Abstract
Grain shape is an important trait for improving rice yield. A number of quantitative trait loci (QTLs) for this trait have been identified by using primary F2 mapping populations and recombinant inbred lines, in which QTLs with a small effect are harder to detect than they would be in advanced generations. In this study, we developed two advanced mapping populations (chromosome segment substitution lines [CSSLs] and BC4F2 lines consisting of more than 2000 individuals) in the genetic backgrounds of two improved cultivars: a japonica cultivar (Koshihikari) with short, round grains, and an indica cultivar (IR64) with long, slender grains. We compared the ability of these materials to reveal QTLs for grain shape with that of an F2 population. Only 8 QTLs for grain length or grain width were detected in the F2 population, versus 47 in the CSSL population and 65 in the BC4F2 population. These results strongly suggest that advanced mapping populations can reveal QTLs for agronomic traits under complicated genetic control, and that DNA markers linked with the QTLs are useful for choosing superior allelic combinations to enhance grain shape in the Koshihikari and IR64 genetic backgrounds.
Keywords: chromosome segment substitution lines (CSSLs), advanced mapping population, quantitative trait loci (QTL), grain shape, Oryza sativa L
Introduction
The agronomic traits of rice are generally controlled by multiple quantitative trait loci (QTLs; Yamamoto et al. 2009, Yano and Sasaki 1997). Identification of QTLs that control grain production and resistance to biotic and abiotic stresses is a primary step in efforts to enhance rice cultivars by incorporating beneficial QTL alleles into superior genetic backgrounds. Grain shape in rice is an important trait, and QTL alleles that increase grain size and to control shape are a target in allele mining studies with the goal of increasing yield (Huang et al. 2013). Grain shape in Asian cultivated rice is diverse, and extensive research has been conducted to identify QTLs controlling it. More than 100 papers have reported nearly 200 QTLs for grain length and grain width (reviewed by Huang et al. 2013), of which 15 have been mapped on a fine scale (Bai et al. 2010, Guo et al. 2009, Li et al. 2004, Qiu et al. 2012, Shao et al. 2012, Wan et al. 2006) and 16 have been cloned including GS3 (Fan et al. 2006, Mao et al. 2010), GW2 (Song et al. 2007), GW5/qSW5 (Shomura et al. 2008, Weng et al. 2008), GS5 (Li et al. 2011), TGW6 (Ishimaru et al. 2013), and GW6a (Song et al. 2015). Although this progress has encouraged marker-assisted breeding to target these QTLs and genes in rice, those studies focused mainly on QTLs with large effects, and the information available on QTL alleles is not currently sufficient for fine-tuning grain shape in practical breeding programs.
A drastic increase in grain size usually does not increase grain productivity proportionally, owing to decreases in both grain filling and grain quality as result of imbalances between the sink and source potentials (Peng et al. 2008, Takai et al. 2013, Takita 1983). Therefore, grain shape should be improved by using appropriate QTL alleles to maintain an appropriate balance between sinks and sources and thus to allow an increase in grain yield. Thus, alleles that govern various effects on grain size are needed for efficient improvement of rice, since this would provide a “tool-box” of QTL alleles that provide an appropriate grain shape in genotypes with diverse source capacities.
Most QTL mapping studies in rice use primary mapping populations such as F2 populations, recombinant inbred lines (RILs), or backcross inbred lines (BILs), and thus might not detect QTLs with small effects owing to the lower detection power of using such early generations (Yamamoto et al. 2009). In contrast, using advanced mapping populations can overcome this problem because their more uniform genetic background increases the ability to detect even QTLs with small effects (Tanksley and Nelson 1996, Yamamoto et al. 2009). Among these populations, chromosome segment substitution lines (CSSLs) let us detect QTLs distributed throughout the genome with high sensitivity and by using a smaller number of plants than would be required in analyses using F2 populations or RILs (Abe et al. 2013, Ando et al. 2008, Ebitani et al. 2005, Hori et al. 2010, Ishikawa et al. 2005, Kubo et al. 2002, Murata et al. 2014, Takai et al. 2009). Through the use of such populations, more than 40 QTLs have been detected that are associated with yield potential in rice, a complex trait whose evaluation requires much time and labor (Takai et al. 2014). Takai et al. (2014) also confirmed the importance of maintaining the sink–source balance in efforts to increase yield, and noted that this balance fluctuated in response to the nature of introgression of the donor’s genome.
The size of the donor’s chromosome segments in the respective CSSLs is generally not small enough to define the position of QTLs within a smaller chromosomal region relative to other mapping populations, so genetic mapping using CSSLs sometimes fails to assign a position to some QTLs (Ebitani et al. 2005). However, an advanced mapping population in which small chromosomal regions have segregated in a highly homogeneous background represents a promising alternative to locate such QTLs (Paterson et al. 1990). Recently, a genome-wide genetic analysis using such populations was conducted to uncover the genetic architecture underlying rice’s heading date (K. Hori et al., unpublished). In that study, 255 QTLs throughout the rice genome were detected by using 11 cross combinations, of which 127 were detected in regions that differed from those of the 650 previously identified QTLs for heading date that are listed in public databases (Yonemaru et al. 2010, Youens-Clark et al. 2011). The evidence strongly suggests that this type of advanced mapping population is useful to detect naturally occurring variations that underlie agronomic traits, including QTLs with small effects.
To enhance our understanding of the genetic control of grain shape in rice, we performed a QTL analysis for grain shape (i.e., grain length and width) in a cross between a japonica rice cultivar with short, round grains (Koshihikari) and an indica cultivar (IR64) with long, slender grains in three mapping populations: an F2 primary mapping population and reciprocal CSSL and BC4F2 populations as the advanced mapping populations. On the basis of the results, we compare the QTL detection power among these populations and comprehensively review the genetic control of grain shape in this japonica × indica cross.
Materials and Methods
Plant materials
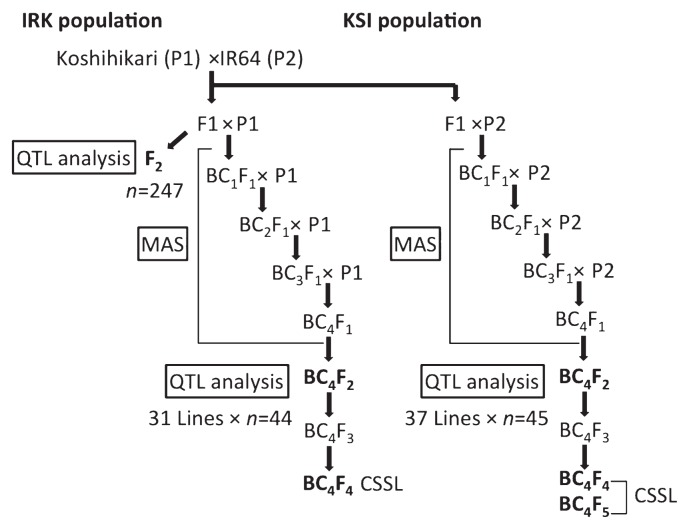
We used three Koshihikari × IR64 populations in this study: an F2 population and advanced reciprocal CSSL and BC4F2 populations (Fig. 1). We began with an F1 population derived from the cross between Koshihikari and IR64 (IRGC66970), and repeatedly backcrossed the progeny with either Koshihikari or IR64 to produce BC4F1 plants. We conducted a whole-genome survey using 150 simple sequence repeat (SSR) markers in each backcrossed generation (BC1F1 to BC4F1) to select target chromosome segments in each CSSL and to minimize non-target chromosome segments from the donor, then selected plants that are homozygous for the target chromosome segments in the self-pollinated progeny (Supplemental Table 1). From BC4F4 or BC4F5 plants, we selected 42 CSSLs with a Koshihikari genetic background (IRK-CSSL) and 40 with an IR64 genetic background (KSI-CSSL), and grew the plants with their parents in 2011 (Fig. 1).
