Abstract
Epidemiological studies evaluate multiple exposures, but the extent of multiplicity often remains non-transparent when results are reported. There is extensive debate in the literature on whether multiplicity should be adjusted for in the design, analysis, and reporting of most epidemiological studies, and, if so, how this should be done. The challenges become more acute in an era where the number of exposures that can be studied (the exposome) can be very large. Here, we argue that it can be very insightful to visualize and describe the extent of multiplicity by reporting the number of effective exposures for each category of exposures being assessed, and to describe the distribution of correlation between exposures and/or between exposures and outcomes in epidemiological datasets. The results of new proposed associations can be placed in the context of this background information. An association can be assigned to a percentile of magnitude of effect based on the distribution of effects seen in the field. We offer an example of how such information can be routinely presented in an epidemiological study/dataset using data on 530 exposure and demographic variables classified in 32 categories in the National Health and Nutrition Examination Survey (NHANES). Effects that survive multiplicity considerations and that are large may be prioritized for further scrutiny.
Observational epidemiological studies almost always measure multiple correlated variables in their populations, for example, individual nutrient intake measurements collected through food frequency questionnaires or panels of biomarkers. Furthermore, newer high-throughput measurement of hundreds (to thousands) of non-genetic, environmental variables in individuals can result in an expansion of exposure variables available for epidemiological studies (table 1).1–10 The promise of assessing personal exposomes—the totality of exposure load occurring from birth to death11—includes scaling up the number of environmental exposures measured in individuals to enable a data-driven search for putatively novel exposures associated with disease or other exposures and outcomes of interest through large-scale analyses such as the environment-wide association studies.12–19 One promising technology to scale up the ascertainment of ‘endogenous’ exposures includes metabolomics (table 1) in which hundreds to thousands of small-molecule metabolites are ascertained in human tissue.2,4,20 Existing epidemiological cohorts have begun to ascertain hundreds of variables of these endogenous exposures such as metabolites and lipids5,21,22 while smaller studies have ascertained on the order of thousands of chemical analytes.1 Of course, the raw number of exposure-related variables that can be measured are still far lower than what is seen in current day genome-wide association studies and genome sequencing, which assess millions of genetic variants simultaneously. However, the multiplicity burden on the exposome side is already impressive and is likely to get even greater with the advent of new platforms, including sensors that allow continuous streaming of personalised exposure signals.
Table 1.
Emerging technologies to measure multiple environmental/non-genetic variables
| Measurement platform type | Number of variables | Example references |
|---|---|---|
| Metabolomics (small molecules) | 10–1000s | 1,2,4,5 |
| Transcriptomics/proteomics (mRNA/protein) | 30–50 000 | 3,6 |
| Nutrient questionnaires | 100s | 7 |
| Automated dietary intake sensors | 100s | 8 |
| Physical activity/behaviour sensors (iPhone/smart devices) | 10–100s | 9,10 |
Testing multiple variables for associations with other exposures and outcomes multiply the prospects of making interesting discoveries. At the same time, this multiplicity can lead to more false positives (due to type 1 error).23 There is a need to take this multiplicity into consideration in designing, analysing, interpreting and communicating epidemiological results in a transparent manner.
Transparency about multiplicity goes beyond the issue of whether statistical inferences should account/adjust for multiplicity or not. The debate of whether p values or other statistical inferences should be corrected for multiple comparisons has been a long one. Arguments have been expressed in the past that there is no need to adjust for multiple comparisons in observational epidemiology.24 This has been a dominant view to-date for most applications of traditional epidemiology. These arguments are stronger when explicit, unique hypotheses are tested that have been prespecified to be of primary interest. However, such prespecification is often non-transparent. In the absence of public preregistration25 of protocols and hypotheses, claims of prespecification can even be dubious and may meet with some healthy scepticism. Therefore, other investigators have highlighted the need for more stringency, for example, by using routinely more stringent thresholds and avoiding focus on statistical significance.26 At the other extreme, for agnostic genomic epidemiological studies, adjusting for the entire multiplicity of genomic comparisons has become standard practice.27 Increasingly, epidemiological studies of exposures will be dealing with numerous variables. Being explicit and transparent about the extent of multiplicity may be important for other scientists to understand the background/context against which the result is reported.
