Abstract
The large-scale organization of the brain has features of complex networks that can be quantified using network measures from graph theory. However, many network measures were designed to be calculated on binary graphs, whereas functional brain organization is typically inferred from a continuous measure of correlations in temporal signal between brain regions. Thresholding is a necessary step to use binary graphs derived from functional connectivity data. However, there is no current consensus on what threshold to use, and network measures and group contrasts may be unstable across thresholds. Nevertheless, whole-brain network analyses are being applied widely with findings typically reported at an arbitrary threshold or range of thresholds. This study sought to evaluate the stability of network measures across thresholds in a large resting state functional connectivity dataset. Network measures were evaluated across absolute (correlation-based) and proportional (sparsity-based) thresholds, and compared between sex and age groups. Overall, network measures were found to be unstable across absolute thresholds. For example, the direction of group differences in a given network measure may change depending on the threshold. Network measures were found to be more stable across proportional thresholds. These results demonstrate that caution should be used when applying thresholds to functional connectivity data and when interpreting results from binary graph models.
Keywords: network analysis, graph theory, functional connectivity, resting state, threshold
Introduction
The human brain is a large-scale system of functionally connected brain regions. This system can be modeled as a network, or graph, by dividing the brain into a set of regions, or “nodes,” and quantifying the strength of the connections between nodes, or “edges,” as the temporal correlation in their patterns of activity. Network analysis, a part of graph theory, provides a set of summary statistics that can be used to describe complex brain networks using a reduced number of observations that can be meaningfully compared between groups and/or related to behavior (Rubinov & Sporns, 2010). Recent studies have used this approach to characterize network properties as they relate to sex (Tian et al., 2011), age (Meunier et al., 2009), and cognition (Dosenbach et al., 2007), as well as in conditions such as addiction (Chanraud et al., 2011), Alzheimer’s disease (Supekar et al., 2008), schizophrenia (Alexander-Bloch et al., 2013; Bassett et al., 2008), and others (Bassett and Bullmore, 2006).
Network analyses of functional connectivity data are commonly based on the blood oxygen level-dependent (BOLD) signal in functional magnetic resonance imaging (fMRI), but can also be derived from electroencephalography (EEG) or magnetoencephalography (MEG). For example, network analysis of fMRI data was used to show that more efficient global information processing (characteristic path length) in resting state was related to high intelligence quotient (IQ; van den Heuvel et al., 2009b). Another study used network analysis of MEG data to show that functional integration (characteristic path length) and functional segregation (clustering coefficient) were decreased in Alzheimer’s disease (Stam et al., 2009).
However, a central challenge of applying network analysis to functional connectivity data is that many network measures were designed to be calculated on binary graphs in which connections are either present or not, whereas temporal correlations in fMRI signal are continuous from −1 to 1. The typical approach is to: (1) define n non-overlapping nodes across the brain using an anatomical atlas or a functional parcellation method where nodes have voxels with similar time courses; (2) estimate the network by computing the entries of the n-by-n matrix representing the functional connections between node pairs, either by linear association such as correlation or by some other nonlinear measure such as mutual information; and (3) apply a threshold to produce an n-by-n binary adjacency matrix representing the network edges and to remove weak connections (Bullmore and Sporns, 2009; Simpson et al., 2013). Thus a threshold is commonly applied to construct a binary graph from functional connectivity data.
The use of thresholded binary graphs is attractive because it facilitates the calculation of many network measures and reduces the computational burden of analyzing the graph. An alternative approach is to use weighted graphs that are not binary but instead allow edges to carry some sort of continuous weight value. With weighted graphs, all edges remain in the graph and any node is connected to every other node. However, some have argued that weighted graphs are not reasonable biological structures because brain regions have sparse anatomical connections only to other specific brain regions (Sporns, 2011). Weighted graphs are also less computationally efficient, especially in the analysis of large-scale networks such as voxel-based functional connectivity networks (Telesford et al., 2011); the overwhelming number of connections makes it difficult to extract meaningful information (Serrano et al., 2009).
To define binary graphs, the applied threshold is typically absolute (correlation-based) or sparsity-based (proportional). Each approach has advantages and disadvantages. Absolute thresholds set a value for the correlation coefficient between node pairs, above which they are considered connected and below which they are not. Proportional thresholds utilize a set percentage of the strongest connections (edges), such as the top 10% of correlation values in the network. For group comparisons, using a proportional threshold ensures that the networks in each group have the same number of nodes, or network size, and the same number of edges, or global degree. This allows for more meaningful comparisons of other network measures that rely on degree. However, proportional thresholds do not take into account absolute differences in correlation values, therefore information about overall group differences may be lost. Absolute thresholds retain this information, but may result in networks with different size or degree, or in a network that is connected in one group but disconnected in the other group. While a difference in node-connectedness between groups may be informative, it confounds the comparison of graph measures that vary significantly with degree (Alexander-Bloch et al., 2010). Moreover, absolute thresholds may be too large for low-average connectivity networks or too small for high-average connectivity networks, thus eliminating strong and significant connections or overemphasizing weak connections (van Wijk et al., 2010).
There is also no consensus in the literature as to what specific threshold should be used. A large range of absolute thresholds have been applied, from a correlation coefficient of r=0.1 (Buckner et al., 2009) to r=0.8 (Tomasi and Volkow, 2010). Likewise a range of proportional thresholds have been reported, from 5–40% (e.g., Fornito et al., 2010). In attempts to show that results are not sensitive to the choice of threshold, findings are often reported across a narrow range of thresholds (e.g., Cole et al., 2013, top 2–10%; van den Heuvel et al., 2009a, r=0.3–0.5). However, this can lead to incomplete results or even misleading results if network properties are unstable across a larger range of thresholds, for example if there is a reversal of group differences in a network measure across thresholds (e.g., Scheinost et al., 2012). Lastly, even if a canonical threshold was determined and agreed upon, different preprocessing decisions such as whether to use global signal regression (GSR) may shift the distribution of correlations (Murphy et al., 2009), leading to binary graphs that are not comparable across studies.
Therefore, this study sought to characterize the (in)stability of network measures across thresholds. A large resting-state fMRI dataset was used to measure network properties across the full range of absolute and proportional thresholds. In addition, the effects of GSR on network measures across thresholds were tested in order to highlight how preprocessing decisions can influence the (in)stability of network measures.
Methods
Participants
One hundred right-handed individuals participated in the study. All participants provided written informed consent in accordance with the Yale Human Investigations Committee at the Yale University School of Medicine. The analyses included 50 males (age 35 ± 10 years) and 50 females (age 34 ± 12 years). 99 of these were from a prior dataset of 103 subjects (Scheinost et al., 2015), 4 of whom were not included because they did not complete all 8 runs, and 1 additional participant was scanned in order to have equal sex groups. A subset of the current sample was grouped by age as younger participants (age range 18–25 years, mean age 22 ± 3 years) and older participants (age range 44–66 years, mean age 51 ± 6 years). Age groups were selected as the youngest and oldest participants matched by gender, and age groups were selected to have participants matched by age and gender. Younger participants included 10 males (age range 18–25 years, mean age 23 ± 3 years) and 10 females (age range 18–25 years, mean age 22 ± 2 years). Older participants included 10 males (age range 44–66 years, mean age 51 ± 6 years) and 10 females (age range 44–63 years, mean age 51 ± 6 years).