Fig. 1.
Development of the plant materials used in this study. CSSL, chromosome segment substitution line; MAS, marker-assisted selection; QTL, quantitative trait locus.
To further dissect grain shape, we grew 31 BC4F2 populations developed with Koshihikari as the recurrent parent (IRK-BC4F2), 37 BC4F2 populations developed with IR64 as the recurrent parent (KSI-BC4F2) and their parents in 2012 (Fig. 1). We used 44 plants for each IRK-BC4F2 line and 45 plants for each KSI-BC4F2 line in our genotyping and trait measurements. As a reference primary mapping population for QTL detection, we grew 247 F2 plants and their parents in 2013. All plants were grown in an experimental field at the National Institute of Agrobiological Sciences, Tsukuba, Japan (36.03°N, 140.11°E). Basal fertilizer was applied at a rate of 56 kg/ha N, 56 kg/ha P, and 56 kg K/ha. Month-old seedlings were transplanted in mid-May at one seedling per hill in plots with a double row for each line, at 18 cm between plants and 36 cm between rows. Rice grains were harvested in September and were air-dried before analysis.
Measurement of grain shape
We scanned approximately 250 per individual at 600 dpi on a GTX-820 scanner (Seiko Epson, Inc., Suwa, Japan). We measured the grain length (GL) and grain width (GW) of the seeds in the images using the “SmartGrain” grain shape analysis software (Tanabata et al. 2012). The “awn removal” function in the software was set at “intermediate”, and other parameters used the default values. To calculate the mean and standard deviation (SD) of the grain length and grain width parameters for each population or line, we excluded any grains whose outlines lay outside the 95% confidence interval.
DNA extraction and marker analysis
Fresh leaves were harvested from field-grown plants, and total DNA was extracted from 1- to 3-cm sections of the fresh leaves by using the method of Takeuchi et al. (2008) for genotyping of the CSSLs and by crushing the tissue in 30 μL 0.5 M NaOH and diluting it in 120 μL of 1 M Tris·HCl (pH 8.0) for the F2 and BC4F2 populations. We used a total of 129 SSR markers and a single-nucleotide polymorphism (SNP) marker for GS3 (IDGS3_001_3), with an average marker interval of 2.7 Mb (Supplemental Table 1), to detect QTLs in the F2 and BC4F2 populations. We added 97 SSR or insertion/deletion markers to the chromosomal regions around QTLs to determine their positions in IRK-BC4F2, and 100 for KSI-BC4F2 (Supplemental Table 1). The genotypes of the CSSLs, which we characterized by using 271 SNP markers (Supplemental Tables 2, 3), were determined by using 384-plex SNPs for the Illumina GoldenGate BeadArray technology platform (Illumina, Inc., San Diego, CA, USA) based on previously reported information (Ebana et al. 2010).
Substitution mapping of QTLs in CSSLs
We used Student’s t-test in Microsoft Excel 2010 (Microsoft, Redmond, WA, USA) to identify mean values for the grain-size parameters that differed significantly between the CSSLs and the recurrent parents, with significance at P < 0.05 (two-tailed). A CSSL whose mean for a grain size parameter differed significantly from the recurrent parent by the t-test was judged to carry one or more putative QTLs in one or more regions of the donor’s chromosome segments in that CSSL. When the t-test was not significant, the CSSL was judged to lack a QTL in its donor’s chromosome segments. Putative QTLs were assigned as described by Paterson et al. (1990) and Ishikawa et al. (2005). In brief, a small chromosomal region that carries one or more QTLs was defined on the basis of the association among RILs between the phenotype and genotype within a certain chromosomal region. When a CSSL differed significantly from another CSSL that carried a single putative QTL and the recurrent parent for a trait, this line was judged to carry additional putative QTLs. Accordingly, putative QTLs were assigned to maximize the number of CSSLs whose phenotype could be explained. As a result, CSSLs whose phenotype differed significantly from the recurrent parent but for which the putative QTL’s region was not supported by some other CSSLs were judged to be “not assigned”.
QTL analysis in the F2 and BC4F2 populations
We constructed a genetic map using MAPMAKER/EXP v. 3.0 software (Lander et al. 1987). QTL analysis for grain length and grain width was performed using version 2.5 of QTL Cartographer software (Basten et al. 2005), and the threshold was obtained by using 1000 permutations. QTLs were named according to the nomenclature rules of McCouch et al. (1997). Since the marker interval in the primary QTL analysis was 2.7 Mb, QTLs with a similar additive effect that were located less than 2.7 Mb apart were judged to be a single QTL.
Results
Variation of grain length and grain width in the F2 population
To clarify phenotypic variation of seed shape in F2 of the cross between Koshihikari and IR64, grain length and grain width in the 247 F2 plants and their parents were measured. The mean grain length and grain width of Koshihikari grown in 2013 were 7.15 ± 0.35 mm (mean ± SD) and 3.48 ± 0.17 mm, respectively. Those of IR64 grown in 2013 were 9.64 ± 0.51 mm and 2.54 ± 0.16 mm, respectively. The difference in seed shape in these two cultivars was apparent in the two traits. Grain lengths in the F2 population ranged from 6.81 to 9.34 mm and averaged 8.17 ± 0.38 mm, and grain widths ranged from 2.34 to 3.36 mm and averaged 2.86 ± 0.17 mm (Supplemental Fig. 1A, 1B). The distribution of these two traits in the F2 population was continuous between the values of the parents except for a gap at a grain length of 7.1 mm.
QTLs for grain length and grain width in the F2 population
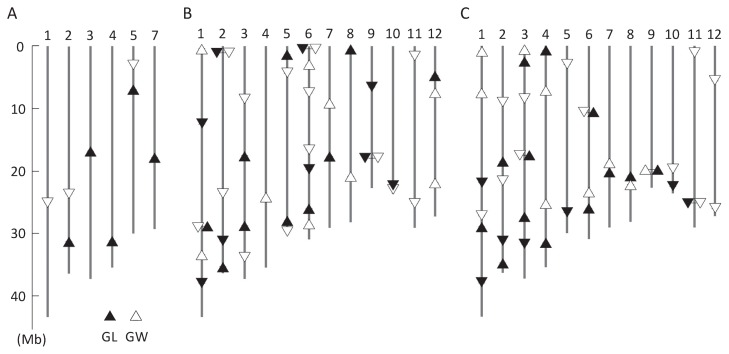
To identify QTLs for grain length and grain width in a primary mapping population, QTL analysis was performed in the F2 population of the cross between Koshihikari and IR64. Five QTLs whose IR64 allele increased grain length and three whose IR64 allele decreased grain width were detected, as expected from the trait values of the parents and the distribution in the F2 population (Fig. 2A, Supplemental Fig. 1). The number of QTLs detected is comparable to that in the previous studies using the F2 (1 to 32, 8.7 on average) (Huang et al. 2013). A QTL for grain length on chromosome 3 accounted for 40% of the phenotypic variation and a QTL for grain width on chromosome 5 accounted for 24% of the phenotypic variation, but the other QTLs had small effects, and each accounted for less than 10% of the phenotypic variation (Table 1).
