Abstract
Here, we report the first detection of a Klebsiella pneumoniae carbapenemase 2 (KPC-2)-producing Klebsiella pneumoniae strain belonging to sequence type 833 (ST833), collected in an Italian hospital from a patient coming from South America. Its blaKPC determinant was carried by a ColE1 plasmid, pKBuS13, that showed the Tn4401b::blaKPC-2 transposon inserted into the regulatory region of an Xer site-specific recombination locus. This interfered with the correct resolution of plasmid multimers into monomers, lowering plasmid stability and leading to overestimation of the number of plasmids harbored by a single host cell. Sequencing of the fragments adjacent to Tn4401b detected a region that did not have significant matches in databases other than the genome of a carbapenem-resistant Escherichia coli strain collected during the same year at a hospital in Boston. This is interesting in an epidemiologic context, as it suggests that despite the absence of tra genes and the instability under nonselective conditions, the circulation of pKBuS13 or of analogous plasmids might be wider than reported.
INTRODUCTION
During the last decade, Klebsiella pneumoniae strains producing K. pneumoniae carbapenemase (KPC) enzymes have become a matter of great concern, as they are often susceptible to only a few antibiotics, cause high mortality among patients with bloodstream infections, and are increasingly being reported worldwide (1).
KPC-type beta-lactamases include 22 variants (http://www.lahey.org/Studies/other.asp) that have been detected in a large number of K. pneumoniae lineages. Among them, KPC-2 and KPC-3 are predominant and largely disseminated worldwide by strains belonging to the clonal complex 258 (CC258), including the sequence type 258 (ST258) lineage defined by multilocus sequence typing (MLST) and its single-locus variants (e.g., ST11, ST437, and ST512) (2–6). Dissemination of blaKPC genes is fueled by their association with Tn4401, a 10-kb Tn3-like element that has been detected on plasmids belonging to different incompatibility groups (FII, N, L/M) and of different sizes (10 to 170 kb) (7).
In Italy, KPC-producing K. pneumoniae (KPC-K. pneumoniae) strains have increasingly been reported since 2009 (8). Most of them belong to the globally spread ST258 and ST512 clones, but some isolates of different STs (ST101 and ST307) have been detected too (9).
In the present work, we report the isolation in the Trieste area (northeast Italy) of a KPC-K. pneumoniae strain belonging to ST833 from the blood culture of a patient coming from a Venezuelan hospital. ST833 is a single locus variant of ST11, which has recently been described as one of the lineages responsible for dissemination of blaKPC determinants carried on different plasmids in Latin America (2, 10, 11). To our knowledge, this is the first finding of a KPC-K. pneumoniae strain belonging to ST833. In addition we describe its blaKPC-2-carrying plasmid, which shows interesting features in an epidemiologic context.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The carbapenem-resistant strain K. pneumoniae KBu-1 was recovered from the blood culture of a 3-year-old patient coming from Venezuela and admitted to the Trieste Pediatric Hospital to undergo marrow transplantation. Both identification and antimicrobial susceptibility were determined by Vitek 2 (bioMérieux, Marcy l'Etoile, France). Extended-spectrum beta-lactamase (ESBL) production was further investigated by the Etest method (AB Biodisk, Solna, Sweden). Detection of carbapenemase production was performed by disc diffusion synergy test (Rosco Diagnostica, Taastrup, Denmark).
Escherichia coli J53 (met-63 pro-22 Rifr) and J62 (lac-28 proC23 his-51 trp-30 Rifr) were used as recipients for conjugation experiments.
E. coli JM101 [supE thi Δ(lac-proAB) F′(lacIq lacZΔM15 traD36 proAB+)] was used as a recipient for electroporation of plasmid DNA isolated from K. pneumoniae KBu-1 and for plasmid DNA preparation for further studies (DNA sequencing, restriction analysis).
Bacteria were grown in Luria-Bertani (LB) medium, supplemented with 100 μg/ml rifampin, 100 μg/ml ampicillin, or 10 μg/ml imipenem when required.