Environmental exposure variables are often densely correlated.14,28,29 We argue that the density of the correlations in the variables of interest is also useful to convey transparently in any epidemiological study. New proposed correlations (or other measures of effect size) may need to be interpreted differently depending on what correlations (or other effect sizes) prevail, on average, in the field. For example, suppose that a new correlation is identified between two variables and its absolute magnitude is r=0.2 (and it is highly statistically significant at p<0.0001). This new correlation of r=0.2 may need to be interpreted differently if correlations seen for these type of exposure variables are usually very strong (eg, r>0.5 on average) or null or very small (eg, r<0.05) on average.
We offer here an example of how an epidemiological study could convey this essential background information: how many exposures it has measured and what are the typical correlation patterns between exposures against which new results may be placed to gauge relative importance. We use data from the National Health and Nutrition Examination Survey (NHANES) from 2003 to 2004.30 NHANES has captured different domains/categories of exposures, such as urine/serum biomarkers of nutrients, pesticides, hydrocarbons, infectious agents, as well as self-reported indicators of behaviour, such as smoking, physical activity and nutrient intake, and also socioeconomic and demographic variables (table 2). We defined categories of exposures/variables based on documented chemical relatedness (eg, polychlorinated biphenyls compounds, phenols or hydrocarbons), dietary relevance (eg, serum nutrients or self-reported diet questionnaire) or behaviour relatedness (eg, physical activity, smoking or pharmaceutical drug intake), and assay type ascertained from the NHANES (eg, serum or urine-based mass spectrometry).
Table 2.
Number of variables in each of the 32 exposure, demographics or behaviour categories
| Category (measurement type) | M | Meff | Mdiff | p Value |
|---|---|---|---|---|
| Polychlorinated biphenyls (s) | 38 | 23.79 | 14.21 | 0.002 |
| Dioxins (s) | 7 | 4.90 | 2.10 | 0.01 |
| Acrylamide (s) | 2 | 1.44 | 0.56 | 0.03 |
| Organochlorine pesticides (s) | 13 | 9.95 | 3.05 | 0.005 |
| Polybrominated ethers (s) | 11 | 8.47 | 2.53 | 0.006 |
| Furans (s) | 10 | 7.99 | 2.01 | 0.006 |
| Perchlorate (u) | 2 | 1.66 | 0.34 | 0.03 |
| Smoking behaviour (s/sr) | 14 | 11.65 | 2.35 | 0.004 |
| Phthalates (u) | 13 | 10.85 | 2.15 | 0.005 |
| Food component recall (sr) | 74 | 63.92 | 10.08 | 0.0008 |
| Phytoestrogens (u) | 6 | 5.22 | 0.78 | 0.01 |
| Hydrocarbons (u) | 21 | 18.47 | 2.53 | 0.003 |
| Nutrients (s) | 29 | 26.32 | 2.68 | 0.002 |
| Volatile compounds (s) | 38 | 34.60 | 3.40 | 0.001 |
| Physical fitness (sr*) | 3 | 2.78 | 0.22 | 0.02 |
| Diakyl metabolites (u) | 6 | 5.59 | 0.41 | 0.009 |
| Socioeconomics (SES) (sr) | 9 | 8.49 | 0.51 | 0.006 |
| Demographics (sr) | 8 | 7.55 | 0.45 | 0.007 |
| Polyflourochemicals (s) | 12 | 11.39 | 0.61 | 0.004 |
| Phenol pesticides (u) | 7 | 6.65 | 0.35 | 0.008 |
| Heavy metals (s/u) | 29 | 27.68 | 1.32 | 0.002 |
| Bacterial infection (s/u) | 33 | 32.19 | 0.81 | 0.002 |
| Viral infection (s) | 16 | 15.72 | 0.28 | 0.003 |
| Phenols (u) | 3 | 2.97 | 0.03 | 0.02 |
| Atrazine-like pesticides (u) | 6 | 5.98 | 0.02 | 0.008 |
| Hormone (s) | 2 | 1.99 | 0.01 | 0.03 |
| Pharmaceutical (sr) | 107 | 106.78 | 0.22 | 0.0005 |
| Organophosphate pesticides (u) | 4 | 4.00 | 0.00 | 0.01 |
| Carbamate pesticides (u) | 4 | 4.00 | 0.00 | 0.01 |
| Melamine (u) | 1 | 1.00 | 0.00 | 0.05 |
| Chloroacetanilide pesticides (u) | 1 | 1.00 | 0.00 | 0.05 |
| Pyrethyroid pesticides (u) | 1 | 1.00 | 0.00 | 0.05 |
| Total | 530 | 476.0 | 54.0 | 0.0001 |
M, number of variables in category; Meff, effective number of variables after taking into account correlation; Mdiff, difference between M and Meff; p value, indicative correlation-adjusted Bonferroni p value threshold (0.05/Meff).