Fig. 2.
Chromosome locations of the QTLs for grain length and grain width detected in this study. (A) F2 population (Koshihikari × IR64), (B) IRK-BC4F2 population (see Fig. 1 for details), (C) KSI-BC4F2 (see Fig. 1 for details). Upward-pointing triangles (△, ▲) indicate that the IR64 alleles increase the parameter value, whereas downward-pointing triangles (▽, ▼) indicate that the IR64 alleles decrease the parameter value.
Table 1.
QTLs for grain length and grain width identified in an F2 population and in the BC4F2 populations
| Chr. | Postion (Mb) | QTL name | Additive effect (BC4F2)e | Additive effect (F2)e | R2 (%) (F2)f | |
|---|---|---|---|---|---|---|
|
| ||||||
| IRK | KSI | |||||
| 1 | 12.49 | qGL1-1 | 0.102 | – | – | – |
| 1 | 21.90 | qGL1-2 | – | 0.083 | – | – |
| 1 | 28.90 | qGL1-3 | −0.117 | −0.156 | – | – |
| 1 | 38.01 | qGL1-4c | 0.116 | 0.258 | – | – |
| 1 | 0.30–0.75a | qGW1-1 | −0.055 | −0.054 | – | – |
| 1 | 7.40 | qGW1-2 | – | −0.014 | – | – |
| 1 | 25.07b | (W) | – | – | 0.060 | 4.9 |
| 1 | 27.19–28.90a | qGW1-3 | 0.026 | 0.059 | – | – |
| 1 | 33.48 | qGW1-4c | −0.015 | – | – | – |
| 2 | 0.98 | qGL2-1 | 0.076 | – | – | – |
| 2 | 18.46 | qGL2-2 | – | −0.168 | – | – |
| 2 | 31.20 | qGL2-3 | 0.080 | 0.091 | – | – |
| 2 | 31.20b | (L) | – | – | −0.020 | 3.4 |
| 2 | 34.69–35.26a | qGL2-4c | −0.023 | −0.236 | – | – |
| 2 | 0.98 | qGW2-1 | 0.064 | – | – | – |
| 2 | 8.98 | qGW2-2 | – | 0.052 | – | – |
| 2 | 21.52–23.57a | qGW2-3 | 0.099 | 0.058 | 0.074 | 8.5 |
| 3 | 2.45 | qGL3-1 | – | −0.161 | – | – |
| 3 | 17.42 | qGL3-2 | −0.304 | −0.347 | −0.315 | 40.2 |
| 3 | 27.32–28.78a | qGL3-3 | −0.102 | −0.131 | – | – |
| 3 | 31.74 | qGL3-4c | – | 0.162 | – | – |
| 3 | 0.54 | qGW3-1c | – | −0.038 | – | – |
| 3 | 8.39 | qGW3-2 | 0.028 | 0.057 | – | – |
| 3 | 17.42 | qGW3-3 | – | 0.071 | – | – |
| 3 | 33.80 | qGW3-4d | 0.044 | – | – | – |
| 4 | 0.69 | qGL4-1 | – | −0.105 | – | – |
| 4 | 31.56 | qGL4-2 | – | −0.176 | −0.127 | 4.4 |
| 4 | 7.10 | qGW4-1d | – | −0.054 | – | – |
| 4 | 24.24–25.22a | qGW4-2 | −0.022 | −0.036 | – | – |
| 5 | 1.23 | qGL5-1 | −0.159 | – | – | – |
| 5 | 6.83b | (L) | – | – | −0.124 | 4.5 |
| 5 | 26.65 | qGL5-2c | – | 0.173 | – | – |
| 5 | 28.02 | qGL5-3 | −0.092 | – | – | – |
| 5 | 2.89–4.26a | qGW5-1 | 0.150 | 0.163 | 0.127 | 24.4 |
| 5 | 29.54 | qGW5-2c | 0.019 | – | – | – |
| 6 | 0.38 | qGL6-1c | 0.093 | – | – | – |
| 6 | 10.65 | qGL6-2 | – | −0.384 | – | – |
| 6 | 19.70 | qGL6-3c | 0.069 | – | – | – |
| 6 | 26.00 | qGL6-4 | −0.096 | −0.127 | – | – |
| 6 | 0.38 | qGW6-1c | 0.026 | – | – | – |
| 6 | 2.85 | qGW6-2c | −0.041 | – | – | – |
| 6 | 7.41 | qGW6-3 | 0.027 | – | – | – |
| 6 | 10.65 | qGW6-4 | – | 0.066 | – | – |
| 6 | 16.58 | qGW6-5 | 0.037 | – | – | – |
| 6 | 23.34 | qGW6-6 | – | −0.044 | – | – |
| 6 | 28.47 | qGW6-7 | −0.039 | – | – | – |
| 7 | 17.69–20.11a | qGL7 | −0.105 | −0.134 | −0.157 | 7.6 |
| 7 | 9.08 | qGW7-1 | −0.052 | – | – | – |
| 7 | 18.66 | qGW7-2 | – | −0.031 | – | – |
| 8 | 0.42 | qGL8-1 | −0.078 | – | – | – |
| 8 | 20.82 | qGL8-2c | – | −0.112 | – | – |
| 8 | 20.82–22.21a | qGW8 | −0.035 | −0.044 | – | – |
| 9 | 6.59 | qGL9-1 | 0.051 | – | – | – |
| 9 | 18.04 | qGL9-2 | 0.072 | – | – | – |
| 9 | 19.79 | qGL9-3c | – | −0.025 | – | – |
| 9 | 18.04 | qGW9-1d | 0.037 | – | – | – |
| 9 | 19.79 | qGW9-2d | – | −0.034 | – | – |
| 10 | 22.43 | qGL10c | 0.116 | 0.082 | – | – |
| 10 | 19.69 | qGW10-1 | – | 0.045 | – | – |
| 10 | 22.83 | qGW10-2 | 0.025 | – | – | – |
| 11 | 25.13 | qGL11d | – | 0.134 | – | – |
| 11 | 1.08–1.46a | qGW11-1 | 0.028 | 0.060 | – | – |
| 11 | 25.13 | qGW11-2 | 0.043 | 0.018 | – | – |
| 12 | 4.75 | qGL12 | −0.058 | – | – | – |
| 12 | 5.51 | qGW12-1d | – | 0.031 | – | – |
| 12 | 7.57 | qGW12-2c | −0.018 | – | – | – |
| 12 | 22.02 | qGW12-3c | −0.050 | – | – | – |
| 12 | 26.00 | qGW12-4c | – | 0.047 | – | – |
The positions of the QTLs in the two BC4F2 populations fell within 2.7 Mb of each other, and they were judged to be the same QTL.