Imaging Parameters
Data were acquired on two identical Siemens 3T Tim Trio MRI scanners, including a localizer, followed by a low-resolution sagittal scan for slice alignment and a T1-weighted axialoblique scan using a conventional spin-echo imaging sequence parallel to the AC-PC (25 slices, TR = 420 ms, TE = 11 ms; bandwidth = 130 Hz/pixel, flip angle = 90°, slice thickness = 6 mm; FOV = 200×200 mm, matrix = 256×256). Resting-state functional data was then obtained at the same slice locations using a T2*-weighted gradient-recalled single shot echo-planar imaging sequence (TR = 1550 ms, TE = 30 ms, flip angle = 80°, FOV = 220×220 mm, matrix = 64×64, slice thickness = 6 mm). Participants were instructed to rest with their eyes open, not to think of anything in particular, and not to fall asleep. There were 8 functional runs of 240 volumes each. The first 6 volumes were discarded to ensure the magnetization had reached steady state. Following the functional runs a high-resolution anatomical scan was obtained using a magnetization-prepared rapid gradient-echo (MPRAGE) imaging sequence (TR = 2530 ms, TE = 2.77 ms, TI = 1100 ms, flip angle = 7°, resolution = 1×1×1 mm).
Connectivity Preprocessing
Images were corrected for slice timing with sinc interpolation and realigned for motion correction using SPM5 (http://www.fil.ion.ucl.ac.uk/spm/software/spm5). In order to preserve the boundaries of functional nodes, no spatial smoothing was applied. All further preprocessing steps used BioImage Suite (www.bioimagesuite.org; Joshi et al., 2011). Several covariates of no interest were regressed from the data including linear and quadratic drift, six rigid-body motion parameters, mean white matter signal, and mean cerebrospinal fluid (CSF) signal. To test the effects of global signal regression (GSR), in an identical dataset the mean overall global signal was regressed. All images were temporally smoothed using a zero mean unit Gaussian filter with an approximate cutoff frequency of 0.12 Hz.
Network Construction
First, 278 nodes were defined by warping the functional brain atlas (http://www.nitrc.org/frs/download.php/5785/shenetal_neuroimage2013_funcatlas.zip; Shen et al., 2010; Shen et al., 2013) into single subject space via a concatenation of linear and nonlinear registrations: The functional images were linearly registered to the T1 axial-oblique (2D anatomical) images. The 2D anatomical images were linearly registered to the MPRAGE (3D anatomical) images. The 3D anatomical images were non-linearly registered to the template brain. All transformations were calculated and combined into a single transform and inverted, in order to reduce interpolation error by warping the atlas to an individual brain with only one transformation. All transformations were estimated using the intensity-based registration algorithms in BioImage Suite.
For each participant, pairwise correlation coefficients were computed for the time courses of each pair of nodes with data from all runs. To test the stability of network measures across runs, pairwise correlation coefficients were computed for the time courses of each pair of nodes with data from each of the 8 runs. The correlation values were normalized to z-scores using the Fisher transformation, resulting in a 278×278 connectivity matrix. Because the matrices are Fisher-transformed correlations, no matrices have zero surviving edges. The connectivity matrices were then converted into a binary adjacency matrix by thresholding using Matlab (www.mathworks.org). Negative correlations were not included in the calculation of binary networks. An absolute threshold (τr) was applied from τr=0–1 at 0.01 intervals. Using a Matlab function from Brain Connectivity Toolbox (https://sites.google.com/site/bctnet; Rubinov and Sporns, 2010), a proportional threshold (τp) was applied from τp=0.01–0.99 at 0.01 intervals.
Comparing Network Measures
Network properties were calculated using Matlab functions from Brain Connectivity Toolbox, and included the commonly reported network-level measures: characteristic path length, global efficiency, transitivity, and modularity; and node-level measures: clustering coefficient, local efficiency, degree, betweenness centrality, and participation coefficient. Definitions are provided in Table 1 (also see: Rubinov and Sporns, 2010). For network measures based on distance, such as characteristic path length, disconnected nodes are removed from the calculation. For network measures based on the adjacency matrix, such as degree, disconnected nodes are assigned a value of 0 (Rubinov and Sporns, 2010).
Table 1.
Network measures (Bassett and Bullmore, 2006; Rubinov and Sporns, 2010).
| Network-level measures |
|---|
| Characteristic path length (L) is the average shortest path length (i.e., the minimal number of edges that form a direct connection between two nodes) between all pairs of nodes in the network and is a measure of functional integration. A short L indicates a more compact network and more efficient global information processing. |
| Global efficiency (Eglob) is the average inverse shortest path length between all pairs of nodes in the network and is a measure of functional integration, and represents the functional efficiency of brain networks for information transmission between multiple parallel paths. |
| Transitivity (T) is the ratio of triangles to triplets in the network, and was a measure of clustering or functional segregation. |
| Modularity (M) quantifies how well the network can be subdivided into non-overlapping groups of nodes or modules and is a measure of functional segregation. |
| Node-level measures |
| Clustering coefficient (C) is a measure of the number of edges between a node’s nearest neighbors or the fraction of triangles around a node, and is a measure of functional segregation. High C represents clustered connectivity at the node. |
| Local efficiency (Eloc) is the global efficiency computed on node neighborhoods, is related to C, and is a measure of functional segregation. |
| Degree (d) is the number of edges connected to a node, and provides information related to the centrality of a node by determining nodes with a large number of connections. |
| Betweenness centrality (CB) is the fraction of all the shortest paths in the network that contain a given node. Nodes with high CB participate in a large number of shortest paths and may connect distinct parts of the network. |
| Participation coefficient (P) is a measure of the inter-modular connections of nodes and indicates centrality. |
To visualize the stability of network measures, a 'survival curve' was plotted to describe how the network measure changes with connection threshold, i.e., survival as a function of threshold. The network measure value can be estimated as a single point on the survival curve. Therefore, the shape of this curve can be used to provide a description of network measure stability across thresholds, for example a monotonic curve would indicate stability, whereas a U-shape would indicate instability. Survival curves are displayed for a range of thresholds from less to more conservative: 0–1 for absolute; 0.99–.01 for proportional thresholds. To test network measure stability across thresholds, independent t-tests were used to compare measures between sex or age groups at each threshold. Reversals in the direction of significant group differences across thresholds (‘sign reversals’) were also used as an index of instability. In addition, correlations between each measure and age across all 100 participants were computed. To test network measure stability across runs, repeated measures analyses of variance were used to test for an effect of run at each threshold. All analyses were performed on graphs derived with and without GSR. Statistical analyses were performed using SPSS 21 (http://www-01.ibm.com/software/analytics/spss) or Matlab, and a significance level of p<.05. All reported findings are significant at this level (specific thresholds are reported in Table 2–Table 3). Several example nodes are described.
Table 2.
Summary of significant group differences across thresholds.