Antibiotic susceptibility profiles of all strains were evaluated according to the guidelines of the CLSI using Sensititre plates produced by Trek Diagnostic Systems (Westlake, OH, USA) and, in the case of imipenem, meropenem, and ceftazidime, by standard microdilution method (12). Antimicrobial agent powders were obtained from Sigma Chemical Co. (St. Louis, MO, USA).
PCR amplification and DNA sequencing.
Molecular confirmation was performed by PCR assays for the ESBL genes (blaTEM, blaSHV, blaCTX-M, and blaOXA-9) and for the carbapenemase genes (blaIMP, blaVIM, blaNDM, blaOXA-48, and blaKPC). Specific primers, used to amplify the blaKPC determinant and other resistant genes, are listed in the supplemental material (see Table S1 in the supplemental material). PCRs were performed, as previously described (13–17), directly on 2 to 3 colonies picked from a pure culture. blaTEM, blaSHV, blaCTX-M, and blaKPC amplicons were entirely sequenced to identify the allelic form.
Sequencing reactions were carried out at a commercial sequence facility (BMR Genomics, Padua, Italy).
The region upstream of blaKPC was amplified and sequenced with the primers 3098U and KPC-Rev (see Table S1 in the supplemental material) to identify the isoform of Tn4401.
The region of the plasmid outward transposon Tn4401 was amplified using the Expand long template PCR system (Roche Molecular Biochemicals, Mannheim, Germany) and two outward-directed primers (EcoRIout and 141R-6). For determination of the sequence of the fragment adjacent to transposon Tn4401, primer walking was carried out with primers Bu13-1 and Bu13-2, designed from sequences obtained with EcoRIout and 141R-6.
Multilocus sequence typing (MLST) was performed according to the protocol described on the K. pneumoniae MLST website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
Conjugation experiments.
Direct transfer of carbapenem resistance into E. coli strains J53 and J62 was attempted by a filter-mating procedure (18). Transconjugant selection was performed on LB agar supplemented with rifampin and imipenem.
Molecular investigations.
Plasmid DNA from K. pneumoniae KBu-1 was extracted by the alkaline lysis method (19) and electroporated into E. coli JM101 using a Gene Pulser apparatus (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions. Transformants were selected on LB agar plus ampicillin and analyzed by PCR for the presence of all the bla genes previously detected in the donor strain.
The plasmid profile was analyzed after S1 nuclease (Roche) digestion (20 U enzyme in each sample) on crude plasmid extract (30 min at 37°C) and on DNA extracted from cells embedded in agarose plugs (20) (1 h at 37°C), followed by separation on agarose gel electrophoresis using different running conditions: (i) 20 V for 20 h on 1% agarose gel and (ii) pulsed-field gel electrophoresis (PFGE) on 0.8% agarose gel with a CHEF-DR III apparatus (Bio-Rad) at 14°C and 6 V/cm for 13 h by using pulse times from 1 to 10 s. Separated DNA was hybridized with a digoxigenin (DIG)-labeled blaKPC-specific probe, obtained by amplification of an internal fragment of blaKPC with primers KPC-F and KPC-R (21) in the presence of 70 μM DIG-11-dUTP (Roche) after capillary blotting onto Hybond-N-positive (N+) membranes (Amersham Biosciences, Piscataway, NJ).
Plasmid restriction analysis was carried on with BamHI, HindIII, PstI, and SacI restriction enzymes according to the manufacturer's instructions (New England BioLabs, Mississauga, Ontario, Canada) followed by separation on 0.8% agarose gel.
The 13-kb band recognized by the blaKPC-specific probe was extracted from low-melting agarose by GELase digestion (Epicentre, Madison, WI, USA) and electroporated into E. coli JM101.
Stability assay.
Evaluation of the number of plasmid-free cells among bacteria grown under nonselective conditions was carried out as described by Tolmasky et al. (22). Each test was replicated three times.
Comparative analysis.
