI.23-06-02131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin LJ, Liu Z, Chen K, Price AC, Pan Y, Swaby JA, Golden WC. Motor neuron degeneration in amyotrophic lateral sclerosis mutant superoxide dismutase-1 transgenic mice: mechanisms of mitochondriopathy and cell death. J Comp Neurol. 2007;500:20–46. doi: 10.1002/cne.21160. [DOI] [PubMed] [Google Scholar]
- 54.Illuzzi J, Yerkes S, Parekh-Olmedo H, Kmiec EB. DNA breakage and induction of DNA damage response proteins precede the appearance of visible mutant huntingtin aggregates. J Neurosci Res. 2009;87:733–747. doi: 10.1002/jnr.21881. [DOI] [PubMed] [Google Scholar]
- 55.Pelegrí C, Duran-Vilaregut J, del Valle J, Crespo-Biel N, Ferrer I, Pallàs M, Camins A, Vilaplana J. Cell cycle activation in striatal neurons from Huntington’s disease patients and rats treated with 3-nitropropionic acid. Int J Dev Neurosci. 2008;26:665–671. doi: 10.1016/j.ijdevneu.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 56.Devine MJ, Plun-Favreau H, Wood NW. Parkinson’s disease and cancer: two wars, one front. Nat Rev Cancer. 2011;11:812–823. doi: 10.1038/nrc3150. [DOI] [PubMed] [Google Scholar]
- 57.Höglinger GU, Breunig JJ, Depboylu C, Rouaux C, Michel PP, Alvarez-Fischer D, Boutillier AL, Degregori J, Oertel WH, Rakic P, et al. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proc Natl Acad Sci USA. 2007;104:3585–3590. doi: 10.1073/pnas.0611671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mandel SA, Fishman T, Youdim MB. Gene and protein signatures in sporadic Parkinson’s disease and a novel genetic model of PD. Parkinsonism Relat Disord. 2007;13 Suppl 3:S242–S247. doi: 10.1016/S1353-8020(08)70009-9. [DOI] [PubMed] [Google Scholar]
- 59.Kim D, Tsai LH. Linking cell cycle reentry and DNA damage in neurodegeneration. Ann N Y Acad Sci. 2009;1170:674–679. doi: 10.1111/j.1749-6632.2009.04105.x. [DOI] [PubMed] [Google Scholar]
- 60.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 61.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 62.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 64.Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 66.Hansen JC, Wolffe AP. Influence of chromatin folding on transcription initiation and elongation by RNA polymerase III. Biochemistry. 1992;31:7977–7988. doi: 10.1021/bi00149a032. [DOI] [PubMed] [Google Scholar]
- 67.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 68.Humpal SE, Robinson DA, Krebs JE. Marks to stop the clock: histone modifications and checkpoint regulation in the DNA damage response. Biochem Cell Biol. 2009;87:243–253. doi: 10.1139/O08-109. [DOI] [PubMed] [Google Scholar]
- 69.House NC, Koch MR, Freudenreich CH. Chromatin modifications and DNA repair: beyond double-strand breaks. Front Genet. 2014;5:296. doi: 10.3389/fgene.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Polo SE. Reshaping Chromatin after DNA Damage: The Choreography of Histone Proteins. J Mol Biol. 2015;427:626–636. doi: 10.1016/j.jmb.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathis GA, Althaus FR. Isolation of 8-methoxypsoralen accessible DNA domains from chromatin of intact cells. Cell Biol Toxicol. 1990;6:35–45. doi: 10.1007/BF00135025. [DOI] [PubMed] [Google Scholar]
- 72.Frost B, Hemberg M, Lewis J, Feany MB. Tau promotes neurodegeneration through global chromatin relaxation. Nat Neurosci. 2014;17:357–366. doi: 10.1038/nn.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arendt T, Holzer M, Stöbe A, Gärtner U, Lüth HJ, Brückner MK, Ueberham U. Activated mitogenic signaling induces a process of dedifferentiation in Alzheimer’s disease that eventually results in cell death. Ann N Y Acad Sci. 2000;920:249–255. doi: 10.1111/j.1749-6632.2000.tb06931.x. [DOI] [PubMed] [Google Scholar]
