Abstract
Denitrification is an important process in the global nitrogen cycle. The genes encoding NirK and NirS (nirK and nirS), which catalyze the reduction of nitrite to nitric oxide, have been used as marker genes to study the ecological behavior of denitrifiers in environments. However, conventional polymerase chain reaction (PCR) primers can only detect a limited range of the phylogenetically diverse nirK and nirS. Thus, we developed new PCR primers covering the diverse nirK and nirS. Clone library and qPCR analysis using the primers showed that nirK and nirS in terrestrial environments are more phylogenetically diverse and 2–6 times more abundant than those revealed with the conventional primers. RNA- and culture-based analyses using a cropland soil also suggested that microorganisms with previously unconsidered nirK or nirS are responsible for denitrification in the soil. PCR techniques still have a greater capacity for the deep analysis of target genes than PCR-independent methods including metagenome analysis, although efforts are needed to minimize the PCR biases. The methodology and the insights obtained here should allow us to achieve a more precise understanding of the ecological behavior of denitrifiers and facilitate more precise estimate of denitrification in environments.
Introduction
Denitrification is a microbial dissimilatory process where nitrate and nitrite are reduced stepwise to gaseous compounds, such as nitric oxide (NO), nitrous oxide (N2O) and dinitrogen (N2) (Knowles, 1982; Zumft, 1997). Denitrification causes nitrogen (N) loss from natural ecosystems and agricultural fields, as well as N removal during wastewater treatment (Tiedje, 1988; Conrad, 1996; Martin et al., 1999; Bouwman et al., 2002), and it can also facilitate the anaerobic degradation of organic pollutants (Leahy and Olsen, 1997). One of the end products of denitrification, N2O, is a potent greenhouse gas and ozone-depleting substance (Davidson, 2009; Ravishankara et al., 2009). Therefore, denitrification has attracted much attention from the scientific, agricultural and industrial fields for a long time. Recent studies have demonstrated that ecological traits such as the diversity, abundance and ability to produce N2O/N2 of denitrifying microbial communities could reflect denitrification in environments (Philippot et al., 2009; Moralesl et al., 2010; Wei et al., 2014); thus, the importance of a precise understanding of these traits is widely recognized by many fields where major research efforts are ongoing.
The reduction of nitrite to NO is catalyzed by two structurally different but functionally equivalent nitrite reductases, that is, copper-containing reductase (NirK) and cytochrome cd1-containing reductase (NirS) (Hochstein and Tomlinson, 1988; Cutruzzola et al., 2001; Sakurai and Kataoka, 2007), and this is a key step in the denitrification process because dissolved N is converted into gaseous N during this step. Taxonomically diverse microorganisms have the ability to denitrify (Tiedje, 1994), and phylogenies based on the nir and 16S rRNA genes are incongruent (Jones et al., 2008). Therefore, nirK and nirS are used as marker genes to study the ecological behavior of denitrifying microorganisms in environments. Many attempts have been made to design and modify the polymerase chain reaction (PCR) primers required to amplify nirK and nirS (Braker et al., 1998; Hallin and Lindgren, 1999; Braker et al., 2000; Michotey et al., 2000; Throbäck et al., 2004). These approaches were conducted based on the available nirK and nirS sequences from cultivable denitrifying bacterial strains, most of which belong to the classes of alpha-, beta- and gamma-proteobacteria.
Recent developments in the genome analysis of cultured and uncultured strains have shown that many diverse microorganisms possess nirK or nirS, including the phyla Nitrospirae, Actinobacteria, Bacteroidetes, Spirochetes, Chloroflexi and Firmicutes, and even the phyla Euryarchaeota and Crenarchaeota in Archaea (Cantera and Stein, 2007; Nolan et al., 2009; Bartossek et al., 2010; Moir, 2011; Mardanov et al., 2012; Murawska et al., 2013; Nishizawa et al., 2013 ). Furthermore, as demonstrated by Jones et al. (2008), these nirK and nirS sequences were not considered during the design and modification of the nirK and nirS primers described above. However, even after the report of Jones et al. (2008), the conventional primers are still widely used to illustrate the ecological traits of denitrifying microorganisms in the environment (for example, Katsuyama et al., 2008; Yoshida et al., 2012; Li et al., 2014). This suggests that the diversity, abundance and functional importance of denitrifying microorganisms in the environment are underestimated at present.
Thus, the objective of this study was to determine the overall diversity, abundance and functional importance of denitrifying microorganisms in environments. To achieve this objective, we performed a phylogenetic analysis of the sequence diversity of all currently available nirK and nirS sequences, designed multiple primer sets to cover these diverse nirK and nirS, examined the diversity, abundance and distribution of nirK and nirS in various terrestrial environments using the newly designed primers, and assessed the presence of microorganisms with previously unconsidered nirK or nirS using cropland soil.
Materials and methods
Selection, alignment and analysis of the sequences of NirK and NirS genes
The full-length nucleotide sequences of nirK and nirS were obtained from the Kyoto Encyclopedia of Genes and Genomes Repository (http://www.genome.jp/kegg) and the Functional Gene Repository (http://fungene.cme.msu.edu/index.spr). All of the reference sequences were downloaded in June 2012. Thus, genomes of some of the denitrifying microorganisms that have been deposited in database after the month of download are not included in this study, such as nirK of Bacillus thuringiensis in Firmicutes (Murawska et al., 2013) and nirS of Pyrobaculum sp. in Archaea (Mardanov et al., 2012). The nirK and nirS sequences were aligned by amino acids using ClustalW2 (Larkin et al., 2007). The homologs of the copper center type 1 domain in nirK, which contained a type 1 copper ligand (that is, two His and one Met) and a type 2 copper ligand (that is, two His), were retained for analysis (Supplementary Figures S1a and S1b). The homologs of the cytochrome d1 domain in nirS, which bound and reduced the nitrite substrate, were retained for analysis (Supplementary Figures S1c and S1d). The phylogenetic trees based on the amino-acid sequences were generated using MEGA 5 with the maximum likelihood algorithm (Tamura et al., 2011), and node support was determined using 500 bootstrap replicates.
