Abstract
Ras GTPase is a molecular switch controlling a number of cellular pathways including growth, proliferation, differentiation, and apoptosis. Recent reports indicated that Ras undergoes dimerization at the membrane surface through protein-protein interactions. If firmly established this property of Ras would require profound reassessment of a large amount of published data and modification of the Ras signaling paradigm. One proposed mechanism of dimerization involves formation of salt bridges between the two GTPase domains (G domains) leading to formation of a compact dimer as observed in Ras crystal structures. In this work, we interrogated the intrinsic ability of Ras to self-associate in solution by creating conditions of high local concentration through irreversibly tethering the two G domains together at their unstructured C-terminal tails. We evaluated possible self-association in this inverted tandem conjugate via analysis of the time-domain fluorescence anisotropy and NMR chemical shift perturbations. We did not observe the increased rotational correlation time expected for the G domain dimer. Variation of the ionic strength (to modulate stability of the salt bridges) did not affect the rotational correlation time in the tandem further supporting independent rotational diffusion of two G domains. In a parallel line of experiments to detect and map weak self-association of the G domains, we analyzed NMR chemical shifts perturbations at a number of sites near the crystallographic dimer interface. The nearly complete lack of chemical shift perturbations in the tandem construct supported a simple model with the independent G domains repelled from each other by their overall negative charge. These results lead us to the conclusion that self-association of the G domains cannot be responsible for homodimerization of Ras reported in the literature.
Introduction
Small monomeric GTPases of Ras superfamily operate as molecular switches in multiple regulatory and signaling cascades (1–3). Ras and its closest homologs are involved in cell signaling cascades controlling mainly growth and proliferation but also involved in differentiation and apoptosis (4–6). Ras contains a soluble N-terminal GTPase domain (G domain) and the posttranslationally lipidated C-terminal tail. Addition of lipids localizes Ras to the membrane surface, which is a required condition for function (7–9).
In the current paradigm, Ras and its homologs function strictly as monomers in contrast to the mechanism of GTPases activated by dimerization (3,10,11). In solution, dimerization of the Ras G domain (lacking C-terminal tail and lipidation) has never been reported; yet, recent studies of the full-length Ras mimics attached to the membranes indicated that the dimerization might be possible at the membrane surface (12).
The earliest proposal that Ras functions at the membrane in an oligomeric form came from observations of radiation inactivation (target size analysis) (13). In the later cross-linking study by Inouye and others (14), Ras dimers were proposed to form on liposomes and facilitate activation of Ras effector Raf-1. Recently, Güldenhaupt analyzed 71 crystal structures of Ras G domains in the Protein Data Bank and pointed out that as many as 50 of them feature an extensive conserved crystal contact between two adjacent Ras molecules forming a conserved crystallographic dimer (15). Based on this observation, the authors proposed a structural model of N-Ras dimers at the membrane surface supported by their experimental Infra-Red polarization data and Förster resonance energy transfer measurements. Fig. S1 in the Supporting Material, top panel, illustrates these dimers as observed in Ras crystals. The extensive crystal contact between two G domains from the neighboring crystallographic units (red and blue in Fig. S1) involves helices 4 and 5 and β2–3 loop. Güldenhaupt and coworkers noted that the this protein-protein interface is mediated by a set of salt bridges involving residues D47, E49, H131, K135, D154, R161, and R164 in both G domains (see Fig. 5 in (15)). These residues are identical or conserved amino acids in all three human Ras isoforms (Fig. S1, bottom panel) indicating that this protein-protein interface is not isoform specific. Most recently, Lin reported observation of the dimeric species of H-Ras detected via the single-molecule tracking and step photobleaching analysis in fluorescence microscopy of supported lipid bilayers (16). Basing their argument on a limited mutational analysis (Y64A substitution) and relative independence of diffusion coefficients on the lipidation pattern (one versus two lipid anchors), the authors arrived at a conclusion that the dimer formation is mediated by protein interactions and does not require lipid anchor clustering (16).
In summary, one plausible interpretation of the reported data implies Ras dimerization through the direct contact of N-terminal G domains of two Ras molecules as exemplified by the crystallographic dimers. In both reports (15,16), the membrane is thought to play a passive role of a diffusional restraint—preventing translational diffusion in the direction normal to the membrane plane and increasing local concentration of the G domains. If this hypothesis is correct one should be able to stimulate formation of Ras dimers by merely sequestering G domains close enough in space to remove entropic penalty of translational diffusion. To test this hypothesis, we created tandem H-Ras constructs via joining two G domains with flexible linkers of variable lengths. We specifically assessed contribution of the salt-bridge formation to the dimerization (15) by varying the ionic strength in the samples. Measurements of rotational diffusion (via time-domain fluorescence anisotropy) and NMR chemical shift perturbation analysis enabled probing the intrinsic propensity of G domains for dimerization. Based on our observations we concluded that the G domains, when brought into close proximity, do not interact to any significant extent and do not form a dimeric species. Consequently, Ras dimerization reports by Güldenhaupt (15) and Lin (16) cannot be explained by tendency of Ras molecules to self-associate via their conserved G domains; instead, the Ras-Ras dimerization models must include direct interaction with the phospholipids (beyond mere tethering of the G domain to the surface of the membrane).
Materials and Methods
Protein constructs
The untagged genetic construct encoding for the wild-type H-Ras, residues 1–166, was engineered in our earlier work (17). The full-length gene of the wild-type H-Ras (a kind gift of Dr. Robert Deschenes, University of South Florida) was subcloned into the pET vector (EMD Millipore, Billerica, MA) for untagged expression. To create an expression construct for the truncated H-Ras ending with the cysteine 181 (residues 1–181), we introduced a stop codon in place of the methionine 182. An additional mutation, C118S, was introduced to remove the only exposed cysteine on the G domain, which may spuriously react with maleimido groups of the cross-linkers (see next section). The C118S mutation has been shown not to have any adverse effect on H-Ras function (18). All mutagenesis steps were performed using the QuikChange Site-Directed Mutagenesis Kit (Life Technologies, Grand Island, NY). The C118S H-Ras protein construct, residues 1–181, (Ras181 in the following text) and the wild-type H-Ras, residues 1–166, (Ras166 in the following text) were expressed and purified using a published protocol (17). The Ras181 protein was expressed in a minimal medium with uniform 15N-labeling to enable both fluorescence and NMR measurements on the protein originating from the same preparation.