The positions of the QTLs in the F2 population fell >2.7 Mb from that in the two BC4F2 populations, but they were judged to be the same on the basis of their additive effects.
The QTLs that were not detected in the F2 or CSSL populations.
The QTLs that were detected only in the BC4F2 population of the same genetic background.
Additive effect of Koshihikari allele on grain length or grain width.
Percentage of phenotypic variance explained by QTL.
Genomic constitution of CSSLs
To determine grain shape in advanced mapping populations of the same cross combination, 42 CSSLs with a Koshihikari genetic background (IRK-CSSL) and 40 with an IR64 genetic background (KSI-CSSL) were developed and their genomic constitution was characterized. Each of the 42 IRK-CSSLs contained at least one large substituted segment of a particular target chromosomal region from IR64 and additional small segments in non-target regions in the Koshihikari genetic background (Supplemental Table 2). If we assume for simplicity that each recombination occurred midway between the two surrounding markers, the size of the target IR64 chromosomal segments in each CSSL ranged from 1.3 to 22.3 Mb and averaged 11.1 Mb (Supplemental Table 2). Although several non-target regions from IR64 (a mean size of 2.2 Mb) and heterozygous regions (a mean size of 0.8 Mb) remained, each CSSL contained approximately 95.9% of the Koshihikari genome (Supplemental Table 2). Similarly, each of the 40 KSI-CSSLs contained at least one large substituted segment of a particular target chromosomal region from Koshihikari and additional small segments in non-target regions in the IR64 genetic background (Supplemental Table 3). The size of the target Koshihikari chromosomal segments in each CSSL ranged from 2.9 to 27.8 Mb and averaged 12.3 Mb, with several non-target regions from Koshihikari (a mean of 2.3 Mb) and heterozygous regions (a mean of 2.6 Mb), and each CSSL contained approximately 95.4% of the IR64 genome (Supplemental Table 3).
QTL detection in the CSSLs
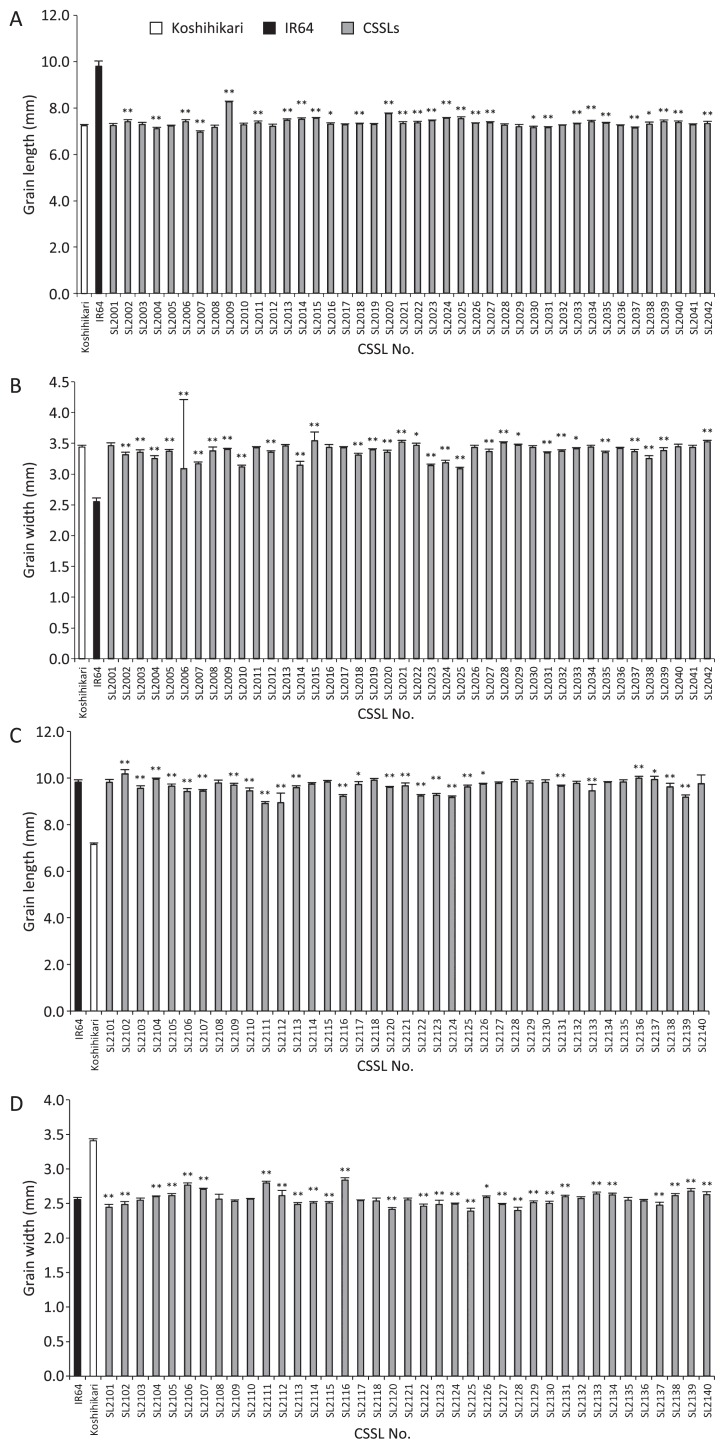
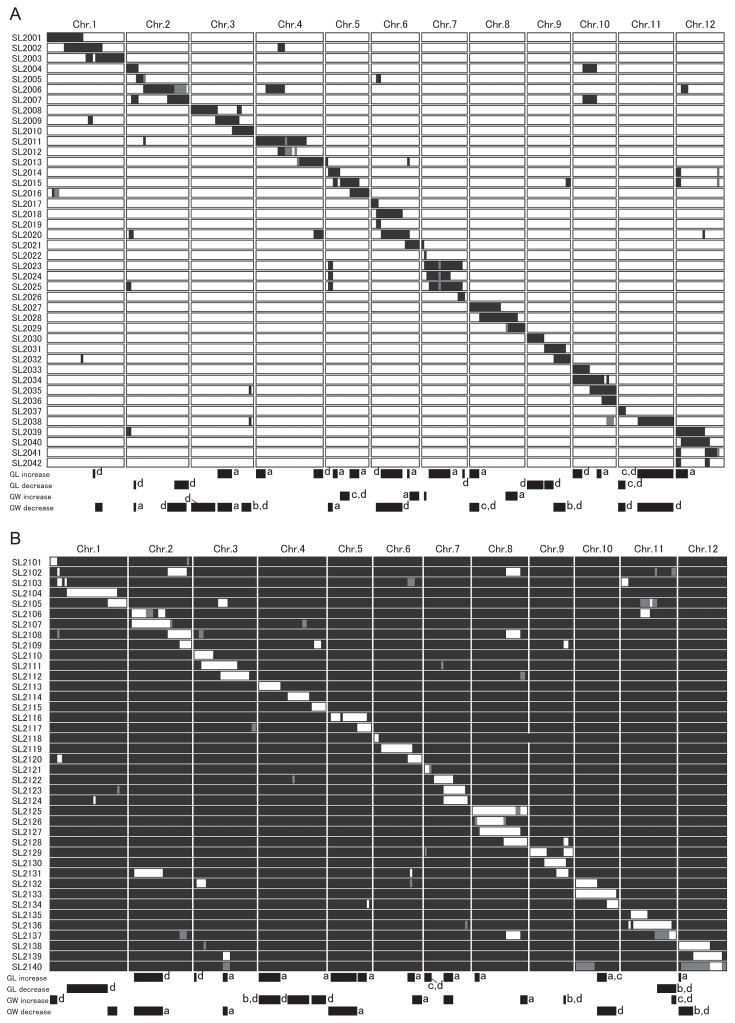
To identify putative QTLs and their chromosomal locations in the IRK-CSSLs and KSI-CSSLs developed in the present study, grain length and grain width in these CSSLs and their parents were measured and substitution mapping for these traits was performed. In the IRK-CSSLs, grain lengths ranged from 6.95 to 8.27 mm, versus 7.24 mm in Koshihikari and 9.80 mm in IR64 in 2011 (Fig. 3A). Grain widths ranged from 3.09 to 3.55 mm, versus 3.44 mm in Koshihikari and 2.55 mm in IR64 (Fig. 3B). Twenty-nine IRK-CSSLs differed significantly from ‘Koshihikari’ in grain length (Fig. 3A), and 31 differed in grain width (Fig. 3B). Putative QTLs were assigned at 20 chromosomal regions for grain length and 16 for grain width (Fig. 4A). We could not assign putative QTLs in one line (SL2022) that differed significantly from Koshihikari in grain length and in four lines (SL2012, SL2033, SL2039, and SL2042) that differed in grain width (Fig. 4A). Seven CSSLs carried multiple putative QTLs for either grain length or grain width (Fig. 4A, footnote c).
