Abstract
Background
Pneumococcal serotype identification is essential to monitor pneumococcal vaccine effectiveness and serotype replacement. Serotyping by conventional serological methods are costly, labour-intensive, and require significant technical expertise. We compared two different molecular methods to serotype pneumococci isolated from the nasopharynx of South African infants participating in a birth cohort study, the Drakenstein Child Health Study, in an area with high 13-valent pneumococcal conjugate vaccine (PCV13) coverage.
Methods
A real-time multiplex PCR (rmPCR) assay detecting 21 different serotypes/-groups and a sequetyping assay, based on the sequence of the wzh gene within the pneumococcal capsular locus, were compared. Forty pneumococcal control isolates, with serotypes determined by the Quellung reaction, were tested. In addition, 135 pneumococcal isolates obtained from the nasopharynx of healthy children were tested by both serotyping assays and confirmed by Quellung testing. Discordant results were further investigated by whole genome sequencing of four isolates.
Results
Of the 40 control isolates tested, 25 had a serotype covered by the rmPCR assay. These were all correctly serotyped/-grouped. Sequetyping PCR failed in 7/40 (18%) isolates. For the remaining isolates, sequetyping assigned the correct serotype/-group to 29/33 (88%) control isolates. Of the 132/135 (98%) nasopharyngeal pneumococcal isolates that could be typed, 69/132 (52%) and 112/132 (85%) were assigned the correct serotype/-group by rmPCR and sequetyping respectively. The serotypes of 63/132 (48%) isolates were not included in the rmPCR panel. All except three isolates (serotype 25A and 38) were theoretically amplified and differentiated into the correct serotype/-group with some strains giving ambigous results (serotype 13/20, 17F/33C, and 11A/D/1818F). Of the pneumococcal serotypes detected in this study, 69/91 (76%) were not included in the current PCV13. The most frequently identified serotypes were 11A, 13, 15B/15C, 16F and 10A.
Conclusion
The rmPCR assay performed well for the 21 serotypes/-groups included in the assay. However, in our study setting, a large proportion of serotypes were not detected by rmPCR. The sequetyping assay performed well, but did misassign specific serotypes. It may be useful for regions where vaccine serotypes are less common, however confirmatory testing is advisable.
Introduction
The pneumococcus (Streptococcus pneumoniae) is a common cause of invasive disease and respiratory tract infections including bloodstream infections, meningitis, pneumonia and otitis media [1–3]. Patients at risk include those at the extremes of age and the immunocompromised, particularly those affected by cell-mediated immune deficiencies. Colonisation of the nasopharynx with a homologous strain of pneumococci precedes the development of invasive and respiratory tract disease [2,4,5]. Serotyping of the pneumococcal polysaccharide capsule, the immunogenic component of current vaccines, remains the cornerstone of strain characterization. To date, more than 90 capsular serotypes have been described and new ones continue to be described [6,7]. Multiple pneumococcal serotypes can colonize the nasopharynx successively over long period of time, or at any one time [8–10]. Invasive disease is commonly regarded as resulting from a single serotype. Public health programs employ serotype prevalence data from invasive disease to assist vaccine selection. Regular surveillance is required, and relies mostly on phenotypic serotyping methods, most notably the Quellung method developed in 1902 [11]. The antiserum utilised in this assay is costly, methods employed are labour intensive, and require significant technical expertise and experience.
More practical, higher throughput typing techniques are required for expanding public health laboratory services in many areas of the world to support growing disease control programs and epidemiological surveillance. Emerging technologies include alternative culture-based phenotypic methods such as latex agglutination, dot blot ELISA and microbead assays [10,12,13]. While the newer phenotypic methods all have their distinct benefits and often surpass Quellung in terms of rapidity and cost, some of the methods require sophisticated and expensive instruments.
Promising genotypic typing methods that target serotype-specific regions of the cps genes have been developed including multiplex Polymerase Chain Reaction (PCR) with subsequent agarose gel electrophoresis [14–16]; restriction fragment length polymorphism (PCR-RFLP) [17]; automated fluorescent capillary electrophoresis (FAF-mPCR) [18]; electrospray ionization mass spectrometry (PCR/ESI-MS) [19]; reverse line blot hybridization assay (mPCR/RLB) [20] and real-time multiplex PCR (rmPCR) [21] including the recently described nanofluidic rmPCR [22]. PCR with subsequent target detection is prone to amplicon contamination and is more labour intensive than rmPCR. rmPCR obviates the need for amplicon manipulation, is highly sensitive, fast and less labour intensive. PCR assays do not require viable isolates and have the potential to detect multiple serotypes simultaneously [21,23–26]. More recently sequetyping, a sequence-based typing method, has been described [27]. There are currently no published head-to-head comparisons of the accuracy of the sequetyping vs. multiplex PCR approaches. Given the heterogeneity and recombinogenic nature of pneumococci, capsular typing tools which infer type from DNA sequence, including target enrichment-based next generation sequencing (NGS) and whole genome sequencing (WGS) [28] are attractive newer methods to complement the molecular typing methods discussed above and may also aid in resolving discrepant phenotypic and genotypic findings.
Materials and Methods
Assay validation
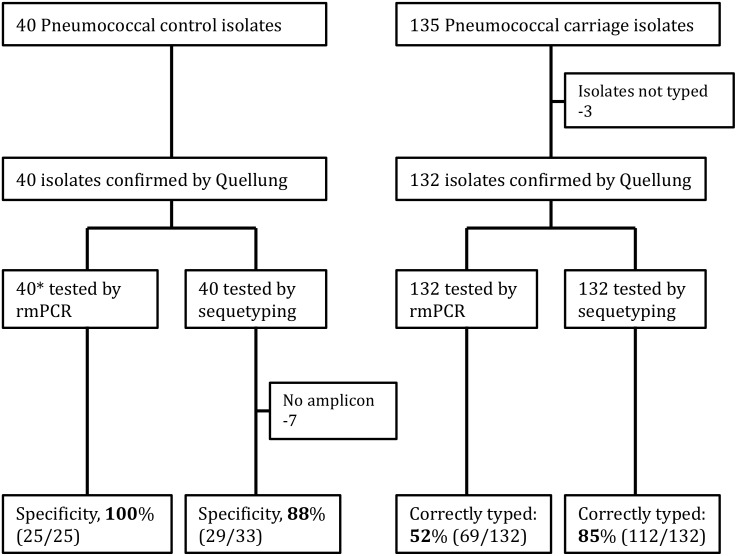
Isolates comprised 40 Quellung-typed control strains, Fig 1, (kindly donated by Dr. Anne von Gottberg, Centre for Respiratory Diseases and Meningitis (CRDM), National Institute for Communicable Diseases (NICD), South Africa [29]). These isolates were transported on Dorset egg medium [30], subcultured onto Columbia blood agar base with 2% agar, 5% horse blood and 4 μg/mL gentamicin media (CAG) upon receipt (Green point Media Laboratory of the National Health Laboratory Service, Cape Town, South Africa) and incubated at 37°C in 5% CO2 overnight. The resulting colonies were inoculated into in 1 ml skim milk-tryptone-glucose-glycerol (STGG) transport medium frozen at -80°C for batch processing.
