Abstract
Vinculin is an abundant protein found at cell-cell and cell-extracellular matrix junctions. In muscles, a longer splice-isoform of vinculin, metavinculin, is also expressed. The metavinculin-specific insert is part of the C-terminal tail domain, the actin-binding site of both isoforms. Mutations in the metavinculin-specific insert are linked to heart disease such as dilated cardiomyopathies. Vinculin tail domain (VT) both binds and bundles actin filaments. Metavinculin tail domain (MVT) binds actin filaments in a similar orientation but does not bundle filaments. Recently, MVT was reported to sever actin filaments. In this work, we asked how MVT influences F-actin alone or in combination with VT. Cosedimentation and limited proteolysis experiments indicated a similar actin binding affinity and mode for both VT and MVT. In real time TIRF microscopy experiments MVT’s severing activity was negligible. Instead, we found that MVT binding caused a two-fold reduction in F-actin’s bending persistence length and increased susceptibility to breakage. Perhaps MVT allows the load of muscle contraction to act as a signal to reorganize actin filaments. Using mutagenesis and site-directed labeling with fluorescence probes, we determined that MVT alters actin interprotomer contacts and dynamics, which presumably reflect the observed changes in bending persistence length. Finally, we found that MVT decreases the density and thickness of actin filament bundles generated by VT. Altogether, our data suggest that MVT alters actin filament flexibility and tunes filament organization in the presence of VT. Both of these activities are potentially important for muscle cell function.
Introduction
Vinculin is an essential 116 kDa adhesion protein of cell-matrix and cell-cell junctions 1,2. It contains largely helical head and tail domains connected by a flexible proline-rich linker 3. The head and tail domains interact in an auto-inhibitory fashion, occluding binding to a large number of potential ligands 3-6. Many of vinculin’s ligands are adhesion proteins and/or cytoskeletal proteins. Through these interactions, vinculin anchors the actin cytoskeleton to membrane adhesion structures, making it an important player at these sites.
The vinculin tail (VT) is sufficient to bind and bundle actin filaments. Crystal structures reveal that VT is composed of five alpha-helices (H1-H5) connected by short loops in an antiparallel bundle configuration similar to that seen in exchangeable apolipoproteins 3. The helices are flanked by a so-called N-terminal strap and C-terminal arm, which interact with each other and a subset of the VT helices. The VT can dimerize in a number of configurations, one of which relies on a cryptic binding site that is revealed upon binding to actin and results in bundling 7,8.
Metavinculin, a larger splice variant of vinculin, is muscle specific. In humans, alternative splicing introduces a 68 amino acid acidic insert after residue 915 in the C-terminal half of vinculin 9,10. A recent crystal structure of full length metavinculin 11 revealed that the sole structural difference between the two isoforms is the replacement of vinculin tail’s H1 α-helix with a new α-helix (referred to as H1’) formed by the C-terminus of the MVT insert. The preceding N-terminal region of the MVT insert is disordered in the crystal structure. When MVT-decorated filaments are compared with VT-decorated filaments by electron microscopy, an extra density away from the binding interface is apparent and attributed to MVT’s acidic insert 12.
Metavinculin colocalizes with vinculin at adhesive structures, such as dense plaques in smooth muscles, intercalated discs in cardiomyocytes, and costameres in skeletal muscles 13-15. In smooth muscle, metavinculin can account for up to half of the total vinculin pool 13,16. Importantly, metavinculin expression was shown to increase with muscle differentiation, and be specific to skeletal muscle fiber subtype, pointing to a role in contractility 16,17. Moreover, deficiency or reduction in metavinculin expression levels are linked to dilated cardiomyopathy 18 or plaque formation, respectively, during human coronary arteriosclerosis 19.
Several lines of evidence suggest that binding of VT and MVT to actin results in distinct filament organization. The actin binding and bundling activity of VT is well characterized in vitro 8,11,12,17,20-23. MVT has been shown to induce a fine meshwork of actin filaments, distinct from VT induced bundles 11,17,22,24. A trivial explanation is that MVT does not dimerize like VT, preventing bundling 11,22. However, MVT was also found to sever actin filaments 12. Tissue analysis from patients with mutations in the MVT insert revealed a disrupted organization of intercalated discs and actin filaments 24,25. Together these data suggest that metavinculin influences actin organization and mechano-transduction properties though the mechanism remains unclear.
Because metavinculin and vinculin are expressed at varying levels relative to one another in the same cells, we set out to understand the unique actin organization properties of MVT in the absence and presence of VT. We initially focused on the recently reported severing activity. However, we found that MVT does not actively sever actin filaments. We show here that MVT alters the mechanical properties of F-actin, rendering filaments partially decorated with MVT prone to fragmentation. Given that metavinculin is specifically expressed in muscle cell, the induced change in actin filament mechanics could be a mechanism to sense load. When examining the combined effects of the two vinculin tail isoforms on actin network formation, we observed a greater loss of bundling than would be predicted by a simple competition model. We propose that metavinculin acts as a regulator of actin filament mechanics and of architecture induced by vinculin.
Results
Comparison of binding between vinculin tail isoforms and actin filaments
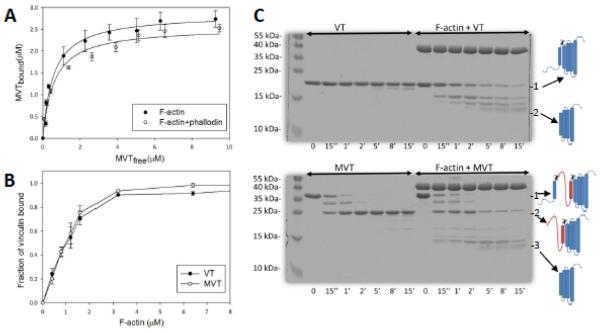
We measured the binding affinity of VT and MVT for actin filaments using high-speed cosedimentation assays. Unless otherwise indicated, experiments were performed with rabbit skeletal muscle actin. Both VT (residues 879-1066) and MVT (residues 879-1134) bind to F-actin with 1:1 stoichometry and approximately the same binding affinity (Kd MVT = 0.6+/−0.2 μM, Kd VT = 0.5+/−0.1 μM) suggesting that the acidic insert does not alter MVT’s binding to F-actin (Fig. 1). Phalloidin does not alter MVT’s affinity for F-actin (Fig. 1a). The findings are comparable to the dissociation constant reported for a slightly longer construct of VT (residues 811-1066) and F-actin, ~0.6-0.8 μM 21,26.
Figure 1. Comparison of binding between vinculin isoforms and actin filaments.
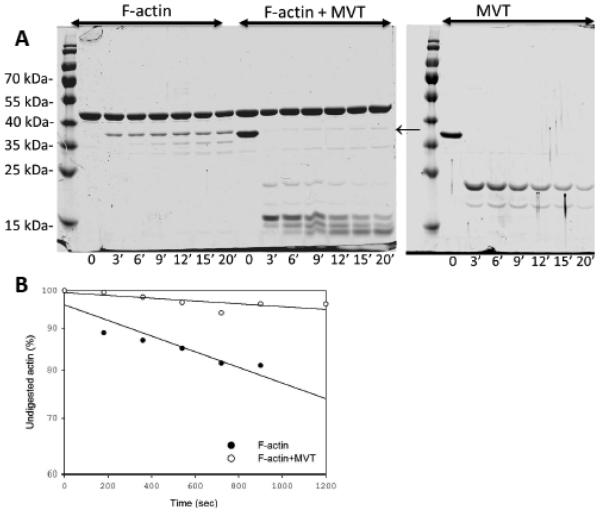
A) Binding of MVT to F-actin. F-actin (2.5 μM) or F-actin stabilized with equimolar phalloidin was incubated with increasing amounts of MVT. Bound MVT was determined by centrifugation and SDS-PAGE analysis. Solid line represents the best fit to the average values from three independent experiments. Average Kd of MVT for F-actin was 0.56 ± 0.09 μM and for F-actin-phalloidin was 0.6 ± 0.1 μM. B) Binding of F-actin to VT or MVT. VT (2.0 μM) or MVT (2.0 μM) was incubated with increasing amounts of phalloidin stabilized F-actin. Affinities are similar for the two vinculin isoforms. C) F-actin binding increases the proteolytic access of VT and MVT’s N-terminal helices. 10 μM MVT or VT alone or bound to 10 μM F-actin were digested with α-chymotrypsin at an enzyme to protein mass ratio of 1:300 (α-chymotrypsin:actin) for the indicated time intervals. Reaction aliquots were taken at the indicated time points and were analyzed with 12% SDS-PAGE gels. The molecular masses of digested peptides in aliquots of VT (15 min), MVT (1 min), F-actin+VT (18 min) and F-actin+MVT (21 min) were analyzed by MALDI-TOF mass spectrometry. The cartoon on the right illustrates the full length VT (“1”) and MVT (“1”) with expected cleavage sites and their major cleavage products (“2” for VT, and “2 and 3” for MVT). The MVT insertion is rapidly digested in the presence or absence of actin. F-actin accelerates digestion of the vinculin isoforms in a similar manner.