Fig. 3.
Grain length and grain width in two sets of CSSLs: (A, B) IRK-CSSLs, with IR64 introgression in a Koshihikari background; (C, D) KSI-CSSLs, with Koshihikari introgression in an IR64 background. Values are means ± SD. Bars labeled with asterisks differed significantly from the means for (A, B) Koshihikari and (C, D) IR64: *, P < 0.05; **, P < 0.01 (two-tailed t-test). Data for SL2119 were excluded from this figure because this line headed too late for us to harvest seeds under natural field conditions.
Fig. 4.
(A) Graphical genotypes and positions of the putative QTLs in the IRK-CSSLs, with IR64 introgression in a Koshihikari background. Black regions, homozygous for IR64 alleles; white, homozygous for Koshihikari alleles; gray, heterozygous. (B) Graphical genotypes and positions of the putative QTLs in the KSI-CSSLs, with Koshihikari introgression in an IR64 background. Black regions, homozygous for IR64 alleles; white, homozygous for Koshihikari alleles; gray, heterozygous. a, Putative QTLs that were detected in both the IRK- and KSI-CSSLs; b, putative epistatic QTLs that were not detected in the other genetic background in the CSSLs or in the BC4F2 population; c, putative QTLs that were not detected in the BC4F2 population. d, putative QTLs that were detected in one set of CSSLs; Chr., chromosome number; GL, grain length; GW, grain width.
In the KSI-CSSLs, grain lengths ranged from 8.91 to 10.18 mm, versus 7.15 mm in Koshihikari and 9.83 mm in IR64 (Fig. 3C). Grain widths ranged from 2.39 to 2.84 mm, versus 3.41 mm in Koshihikari and IR64 in 2.56 mm (Fig. 3D). Twenty-six KSI-CSSLs differed significantly from IR64 in grain length (Fig. 3C), and 29 differed in grain width (Fig. 3D). Putative QTLs were assigned at 14 chromosomal regions for grain length and 15 for grain width (Fig. 4B). We could not assign putative QTLs in one line (SL2109) that differed significantly from IR64 in grain length and in two lines (SL2126 and SL2127) that differed in grain width (Fig. 4B). Three CSSLs were judged to be carrying multiple putative QTLs for either grain length or grain width (Fig. 4B, footnote c).
Nine chromosomal regions that carried putative QTLs for grain length were shared in the two sets of CSSLs (Fig. 4A, 4B, footnote a), but another 16 putative QTLs were found in only one of the sets (Fig. 4A, 4B, footnote d). Five chromosomal regions that carried putative QTLs for grain width were shared in the two sets of CSSLs (Fig. 4A, 4B, footnote a), but another 17 putative QTLs were only found in one of the sets (Fig. 4A, 4B, footnote d). In total, 47 putative QTLs for the two traits, nearly six times that in our analysis using the F2 population, were identified and assigned a location in the two sets of CSSLs (Fig. 4A, 4B).
Variation of grain length and grain width in the BC4F2 populations
To further clarify grain shape in another advanced mapping population, grain length and grain width were measured in 31 BC4F2 populations developed with Koshihikari as the recurrent parent (IRK-BC4F2) and 37 BC4F2 populations developed with IR64 as the recurrent parent (KSI-BC4F2). The mean grain length and grain width of Koshihikari grown in 2012 were 7.03 ± 0.32 mm and 3.41 ± 0.16 mm, respectively. Those of IR64 grown in 2012 were 9.49 ± 0.51 mm and 2.50 ± 0.16 mm, respectively. Grain length in IRK-BC4F2 ranged from 6.55 to 8.65 mm and averaged 7.12 ± 0.21 mm, whereas that in KSI-BC4F2 ranged from 8.27 to 10.54 mm and averaged 9.35 ± 0.28 mm (Supplemental Fig. 1C, 1E). Grain width in IRK-BC4F2 ranged from 2.97 to 3.82 mm, and averaged 3.38 ± 0.09 mm, whereas that in KSI-BC4F2 ranged from 2.07 to 3.10 mm, and averaged 2.52 ± 0.10 mm (Supplemental Fig. 1D, 1F). The range of variation of both traits in the BC4F2 populations was smaller than that in the F2 population, and their averages were closer to those of the recurrent parents than in the F2 population.
QTLs for grain length and grain width in the BC4F2 populations
QTL analysis for grain length and grain width was performed in each of 31 IRK-BC4F2 and 37 KSI-BC4F2 populations and the number of QTLs and their positions and effects in BC4F2 were compared with those in other mapping populations. In IRK-BC4F2, we detected QTLs in 23 out of 31 populations (Supplemental Table 4): 19 for grain length and 23 for grain width (Fig. 2B). In KSI-BC4F2, we detected QTLs in 26 out of 37 populations (Supplemental Table 5): 20 for grain length and 21 for grain width (Fig. 2C). We detected a total of 65 QTLs for the two traits in the two populations (Table 1). This number is 8 times that in our analysis using the F2 and 1.4 times that in our analysis using the CSSLs.