| Absolute thresholds | Proportional thresholds | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Metric | No GSR |
τr | GSR | τr | No GSR |
τp | GSR | τp | ||
| Sex group differences | L | F | 0.34–0.82 | F | 0.04–0.09, 0.24–0.57 | M | 0.28–0.38 | M | 0.03, 0.05–0.38 | |
| M | 0.92–1 | M | 0.73–0.75, 0.82–0.88, 0.94–0.98 | |||||||
| Eglob | M | 0.37–0.98 | M | 0.03–0.95, 0.97 | F | 0.04–0.27 | ||||
| T | M | 0.21–0.42 | M | 0–0.34 | M | 0.04–0.99 | ||||
| F | 0.64–1 | F | 0.63–0.89 | |||||||
| M | M | 0–0.02, 0.92–0.93, 0.96–0.97 | M | 0.06–0.99 | ||||||
| F | 0.41, 0.66, 0.69 | |||||||||
| Node | ||||||||||
| 15 | C | M | 0.08, 0.1–0.29, 0.31–0.33 | M | 0.02–0.25 | M | 0.15, 0.23–0.24, 0.29–0.5 | M | 0.01, 0.08–0.39, 0.41 | |
| F | 0.59–0.75, 0.77, 0.79, 0.9 | F | 0.57–0.58, 0.61–0.65 | |||||||
| 272 | Eloc | M | 0.03–0.07, 0.1–0.24 | M | 0–0.17 | M | 0.71–0.82, 0.86, 0.88–0.93 | M | 0.17–0.99 | |
| F | 0.71–0.73 | F | 0.63 | |||||||
| 249 | d | M | 0.05–0.25 | F | 0.01–0.05 | F | 0.01–0.05 | |||
| F | 0.49–0.71 | M | 0.3–0.36 | |||||||
| 104 | CB | F | 0–0.70, 0.74–0.75, 0.81 | F | 0.02–0.16, 0.18, 0.21–0.29 | F | 0.03–0.05, 0.07–0.99 | M | 0.01 | |
| M | 0.96, 0.98 | M | 0.64, 0.66–0.75, 0.81–0.89 | F | 0.11, 0.13, 0.28–0.39 | |||||
| 20 | P | F | 0.07–0.08, 0.17, 0.19- 0.25, 0.4, 0.53–0.54 |
F | 0.18, 0.21–0.37, 0.39, 0.41–0.45, 0.48 |
M | 0.02 | M | 0.01 | |
| M | 0.67, 0.69, 0.93 | M | 0.71, 0.73, 0.77, 0.8, 0.82–0.83 | F | 0.09–0.1, 0.16, 0.2–0.23, 0.25, 0.27–0.36, 0.38–0.42, 0.44- 0.48, 0.54–0.71, 0.77–0.81 |
F | 0.06–0.18, 0.28, 0.3, 0.32–0.33 |
|||
| Age group differences | L | younger | 0.69, 0.71–0.72, 0.75–0.76, 0.89, 0.91–0.92 |
|||||||
| Eglob | ||||||||||
| T | ||||||||||
| M | younger | 0.98 | younger | 0.52, 0.54, 0.68, 0.70, 0.76, 0.83, 0.88 |
younger | 0.03–0.09, 0.12 | younger | 0.05–0.06 | ||
| Node | ||||||||||
| 174 | C | older | 0.29 | older | 0.04–0.29, 0.31 | older | 0.28–0.39, 0.41, 0.43, 0.45- 0.46, 0.48–0.49, 0.51–0.53, 0.57–0.58 |
older | 0.16, 0.18–0.39 | |
| younger | 0.76–0.79 | younger | 0.63–0.69 | |||||||
| 128 | Eloc | older | 0.33–0.34, 0.4 | younger | 0.64, 0.66–0.69 | older | 0.28–0.31 | |||
| younger | 0.66 | |||||||||
| 199 | d | younger | 0–0.11 | older | 0.23–0.28, 0.34–0.37, 0.39–0.46 | older | 0.04–0.05, 0.14 | older | 0.04–0.08 | |
| older | 0.55–0.65 | younger | 0.63–0.99 | |||||||
| 26 | CB | older | 0.16–0.25, 0.36 | older | 0.1 | older | 0.17, 0.19–0.22, 0.34–0.57 | younger | 0.03 | |
| younger | 0.75–0.76 | younger | 0.61 | |||||||
| 62 | P | younger | 0.01–0.1, 0.19–0.32 | younger | 0–0.07 | younger | 0.01, 0.28–0.37, 0.39–0.41, 0.52–0.53, 0.75–0.76, 0.78–0.8, 0.82–0.83, 0.85, 0.88–0.89 |
younger | 0.25, 0.32, 0.35–0.99 | |
| older | 0.77, 0.93 | |||||||||
L – characteristic path length; Eglob – global efficiency; T – transitivity, M – modularity, C – clustering coefficient, Eloc – local efficiency, d – degree, CB – betweenness centrality, P – participation coefficient; τr – absolute threshold, τp – proportional threshold; all results significant at p<.05.
Table 3.
Summary of significant effects of run across thresholds.
| Effect of run | Absolute thresholds (τr) | Proportional thresholds (τp) | |||
| Metric | No GSR | GSR | No GSR | GSR | |
| L | 0.37–0.56 | 0.16–0.62, 0.93 | 0.06–0.53, 0.55, 0.59, 0.64, 0.82–0.92 | 0.02–0.03 | |
| Eglob | 0.54–0.86, 0.97–0.98 | 0.16–0.78 | 0.03–0.37, 0.82–0.91 | ||
| T | 0–0.47, 0.91–1 | 0.14–0.99 | |||
| M | 0.65, 0.69, 0.71 | 0.56, 0.71, 0.76, 0.78, 0.85 | 0.05 | ||
L – characteristic path length; Eglob – global efficiency; T – transitivity, M – modularity; all results significant at p<.05.
Results
Network-Level Measures
Characteristic path length (L)
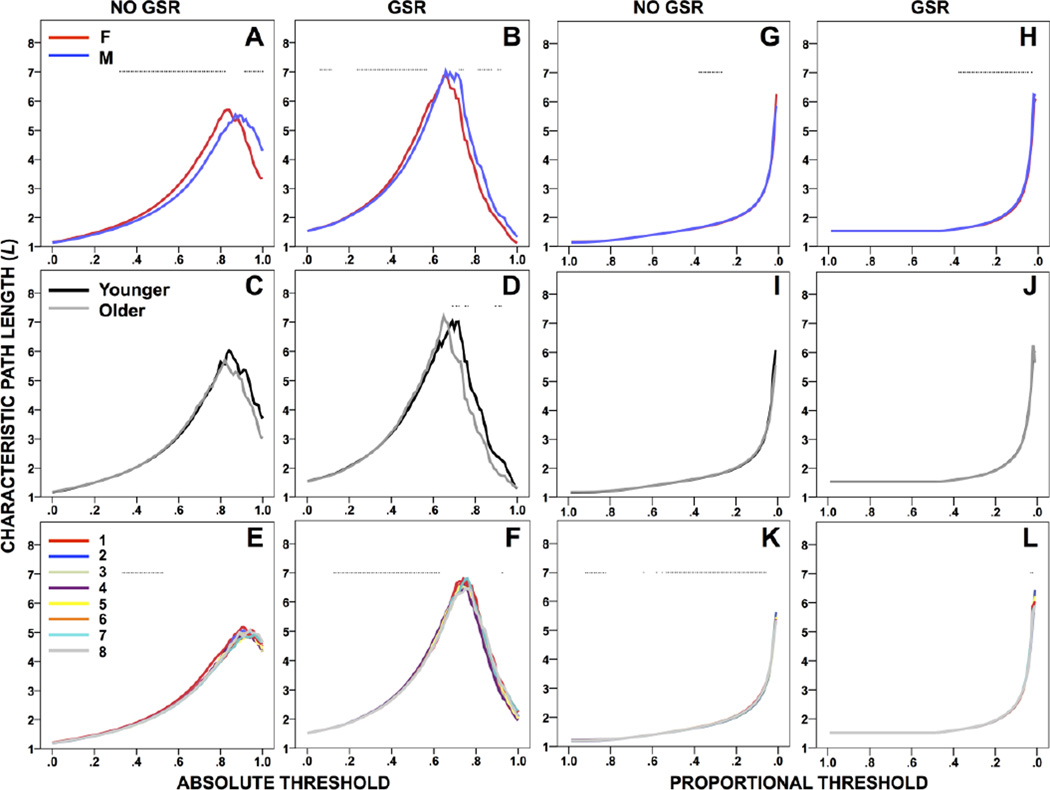
Absolute threshold
The survival curve for L has an inverted V-shape without GSR, with the maximum shifted to a lower threshold with GSR (Figure 1A–F). L was longer for females or males (depending on threshold) with and without GSR (Figure 1A–B). L did not differ by age without GSR; with GSR, L was longer for younger participants and was negatively correlated with age (Figure 1C–D, S1). The shape of the survival curve for L was consistent across runs, though an effect of run was found with and without GSR (Figure 1E–F).
Figure 1.
Characteristic path length (L) across absolute (A–F) and proportional (G–L) thresholds, compared between sex groups (top), age groups (middle), and across runs (bottom), without and with GSR. *p<.05 (Table 2).