Denotes quantitative assessment of physical activity (VO2Max) was also measured.
s, serum; sr, self-report; u, urine.
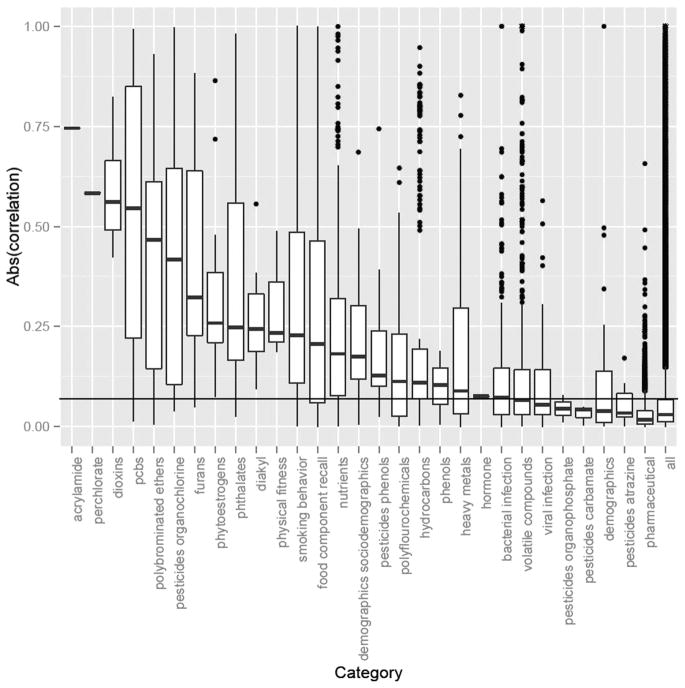
Table 2 shows the number of exposures assessed for each of the 32 categories. Given the substantial correlation between some variables, the number of independent comparisons is somewhat smaller than the number of variables assessed. To correct for this, we have adopted here a method used in genetic association studies to estimate the number of effective variables that are present after taking the between-variable correlation into account.31 For example, 38 polychlorinated biphenyl analytes were assessed, but after taking into account their correlation, this is equivalent to 24 ‘independent’ variables (table 2). This adjustment matters most where the correlations are largest (eg, polychlorinated biphenyls, dioxins, furans) (figure 1). We also show indicatively what p value thresholds would be used, if one wanted to correct for the number of independent variables in each category. If we assume all 530 variables were independent, correcting for 530 comparisons would result in a p value threshold of 0.05/530=0.0001. We do not intend to settle here the debate of whether p values should be avoided, left uncorrected, corrected for the effective number of variables in the category of interest or whether a different method (eg, false discovery rate using the ‘step-down’32 or permutation-based approach33) should be adopted. However, presenting the number of variables and effective variables (M and Meff, respectively) will be useful in helping other scientists and readers to understand the extent of multiplicity at hand behind an epidemiological dataset that has been analysed to obtain some reported results. Further, reporting of such information can be mandated by journal editors just as authorship and participant roles are documented prior to submission of a manuscript.
Figure 1.
Within-category absolute value of correlations (Spearman or biserial). Boxplot illustrates IQR (25–75th percentile) with median in the middle of the box. Only one pair of correlations was estimated for acrylamide and perchlorate exposures. Outliers outside of the 99th percentile are shown in actual points. We estimated significance of correlations through ‘permutation’ testing, randomly shuffling exposures to estimate correlation thresholds under the null distribution of no correlation. The horizontal black line denotes the 95th percentile of all ‘permuted’ correlations. ‘all’: denotes correlation across all pairs of variables available.