Preparation of the inverted tandem conjugate
To prepare the inverted tandem conjugates of Ras181, we used cross-linking of C-terminal cysteines with bis-maleimide-polyethyleneglycols with two and 11 ethyleneglycol units (Ras-2-Ras and Ras-11-Ras in Fig. S2A). The cross-linkers were BM(PEG)2 (Thermo Fisher Scientific, Waltham, MA) and BM(PEG)11 (Conju-Probe, San Diego, CA). The BM(PEG)2 creates an ∼1.5 nm spacer, whereas the BM(PEG)11 ends are separated by ∼5 nm. The polyethyleneglycol spacer is hydrophilic and inherently flexible. The maleimide moiety irreversibly reacts with sulfhydryl groups at neutral pH with high specificity (19). To ensure that the cysteine 181 in Ras181 is fully reduced before the conjugation reactions, we added DTT to 5 mM and incubated the protein sample in the desiccator under vacuum for 30 min at room temperature. To completely remove DTT from the protein sample, we injected the protein solution into the XK16/40 size-exclusion column packed with Ultrogel Aca54 (Sigma, St. Louis, MO) equilibrated with the reaction buffer containing 20 mM HEPES pH 7.2, 1 mM MgCl2, and 100 mM NaCl. The protein sample eluted as two peaks with the maxima at 22 and 34 ml. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed that the second peak contains Ras181, whereas the first peak consisted of residual contaminating proteins. The Ras181 peak was concentrated to 46 μM using Centriprep YM-3 centrifugal filters (EMD Millipore). At this stage, ∼1/3 of the total protein was set aside for preparation of Ras181 samples for fluorescence and NMR spectroscopy. The rest of the Ras181 concentrate was mixed with the cross-linkers in 3:1 ratio (protein: BM(PEG)); the protein excess over the cross-linker ensured complete use of the cross-linker. Reactions with each cross-linker were stopped after 24 h at room temperature by the addition of β-mercaptoethanol. The inverted tandem conjugates, Ras181-BM(PEG)2-Ras181 (Ras-2-Ras) and Ras181-BM(PEG)11-Ras181 (Ras-11-Ras), were isolated from the reaction mixtures using the Ultrogel Aca54 size-exclusion column. The representative size-exclusion chromatography profile of the reaction mixture and its SDS-PAGE analysis are shown in Fig. S2, B and C; the shaded fractions were pooled and concentrated to obtain the purified Ras-2-Ras conjugate sample. Fig. S2D shows the final purity of all samples and confirms that contamination of conjugates with Ras181 monomers did not exceed 5%.
The molecular mass of the uniformly 15N-labeled Ras-2-Ras conjugate has been determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to be 41,700 ± 30 Da, which is consistent with the theoretical value of 41,668 Da. The 15N-labeled Ras-11-Ras conjugate exhibited a molecular mass of 42,229 Da with the theoretical value of 42,205 Da. The electrophoretic mobilities of Ras conjugates are anomalous—in agreement with the anomalous electrophoretic mobility of the Ras monomer due to its acidic pI of 5.0 (calculated using the ExPASy ProtParam tool (20,21)). The final yield of the Ras-2-Ras conjugate in this procedure was 3 mg; Ras-11-Ras yield was significantly lower. Protein concentrations were determined using Bradford assay (Thermo Fisher Scientific). The pure Ras181, Ras-2-Ras, and Ras-11-Ras conjugate preparations were dialyzed in a 3.5 kDa molecular weight cutoff (MWCO) dialysis bags against the working buffer containing 20 mM Hepes pH 7.2, 5 mM MgCl2, 1 mM DTT with 1.5 mM NaN3 added as an antibacterial agent.
Preparation of protein samples for fluorescence measurements
To prepare for the nucleotide exchange, the Ras181 and Ras conjugate samples were concentrated to 50 μM (in units of Ras monomers). The GDP nucleotide associated with the GTPase site in Ras was replaced with the (2′-(or-3′)-O-(N-methylanthraniloyl) guanosine 5′-diphosphate (mant-GDP; Life Technologies) using the EDTA-assisted method. In brief, to increase the rate of spontaneous nucleotide exchange (22), the magnesium ions in the protein samples were chelated with EDTA added to 6 mM along with the 0.8 mM mant-GDP and additional 10 mM DTT. The reaction mixtures were incubated for 1 h at room temperature, and further separated using the illustra NAP-5 Columns (GE Healthcare Bio-Sciences, Pittsburgh, PA) packed with G-25 size-exclusion resin and equilibrated with the working buffer. Protein concentration was determined by Bradford assay to be 0.34 mg/ml (16 μM) for Ras181, 0.26 mg/ml (12 μM monomers) for Ras-2-Ras, and 0.11 mg/ml (5 μM monomers) and 0.40 mg/ml (18 μM) for two separate preparations of Ras-11-Ras. Adjustment of salt content in both fluorescence and NMR samples (next section) was done by direct addition of calculated aliquots of 5 M NaCl solution. The effect of ionic strength on pH was tested by addition of an equivalent amount of NaCl to the buffer and found to be negligible: pH was reduced by as little as 0.03 pH units in 300 mM NaCl.
Preparation of protein samples for NMR measurements
The 10% D2O was added to protein solutions to allow for spectrometer locking, and protein samples were concentrated to ∼150 μM (Ras181) and 300 μM (Ras-2-Ras conjugate; in moles of Ras monomers); obtained quantity of Ras-11-Ras conjugate was not sufficient for NMR experiments. The protein samples were thoroughly degassed (1 h under vacuum in the desiccator) and loaded into Shigemi tubes.