- 74.Arendt T. Alzheimer’s disease as a disorder of mechanisms underlying structural brain self-organization. Neuroscience. 2001;102:723–765. doi: 10.1016/s0306-4522(00)00516-9. [DOI] [PubMed] [Google Scholar]
- 75.Pan L, Penney J, Tsai LH. Chromatin regulation of DNA damage repair and genome integrity in the central nervous system. J Mol Biol. 2014;426:3376–3388. doi: 10.1016/j.jmb.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci. 2009;10:100–112. doi: 10.1038/nrn2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coppedè F, Migliore L. DNA damage and repair in Alzheimer’s disease. Curr Alzheimer Res. 2009;6:36–47. doi: 10.2174/156720509787313970. [DOI] [PubMed] [Google Scholar]
- 78.Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, Ahanonu B, Pao PC, Qiu Y, Zhao Y, et al. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci. 2013;16:1008–1015. doi: 10.1038/nn.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fishel ML, Vasko MR, Kelley MR. DNA repair in neurons: so if they don’t divide what’s to repair? Mutat Res. 2007;614:24–36. doi: 10.1016/j.mrfmmm.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 80.Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci. 2013;16:1383–1391. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weissman L, Jo DG, Sørensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA. Defective DNA base excision repair in brain from individuals with Alzheimer’s disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007;35:5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ly CV, Verstreken P. Mitochondria at the synapse. Neuroscientist. 2006;12:291–299. doi: 10.1177/1073858406287661. [DOI] [PubMed] [Google Scholar]
- 83.Martin LJ. Biology of mitochondria in neurodegenerative diseases. Prog Mol Biol Transl Sci. 2012;107:355–415. doi: 10.1016/B978-0-12-385883-2.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 85.Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakaimal S, Nair KS. Decline in skeletal muscle mitochondrial function with aging in humans. Proc Natl Acad Sci USA. 2005;102:5618–5623. doi: 10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chistiakov DA, Sobenin IA, Revin VV, Orekhov AN, Bobryshev YV. Mitochondrial aging and age-related dysfunction of mitochondria. Biomed Res Int. 2014;2014:238463. doi: 10.1155/2014/238463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chaturvedi RK, Flint Beal M. Mitochondrial diseases of the brain. Free Radic Biol Med. 2013;63:1–29. doi: 10.1016/j.freeradbiomed.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 88.Borgesius NZ, de Waard MC, van der Pluijm I, Omrani A, Zondag GC, van der Horst GT, Melton DW, Hoeijmakers JH, Jaarsma D, Elgersma Y. Accelerated age-related cognitive decline and neurodegeneration, caused by deficient DNA repair. J Neurosci. 2011;31:12543–12553. doi: 10.1523/JNEUROSCI.1589-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vijg J, Suh Y. Genome instability and aging. Annu Rev Physiol. 2013;75:645–668. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- 90.Frank CL, Tsai LH. Alternative functions of core cell cycle regulators in neuronal migration, neuronal maturation, and synaptic plasticity. Neuron. 2009;62:312–326. doi: 10.1016/j.neuron.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang Z, Zang K, Reichardt LF. The origin recognition core complex regulates dendrite and spine development in postmitotic neurons. J Cell Biol. 2005;170:527–535. doi: 10.1083/jcb.200505075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Y, Kim AH, Bonni A. The dynamic ubiquitin ligase duo: Cdh1-APC and Cdc20-APC regulate neuronal morphogenesis and connectivity. Curr Opin Neurobiol. 2010;20:92–99. doi: 10.1016/j.conb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li J, Han YR, Plummer MR, Herrup K. Cytoplasmic ATM in neurons modulates synaptic function. Curr Biol. 2009;19:2091–2096. doi: 10.1016/j.cub.