Fig 1. Flow chart showing the pneumococcal isolates included in the study.
*Of the 40 isolates that were tested by rmPCR, only 25 were included as part of the rmPCR targets.
Subsequently, 135 pneumococcal isolates (Fig 1) were cultured from nasopharyngeal (NP) swabs that were collected from 83 healthy infants by employing nylon flocked swabs (Copan Italia, Brescia, Italy). Infants were recruited between May 2012 and September 2013 as part the Drakenstein Child Health Study (DCHS), a South African birth cohort study [31]. NP swabs were collected employing the World Health Organization protocol for pneumococcal carriage studies. Briefly, the collected NP swabs were immediately placed into 1 ml STGG, transported on ice to the laboratory and frozen at -80°C for batch processing. After thawing, STGG samples were vortexed for 15 s before a 10 μl aliquot was inoculated onto Columbia blood agar base with 2% agar, 5% horse blood (BA) plates and incubated at 37°C in 5% CO2 overnight. Presumptive pneumococcal isolates were identified by colony morphology, α-hemolysis and ethylhydrocupreine (optochin) disk susceptibility (Oxoid, Basingstoke, UK) as previously described [32–34].
Nucleic acid extraction
Prior to rmPCR and sequetyping, all isolates were subjected to nucleic acid extraction employing a heat lysis method as previously described [35]. Briefly, a sweep of pneumococcal colonies was obtained from primary BA plates that were inoculated with thawed STGG aliquots containing either pneumococcal control strains or carriage isolates. The colony sweeps were resuspended in 100 μl of phosphate-buffered saline, pH 7.4 (PBS; Sigma-Aldrich, St. Louis, MI) thereafter heated at 95°C for 5 min. The supernatant containing genomic DNA (gDNA) was ten-fold serially diluted in PBS before nucleic acid amplification.
Real-time multiplex PCR
The rmPCR, designed as a 7x3-plex that targets 21 serotypes. We used the multiplex scheme for an African region, based on a relatively limited sample observed [21]. This assay targets all the pneumococcal serotypes included in PCV13 and 8 other additional serotypes/-groups (PCV13 serotypes: 1, 3, 4, 5, 6A/6B, 7F/7A, 9V/9A, 14, 18C/18A/18B/18C, 19A, 19F, 23F, 23A; The 8 additional serotypes/-groups include: 2, 6C/6D, 11A/11D, 12B/2F/46, 15A/15F, 16F, 22F, 33A/33F). Briefly, the PCR reaction comprised 12.5 μl of 2X SensiFAST Probe No-ROX One-Step master mix (Bioline, Taunton, MA), primers and probes for serotype as described by Pimenta et al, 5 μl gDNA (diluted 1:1000) and nuclease/RNase-free water (Applied Biosystems, Irving, CA) for a final reaction volume of 25 μl (S1 Table). The thermal cycling conditions consisted of initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min employing a CFX96 Touch Real-Time PCR amplification system (Bio-Rad Laboratories, Hercules, CA)
Sequetyping
The assay was performed as previously described [27] with minor modification: the PCR reaction comprised 12.5 μl of 2X KAPA Taq Ready Mix (KAPA Biosystems, Boston, MA), 1 μl of primer mix, 2 μl gDNA (diluted 1:10), 8.5 μl nuclease/RNase-free H2O (Applied Biosystems) in a final volume of 25 μl (S2 Table). Thermal cycling consisted of an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 65°C for 30 s, and extension at 72°C for 90 s employing an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems). The PCR products were separated by electrophoresis in 1.5% agarose gel (SeaKem LE Agarose; Lonza, Rockland, ME) for 45 min at 80 V in a 1X Tris-acetate EDTA buffer. Ethidium bromide-stained DNA products were visualized under UV illumination and sized by using a 1–kb DNA molecular size marker (HyperLadderv1-kb; Bioline).
PCR products were prepared for sequencing employing Exo-SAP IT (Affymetrix, Maumee, OH) according to the manufacturer’s instructions. Prepared amplicons were submitted for cycle sequencing employing the BigDye Sequence Terminator kit V3.1 (Applied Biosystems) and analysed on an ABI 3500 XL Genetic Analyzer (Appplied Biosystems) by Inqaba Biotech (Inqaba Biotechnical Industries [Pty] Ltd, Pretoria, South Africa). Sequencing was performed in both directions using forward (cps1), 5’-GCA ATG CCA GAC AGT AAC CTC TAT-3’, and reverse (cps2), 5’-CCT GCC TGC AAG TCT TGA TT-3’ primers.
DNA sequences obtained were assembled and edited using DNA Baser Sequence Assembler v4 (www.DnaBaser.com). The consensus sequences were used to interrogate the GenBank database (http://www.ncbi.nlm.nih.gov/blast/) and assign a serotype using the criteria as per protocol [27]. Briefly, the serotype of the wzh nucleotide sequence from GenBank with the highest BLAST bit score was assigned, provided that sequence identity was >98% with the query amplicon nucleotide sequence. To automate the above process, a Java-based program, Sequetyper (available at http://www.gematics.com/sequetyper.html) was developed and validated to automatically analyse and determine the pneumococcal serotype based on interrogation of GenBank with the input forward and reverse sequences of the generated wzh amplicon. This application is suitable for high-throughput analysis of sequetyping data (S1 Fig).