EM reconstruction of MVT in complex with F-actin 12 shows an extra density away from the binding interface, which was attributed to MVT’s acidic insert. One consequence of such an arrangement would be solvent exposure of the insert and susceptibility to proteolytic digestion. To test this scenario we digested F-actin-MVT and F-actin−VT complexes with α-chymotrypsin as previously done for F-actin−VT 3. Consistent with Bakolitsa et al., (2004) we found that in the absence of F-actin, VT was quite resistant to proteolysis, with a minor cleavage product observed at 15 minutes (Fig. 1C). Using MALDI-TOF mass spectrometry, we identified the cleaved site to be the C-terminal arm of VT after residue F1054. In contrast, VT was readily cleaved in the presence of F-actin. The major cleavage site was between residues W912 and S913 at the linker connecting helix 1 to helix 2. The acidic insert of MVT is positioned between these two helices. Interestingly, the insert was shown to allow the formation of a helix, H1’, with an identical amino acid sequence to the C-terminus of original helix, H1. We found that cleavage between W912 and S913 was targeted by chymotrypsin in MVT alone (Fig. 1c). This site was also targeted rapidly when MVT was bound to F-actin. This result confirms that the W912/S913 site is solvent exposed as predicted based on EM reconstruction and crystal structures11,12. Similar to our results with VT-F-actin, a second chymotrypsin target site became more exposed in MVT when it was bound to F-actin. We confirmed that this cleavage site was between W980 and S981 of helix H1’ in MVT. Notably, F-actin bound MVT and VT were digested with chymotrypsin at similar rates to yield the same protease resistant fragment with an average mass of 15.5 kDa (Fig. 1c). Therefore, our proteolysis data support the structural data that suggest that the core binding interface between actin and vinculin is preserved in both VT and MVT 8,12. This is also consistent with the finding that VT and MVT have approximately the same binding affinity for F-actin (Fig. 1b).
Real time severing assays
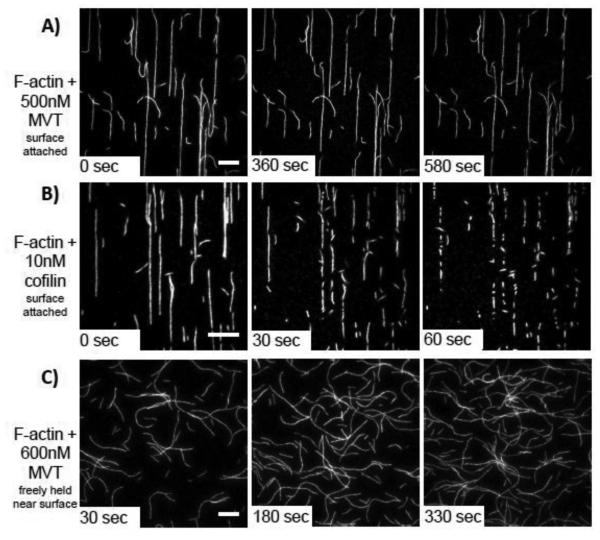
While actin filament bundling by vinculin is well studied, little is known about actin severing by MVT. To gain more insight into this activity, we used TIRF microscopy to observe MVT-induced severing in real time. We attached actin filaments (labeled with 0.5% biotin and 20% Cy3b) to streptavidin coated cover slips and then added MVT at varying concentrations. To our surprise, observable severing events in the presence of MVT were rare. Janssen et al.12 reported that severing activity was greatest at half binding density. Therefore, we added 0.5 µM MVT, which should correspond to a half binding density assuming 5-15 nM F-actin on the surface. Under these conditions only a few breaks were observed over 10 minutes (Fig. 2a, Supplementary Movie 1). Under similar conditions, a known severing protein, yeast cofilin (10 nM), resulted in many break points in as little as 1 minute (Fig. 2b, Supplementary Movie 2). To rule out the possibility that the filaments become saturated with MVT and then resistant to severing, we tested a range of MVT concentrations (50 nM-2 μM, data not shown). Apparent severing events were observed at very low frequency, if at all, regardless of the concentration of MVT added.
Figure 2. MVT does not actively sever actin filaments.
A-B) 1μM Mg-ADP-F-actin (20% Cy3b-labeled, 0.5% Biotin-Peg2-labeled) was attached to biotin-PEG on functionalized coverslips through streptavidin cross-links. Once an initial image was acquired (0 sec), the indicated proteins were added and movies were acquired. A) Very few breaks were observed when MVT was added (see also in Supplementary Movie 1). B) Cofilin rapidly severs filaments under the same conditions (see also in Supplementary Movie 2). Scale bars = 10 μm. Cofilin images are shown at a higher magnification to better visualize the large number of severing events. C) F-actin (20% Cy3b-labeled) was diluted to 25 nM in the presence of 0.6 μM MVT (binding density = 0.5) and immediately added to chambers treated with 1% Pluronic F-127 and 0.5 mg/ml casein. In this experiment, the filaments are not covalently attached to the surface. No cuts were detected. Scale bar = 20 μm.
Because surface contacts can influence F-actin severing in a number of ways, we also performed experiments in which we held filaments near the coverslip without covalent attachments. We pre-treated our flow cells with Pluronic F-127, and then incubated the sample chamber with casein. Similar set ups have been shown to be effective in blocking protein adsorption in single-molecule experiments and allowing visualization of more freely moving actin and microtubule filaments by TIRF 27-29. In this assay, we mixed MVT and F-actin before adding them to the flow cell in order to know the exact ratios and consequently the binding densities of proteins in a given experiment. Despite both of these changes, we did not observe severing of F-actin partially decorated with MVT (Fig. 2c, Supplementary Movie 3). These results led us to question whether or not MVT is an actual severing protein.
Because significant shortening of filaments by MVT was observed by Janssen et al. 12, we reproduced their two-step seeded actin assembly assays. In these assays, the rate of assembly is directly proportional to the amount of free barbed ends or the number of “seeds”. The number of seeds increases if filaments are severed. To create seeds, we pre-incubated F-actin (2 μM) with increasing amounts of MVT. We then added actin monomers and monitored the rate of assembly. Like Janssen et al. 12, we observed a biphasic pattern in actin polymerization rates which was maximal when F-actin was incubated with MVT at half binding density (Supplementary Fig. 1a and 1b). Interpretation of this assay relies on the assumption that MVT is not changing the nucleation and/or elongation rates of actin polymerization. We confirmed this in real time polymerization assays using TIRF microscopy (Supplementary Fig. 1c and 1d). Therefore, we concluded that the dose dependent change we observed in the seeded elongation assay is indeed a consequence of increased barbed ends and that our MVT behaves similar to that used by Janssen et al12.
We next used direct observation of filaments incubated with MVT. We incubated fluorescently labeled F-actin with MVT and analyzed the average filament length (Lavg) at steady state as a function of MVT binding density using fluorescence microscopy (Supplementary Fig. 1e and 1f). Consistent with Janssen et al. and our seeded elongation assay, these results showed that MVT-mediated severing demonstrates a biphasic dependency where maximal severing activity is at binding densities close to 0.5 (molar ratio of 1:1 for actin/MVT). Thus we needed to resolve the discrepancy between the bulk and single filament assays.
Equilibrium severing assays
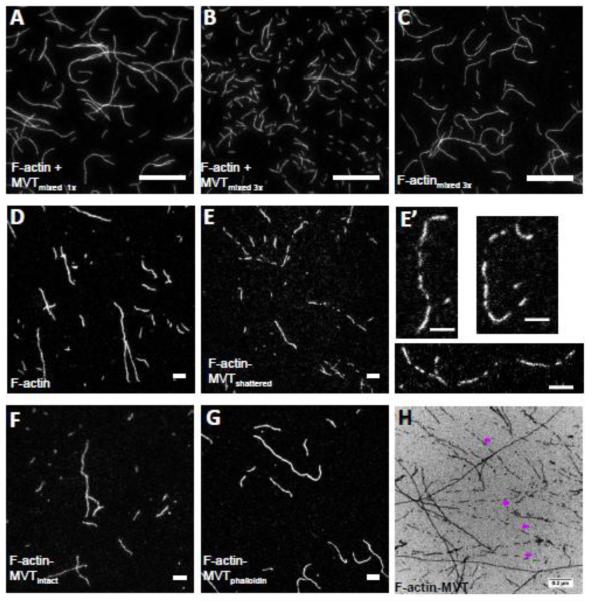
Two major differences exist between TIRF severing assays and barbed end elongation or steady state assays that could influence severing activity: surface interactions and shear forces. We used wide bore pipette tips whenever we handled actin filaments to minimize shear forces but these forces cannot be completely eliminated. In order to ask whether MVT bound filaments are more susceptible to shear forces (such as those introduced when pipetting), we measured filament lengths when F-actin and MVT were mixed and pipetted up and down once or three times. Because apparent severing activity was greatest at half binding density, we continued to use this condition. MVT-decorated filaments were noticeably shorter when mixed three times compared to once (Lavg of 4.3 ± 0.3 μm vs. 16 ± 2 μm) (Fig. 3a vs. b). Notably, control filaments (no MVT) mixed three times were also long (Lavg of 11.5 ± 1.2 μm) (Fig. 3c). Therefore, we propose that MVT destabilizes actin filaments but is not a severing protein like cofilin or gelsolin.
Figure 3. Partial decoration of F-actin with MVT leads to filament fragmentation during sample handling.
A-C) F-actin (1 μM, 20% Cy3b-labeled) was diluted to 25 nM in the presence (A and B) and absence (C) of 0.6 μM MVT (binding density ~0.5) and added to chambers treated with 1% pluronic acid and 0.5 mg/ml casein. Samples with MVT and pipetted three times before addition to the chamber were shorter than those (B) with MVT but pipetted only one time or (C) control samples pipetted three times. Scale bar = 20 μm. D-G) F-actin (2 μM, 20% Cy3b-labeled) was incubated for 2 hours with either 1.6 μM MVT (binding density = 0.5) or MVT storage buffer. Actin samples were diluted to 5 nM in F-buffer supplemented with 100 mM BME containing 0.6 μM MVT in order to maintain a binding density of 0.5 with or without phalloidin. The diluted solutions were spotted onto a glass slide and a poly-L-lysine coated coverslip was placed on top. Scale bars = 5 μm. Representative images are shown for (D) control filaments, (E-F) F-actin-MVT diluted in MVT buffer (different fields from the same sample where filaments appeared fragmented and intact, respectively), and (G) F-actin-MVT diluted in phalloidin buffer. (E’) An enlarged image of fragmented filaments prepared as in (E). See filament length distribution in Supplementary Figure 2. H) Electron micrograph of F-actin (2.5 μM) incubated with equimolar MVT. Pre-polymerized actin was mixed with storage buffer (control not shown) or MVT (H) and spotted on carbon EM grids. As seen in fluorescent images, actin filaments were often fragmented in the presence of MVT as indicated by purple arrows.