Primer design for the detection of diverse NirK and NirS genes
We designed forward primers for nirK, which can anneal with the sequences around the conserved methionine from the type 1 copper ligand, and reverse primers, which can anneal with sequences that contain conserved histidine from the type 2 copper ligand (Supplementary Figures S1a and S1b). We also designed forward primers for nirS, which can anneal with sequences around conserved glycine, and reverse primers, which can anneal with the sequences that contain two consecutive conserved glycines (Supplementary Figures S1c and S1d). Several sets of degenerate primers specific to the nirK and nirS sequences in each cluster were designed using the COnsensus-DEgenerate Hybrid Oligonucleotide Primers (CODEHOP) algorithm (Rose et al., 1998). The degenerate primers of each nirK and nirS cluster consisted of a 3′ degenerate core region with an 11-bp length across four highly conserved amino-acid codons and a 5′ consensus non-degenerate clamp region with an 11- to 14-bp length (Supplementary Figures S1 and S2, Supplementary Table S1). In particular, the sequence of widely used conventional primers that had been designed to amplify the nirK and nirS in Cluster I showed mismatches with the sequences described in Supplementary Tables S2. Therefore, we designed new primers for the nirK and nirS in Cluster I. The 3′ end amino acid of the newly designed forward primer for nirK in Cluster I was a highly conserved proline, which was different from the non-conserved end amino acid (that is, lysine or arginine) of the conventional primer F1aCu (Supplementary Figure S1a) (Hallin and Lindgren, 1999). The 3′ end amino acid of the newly designed reverse primer for nirK in Cluster I was a highly conserved histidine, which was different from the non-conserved end amino acid (that is, serine, asparagine or threonine) of the widely used primer R3Cu (Supplementary Figure S1b) (Hallin and Lindgren, 1999). The 3′ end amino acid of the newly designed reverse primer for nirS in Cluster I was a highly conserved leucine, which was different from the non-conserved end amino acid (that is, isoleucine, valine or leucine) of the conventional primer R3cd (Supplementary Figure S1d) (Throbäck et al., 2004).
Validation of the designed primers using the denitrifying strains
Eighteen bacterial strains obtained from the culture collection of the Japan Collection of Microorganisms (JCM, Tsukuba, Ibaraki, Japan) or the Biological Resource Center (NBRC, Kisarazu, Chiba, Japan), which were expected to belong to each cluster in the phylogenetic trees (Figure 1) according to their 16S rRNA-based taxonomy, were used to validate the coverage and specificity of all designed primer candidates (Supplementary Table S2). In addition, forty-four strains of denitrifying bacteria, which had been isolated from rice field soil in Niigata, Japan (Ashida et al., 2010; Nishizawa et al., 2012; Nishizawa et al., 2013), were used (Supplementary Table S2). The denitrification abilities of all bacterial strains were analyzed using 15N-labeled NaNO3 (99.5 atom%-15N, SI Sciences, Japan) and a gas chromatography-mass spectrometry (GC–MS) system (GCMS-QP2010Plus, Shimadzu, Kyoto, Japan) as previously described (Isobe et al., 2011). Genomic DNA was extracted from the single colonies as previously described (Wei et al., 2014). Genomic DNA from a non-denitrifying bacterium, Geobacillus kaustophilus, which does not possess the nirK or nirS, was used as a negative control.
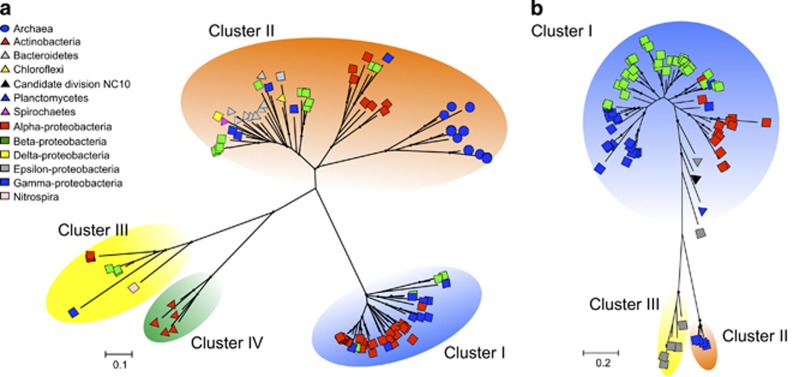
Figure 1.
Unrooted maximum likelihood phylogeny of partial nirK (a) and nirS (b) amino-acid sequences obtained from genomes. The nirK type 1 copper center and nirS cytochrome d1 heme were detected for each cluster. Bootstrap values (500 replicates) >70% are denoted by dots above the branches, and the branch lengths correspond to sequence differences, which are indicated by the scale bar. Symbols on tree tips indicate the taxonomic affiliations of reference sequences.
PCR conditions were optimized for nirK and nirS in each cluster that used the strains. All reactions were performed as described in Supplementary Table S1. The PCR product purification, cloning and sequencing were performed as described elsewhere (Wei et al., 2014). According to the obtained nirK or nirS sequence, each denitrifier isolate was determined to belong to any cluster of nirK or nirS except for nirS Cluster III. On the basis of these results, the best primers were selected, and their sequences and conditions for conventional PCR and quantitative PCR (qPCR) were optimized as shown in Supplementary Table S1.
Diversity of NirK and NirS genes in various environments
Clone library analyses were performed using the environmental samples to determine the diversity of the nirK and nirS sequences in various environments. We used cropland soil (Takahashi et al., 1991), rice paddy soil (Itoh et al., 2013), forest soils (Urakawa et al., 2015) and lake sediment (Abe et al., 2000) as described below. All soil samples were collected in triplicate at 5–10 cm depth from each experimental site, and lake sediment was sampled in triplicate at 10 cm depth from the sediment surface using an Ekman sampler. Environmental DNA was extracted using an ISOIL kit (Nippon Gene, Toyama, Japan). The PCR conditions were the same as previously described except that the MgCl2 concentration was reduced to 2.0 mM. Three hundred fifty-two sequences of nirK and 228 sequences of nirS were obtained after the removal of poor-quality reads (that is, low-quality base calling and frame-shift errors) and potential chimeric sequences. The sequences from each library were then clustered into operational taxonomic units with 3% differences using the Mothur program (Schloss et al., 2009). The final set of 195 sequences for nirK and 102 sequences for nirS was aligned by translating them to amino-acid sequences as previously described, and the amino-acid sequences from the selected microbial genomes and denitrifying cultured collections were used as a reference alignment.
Abundance and distribution of NirK and NirS genes in various environments
The abundance of nirK, nirS and 16S rRNA genes in the environmental samples was determined by qPCR. Various environmental samples from terrestrial habitats were used, including a cropland soil (Andosol) applied with an organic fertilizer (Wei et al., 2014), a cropland soil (Gray Lowland Soil, Eutric Fluvisol) applied with an organic or urea fertilizer (Takahashi et al., 1991), paddy soils from flooded and non-flooded seasons (Gray Lowland Soil, Eutric Fluvisol) (Itoh et al., 2013), two natural forest soils (Brown Forest soil and Andosol) (Urakawa et al., 2015), a planted forest soil (Brown Forest soil) (Oda et al., 2009) and a lake sediment (Abe et al., 2000).