Fluorescence spectroscopy experiments
The time-dependent polarization anisotropy measurements were performed using the QM40 QuantaMaster system manufactured by Photon Technology International (HORIBA Scientific, Edison, NJ) equipped with PicoMaster 1 time-correlated single-photon counting (TCSPC). The pulsed excitation was provided by 365 nm LED; the emission was detected at 440 nm with either 5 or 24 nm slit widths. Temperature of the sample was controlled using Peltier-based Turret 400 (Quantum Northwest, Shoreline, WA). The G-factor at 440 nm was measured using the steady-state Xenon lamp excitation. The polarized fluorescence decays were recorded with the motorized Glan-Thompson polarizers and the emission slits at 24 nm. The typical TCSPC counting rate was kept below 2%. The instrument response functions were recorded using a solution of a generic scatterer at 1–2% TCSPC counting rate. In all measurements, we used 30–60 min acquisition time per one emission polarizer orientation. Experiments were repeated multiple times for averaging. The polarized fluorescence decays were recorded for three sample temperatures (20, 25, and 37°C) and three concentrations of NaCl in solutions (0, 150, and 300 mM). The polarized decays of Ras166 were recorded at 20°C, 0 mM NaCl in the same buffer.
Fluorescence anisotropy decays were analyzed using AniFit software (kindly shared by Søren Preus; available from www.fluortools.com). The software performed global fitting of parallel and perpendicular components to optimize parameters in the intensity and anisotropy decay laws. In the first step of the optimization algorithm the isotropic fluorescence decay is reconstructed and fit with the multiexponential law. Next, the anisotropy decay law is assumed and the fitted isotropic decay is split into parallel and perpendicular components that are further reconvoluted with the instrument response function to simulate the polarized decays. Finally, these simulated decays are compared to the experimental decays obtained with the parallel and perpendicular orientations of the emission polarizer. To compensate for a small contribution of scattered light in the earliest times of the decay, the instrument response function is added to the simulated data with an adjustable coefficient. Multiple rounds of optimization resulted in the best-fit parameters of intensity and anisotropy decay laws along with the 95% confidence intervals estimated from the Jacobian matrix. The fits were performed with either single- or double-exponential laws of anisotropy decay and did not yield a significant difference in the slow rotational correlation times. Therefore, the single-exponential anisotropy decay with a contribution from scattered light was used for analysis of all polarized decays.
NMR spectroscopy experiments
Two-dimensional 15N-1H heteronuclear single-quantum correlation (HSQC) NMR spectra were obtained using an Agilent Technologies VNMR-S spectrometer (Santa Clara, CA) with the Cold Probe operating at the magnetic field strength of 14.1 Tesla corresponding to 600 MHz Larmor frequency of 1H nuclear spins. Spectral processing was performed using NMRPipe (23) and Sparky (24). The NMR signal assignment of the G-domain residues in Ras181 and Ras-2-Ras conjugate was achieved by analyzing spectral overlays with the HSQC spectra of the wild-type H-Ras sample, residues 1–166, complexed with GDP (recorded in the identical buffer conditions at 20°C and assigned in our earlier work (25)). Averaged chemical shift differences between Ras181 and Ras-Ras samples, Δav, were calculated according to the following equation (26):
where δ stands for chemical shifts of the 1H (δHN) or 15N (δN) in the amide group in Ras181 and Ras-2-Ras spectra; the factor of 5 is a weight reflecting greater relative sensitivity of nitrogen-15 chemical shift on the ppm scale (26).
Control of Ras-Ras sample stability
To confirm that the Ras conjugates did not hydrolyze in the course of experiments, we performed SDS-PAGE analysis of the protein samples after the measurements were completed. Fig. S3 shows unchanged molecular masses indicating that Ras conjugates remained intact.
Results
Experimental strategy to detect dimerization
The goal of this study was to evaluate probability of direct binding of two G domains to form a dimer under physiologically relevant solution conditions. Specific self-association of the G domains would have two major consequences for biophysical properties of the sample. First, rotational diffusion would slow down due to doubling of the mass and increased molecular dimensions. Second, formation of an extensive dimer interface would change environment of a significant number of atoms in the vicinity of such an interface. To detect these effects, we evaluated the rotational correlation time of different Ras constructs through the time-dependent fluorescence anisotropy measurements, and investigated the possible dimer interface by analyzing the NMR chemical shift perturbations.
Protein models
To sensitively isolate effects due to dimer formation from influences of overall protein structure and solution environment, we prepared a set of three protein models shown in Fig. S2A. The isolated G domain of H-Ras, residues 1–166, (Ras166) serves as a reference for rotational diffusion and chemical shifts of the G domain. Ras166 was reported to be strictly monomeric in solution in a number of studies (17,27–29). The elongated H-Ras construct, residues 1–181, (Fig. S2A, Ras181) ending at the site of the first lipid modification, Cys-181 (30), represents a portion of Ras polypeptide exposed to the cytosol in vivo. The Ras181 is identical to the H-Ras construct used for conjugation to the membranes by a number of research groups to mimic the native state of Ras (16,31,32). In our study, Ras181 serves to assess the effect of the unstructured C-terminal tail, residues 167–181, on the rotational diffusion and chemical shifts of the G domain (cysteine 181 remains reduced and unlipidated in the experiments).
Protein-protein interactions are enhanced at high concentrations due to the bimolecular nature of the binding reaction. To create high local concentration of the G domain, we tethered two Ras181 proteins through their Cys-181 side chains using the bis-maleimido cross-linkers BM(PEG)2 and BM(PEG)11 (producing Ras-2-Ras and Ras-11-Ras constructs, respectively). The resulting inverted-tandem conjugates restrict a pair of G domains in the close proximity of each other connected by a flexible unstructured chain comprising the residues 173–181 of two Ras181 molecules and the cross-linker (the helix 5 is known to extend to residue 172 (18,29,33)). The C-termini of the two G domains in the crystallographic dimer are oriented in a V-shaped fashion toward one side of the dimer (Fig. S2A). The Ras-Ras conjugates with both 1.5 and 5 nm linker lengths allow for the crystallographic dimer conformation as well as other possible interaction modes. The linkage of cysteine side chains with bis-maleimido cross-linkers is irreversible (19); in addition, the Ras conjugates were shown to remain intact throughout all the experiments (Fig. S3).