To further assess whether or not MVT could sever, we revisited the steady state assay. We took two approaches in sample preparation, either diluting samples into more MVT to maintain constant binding density or diluting samples into phalloidin to capture the length distribution before adding the sample to the slide. When samples were diluted in MVT we observed sometimes two distinct populations of filaments on the microscope slides. Namely, there were regions with short filaments (Fig.3e and e’) and regions with longer filaments (Fig.3f and Supplementary Fig. 2). Many of the short filaments were aligned as if they were the product of a single filament, which shattered (Fig. 3e’). We observed the same bimodal length distribution by fluorescence microscopy and electron microscopy (Fig. 3h). The short filaments were absent when the same MVT-F-actin solution was diluted into phalloidin (Fig. 3g), which is well documented to stabilize F-actin. The average filament length, when diluted into phalloidin, (Lavg of 7.5 ± 0.6 μm) was comparable to the control (Lavg of 9.4 ± 0.7 μm) (Fig.3d vs 3g and Supplementary Fig. 2). Thus, in all cases where MVT decorated filaments were pipetted and/or placed on a slide, the filaments were shorter than controls. In contrast, when filaments were stabilized with phalloidin they were approximately the length of control filaments. Vinculin and metavinculin may encounter both muscle actin and non-muscle actin. Because actin binding proteins can be sensitive to actin isoforms we repeated some experiments with actin purified from yeast as a surrogate for non-muscle actin. Yeast actin is well known to have cytoplasmic actin-like properties, in terms of dynamics and polymerization, in comparison to sarcomeric actin. We asked whether wild type yeast actin filaments were more susceptible to fragmentation when half saturated (v = 0.5) with MVT. Indeed they were (Supplementary Figure 3a vs. 3b). Incubation of yeast actin with MVT but no phalloidin resulted in filaments with a shorter average length and many aligned short filaments that appear to come from the breakage of a single longer actin filament (Supplementary Figure 3b’). These observations, along with the real time severing assays, led us to hypothesize that MVT does not sever but instead destabilizes filaments.
In light of these conclusions we revisited the two-step seeded actin assembly assays. We redesigned it to minimize shear stress that the actin filaments experience from pipetting. Specifically, we allowed F-actin and MVT to incubate for 30 minutes in the multiwell plate where measurements were made. We then gently added buffer and monomeric actin and mixed by stirring, not pipetting. In this case, MVT did not cause increased barbed end production (Supplementary Fig. 1e and 1f). We used cofilin as a positive control to demonstrate that the mixing was sufficient. Together, these data support our revised understanding of MVT: it does not sever but does destabilize filaments leaving them prone to fragmentation.
MVT binding alters F-actin mechanics
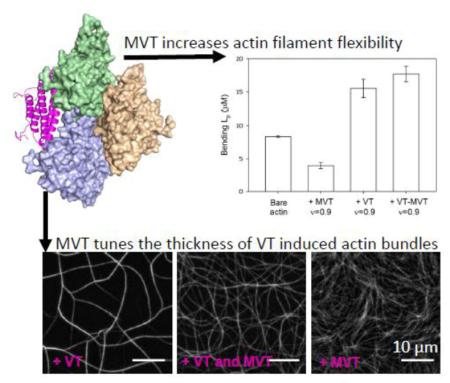
The above conclusion on filament destabilization by MVT led us to measure the flexural rigidity of filaments decorated with MVT. We determined the bending persistence length (Lp) of MVT-decorated Alexa488-labeled filaments from a two-dimensional cosine correlation of segment tangent angles 30,31. MVT-decorated actin filaments (v = 0.9) are ~2-fold more compliant in bending than bare filaments, as indicated by the persistence length values of 3.9 ± 0.5 μm and 8.4 ± 0.2 μm, respectively (Fig. 4). These results raise the possibility that fragmentation of filaments partially decorated with MVT may arise from mechanical heterogeneities introduced by MVT. The observation that apparent severing peaks when filaments are half-saturated (v = 0.5; Supplementary Fig. 1b), suggests that fragmentation occurs at or near boundaries between bare and decorated segments.
Figure 4. MVT binding alters F-actin mechanics.
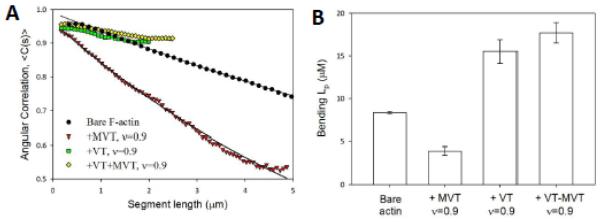
A) Flexural rigidities of bare, MVT−, VT−, and VT+MVT-decorated actin filaments (All binding densities = 0.9). Solid lines represent best fits of the two-dimensional average cosine angular correlation, <C(s)>, along filament segment lengths (x) (<C(s)> = A*exp(−x/2 Lp)). B) Bending Lp calculated from the best fit of angular correlation data as shown in (A). Lp of bare actin: 8.35 ± 0.17μm, MVT-decorated actin: 3.92 ± 0.46μm, VT-decorated actin: 15.5 ± 1.4μm, MVT/VT-decorated actin: 17.7 ± 1.2μm. Uncertainty corresponds to the standard error of the fit, n = 50-200 filaments/condition.
MVT binding modulates actin-actin contacts
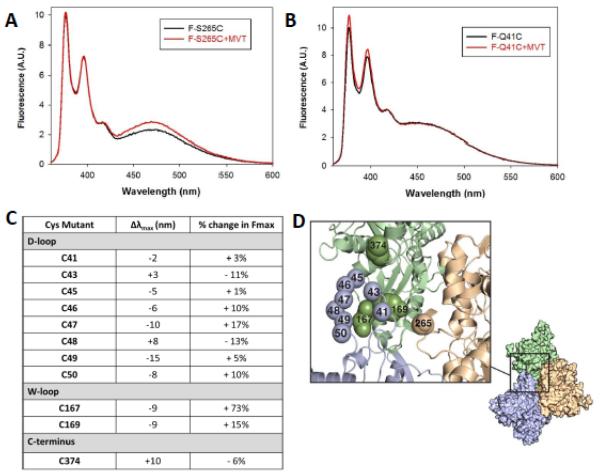
Next, we probed interprotomer contacts to study structural changes caused by MVT using yeast actin mutants. The fact that yeast actin was susceptible to fragmentation at half binding densities similar to alpha skeletal muscle actin (Supplementary Fig. 3a-c) suggests that yeast actin mutants reliably report MVT induced changes in actin. Therefore, we focused on understanding the MVT induced changes in dynamic actin loops (H- (a.a. 262-274), D-(a.a. 40-50), W- (a.a. 167-170) and the C-terminus 32-35. First, we asked whether MVT caused rearrangements in lateral and/or longitudinal interprotomer contacts. To do so, we measured fluorescence excimer formation by pyrene labeled cysteine pairs in mutant yeast actins36: lateral actin proximity is reported by pyrene labels at Cys265 and Cys374 (referred to as S265C because this residue is mutated)33,37; longitudinal proximity is reported by pyrene bound to Cys41 and Cys374 (Q41C) 32. In both cases, upon polymerization, two pyrene rings from neighboring protomers stack to form an excimer band. Any changes in the amplitude of these excimer spectra are likely to reflect a change in the plasticity or the position of interprotomer loops in F-actin caused by MVT. We observed that MVT binding enhances the pyrene excimer emission of S265C actin (~25% increase in excimer emission maxima, Fig. 5a). However, MVT had essentially no effect on the excimer emission of Q41C actin (Fig. 5b). The simplest interpretation of these data is that MVT binding predominantly affects the lateral interactions between actin filament strands.
Figure 5. MVT binding predominantly affects the lateral interactions between actin protomers.
Pyrene emission spectra of 8 μM (A) F-S265C or (B) F-Q41C actin with or without 8μM MVT. Spectra were recorded between 360–600 nm, with the excitation wavelength set at 344 nm. The pyrene excimer emission maxima was enhanced by MVT addition to S265C while it was unaffected in Q41C filaments. C) Relative differences in fluorescence amplitude (Fmax) and emission wavelength maxima (Dlmax) caused by addition of MVT (5 μM) to actin filaments (5 μM) labeled with prodan at the indicated residues. Fluorescence emission spectra of prodan were recorded between 400–650 nm, with the excitation wavelength set at 385 nm. D) A three subunit Fujii model69 of the actin filament is shown 70. The spheres indicate the locations of amino acids that were mutagenized and probed in A-C. Proximity of these residues to MVT is modeled in Supplementary Figure 4.
Next, we used prodan-labeled yeast actin with cysteine mutants introduced in the D-loop, W-loop, and C-terminus. These mutants were previously used to probe direct and/or allosteric changes in F-actin induced by myosin binding 38. Addition of equimolar or more MVT to prodan labeled filaments revealed spectral changes at each residue tested (Fig. 5c), while the magnitude of spectral changes varied among the positions tested. We observed that the prodan located on C-terminal residues of the D-loop (residues 47 to 50) had more pronounced changes in quantum yields and emission maxima compared to the N-terminal residues of the D-loop (residues 41 to 46). Mutant C167 in the W-loop showed the most dramatic change in spectral properties with 70% increase in quantum yield. This large change at C167 may arise from direct binding of MVT in the vicinity of the W-loop as this site was modeled to be at the binding interface between VT and F-actin by mutagenesis, molecular dynamics and EM reconstruction 8,39. Spectral changes in probes positioned away from the predicted binding interface suggest that some allosteric conformational changes in F-actin loops are induced by MVT binding (Fig. 5d and Supplementary Fig. 4).