Quellung testing
Pneumococcal control and nasopharyngeal isolates were submitted to CRDM for quellung testing using specific anti-sera (Statens Serum Institut, Copenhagen, Denmark). Serotype 6C was distinguished from serotype 6A by PCR [36], while serotype 25A and 38 were undistinguishable and hence reported as 25A/38 [37].
Next-generation sequencing
Three pneumococcal carriage isolates serotyped as 16F by Quellung and rmPCR but identified as 9V by sequetyping were subjected to WGS. The 3 discordant isolates as well as a control strain identified as 9V by Quellung, sequetyping and rmPCR were also included. Briefly, gDNA was isolated with a Wizard Genomic DNA Purification Kit (Promega Corporation, Fitchburg, WI) according to the manufacturer's instructions. The gDNA quality was assessed using the Qubit Fluorometer (Life Technologies, Carlsbad, CA), the NanoDrop ND-1000 (Life Technologies) and agarose gel electrophoresis used to determine absolute concentration, polyphenolic/polysaccharide/chaotropic salt contamination and gDNA integrity respectively. Quantified gDNA was submitted to the Centre for Proteomics and Genomic Research (CPGR) for WGS. Briefly, sequencing libraries were generated using the Nextera XT DNA Sample Prep Kit (Illumina, San Diego, CA) and the libraries were indexed according to the dual-bar cording protocol (with i7 and i5 primers) using the Nextera XT Index Kit (Illumina). Libraries were then normalized, pooled, and a 5% PhiX control added before sequencing with the Illumina MiSeq Reagent Kit v2 (500 cycle) on the Illumina MiSeq system.
De novo sequence assembly
The quality of the output sequence data was assessed using FastQC [38] and sequencing adapters were trimmed using Trimmomatic [39]. The 3'-end nucleotides with PHRED scores below 20 were trimmed using the fastx_trimmer tool of FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit) [40]. The sequence data was then assembled de novo using SPAdes v3.0.0 assembler [41]. Draft genome assemblies were annotated individually using RAST (Rapid Annotation using Subsystem Technology) [42]. The contigs containing putative cps regions were identified through the standalone blastall homology searches against the 16F (Accession: CR931668) and 9v (Accession: CR931648) annotated reference genomes and then extracted to a separate file using a shell command based on SAMtools [43]. These contigs were then aligned and visual representation of the alignments was performed using the Artemis Comparison Tool (ACT) v6 and WebACT [44].
Data analysis
Results of the two molecular serotyping assays up to the serogroup level were compared with serotyping results obtained by Quellung testing. In cases of discordance between the two molecular serotyping assays, the results were confirmed by Quellung testing. Serotype distribution was determined based on Quellung results. Where more than one isolate was tested from the same child, isolates of the same serotype were included only once in the analysis.
Ethical consideration
Ethical approval was obtained from the Human Research Ethics Committee of the Faculty of Health Sciences, University of Cape Town (HREC ref: 062/2011) and the Western Cape Provincial Child Health Research committee. Mothers provided written informed consent at enrolment.
Results
Real-time multiplex PCR
Of the 40 pneumococcal control isolates subjected to rmPCR, 25 isolates yielded a positive signal; 15 isolates failed to yield detectable amplification signal. Of the 25 rmPCR positive isolates, results were all (25/25) concordant with Quellung confirmed serotypes (Table 1).
Table 1. Concordance of molecular serotyping results of pneumococcal control strains.
| Serotype | rmPCR a | Sequetyping |
|---|---|---|
| 1 | 1 | 1 |
| 2 | 2 | No IDb |
| 3 | 3 | 3 |
| 4 | 4 | 4 |
| 5 | 5 | 5 |
| 6A | 6A/6B | 6A |
| 6C | 6C/6D | 6C/6D |
| 7F | 7F/7A | 7F/7A |
| 9V | 9V/9A | 9V |
| 11A | 11A | Negc |
| 12B | 12F/12A/12B/44/46 | 12B |
| 12F | 12F/12A/12B/44/46 | Neg |
| 14 | 14 | 14 |
| 15A | 15A/15F | 15A |
| 15F | 15A/15F | 15F |
| 16F | 16F | 9V d |
| 18C | 18C/18A/18B/18F | 18B d |
| 19A | 19A | Neg |
| 19F | 19F | 19F |
| 22F | 22F/22A | Neg |
| 23A | 23A | 23A |
| 23F | 23F | Neg |
| 33A | 33F/33A/37 | 33A/33F/35A |
| 33F | 33F/33A/37 | 33A/33F/35A |
| 46 | 12F/12A/12B/44/46 | 12A |
| 7C | N/A* | 7C |
| 8 | N/A* | 8 |
| 9N | N/A* | 9N |
| 10A | N/A* | 10A |
| 10F | N/A* | 10C/10F |
| 11B | N/A* | Neg |
| 15B | N/A* | 15B/15C |
| 21 | N/A* | Neg |
| 23B | N/A* | 23B |
| 24B | N/A* | 24B |
| 27 | N/A* | 27 |
| 28A | N/A* | 28A |
| 31 | N/A* | 31 |
| 35B | N/A* | 35B/35C |
| 35C | N/A* | 33A/33F/35C |
armPCR: real-time Multiplex PCR.
N/A* = serotype not included in the rmPCR panel.
No ID b. = sequence identity of ≤98% with sequences in GenBank.
Negc = negative sequetyping PCR result.
dThe Sequetyping assay mistyped serotype 18C as 18B.
Sequetyping
Of the 40 pneumococcal control isolates subjected to sequetyping, 33 isolates yielded single amplicons of ~ 1,061 bp; 7 isolates failed to yield detectable amplicons. Sequence analysis yielded 29/33 (88%) sequetype-Quellung concordant results (Table 1). Of the four discordant results, Quellung 16F was identified as 9V by sequetyping, Quellung 46 was identified as 12A by sequetyping while Quellung 18C was identified as 18B. The fourth discordant isolate yielded no match (>98%) when submitted to GenBank and is considered novel (GenBank submission accession number: BankIt1792036). The wzh PCR was negative in 7/40 (18%) control isolates tested, which consequently could not be sequetyped. Detailed results are provided in Table 1.