Environmental DNA was extracted as previously described. The qPCR was conducted using a StepOne real-time PCR system (Applied Biosystems, Warrington, UK) and a KOD SYBR qPCR Mix kit, which was suitable for the degenerate primers that had long sequences and high Tm values. The quantification of the 16S rRNA gene was performed with the primers 357F/520R as previously described (Itoh et al., 2013). The quantification of nirK and nirS in each cluster was performed with the designed primers. We also utilized the widely used conventional primers F1aCu/R3Cu for nirK in Cluster I and cd3aF/R3cd for nirS in Cluster I for comparison. All reactions of qPCR were performed as described in Supplementary Table S1.
The standard curves in the qPCR analyses of the 16S rRNA, nirK and nirS genes were generated using linearized plasmids that contained the following: the cloned 16S rRNA gene from Pseudomonas stutzeri JCM-5965; nirK from Ochrobactrum anthropi JCM21032 in Cluster I, which can be amplified by the designed primers nirKC1F and nirKC1R, as well as by the widely used primers F1aCu/R3Cu; nirK from Azospirillum lipoferum NBRC-1022290 in Cluster II; nirK from Actinosynnema mirum NBRC-10460 in Cluster III; nirK from Nitrobacter winogradskyi NBRC-14297 in Cluster IV; nirS from Denitratisoma oestradiolicum JCM12830 in Cluster I, which can be amplified by the designed primers nirSC1F and nirSC1R, as well as the widely used conventional primers cd3aF andR3cd; and nirS from Methylomonas koyamae NBRC-105905 in Cluster II. The absence of the PCR inhibitors in soil DNA was confirmed by mixing a known amount of standard DNA with environmental DNA in a qPCR reaction. The amplification efficiencies, R2 of the standard curve and Tm value of the melting curve in the qPCR assay for the 16S rRNA gene, nirK in Clusters I–IV and nirS in Clusters I and II were estimated as shown in Supplementary Table S1.
Response of NirK and NirS genes under denitrification-induced conditions
The expression levels of nirK and nirS in soil were analyzed using the soil microcosm under denitrification-induced conditions. We used the Gray Lowland Soil (Eutric Fluvisol) of cropland in Niigata, Japan. This soil is the same as the soil from which forty-four bacterial strains have previously been isolated (Supplementary Table S2). Forty grams of the non-fertilized soil were placed in 80-ml glass bottles, mixed with 0.5 g of granular organic fertilizers and incubated for 20 days as previously described (Wei et al., 2014). The soil without fertilizer was also incubated as a control. The N2O flux was measured every 2 days, and the highest N2O emission rate was observed on the eighth day. The soil RNA and DNA were extracted on the eighth day using an RNA PowerSoil Total RNA Isolation Kit and DNA Elution Accessory Kit (MO BIO Laboratories, Carlsbad, CA, USA). Total RNA and DNA were extracted from 1.2 g of soil according to the manufacturer's protocol. The concentrations of the extracted RNA and DNA were determined using the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Digestion of the residual DNA in RNA solution was performed using the Ambion TURBO DNA-free Kit (Life Technologies). RNA was transcribed into complementary DNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The absence of residual DNA was confirmed in the PCR without reverse transcription. The DNA and cDNA of 16S rRNA gene and nirK and nirS in soils were quantified as previously described. DNA-based clone library analyses of nirK and nirS in the soil on the eighth day after fertilization were simultaneously performed as previously described.
Nucleotide sequence accession numbers
The nucleotide sequences of partial nirK and nirS from the environmental samples in this study have been deposited in the DDBJ/EMBL/GenBank databases with accession numbers AB936839 to AB937093 and AB937560 to AB937661, respectively. The nucleotide sequences of partial nirK and nirS from the denitrifying isolates have been deposited in the databases with accession numbers AB937662 to AB937717.
Results
Phylogeny of NirK and NirS genes
Ninety-seven full-length nirK sequences that belong to the bacterial phyla Actinobacteria, Bacteroidetes, Chloroflexi, Nitrospirae, Proteobacteria and Spirochetes and the archaeal phyla Euryarchaeota were obtained from a public database after selecting a representative sequence with high similarity (>99% of amino-acid sequence) from one species. We constructed a phylogenetic tree based on the sequences for the electron entry site (including type I copper center; 480-560 bp) in NirK. All widely used conventional primer sets, such as F1aCu/R3Cu and nirK2F/nirK5R (Braker et al., 1998; Hallin and Lindgren, 1999), were also designed to amplify the sequence for this site. The tree was divided into four clusters (that is, Clusters I–IV) with high bootstrap support (>70%) (Figure 1a; see Supplementary Figure S2 for species names). We determined that the sequences that can be amplified with the conventional primers are located in Cluster I, which contains nirK from the class alpha-, beta- and gamma-proteobacteria, but not in Clusters II–IV (Supplementary Figures S1 and S2). Cluster II contains nirK from the class alpha-, beta-, gamma-, delta- and epsilon-proteobacteria, and the phyla Bacteroidetes, Chloroflexi and Spirochetes. The nirK from the halophilic archaeal family, Halobacteriaceae, formed a distinct clade, but it had similar sequences in the primer region to all bacterial phyla in Cluster II (Supplementary Figure S1). Cluster III consisted entirely of the nirK from the phylum Actinobacteria. Cluster IV consisted entirely of nirK from nitrifiers, including the genera Nitrospira, Nitrosococcus, Nitrosomonas and Nitrobacter; however, additional nirK sequences from other nitrifiers were also assembled in Clusters I and II (Supplementary Figure S1).
Seventy-five full-length nirS sequences that belong to the bacterial phyla Bacteroidetes, NC10, Planctomycetes and Proteobacteria were obtained with the same approach as the nirK sequences. We constructed the phylogenetic tree based on the sequences of the catalytic site (including cytochrome heme d1; 530–570 bp) in NirS. All widely used conventional primers, such as cd3aF/R3cd and nirS2F/nirS4R (Braker et al., 1998); Michotey et al., 2000; Throbäck et al., 2004, were also designed to amplify the sequence of this site. The tree was divided into three clusters (that is, Clusters I–III) with high bootstrap support (>87%) (Figure 1b; see Supplementary Figure S2 for species names). We determined that the sequences that can be amplified with conventional primers are located in Cluster I, but not in Clusters II and III (Supplementary Figures S1 and S2). Cluster I contained the nirS from the class alpha-, beta-, gamma- and epsilon-proteobacteria, and the phyla Bacteroidetes, Planctomycetes and NC10. Cluster II consisted entirely of the nirS from methane-oxidizing bacteria, including the genera Methylobacter, Methylomonas and Methylomicrobium. Cluster III consisted entirely of the epsilon-proteobacterial nirS from deep-sea sediments in hydrothermal fields. These phylogenetic analyses indicate that nirK and nirS are distributed among taxonomically diverse microorganisms, and a considerable proportion (that is, primarily nirK in Clusters II, III and IV and nirS in Clusters II and III, Figure 1) cannot be detected by the widely used conventional primers; thus, they represent the previously unconsidered nirK and nirS sequences.