Next we asked whether MVT increases the exposure of actin’s D-loop using limited proteolysis with subtilisin. Cofilin, which also decreases actin filament persistence length 30,40,41, increases the proteolytic exposure of actin’s D-loop 34 .In contrast, decoration by MVT inhibits subtilisin cleavage of actin’s D-loop between residues 47 and 48 compared to actin alone (Fig. 6). F-actin bound MVT was cleaved to yield a lower molecular weight product (above 15 kDa) compared to MVT alone at early time points of the reaction. This is consistent with our limited digest findings with chymotrypsin (Fig. 1c) suggesting that the N-terminal helices of MVT undergo a conformational change upon actin binding and become more exposed to the solvent. We note that this remaining fragment of MVT, which is comparable molecular weight to the protease resistant core of MVT observed upon digestion with chymotrypsin, is also sufficient to protect F-actin from subtilisin cleavage.
Figure 6. Limited proteolysis of MVT decorated F-actin by subtilisin.
A) 10 μM F-actin, 10 μM MVT, and 10 μM F-actin+MVT were all digested with the same concentration of subtilisin, based on enzyme to protein mass ratio of 1:50 (subtilisin:F-actin). Reaction aliquots were taken at the indicated time points and were analyzed by 12% SDS/PAGE gels. B) Digestion of actin monomer was analyzed by densitometry of intact actin monomer band. MVT protects actin’s D-loop from proteolytic cleavage by subtilisin.
In summary, the more pronounced changes we detected were in lateral interactions. We propose that they account for, or at least contribute to, the decrease in persistence length we measured in the presence of MVT.
MVT and the F-actin bundling activity of VT
In order to gain insight into the effects of vinculin isoforms on actin filament mechanics, we also measured the Lp of VT decorated filaments. In the samples containing only VT, we observed primarily large (10-20 μm long) bundles but a population of single filaments was also evident. We analyzed these single filaments. In contrast to MVT, VT decoration resulted in longer bending persistence length (Lp of 15±1 μm, Fig. 4). Due to the potent bundling activity of VT, we could not measure its effects on actin-actin contacts, as we did for MVT. Therefore, we cannot determine which, if any, of the structural changes we observed are unique to MVT. However, the difference in persistence length supports the idea that MVT and VT influence actin filaments in distinct manners.
Given that metavinculin is always expressed in conjunction with vinculin, studying the combination of both isoforms may be more relevant for understanding their roles. We asked how a 1:1 mixture of the two tail isoforms affects the mechanical properties of actin filaments. As in the samples containing only VT, we observed some bundles as well as short single filaments. Analysis of the single filaments showed a minor increase in the persistence length (18±1 μm) compared to VT alone (Fig. 4). Therefore, we conclude that the filaments co-decorated with MVT and VT are very similar to the VT-decorated filaments in terms of mechanics (Fig. 4). However, the local ratios of MVT to VT may vary between 0-1 so our results with the 1:1 mixture reflect an ensemble average and may not reflect the local mechanical properties of filaments decorated with vinculin tail isoforms.
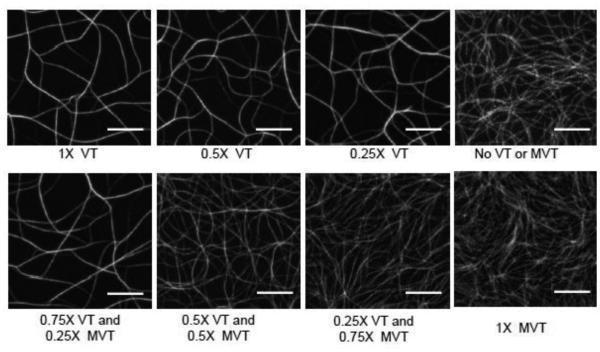
To further elucidate the functional role of MVT in actin organization and the consequences of having both isoforms in a given cell, we studied the combination of proteins in bundling assays using TIRF microscopy. Handling VT-induced actin bundles was challenging and we often observed the breakage of bundles when we transferred them from test tubes to cover slips for visualization by fluorescence microscopy. Therefore, we employed real time bundling assays in which we functionalized cover slips with a low density of biotin-HMM to allow for flexible tethering of F-actin to the surface. We then added monomeric actin in the presence of VT and/or MVT and observed formation of F-actin bundles that were not perturbed by pipetting. Addition of VT, even at low fractions compared to actin, resulted in the formation of heavily bundled F-actin networks (Fig. 7, top row) as expected based on previous reports 8,11,12,17,20-23. In contrast, the presence of MVT did not result in bundling (Fig. 7, bottom row 1X MVT) also agreeing with previous results 11,17,22,24. Next, we prepared samples with varying ratios of MVT:VT. We observed that increasing the MVT:VT ratio resulted in a reduction in bundling density and thickness. At a ratio of 1:1 (MVT:VT), the organization of the actin network was visibly different: The fields were denser, presumably reflecting a decrease in bundling, and the bundles were thinner as judged by the intensity levels of individual bundles (Fig. 7) 42. We found that the average and maximum numbers of F-actin filaments in bundles decreased ~2-fold when equimolar MVT was mixed with VT (Supplementary Figure 5). At a ratio of 3:1 (MVT:VT), the actin looked more like MVT alone, despite this concentration of VT being sufficient to bundle when alone (Fig. 7, top row 0.25X VT). It appears that MVT binding inhibits VT binding or competes with VT’s ability to bundle F-actin.
Figure 7. MVT attenuates and alters the architecture of VT induced F-actin bundle networks.
Monomeric actin (1.3 μM = 1X, 15% Cy3b) and MVT and/or VT were mixed at the indicated molar ratios with polymerizing TIRF F-buffer (pH 7.4) and add to the sample chamber. The resulting actin network was visualized in real time or at steady state in flow chambers functionalized with biotin-HMM. Images shown were taken after 30 minutes of incubation at 24 °C. Scale bars = 10 μm.
MVT and VT colocalizes with the intercalated discs and costameres of cardiac muscle cells, suggesting that these proteins may interact with the sub-sarcolemmal cytoplasmic actin and/or sarcomeric muscle actin. Therefore, we repeated our experiments with yeast actin. VT induced thick actin bundles whereas MVT induced a fine mesh of actin filaments that was noted in prior work (Supplementary Fig. 6). The reduction in bundle thickness and density was obvious at a ratio of 1:1 (VT:MVT). Therefore, our microscopy observations are similar with both yeast and alpha skeletal muscle actin.
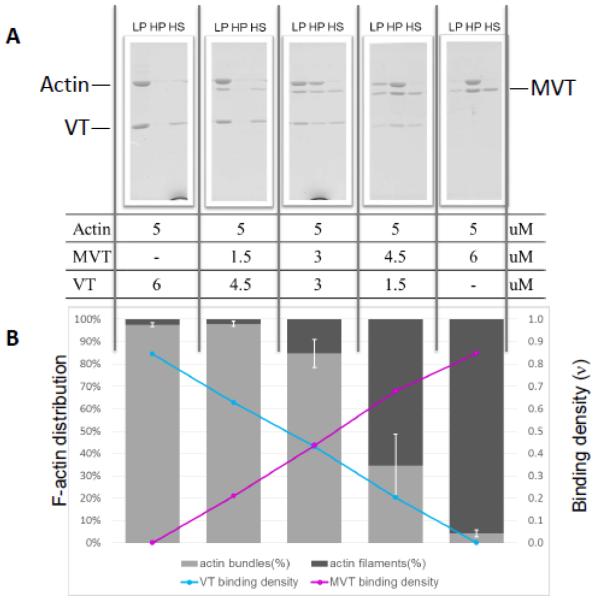
In order to analyze quantitatively what we observed in TIRF bundling assays, we turned to differential co-sedimentation assays. Consistent with our measurements showing that MVT and VT have similar affinities for actin, the amount of MVT and VT bound to filaments, reflects their input (Fig. 8). This observation suggests that there is a simple binding competition between these proteins for actin filaments. These data also agreed with our TIRF microscopy observations, showing that addition of MVT decreased VT’s bundling activity in a concentration dependent manner (Fig. 8). Noticeably, when MVT was added, bundling was reduced more than would be expected based on the VT binding densities measured (Supplementary Fig. 7). For example a binding density of ~0.2 VT is sufficient to bundle 80-90% of the F-actin in the absence of MVT. However, when conditions resulted in 20% VT and 60% MVT only ~30% of the filaments were bundled (Fig 8 and Supp. Fig. 7). Similar results were obtained using yeast actin filaments (Supplementary Fig.8). Therefore, the loss of bundling we observed in the presence of MVT is greater than would be predicted by a simple competition model and suggest that MVT may “poison” VT’s ability to bundle as discussed below.
Figure 8. MVT reduces the F-actin bundling activity of VT.
F-actin and vinculins were mixed at the indicated molar ratios and incubated for 1 hour at 24 °C. Samples were subjected to differential centrifugation to separate fractions with actin bundles (LP, low speed pellet) or filaments (HP, high speed pellet). Unbound vinculins were in the high speed supernatant fraction (HS). A) Fractions were analyzed on SDS-PAGEs and B) subjected to densitometry analysis to determine the percentage of actin filaments in bundles and the binding densities of VT and MVT on F-actin (sum of LP+HP). Increasing molar ratios of MVT to VT were found to decrease VT’s bundling activity as seen in Figure 7.
Discussion
Metavinculin is a longer splice-isoform of vinculin that is specifically expressed in mature muscle cells 13-17,43. Several lines of evidence suggest that metavinculin serves a more specialized function in mechano-transduction than vinculin. For example, metavinculin expression correlates with the contractility of muscle cells and its expression is developmentally regulated 15,44-46. Additionally, low expression levels of metavinculin 18,19 and mutations in the acidic insert of MVT 24,25 have been linked to cardiomyopathies and atherosclerosis. However, despite current efforts, we do not fully understand MVT’s function. Recent work by Janssen et al. 12 proposed that MVT severs F-actin. This was an intriguing finding, which suggested that the 68 a.a. acidic insert can switch the function of a well-known adhesion protein, vinculin, from bundling to severing. Therefore, we initially aimed to learn more about the severing activity of MVT. Instead, we determined that F-actin partially decorated with MVT has a shorter bending persistence length and is more prone to breakage. A recent theoretical study is consistent with this outcome 47.