Figure 1 shows the magnitude of the absolute correlation coefficients across each category of variables for 29 of the 32 categories that had two or more variables. These correlations are Spearman rank for continuous variables and biserial correlations for binary variables. As documented, most categories had correlated exposures; however, most correlations were not very large (r<0.5 in absolute values). Serum measures of persistent pollutants such as polychlorinated biphenyls, acrylamide, dioxins and organochlorine pesticides had the highest average correlations in their respective categories. Further, dietary measures, such as serum levels of nutrients and self-reported dietary factors, were also densely but modestly correlated with one another, perhaps reflecting the phenomenon of cluster of nutrients that are consumed together. Epidemiological associations may be better understood in the context of other associations in the same field where similar variables are involved. For example, an absolute correlation coefficient of 0.15 between two new nutritional variables is probably not that noteworthy, since the average absolute correlation between two nutritional variables exceeds that value.
The same principle can also be extended to the evaluation of exposures with specific outcomes of interest. For example, in NHANES the median absolute correlation coefficient between the serum nutrients and low-density lipoprotein (LDL)-cholesterol is 0.17. A new proposed correlation between a nutrient and LDL-cholesterol with correlation coefficient less than 0.17 may not be particularly noteworthy, regardless of its level of statistical significance, since 0.17 is around the average absolute correlation that is seen for the average nutrient. Conversely, the median absolute correlation between the nutritional variables and diabetes mellitus (defined as serum blood glucose greater or equal to 126 mg/dL34) is 0.07. Thus, a correlation of 0.17 between a nutrient and diabetes mellitus may be noteworthy since it is greater than two times the size of the typical correlation seen for this field.
One way to place a new proposed association in context is to state what the percentile of its estimated effect is as compared with other associations in the same field using exposure variables and/or outcomes of the same family/type of variables (its ‘relative importance’). In the example given above using correlation coefficients, a correlation of 0.17 with LDL-cholesterol would be at the 50th percentile, while a correlation of 0.17 with diabetes would be at 97th percentile. While we use correlation coefficients for computational convenience, other metrics of effect may be used (eg, standardised effects or regression slopes per unit of exposure) and these metrics are typically easily interchangeable, for example, correlation coefficients may be transformed to an equivalent standardised effect (=0.5ln((1 +r)/(1 − r))). For example, a correlation of 0.8 would be equivalent to a standardised effect of 1.1 (a change in 1 SD of one exposure is associated with a 1.1 unit change of the other exposure or outcome of interest). Correlations of 0.17 and 0.07, respectively, are equivalent to 0.17 and 0.07 units of standardised effects. Standardised effects may also be converted to equivalents of ORs.35 Understanding the relative effect sizes may be important for planning follow-up investigations and eventually (for those correlations that have additional causal support) considering what the health policy may be, if any.
Of course, one cannot stress enough that causality cannot be discerned based alone on the level of statistical significance (no matter whether and how it is corrected or not) or magnitude of correlation (or other effect metric) in observational studies. However, we argue that the extra transparency of presenting the multiplicity inherent in an epidemiological dataset and of placing the effect size against the distribution of typical effects seen in the same field might be insightful and mitigate chances for false positive reporting. Many epidemiological datasets are used in reporting dozens and hundreds of separate papers on diverse associations. A report of the data multiplicity and correlation profile of each dataset can be used to inform all of these papers. This profile can be added in a dataset registration record that describes in public view the overall dataset and study design.
Multiplicity can also be induced by the procedure in which analysts produce findings such as different study designs and even choice of adjustment variables that are used to correct for confounding. Confounding is a major hindrance to assessing causality. In this regard, efforts for causal modelling (eg, via directed acyclic graphs36) exist, but a stumbling block is how the structure of the directed acyclic graph is to be chosen. Often, this choice is achieved through a combination of some a priori knowledge and biological plausibility (or biological speculation) and through examining statistical associations (eg, correlations) between arbitrary variables.37 By considering the correlations between all variables (and the effective number of tests), analysts may prioritise for further consideration adjustments for the strongest correlates with their outcome and exposures of interest. These correlates might have been missed by traditional epidemiological thinking that relies on arbitrarily picking potential adjustment variables. Different teams of epidemiologists may think of adjusting for very different sets of variables, even when the same exposure and outcome are assessed across their studies. This is routinely demonstrated in systematic reviews and meta-analyses of epidemiological studies where the set of adjusting factors is almost always markedly different across the combined investigations. Part of this divergence may be due to the fact that not all studies measure the very same confounders. However, even for routinely collected variables, clearly there is large subjectivity among investigators on how they use them. This creates a situation where almost any result can be achieved that would conform to the original expectations of the traditional epidemiologist, provided a suitable set of adjustments and a self-justifying causal model is adopted. This flexibility can transform epidemiology into a champion field for subjectivity, allegiance bias and confirmation bias.