The nucleotide and protein sequences were analyzed using the blastn, blastp, and bl2seq algorithms available at the National Center of Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Direct and tandem repeats were detected using the Tandem Repeats Finder software, version 4.07b (23).
Nucleotide sequence accession numbers.
The regions of pKBuS13 sequenced in this work have been deposited in GenBank under the accession numbers KM076933, KM076934, and KM076935.
RESULTS AND DISCUSSION
Isolation and molecular characterization of KBu-1.
In May 2012, a 3-year-old patient coming from Venezuela was admitted to the Trieste Children's Hospital IRCCS Burlo Garofolo to undergo bone marrow transplantation. A culture of a surveillance rectal swab detected different multidrug-resistant organisms: extended spectrum beta-lactamase (ESBL)-producing Escherichia coli, vancomycin-resistant Enterococcus faecium (VRE), and K. pneumoniae resistant to all beta-lactams, with MICs for imipenem and meropenem of ≥16 μg/ml. Unfortunately, at a later stage, the patient became neutropenic, developed a severe K. pneumoniae sepsis, and died. Further analysis revealed identical features to the previous isolate: (i) they showed the same antibiotype (Table 1); (ii) they were positive for carbapenemase production, and screening by PCR revealed the presence of the blaKPC gene and was negative for other carbapenemase determinants; and (iii) ESBL production was not detected by Vitek 2 and resulted nondeterminable by Etest, as MIC values were above the test ranges; further analysis by PCR and sequencing of the amplicons revealed the presence of the blaCTX-M-1, blaTEM-1b, and blaSHV-11 genes while blaOXA-9 was not detected.
TABLE 1.
Antimicrobial susceptibility patterns of K. pneumoniae KBu-1, the E. coli JM101 recipient, and the E. coli JM101 transformants
| Antimicrobial agent(s) | MICa (μg/ml) for: |
||
|---|---|---|---|
| K. pneumoniae KBu-1 | E. coli JM101 | E. coli JM101 transformantsb | |
| Imipenemc | 512 | 0.25 | 4 |
| Meropenemc | 512 | 0.03 | 4 |
| Ceftazidimec | 64 | 0.12 | 8 |
| Amoxicillin-clavulanic acid | >8 | 4 | >8 |
| Ampicillin-sulbactam | >32 | ≤8 | >32 |
| Cefepime | >32 | ≤1 | 2 |
| Cefotaxime | >4 | ≤0.06 | 4 |
| Piperacillin-tazobactam | >128 | ≤2 | 128 |
| Amikacin | ≤4 | ≤4 | ≤4 |
| Gentamicin | ≤1 | ≤1 | ≤1 |
| Colistin | ≤0.5 | ≤0.5 | ≤0.5 |
| Nitrofurantoin | >64 | ≤32 | ≤32 |
| Tigecycline | 1 | 0.25 | 0.25 |
| Trimethoprim-sulfamethoxazole | >4 | ≤0.5 | ≤0.5 |
| Ciprofloxacin | >2 | ≤0.06 | ≤0.06 |
| Levofloxacin | >4 | ≤1 | ≤1 |
Reported MIC values were determined by Sensititre plates (Trek Diagnostic Systems) with the exception of those of imipenem, meropenem, and ceftazidime.
E. coli JM101 transformed with the entire K. pneumoniae KBu-1 plasmid content and with the 13-kb band alone displayed the same susceptibility profile.
For these antibiotics, the CLSI standard microdilution method was used (12) in order to obtain a more precise evaluation.
This KPC-K. pneumoniae isolate remained a unique one, thanks to strict infection control procedures (segregation and barrier nursing) adopted for patient management: cultures of rectal swabs of all the patients recovered in the same unit gave negative results.
The isolate was named KBu-1 and was further characterized at the molecular level.