New primer design and its validation using denitrifying strains
We designed seven sets of primers that can potentially amplify nirK or nirS located in each of the clusters shown in Figure 1 (nirKC1F/nirKC1R, nirKC2F/nirKC2R, nirKC3F/nirKC3R and nirKC4F/nirKC4R for Clusters I–IV in the phylogenetic tree of nirK and nirSC1F/nirSC1R, nirSC2F/nirSC2R and nirSC3F/nirSC3R for Clusters I-III in the phylogenetic tree of nirS, Supplementary Table S1). The amplified position and expected amplified fragment size (∼430–468 bp for nirK and 410–420 bp for nirS) were approximately the same as the widely used conventional primers (for example, F1aCu/R3Cu or nirK2F/nirK5R for nirK and cd3aF/R3cd for nirS) (Supplementary Figures S1 and S2). This enables the comparative analysis of the nir sequences obtained via the newly designed primers with the massive store of nir sequences previously obtained using the conventional primers.
All primer sets successfully generated single amplified fragments with the expected size from the genomic DNA of the denitrifying bacteria obtained from the culture collection and isolates from the rice paddy soil. A sequence analysis of the amplified fragments confirmed that each primer set amplified the nirK or nirS belonging to each target cluster. No amplification product was generated from non-denitrifying bacteria (Supplementary Table S2). For comparison, the amplification of nirK and nirS from the denitrifier strains using the most widely used conventional primers (F1aCu/R3Cu for nirK and cd3aF/R3cd for nirS) was tested. These primers amplified the nirK and nirS in Cluster I, but failed to amplify them in the other Clusters. Non-specific amplifications were also observed (Supplementary Tables S2). These results indicate the superior coverage and specificity of the newly designed primer sets compared with the conventional primers.
Diversity of NirK and NirS genes in various environments
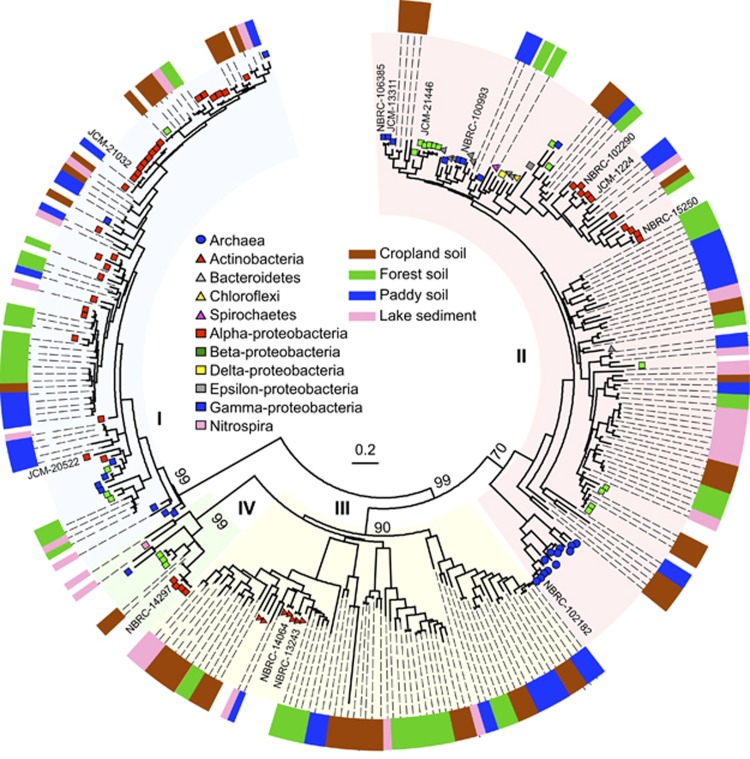
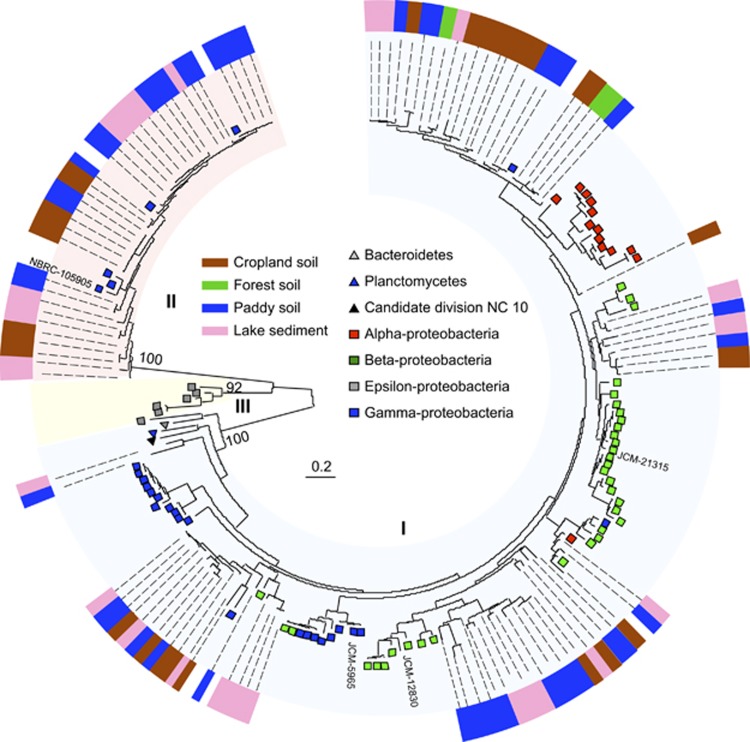
To examine the diversity of nirK and nirS in the environment, including previously unconsidered nirK and nirS, we performed a DNA-based clone library analysis using the newly designed primers. The nirK clones in Clusters I–IV and the nirS clones in Clusters I and II were obtained from all environmental samples examined, that is, cropland, rice paddy, forest soils and lake sediment (Figures 2 and 3) (Supplementary Table S3). The nirS clones in Cluster III, which contained the entire nirS sequence from deep-sea sediments in hydrothermal fields (Takai et al., 2006), were not obtained from these environmental samples.
Figure 2.
Maximum likelihood phylogeny of the four clusters of nirK amino-acid sequences obtained from the environmental samples listed in Supplementary Table S3 and the reference sequences from the genomes and denitrifying strains listed in Supplementary Table S2. Bootstrap values (500 replicates) of each cluster are denoted above the branches. Non-coded and coded symbols on tree tips indicate the taxonomic affiliations of the reference sequences from genomes and denitrifying strains, respectively. The outer color strip shows the source of environmental clones. The branch lengths correspond to sequence differences, which are indicated by the scale bar.
Figure 3.