Fluorescence anisotropy analysis
Comparison of the rotational correlation time expected for a Ras dimer with the measured times for the Ras conjugates could help assess a degree of dimerization of G domains under conditions of a high local concentration. To improve sensitivity to dimerization, we used a series of solution conditions with increasing ionic strength to disrupt salt bridges at the crystallographic dimer interface (15) and thus favor the monomeric species. Our fluorescence measurements were performed at low protein concentrations (≤18 μM) ensuring that Ras166 and Ras181 served as truly monomeric controls. Selection of the fluorophore was also critical because most fluorophores are lipophilic and may drive weak self-association of the fluorophore-tagged molecules. Generally, to reduce the self-association propensity one needs a smaller fluorophore. In this study we used mant-GDP, the guanine diphosphate modified with N-methylanthraniloyl group (mant) at the position of 2′ or 3′ of ribose (34). Mant group is a small ultraviolet-range fluorophore (λex = 360 nm; λem = 440 nm) minimizing a chance of fluorophore-driven association and causing little perturbation to Ras functions (35). We performed all experiments with the diphosphate nucleotide because both dimerization reports from Lin (16) and Güldenhaupt (15) involved Ras-GDP, and the crystallographic dimers are observed in Ras complexed with either GDP or GTP mimics. Representative time-domain fluorescence decays are shown in Fig. S4 and all fitted rotational correlation times are given in the Table S1.
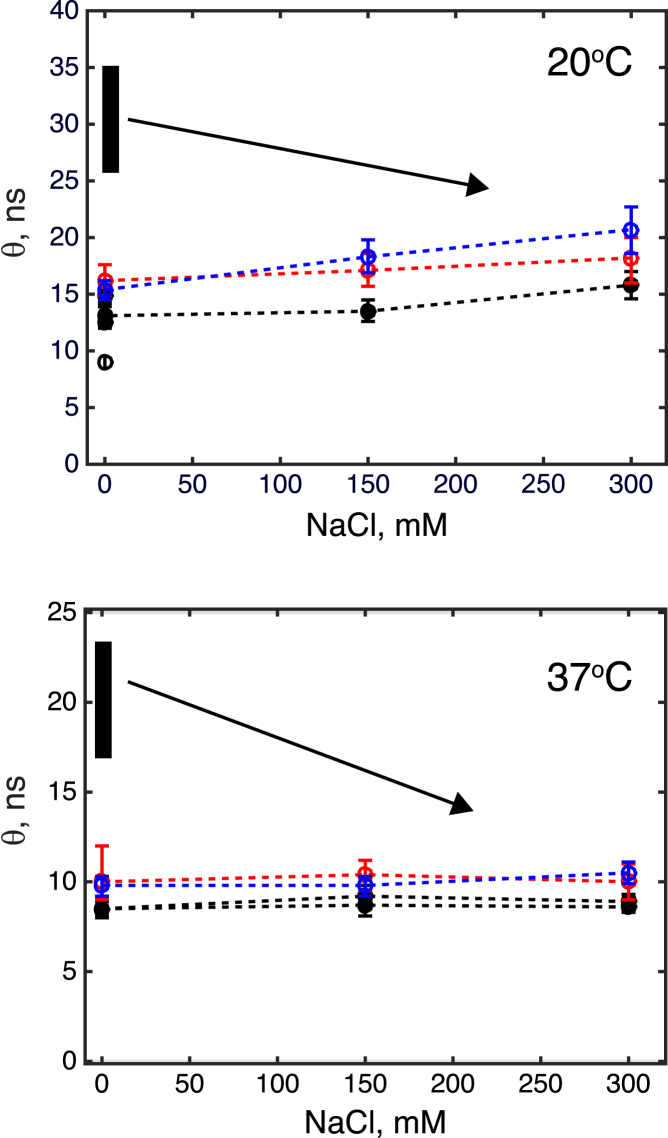
Fig. 1 shows dependence of the rotational correlation times of Ras181 and Ras conjugates on the NaCl concentration. The rotational correlation time of Ras181 is increased relatively to the isolated G domain Ras166 likely due to the extended helix 5 (18) (truncated in Ras166 construct) and unstructured C-terminal residues 173–181 producing additional hydrodynamic drag (36). With the addition of salt we observe little to no change in the rotational correlation time of Ras181 (black circles); a weak upward trend at 20°C may be attributed to the nonspecific effects such as increased viscosity of the solution and a larger size of the solvation shell of the G domain due to presence of ions. The joining of two Ras181 molecules to form Ras-2-Ras and Ras-11-Ras inverted-tandem conjugates (red and blue circles, respectively) results in a modest increase in the rotational correlation times. Increasing ionic strength affects Ras-2-Ras and Ras-11-Ras similar to Ras181.
Figure 1.
Effect of ionic strength on rotational correlation times of the G domains in Ras181 (black circles), Ras-2-Ras (red circles), and Ras-11-Ras (blue circles) at 20°C (top) and 37°C (bottom). Dashed lines connect data points for the same protein sample to guide the eye. The correlation time of a monomeric G domain Ras166 at low salt and 20°C is shown in the top panel with an open circle. Correlation times measured with two independent preparations of Ras181 at 37°C (black circles) are shown separately to demonstrate reproducibility of the measurements. Error bars represent 95% confidence intervals. Vertical black bars show the expected range of rotational correlation times for the Ras conjugates if the G domains formed tight dimers at low salt condition (see Supporting Material for details of this estimate). The arrows indicate anticipated reduction of the dimer correlation time upon increasing the ionic strength. To see this figure in color, go online.
The first test for the dimer formation is to compare the correlation times for Ras conjugates with the expected rotational correlation times of the tight dimer of Ras G domains. We estimated that the dimeric species should exhibit correlation times 2.0- to 2.7-fold greater than the values observed for the monomer (see Supporting Material). Considering the correlation time of Ras181 at 20°C of 13 ns at the lowest ionic strength (most favorable for the salt-bridge formation), the dimerized Ras-Ras conjugate should exhibit correlation times from 26 to 35 ns. Similar calculation for 37°C produces expected correlation times of 17 to 23 ns for the dimer. These expected ranges are shown as vertical black bars in Fig. 1—both far outside the error range of the corresponding measured values for Ras181 (solid black circles) at 0 mM NaCl.