Carriage isolates
Of 135 pneumococcal isolates tested, 132 (98%) were assigned a serotype/-group by the Quellung reaction (Tables 2 and 3). Three (3) isolates could not be typed by either Quellung or molecular methods. A total of 69 (52%) isolates were assigned a serotype covered by the rmPCR assay. Of these, the rmPCR assay assigned the correct serotype to all 69 isolates (Tables 2 and 3).
Table 2. Carriage isolate serotyping method concordance.
| Serotype (n) a | rmPCR (n) b | Sequetyping (n) | Remarks |
|---|---|---|---|
| 1 (1) | 100% (1) | 100% (1) | |
| 3 (1) | 100% (1) | 100% (1) | |
| 4 (1) | 100% (1) | 100% (1) | |
| 6A (2) | 100% (2) | 50% (1) | 2 rmPCR as 6A/6B; 1 sequetyped as 9V |
| 6B (11) | 100% (11) | 100% (11) | 11 rmPCR as 6A/6B |
| 6C (1) | 100% (1) | 100% (1) | 1 rmPCR as 6C/6D; 1 sequetyped as 6C/6D |
| 10A/11A (1) | 100% (1) | 100% (1) | 1 rmPCR as 11A/11D; 1 sequetyped as 10A |
| 11A (8) | 100% (8) | 75% (6) | 8 rmPCR as 11A/11D; 6 sequetyped as 11A/11D/18F; 2 no amplicon in sequetyping |
| 14 (5) | 100% (5) | 100% (5) | |
| 15A (10) | 100% (10) | 100% (10) | 10 rmPCR as 15A/15F |
| 16F (8) | 100% (8) | 63% (5) | 3 sequetyped as 9V |
| 17F/1 ¥ (2) | 100% (2) | 100% (2) | 2 rmPCR as 1; 2 sequetyped as 1 |
| 18C (3) | 100% (3) | 33% (1) | 3 rmPCR as 18A/18B/18C/18F; 1 sequetyped as 18B, 2 sequetyped with low identity score |
| 19A (8) | 100% (8) | 75% (6) | 2 no amplicon in sequetyping |
| 19F (5) | 100% (5) | 100% (5) | |
| 22F (2) | 100% (2) | 100% (2) | 2 rmPCR as 22A/22F; 2 sequetyped as 22A/22F |
| 7C (1)* | N/A | 100% (1) | |
| 9N (4)* | N/A | 100% (4) | |
| 10A (8)* | N/A | 100% (8) | |
| 13 (14)* | N/A | 93% (13) | 1 sequetyped as 15B; 13 sequetyped as 13/20 |
| 15B (7)* | N/A | 100% (7) | 7 sequetyped as 15B/15C |
| 15C (7)* | N/A | 100% (7) | 7 sequetyped as 15B/15C |
| 17F (3)* | N/A | 33% (1) | 2 sequetyped as 33C |
| 19B (2)* | N/A | 100% (2) | 1 sequetyped as 19C, 1 sequetyped as 19F |
| 21 (5)* | N/A | 100% (5) | |
| 25A/38 (3)* c | N/A | Neg d | 3 no amplicon in sequetyping |
| 35A (9)* | N/A | 89% (8) | 8 sequetyped as 33A/33F/35A, 1 sequetyped as 13/20 |
| OMNI NEG (3) | Neg | Neg | |
| Total (132) | 69 | 112 |
aThe numbers in closed brackets indicate the correct identification of a Quellung-confirmed serotype by the rmPCR and sequetyping assays;
brmPCR: real-time multiplex PCR;
* = serotypes not included in rmPCR assay.
¥ Mixed serotypes detected;
Negd = negative sequetyping PCR result;
cSerotype 25A and 38 were undistinguishable by Quellung and hence reported as 25A/38 [37].
Table 3. Summary of molecular serotyping results of pneumococcal nasopharyngeal isolates from healthy children compared with the serotype determined by the Quellung reaction.
| rmPCR a , correctly serotyped b | Sequetyping, correctly serotyped b | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| Yes | No | Negative c | Yes | No | Negative c | |||
| Quellung | Typeable | 69 | 0 | 63 ¥ | 112 | 13 | 7 | 132 |
| nontypeable | 0 | 0 | 3 | 0 | 0 | 3 | 3 | |
| Total | 69 | 0 | 66 | 112 | 13 | 10 | 135 | |
a rmPCR: real-time multiplex PCR.
b isolates typed correctly to the serogroup level compared with phenotypic Quellung reaction results.
c Negative: no amplification.
¥ all serotypes not covered by the rmPCR panel.
Of the 135 pneumococcal nasopharyngeal isolates that were sequetyped, 125 isolates yielded single amplicons of ~ 1,061 bp. A correct serotype/-group was determined in 112 (85%) of the 132 nasopharyngeal isolates. The partial wzh sequence of 2/3 Quellung 18C isolates did not match any of the pneumococcal wzh sequences in GenBank with >98% identity while the third was determined as 18B. The wzh PCR was negative for seven isolates of which three were serotype 25A/38, two were serotype 11A and two were serotype 19A, as confirmed by Quellung testing. Consistent misidentifications by sequetyping, occurring in more than one isolate, were observed for serotype 16F (two isolates sequetyped as 9V) and for serotype 17F (two isolates sequetyped as 33C).
Fig 2 shows the serotype distribution of the pneumococcal isolates, excluding duplicate isolates of the same serotype from the same infant. The most frequently identified serotypes were 11A (9 infants), 13 (8 infants), 15B, 15C (both 7 infants), 16F and 10A (both 6 infants). Of the 91 isolates (the total number of isolates when calculating each serotype only once per child), 22 (24%) were serotypes included in PCV13 while 69 (76%) serotypes were not.
Fig 2. Serotype distribution of nasopharyngeal pneumococcal isolates.
The figure includes serotypes detected from the Drakenstein Child Health Study, determined by Quellung reaction, excluding duplicate serotypes from the same infant. Blue = serotypes included in PCV13; Red = serotypes not included in PCV13. Green = non-typable isolates.