Proportional threshold
The survival curve for L increases smoothly without GSR, or is flat and then increases smoothly with GSR (Figure 1G–L). L was longer for males with and without GSR (Figure 1G–H). L did not differ between age groups without GSR, yet was negatively or positively correlated with age; with GSR, L did not differ between age groups, yet was negatively correlated with age (Figure 1I–J, S1). The shape of the survival curve for L was consistent across runs, though an effect of run was found with and without GSR (Figure 1K–L).
Global efficiency (Eglob)
Absolute threshold
The survival curve for Eglob is monotonic and decreases without GSR, with the minimum shifted to a lower threshold with GSR (Figure S2A–F). Eglob was higher for males with and without GSR (Figure S2A–B). Eglob did not differ by age with or without GSR (Figure S2C–D, S1). The shape of the survival curve for Eglob was consistent across runs, though an effect of run was found with and without GSR (Figure S2E–F).
Proportional threshold
The survival curve for Eglob is monotonic and decreases without GSR, or is flat and then decreases with GSR (Figure S2G–L). Eglob did not differ between sex groups without GSR; with GSR, Eglob was higher for females (Figure S2G–H). Eglob did not differ between age groups without GSR, yet was negatively correlated with age; with GSR, Eglob did not differ between age groups, yet was positively correlated with age (Figure S2I–J, S1). The shape of the survival curve for Eglob was consistent across runs, though an effect of run was found without GSR (Figure S2K–L).
Transitivity (T)
Absolute threshold
The survival curve for T is U-shaped without GSR, with the minimum shifted to a lower threshold and J-shaped with GSR (Figure S3A–F). T was higher for males or females with and without GSR (Figure S3A–B). T did not differ between age groups without GSR, yet was positively correlated with age; with GSR, T did not differ by age (Figure S3C–D, S1). The shape of the survival curve for T was consistent across runs, though an effect of run was found without GSR (Figure S3E–F).
Proportional threshold
The survival curve for T is monotonic and decreases without GSR, or is flat and then decreases with GSR (Figure S3G–L). T did not differ between sex groups without GSR; with GSR, T was higher for males (Figure S3G–H). T did not differ between age groups without GSR, yet was positively correlated with age; with GSR, T did not differ by age (Figure S3I–J, S1). The shape of the survival curve for T was consistent across runs, though an effect of run was found without GSR (Figure S3K–L).
Modularity (M)
Absolute threshold