The second test for the crystallographic dimer formation employs variation of the ionic strength because the dimerization interface (15) features multiple salt bridges. Long-range electrostatic interactions are strongest at a low ionic strength, whereas the high ionic strength disrupts salt bridges due to ionic shielding. Therefore, one would expect the rotational correlation time of the G domain in Ras conjugates to be greatest at the low salt concentration (due to enhanced stability of G domain dimers) and to gradually reduce with increasing ionic strength (due to a gradual shift of populations toward dissociated independently tumbling G domains in Ras conjugates). This anticipated trend is schematically illustrated by arrows in Fig. 1. Contrary to these expectations, we observe the variation of rotational correlation times of the G domains in Ras conjugates closely tracking the trends displayed by the monomeric Ras181 construct.
NMR chemical shift perturbations
NMR spectroscopy is particularly suitable for detecting and mapping weak interactions between proteins because binding events often lead to perturbation of resonance frequencies of multiple nuclear spins, which are simultaneously detected in the NMR spectrum (37). In addition, the isotopic labeling required for NMR detection is nonperturbing (relative to the effect of extrinsic labeling in fluorescence measurements) thus further reducing chances of artifacts.
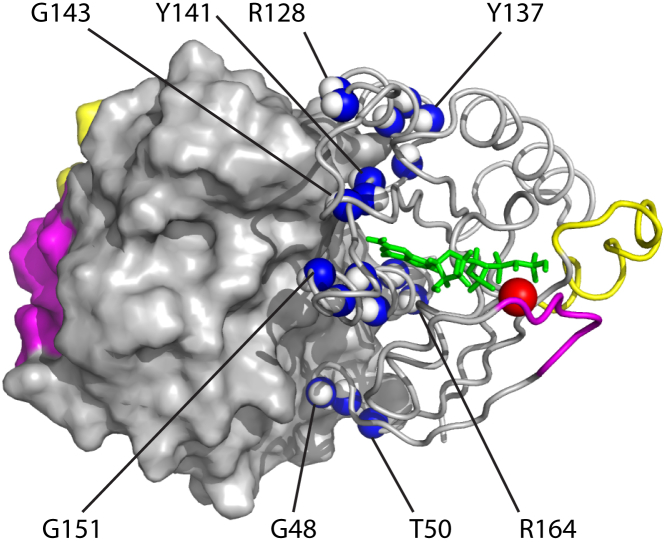
Examination of the dimer structure observed in Ras crystals indicates that many nuclear spins in the G domain are likely to have their magnetic environments significantly altered due to 1) desolvation of the protein surface upon the dimer formation, 2) establishing of the new van der Waals contacts, and 3) formation of electrostatic pairs (salt bridges) between the monomers. The amide groups of ∼26 amino acid residues lie within 5 Å from the surface of the neighboring G domain in the dimer structure or have their respective side-chains involved in the interaction. Out of these sensor residues, we were able to assign as many as 20 1H-15N amide HSQC peaks in the Ras181 and Ras-2-Ras NMR spectra. Localization of these amide groups in the Ras crystallographic dimer is shown in Fig. 2—with the dimer surface well covered by the NMR probes the dimerization event should be difficult to miss.
Figure 2.
Localization of the assigned 1H and 15N amide nuclear spins expected to experience chemical shift perturbation due to formation of the Ras dimer. Blue and white spheres indicate amide groups of the following residues: G48, E49, T50, S127, R128, R135, S136, Y137, I139, Y141, I142, E143, G151, D154, A155, Y157, T158, R161, and E162 (some are labeled for a visual guidance). One G domain is shown as a cartoon; another—with the van der Waals surface. Switch I, magenta; switch II, yellow; GDP, green sticks; and active site magnesium ion, a red sphere. To see this figure in color, go online.
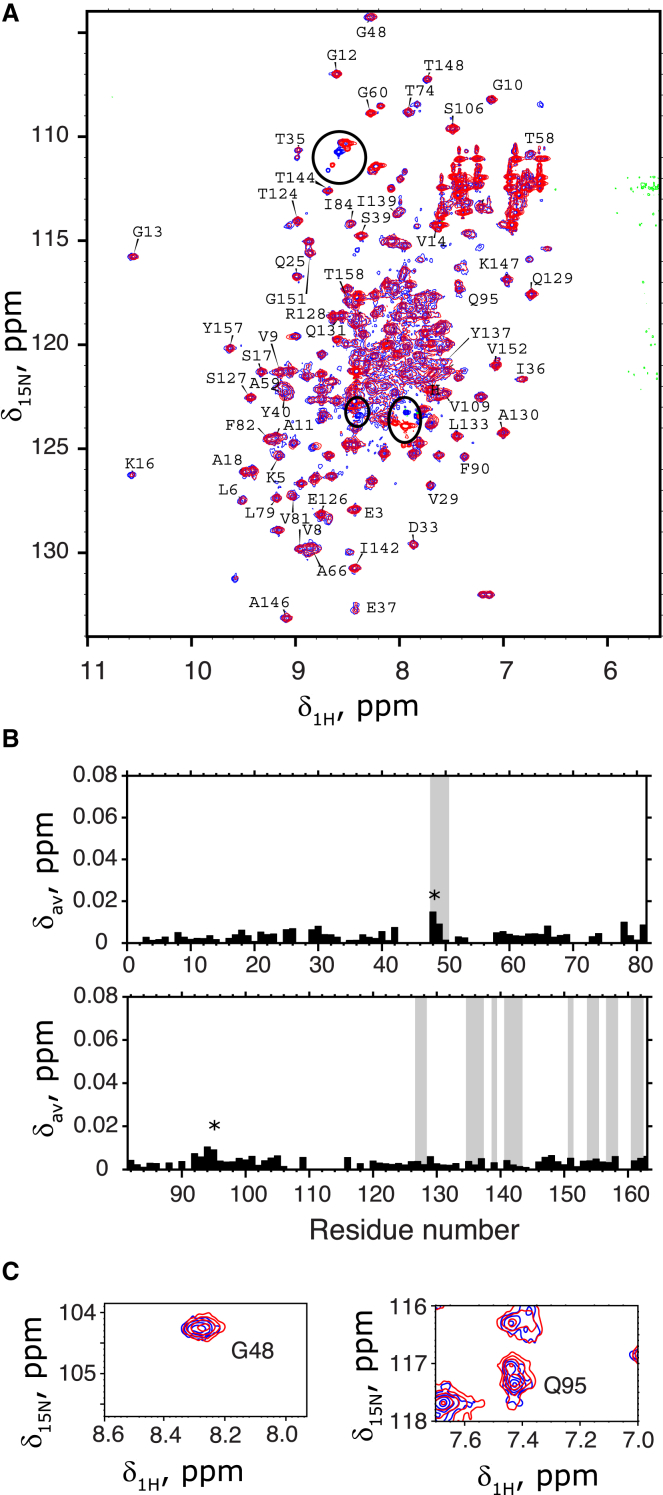
To measure chemical shifts of amide nitrogen-15 and proton nuclear spins, we recorded 1H-15N HSQC spectra of Ras181 and Ras-2-Ras at 20 and 25°C in the buffer conditions and ionic strengths matching the ones employed in our fluorescence anisotropy measurements. An overlay of Ras181 and Ras-2-Ras spectra at low ionic strength (to favor crystallographic dimer formation) is shown in panel A of Fig. 3, whereas panel B summarizes chemical shift differences for the residues of the G domain. A typical averaged chemical shift change due to association of polypeptides is expected to be in excess of 0.05 ppm up to 0.2–0.3 ppm (for example, see (26)); in our data, the maximum difference between Ras181 and Ras-2-Ras amounts to 0.015 at G48. Panel C illustrates two examples of peaks in the G domain experiencing the largest chemical shift change suggesting the source of these variations being the random noise in the line shape. Insignificant chemical shift differences between the G domain amide resonances in Ras181 and Ras-2-Ras indicate that no detectable dimerization takes place in the inverted tandem conjugate. Increasing the salt content to 300 mM did not induce any additional chemical shift differences between Ras181 and Ras-2-Ras spectra (Fig. S5), which could be anticipated if G domains in the Ras conjugate remained unassociated at all ionic strengths.
Figure 3.