Next-generation sequencing
A total of 14.3 million paired-end sequence reads (2 x 250) were obtained for the four samples as shown in Table 4. The quality control steps used preserved the sequence number though reducing the sequence read length to 230 forward, and 120 reverse (230–120 fr) respectively.
Table 4. NGS data and assembly metrics.
| Isolate ID | Paired sequence reads | Number of contigs | N50 (Kb) | Draft genome size (Mb) | Sequencing coverage |
|---|---|---|---|---|---|
| 9v | 1 688 340 | 52 | 57.1 | 2.1 | 144x |
| 16f | 4 621 214 | 47 | 68.1 | 2.1 | 368x |
| 103347 | 2 226 252 | 62 | 77.3 | 2.1 | 184x |
| 103385 | 5 773 884 | 56 | 82.3 | 2.1 | 437x |
One of pneumococcal control strains and two DCHS strains were serotyped as 16F in Quellung, but mistyped as 9V by sequetyping. A comparison of the cps gene loci showed that the wzh sequence of all the three queried 16F strains was entirely 9V-like (Fig 3). This is in contrast to the rest of their cps loci: which in terms of structural gene organization as well as specific sequence of these genes were entirely 16F-like. Comparative genome analysis of the annotated gene structure showed a marked clustering for the other three queried 16F serotypes and were all significantly different from the 9V reference (Fig 4). MLST loci of these 16F strains showed a shared a 16F-like-MLST-type, except the third strain (103385) which had a unique glutamate dehydrogenase gene (gdh) allele.
Fig 3. Similarity of 16F-like capsular polysaccharide (cps) gene loci.
Sequences from pneumococci serotyped as 16F Quellung but sequetyped as 9V was compared to reference 9V (CR931648) and 16F (CR931668) cps sequences. Artemis Comparison Tool (ACT) was used to generate and view gene homology. The top lines represent the forward and reverse strand of a serotype 9v reference, the middle lines represent the queried 16F strain and the bottom lines shows the 16F reference. The portion of the wzh gene that is amplified by the sequetyping assay is shown by the blue rectangle. The clear blocks below the blue box shows regions were the genes that are not similar. BLASTN matches are shown as red bands between sequences, indicating the degree of similarity between the sequences.
Fig 4. Comparative genome analysis of pneumococcal serotypes 16F and 9V genetic background.
When the sequence identities of all four genomes were compared using RAST(Rapid Annotation using Subsystem Technology), the genome backbone of all three 16F (103347 and 103385 from this study and a 16F control strain) were mostly identical but divergent from 9V. The colour codes represent how close or divergent the genomes are. Therefore, similar genome backgrounds will have similar colours.
Discussion
To identify a rapid high throughput molecular serotyping assay, rmPCR was compared to sequetyping, in the first place using a panel of 40 control isolates. rmPCR is designed to detect and identify 21 serotypes including all serotypes/-groups in PCV13, all of which were included in our analysis. Concordance with Quellung was 100% (25/25) for those control isolates included in the rmPCR panel. Sequetyping is designed to identify up to 46 different serotypes/-groups, concordance with Quellung for the 40 control strains was 88% (29/33), with failure of sequetyping PCR for 7 strains.
Amongst the pneumococcal carriage isolates tested, the correct serotype/-group could be assigned to 52% (69/132) and 85% (112/132) by rmPCR and sequetyping respectively. Of these, 63/132 isolates were not included in the rmPCR panel. However, the sequetyping assay was theoretically expected to amplify and differentiate all except three isolates (Serotypes 25A and 38) into the correct serotype/-group with some strains giving ambiguous results (serotype 13/20, 17F/33C, and 11A/D/1818F). For those serotypes included in the rmPCR assay, there was good agreement between the results across all three assays. The high number of negative results from rmPCR amongst nasopharyngeal isolates was not surprising since this assay is likely to be less useful in areas where pneumococcal conjugate vaccines have been implemented resulting in serotype replacement which may arise as a result of either serotype unmasking or capsular switching. Data from the United States on invasive pneumococcal isolates showed a decline in serotypes included in the rmPCR assay from 92% (3812/4106) prior to PCV7 implementation to 79% (2939/3708) after PCV7 roll out and a further decrease to 74% (2581/3480) post PCV13 implementation (Unpublished US Active Bacterial Core surveillance data). Amongst our small cohort, non-vaccine serotypes 11A, 13, 15B, 15C, 16F and 10A were the most prevalent serotypes identified. Similarly, data from a number of other post PCV13 surveillance studies have reported serotypes 11A, 15A/B/C, 16, but also 22F, 21 and 34 as prevalent non-vaccine serotypes [45–51]. The original rmPCR protocol [21] did not make reference to any internal control in the assay set up. However, as part of our assay set-up and validation, we screened all the samples with a 16S rRNA PCR to check for inhibition and subjected all rmPCR negative samples to cpsA PCR to check the integrity of the capsulation locus, although not applicable for serotypes 14, 25, 35A and 38 [52].
The broad range of serotypes that are theoretically detectable by sequetyping is a major advantage of this technique. It is not clear why, in this study, amplification failed for 7/40 (18%) control strains. Interrogation of published gene sequences for these serotypes indicated that these serotypes should generate PCR products with the protocol used here [27]. PCR inhibition was excluded based on successful lytA PCR in all 7 strains. Interestingly, four of the nasopharyngeal cariage isolates that failed to amplify during sequetyping were serotypes for which a similar problem was encountered when sequetyping the control isolates (serotypes 11A and 19A). The remaining three sequetype-negative isolates were of serotype 25A/38 which were expected to be non-amplifiable because of absence of the reverse primer binding site in the wzd gene [6,27,53]. Sequetyping misidentified three control strains (for which Quellung and rmPCR were concordant). The sequence obtained from wzh amplicon of serotype 2 did not match any of the sequences in GenBank with >98% sequence identity. The original study describing sequetyping did not test this serotype although their insilico analysis had predicted that the primer sets should be able to amplify serotype 2 [27]. Misidentification of the serotype 46 isolate as 12A is explained by high relatedness between these serotypes as their cps gene clusters are almost identical [6]. Based on our observation of mistyping the 18C PCV13 serotype as 18B by sequetyping, it may be warranted to confirm all 18B results by Quellung. Misidentification of 17F isolates as 33C was predicted by the original sequetyping paper as these serotypes cannot be distinguished based on their wzh sequence [27].