Maximum likelihood phylogeny of the three clusters of nirS amino-acid sequences obtained from the environmental samples listed in Supplementary Table S3 and the reference sequences from the genomes and denitrifying strains listed in Supplementary Table S2. Bootstrap values (500 replicates) of each cluster are denoted above the branches. Non-coded and coded symbols on tree tips indicate the taxonomic affiliations of the reference sequences from genomes and denitrifying strains, respectively. The outer color strip shows the source of environmental clones. The branch lengths correspond to sequence differences, which are indicated by the scale bar.
A comparative sequence analysis of the obtained environmental clones with the nir sequences of known denitrifier strains in the public database was conducted. The nirK sequences of all clones in Cluster I showed the highest similarities (>70% of amino-acid sequence) with those of the denitrifiers that belong to class alpha-, beta- and gamma-proteobacteria. The nirK sequences of 90, 13 and 5 clones of a total of 112 clones in Cluster II exhibited the highest similarities (64–99%, 68–87% and 72–76%, respectively) with those of the denitrifiers that belong to the phyla Proteobacteria, Bacteroidetes and Spirochetes, respectively. The other four clone sequences did not exhibit similarities with those of the known denitrifier strains or genomes. The nirK sequences from 34 of a total of 103 clones in Cluster III showed the highest similarity (60–90%) with those of the phylum Actinobacteria. The remaining clones did not show substantial similarity (<50%) with those of any of the known Actinobacterial strains or genomes. Moreover, 26 sequences formed an unexpected clade distant to Cluster III. However, these sequences were amplified with the primers for Cluster III; thus, we included this clade in Cluster III (Figure 2). The nirK sequences of all 31 clones in Cluster IV showed the highest similarities (60–90%) with those from the nitrifying genera, including Nitrobacter, Nitrosomonas, Nitrosococcus and Nitrospira.
The nirS sequences from 96 of 99 clones in Cluster I showed the highest similarities (71–95%) with those of the denitrifiers that belong to class alpha-, beta- and gamma-proteobacteria. The other three clone sequences showed the highest similarity (77–80%) with those from the phylum Chloroflexi. The nirS sequences of all 65 clones in Cluster II showed the highest similarities (82–99%) with those from the methane oxidizing genera Methylomicrobium, Methylobacter and Methylomonas.
Abundance of NirK and NirS genes in various environments
We performed a quantitative PCR study using the newly designed primers to examine the abundance and distribution of nirK and nirS sequences, including previously unconsidered sequences, in various terrestrial environmental samples: cropland soils with different soil types or fertilizations, rice paddy soils in water-flooding and non-flooding seasons, natural and planted forest soils and a lake sediment. For comparison, nirK and nirS in Cluster I were quantified using the widely used conventional primers F1aCu/R3Cu for nirK and cd3aF/R3cd for nirS. The abundance of the nirK and nirS in each cluster was normalized by the abundance of the 16S rRNA gene to standardize the unit.
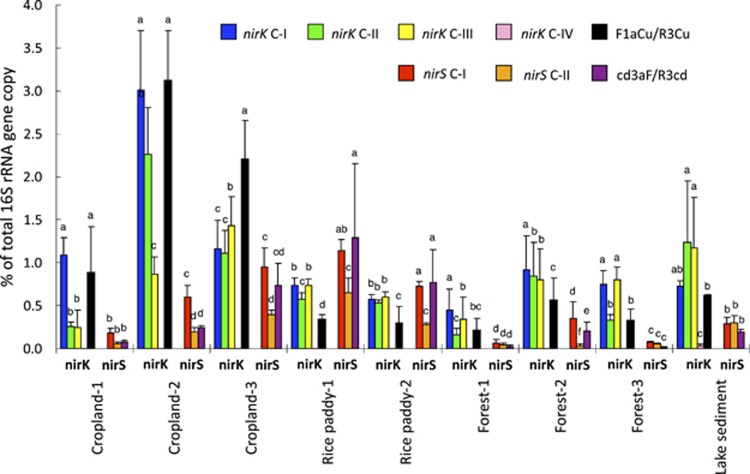
First, we determined that the nirK sequence in Clusters I–IV and the nirS sequence in Clusters I–II were present in abundance in all environmental samples (Figure 4). The nirS sequence in Cluster III, which contained only epsilon-proteobacteria nirS sequences from hydrothermal regions, was not detected in the tested samples. Previous studies also showed that the abundance of epsilon-proteobacteria was quite low in soils and sediments, as examined by their 16S rRNA and nosZ genes (Roesch et al., 2007; Jones et al., 2013). Many studies have used the conventional primer sets F1aCu/R3Cu and cd3aF/R3cd for the quantification of nir in environments, despite the possibility of nonspecific amplification previously described. The abundances of nirK and nirS in Cluster I that were quantified by the newly designed primers were similar to or higher than those quantified by the conventional primers, with the exception of nirK in Cropland-3. The total abundances of nirK in Clusters I–IV and nirS in Clusters I–II quantified with the newly designed primers were ∼2–6 times larger compared with the primers F1aCu/R3Cu and cd3aF/R3cd (Supplementary Table S4).
Figure 4.
Relative abundance of each nirK and nirS cluster of gene copies from the different environmental samples listed in Supplementary Table S4, calculated as a proportion of the total number of bacterial 16S rRNA gene copies (for the mean and s.d. of replicates by site, see Supplementary Tables S4). Relative abundance of nirK cluster IV and nirS cluster I genes detected by the widely used primer sets F1aCu/R3Cu and cd3aF/R3cd are shown as a reference. Different small letters indicate the significant difference among nir clusters in each environmental sample according to the least significant difference test at P<0.05.
The distribution of nirK and nirS differed in the terrestrial environments (Figure 4). For example, cropland soils (Fluvisols) that received organic fertilization (Cropland-2) had more nirK in Clusters I and II but less nirS in Clusters I and II compared with cropland soils (Fluvisols) that received urea fertilization (Cropland-3). Water flooding increased nirK and nirS in paddy soils (Rice paddy-1 and 2). In addition, the abundance of nirS in Cluster II, which contains nirS sequences only from methane oxidizers, was high in rice paddy soils (10–17% of total nir gene) but low in cropland, forest soil and lake sediment (1–3% of total nir gene), which indicated a higher abundance or contribution to the denitrification of methane oxidizers in rice paddy soil. The total abundance of nirK fragments (Clusters I, II, III and IV) was highest in cropland soils (Fluvisols), whereas the total abundance of nirS fragments (Clusters I and II) was highest in flooded paddy soils (Fluvisols).