Among the assays used in this study, TIRF experiments provide direct evidence of severing in real time. However, surface effects and protein labeling required to visualize samples can have unpredictable results on actin filaments, the potential severing protein and thereby the rate of severing (e.g. Pavlov et al. 48). Solution experiments can avoid most of these pitfalls. The surface chemistry effects are minimized but actin still has to be labeled, to be detected. We also used two categories of solution experiment, measurement of filament lengths after treatment and a functional assay to measure the number of barbed ends in solution. Both of these assays suffer from the same weakness, damaging forces. In both cases, these forces are created by pipetting, even with the best pipetting practices. Slide preparation can further perturb the sample. We sought to overcome these problems in two ways: 1) By adding phalloidin before applying the filaments to a slide, we capture the filament length distribution and protect against shearing. For practical reasons, we pipetted the MVT-treated sample once, when diluting it into a solution with phalloidin. This presumably explains why the phalloidin-stabilized filaments are slightly shorter than those of the control sample. 2) We designed our seeded elongation assay such that the MVT + F-actin solution was not pipetted. We added actin monomers to the sample and stirred gently. Given that the results from these two assays indicated that MVT does not actively sever, we also believe that our TIRF assay faithfully reports MVT’s behavior. The fact that yeast actin and rabbit muscle actin were affected similarly by MVT, combined with the mechanical and structural assays, we conclude that MVT changes interprotomer contacts in such a way that a filament is prone to breaking. The fact that metavinculin is specifically expressed in muscle cells and at varying amounts depending on stages of development or muscle fiber type, leads us to propose that metavinculin contributes to reorganization of F-actin in these cells in a manner that is sensitive to contractility.
MVT tunes the architecture and density of VT induced actin bundles
We performed experiments on the combined effects of VT and MVT because the full length proteins co-localize to muscular adhesion sites 13,43 and are proposed to work together 17. Intriguingly, the bundling architecture and density of actin networks formed in the presence of both VT and MVT was significantly different from either VT or MVT alone (Fig. 8). We found that VT and MVT binding to actin is indistinguishable. Our co-sedimentation results demonstrated that MVT and VT bind actin filaments in ratios that reflect their input ratio, which suggests that there is a simple binding competition between the two. However, the reduction in bundling we observed in the presence of both VT and MVT could not be solely due to the binding competition between the two isoforms.
The mechanism by which MVT poisons VT bundling activity is not clear at this point. MVT and VT may hetero-dimerize. There is prior work showing MVT and VT can heterodimerize in the presence of phospholipids11,17.. This hetero-dimerization would compete with VT homo-dimerization. However, in the case of VT, dimerization in the presence of phospholipids is distinct from the dimerization required for actin crosslinking. Further, if MVT and VT heterodimerized while bound to actin, the ratios of MVT and VT in our cosedimentation assays would probably not reflect the input. Thus we do not favor a model in which the two vinculin tails tightly heterodimerize. One possibility is that the MVT insert weakly interacts with the C-terminal hairpin 49 of a neighboring VT, when bound to actin, preventing VT homo-dimerization. Alternatively, or perhaps additionally, filaments decorated by MVT could repel one another due to the negative charges introduced by the acidic insert. This would also reduce the probability and frequency of VT dimerization between individual actin filaments, resulting in a network composed of more, thinner bundles.
The organization of actin filaments is likely to play a role in force generation or attachment strength at the adhesion sites. The fact that the single filaments with dual decoration have similar bending persistence lengths to VT bound filaments suggests that MVT may adjust bundle thickness and branching without altering mechanical properties. Moreover, such organization of a network of filaments with more, thinner and/or looser bundles may allow faster reorganization of the actin cytoskeleton while maintaining mechanically strong adhesion sites. Thick, tight bundles are resistant to tailoring by actin binding proteins such as cofilin, gelsolin, and Arp2/3 complex and we note that the proline rich domain in both metavinculin and vinculin possess binding sites for a number of actin binding proteins such as profilin, Arp2/3 complex, and N-Wasp 50.
Alternatively, if there are local binding density differences of VT and MVT, the differential filament compliance that we present here may serve to direct cues for the binding of other partners at the adhesion sites. If so, the initial filament binding to MVT may influence subsequent binding events and result in a distinct network architecture. This would be important for regulating the contractile force distribution. The main binding partners of vinculins at the adhesion sites are mechanosensor proteins including integrins, talin, paxillin, and alpha-actinin, supporting this conclusion 51.
MVT provides insight into severing by cofilin
Although MVT is not an active severing factor, our studies provide insight into cofilin-like severing. Given that MVT decreases the Lp of actin filaments by almost 2-fold and causes maximal susceptibility to fragmentation at half binding density, we speculate that mechanical discontinuities are introduced along partially-decorated filaments, which can lead to breakage. Similar discontinuities have been proposed to contribute to filament severing by the ADF/cofilin family of proteins 30,41,52,53. Therefore, we asked how ADF/cofilin and MVT interactions with actin differ such that one actively severs and the other does not. ADF/cofilins bind F-actin with high affinity and positive cooperativity, forming clusters on filaments even at low binding densities 30,41,52-58. Our binding isotherms did not identify cooperativity in binding of MVT to actin which suggests that MVT - unlike cofilins 52 - may not form large clusters upon binding to F-actin. In general, the presence of clusters would be expected to amplify the structural changes at the actin interprotomer contact regions believed to be responsible for filament destabilization and fragmentation. Mechanical discontinuities in the F-actin-MVT complex may be less pronounced than those found in F-actin-cofilin. This would result in less spontaneous fracture due to thermal fluctuations but we do not believe that this is sufficient to explain the difference in activity between MVT and ADF/cofilin.
Like MVT, many ADF/cofilins reduce the persistence length of actin filaments. Depending on the isoform used, the change is as great as four fold, and the magnitude of change is correlated with the severing activity 30,40,41. Unlike MVT, ADF/cofilins significantly disrupt the interactions between actin interprotomer loops. Specifically interactions of actin’s subdomains 1 and 2 are disrupted by an increase in the plasticity of the D-loop upon cofilin binding 32,33,35,54,59-61. This structural change accounts for the altered tilt and twist of cofilin decorated filaments observed by EM 60,61, Cryo-EM 59 and AFM 40. No such tilt and twist in actin filaments decorated with MVT is observed by EM 7,12. This is consistent with our finding that the D-loop and other structures are only mildly altered when actin is decorated with MVT. This implies that a combination of mechanical and strong structural changes, such as twisting, are required for efficient severing of actin filaments by thermal fluctuations as proposed by Elam et al (2013) and Kang et al., (2014).
The vertebrate cofilin, h-Cof1, provides a good complementary example: h-Cof1 does not change the bending persistence length of yeast actin filaments and, as one might predict, only weakly severs these filaments 41,62. However, h-Cof1 does change the twist of yeast actin filaments as observed by EM (unpublished data). Thus inducing twist is not sufficient for severing. Therefore, our results with MVT and these observations with h-Cof1 argue strongly that both mechanical and structural changes are necessary for severing by thermal fluctuations.
Conclusions
We present evidence that MVT binding changes the mechanical properties of actin filaments with only mild effects on the structure of the filament. This change may make filaments prone to fragmentation due to mechanical load, which could be critical in developing and mature muscle. We also found that MVT tunes the density and thickness of actin filament bundles generated by VT. Interestingly, MVT alters VT driven filament architecture but does not change the mechanical properties of VT decorated filaments. Local density differences between metavinculin and vinculin could result in different mechanical properties of F-actin. In the future, it will be important to learn whether full-length metavinculin and vinculin are differentially regulated and how these proteins are organized in adhesion structures to know which role of metavinculin dominates. Of particular interest is the role metavinculin plays in the heart given its link to cardiac disease.
Methods
Protein expression and purification
Rabbit skeletal muscle α-actin was purified as described 63 and gel filtered using a HiLoad 16/60 Superdex 200 column (GE Healthcare; column buffer “A”: 2 mM Tris (pH 8.0), 0.5 mM dithiothreitol (DTT), 0.25 mM TCEP, 0.2 mM ATP, and 0.2 mM CaCl2).
Yeast (Saccharomyces cerevisiae) actin, wild type (WT) and mutant strains, Q41C/C374S (C41), I43C/C374S (C43), V45C/C374S (C45), G46C/C374S (C46), M47C/C374S (C47), G48C/C374A (C48), Q49C/C374A (C49), K50C/C374A (C50), A167C/C374A (C167), F169C/C374A (C169), Q41C, and S265C were purified by a DNase I affinity chromatography and actin cycling procedure as described 64. After cycling, proteins were stored in 10 mM HEPES (pH 7.4), 1 mM DTT, 0.2 mM ATP, and 0.2 mM CaCl2 on ice.
Human metavinculin tail (MVT, residues 879-1134) with no affinity tags and vinculin tail (VT, residues 879–1,066) with an N-terminal 6-His tag were expressed in MMI medium containing 0.5 mM IPTG at 18°C overnight in Escherichia coli strain BL21(DE3). Cell pellets were frozen in liquid nitrogen and stored at −80°C. Before purification, cell pellets were thawed and resuspended in 20 mM MOPS, pH 7.0, 20 mM NaCl, 1 mM EDTA, 0.5% Triton X100, 0.2 mM PMSF, and 5mM BME and lysed using a microfluidizer (Microfluidics, Newton, MA). The lysate was cleared by centrifugation at 20,000 × g for 25 min, and the supernatant was loaded onto a 5 mL HiTrap SP column (GE healthcare) pre-equilibrated with 10 mM MOPS, pH 7.0, 0.2 mM PMSF, and 1mM DTT. HiTrap SP bound MVT or VT was eluted with a NaCl gradient. For MVT purification, the purest fractions were pooled and further purified by HiLoad 16/60 Superdex 75 column or 26/60 Sepharacyl 100 (GE Healthcare; column buffer: 10 mM HEPES (pH 7.5), 150mM KCl, 1 mM DTT, 0.2 mM PMSF). After this step, if further purification was needed, the gel filtered fractions were purified on a MonoQ column (buffer: 10 mM TRIS (pH 8.0), 1 mM DTT, 0.2 mM PMSF, up to 0.5 M NaCl gradient). Purified MVT was dialyzed and stored in MVT storage buffer (10 mM HEPES (pH 7.5), 150mM KCl, 1 mM DTT, 0.2 mM PMSF).