Lack of significant chemical shift differences between amide resonances of the G domain in Ras181 and Ras-2-Ras. (A) Overlay of 15N-1H HSQC NMR spectra for Ras-2-Ras (red) onto the Ras181 (blue) at low ionic strength at 20°C. Peak assignments are shown for resonances of the G domain; labels in crowded regions were removed for clarity. Signals from the C-terminal peptide affected by the conjugation reaction are indicated by black ovals. (B) Averaged chemical shift differences, Δav, plotted versus the residue number in the G domain. The C-terminal extensions were not included in analysis. Residues at the dimer interface (their NH groups indicated by spheres in Fig. 2) are indicated as shaded areas. Intervals without black bars correspond to gaps in the assignment or unresolved spectral overlap. (C) Enlarged spectral views of the two peaks indicated with an asterisk in (B).To see this figure in color, go online.
Discussion
In this work, we tested a hypothesis that protein-protein interactions between G domains of Ras molecules are prominent enough to drive specific dimerization of Ras as suggested in recent reports (15,16). The key approach in our study was to create conditions of high local concentration of G domains to favor their self-association thus revealing their intrinsic dimerization propensity. In the native membrane-bound state, this propensity would be enhanced by the increased local concentration due to membrane attachment (38,39) and further modulated by direct protein-lipid interactions (preferred orientations (32)). In our reductionist approach, we focused on the self-association driven solely by the G domains (in absence of the membrane) because crystallographic dimers noted by Güldenhaupt (15) occur in absence of the membrane in the crystal; in addition, Ras dimerization reported by Lin (16) did not depend on the number of lipid anchors at the C-terminal peptide suggestive of a passive role of the membrane in the self-association process.
Biomolecular interactions typically include favorable enthalpic component (formation of noncovalent bonds) and unfavorable entropic component due to reduction of translational freedom of binding partners. We reasoned that tethering the two G domains using a flexible irreversible linker should reduce the entropic penalty of dimerization thus increasing the thermodynamic self-association constant. Flexibly tethering the binding partners together to reveal weak binding modes and trap transient protein-protein interactions is an established approach to produce stable protein-protein complexes and protein dimers suitable for detailed interrogation by structural and biophysical methods (for review, see (40)). For example, transient interaction between T cell receptor and peptide/major histocompatibility complex was effectively stabilized to allow crystallization by connecting the binding partners with the peptide linker (41); in another example, the native dimeric structure HIV protease was stabilized by preparation of a tethered dimer, which helped reveal a crucial contribution of dimerization to the enzyme function (42). In all cases, it is important that the linker provides for sufficient conformational freedom to the binding partners to assume productive mutual orientations.
To ensure that the tethering method in our study does not restrict available mutual orientations of the G domains, we cross-linked two molecules of Ras, residues 1–181, at their C-terminal cysteine 181 distanced from the folded G domain by approximately eight-residue-long C-terminal peptide, which lacks any stable structure (3,18). To make our experiments further less sensitive to the specific structure of the cross-linker, we created two versions of the Ras-Ras conjugate using flexible polyethylene glycols spacers of two different lengths: 1.5 and 5 nm (Fig. S2A). Comparative analysis of monomeric and conjugated Ras preparations established that the Ras G domain is not capable of forming a stable dimer via the direct protein-protein interaction in a temperature range from 20 to 37°C both in the absence and presence of electrolytes (up to 300 mM NaCl) at the cytosolic pH 7.2.
Consideration of protein electrostatics lends further support to the argument against the direct contact of G domains in Ras dimers reported in the literature. The overall charge of the G domain is negative at the cytosolic pH because the theoretical pI of Ras166 polypeptide is 4.9 (the overall charge will become even more negative if the charges of the GDP phosphates are considered). Therefore, the translational diffusion of G domains may be significantly biased by their mutual repulsion thus favoring the monomeric state. Similar electrostatic repulsion is expected to keep the G domain at a distance from the membrane surface because the inner leaflet of the plasma membrane is also strongly negatively charged (43).