The survival curve for M is S-shaped without GSR, with the maximum shifted to a lower threshold and inverted J-shaped with GSR (Figure S4A–F). M did not differ between sex groups without GSR; with GSR, M was higher for males or females (Figure S4A–B). M was higher for younger participants without GSR and negatively correlated with age; with GSR, M was higher for younger participants and negatively correlated with age (Figure S4C–D, S1). The shape of the survival curve for M was consistent across runs, though an effect of run was found with and without GSR (Figure S4E–F).
Proportional threshold
The survival curve for M increases smoothly to a final sharp peak without GSR, or is flat and then increases to a sharp peak with GSR (Figure S4G–L). M did not differ between sex groups without GSR; with GSR, M was higher for males (Figure S4G–H). M was higher for younger participants without GSR and negatively correlated with age; with GSR, M was higher for younger participants and negatively correlated with age (Figure S4I–J, S1). The shape of the survival curve for M was consistent across runs, though an effect of run was found with GSR (Figure S4K–L).
Node-Level Measures
Clustering coefficient (C)
Absolute threshold
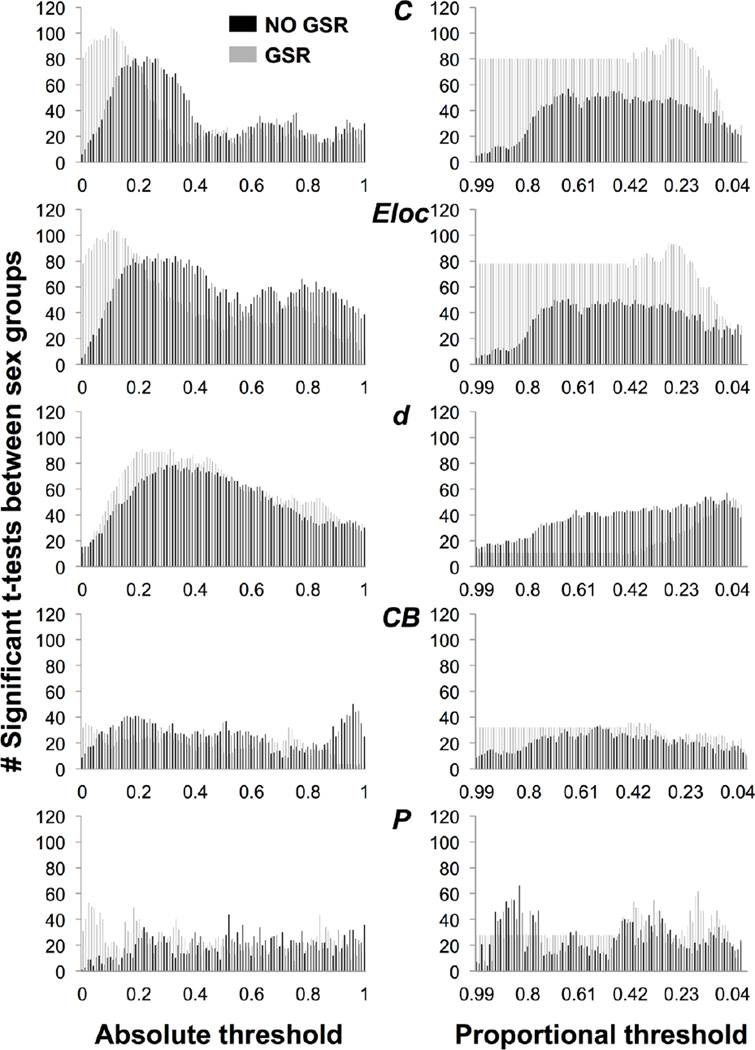
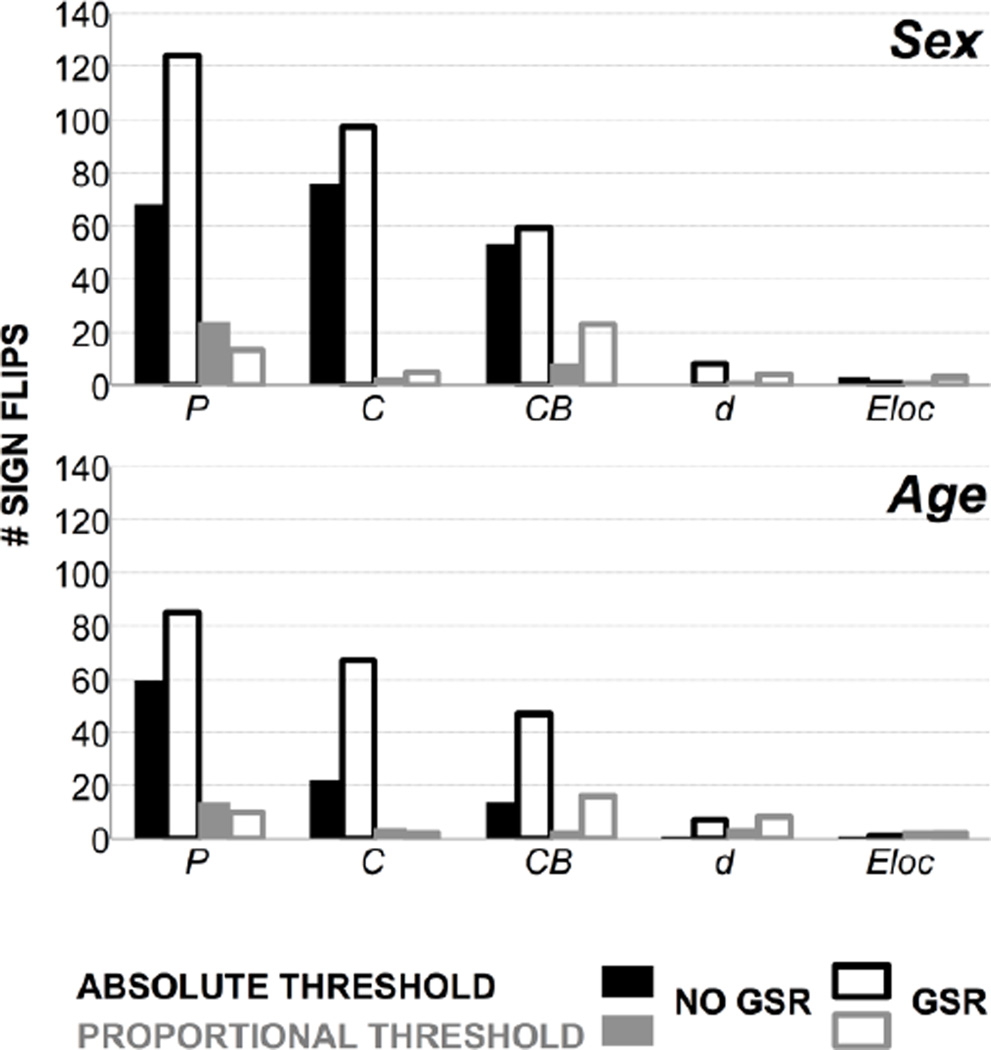
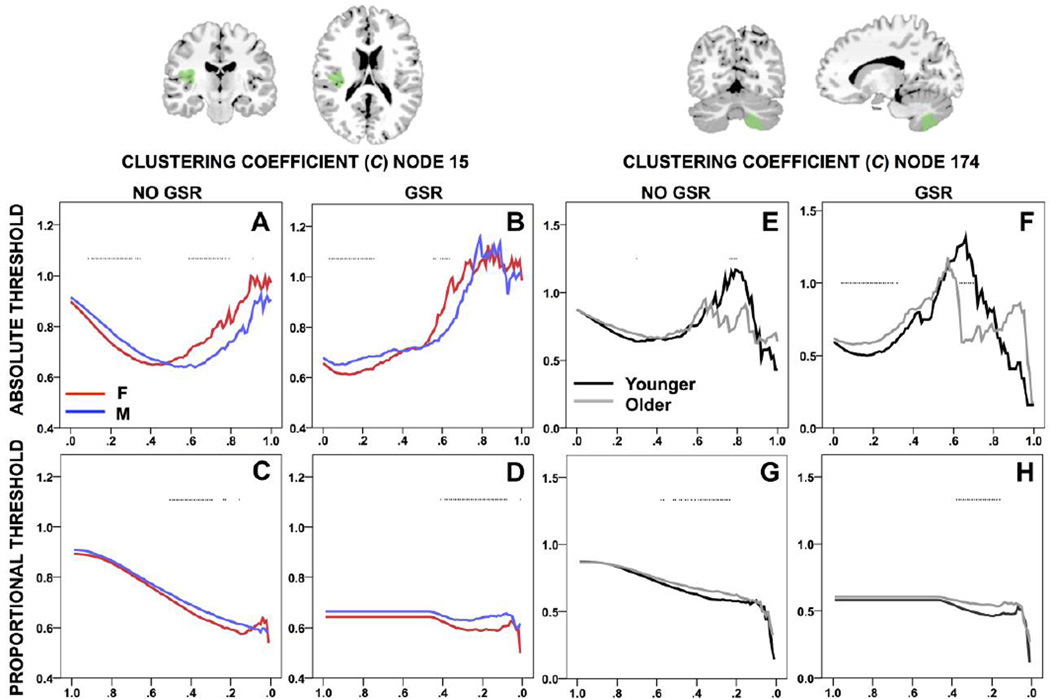
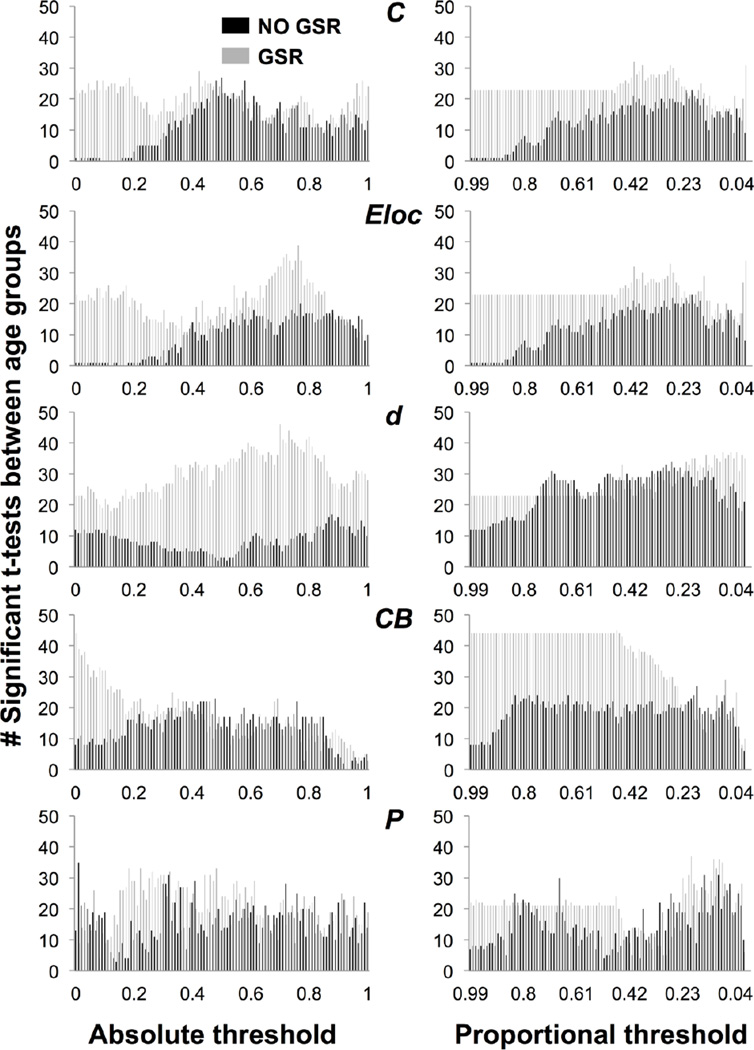
The number of nodes showing a difference in C between sex groups is skewed toward low thresholds without GSR, with the peak shifted toward lower thresholds with GSR (Figure 2). Seventy-six nodes showed sign reversals between sex groups without GSR; 97 nodes showed sign reversals with GSR (Figure 4). The survival curve for C is typically U- or S-shaped at a given node. At node 15, C was higher for males or females with and without GSR (Figure 5A–B). The number of nodes showing a difference in C between age groups is minimal at low thresholds then increases without GSR, and higher at low thresholds and variable with GSR (Figure 3). Twenty-two nodes showed sign reversals between age groups without GSR; 67 nodes showed sign reversals with GSR (Figure 4). At node 174, C was higher for older or younger participants with and without GSR (Figure 5E–F).
Figure 2.
Number of nodes showing a significant t-test between sex groups for clustering coefficient (C), local efficiency (Eloc), degree (d), betweenness centrality (CB), and participation coefficient (P), across absolute and proportional thresholds, without GSR (black) and with GSR (gray).
Figure 4.
Number of “sign reversals” in the direction of significant differences for sex and age groups, for participation coefficient (P), clustering coefficient (C), betweenness centrality (CB), degree (d), and local efficiency (Eloc), across absolute and proportional thresholds, without and with GSR.
Figure 5.
Clustering coefficient (C) compared between sex groups at node 15, and between age groups at node 174, across absolute and proportional thresholds, without and with GSR. *p<.05 (Table 2). Node displayed (green) overlaid onto MNI brain.