We found one control strain and three nasopharyngeal strains that were serotyped as 16F by Quellung, but sequetyped as 9V. Even though the wzh sequences of the queried 16F was entirely 9V-like, the serotype specific wzy/wzx genes are entirely 16F-like. Based on analysis of the core genome, the 16F control strain was identified as sequence type (ST) 5326, one of the nasopharyngeal isolates was identified as ST4088, while the other nasopharyngeal isolates was a new ST, which was a single-locus variant of ST5326. These sequence types are all commonly associated with serotype 16F [44]. Therefore, our strains seem to be 16F strains in almost every sense, they only have a 9V-wzh gene. This structural difference is not expected to have occurred as a result of vaccination, because none of the currently used vaccine formulations include serotype 16F and the exchanged 9V gene does not result in a modified phenotype. In practice, in our setting, each isolate with a 9V sequetype result should be investigated further.
Both molecular assays are able to type many pneumococcal strains only to the serogroup level. Discrimination of individual serotypes within a serogroup may be important for more detailed assessment of carriage and vaccine effectiveness. When selecting a serotyping method, test characteristics other than accuracy may also be relevant. The sequetyping assay, which involves a single amplification step, is inexpensive compared with the rmPCR assay, which is labour intensive, includes many costly PCR probes and is constrained by the limited multiplexing options of real-time PCR. Interpretation of the sequetyping results is based on the publically available GenBank database. An advantage of this is its free accessibility, but the uncontrolled and changing nature of this database could be a risk for the assignment of serotypes. Our automated ‘sequetyper’ application makes analysis of the relevant sequence data for sequetyping rapid and simple. A significant disadvantage of sequetyping is that the targeted wzh gene is not serotype-specific and does itself not determine serotype—the results are inferred based on association. It is entirely feasible therefore (as we found here) that for specific serotypes and in particular populations of pneumococci that this association may not correctly predict serotype. The technique is therefore likely only useful for typing pneumococci from populations of pneumococci where such association has been confirmed using another typing technique. In our case this would mean confirming serotype for a smaller subset such as serotype 9V, 13, 20 and serogroup 33. The CDC Streptococcal laboratory has recently provided an update of a conventional multiplex PCR assay (not available at the time of this study) that utilises 41 serotype-specific primer sets to detect upto 70 different pneumococcal serotypes (http://www.cdc.gov/streplab/downloads/pcr-oligonucleotide-primers.pdf). The basis/methodology for this assay is similar to the rmPCR employed here although less costly. The benefits of deducing more than 70 serotypes by this assay needs to be weighed against sensitivity and risk of amplicon contamination.
In conclusion, sequetyping is a useful technique for large scale molecular serotyping of pneumococcal strains, particularly post-PCV introduction, because of the broad range of non-vaccine serotypes that can be detected, low cost and ease of use. Our results suggest the need for an extended and carefully curated database of serotype-specific sequence data, which will increase the accuracy and expand the serotype coverage of the sequetyping method. However, given the potential for gene exchange that could result in false assignment of serotype by sequetyping, it is necessary to confirm serotype assignment using a different method. This may still be cost-saving as it would involve, for example, testing only the specific serotype assigned by serotyping, using the Quellung method, in most instances. The rmPCR assay, ideally extended to include more serotypes is reliable but cost, time required to perform testing, and currently restricted serotype coverage may limit its widespread application for large epidemiological studies. In the future it is likely that WGS will be increasingly used as a tool for serotype inference. WGS has many advantages, in that additional information (such as multi-locus sequence type and antimicrobial resistance) can be inferred from the same dataset without additional testing, and that serotype can be definitively assigned. As sequence costs decline further, bioinformatic pipelines are increasingly automated and the technology is more widely available in low-resource settings it is likely that WGS will replace conventional typing tools for pneumococci.
Supporting Information
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Anne von Gottberg, Linda de Gouveia, Mushal Ali and the staff of the Centre for Respiratory Diseases and Meningitis (CRDM), National Institute for Communicable Diseases (NICD) of the National Health Laboratory Service for training, sharing of standard operating procedures, supplying control isolates and performing phenotypic serotyping. We thank the clinical research staff involved in the Drakenstein Child Lung Health Study for collection of samples and the children and parents for participating in the study. We further wish to thank Mamadou Kaba, Charmaine Barthus, Widaad Zemanay, Layla Hendricks, Nchimunya Hapeela, Whitney Barnett and the rest of the Drakenstein Child Lung Health Study team for their help and technical assistance. We thank the Western Health Department and the staff at Paarl hospital, Mbekweni and TC Newman clinics for their support of the study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded in part by grants from the Bill and Melinda Gates Foundation Global Health Grant (OPP1017641) and H3Africa (1U01 HG006961-01). Felix S. Dube is supported by the National Research Foundation of South Africa; S.P. van Mens received funding from the St Antonius Research Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wang H, Liddell CA, Coates MM, Mooney MD, Levitz CE, Schumacher AE, et al. (2014) Global, regional, and national levels of neonatal, infant, and under-5 mortality during 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bogaert D, De Groot R, Hermans PWM (2004) Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4: 144–154. [DOI] [PubMed] [Google Scholar]