Sequencing of the blaKPC amplicon and of the genes used to determine the MLST group of the isolate revealed that it harbored the blaKPC-2 gene and belonged to ST833 (allelic profile 3-3-1-1-1-1-12). To our knowledge, this is the first report of a KPC-K. pneumoniae strain belonging to ST833. It differs for a single point mutation from the ST11 lineage (370 C→G in the tonB allele, leading to the amino acid substitution 121 P→A) and belongs to CC258, which is considered of special concern as it gathers the most common lineages spread worldwide (4–6), including South America (2, 3, 11). The report of the SENTRY Antimicrobial Surveillance Program on strains collected from different South American hospitals during 2010 confirmed the expansion of CC258 in this area and particularly of strains belonging to ST11, mostly detected in Brazil (10).
Unfortunately, no data are available about STs circulating in Venezuela, as none of the hospitals were part of the study in 2010, although the circulation of the blaKPC determinant in Venezuelan hospitals is documented (24, 25).
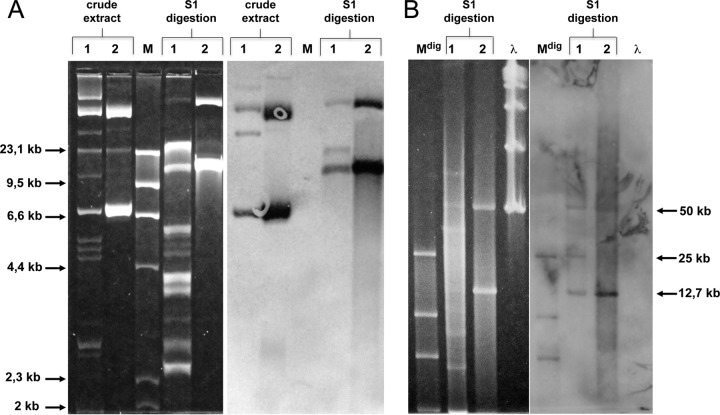
Plasmid extraction followed by S1 digestion revealed at least 12 bands of various sizes (ranging from 3 to 80 kb), three of which (approximately 13, 25, and 50 kb) were recognized by an internal probe for the blaKPC gene (Fig. 1). All attempts to transfer resistance to imipenem by conjugation from KBu-1 to E. coli J53 Rifr and to E. coli J62 Rifr were unsuccessful. However, when the plasmid mixture was electroporated into E. coli JM101, transformants carrying both the 13-kb and the 50-kb plasmids were obtained (Fig. 1). The same result was achieved when we electroporated the 13-kb band alone, extracted from low-melting agarose (see Fig. S1 in the supplemental material). Analysis by PCR on plasmid DNA from E. coli JM101 transformants revealed the presence of the blaKPC determinant, while blaCTX, blaSHV, and blaTEM were not detected. The 13-kb plasmid was named pKBuS13 and was further investigated.
FIG 1.
Hybridization with a blaKPC probe of plasmid DNA separated on agarose gel electrophoresis. (A) Plasmid extract from K. pneumoniae KBu-1 and from E. coli JM101 transformed with KBu-1 plasmid content was run at 20 V for 20 h before and after S1 nuclease digestion. (B) Fragments higher than 30 kb obtained by S1 digestion were better separated on PFGE, switch 1–10 s for 13 h. Lane 1, KBu-1 plasmid content; lane 2, plasmid extraction from E. coli JM101 transformed with KBu-1 plasmid content; lane M, Molecular Weight Marker II (Roche); lane Mdig, digoxigenin-labeled Molecular Weight Marker II (Roche); lane λ, λ ladder (New England BioLabs).
Sequence analysis of plasmid pKBuS13.
Besides the spread of few strain lineages, the worldwide dissemination of the blaKPC-2 determinant is favored by its location on the Tn4401 transposon, a Tn3-like element that supports replicative transposition and has been found inserted at different loci on a broad variety of plasmids (7, 11).
Most of the KPC-K. pneumoniae strains circulating in South America carry the blaKPC-2 determinant on the Tn4401b variant of Tn4401, located on plasmids of variable size (20 to 300 kb) and belonging to different incompatibility groups (IncFII, IncL/M, and IncN) (2, 3, 11).