Mazhab-Jafari and coworkers recently investigated Ras homolog Rheb GTPase chemically linked to the membrane surface (lipid nanodisc) via C-terminal cysteine side chain and reported that Rheb retained the same three-dimensional structure and only transiently associated with the nanodisc surface (32). In their paramagnetic relaxation enhancement NMR experiments the authors identified that there are two preferred orientations of the G domain of Rheb at a time when it contacts the lipid surface, and proposed that these preferred orientations may enhance specific protein-protein interactions. Yet, Mazhab-Jafari and coworkers stressed that these pre-oriented membrane-bound states were minor conformations of the protein-nanodisc complex with a majority of the protein exhibiting a high degree of freedom (dynamics) despite being tethered to the membrane (32). These observations are in line with the report by Werkmüller and coworkers that rotational diffusion of N- and K-Ras was only modestly retarded by tethering these proteins to large liposomes (44).
It is possible that preferred orientations of Ras induced by the membrane proximity enhance the hypothetical weak propensity of G domains to form dimers. However, thermodynamics requires such preoriented states to constitute the majority of the protein population to contribute significant Gibbs energy to the further binding process. In reports from both Mazhab-Jafari and Werkmuller the nearly free diffusion of G domains near the membrane surface was consistent with a small minority of G domains being in direct contact with the membrane implying that their contribution to the overall sample properties was respectively small.
In addition to this analysis, we would like to argue that although preferred orientations may certainly improve productive interactions, capability for noncovalent bonding between two G domains must exist in the first place. As such it should be detectable in other settings, for example, by creating high local concentration in the absence of the membrane surface. If the interaction remains only transient even in such a favorable thermodynamic condition one could rightfully doubt biological significance of this interaction mode.
Concluding our analysis we need to note that there are other studies of Ras tethered to the membrane surfaces that did not require dimerization to explain experimental observations. One example is the work published nearly at the same time with Güldenhaupt’s dimerization report (15) where Kapoor and coworkers (45) investigated N-Ras (and K-Ras) tethered to the supported phospholipid bilayers. The authors, however, did not detect the same signatures in the infrared spectra that made Güldenhaupt propose dimerization in their samples. It is important to note that, in the previously mentioned studies, Ras constructs were anchored or chemically tethered to the membranes of variable lipid composition, which may be one of the sources of reported differences.
In summary, we demonstrated that the protein-protein interactions between H-Ras G domains alone cannot stabilize G domain dimers and the crystallographic dimer does not form in solutions. High sequence identity between H-, N-, and K-Ras G domains allows for extending these conclusions to all three human Ras isoforms. We suggest that in all cases in vivo and in vitro where dimerization of Ras is suspected one should focus on other possible causes such as the direct interaction of G domains with the lipid bilayer, involvement of lipid anchors of the C-terminal peptides, or membrane-induced conformational changes in the G-domain structure.
Author Contributions
E.A.K., A.R.G., and E.L.K. designed and performed research; A.R.G., and E.L.K. performed analysis; E.L.K. wrote the article.
Acknowledgments
The authors are grateful to Dr. Alemayehu Gorfe and Dr. Qadir Timerghazin for helpful discussions.
E.L.K. acknowledges Committee on Research (COR) Summer Faculty Fellowship 2014 from Marquette University.
Editor: Elizabeth Rhoades.
Footnotes
Elizaveta A. Kovrigina and Azamat R. Galiakhmetov contributed equally to this work.
Supporting Discussion, six figures, and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00726-2.
Supporting Citations
References (46–52) appear in the Supporting Material.
Supporting Material
References
- 1.Takai Y., Sasaki T., Matozaki T. Small GTP-binding proteins. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 2.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci. STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wittinghofer A., Vetter I.R. Structure-function relationships of the G domain, a canonical switch motif. In: Kornberg R.D., Raetz C.R.H., Rothman J.E., Thorner J.W., editors. Annual Review of Biochemistry. Annual Reviews; Palo Alto, CA: 2011. pp. 943–971. [DOI] [PubMed] [Google Scholar]
- 4.Lowy D.R., Willumsen B.M. Function and regulation of ras. Annu. Rev. Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 5.Vojtek A.B., Der C.J. Increasing complexity of the Ras signaling pathway. J. Biol. Chem. 1998;273:19925–19928. doi: 10.1074/jbc.273.32.19925. [DOI] [PubMed] [Google Scholar]
- 6.Feig L.A., Buchsbaum R.J. Cell signaling: life or death decisions of ras proteins. Curr. Biol. 2002;12:R259–R261. doi: 10.1016/s0960-9822(02)00787-x. [DOI] [PubMed] [Google Scholar]
- 7.Groves J.T., Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prior I.A., Hancock J.F. Ras trafficking, localization and compartmentalized signalling. Semin. Cell Dev. Biol. 2012;23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahearn I.M., Haigis K., Philips M.R. Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 2012;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böhme S., Meyer S., Klare J.P. Stabilization of G domain conformations in the tRNA-modifying MnmE-GidA complex observed with double electron electron resonance spectroscopy. J. Biol. Chem. 2010;285:16991–17000. doi: 10.1074/jbc.M109.096131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zent E., Wittinghofer A. Human septin isoforms and the GDP-GTP cycle. Biol. Chem. 2014;395:169–180. doi: 10.1515/hsz-2013-0268. [DOI] [PubMed] [Google Scholar]
- 12.Santos E. Dimerization opens new avenues into Ras signaling research. Sci. Signal. 2014;7:pe12. doi: 10.1126/scisignal.2005318. [DOI] [PubMed] [Google Scholar]
- 13.Santos E., Nebreda A.R., Kempner E.S. Oligomeric structure of p21 ras proteins as determined by radiation inactivation. J. Biol. Chem. 1988;263:9853–9858. [PubMed] [Google Scholar]
- 14.Inouye K., Mizutani S., Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem. 2000;275:3737–3740. doi: 10.1074/jbc.275.6.3737. [DOI] [PubMed] [Google Scholar]
- 15.Güldenhaupt J., Rudack T., Gerwert K. N-Ras forms dimers at POPC membranes. Biophys. J. 2012;103:1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin W.-C., Iversen L., Groves J.T. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. USA. 2014;111:2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connor C., Kovrigin E.L. Global conformational dynamics in ras. Biochemistry. 2008;47:10244–10246. doi: 10.1021/bi801076c. [DOI] [PubMed] [Google Scholar]
- 18.Thapar R., Williams J.G., Campbell S.L. NMR characterization of full-length farnesylated and non-farnesylated H-Ras and its implications for Raf activation. J. Mol. Biol. 2004;343:1391–1408. doi: 10.1016/j.jmb.2004.08.106. [DOI] [PubMed] [Google Scholar]
- 19.Smyth D.G., Blumenfeld O.O., Konigsberg W. Reactions of N-ethylmaleimide with peptides and amino acids. Biochem. J. 1964;91:589–595. doi: 10.1042/bj0910589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasteiger E., Gattiker A., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasteiger E., Hoogland C., Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press; NY: 2005. pp. 571–607. [Google Scholar]
- 22.Hall A., Self A.J. The effect of Mg2+ on the guanine nucleotide exchange rate of p21N-ras. J. Biol. Chem. 1986;261:10963–10965. [PubMed] [Google Scholar]
- 23.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 24.Goddard, T. D., and D. G. Kneller. SPARKY 3; http://www.cgl.ucsf.edu/home/sparky/.