- 3. Rudan I, O’Brien KL, Nair H, Liu L, Theodoratou E, Qazi S, et al. (2013) Epidemiology and etiology of childhood pneumonia in 2010: estimates of incidence, severe morbidity, mortality, underlying risk factors and causative pathogens for 192 countries. J Glob Health 3: 010401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coles CL, Sherchand JB, Khatry SK, Katz J, Leclerq SC, Mullany LC, et al. (2009) Nasopharyngeal carriage of S. pneumoniae among young children in rural Nepal. Trop Med Int Health 14: 1025–1033. 10.1111/j.1365-3156.2009.02331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antonio M, Dada-Adegbola H, Biney E, Awine T, O’Callaghan J, Pfluger V, et al. (2008) Molecular epidemiology of pneumococci obtained from Gambian children aged 2–29 months with invasive pneumococcal disease during a trial of a 9-valent pneumococcal conjugate vaccine. BMC Infect Dis 8: 81 10.1186/1471-2334-8-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, et al. (2006) Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2: 0262–0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calix JJ, Porambo RJ, Brady AM, Larson TR, Yother J, Abeygunwardana C, et al. (2012) Biochemical, Genetic, and Serological Characterization of Two Capsule Subtypes among Streptococcus pneumoniae Serotype 20 Strains: DISCOVERY OF A NEW PNEUMOCOCCAL SEROTYPE. J Biol Chem 287: 27885–27894. 10.1074/jbc.M112.380451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brugger SD, Frey P, Aebi S, Hinds J, Mühlemann K (2010) Multiple colonization with S. pneumoniae before and after introduction of the seven-valent conjugated pneumococcal polysaccharide vaccine. PLoS One 5: e11638 10.1371/journal.pone.0011638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rivera-Olivero IA, Blommaart M, Bogaert D, Hermans PWM, de Waard JH (2009) Multiplex PCR reveals a high rate of nasopharyngeal pneumococcal 7-valent conjugate vaccine serotypes co-colonizing indigenous Warao children in Venezuela. J Med Microbiol 58: 584–587. 10.1099/jmm.0.006726-0 [DOI] [PubMed] [Google Scholar]
- 10. Turner P, Hinds J, Turner C, Jankhot A, Gould K, Bentley SD, et al. (2011) Improved detection of nasopharyngeal cocolonization by multiple pneumococcal serotypes by use of latex agglutination or molecular serotyping by microarray. J Clin Microbiol 49: 1784–1789. 10.1128/JCM.00157-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Austrian R (1960) The quellung reaction, a neglected microbiologic technique. Mt Sinai J Med 43: 699–709. [PubMed] [Google Scholar]
- 12. Roca A, Hill PC, Townend J, Egere U, Antonio M, Bojang A, et al. (2011) Effects of community-wide vaccination with PCV-7 on pneumococcal nasopharyngeal carriage in the Gambia: a cluster-randomized trial. PLoS Med 8: e1001107 10.1371/journal.pmed.1001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bronsdon MA, O’Brien KL, Facklam RR, Whitney CG, Schwartz B, Schwartz B, et al. (2004) Immunoblot method to detect Streptococcus pneumoniae and identify multiple serotypes from nasopharyngeal secretions. J Clin Microbiol 42: 1596–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richter SS, Heilmann KP, Dohrn CL, Riahi F, Diekema DJ, Doern GV (2013) Evaluation of pneumococcal serotyping by multiplex PCR and quellung reactions. J Clin Microbiol 51: 4193–4195. 10.1128/JCM.01876-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brito DA, Ramirez M, de Lencastre H (2003) Serotyping Streptococcus pneumoniae by multiplex PCR. J Clin Microbiol 41: 2378–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pai R, Gertz RE, Beall B (2006) Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol 44: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Batt SL, Charalambous BM, McHugh TD, Martin S, Gillespie SH (2005) Novel PCR-restriction fragment length polymorphism method for determining serotypes or serogroups of Streptococcus pneumoniae isolates. J Clin Microbiol 43: 2656–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Selva L, del Amo E, Brotons P, Muñoz-Almagro C (2012) Rapid and easy identification of capsular serotypes of Streptococcus pneumoniae by use of fragment analysis by automated fluorescence-based capillary electrophoresis. J Clin Microbiol 50: 3451–3457. 10.1128/JCM.01368-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Massire C, Gertz RE, Svoboda P, Levert K, Reed MS, Pohl J, et al. (2012) Concurrent serotyping and genotyping of pneumococci by use of PCR and electrospray ionization mass spectrometry. J Clin Microbiol 50: 2018–2025. 10.1128/JCM.06735-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O’Sullivan MVN, Zhou F, Sintchenko V, Kong F, Gilbert GL (2011) Multiplex PCR and reverse line blot hybridization assay (mPCR/RLB). J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pimenta FC, Roundtree A, Soysal A, Bakir M, du Plessis M, Wolter N, et al. (2013) Sequential triplex real-time PCR assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J Clin Microbiol 51: 647–652. 10.1128/JCM.02927-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dhoubhadel BG, Yasunami M, Yoshida L-M, Thi HAN, Thi THV, Thi TAN, et al. (2014) A novel high-throughput method for molecular serotyping and serotype-specific quantification of Streptococcus pneumoniae using a nanofluidic real-time PCR system. J Med Microbiol 63: 528–539. 10.1099/jmm.0.071464-0 [DOI] [PubMed] [Google Scholar]
- 23. Azzari C, Moriondo M, Indolfi G, Cortimiglia M, Canessa C, Becciolini L, et al. (2010) Realtime PCR is more sensitive than multiplex PCR for diagnosis and serotyping in children with culture negative pneumococcal invasive disease. PLoS One 5: e9282 10.1371/journal.pone.0009282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blaschke AJ (2011) Interpreting assays for the detection of Streptococcus pneumoniae . Clin Infect Dis 52 Suppl 4: S331–S337. 10.1093/cid/cir048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moore CE, Sengduangphachanh A, Thaojaikong T, Sirisouk J, Foster D, Phetsouvanh R, et al. (2010) Enhanced determination of Streptococcus pneumoniae serotypes associated with invasive disease in Laos by using a real-time polymerase chain reaction serotyping assay with cerebrospinal fluid. Am J Trop Med Hyg 83: 451–457. 10.4269/ajtmh.2010.10-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Resti M, Moriondo M, Cortimiglia M, Indolfi G, Canessa C, Becciolini L, et al. (2010) Community-acquired bacteremic pneumococcal pneumonia in children: diagnosis and serotyping by real-time polymerase chain reaction using blood samples. Clin Infect Dis 51: 1042–1049. 10.1086/656579 [DOI] [PubMed] [Google Scholar]