Some traditional epidemiologists who have heard us suggest agnostic approaches for assessments of the exposome take a defensive stance claiming that most epidemiology is (and should be) testing specific focused a priori hypotheses. For example, a reviewer of the original submission of this paper felt that ‘it probably would be extremely complicated and finally useless to produce the same approach with a study such as European Prospective Investigation into Cancer and Nutrition (EPIC). Many environmental studies do have solid a priori hypotheses’. However, a perusal of PubMed and of the EPIC website shows that EPIC (a classic paradigm of a cohort that claims to have prior hypotheses) has already published over a thousand papers and the same applies to other leading cohorts such as the Nurses’ Health Study. While each paper usually does test only a few hypotheses of associations, cumulatively this cohort-specific publication corpus results in investigating several thousands of hypotheses in the very same single cohort dataset. We think it is better to adapt a systematic all-inclusive approach to the data rather than try to separate arbitrarily and one-at-a-time the thousands of ‘solid a priori’ hypotheses from the thousands that no one has happened to pick—until now.
Finally, a caveat is also needed about the meaning and implications of large effects. Large effects are considered to have increased credibility in evidence appraisal schemes such as GRADE.38 Nevertheless, even a very large effect at the top percentile of a field may sometimes reflect an error (‘too good to be true’) or an inflated effect (due to the winner’s curse).39 Thus, it would require careful examination and replication in additional epidemiological studies. Effects that remain at the top percentiles across multiple studies may be worth prioritising for further corroboration with other types of biological or experimental evidence. One would need to consider also whether this variable that has the top effect is not strongly correlated with other variables or has strong correlations with many other variables.14 Finally, this approach does not negate also the possibility that many genuine effects, including those that reflect causal relationships, may be small or even tiny.40 Regardless, deciding what is large, small or tiny, and what is significant or not may benefit from knowing the profile of the relevant correlation matrix of studied exposures, its inherent multiplicity and the size of average effects that circulate in the field.
Acknowledgments
Funding Chirag J Patel is supported by a NIH National Institute of Environmental Health Sciences (K99 ES023504) and a PhRMA Foundation informatics fellowship. The Meta-Research Innovation Center at Stanford is supported by a grant by the Laura and John Arnold Foundation.
Footnotes
Contributors CJP and JPAI wrote the manuscript.
Provenance and peer review Commissioned; externally peer reviewed.
References
- 1.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappaport SM, Barupal DK, Wishart D, et al. The blood exposome and its role in discovering causes of disease. Environ Health Perspect. 2014 doi: 10.1289/ehp.1308015. http://dx.doi.org/10.1289/ehp.1308015. [DOI] [PMC free article] [PubMed]
- 3.Idaghdour Y, Czika W, Shianna KV, et al. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat Genet. 2010;42:62–7. doi: 10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athersuch TJ. The role of metabolomics in characterizing the human exposome. Bioanalysis. 2012;4:2207–12. doi: 10.4155/bio.12.211. [DOI] [PubMed] [Google Scholar]
- 5.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spira A, Beane J, Shah V, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA. 2004;101:10143–8. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willett WC, Sampson L, Stampfer MJ, et al. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol. 1985;122:51–65. doi: 10.1093/oxfordjournals.aje.a114086. [DOI] [PubMed] [Google Scholar]
- 8.Sun M, Fernstrom JD, Jia W, et al. A wearable electronic system for objective dietary assessment. J Am Diet Assoc. 2010;110:45–7. doi: 10.1016/j.jada.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerr J, Duncan S, Schipperijn J. Using global positioning systems in health research: a practical approach to data collection and processing. Am J Prev Med. 2011;41:532–40. doi: 10.1016/j.amepre.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 10.Madan A, Cebrian ME, Lazer D, et al. Social Sensing to Model Epidemiological Behavior Change. Proceedings of ACM Ubicomp; 2010. [Google Scholar]