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Odajima J, Wills ZP, Ndassa YM, Terunuma M, Kretschmannova K, Deeb TZ, Geng Y, Gawrzak S, Quadros IM, Newman J, et al. Cyclin E constrains Cdk5 activity to regulate synaptic plasticity and memory formation. Dev Cell. 2011;21:655–668. doi: 10.1016/j.devcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang J, Cicero SA, Wang L, Romito-Digiacomo RR, Yang Y, Herrup K. Nuclear localization of Cdk5 is a key determinant in the postmitotic state of neurons. Proc Natl Acad Sci USA. 2008;105:8772–8777. doi: 10.1073/pnas.0711355105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jackson WS, Borkowski AW, Watson NE, King OD, Faas H, Jasanoff A, Lindquist S. Profoundly different prion diseases in knock-in mice carrying single PrP codon substitutions associated with human diseases. Proc Natl Acad Sci USA. 2013;110:14759–14764. doi: 10.1073/pnas.1312006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang H, Tian C, Xu Y, Xie WL, Zhang J, Zhang BY, Ren K, Wang K, Chen C, Wang SB, et al. Abortive cell cycle events in the brains of scrapie-infected hamsters with remarkable decreases of PLK3/Cdc25C and increases of PLK1/cyclin B1. Mol Neurobiol. 2013;48:655–668. doi: 10.1007/s12035-013-8455-1. [DOI] [PubMed] [Google Scholar]
- 98.Seeburg DP, Pak D, Sheng M. Polo-like kinases in the nervous system. Oncogene. 2005;24:292–298. doi: 10.1038/sj.onc.1208277. [DOI] [PubMed] [Google Scholar]
- 99.Jalland CM, Benestad SL, Ersdal C, Scheffler K, Suganthan R, Nakabeppu Y, Eide L, Bjørås M, Tranulis MA. Accelerated clinical course of prion disease in mice compromised in repair of oxidative DNA damage. Free Radic Biol Med. 2014;68:1–7. doi: 10.1016/j.freeradbiomed.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 100.Thackray AM, Di Y, Zhang C, Wolf H, Pradl L, Vorberg I, Andréoletti O, Bujdoso R. Prion-induced and spontaneous formation of transmissible toxicity in PrP transgenic Drosophila. Biochem J. 2014;463:31–40. doi: 10.1042/BJ20140129. [DOI] [PubMed] [Google Scholar]
- 101.Thackray AM, Zhang C, Arndt T, Bujdoso R. Cytosolic PrP can participate in prion-mediated toxicity. J Virol. 2014;88:8129–8138. doi: 10.1128/JVI.00732-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thackray AM, Muhammad F, Zhang C, Di Y, Jahn TR, Landgraf M, Crowther DC, Evers JF, Bujdoso R. Ovine PrP transgenic Drosophila show reduced locomotor activity and decreased survival. Biochem J. 2012;444:487–495. doi: 10.1042/BJ20112141. [DOI] [PubMed] [Google Scholar]
- 103.Thackray AM, Muhammad F, Zhang C, Denyer M, Spiropoulos J, Crowther DC, Bujdoso R. Prion-induced toxicity in PrP transgenic Drosophila. Exp Mol Pathol. 2012;92:194–201. doi: 10.1016/j.yexmp.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 104.Chiesa R, Harris DA. Prion diseases: what is the neurotoxic molecule? Neurobiol Dis. 2001;8:743–763. doi: 10.1006/nbdi.2001.0433. [DOI] [PubMed] [Google Scholar]
- 105.Aguzzi A, Sigurdson C, Heikenwaelder M. Molecular mechanisms of prion pathogenesis. Annu Rev Pathol. 2008;3:11–40. doi: 10.1146/annurev.pathmechdis.3.121806.154326. [DOI] [PubMed] [Google Scholar]
- 106.Borchelt DR, Taraboulos A, Prusiner SB. Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J Biol Chem. 1992;267:16188–16199. [PubMed] [Google Scholar]
- 107.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner SB. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, Anderson RG, Taraboulos A, Prusiner SB. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci USA. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thackray AM, Andréoletti O, Bujdoso R. Bioassay of plasma from prion-infected sheep in ovine PrP transgenic Drosophila. Meeting abstract, Prions: Epigenetics and Neurodegenerative Diseases. Trieste, Italy; 2014. [Google Scholar]