On the assumption that the KBu-1 isolate carried the blaKPC determinant inside Tn4401, we investigated the variable region of the transposon located upstream of blaKPC. As expected, amplification and sequencing of this region yielded the typical structure of the Tn4401b variant, without the deletions of 100 or 200 bp detected in the Tn4401 or Tn4401a isoforms.
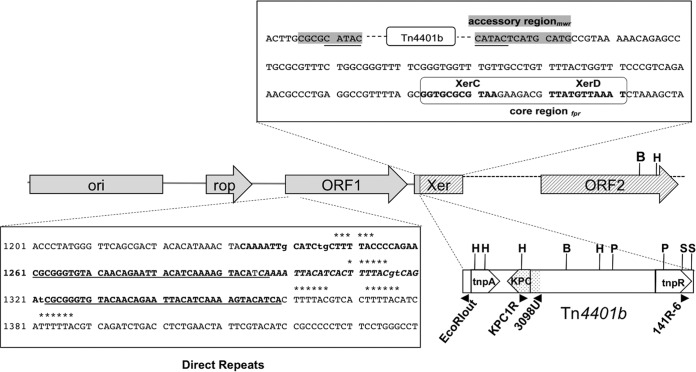
The region of pKBuS13 adjacent to Tn4401b was amplified using outward-directed primers, and the 2,700-bp amplicon was fully sequenced. The location of genes and genetic structures identified by comparative analysis is shown in Fig. 2.
FIG 2.
Genetic map of relevant region of pKBuS13. Genes, ORFs, and genetic structures in the regions adjacent to Tn4401b are shown. The 1,605-bp region upstream the tnpA side (GenBank accession number KM076933) is shaded gray; the 1,118-bp region downstream of the tnpR-side (GenBank accession number KM076935) is crosshatched. Tn4401b is drawn schematically, not to scale, indicating the tnpA and tnpR genes located at the boundaries. The region evidenced by dots was verified by sequencing (GenBank accession number KM076934). The position of some primers used in this work and the sites of the enzymes used for restriction analysis are shown (B, BamHI; H, HindIII; P, PstI; S, SacI). Two sequences are enlarged: above, the Xer site with the mwr locus (interrupted by the Tn4401b insertion) in the accessory region shown in the gray box, the 5-bp duplication resulting from transposon insertion underlined, and the fpr locus in the core region boxed with the binding sites for XerC and XerD shown in bold; below, the sequence containing the direct repeats (DR) identified inside ORF1, with the different DR motifs marked as follows: ******, 6-bp motif (5 repeats); underlined, 37-bp stretches separated by 25 bp; bold, tandem repeat identified by the tandem repeat finder software (23): two 61-bp stretches separated by one T. It is an imperfect DR, with three mismatches (lowercase) compared with the consensus sequence CGCGGGTGTACAACAGAATTACATCAAAAGTACA.
The 1,605-bp region adjacent to the tnpA side of Tn4401b contained two genes responsible for replication (ori p15A) and control of the copy number (rop) of plasmids belonging to the ColE1 family. In addition, an open reading frame (ORF1) containing different direct repeats was found (Fig. 2).
The insertion site of Tn4401 looked peculiar, as it was inserted quite inside an Xer site-specific recombination locus. This locus, involved in the resolution of plasmid multimers (26), usually consists of a core region containing the binding sites for two recombinases (XerC and XerD) and an accessory region, which provides the binding sites for the specific accessory proteins needed for the regulation of the entire process. Different core recombination sites have been described (mwr, psi, cer, dif, dxs, and fpr), which work with different efficiency and are regulated by different accessory proteins (27, 28). Two of them, mwr and fpr, are osmoregulated: that is, at high salt concentrations their recombination efficiency is lower than that required for multimers resolution. These sites have been detected so far only on two natural plasmids, pJHCMW1 (22) and pFPTB1 (29), in a Salmonella enterica serovar Typhimurium isolate and in a K. pneumoniae isolate, respectively (22, 29), each carrying a transposon inserted about 20 bp downstream of Xer. It has been postulated that multimer resolution of these plasmids is provided by the transposon resolvase besides the Xer system, suggesting that they form a group of plasmids whose stability is significantly enhanced by transposon acquisition (28). pKBuS13 is, to our knowledge, the third natural plasmid belonging to this group. However, its Xer recombination system is probably ineffective because the fpr site is the less efficient among those detected in the core region (28), and, most importantly, its accessory region is broken by Tn4401b insertion. Xer system inactivity is supported by two observations: (i) under nonselective conditions, both K. pneumoniae KBu-1 and E. coli JM101 lost pKBuS13 at approximately the same rate of pUC19, which lacks an Xer recombination site and is randomly partitioned during cell division (Fig. 3); and (ii) plasmid stability did not increase in the absence of NaCl (data not shown).