- 25.O’Connor C., Kovrigin E.L. Characterization of the second ion-binding site in the G domain of H-Ras. Biochemistry. 2012;51:9638–9646. doi: 10.1021/bi301304g. [DOI] [PubMed] [Google Scholar]
- 26.Grzesiek S., Stahl S.J., Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 27.Campbell-Burk S.L., Carpenter J.W. Refolding and purification of ras proteins. Methods Enzymol. 1995;255:3–13. doi: 10.1016/s0076-6879(95)55003-8. [DOI] [PubMed] [Google Scholar]
- 28.Kraulis P.J., Domaille P.J., Laue E.D. Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- 29.Ito Y., Yamasaki K., Miyazawa T. Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry. 1997;36:9109–9119. doi: 10.1021/bi970296u. [DOI] [PubMed] [Google Scholar]
- 30.Hancock J.F., Magee A.I., Marshall C.J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 31.Gureasko J., Galush W.J., Kuriyan J. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 2008;15:452–461. doi: 10.1038/nsmb.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazhab-Jafari M.T., Marshall C.B., Ikura M. Membrane-dependent modulation of the mTOR activator Rheb: NMR observations of a GTPase tethered to a lipid-bilayer nanodisc. J. Am. Chem. Soc. 2013;135:3367–3370. doi: 10.1021/ja312508w. [DOI] [PubMed] [Google Scholar]
- 33.Muto Y., Yamasaki K., Yokoyama S. Sequence-specific 1H and 15N resonance assignments and secondary structure of GDP-bound human c-Ha-Ras protein in solution. J. Biomol. NMR. 1993;3:165–184. doi: 10.1007/BF00178260. [DOI] [PubMed] [Google Scholar]
- 34.Lenzen C., Cool R.H., Hall A. Analysis of intrinsic and CDC25-stimulated guanine nucleotide exchange of p21ras-nucleotide complexes by fluorescence measurements. Methods Enzymol. 1995;255:95–109. doi: 10.1016/s0076-6879(95)55012-7. [DOI] [PubMed] [Google Scholar]
- 35.Ahmadian M.R., Wittinghofer A., Herrmann C. Fluorescence methods in the study of small GTP-binding proteins. Methods Mol. Biol. 2002;189:45–63. doi: 10.1385/1-59259-281-3:045. [DOI] [PubMed] [Google Scholar]
- 36.Hazlett T.L., Moore K.J., Eccleston J.F. Solution dynamics of p21ras proteins bound with fluorescent nucleotides: a time-resolved fluorescence study. Biochemistry. 1993;32:13575–13583. doi: 10.1021/bi00212a025. [DOI] [PubMed] [Google Scholar]
- 37.Wemmer D.E., Williams P.G. Use of nuclear magnetic resonance in probing ligand-macromolecule interactions. Methods Enzymol. 1994;239:739–767. doi: 10.1016/s0076-6879(94)39028-2. [DOI] [PubMed] [Google Scholar]
- 38.McLaughlin S., Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem. Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 39.Kuriyan J., Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- 40.Reddy Chichili V.P., Kumar V., Sivaraman J. Linkers in the structural biology of protein–protein interactions. Protein Sci. 2013;22:153–167. doi: 10.1002/pro.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinherz E.L., Tan K., Wang J. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y.S., Yin F.H., Kettner C.A. Stability and activity of human immunodeficiency virus protease: comparison of the natural dimer with a homologous, single-chain tethered dimer. Proc. Natl. Acad. Sci. USA. 1990;87:9660–9664. doi: 10.1073/pnas.87.24.9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stace C.L., Ktistakis N.T. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta. 2006;1761:913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Werkmüller A., Triola G., Winter R. Rotational and translational dynamics of ras proteins upon binding to model membrane systems. ChemPhysChem. 2013;14:3698–3705. doi: 10.1002/cphc.201300617. [DOI] [PubMed] [Google Scholar]
- 45.Kapoor S., Weise K., Winter R. The role of G-domain orientation and nucleotide state on the Ras isoform-specific membrane interaction. Eur. Biophys. J. 2012;41:801–813. doi: 10.1007/s00249-012-0841-5. [DOI] [PubMed] [Google Scholar]
- 46.Schrodinger, LLC. 2010. The PyMOL Molecular Graphics System, Version 1.3r1.
- 47.Larkin M.A., Blackshields G., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 48.Kawski A. Fluorescence anisotropy: theory and applications of rotational depolarization. Crit. Rev. Anal. Chem. 1993;23:459–529. [Google Scholar]
- 49.Lakowicz J.R. Springer; NY: 2010. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 50.Chuang T.J., Eisenthal K.B. Theory of fluorescence depolarization by anisotropic rotational diffusion. J. Chem. Phys. 1972;57:5094–5097. [Google Scholar]
- 51.Small E.W., Isenberg I. Hydrodynamic properties of a rigid molecule: rotational and linear diffusion and fluorescence anisotropy. Biopolymers. 1977;16:1907–1928. doi: 10.1002/bip.1977.360160907. [DOI] [PubMed] [Google Scholar]
- 52.Lide D.R., editor. CRC Handbook of Chemistry and Physics. Taylor and Francis; New York: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.