Involvement of microorganisms with NirK and NirS genes in the denitrification in soil
We performed RNA- and culture-based analyses using a soil microcosm system to examine the involvement in denitrification and taxonomic positions of microorganisms with nirK or nirS detected using the newly designed primers. First, we performed gene expression analyses of nirK and nirS in the soil microcosm. The soil was incubated in a vial after the application of the N fertilizer to induce denitrification. Control soil without fertilizer application was also prepared. N2O emissions due to denitrification peaked at 8 days after the application of the fertilizer (Supplementary Figure S3). The DNA and RNA transcripts of nirK and nirS were quantified in both soils using the newly designed primers (Supplementary Table S4), and their abundances were normalized based on the 16S rRNA gene abundance (Table 1). We found that nirK in Clusters I–III and nirS in Clusters I–II were abundant in both soils, and the abundance of nirK in Clusters I–II was higher in the denitrification-induced soil compared with the control soil. The nirK transcripts in Clusters I–III and the nirS transcripts in Cluster I were detected in both soils, and the abundances of these transcripts were higher in the denitrification-induced soil compared with the control soil (Table 1).
Table 1. The relative abundance of the nitrite reductase gene and transcript responsible for N2O emission in a soil microcosm.
| Treatmentsa | nirK in Cluster Ib | nirK in Cluster II | nirK in Cluster III | nirK in Cluster IV | Total nirK | nirS in Cluster I | nirS in Cluster II | Total nirS | Total Nitrite reductase gene | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | NF-8 | 0.506 | 0.717 | 2.191 | 0.027 | 3.441 | 1.863 | 0.498 | 2.36 | 5.802 |
| OF-8 | 5.631 | 5.272 | 1.282 | 0.014 | 12.199 | 1.344 | 0.275 | 1.619 | 13.818 | |
| Transcript | NF-8 | 0.002 | 0.003 | 0.001 | —c | 0.006 | — | — | — | 0.006 |
| OF-8 | 0.010 | 0.049 | 0.002 | — | 0.062 | 0.001 | — | 0.001 | 0.063 |
NF-8 and OF-8 represent the control and organic fertilized soil on the eighth day of N2O flux in the soil microcosm, which corresponds to the curve in Supplementary Figure S3.
Relative abundance of the nitrite reductase gene or transcript copies calculated as a percentage of the total bacterial 16S rRNA gene copies (for the mean and s.d. of replicates by treatment see Supplementary Table S4).
Not detected.
We analyzed the homology between the nirK and nirS sequences obtained from the soil microcosm study (Supplementary Table S3) and those from the denitrifying strains (Supplementary Table S2), which had been isolated previously from the same soil using a single-cell isolation method (Ashida et al., 2010). The strains were isolated in conditions where the activity and growth of denitrifiers were enhanced (Ashida et al., 2010; Nishizawa et al., 2012); thus, they were considered to be the dominant and active denitrifying populations in the soil. Among the 44 denitrifying isolates, 24 isolates possessed nirK or nirS that could be detected using the newly designed primers, but they could not be detected with the conventional primers (Supplementary Table S2). The nir sequences of 45 of 111 soil clones obtained from the microcosm experiment that belonged to Clusters I–III for nirK and Cluster I for nirS shared high homology (>80% of the amino-acid sequence) with the sequences of the denitrifying isolates (Supplementary Figure S4). The nir sequences from 26 of these 45 clones shared high homology with the nir sequences of the denitrifying isolates that could not be detected with the conventional primers, which included nirK of Enterobacter sp. and Sinorhizobium sp. in Cluster I; nirK of Ralstonia sp., Curvibacter sp., Wautersia sp. and Yersinia sp. in Cluster II; nirK of Streptomyces sp. and Micromonospora sp. in Cluster III; and nirS of Dechloromonas sp. in Cluster I. All of these denitrifying isolates exhibited a high capacity to produce N2O as the end product of denitrification (that is, the ratio of N2O production relative to N2O and N2 production was >80%).
Discussion
Previous studies utilized conventional primer sets to analyze nirK and nirS of denitrifiers that belonged primarily to alpha-, beta-, and gamma-proteobacteria from various environmental samples (Braker et al., 2000; Heylen et al., 2006; Smith et al., 2007; Ishii et al., 2011; Palmer et al., 2012). However, our phylogenetic analysis of the available nirK and nirS showed that a greater diversity of microorganisms possess nirK or nirS, including Actinobacteria, Bacteroidetes, Chloroflexi, Nitrospirae, Spirochetes, Planctomycetes, NC10, and even the archaeal phylum Euryarchaeota (Figure 1). Their nirK and nirS were not detected using the conventional primers (Supplementary Figures S1 and S2). The clone library analysis verified that the previously unconsidered nirK and nirS are distributed in many terrestrial environments, including cropland and paddy soils, natural and planted forest soils, and lake sediments (Figures 2 and 3). We also found that the microorganisms with nirK or nirS were 2–6 times more abundant than those determined using conventional primer sets in all of the terrestrial environments tested (Figure 4). These results strongly suggest that denitrifying microorganisms with nirK or nirS are more diverse and abundant than considered previously, and they are widely distributed in terrestrial environments.
The distributions of nirK and nirS differed in the terrestrial environments that we tested (Figure 4). Habitat-specific factors may have a crucial effect on the abundance and diversity of denitrifying microorganisms with nirK or nirS in different environments as suggested by Jones and Hallin (2010). In particular, the high abundance of nirS compared with nirK in rice paddy soils, where denitrification is generally active, may be attributed to the high availability of soluble iron (Yamazaki et al., 1995; Einsle et al., 2002). Soluble iron is used for the production of NirS, but it is generally limited by the insolubility of iron under aerobic conditions above pH 4 (Kraemer 2004). However, soluble iron can be seasonally available in paddy field soils because of water flooding. In contrast, Enwall et al. (2010) noted that copper availability might be a strong driving factor that shapes the abundance of nirK because NirK is a multicopper protein. The different distributions of nirK and nirS in each cluster also highlight the different physiological features of the total denitrifying communities in these environments as the results indicated a higher abundance or contribution to the denitrification of methane oxidizers in rice paddy soil. The different distributions also may affect the specific rates or end products of denitrification. Determination of the relationships between the phylogenies of nirK and nirS and the physiological features or responses to environmental gradients of denitrifying microorganisms will be important for understanding the spatial and temporal dynamics of denitrification (Philippot et al., 2009; Moralesl et al., 2010).
RNA- and culture-based analyses using a cropland soil also suggested that previously unconsidered microorganisms that possess nirK and nirS are responsible for N2O emissions. We performed RNA-based analyses to investigate their involvement in denitrification because microorganisms with nirK and nirS generally have life strategies other than denitrification, such as oxygen respiration, nitrification and methane oxidation. The abundances of transcripts of nirK in Clusters I–III and nirS in Cluster I were clearly higher in the denitrification-induced soil compared with the non-induced soil (Table 1). In addition, we found that many soil clones with nirK in Clusters I–III and nirS in Cluster I shared similar sequences with nirK or nirS, which could not be detected by the conventional primers, of the denitrifying strains isolated from the same soil (Supplementary Table S2 and Supplementary Figure S4). These strains exhibited a high capacity to produce N2O as the end product of denitrification. We also showed that the microorganisms in the soil microcosm tested were affiliated to Enterobacter sp., Sinorhizobium sp., Ralstonia sp., Curvibacter sp., Wautersia sp., Yersinia sp., Streptomyces sp., Micromonospora sp. and Dechloromonas sp.
