Summary
There is an urgent need to understand the molecular mechanisms by which sustained exposure to low-dose environmental chemical mixtures promotes carcinogenesis. This Halifax Project review specifically examines the effects of environmental chemicals on the cancer hallmark of evading growth suppression.
Abstract
As part of the Halifax Project, this review brings attention to the potential effects of environmental chemicals on important molecular and cellular regulators of the cancer hallmark of evading growth suppression. Specifically, we review the mechanisms by which cancer cells escape the growth-inhibitory signals of p53, retinoblastoma protein, transforming growth factor-beta, gap junctions and contact inhibition. We discuss the effects of selected environmental chemicals on these mechanisms of growth inhibition and cross-reference the effects of these chemicals in other classical cancer hallmarks.
Introduction
The normal cell cycle contains multiple checkpoints and molecular pathways that suppress cellular proliferation and growth in response to DNA-damaging agents or harmful stimuli. A hallmark of cancer is the ability to evade these growth-inhibitory signals. This loss of response to growth inhibition can occur as a result of sustained exposure to specific environmental chemicals. In this review, we will examine key molecular and cellular mechanisms through which cancers evade growth suppression. Specifically, we will examine the mechanisms by which cancer cells evade the growth-inhibitory signals of p53, the retinoblastoma protein, transforming growth factor-beta (TGF-β), gap junctions and contact inhibition.
We have organized the review article to discuss each mechanism of growth inhibition individually. At the end of each section, we discuss selected environmental chemicals that have been reported to disrupt that specific mechanism of growth inhibition. Thus, our manuscript contains ‘mini-reviews’ of specific mechanisms of growth suppression and the influence of selected environmental chemicals on these mechanisms. We then examine if the selected environmental chemicals affect multiple mechanisms within the hallmark of escaping growth inhibition. Finally, we review whether these chemicals affect other classical hallmarks of cancer. The overall goal is to understand the molecular mechanisms through which cancer cells evade growth inhibition in response to specific environmental chemicals.
P53
The p53 tumor suppressor is a tetrameric nuclear transcription factor (1,2). Almost 50% of all cancers harbor p53 mutations, although the frequency varies with tumor type. For example, although p53 is mutated in up to 70% of lung cancers, the frequency drops to around 10% for leukemias (http://p53.free.fr/Database/p53_cancer_db.html). Ninety percent of somatic p53 mutations occur in the DNA-binding domain (http://p53.iarc.fr/TP53SomaticMutations.aspx). This prevents p53-dependent transactivation, as the mutant can act in a dominant-negative manner by oligomerization with wild-type p53 (3). Genetic evidence shows that p53 activity is controlled by two homologous negative regulators, MDM2 and MDM4 (also known as MDMX). Both MDM proteins have amino-terminal hydrophobic pockets that bind to an alpha-helical region of p53, leading to inhibition of p53 transcription factor functions (4,5). MDM2 also has a carboxy-terminal RING domain that recruits E2 ubiquitin-conjugating enzymes. Thus, MDM2 has intrinsic ubiquitin-ligase activity that leads to the degradation of p53. Although MDMX also has the C-terminal RING domain, it cannot promote ubiquitination or degradation of p53 on its own. However, there is genetic evidence that the RING domains of both MDMX and MDM2 are required for the in vivo suppression of p53 activity (6,7). In addition, MDMX can stimulate the ubiquitin-ligase activity of MDM2 through RING-RING-mediated MDM2/MDMX hetero-oligomerization (8). Thus, a model has been proposed in which the MDM2/MDMX complex serves as an ‘optimal’ p53 inhibitor in certain contexts. Importantly, both MDM2 and MDMX are bonafide oncogenes, and clinical trials are testing small molecule antagonists of their interaction with p53 (9).