Figure 3.
Number of nodes showing a significant t-test between age groups for clustering coefficient (C), local efficiency (Eloc), degree (d), betweenness centrality (CB), and participation coefficient (P), across absolute and proportional thresholds, without GSR (black) and with GSR (gray).
Proportional threshold
The number of nodes showing a difference in C between sex groups is minimal at high thresholds then increases without GSR, and with GSR is flat and then peaks (Figure 2). Three nodes showed sign reversals between sex groups without GSR; 5 nodes showed sign reversals with GSR (Figure 4). The survival curve for C typically decreases or is stable and then drops off. At node 15, C was higher for males with and without GSR (Figure 5C–D). The number of nodes showing a difference in C between age groups is minimal at high thresholds and variable without GSR, and with GSR is flat and then peaks (Figure 3). Four nodes showed sign reversals between age groups without GSR; 2 nodes showed sign reversals with GSR (Figure 4). At node 174, C was higher for older participants with and without GSR (Figure 5G–H).
Local efficiency (Eloc)
Absolute threshold
The number of nodes showing a difference in Eloc between sex groups is skewed toward low thresholds without GSR, with the peak shifted toward lower thresholds with GSR (Figure 2). Three nodes showed sign reversals between sex groups without GSR; 1 node showed a sign reversal with GSR (Figure 4). The survival curve for Eloc is typically non-monotonic with a downward trend. At node 272, Eloc was higher for males or females with and without GSR (Figure S5A–B). The number of nodes showing a difference in Eloc between age groups is minimal at high thresholds then increases without GSR, and is higher at low thresholds and then peaks with GSR (Figure 3). One node showed a sign reversal between age groups with or without GSR (Figure 4). At node 128, Eloc was higher for older or younger participants without GSR, and higher for younger participants with GSR (Figure S5E–F).
Proportional threshold
The number of nodes showing a difference in Eloc between sex groups is minimal at high thresholds then increases without GSR, and with GSR is flat and then peaks (Figure 2). Two nodes showed sign reversals between sex groups without GSR; 3 nodes showed sign reversals with GSR (Figure 4). The survival curve for Eloc is typically nonmonotonic and decreasing or is flat with a peak and drop-off. At node 272, Eloc was higher for males with and without GSR (Figure S5C–D). The number of nodes showing a difference in Eloc between age groups is minimal at high thresholds then increases without GSR, and with GSR is flat and then peaks (Figure 3). Three nodes showed sign reversals between age groups without GSR; 2 nodes showed sign reversals with GSR (Figure 4). At node 128, Eloc was higher for older participants without GSR (Figure S5G–H).
Degree (d)
Absolute threshold
The number of nodes showing a difference in d between sex groups is skewed toward low thresholds without GSR, with the peak shifted toward a slightly lower threshold with GSR (Figure 2). No nodes showed a sign reversal between sex groups without GSR; 8 nodes showed sign reversals with GSR (Figure 4). The survival curve for d is typically monotonic and decreasing. At node 249, d did not show a difference between sex groups without GSR, but was higher for males or females with GSR (Figure S6A–B). The number of nodes showing a difference in d between age groups is variable without GSR, and is higher and skewed toward high thresholds with GSR (Figure 3). One node showed a sign reversal between age groups without GSR; 7 nodes showed sign reversals with GSR (Figure 4). At node 199, d was higher for younger or older participants without GSR, and higher for older participants with GSR (Figure S6E–F).
Proportional threshold
The number of nodes showing a difference in d between sex groups increases without GSR, and with GSR is flat and then increases to peak (Figure 2). Two nodes showed sign reversals between sex groups without GSR; 4 nodes showed sign reversals with GSR (Figure 4). The survival curve for d is typically monotonic and decreasing or flat and then decreasing. At node 249, d was higher for females without GSR, and higher for females or males with GSR (Figure S6C–D). The number of nodes showing a difference in d between age groups was variable and increasing without GSR, and with GSR is flat and then peaks (Figure 3). Four nodes showed sign reversals between age groups without GSR; 8 nodes showed sign reversals with GSR (Figure 4). At node 199, d was higher for older or younger participants without GSR, and higher for older participants with GSR (Figure S6G–H).
Betweenness centrality (CB)
Absolute threshold
The number of nodes showing a difference in CB between sex groups is low and variable with or without GSR (Figure 2). Fifty-three nodes showed sign reversals between sex groups without GSR; 59 nodes showed sign reversals with GSR (Figure 4). The survival curve for CB is typically non-monotonic with an inverted V-shape. At node 104, CB was higher for females or males with and without GSR (Figure S7A–B). The number of nodes showing a difference in CB between age groups was highest at moderate thresholds without GSR, and skewed toward low thresholds with GSR (Figure 3). Fourteen nodes showed sign reversals between age groups without GSR; 47 nodes showed sign reversals with GSR (Figure 4). At node 26, CB was higher for older or younger participants with and without GSR (Figure S7E–F).
Proportional threshold
The number of nodes showing a difference in CB between sex groups is low and variable without GSR, and with GSR is flat and then peaks (Figure 2). Eight nodes showed sign reversals between sex groups without GSR; 23 nodes showed sign reversals with GSR (Figure 4). The survival curve for CB typically increases to a sharp peak and then drops off. At node 104, CB was higher for females without GSR, and higher for males or females with GSR (Figure S7C–D). The number of nodes showing a difference in CB between age groups is variable without GSR, and with GSR is higher and flat and then peaks and decreases (Figure 3). Three nodes showed sign reversals between age groups without GSR; 16 nodes showed sign reversals with GSR (Figure 4). At node 26, CB was higher for older participants without GSR, and higher for younger participants with GSR (Figure S7G–H).
Participation coefficient (P)
Absolute threshold
The number of nodes showing a difference in P between sex groups is low and variable with and without GSR (Figure 2). Sixty-eight nodes showed sign reversals between sex groups without GSR; 124 nodes showed sign reversals with GSR (Figure 4). The survival curve for P is typically non-monotonic with a downward trend. At node 20, P was higher for females or males with or without GSR (Figure S8A–B). The number of nodes showing a difference in P between age groups is variable with and without GSR (Figure 3). Sixty nodes showed sign reversals between age groups without GSR; 85 nodes showed sign reversals with GSR (Figure 4). At node 62, P was higher for younger or older participants without GSR, and higher for younger participants with GSR (Figure S8E–F).
Proportional threshold
The number of nodes showing a difference in P between sex groups is variable without GSR, and with GSR is flat and then peaks (Figure 2). Twenty-four nodes showed sign reversals between sex groups without GSR; 13 nodes showed sign reversals with GSR (Figure 4). The survival curve for P is typically non-monotonic with an inverted J-shape or flat and then J-shaped. At node 20, P was higher for males or females with and without GSR (Figure S8C–D). The number of nodes showing a difference in P between age groups is variable without GSR, and with GSR is flat and then drops off and then peaks (Figure 3). Fourteen nodes showed sign reversals between age groups without GSR; 10 nodes showed sign reversals with GSR (Figure 4). At node 62, P was higher for younger participants with and without GSR (Figure S8G–H).
Discussion
Network measures were found to be unstable in a number of ways, particularly across absolute thresholds. Few of the survival curves were monotonic across absolute thresholds (e.g., Figure 1A–F); this can make it difficult to parameterize the curve with a simple function (as in Scheinost et al., 2012). Most of the network measures showed “sign reversals” in the direction of group differences across absolute thresholds (Figure 4). Finally, the number of nodes showing a group difference in a given network measure across absolute thresholds was typically variable or skewed, i.e., not flat (Figure 2–Figure 3).