FIG 3.
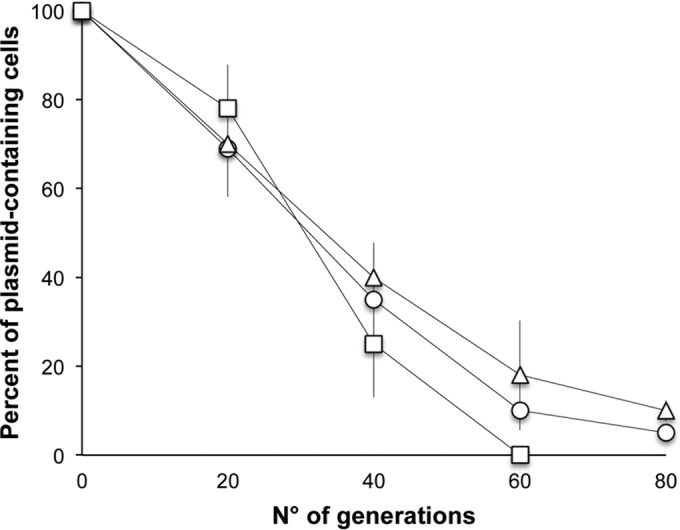
Stability of pKBuS13 in K. pneumoniae KBu-1 (circles) and in E. coli JM101 (triangles). Plasmid pUC19 carried by E. coli JM101 (squares) was used as a control, as it lacks a Xer recombination site and is randomly partitioned during cell division. Plasmid content of strains cultured under nonselective conditions for the indicated number of generations was analyzed. The graph shows the means of three independent experiments plus or minus the standard deviations.
The low stability of pKBuS13 proves that the activity of the transposon resolvase alone is not sufficient to stabilize this plasmid, suggesting that the level of dimer resolution needed for stabilization may be achieved by the cooperation between the Xer system and the transposon resolvase, and therefore they are both necessary.
These results suggested that the two plasmids detected in the E. coli recipient were the monomeric and tetrameric forms of pKBuS13, a hypothesis that was confirmed by restriction analysis with four different enzymes (BamHI, HindIII, SacI, and PstI), of plasmids extracted from E. coli JM101, which always gave the pattern expected for pKBuS13 (Fig. 4).
FIG 4.
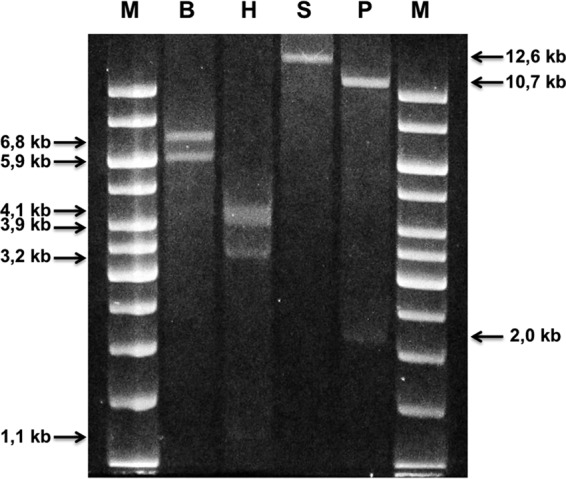
Restriction analysis of pKBuS13 separation on 0.8% agarose gel electrophoresis of pKBuS13 extracted from the E. coli JM101 recipient and digested with BamHI (lane B), HindIII (lane H), SacI (lane S), and PstI (lane P). Lane M, GeneRuler 1-kb DNA ladder (Thermo Scientific).