- 11.Rappaport SM, Smith MT. Environment and disease risks. Science. 2010;330:460–61. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS ONE. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel CJ, Cullen MR, Ioannidis JP, et al. Systematic evaluation of environmental factors: persistent pollutants and nutrients correlated with serum lipid levels. Int J Epidemiol. 2012;41:828–43. doi: 10.1093/ije/dys003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel CJ, Ioannidis JPA. Studying the elusive environment in large-scale. J Am Med Assoc. 2014 doi: 10.1001/jama.2014.4129. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel CJ, Rehkopf DH, Leppert JT, et al. Systematic evaluation of environmental and behavioural factors associated with all-cause mortality in the United States National Health and Nutrition Examination Survey. Int J Epidemiol. 2013;42:1795–810. doi: 10.1093/ije/dyt208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel CJ, Yang T, Hu Z, et al. Investigation of maternal environmental exposures in association with self-reported preterm birth. Reprod Toxicol. 2013;45C:1–7. doi: 10.1016/j.reprotox.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzoulaki I, Patel CJ, Okamura T, et al. A nutrient-wide association study on blood pressure. Circulation. 2012;126:2456–64. doi: 10.1161/CIRCULATIONAHA.112.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall MA, Dudek SM, Goodloe R, et al. Environment-wide association study (EWAS) for type 2 diabetes in the Marshfield Personalized Medicine Research Project Biobank. Pac Symp Biocomput. 2014:200–11. [PMC free article] [PubMed] [Google Scholar]
- 19.Lind PM, Riserus U, Salihovic S, et al. An environmental wide association study (EWAS) approach to the metabolic syndrome. Environ Int. 2013;55:1–8. doi: 10.1016/j.envint.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 20.Rappaport SM. Biomarkers intersect with the exposome. Biomarkers. 2012;17:483–9. doi: 10.3109/1354750X.2012.691553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee EP, Cheng S, Larson MG, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121:1402–11. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Townsend MK, Clish CB, Kraft P, et al. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin Chem. 2013;59:1657–67. doi: 10.1373/clinchem.2012.199133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ioannidis JPA. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–6. [PubMed] [Google Scholar]
- 25.Dal-Re R, Ioannidis JP, Bracken MB, et al. Making prospective registration of observational research a reality. Sci Transl Med. 2014;6:224cm1. doi: 10.1126/scitranslmed.3007513. [DOI] [PubMed] [Google Scholar]
- 26.Sterne JA, Davey Smith G. Sifting the evidence-what’s wrong with significance tests? BMJ. 2001;322:226–31. doi: 10.1136/bmj.322.7280.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson TA, Manolio TA. How to interpret a genome-wide association study. J Am Med Assoc. 2008;299:1335–44. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 28.Ioannidis J, Loy EY, Poulton R, et al. Researching genetic versus nongenetic determinants of disease: a comparison and proposed unification. Sci Transl Med. 2009;1:8. doi: 10.1126/scitranslmed.3000247. [DOI] [PubMed] [Google Scholar]
- 29.Smith GD, Lawlor DA, Harbord R, et al. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;4:e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Centers for Disease Control and Prevention (CDC), National Center for Health Statistics (NCHS) National health and nutrition examination survey data. Hyattsville, MD: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; 2003–2004. [Google Scholar]
- 31.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 33.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2014;37(Suppl 1):S81–90. doi: 10.2337/dc14-S081. [DOI] [PubMed] [Google Scholar]
- 35.Chinn S. A simple method for converting an odds ratio to effect size for use in meta-analysis. Stat Med. 2000;19:3127–31. doi: 10.1002/1097-0258(20001130)19:22<3127::aid-sim784>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 36.Greenland S, Pearl J, Robins JM. Causal diagrams for epidemiologic research. Epidemiology. 1999;10:37–48. [PubMed] [Google Scholar]
- 37.Hernan MA, Hernandez-Diaz S, Werler MM, et al. Causal knowledge as a prerequisite for confounding evaluation: an application to birth defects epidemiology. Am J Epidemiol. 2002;155:176–84. doi: 10.1093/aje/155.2.176. [DOI] [PubMed] [Google Scholar]
- 38.Guyatt GH, Oxman AD, Vist GE, et al. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ. 2008;336:924–6. doi: 10.1136/bmj.39489.470347.AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ioannidis JPA. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–8. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 40.Siontis GC, Ioannidis JP. Risk factors and interventions with statistically significant tiny effects. Int J Epidemiol. 2011;40:1292–307. doi: 10.1093/ije/dyr099. [DOI] [PubMed] [Google Scholar]