Eglob and M were the most stable network-level measures across absolute thresholds. Eglob is defined as the average inverse shortest path length (Rubinov and Sporns, 2010) and is therefore related to L The current findings suggest that Eglob may be a more stable measure of functional integration than L when using absolute thresholds. Further, although Eglob exhibited a monotonic survival curve across absolute thresholds (Figure S2A–F), the curve for Eloc was typically non-monotonic (Figure S5A–B, E–F). This suggests that nodes may contribute to Eglob differently across absolute thresholds. The most stable node-level measures were d and Eloc, with few to no nodes showing sign reversals in these measures across absolute thresholds (Figure 4). Only one node showed a sign reversal in d across absolute thresholds (node 199, Figure S6E).
The stability of network measures was improved using proportional thresholds. The survival curves for network measures across proportional thresholds had more smoothly varying shapes. For example, the inverted V-shaped curves of L across absolute thresholds were not found (Figure 1). Although a few network measures showed sign reversals across proportional thresholds (Figure 4), in each instance, the reversal occurred at one or a few extreme thresholds. For example, at node 20, P was higher for males at τp=0.01–0.02, but higher for females at virtually every other threshold (Figure S8C–D). The number of nodes showing a group difference in a network measure across proportional thresholds was also less variable or more flat (Figure 2–Figure 3). These findings suggest that network measures are more stable across proportional than absolute thresholds. The application of proportional thresholds has become more common in graph theoretical analyses of human brain networks (e.g., Achard and Bullmore, 2007; Zhang et al., 2011). However, these findings demonstrate that the outcome of network analyses can differ depending on whether absolute or proportional thresholds are applied. For example, Eglob was found to be greater for males than females across absolute thresholds, but greater for females than males across proportional thresholds, when the same preprocessing method was used (Figure S2B, H).
The instability of network measures at very high or very low thresholds is expected and is related to the calculation of these measures as the graph becomes more or less connected. The current findings demonstrate, however, that network measure instability is not restricted to extreme thresholds, but occurs within reasonable, commonly applied ranges. For instance, L decreases at high absolute thresholds (and also at very low proportional thresholds <.01; not displayed) because many nodes no longer have paths between them that survive threshold, thereby decreasing the average shortest path length. However, the instability of L was not restricted to high absolute thresholds (Figure 1). With GSR, L was greater for females at reportable, low thresholds (r=0.24–0.57), but greater for males at reportable, high thresholds (r=0.73–0.75, 0.82–0.88), and published studies have reported findings at absolute thresholds ranging from r=0.1–0.8 (Buckner et al., 2009; Tomasi and Volkow, 2010). For network measures showing sign reversals in the direction of group differences across absolute thresholds, most showed sign reversals within the typically reported range (Table 2). Proportional thresholds were found to improve network measure stability, and this was indicated by all sign reversals across proportional thresholds occurring at one or a few extreme thresholds, as described above.
Network measures were also found to be somewhat unstable within subjects across the eight resting-state fMRI runs. Only network-level measures were tested across runs. Although the shapes of the survival curves were consistent across runs (e.g., Figure 5E–F, K–L), indicating relative stability, there was nevertheless a significant effect of run on every network measure at some threshold, depending on the type of threshold applied and the preprocessing method.
Finally, these findings demonstrate an impact of preprocessing with GSR, but did not reveal a consistent effect of GSR on the stability of network measures. The survival curves were typically shifted by GSR across absolute or proportional thresholds. GSR impacted group differences in a number of ways: (a) group differences were only found with GSR (e.g., Eglob, T, M; proportional threshold; sex); (b) group differences found without GSR were no longer found with GSR (e.g., Eloc; node 128; proportional threshold; age); (c) sign reversals found without GSR resolved to one direction of group differences with GSR (e.g., d; node 199; absolute or proportional threshold; age); and (d) at worst, the direction of group differences found without GSR reversed with GSR (e.g., CB; proportional threshold; age). Across absolute thresholds, with GSR, all network measures showed a sign reversal by sex with the exception of Eglob. Across proportional thresholds, d, CB, and P showed sign reversals. These findings suggest that GSR does not improve network measure stability in any consistent way. It is possible that in some instances, the group differences found with GSR are a correct representation of the data. GSR may facilitate observations of true physiological relationships in the brain (Fox et al., 2009) and can help mitigate artifacts due to motion (Yan et al., 2013) or variations in carbon dioxide (Wise et al., 2004). However, in the current findings, it is not possible to determine whether GSR reveals true group differences, and overall, GSR was not found to stabilize network measures across thresholds. Alternative solutions to account for global signal fluctuations include physiological noise removal using independent components (Griffanti et al., 2014) or principal components analyses (e.g, CompCor; Behzadi et al., 2007).
In summary, network analyses enable characterization of the large-scale organization of the brain from fMRI connectivity data, however, a threshold is commonly applied to infer network structure from the data, and the current findings, and other studies (e.g., Scheinost et al., 2012), have shown that network measures are unstable across thresholds. Here we provide additional evidence that network measures are unstable across runs and can change with different preprocessing pipelines. As there is no biologically principled way to set a threshold for a brain network, thresholds remain an ‘educated guess’ (Van Dijk et al., 2010). The current findings demonstrate that the choice of thresholding approach can dramatically impact results, including even reversing the direction of group differences in a network measure.
Recent studies have reported findings across a range of thresholds. This is useful for a more complete understanding of the data. Other approaches to thresholding were not tested in the current study, such as weighted graphs, in which edges carry some continuous weight value, such as related to the strength or effectiveness of connections, or the distance between nodes (Reijneveld et al., 2007). However, it is not straightforward how best to set weights that reflect the relative strengths of connections (Fornito et al., 2010). It has also been suggested that weighted graphs are not reasonable biological structures because in weighted graphs all brain regions are connected to all other brain regions (Sporns, 2011). However, unlike measures of anatomical connectivity which are considered to reflect direct connections, functional connectivity measures reflect both direct (monosynaptic) and indirect (polysynaptic) connections (Buckner et al., 2013), and therefore weighted graphs in which all nodes are connected may still reflect a reasonable representation of brain networks. Methods have improved for modeling weighted brain networks (e.g., Latora and Marchiori, 2003), and the weighted approach has been used to characterize network properties as they relate to sex and age (Gong et al., 2009), among others. Additional methods can be used that integrate network measures over a full range of thresholds and, thus, are not sensitive to threshold effects (Ginestet et al., 2011; Scheinost et al., 2012). One approach is to parameterize the distribution of a network measure calculated over all thresholds (Scheinost et al., 2012). These parameters can then be used to compare connectivity between groups (e.g., Garrison et al., 2014), or to relate connectivity to behavior or other variables (e.g., Mitchell et al., 2013). A second approach is to use Monte Carlo sampling to integrate network measures over a range of proportional thresholds for both weighted and un-weighted networks (Ginestet et al., 2011).
Conclusion
Network analyses are useful for summarizing large-scale brain organization, to relate features of network topology to behavior and cognition, and to examine changes in these features in clinical populations such as individuals with neurological and psychiatric disorders. However, caution must be used when conducting and interpreting network analyses. As studies work to elucidate the organization of functional brain networks in these and other contexts, the choice of thresholding approach must be carefully motivated because the network structure inferred largely determines the neurobiological interpretation (Rubinov and Sporns, 2010).
Supplementary Material
Highlights.
Network measures were found to be unstable across absolute thresholds.
For instance, the direction of significant group differences can reverse across thresholds.
Network measures were found to be more stable across proportional thresholds.
Caution should be used when applying thresholds to functional connectivity data.