The 1,118-bp region located downstream of the tnpR side of the transposon carried an unknown ORF2 that retrieved a single match in the database: a fragment of the genome of a carbapenem-resistant E. coli strain, named BIDMC43b (GenBank accession number JAPE01000031), detected in a blood culture from a hospital in Boston in December 2012. E. coli BIDMC43b is part of the Carbapenem Resistance Initiative, an epidemiologic study currently in progress at the Broad Institute of MIT and Harvard (broadinstitute.org). Its entire genome has been sequenced by a shotgun approach and is now at the scaffold assembly level, so little information is yet available (January 2015). The same strain carries a Tn4401b too, although in a different region of the genome (GenBank accession number JAPE01000025), so the hypothesis that pKBuS13 might have originated by genomic rearrangements in this strain (or in an analogous one) should be taken into account.
In conclusion, pKBuS13 is a small plasmid carrying only one resistance determinant, and it is not self-transmissible by conjugation as it does not contain tra genes (although its mobilization in the presence of a helper plasmid cannot be excluded), so it might be considered unimportant for dissemination of antibiotic resistance. Nevertheless, the finding that part of its sequence did not have significant matches in the database other than the genome of a carbapenem-resistant E. coli strain detected very far from Italy and from South America is interesting for epidemiologic studies, as it might mirror a wider distribution of this kind of plasmids than that reported. Moreover, the finding that it is carried by a strain that hosts many different plasmids (Fig. 1A), along with the ability of Tn4401 to undergo replicative transposition, agrees with the report that many different blaKPC-carrying genetic platforms are circulating in Latin America (2) and represents a particularly worrisome circumstance.
The plasmid instability described for pKBuS13 is a peculiar feature that shows both positive and negative aspects. In the clinic, the detection of unstable plasmids might be considered less alarming compared to that of other plasmids, as their spread might be considered containable provided that appropriate antibiotic control policies were adopted. However, researchers that study the epidemiology of resistance determinants should take into account this property, as it might lead to overestimation of the number of plasmids harbored by clinical isolates.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the University of Trieste (grant FRARizzo 2013).
We are grateful to D. Pizzo for the experiments carried out during his thesis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04543-14.
REFERENCES
- 1.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade LN, Curiao T, Ferreira JC, Longo JM, Climaco EC, Martinez R, Bellissimo-Rodrigues F, Basile-Filho A, Evaristo MA, Del Peloso PF, Ribeiro VB, Barth AL, Paula MC, Baquero F, Canton R, Darini AL, Coque TM. 2011. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother 55:3579–3583. doi: 10.1128/AAC.01783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomez SA, Pasteran FG, Faccone D, Tijet N, Rapoport M, Lucero C, Lastovetska O, Albornoz E, Galas M, Melano RG, Corso A, Petroni A. 2011. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin Microbiol Infect 17:1520–1524. doi: 10.1111/j.1469-0691.2011.03600.x. [DOI] [PubMed] [Google Scholar]
- 4.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leavitt A, Carmeli Y, Chmelnitsky I, Goren MG, Ofek I, Navon-Venezia S. 2010. Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob Agents Chemother 54:3002–3006. doi: 10.1128/AAC.01818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. 2011. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 66:307–312. doi: 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- 7.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giani T, D'Andrea MM, Pecile P, Borgianni L, Nicoletti P, Tonelli F, Bartoloni A, Rossolini GM. 2009. Emergence in Italy of Klebsiella pneumoniae sequence type 258 producing KPC-3 carbapenemase. J Clin Microbiol 47:3793–3794. doi: 10.1128/JCM.01773-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R, Pantosti A, Pagani L, Luzzaro F, Rossolini GM. 2013. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey. Euro Surveill 18(22):pii: 20489 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20489. [PubMed] [Google Scholar]