For VT purification, the fractions from HiTrap SP column chromatography were pooled and dialyzed into thrombin cleavage buffer (10 mM TRIS (pH 8.0), 100 mM KCl, 2.5 mM CaCl2) and the N-terminal His tag was cleaved with 1 unit of human thrombin per 10-15 mg total protein (Novagen, USA) for 3-4 hours at 4°C. Affinity tag-free VT was further purified on a MonoS column (buffer: 10 mM MOPS (pH 7.0), 1 mM DTT, 0.2 mM PMSF, up to 0.5M NaCl gradient) and dialyzed into MVT storage buffer. Protein concentrations were determined using extinction coefficients 17790 M−1cm−1 and 24980 M−1cm−1 for VT and MVT, respectively. 20-25 uL aliquots were flash frozen in liquid nitrogen and stored at −80°C.
Yeast cofilin was expressed and purified as previously described 32.
Cosedimentation Experiments
Cosedimentation experiments for determining the affinity between MVT and F-actin were performed by incubating 2.5 μM F-actin in KMEH buffer (10 mM HEPES (pH 7.4), 0.4 mM EGTA, 50 mM KCl, 1 mM MgCl2) with increasing concentrations of MVT (0-12 μM) for 45 min. The samples were then centrifuged for 25 min at 90,000 rpm in a Beckman TLA100 rotor. Pellets were concentrated by resuspending them in one fourth of the original volume and run on an SDS-PAGE gel along with MVT standards. The amount of bound MVT was analyzed by densitometry analysis. The binding curves were obtained using Systat SigmaPlot.
Comparative cosedimentation assays for MVT and VT binding to F-actin were done by incubating 2.0 μM MVT or VT in KMEH buffer with increasing concentrations of phalloidin stabilized F-actin (0-12.8 μM) for 45 min. The samples were then centrifuged for 25 min at 90,000 rpm in a Beckman TLA100 rotor. The supernatant and pellet fractions were run on SDS-PAGE. The gels were later stained with Sypro-Red (Invitrogen). The fractions of VT or MVT in supernatants versus pellets were analyzed by densitometry using a Pharos FX Plus Molecular Imager with Quantity One software (Bio-Rad).
For differential cosedimentation experiments, in which the bundles and filaments were separated, a two-step centrifugation procedure was followed. First, the samples were prepared by incubating 5 μM F-actin with VT, MVT, or both at various ratios (0-6 μM) for 1 hour at 24°C in KMEH buffer. The bundled fractions were separated by centrifugation for 15 min at 12,000 × g. The resulting supernatant was further centrifuged for 25 min at 90,000 rpm in a Beckman TLA100 rotor to separate the filament fraction. Both pellets and the high speed supernatants were run on SDS-PAGE gels and the percentage of VT, MVT, and actin in each fraction was calculated by densitometry analysis.
Limited Proteolysis
Limited proteolysis of alpha skeletal F-actin, MVT, and their complex with subtilisin were performed as previously described 34. Briefly, 10 μM F-actin, 10 μM MVT, and 10 μM F-actin+MVT were digested with subtilisin at an enzyme to protein mass ratio of 1:50 (subtilisin: -actin). This subtilisin concentration was also used for MVT alone. Digestions took place in 10 mM HEPES (pH 7.4), 0.2 mM ATP, 0.4 mM MgEGTA, 5 mM BME, 2 mM MgCl2, and 50 mM KCl at 25°C. Reaction samples were taken at 0, 3, 6, 9, 12, 15, 20 min and placed in small tubes with PMSF (final concentration 2mM) to stop the digestions. Cleaved peptides were analyzed by running them on 12% SDS-PAGE gels.
Limited proteolysis of 10 μM of MVT, VT, and their complexes with F-actin by alpha-chymotrypsin were performed as described above, but at an enzyme to protein mass ratio of 1:300 (α-chymotrypsin: actin). Reaction samples were taken at 0, 15 s, 1, 2, 5, 8, 15, and 20 mins. Cleaved peptides were analyzed by running them on 12% SDS-PAGE gels. In addition, the molecular masses of digested peptides found in aliquots of VT (15 min), MVT (1 min), F-actin+VT (18 min) and F-actin+MVT (20 min) were analyzed using a Voyager-DE STR MALDI-TOF Mass Spectrometer (Applied Biosystems, Foster City, CA).
Pyrene actin elongation assays using F-actin seeds
We used a microplate based seeded actin assembly assay to quantify the amount of MVT induced F-actin severing. Briefly, seeds were generated by incubating 2 μM F-actin (polymerized overnight in F-buffer containing final concentrations of 1mM MgCl2 and 50 mM KCl) with increasing amounts of MVT (0, 0.25, 0.5, 1.0, 1.5, 2, 3, 4, and, 8 μM) for 30 minutes at 24°C. Next, 5 μM actin (5% pyrene labeled) was incubated for 2 min at 24°C with ME buffer (final concentration, 200 μM ethylene glycol tetraacetic acid [EGTA] and 50 μM MgCl2) to convert Ca-G-actin to Mg-G-actin. Following this, 25 μL seeds and 50 μL of 2.5X KMEH polymerization buffer (final concentration, 10 mM Na-HEPES, pH 7.4, 0.4 mM EGTA, 50 mM KCl, 1 mM MgCl2) were quickly mixed into 50 μL of Mg-G-actin which was pre-distributed into microplate wells. Increase in pyrene fluorescence upon actin polymerization was monitored using a TECAN F200 plate reader with excitation and emission wavelengths set at 365 nm and 407 nm, respectively.
Total Internal Reflection Fluorescence (TIRF) Microscopy
Rabbit alpha-actin was labeled with Cy3b-maleimide and biotin-maleimide as described 65. Cy3b-actin and biotin actin were gel filtered on a 300/10 Superdex 200 column (GE Healthcare; column buffer “A”: 2 mM Tris (pH 8.0), 0.5 mM dithiothreitol (DTT), 0.25 mM TCEP, 0.2 mM ATP, and 0.2 mM CaCl2) and stored on ice. Percentage labeling of actin was calculated from the extinction coefficient for Cy3b (ε561= 130,000 M−1cm−1).
Direct visualization of actin filament growth 66, severing 65 or bundle formation66 was performed as previously described. Briefly, 22 × 22 mm or 22 × 40 mm coverslips were prepared and functionalized with polyethylene glycol (final surface composition 97% methoxy-PEG and 3% biotin-PEG; JenKem Technology, Allen, TX) as described 66. The larger coverslips were used to allow exchange of buffers in flow cells while on the microscope stage. Flow cells were prepared with double-sided tape as spacers between coverslips. Two different TIRF buffers were used. TIRF(pH 7.0) is composed of 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM HEPES, pH 7, 0.2 mM ATP, 50 mM DTT, 0.2% methylcellulose and , 20 mM glucose. TIRF(pH 7.4) is composed of 50 mM KCl, 1 mM MgCl2, 0.4 mM EGTA, 10 mM HEPES pH 7.4, 0.2 mM ATP, 100 mM DTT, and , 20 mM glucose.
For real time actin filament elongation assays, 25 μl of 40 nM (tetramer concentration) streptavidin was flown into the chamber (volume ~15uL) and incubated for 30 s. Unbound streptavidin was washed with TIRF (pH7.0) buffer. Next, Ca-G-actin (final concentration, 0.6 or 1 μM actin, 15% Cy3b labeled, 1% biotin labeled) was converted to Mg-G-actin by incubating it with ME buffer for 2 min at 24°C. Interacting proteins (VT or MVT) were diluted into 2X TIRF (pH7.0) buffer supplemented with glucose oxidase (final concentration 0.25 mg/ml) and catalase (final concentration 0.05 mg/ml). Equal volumes of Mg-G-actin and MVT or VT were mixed (final concentration 1X TIRF (pH7.0)) and added to the flow cells. Filament elongation was monitored in real time using a DMI6000 TIRF microscope (Leica, Wetzlar, Germany) and the elongation rates were analyzed with JFilament 67 plugin in Fiji 68 from the images captured with 10 s intervals.
For actin filament bundling assays, a similar experimental approach was used except that the actin filaments were attached to the surface through biotin-Peg11-HMM as described 66. Mg-G-actin mix (final concentration 1.3μM, 15% cy3b labeled) in TIRF (pH7.0) buffer supplemented with glucose oxidase (final concentration 0.25 mg/ml), catalase (final concentration 0.05 mg/ml), and interacting proteins was prepared as described above. 30 μl of the Mg-G-actin mix was flown into the cells and visualized with a TIRF microscope. For steady state assays, the flow chambers were sealed with VALAP to prevent evaporation and kept at 24°C for 30 minutes before image acquisition.
Real time actin filament severing assays were performed two ways: 1) Flow cells were treated with streptavidin or neutravidin as described above. 1 μM actin filaments (20% Cy3b labeled, and 0.5% biotin labeled) polymerized overnight were incubated in these flow cells for 30 s. Unbound filaments were washed out with 1X TIRF(pH7.4) buffer supplemented with glucose oxidase and catalase and the bound filaments were immediately visualized. Then, yeast cofilin (10 nM) or MVT (50 nM to 2 μM) in 1X TIRF(pH7.4) buffer supplemented with glucose oxidase and catalase was added and images were acquired at 10-20 s time intervals for up to 15 minutes. 2) Flow cells were treated with 1% Pluronic F-127 and 0.5 mg/ml casein and rinsed with 1X TIRF(pH7.4) buffer. F-actin (20% Cy3b-labeled) was diluted to 25 nM in the presence of MVT in 1X TIRF(pH7.4) buffer supplemented with glucose oxidase and catalase and immediately added to chambers treated.