Acknowledgements
We thank our research participants for their time and efforts. This work was funded by grants from the National Institutes of Health, National Institute on Drug Abuse (to KAG: K12DA00167; to DS: T32DA022975); the American Heart Association (to KAG: 14CRP18200010); and the National Science Foundation (to EF: Graduate Research Fellowship).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achard S, Bullmore E. Efficiency and cost of economical brain functional networks. PLoS Comput Biol. 2007;3:e17. doi: 10.1371/journal.pcbi.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch AF, Gogtay N, Meunier D, Birn R, Clasen L, Lalonde F, Lenroot R, Giedd J, Bullmore ET. Disrupted modularity and local connectivity of brain functional networks in childhood-onset schizophrenia. Front Syst Neurosci. 2010;4:147. doi: 10.3389/fnsys.2010.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch AF, Vertes PE, Stidd R, Lalonde F, Clasen L, Rapoport J, Giedd J, Bullmore ET, Gogtay N. The anatomical distance of functional connections predicts brain network topology in health and schizophrenia. Cerebral Cortex. 2013;23:127–138. doi: 10.1093/cercor/bhr388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E. Small-world brain networks. Neuroscientist. 2006;12:512–523. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A. Hierarchical organization of human cortical networks in health and schizophrenia. Journal of Neuroscience. 2008;28:9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Yeo BT. Opportunities and limitations of intrinsic functional connectivity MRI. Nature Neuroscience. 2013;16:832–837. doi: 10.1038/nn.3423. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. Journal of Neuroscience. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Chanraud S, Pitel AL, Pfefferbaum A, Sullivan EV. Disruption of functional connectivity of the default-mode network in alcoholism. Cerebral Cortex. 2011;21:2272–2281. doi: 10.1093/cercor/bhq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Reynolds JR, Power JD, Repovs G, Anticevic A, Braver TS. Multitask connectivity reveals flexible hubs for adaptive task control. Nature Neuroscience. 2013;16:1348–1355. doi: 10.1038/nn.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NU, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RA, Fox MD, Snyder AZ, Vincent JL, Raichle ME, Schlaggar BL, Petersen SE. Distinct brain networks for adaptive and stable task control in humans. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11073–11078. doi: 10.1073/pnas.0704320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Bullmore ET. Network scaling effects in graph analytic studies of human resting-state FMRI data. Front Syst Neurosci. 2010;4:22. doi: 10.3389/fnsys.2010.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. Journal of Neurophysiology. 2009;101:3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison KA, Scheinost D, Constable RT, Brewer JA. BOLD signal and functional connectivity associated with loving kindness meditation. Brain Behav. 2014;4:337–347. doi: 10.1002/brb3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestet CE, Nichols TE, Bullmore ET, Simmons A. Brain network analysis: separating cost from topology using cost-integration. PLoS One. 2011;6:e21570. doi: 10.1371/journal.pone.0021570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, Rosa-Neto P, Carbonell F, Chen ZJ, He Y, Evans AC. Age- and gender-related differences in the cortical anatomical network. Journal of Neuroscience. 2009;29:15684–15693. doi: 10.1523/JNEUROSCI.2308-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L, Salimi-Khorshidi G, Beckmann CF, Auerbach EJ, Douaud G, Sexton CE, Zsoldos E, Ebmeier KP, Filippini N, Mackay CE, Moeller S, Xu J, Yacoub E, Baselli G, Ugurbil K, Miller KL, Smith SM. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. Neuroimage. 2014;95:232–247. doi: 10.1016/j.neuroimage.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Scheinost D, Okuda H, Belhachemi D, Murphy I, Staib LH, Papademetris X. Unified framework for development, deployment and robust testing of neuroimaging algorithms. Neuroinformatics. 2011;9:69–84. doi: 10.1007/s12021-010-9092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latora V, Marchiori M. Economic small world behavior in weighted networks. European Physical Journal. 2003;32:249–263. [Google Scholar]
- Meunier D, Achard S, Morcom A, Bullmore E. Age-related changes in modular organization of human brain functional networks. Neuroimage. 2009;44:715–723. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- Mitchell MR, Balodis IM, Devito EE, Lacadie CM, Yeston J, Scheinost D, Constable RT, Carroll KM, Potenza MN. A preliminary investigation of Stroop-related intrinsic connectivity in cocaine dependence: associations with treatment outcomes. American Journal of Drug and Alcohol Abuse. 2013;39:392–402. doi: 10.3109/00952990.2013.841711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijneveld JC, Ponten SC, Berendse HW, Stam CJ. The application of graph theoretical analysis to complex networks in the brain. Clinical Neurophysiology. 2007;118:2317–2331. doi: 10.1016/j.clinph.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Scheinost D, Benjamin J, Lacadie CM, Vohr B, Schneider KC, Ment LR, Papademetris X, Constable RT. The intrinsic connectivity distribution: a novel contrast measure reflecting voxel level functional connectivity. Neuroimage. 2012;62:1510–1519. doi: 10.1016/j.neuroimage.2012.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinost D, Finn ES, Tokoglu F, Shen X, Papademetris X, Hampson M, Constable RT. Sex differences in normal age trajectories of functional brain networks. Human Brain Mapping. 2015;36:1524–1535. doi: 10.1002/hbm.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano MA, Boguna M, Vespignani A. Extracting the multiscale backbone of complex weighted networks. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6483–6488. doi: 10.1073/pnas.0808904106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Papademetris X, Constable RT. Graph-theory based parcellation of functional subunits in the brain from resting-state fMRI data. Neuroimage. 2010;50:1027–1035. doi: 10.1016/j.neuroimage.2009.12.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Tokoglu F, Papademetris X, Constable RT. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage. 2013;82:403–415. doi: 10.1016/j.neuroimage.2013.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SL, Bowman FD, Laurienti PJ. Analyzing complex functional brain networks: Fusing statistics and network science to understand the brain. Statistics Surveys. 2013;7:1–36. doi: 10.1214/13-SS103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. Networks of the brain. Cambridge, MA: MIT Press; 2011. [Google Scholar]
- Stam CJ, de Haan W, Daffertshofer A, Jones BF, Manshanden I, van Cappellen van Walsum AM, Montez T, Verbunt JP, de Munck JC, van Dijk BW, Berendse HW, Scheltens P. Graph theoretical analysis of magnetoencephalographic functional connectivity in Alzheimer's disease. Brain. 2009;132:213–224. doi: 10.1093/brain/awn262. [DOI] [PubMed] [Google Scholar]
- Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer's disease. PLoS Comput Biol. 2008;4:e1000100. doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesford QK, Simpson SL, Burdette JH, Hayasaka S, Laurienti PJ. The brain as a complex system: using network science as a tool for understanding the brain. Brain Connect. 2011;1:295–308. doi: 10.1089/brain.2011.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Wang J, Yan C, He Y. Hemisphere- and gender-related differences in small-world brain networks: a resting-state functional MRI study. Neuroimage. 2011;54:191–202. doi: 10.1016/j.neuroimage.2010.07.066. [DOI] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND. Functional connectivity density mapping. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9885–9890. doi: 10.1073/pnas.1001414107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Mandl RC, Kahn RS, Hulshoff Pol HE. Functionally linked resting-state networks reflect the underlying structural connectivity architecture of the human brain. Human Brain Mapping. 2009a;30:3127–3141. doi: 10.1002/hbm.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Kahn RS, Hulshoff Pol HE. Efficiency of functional brain networks and intellectual performance. Journal of Neuroscience. 2009b;29:7619–7624. doi: 10.1523/JNEUROSCI.1443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KRA, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic Functional Connectivity As a Tool For Human Connectomics: Theory, Properties, and Optimization. Journal of Neurophysiology. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk BC, Stam CJ, Daffertshofer A. Comparing brain networks of different size and connectivity density using graph theory. PLoS ONE. 2010;5:e13701. doi: 10.1371/journal.pone.0013701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21:1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Yan CG, Cheung B, Kelly C, Colcombe S, Craddock RC, Di Martino A, Li Q, Zuo XN, Castellanos FX, Milham MP. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 2013;76:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang J, Wu Q, Kuang W, Huang X, He Y, Gong Q. Disrupted brain connectivity networks in drug-naive, first-episode major depressive disorder. Biological Psychiatry. 2011;70:334–342. doi: 10.1016/j.biopsych.2011.05.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.