Of these 65 QTLs, 8 (6 for grain length and 2 for grain width) were mapped at the same marker intervals, and another 10 (3 for grain length and 7 for grain width) were mapped within 2.7 Mb of each other between the IRK-BC4F2 and KSI-BC4F2 populations (Table 1, footnote a). Another 17 (9 for grain length and 8 for grain width) were not detected in the F2 or CSSL populations (Table 1, footnote c). Among the 33 putative QTLs that were detected in one set of CSSLs (Fig. 4A, 4B, footnote d), 6 were detected only in the BC4F2 population of the same genetic background (Fig. 4A, 4B, footnote b, Table 1, footnote d), and the other 27 were detected in the BC4F2 of the other genetic background (Table 1). Among the 47 putative QTLs detected in the CSSLs, 7 were not detected in the BC4F2 populations (Fig. 4A, 4B, footnote c). Our analysis confirmed that most of the 65 QTLs detected using BC4F2 were found in both the Koshihikari and IR64 genetic backgrounds.
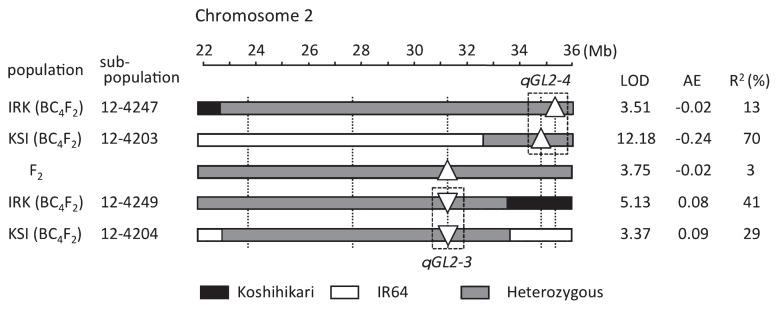
All 8 of the QTLs detected in the F2 population were validated in the BC4F2 populations, although 3 of the BC4F2 QTLs were located more than 2.7 Mb from the corresponding QTL in the F2 population (Table 1, footnote b). For example, in the F2 population, a QTL whose IR64 allele increases grain length (additive effect [AE] = −0.02) was detected at 31.20 Mb on chromosome 2. But in IRK-BC4F2, QTL qGL2-3, with the opposite additive effect (AE = 0.09), was detected at the same position, and qGL2-4, whose additive effect was in the same direction (AE = −0.03) as in the F2 population, was detected at 34.69 Mb. The positions of the two QTLs in IRK-BC4F2 were almost same as those in KSI-BC4F2. Notably, the additive effects of qGL2-4 in the KSI-BC4F2 population (AE = −0.24), in which only qGL2-4 is segregating, was 8 times that in the IRK-BC4F2 population (AE = −0.03) and more than 8 times that in the F2 population (AE = −0.02), in which both qGL2-3 and qGL2-4 are segregating (Fig. 5).
Fig. 5.
Graphical genotypes and positions of the QTLs for grain length that were detected in the region from 30 to 36 Mb on chromosome 2 in the F2 population and in two BC4F2 sub-populations in the genetic background of Koshihikari (IRK) or IR64 (KSI). Black boxes, homozygous for IR64; white, homozygous for Koshihikari; gray, heterozygous. AE, additive effect. Upward-pointing triangles (△) indicate that the IR64 alleles increase values, and downward-pointing triangles (▽) indicate that the IR64 alleles decrease values.
Collectively, BC4F2 population exhibited highest sensitivity in detecting QTLs among three populations, yet our analysis underestimated in the additive effect of some QTLs.
Discussion
Our results clearly indicate that the genetic control of grain shape, an important agronomic trait of rice, is complex, as we found 65 QTLs involved in genetic variation of grain shape in a single cross combination. Extensive genetic research on grain shape previously identified nearly 200 QTLs for grain length and grain width, of which 16 have been cloned in rice (Huang et al. 2013). The two QTLs with major effects detected here, qGL3-2 and qGW5-1, probably correspond to GS3 and GW5/qSW5, respectively (Fan et al. 2006, Mao et al. 2010, Shomura et al. 2008, Weng et al. 2008). The three QTLs (qGL1-4, qGW2-2, and qGW8) were located in genomic regions harboring previously cloned or finely mapped gene loci (Singh et al. 2012, Song et al. 2007, Xie et al. 2006). Nevertheless, some QTLs detected here (qGW1-1, qGL4-1, qGL9-1, qGL12-1, and qGL12-2) appear to be new on the basis of the map locations of the previously reported QTLs (Huang et al. 2013). The allelic effects of these QTLs need to be validated as potential sources of alleles for modulating grain shape. In addition, further screening of QTL alleles for grain shape in other cultivars by using advanced mapping populations and fine-scale genetic mapping would provide a more comprehensive elucidation of the genetic control of grain shape in Asian rice cultivars.
QTL alleles for grain length were co-localized with those for grain width in 18 regions (Table 1). Previous studies detected QTLs for either grain length or grain width in a given chromosomal region, but complementation testing of the cloned genes (GW2, GS3, GS5, and GW6a) revealed pleiotropic effects of single genes for grain length or grain width (Li et al. 2011, Mao et al. 2010, Song et al. 2007, 2015). Thus, some of our examples (i.e., qGL3-2 and qGW3-3, qGL9-2 and qGW9-2, and qGL11 and qGW11-2) might result from pleiotropic effects of single genes, whereas others might represent close linkage of independent genes, because the two QTLs were not located in the same marker interval in a single mapping population (i.e., qGL5-3 and qGW5-2, qGL6-4 and qGW6-6, and qGL10 and qGW10-1). Therefore, fine genetic mapping and functional characterization of the genes that control grain shape will be indispensable for uncovering the complexity of the traits and identifying loci that have been mapped in similar regions in different cross combinations.
Previous studies detected 1 to 32 QTLs (8.7 on average) for grain length and grain width per single cross combination (Huang et al. 2013). This number is comparable to that in our analysis using the F2 population; we therefore believe that QTL mapping using such a primary mapping population cannot reliably detect QTLs with small effects, probably owing to noise from the QTLs with large effects (Yamamoto et al. 2009). In addition to the QTLs with large effects, epistasis between pairs of QTLs with small effects potentially involves complicated genetic control of yield-related traits, including grain shape (Lei et al. 2008, Li et al. 1997, Li et al. 2008). Most of the 65 QTLs in the present study were detected under both the Koshihikari and IR64 genetic backgrounds, and some (qGW5-1, qGL8-2, and qGW8) were located in chromosomal regions that harbored a QTL with an epistatic effect in previous studies. Thus, we hypothesize that a larger proportion of the genetic variation than that expected on the basis of previous studies could be explained by an additive model in the cross combination we used. To further clarify non-additive effects of the QTL alleles, the combined effect of QTL alleles should be evaluated in a homogeneous genetic background by using crosses between near-isogenic lines.