To summarize, our new primers showed that microorganisms with nirK or nirS are phylogenetically more diverse and abundant than previously considered and are involved in denitrification in terrestrial environments. Although denitrifying microorganisms have been studied for a long time in various environments using culture- and PCR-based techniques with nirK and nirS as marker genes, these genes from many of the denitrifying strains that we isolated could not be amplified using the conventional primers. The methodology developed in the present study and the insights obtained should allow us to achieve a more precise understanding of the spatial and temporal dynamics of denitrification and facilitate more precise estimates of denitrification in different environments. It should be noted that the number of well-defined nirK and nirS is increasing. For example, since our collection of nirK and nirS sequences from databases, sequences have been obtained from more diverse microorganisms, such as nirK from Firmicutes (Murawska et al., 2013) and nirS from Crenarchaeota (Mardanov et al., 2012). Some groups of fungi also have nirK (Wei et al., 2015). PCR techniques still have a greater capacity for the deep analysis of target genes, although efforts are needed to minimize the PCR biases. The currently unknown diversity detected using metagenomic analyses and obtained by sophisticated isolation techniques will allow us to design primers with greater coverage.
Acknowledgments
This work was supported by the Institute for Fermentation, Osaka, the Program for Promotion of Basic and Applied Research for Innovation in a Bio-oriented Industry, Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry, a Grants-in-Aid for Scientific Research from the Japanese Society for the Promotion of Science (Nos. 24658133, 24780055, 25252026, 26292085 and 26712015) and the GRENE/Ecoinformatics project from the Ministry of Education, Culture, Sports, Science and Technology, Japan. This work also formed part of the project, assessment and extension of technologies for mitigating greenhouse gas emission from agricultural soils launched by MAFF.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abe DS, Kato K, Adams DD, Terai H, Tundisi JG. Contribution of free-living and attached bacteria to denitrification in the hypolimnion of a mesotrophic Japanese lake. Microbes Environ. 2000;15:93–101. [Google Scholar]
- Ashida N, Ishii S, Hayano S, Tago K, Tsuji T, Yoshimura Y, et al. Isolation of functional single cells from environments using a micromanipulator: application to study denitrifying bacteria. Appl Microbiol Biotechnol. 2010;85:1211–1217. doi: 10.1007/s00253-009-2330-z. [DOI] [PubMed] [Google Scholar]
- Bartossek R, Nicol GW, Lanzen A, Klenk HP, Schleper C. Homologues of nitrite reductases in ammonia-oxidizing archaea: diversity and genomic context. Environ Microbiol. 2010;12:1075–1088. doi: 10.1111/j.1462-2920.2010.02153.x. [DOI] [PubMed] [Google Scholar]
- Bouwman AF, Boumans LJM, Batjes NH. Emissions of N2O and NO from fertilized fields: summary of available measurement data. Global Biogeochem Cycle. 2002;16:1058–1071. [Google Scholar]
- Braker G, Fesefeldt A, Witzel KP. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol. 1998;64:3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braker G, Zhou J, Wu L, Devol AH, Tiedje JM. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl Environ Microbiol. 2000;66:2096–2104. doi: 10.1128/aem.66.5.2096-2104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantera JJ, Stein LY. Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ Microbiol. 2007;9:765–776. doi: 10.1111/j.1462-2920.2006.01198.x. [DOI] [PubMed] [Google Scholar]
- Conrad R. Soil organisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO) Microbiol Rev. 1996;60:609–640. doi: 10.1128/mr.60.4.609-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutruzzola F, Brown K, Wilson EK, Bellelli A, Arese M, Tegoni M, et al. The nitrite reductase from Pseudomonas aeruginosa: Essential role of two active-site histidines in the catalytic and structural properties. PNAS. 2001;98:2232–2237. doi: 10.1073/pnas.041365298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EA. The contribution of manure and fertilizer nitrogen to atmospheric nitrous oxide since 1860. Nat Geosci. 2009;2:659–662. [Google Scholar]
- Einsle O, Messerschmidt A, Huber R, Kroneck PM, Neese F. Mechanism of the six-electron reduction of nitrite to ammonia by cytochrome c nitrite reductase. J Am Chem Soc. 2002;124:11737–11745. doi: 10.1021/ja0206487. [DOI] [PubMed] [Google Scholar]
- Enwall K, Throbäck IN, Stenberg M, Söderström M, Hallin S. Soil resources influence spatial patterns of denitrifying communities at scales compatible with land management. Appl Environ Microbiol. 2010;76:2243–2250. doi: 10.1128/AEM.02197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin S, Lindgren PE. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl Environ Microbiol. 1999;65:1652–1657. doi: 10.1128/aem.65.4.1652-1657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heylen K, Gevers D, Vanparys B, Wittebolle L, Geets J, Boon N, et al. The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ Microbiol. 2006;8:2012–2021. doi: 10.1111/j.1462-2920.2006.01081.x. [DOI] [PubMed] [Google Scholar]
- Hochstein LI, Tomlinson GA. The enzymes associated with denitrification. Annu Rev Microbiol. 1988;42:231–261. doi: 10.1146/annurev.mi.42.100188.001311. [DOI] [PubMed] [Google Scholar]
- Ishii S, Ohno H, Tsuboi M, Otsuka S, Senoo K. Identification and isolation of active N2O reducers in rice paddy soil. ISME J. 2011;5:1936–1945. doi: 10.1038/ismej.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe K, Koba K, Ueda S, Senoo K, Harayama S, Suwa Y. A simple and rapid GC/MS method for the simultaneous determination of gaseous metabolites. J Microbiol Methods. 2011;84:46–51. doi: 10.1016/j.mimet.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Itoh H, Ishii S, Shiratori Y, Oshima K, Otsuka S, Hattori M, et al. Seasonal transition of active bacterial and archaeal communities in relation to water management in paddy soils. Microbes Environ. 2013;28:370–380. doi: 10.1264/jsme2.ME13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Graf DR, Bru D, Philippot L, Hallin S. The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J. 2013;7:417–426. doi: 10.1038/ismej.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Hallin S. Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J. 2010;4:633–641. doi: 10.1038/ismej.2009.152. [DOI] [PubMed] [Google Scholar]