All image analysis was performed using Fiji 68.
Steady state microscopy assays with F-actin
For steady state length distribution measurements, F-actin (20% cy3b) was polymerized in 1x KMEH (pH 7.4) buffer overnight on ice as described above. Next day, F-actin (2 μM) was incubated at 24°C for 2 hours with either 1.6 μM MVT or MVT storage buffer. F-actin samples were diluted to 5 nM in 1x KMEH (pH 7.4) buffer supplemented with 100 mM BME, which contained either the MVT storage buffer or 0.6 μM MVT with or without 50 nM phalloidin. Dilute samples were spotted onto the slide and a cover slip poly-L-lysine coated coverslip coated with 2 mg/ml poly-L-lysine was placed on top of the sample and sealed with nail polish. All pipetting was performed with cut tips as gently as possible. Sample images were acquired using an Andor CCD camera controlled by Leica software on a Leica DMI6000 TIRF microscope. Actin filament lengths were measured using the JFilament 67 plugin in Fiji 68. For electron microscopy, F-actin (2.5 μM) was mixed with buffer or equimolar MVT. Then, 5 μLs of these mixtures were spotted on 400-mesh carbon-coated copper grids coated with formvar films (EM Sciences) as described previously. Briefly, protein mixtures were adsorbed to grids for 60 seconds and stained with 1% uranyl acetate for 45 seconds. Electron microscopy grids were later examined using a JEOL 1200EX-II electron microscope operated at 80 kV. All image analysis were performed using Fiji 68.
Bending stiffness measurements
For bending persistence length measurements, actin was labeled on lysines with Alexa 488-succimidyl ester fluorophore 30,41 for fluorescence microscopy. Ca-ATP-G-actin was converted to Mg-ATP-G-actin with 0.2 mM EGTA and MgCl2 equal to the concentration of G-actin plus 10 μM. Mg-ATP-G-actin was polymerized at room temperature (~22-25°C) by adding 0.1 volume of 10X polymerization KMI6.8 buffer ( 1x KMI6.8: 50 mM KCl, 2 mM MgCl2, 2 mM DTT, 0.2 mM ATP, 10 mM imidazole, pH 6.8).
Alexa-488-labeled actin filaments (2 μM) were equilibrated at room temperature for 1 hr with MVT and/or VT at saturating binding densities and then serially diluted in 1× KMEH buffer (final concentration, 50 mM KCl, 0.4 mM EGTA, 1 mM MgCl2, 10 mM Na-HEPES, pH 7.4,) containing free MVT and/or VT supplemented with 15 mM glucose, 20 μg/mL catalase, and 100 μg/mL glucose oxidase. Samples were added to microscope slides cleaned with absolute ethanol then 0.1 M KOH, followed by extensive rinsing with MilliQ water. Images of MVT and/or VT-decorated Alexa-488-labeled actin filaments were acquired at room temperature (~22-25°C) using an iMic digital objective-based TIRF microscope (Till Photonics) equipped with a 100× objective (Olympus), an iXon 897 EMCCD camera (Andor Technology) and LiveAcquisition software (Till Photonics).
The actin filament bending persistence lengths (Lp) of decorated filaments were determined from a cosine angular correlation analysis of digitized filament images (n = 100-300 individual filaments for each condition) using a custom Matlab script 31.
Fluorescence spectroscopy of prodan and pyrene labeled F-actins
Yeast WT actin (native cysteine 374) and cysteine bearing yeast actin mutants (C41, C43, C45-50, C167, and C169) were labeled with prodan as previously described 38,64. Yeast actins Q41C and S265C were labeled with pyrene maleimide as previously described (4).
Fluorescence emission spectra of prodan labeled F-actins (5 μM) with or without equimolar MVT were recorded between 400–650 nm, with the excitation wavelength set at 385 nm. Fluorescence emission spectra of 200% pyrene labeled Q41C and S265C F-actins (8 μM) in the presence and absence of equimolar MVT were recorded between 360–600 nm, with the excitation wavelength set at 344 nm. In all recordings, a PTI (Photon Technology International) spectrofluorometer was used.
Supplementary Material
Highlights.
1) Metavinculin tail (MVT) decreases actin filament’s bending persistence length
2) MVT makes actin more susceptible to mechanical breakage
3) MVT and VT bind actin with similar affinity and mode
4) MVT reduces VT’s actin bundling activity
Acknowledgments
This work was supported by National Institutes of Health (NIH) NRSA postdoctoral fellowship F32HL119069 to Z.A.O.D., NIH grant (R01GM097348) to E.M.D.L.C., NIH grant (R01GM077190) to E.R. and D.H., and NIH grant (R01GM096133) and Burroughs- Welcome Fund (Career Award in the Biomedical Sciences) to M.E.Q. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Carisey A, Ballestrem C. Vinculin, an adapter protein in control of cell adhesion signalling. Eur J Cell Biol. 2011;90:157–63. doi: 10.1016/j.ejcb.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng X, Nelson ES, Maiers JL, DeMali KA. New insights into vinculin function and regulation. Int Rev Cell Mol Biol. 2011;287:191–231. doi: 10.1016/B978-0-12-386043-9.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakolitsa C, de Pereda JM, Bagshaw CR, Critchley DR, Liddington RC. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–13. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- 4.Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J Biol Chem. 1994;269:12611–9. [PubMed] [Google Scholar]
- 5.Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–60. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–6. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- 7.Thompson PM, Tolbert CE, Campbell SL. Vinculin and metavinculin: oligomerization and interactions with F-actin. FEBS Lett. 2013;587:1220–9. doi: 10.1016/j.febslet.2013.02.042. [DOI] [PubMed] [Google Scholar]
- 8.Janssen ME, Kim E, Liu H, Fujimoto LM, Bobkov A, Volkmann N, Hanein D. Three-dimensional structure of vinculin bound to actin filaments. Mol Cell. 2006;21:271–81. doi: 10.1016/j.molcel.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Gimona M, Small JV, Moeremans M, Van Damme J, Puype M, Vandekerckhove J. Porcine vinculin and metavinculin differ by a 68-residue insert located close to the carboxy-terminal part of the molecule. EMBO J. 1988;7:2329–34. doi: 10.1002/j.1460-2075.1988.tb03076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strasser P, Gimona M, Herzog M, Geiger B, Small JV. Variable and constant regions in the C-terminus of vinculin and metavinculin. Cloning and expression of fragments in E. coli. FEBS Lett. 1993;317:189–94. doi: 10.1016/0014-5793(93)81274-4. [DOI] [PubMed] [Google Scholar]
- 11.Rangarajan ES, Lee JH, Yogesha SD, Izard T. A helix replacement mechanism directs metavinculin functions. PLoS One. 2011;5:e10679. doi: 10.1371/journal.pone.0010679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssen ME, Liu H, Volkmann N, Hanein D. The C-terminal tail domain of metavinculin, vinculin's splice variant, severs actin filaments. J Cell Biol. 2012;197:585–93. doi: 10.1083/jcb.201111046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belkin AM, Ornatsky OI, Glukhova MA, Koteliansky VE. Immunolocalization of meta-vinculin in human smooth and cardiac muscles. J Cell Biol. 1988;107:545–53. doi: 10.1083/jcb.107.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feramisco JR, Smart JE, Burridge K, Helfman DM, Thomas GP. Co-existence of vinculin and a vinculin-like protein of higher molecular weight in smooth muscle. J Biol Chem. 1982;257:11024–31. [PubMed] [Google Scholar]
- 15.Glukhova MA, Kabakov AE, Belkin AM, Frid MG, Ornatsky OI, Zhidkova NI, Koteliansky VE. Meta-vinculin distribution in adult human tissues and cultured cells. FEBS Lett. 1986;207:139–41. doi: 10.1016/0014-5793(86)80027-8. [DOI] [PubMed] [Google Scholar]
- 16.Saga S, Hamaguchi M, Hoshino M, Kojima K. Expression of meta-vinculin associated with differentiation of chicken embryonal muscle cells. Exp Cell Res. 1985;156:45–56. doi: 10.1016/0014-4827(85)90260-5. [DOI] [PubMed] [Google Scholar]
- 17.Witt S, Zieseniss A, Fock U, Jockusch BM, Illenberger S. Comparative biochemical analysis suggests that vinculin and metavinculin cooperate in muscular adhesion sites. J Biol Chem. 2004;279:31533–43. doi: 10.1074/jbc.M314245200. [DOI] [PubMed] [Google Scholar]
- 18.Maeda M, Holder E, Lowes B, Valent S, Bies RD. Dilated cardiomyopathy associated with deficiency of the cytoskeletal protein metavinculin. Circulation. 1997;95:17–20. doi: 10.1161/01.cir.95.1.17. [DOI] [PubMed] [Google Scholar]
- 19.Meyer T, Brink U, Unterberg C, Stohr S, Kreuzer H, Buchwald AB. Expression of meta-vinculin in human coronary arteriosclerosis is related to the histological grade of plaque formation. Atherosclerosis. 1994;111:111–9. doi: 10.1016/0021-9150(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 20.Huttelmaier S, Bubeck P, Rudiger M, Jockusch BM. Characterization of two F-actin-binding and oligomerization sites in the cell-contact protein vinculin. Eur J Biochem. 1997;247:1136–42. doi: 10.1111/j.1432-1033.1997.01136.x. [DOI] [PubMed] [Google Scholar]
- 21.Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–4. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 22.Rudiger M, Korneeva N, Schwienbacher C, Weiss EE, Jockusch BM. Differential actin organization by vinculin isoforms: implications for cell type-specific microfilament anchorage. FEBS Lett. 1998;431:49–54. doi: 10.1016/s0014-5793(98)00723-6. [DOI] [PubMed] [Google Scholar]