- 10.Castanheira M, Costello AJ, Deshpande LM, Jones RN. 2012. Expansion of clonal complex 258 KPC-2-producing Klebsiella pneumoniae in Latin American hospitals: report of the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 56:1668–1669. doi: 10.1128/AAC.05942-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, Gales AC, Venezia SN, Quinn JP, Nordmann P. 2010. Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg Infect Dis 16:1349–1356. doi: 10.3201/eid1609.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Mugnaioli C, Luzzaro F, De Luca F, Brigante G, Perilli M, Amicosante G, Stefani S, Toniolo A, Rossolini GM. 2006. CTX-M-type extended-spectrum beta-lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob Agents Chemother 50:2700–2706. doi: 10.1128/AAC.00068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanson ND, Thomson KS, Moland ES, Sanders CC, Berthold G, Penn RG. 1999. Molecular characterization of a multiply resistant Klebsiella pneumoniae encoding ESBLs and a plasmid-mediated AmpC. J Antimicrob Chemother 44:377–380. doi: 10.1093/jac/44.3.377. [DOI] [PubMed] [Google Scholar]
- 15.Ellington MJ, Kistler J, Livermore DM, Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother 59:321–322. [DOI] [PubMed] [Google Scholar]
- 16.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Smith Moland E, Hanson ND, Herrera VL, Black JA, Lockhart TJ, Hossain A, Johnson JA, Goering RV, Thomson KS. 2003. Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J Antimicrob Chemother 51:711–714. doi: 10.1093/jac/dkg124. [DOI] [PubMed] [Google Scholar]
- 18.Selvaraj G, Iyer VN. 1983. Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol 156:1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning. A laboratory manual. 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 20.Grundmann H, Schneider C, Hartung D, Daschner FD, Pitt TL. 1995. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J Clin Microbiol 33:528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenover FC, Kalsi RK, Williams PP, Carey RB, Stocker S, Lonsway D, Rasheed JK, Biddle JW, McGowan JE Jr, Hanna B. 2006. Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg Infect Dis 12:1209–1213. doi: 10.3201/eid1208.060291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tolmasky ME, Colloms S, Blakely G, Sherratt DJ. 2000. Stability by multimer resolution of pJHCMW1 is due to the Tn1331 resolvase and not to the Escherichia coli Xer system. Microbiology 146:581–589. [DOI] [PubMed] [Google Scholar]
- 23.Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labrador I, Araque M. 2014. First description of KPC-2-producing Klebsiella oxytoca isolated from a pediatric patient with nosocomial pneumonia in Venezuela. Case Rep Infect Dis 2014:434987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcano D, De Jesus A, Hernandez L, Torres L. 2011. Frequency of enzymes associated with reduced sensitivity to beta-lactam antibiotics in enterobacteria isolates, Caracas, Venezuela. Rev Panam Salud Publica 30:529–534. (In Spanish.) [PubMed] [Google Scholar]
- 26.Colloms SD. 2013. The topology of plasmid-monomerizing Xer site-specific recombination. Biochem Soc Trans 41:589–594. doi: 10.1042/BST20120340. [DOI] [PubMed] [Google Scholar]
- 27.Sarno R, McGillivary G, Sherratt DJ, Actis LA, Tolmasky ME. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob Agents Chemother 46:3422–3427. doi: 10.1128/AAC.46.11.3422-3427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran T, Sherratt DJ, Tolmasky ME. 2010. fpr, a deficient Xer recombination site from a Salmonella plasmid, fails to confer stability by dimer resolution: comparative studies with the pJHCMW1 mwr site. J Bacteriol 192:883–887. doi: 10.1128/JB.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquali F, Kehrenberg C, Manfreda G, Schwarz S. 2005. Physical linkage of Tn3 and part of Tn1721 in a tetracycline and ampicillin resistance plasmid from Salmonella Typhimurium. J Antimicrob Chemother 55:562–565. doi: 10.1093/jac/dkh553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.