The case of qGL2-3 and qGL2-4, two QTLs for grain length that lie about 4 Mb apart on chromosome 2 (Fig. 5), provides an important insight into the genetic control of agronomic traits. The position of qGL2-3 in the F2 population might not have been estimated correctly, possibly owing to the heterogeneous genetic background. Even in the analysis using the BC4F2 populations, linkage of the QTL alleles in repulsion decreased our ability to detect QTLs and resulted in underestimation of the additive effect when the two QTLs are segregating (Fig. 5). Six putative QTLs detected in our analysis using CSSLs were not detected in the analysis using the BC4F2 populations for similar or other reasons (Fig. 4A, 4B, footnote c). The cases in which multiple QTL alleles for a trait are linked in coupling might result in overestimation of the effect of a single QTL in primary mapping studies, as has been reported previously (Ashikari et al. 2005, Fukuoka et al. 2012, Song et al. 2015, Yu et al. 2008). From these observations, both a homogeneous background and the number of recombination events in a mapping population are important factors when dissecting agronomic traits that are under complicated genetic control.
By using fewer than 50 genotypes per population, our analysis using CSSLs efficiently assigned a greater number of putative QTLs for grain length and grain width than our analysis using the F2 population. CSSLs have been used to detect QTLs for other traits such as heading date, yield, and preharvest sprouting resistance (Abe et al. 2013, Ando et al. 2008, Ebitani et al. 2005, Hori et al. 2010, Ishikawa et al. 2005, Takai et al. 2009, 2014). Previous studies and the present study together suggest that CSSLs are useful genetic materials for dissecting complex agronomic traits in rice, but the number of recombinations in these lines determines the resolution of the genetic mapping, as was suggested previously (Ebitani et al. 2005, Xu et al. 2010). Koshihikari is a leading cultivar in temperate areas, and IR64 is a leading cultivar in tropical areas. Reciprocal CSSLs from the cross between these cultivars, which are being released by researchers from the Rice Genome Resource Center at the National Institute of Agrobiological Sciences (http://www.rgrc.dna.affrc.go.jp/index.html), will be useful genetic materials for analyzing various agronomic traits to enhance cultivars in both tropical and temperate areas.
We identified a large number of QTL alleles that will be useful for modulating grain shape in the genetic background of the two improved cultivars, Koshihikari and IR64. These QTL alleles mostly had smaller effects than those that have been cloned so far, but will nonetheless be useful for increasing grain yield, because a drastic increase of grain size usually does not increase grain production (Peng et al. 2008, Takai et al. 2013, Takita 1983). Thus, it is necessary to change the traits related to grain production while retaining a balance between sinks and sources. Some QTL alleles that increase the 1000-grain weight are co-localized with QTLs that reduce the number of grains per panicle (Takai et al. 2014). This example suggests that breeders should choose QTL alleles for grain shape that lack negative associations with other agronomic traits or should remove tightly linked undesirable traits to increase grain production in a practical breeding program.
Supplementary Material
Acknowledgments
We are grateful for the technical assistance of the technical staff of the Rice Applied Genomics Research Unit, and for their management of the rice fields at the National Institute of Agrobiological Sciences. This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics-based Technology for Agricultural Improvement, IVG2003). We thank two editors from ELSS, Inc. (http://elss.co.jp/en/) for editing our manuscript before submission.
Literature Cited
- Abe, T., Nonoue, Y., Ono, N., Omoteno, M., Kuramata, M., Fukuoka, S., Yamamoto, T., Yano, M. and Ishikawa, S. (2013) Detection of QTLs to reduce cadmium content in rice grains using LAC23/Koshihikari chromosome segment substitution lines. Breed. Sci. 63: 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, T., Yamamoto, T., Shimizu, T., Ma, X.F., Shomura, A., Takeuchi, Y., Lin, S.Y. and Yano, M. (2008) Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor. Appl. Genet. 116: 881–890. [DOI] [PubMed] [Google Scholar]
- Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., Angeles, E.R., Qian, Q., Kitano, H. and Matsuoka, M. (2005) Cytokinin oxidase regulates rice grain production. Science 29: 741–745. [DOI] [PubMed] [Google Scholar]
- Bai, X., Luo, L., Yan, W., Kovi, M.R., Zhan, W. and Xing, Y. (2010) Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 11: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basten, C.J., Weir, B.S. and Zeng, Z.B. (2005) QTL cartographer, ver. 1.17. Department of Statistics, North Carolina State University, Raleigh, NC. [Google Scholar]
- Ebana, K., Yonemaru, J., Fukuoka, S., Iwata, H., Kanamori, H., Namiki, N., Nagasaki, N. and Yano, M. (2010) Genetic structure revealed by a whole-genome single-nucleotide polymorphism survey of diverse accessions of cultivated Asian rice (Oryza sativa L.). Breed. Sci. 60: 390–397. [Google Scholar]
- Ebitani, T., Takeuchi, Y., Nonoue, Y., Yamamoto, T., Takeuchi, K. and Yano, M. (2005) Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed. Sci. 55: 65–73. [Google Scholar]
- Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., Li, X. and Zhang, Q. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1171. [DOI] [PubMed] [Google Scholar]
- Fukuoka, S., Mizobuchi, R., Saka, N., Suprun, I., Matsumoto, T., Okuno, K. and Yano, M. (2012) A multiple gene complex on rice chromosome 4 is involved in durable resistance to rice blast. Theor. Appl. Genet. 125: 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L., Ma, L., Jiang, H., Zeng, D., Hu, J., Wu, L., Gao, Z., Zhang, G. and Qian, Q. (2009) Genetic analysis and fine mapping of two genes for grain shape and weight in rice. J. Integr. Plant Biol. 51: 45–51. [DOI] [PubMed] [Google Scholar]
- Hori, K., Sugimoto, K., Nonoue, Y., Ono, N., Matsubara, K., Yamanouchi, U., Abe, A., Takeuchi, Y. and Yano, M. (2010) Detection of quantitative trait loci controlling pre-harvest sprouting resistance by using backcrossed populations of japonica rice cultivars. Theor. Appl. Genet. 120: 1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, R., Jiang, L., Zheng, J., Wang, T., Wang, H., Huang, Y. and Hong, Z. (2013) Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci. 18: 218–226. [DOI] [PubMed] [Google Scholar]
- Ishikawa, S., Ae, N. and Yano, M. (2005) Chromosomal regions with quantitative trait loci controlling cadmium concentration in brown rice (Oryza sativa). New Phytol. 168: 345–350. [DOI] [PubMed] [Google Scholar]
- Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., Kashiwagi, T., Ujiie, K., Shimizu, B., Onishi, A.et al. (2013) Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45: 707–711. [DOI] [PubMed] [Google Scholar]