- 23.Wen KK, Rubenstein PA, DeMali KA. Vinculin nucleates actin polymerization and modifies actin filament structure. J Biol Chem. 2009;284:30463–73. doi: 10.1074/jbc.M109.021295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson TM, Illenberger S, Kishimoto NY, Huttelmaier S, Keating MT, Jockusch BM. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–7. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- 25.Vasile VC, Will ML, Ommen SR, Edwards WD, Olson TM, Ackerman MJ. Identification of a metavinculin missense mutation, R975W, associated with both hypertrophic and dilated cardiomyopathy. Mol Genet Metab. 2006;87:169–74. doi: 10.1016/j.ymgme.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Goldmann WH, Guttenberg Z, Tang JX, Kroy K, Isenberg G, Ezzell RM. Analysis of the F-actin binding fragments of vinculin using stopped-flow and dynamic light-scattering measurements. Eur J Biochem. 1998;254:413–419. doi: 10.1046/j.1432-1327.1998.2540413.x. [DOI] [PubMed] [Google Scholar]
- 27.Gurel PS, Ge P, Grintsevich EE, Shu R, Blanchoin L, Zhou ZH, Reisler E, Higgs HN. INF2-mediated severing through actin filament encirclement and disruption. Curr Biol. 2014;24:156–64. doi: 10.1016/j.cub.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gell C, Bormuth V, Brouhard GJ, Cohen DN, Diez S, Friel CT, Helenius J, Nitzsche B, Petzold H, Ribbe J, Schaffer E, Stear JH, Trushko A, Varga V, Widlund PO, Zanic M, Howard J. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 2010;95:221–45. doi: 10.1016/S0091-679X(10)95013-9. [DOI] [PubMed] [Google Scholar]
- 29.Bieling P, Telley IA, Hentrich C, Piehler J, Surrey T. Fluorescence microscopy assays on chemically functionalized surfaces for quantitative imaging of microtubule, motor, and +TIP dynamics. Methods Cell Biol. 2010;95:555–80. doi: 10.1016/S0091-679X(10)95028-0. [DOI] [PubMed] [Google Scholar]
- 30.McCullough BR, Blanchoin L, Martiel JL, De la Cruz EM. Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. J Mol Biol. 2008;381:550–8. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham JS, McCullough BR, Kang H, Elam WA, Cao W, De La Cruz EM. Multi-platform compatible software for analysis of polymer bending mechanics. PLoS One. 2014;9:e94766. doi: 10.1371/journal.pone.0094766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bobkov AA, Muhlrad A, Kokabi K, Vorobiev S, Almo SC, Reisler E. Structural effects of cofilin on longitudinal contacts in F-actin. J Mol Biol. 2002;323:739–50. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- 33.Bobkov AA, Muhlrad A, Shvetsov A, Benchaar S, Scoville D, Almo SC, Reisler E. Cofilin (ADF) affects lateral contacts in F-actin. J Mol Biol. 2004;337:93–104. doi: 10.1016/j.jmb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Muhlrad A, Kudryashov D, Michael Peyser Y, Bobkov AA, Almo SC, Reisler E. Cofilin induced conformational changes in F-actin expose subdomain 2 to proteolysis. J Mol Biol. 2004;342:1559–67. doi: 10.1016/j.jmb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Scoville D, Stamm JD, Altenbach C, Shvetsov A, Kokabi K, Rubenstein PA, Hubbell WL, Reisler E. Effects of binding factors on structural elements in F-actin. Biochemistry. 2009;48:370–8. doi: 10.1021/bi801649j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehrer SS. Pyrene excimer fluorescence as a probe of protein conformational change. Subcell Biochem. 1995;24:115–32. doi: 10.1007/978-1-4899-1727-0_5. [DOI] [PubMed] [Google Scholar]
- 37.Feng L, Kim E, Lee WL, Miller CJ, Kuang B, Reisler E, Rubenstein PA. Fluorescence probing of yeast actin subdomain 3/4 hydrophobic loop 262-274. Actin-actin and actin-myosin interactions in actin filaments. J Biol Chem. 1997;272:16829–37. doi: 10.1074/jbc.272.27.16829. [DOI] [PubMed] [Google Scholar]
- 38.Oztug Durer ZA, Kamal JK, Benchaar S, Chance MR, Reisler E. Myosin binding surface on actin probed by hydroxyl radical footprinting and site-directed labels. J Mol Biol. 2011;414:204–16. doi: 10.1016/j.jmb.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson PM, Tolbert CE, Shen K, Kota P, Palmer SM, Plevock KM, Orlova A, Galkin VE, Burridge K, Egelman EH, Dokholyan NV, Superfine R, Campbell SL. Identification of an actin binding surface on vinculin that mediates mechanical cell and focal adhesion properties. Structure. 2014;22:697–706. doi: 10.1016/j.str.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma S, Grintsevich EE, Phillips ML, Reisler E, Gimzewski JK. Atomic force microscopy reveals drebrin induced remodeling of f-actin with subnanometer resolution. Nano Lett. 2011;11:825–7. doi: 10.1021/nl104159v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCullough BR, Grintsevich EE, Chen CK, Kang H, Hutchison AL, Henn A, Cao W, Suarez C, Martiel JL, Blanchoin L, Reisler E, De La Cruz EM. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101:151–9. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breitsprecher D, Koestler SA, Chizhov I, Nemethova M, Mueller J, Goode BL, Small JV, Rottner K, Faix J. Cofilin cooperates with fascin to disassemble filopodial actin filaments. J Cell Sci. 124:3305–18. doi: 10.1242/jcs.086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thoss F, Dietrich F, Punkt K, Illenberger S, Rottner K, Himmel M, Ziegler WH. Metavinculin: New insights into functional properties of a muscle adhesion protein. Biochem Biophys Res Commun. 2013;430:7–13. doi: 10.1016/j.bbrc.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Belkin AM, Ornatsky OI, Kabakov AE, Glukhova MA, Koteliansky VE. Diversity of vinculin/meta-vinculin in human tissues and cultivated cells. Expression of muscle specific variants of vinculin in human aorta smooth muscle cells. J Biol Chem. 1988;263:6631–5. [PubMed] [Google Scholar]
- 45.Glukhova MA, Frid MG, Koteliansky VE. Developmental changes in expression of contractile and cytoskeletal proteins in human aortic smooth muscle. J Biol Chem. 1990;265:13042–6. [PubMed] [Google Scholar]
- 46.Koteliansky VE, Belkin AM, Frid MG, Glukhova MA. Developmental changes in expression of adhesion-mediating proteins in human aortic smooth muscle. Biochem Soc Trans. 1991;19:1072–6. doi: 10.1042/bst0191072. [DOI] [PubMed] [Google Scholar]
- 47.De La Cruz EM, Martiel J, Blanchoin L. Mechanical heterogeneity favors fragmentation of strained actin filaments. Biophys J. 2015 doi: 10.1016/j.bpj.2015.03.058. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J Mol Biol. 2007;365:1350–8. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen K, Tolbert CE, Guilluy C, Swaminathan VS, Berginski ME, Burridge K, Superfine R, Campbell SL. The vinculin C-terminal hairpin mediates F-actin bundle formation, focal adhesion, and cell mechanical properties. J Biol Chem. 2011;286:45103–15. doi: 10.1074/jbc.M111.244293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–12. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brancaccio M, Hirsch E, Notte A, Selvetella G, Lembo G, Tarone G. Integrin signalling: the tug-of-war in heart hypertrophy. Cardiovasc Res. 2006;70:422–33. doi: 10.1016/j.cardiores.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 52.De La Cruz EM. How cofilin severs an actin filament. Biophys Rev. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elam WA, Kang H, De la Cruz EM. Biophysics of actin filament severing by cofilin. FEBS Lett. 2013;587:1215–9. doi: 10.1016/j.febslet.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J Mol Biol. 2006;356:325–34. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 55.Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. J Mol Biol. 2006;361:257–67. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 56.De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. J Mol Biol. 2005;346:557–64. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 57.De La Cruz EM, Sept D. The kinetics of cooperative cofilin binding reveals two states of the cofilin-actin filament. Biophys J. 2010;98:1893–901. doi: 10.1016/j.bpj.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann AC, Guerin C, Martiel JL, De la Cruz EM, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–8. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schroder GF, Egelman EH. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci U S A. 2011;108:20568–72. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galkin VE, Orlova A, VanLoock MS, Shvetsov A, Reisler E, Egelman EH. ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J Cell Biol. 2003;163:1057–66. doi: 10.1083/jcb.200308144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang H, Bradley MJ, Cao W, Zhou K, Grintsevich EE, Michelot A, Sindelar CV, Hochstrasser M, De La Cruz EM. Site-specific cation release drives actin filament severing by vertebrate cofilin. Proc Natl Acad Sci U S A. 2014;111:17821–6. doi: 10.1073/pnas.1413397111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–71. [PubMed] [Google Scholar]
- 64.Durer ZA, Kudryashov DS, Sawaya MR, Altenbach C, Hubbell W, Reisler E. Structural states and dynamics of the D-loop in actin. Biophys J. 2012;103:930–9. doi: 10.1016/j.bpj.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen CK, Sawaya MR, Phillips ML, Reisler E, Quinlan ME. Multiple forms of Spire-actin complexes and their functional consequences. J Biol Chem. 2012;287:10684–92. doi: 10.1074/jbc.M111.317792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bor B, Vizcarra CL, Phillips ML, Quinlan ME. Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains. Mol Biol Cell. 2012;23:3801–13. doi: 10.1091/mbc.E12-04-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith MB, Li H, Shen T, Huang X, Yusuf E, Vavylonis D. Segmentation and tracking of cytoskeletal filaments using open active contours. Cytoskeleton (Hoboken) 2010;67:693–705. doi: 10.1002/cm.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–8. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 70.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.