- Jones CM, Stres B, Rosenquist M, Hallin S. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol Biol Evol. 2008;25:1955–1966. doi: 10.1093/molbev/msn146. [DOI] [PubMed] [Google Scholar]
- Katsuyama C, Kondo N, Suwa Y, Yamagishi T, Itoh M, Ohte N, et al. Denitrification activity and relevant bacteria revealed by nitrite reductase gene fragments in soil of temperate mixed forest. Microbes Environ. 2008;23:337–345. doi: 10.1264/jsme2.me08541. [DOI] [PubMed] [Google Scholar]
- Kraemer SM. Iron oxide dissolution and solubility in the presence of siderophores. Aquat Sci. 2004;66:3–18. [Google Scholar]
- Knowles R. Denitrification. Microbiol Rev. 1982;46:43–70. doi: 10.1128/mr.46.1.43-70.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leahy JG, Olsen RH. Kinetics of toluene degradation by toluene-oxidizing bacteria as a function of oxygen concentration, and the effect of nitrate. FEMS Microbiol Ecol. 1997;23:23–30. [Google Scholar]
- Li J, Wei G, Wang N, Gao Z. Diversity and distribution of nirK-harboring denitrifying bacteria in the wter column in the Yellow River estuary. Microbes Environ. 2014;29:107–110. doi: 10.1264/jsme2.ME13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardanov AV, Gumerov VM, Slobodkina GB, Beletsky AV, Bonch-Osmolovskaya EA, Ravin NV, et al. Complete genome sequence of strain 1860, a crenarchaeon of the genus Pyrobaculum able to grow with various electron acceptors. J Bacteriol. 2012;194:727–728. doi: 10.1128/JB.06465-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TL, Kaushik NK, Trevors JT, Whiteley HR. Review: Denitrification in temperate climate riparian zones. Water Air Soil Poll. 1999;111:171–186. [Google Scholar]
- Michotey V, Mejean V, Bonin P. Comparison of methods for quantification of cytochrome cd1-denitrifying bacteria in marine samples. Appl Environ Microbiol. 2000;66:1564–1571. doi: 10.1128/aem.66.4.1564-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir JWB. Nitrogen Cycling in Bacteria: Molecular Analysis. Caister Academic Press: Norfolk; 2011. [Google Scholar]
- Moralesl SE, Cosart T, Holben WE. Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J. 2010;4:799–808. doi: 10.1038/ismej.2010.8. [DOI] [PubMed] [Google Scholar]
- Murawska E, Fiedoruk K, Bideshi DK, Swiecicka I. Complete genome sequence of Bacillus thuringiensis subsp. thuringiensis strain IS5056, an isolate highly toxic to Trichoplusia ni. Genome Announc. 2013;1:e0010813. doi: 10.1128/genomeA.00108-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa T, Tago K, Uei Y, Ishii S, Isobe K, Otsuka S, et al. Advantages of functional single-cell isolation method over standard agar plate dilution method as a tool for studying denitrifying bacteria in rice paddy soil. AMB Express. 2012;2:50. doi: 10.1186/2191-0855-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa T, Uei Y, Tago K, Isobe K, Otsuka S, Senoo K. Taxonomic composition of denitrifying bacterial isolates is different among three rice paddy field soils in Japan. Soil Sci Plant Nutr. 2013;59:305–310. [Google Scholar]
- Nolan M, Tindall BJ, Pomrenke H, Lapidus A, Copeland A, Glavina Del Rio T, et al. Complete genome sequence of Rhodothermus marinus type strain (R-10 T) Stand Genomic Sci. 2009;1:283–290. doi: 10.4056/sigs.46736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Asano Y, Suzuki M. Transit time evaluation using a chloride concentration input step shift after forest cutting in a Japanese headwater catchment. Hydrol Process. 2009;23:2705–2713. [Google Scholar]
- Palmer K, Biasi C, Horn MA. Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. ISME J. 2012;6:1058–1077. doi: 10.1038/ismej.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Cuhel J, Saby NP, Chèneby D, Chronáková A, Bru D, et al. Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ Microbiol. 2009;11:1518–1526. doi: 10.1111/j.1462-2920.2009.01879.x. [DOI] [PubMed] [Google Scholar]
- Ravishankara AR, Daniel JS, Portmann RW. Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Kataoka K. Structure and function of type I copper in multicopper oxidases. Cell Mol Life Sci. 2007;64:2642–2656. doi: 10.1007/s00018-007-7183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-source platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Nedwell DB, Dong LF, Osborn AM. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microbiol. 2007;3:3612–3622. doi: 10.1128/AEM.02894-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Chinushi T, Nagumo Y, Nakano T, Ohyama T. Effect of deep placement of controlled release nitrogen fertilizer (coated urea) on growth, yield, and nitrogen fixation of soybean plants. Soil Sci Plant Nutr. 1991;37:223–231. [Google Scholar]
- Takai K, Suzuki M, Nakagawa S, Miyazaki M, Suzuki Y, Inagaki F, et al. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int J Syst Evol Microbiol. 2006;56:1725–1733. doi: 10.1099/ijs.0.64255-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throbäck IN, Enwall K, Jarvis A, Hallin S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol. 2004;49:401–417. doi: 10.1016/j.femsec.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Tiedje JM.1994DenitrifiersIn: Bottomley PS, Angle JS, Weaver RW, (eds). Methods of Soil Analysis: Part 2-Microbiological and Biochemical Properties Soil Science Society of America Press: Madison; 253–254. [Google Scholar]
- Tiedje JM.1988Ecology of denitrification and dissimilatory nitrate reduction to ammoniumIn: Zehnder AJB, (ed). Biology of Anaerobic Microorganisms John Wiley & Sons Press; New York, pp179–244. [Google Scholar]
- Urakawa R, Ohte N, Shibata H, Tateno R, Hishi T, Fukushima K, et al. Biogeochemical nitrogen properties of forest soils in the Japanese archipelago. Ecol Res. 2015;30:1–2. [Google Scholar]
- Wei W, Isobe K, Shiratori Y, Nishizawa T, Ohte N, Otsuka S, et al. N2O emission from cropland field soil through fungal denitrification after surface applications of organic fertilizer. Soil Biol Biochem. 2014;69:157–167. [Google Scholar]
- Wei W, Isobe K, Shiratori Y, Nishizawa T, Ohte N, Ise Y, et al. Development of PCR primers targeting fungal nirK to study fungal denitrification in the environment. Soil Biol Biochem. 2015;81:282–286. [Google Scholar]
- Yamazaki T, Oyanagi H, Fujiwara T, Fukumori Y. Nitrite reductase from the magnetotactic bacterium Magnetospirillum magnetotacticum. A novel cytochrome cd1 with Fe (II): nitrite oxidoreductase activity. Eur J Biochem. 1995;233:665–671. doi: 10.1111/j.1432-1033.1995.665_2.x. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Ishii S, Fujii D, Otsuka S, Senoo K. Identification of active denitrifiers in rice paddy soil by DNA- and RNA-based analyses. Microbes Environ. 2012;27:456–461. doi: 10.1264/jsme2.ME12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.