- Kubo, T., Aida, Y., Nakamura, K., Tunematsu, H., Doi, K. and Yoshimura, A. (2002) Reciprocal chromosome segment substitution series derived from Japonica and Indica cross of rice (Oryza sativa L). Breed. Sci. 52: 319–325. [Google Scholar]
- Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E. and Newberg, L.A. (1987) Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lei, D.Y., Xie, F.M., Xu, J.L. and Chen, L.Y. (2008) QTLs mapping and epistasis analysis for grain shape and chalkiness degree of rice. Chin. J. Rice Sci. 22: 255–260. [Google Scholar]
- Li, J., Thomson, M. and McCouch, S.R. (2004) Fine mapping of a grain-weight quantitative trait locus in the pericentromeric region of rice chromosome 3. Genetics 168: 2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Lu, K., Chen, Z., Mu, T., Hu, Z. and Li, X. (2008) Dominance, overdominance and epistasis condition the heterosis in two heterotic rice hybrids. Genetics 180: 1725–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., Shao, D., Xu, C., Li, X., Xiao, J.et al. (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43: 1266–1269. [DOI] [PubMed] [Google Scholar]
- Li, Z., Pinson, S.R.M., Park, W.D., Paterson, A.H. and Stansel, J.W. (1997) Epistasis for three grain yield components in rice (Oryza sativa L.). Genetics 145: 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, H., Sun, S., Yao, J., Wang, C., Yu, S., Xu, C., Li, X. and Zhang, Q. (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci USA 107: 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch, S.R., Cho, Y.G., Yano, M., Paul, E., Blinstub, M., Morishima, H. and Kinoshita, T. (1997) Report on QTL nomenclature. Rice Genet. Newsl. 14: 11–13. [Google Scholar]
- Murata, K., Iyama, Y., Yamaguchi, T., Ozaki, H., Kidani, Y. and Ebitani, T. (2014) Identification of a novel gene (Apq1) from the indica rice cultivar ‘Habataki’ that improves the quality of grains produced under high temperature stress. Breed. Sci. 64: 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H., DeVerna, J.W., Lanini, B. and Tanksley, S.D. (1990) Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato. Genetics 124: 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, S., Khush, G.S., Virk, P., Tang, Q. and Zoh, Y. (2008) Progress in ideotype breeding to increase rice yield potential. Field Crops Res. 108: 32–38. [Google Scholar]
- Qiu, X., Gong, R., Tan, Y. and Yu, S. (2012) Mapping and characterization of the major quantitative trait locus qSS7 associated with increased length and decreased width of rice seeds. Theor. Appl. Genet. 125: 1717–1726. [DOI] [PubMed] [Google Scholar]
- Shao, G., Wei, X., Chen, M., Tang, S., Luo, J., Jiao, G., Xie, L. and Hu, P. (2012) Allelic variation for a candidate gene for GS7, responsible for grain shape in rice. Theor. Appl. Genet. 125: 1303–1312. [DOI] [PubMed] [Google Scholar]
- Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S. and Yano, M. (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028. [DOI] [PubMed] [Google Scholar]
- Singh, R., Singh, A.K., Sharma, T.R., Singh, A. and Singh, N.K. (2012) Fine mapping of grain length QTLs on chromosomes 1 and 7 in Basmati rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 21: 157–166. [Google Scholar]
- Song, X.J., Huang, W., Shi, M., Zhu, M.Z. and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Song, X.J., Kuroha, T., Ayano, M., Furuta, T., Nagai, K., Komeda, N., Segami, S., Miura, K., Ogawa, D., Kamura, T.et al. (2015) Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. USA 112: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, T., Ohsumi, A., San-oh, Y., Laza, M.R.C., Kondo, M., Yamamoto, T. and Yano, M. (2009) Detection of a quantitative trait locus controlling carbon isotope discrimination and its contribution to stomatal conductance in japonica rice. Theor. Appl. Genet. 118: 1401–1410. [DOI] [PubMed] [Google Scholar]
- Takai, T., Adachi, S., Taguchi-Shiobara, F., Sanoh-Arai, Y., Iwasawa, N., Yoshinaga, S., Hirose, S., Taniguchi, Y., Yamanouchi, U., Wu, J.et al. (2013) A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Sci. Rep. 23: 2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, T., Ikka, T., Kondo, K., Nonoue, Y., Ono, N., Arai-Sanoh, Y., Yoshinaga, S., Nakano, H., Yano, M., Kondo, M.et al. (2014) Genetic mechanisms underlying yield potential in the rice high-yielding cultivar Takanari, based on reciprocal chromosome segment substitution lines. BMC Plant Biol. 14: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, Y., Hori, K., Suzuki, K., Nonoue, Y., Takemoto-Kuno, Y., Maeda, H., Sato, H., Hirabayashi, H., Ohta, H., Ishii, T.et al. (2008) Major QTLs for eating quality of an elite Japanese rice cultivar, Koshihikari, on the short arm of chromosome 3. Breed. Sci. 58: 437–445. [Google Scholar]
- Takita, T. (1983) Breeding of a rice line with extraordinarily large grains as a genetic source for high yielding varieties. JARQ 17: 93–97. [Google Scholar]
- Tanabata, T., Shibaya, T., Hori, K., Ebana, K. and Yano, M. (2012) Smart Grain: High-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 160: 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S.D. and Nelson, J.C. (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 92: 191–203. [DOI] [PubMed] [Google Scholar]
- Wan, X.Y., Wan, J.M., Jiang, L., Wang, J.K., Zhai, H.Q., Weng, J.F., Wang, H.L., Lei, C.L., Wang, J.L., Zhang, X.et al. (2006) QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects. Theor. Appl. Genet. 112: 1258–1270. [DOI] [PubMed] [Google Scholar]
- Weng, J., Gu, S., Wan, X., Gao, H., Guo, T., Su, N., Lei, C., Zhang, X., Cheng, Z., Guo, X.et al. (2008) Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18: 1199–1209. [DOI] [PubMed] [Google Scholar]
- Xie, X., Song, M.H., Jin, F., Ahn, S.N., Suh, J.P., Hwang, H.G. and McCouch, S.R. (2006) Fine mapping of a grain weight quantitative trait locus on rice chromosome 8 using near-isogenic lines derived from a cross between Oryza sativa and Oryza rufipogon. Theor. Appl. Genet. 113: 885–894. [DOI] [PubMed] [Google Scholar]
- Xu, J., Zhao, Q., Du, P., Xu, C., Wang, B., Feng, Q., Liu, Q., Tang, S., Gu, M., Han, B.et al. (2010) Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.). BMC Genomics 11: 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T., Yonemaru, J. and Yano, M. (2009) Towards the understanding of complex traits in rice: Substantially or superficially? DNA Res. 16: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M. and Sasaki, T. (1997) Genetic and molecular dissection of quantitative traits in rice. Plant Mol. Biol. 35: 145–153. [PubMed] [Google Scholar]
- Yonemaru, J., Yamamoto, T., Fukuoka, S., Uga, Y., Hori, K. and Yano, M. (2010) Q-TARO: QTL Annotation Rice Online Database. Rice 3: 194–203. [Google Scholar]
- Youens-Clark, K., Buckler, E., Casstevens, T., Chen, C., Declerck, G., Derwent, P., Dharmawardhana, P., Jaiswal, P., Kersey, P., Karthikeyan, A.S.et al. (2011) Gramene database in 2010: updates and extensions. Nucleic Acids Res. 39: D1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S.W., Yang, C.D., Fan, Y.Y., Zhuang, J.Y. and Li, X.M. (2008) Genetic dissection of a thousand-grain weight quantitative trait locus on rice chromosome 1. Chin. Sci. Bull. 53: 2326–2332. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.