- 27. Leung MH, Bryson K, Freystatter K, Pichon B, Edwards G, Charalambous BM, et al. (2012) Sequetyping: serotyping Streptococcus pneumoniae by a single PCR sequencing strategy. J Clin Microbiol 50: 2419–2427. 10.1128/JCM.06384-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jauneikaite E, Tocheva AS, Jefferies JMC, Gladstone RA, Faust SN, Christodoulides M, et al. (2015) Current methods for capsular typing of Streptococcus pneumoniae . J Microbiol Methods 113: 41–49. 10.1016/j.mimet.2015.03.006 [DOI] [PubMed] [Google Scholar]
- 29. Von Gottberg A, Cohen C, de Gouveia L, Meiring S, Quan V, Whitelaw A, et al. (2013) Epidemiology of invasive pneumococcal disease in the pre-conjugate vaccine era: South Africa, 2003–2008. [DOI] [PubMed] [Google Scholar]
- 30. Wasas AD, Huebner RE, De Blanche M, Klugman KP (1998) Long-term survival of Streptococcus pneumoniae at room temperature on Dorset egg medium. J Clin Microbiol 36: 1139–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Le Roux DM, Myer L, Nicol MP, Zar HJ (2015) Incidence and severity of childhood pneumonia in the first year of life in a South African birth cohort: the Drakenstein Child Health Study. Lancet Glob Heal 3: e95–e103. [DOI] [PubMed] [Google Scholar]
- 32. Satzke C, Turner P, Virolainen-Julkunen A, Adrian PV, Antonio M, Hare KM, et al. (2013) Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 32: 165–179. 10.1016/j.vaccine.2013.08.062 [DOI] [PubMed] [Google Scholar]
- 33. Dube FS, Kaba M, Whittaker E, Zar HJ, Nicol MP (2013) Detection of Streptococcus pneumoniae from Different Types of Nasopharyngeal Swabs in Children. PLoS One 8: e68097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winn WC, Allen SD, Janda WM, Koneman EW, Procop GW, Schreckenberger PC, et al. (2006) Gram-Positive Cocci Part II: Streptococci, Enterococci, and the “Streptococcus-like” Bacteria, p. 672–764 In: Konemans’s Color Atlas and Textbook of Diagnostic Microbiology. 6th ed Wilkins LW and, editor 672–764 p. [Google Scholar]
- 35. Leung MHY, Oriyo NM, Gillespie SH, Charalambous BM (2011) The adaptive potential during nasopharyngeal colonisation of Streptococcus pneumoniae . Infect Genet Evol 11: 1989–1995. 10.1016/j.meegid.2011.09.002 [DOI] [PubMed] [Google Scholar]
- 36. Park IH, Pritchard DG, Cartee R, Brandao A, Brandileone MCC, Nahm MH, et al. (2007) Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae . J Clin Microbiol 45: 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nunes MC, Jones SA, Groome MJ, Kuwanda L, Van Niekerk N, von Gottberg A, et al. (2014) Acquisition of Streptococcus pneumoniae in South African children vaccinated with 7-valent pneumococcal conjugate vaccine at 6, 14 and 40 weeks of age. Vaccine. [DOI] [PubMed] [Google Scholar]
- 38. Andrews S (2011) FastQC a quality-control tool for high-throughput sequence data. [Google Scholar]
- 39. Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7: e30619 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, et al. (2013) Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20: 714–737. 10.1089/cmb.2013.0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jolley KA, Maiden MCJ (2010) BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11: 595 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guevara M, Ezpeleta C, Gil-Setas A, Torroba L, Beristain X, Aguinaga A, et al. (2014) Reduced incidence of invasive pneumococcal disease after introduction of the 13-valent conjugate vaccine in Navarre, Spain, 2001–2013. Vaccine 32: 2553–2562. 10.1016/j.vaccine.2014.03.054 [DOI] [PubMed] [Google Scholar]
- 46. Olarte L, Hulten KG, Lamberth L, Mason EO, Kaplan SL (2014) Impact of the 13-Valent Pneumococcal Conjugate Vaccine on Chronic Sinusitis Associated with Streptococcus pneumoniae in Children. Pediatr Infect Dis J. [DOI] [PubMed] [Google Scholar]
- 47. Kaplan SL, Barson WJ, Lin PL, Romero JR, Bradley JS, Tan TQ, et al. (2013) Early trends for invasive pneumococcal infections in children after the introduction of the 13-valent pneumococcal conjugate vaccine. Pediatr Infect Dis J 32: 203–207. 10.1097/INF.0b013e318275614b [DOI] [PubMed] [Google Scholar]
- 48. Steens A, Bergsaker MAR, Aaberge IS, Rønning K, Vestrheim DF (2013) Prompt effect of replacing the 7-valent pneumococcal conjugate vaccine with the 13-valent vaccine on the epidemiology of invasive pneumococcal disease in Norway. Vaccine 31: 6232–6238. 10.1016/j.vaccine.2013.10.032 [DOI] [PubMed] [Google Scholar]
- 49. Van Hoek AJ, Sheppard CL, Andrews NJ, Waight PA, Slack MPE, Harrison TG, et al. (2014) Pneumococcal carriage in children and adults two years after introduction of the thirteen valent pneumococcal conjugate vaccine in England. Vaccine. [DOI] [PubMed] [Google Scholar]
- 50. Aguiar S, Brito M, Horacio A, Lopes J, Ramirez M, Melo-Cristino J, et al. (2014) Decreasing incidence and changes in serotype distribution of invasive pneumococcal disease in persons aged under 18 years since introduction of 10-valent and 13-valent conjugate vaccines in Portugal, July 2008 to June 2012. Euro Surveill 19. [DOI] [PubMed] [Google Scholar]
- 51. Cohen R, Levy C, Bingen E, Bechet S, Derkx V, Werner A, et al. (2012) Nasopharyngeal carriage of children 6 to 60 months during the implementation of the 13-valent pneumococcal conjugate vaccine. Arch Pediatr 19: 1132–1139. 10.1016/j.arcped.2012.07.013 [DOI] [PubMed] [Google Scholar]
- 52. Da Gloria Carvalho M, Pimenta FC, Jackson D, Roundtree A, Ahmad Y, et al. (2010) Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J Clin Microbiol 48: 1611–1618. 10.1128/JCM.02243-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mavroidi A, Aanensen DM, Godoy D, Skovsted IC, Kaltoft MS, et al. (2007) Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol 189: 7841–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.