Acquisition of p53 mutations (somatic or inherited) and overexpression of MDM2/MDMX are the most direct mechanisms by which the p53 pathway is disabled in cancers (10). However, other mechanisms that limit p53 activity have been described. For example, p53 protein stability is regulated by specific kinase pathways, including the Ataxia-Telangiectasia-mutated (ATM)-checkpoint kinase 2 (Chk2) pathway. This pathway and the Ataxia-Telangiectasia-related (ATR)-Checkpoint kinase 1 (Chk1) pathway are essential for repairing DNA double-strand breaks (11). In normal cells, the presence of double-strand breaks induces phosphorylation of ATM (12,13), which then activates the downstream effector Chk2. Activated Chk2 regulates the S/M checkpoint by phosphorylating and activating multiple substrates, including p53 (11,14). ATM is recognized as an important regulator of p53 checkpoint function, as ATM-deficient cells fail to induce or stabilize p53 protein levels in response to some types of DNA damage, such as ionizing radiation. This may be particularly relevant to environmental carcinogens, as ATM-deficient lymphoblasts exposed to the chemical diepoxybutane are unable to stabilize p53 protein levels to the same extent as cells with wild-type ATM (15). Thus, although many p53-independent functions exist for ATM, disruption of p53 growth-inhibitory checkpoint function is a major mechanism by which ATM-deficient cancer cells fail to respond to DNA-damaging agents, including specific environmental pollutants.
Similar to ATM-Chk2, the liver kinase B1 (LKB1, STK11) gene encodes a 48-kDa kinase that phosphorylates multiple substrates, including p53. A first-hit germ-line mutation in LKB1 occurs in patients born with Peutz–Jeghers Syndrome, which is an autosomal-dominant disorder that confers a 93% risk of developing cancers (16). LKB1 is considered a haploinsufficient tumor suppressor gene, such that additional oncogenic events (e.g., loss of Pten or p53 or activation of K-ras) cooperate with LKB1 loss to promote cancer; biallelic inactivation of LKB1 has also been reported in sporadic cancers (17–19). Similar to ATM-Chk2, LKB1 can promote cell cycle arrest through p53-dependent and independent mechanisms. In particular, LKB1 has been shown to interact with, phosphorylate, and stabilize nuclear p53 (20). P53 phosphorylation by LKB1 occurs on serine 15, which may be mediated by the LKB1 substrate 5′ adenosine monophosphate-activated protein kinase (AMPK), and on C-terminal serine 392 (20). The importance of LKB1-mediated p53 phosphorylation to growth suppression is supported by the finding that an LKB1 kinase-dead mutant no longer phosphorylates p53 and is unable to promote G1 arrest in contrast to wild-type LKB1. In addition, mutant p53 constructs lacking either one of the LKB1 phosphorylation sites are unable to cooperate with wild-type LKB1 to induce G1 arrest (20). P53-dependent growth arrest by LKB1 may be mediated in part by recruitment of the LKB1-p53 complex to the p21 promoter and increased expression of the p21 cdk inhibitor (20). Intriguingly, LKB1 catalytically deficient mutants are not only unable to mediate growth inhibition, but they actually display gain-of-function oncogenic properties. LKB1 mutants are recruited to the CCND1 promoter, resulting in increased cyclin D1 expression and cell cycle progression (21). LKB1 oncogenic mutants are also recruited to the c-myc promoter, increasing expression of this oncogene (22). The oncogenic roles of LKB1 mutants may be particularly relevant in lung cancer cells of patients who are exposed to tobacco smoke, as LKB1 mutations increase susceptibility to carcinogen-induced lung tumors (23). In addition, exposure to cigarette smoke can downregulate levels of LKB1 in lung cancer cells and normal human bronchiolar epithelial cells (24). Thus, environmental pollutants may indirectly alter p53 phosphorylation, stabilization and function by deregulating upstream kinases, such as Chk2 and LKB1.
In addition to effects on upstream regulators of p53, cigarette smoke can directly affect p53. The incidence of p53 mutations in lung cancers of smokers is increased compared to those in nonsmokers (25). There are many carcinogens present in cigarette smoke. One such carcinogen is benzo(a)pyrene, which is specifically implicated in the mutation of p53 (26) (Table 1). Interestingly, benzo(a)pyrene affects multiple levels of checkpoint regulation, not exclusively p53 function. For example, the ATM checkpoint kinase is activated by benzo(a)pyrene in premalignant or malignant esophageal cancers (27). Thus, it is feasible that ATM deficiency may block critical checkpoint activation in response to environmental contaminants, such as tobacco smoke, further increasing the risk of carcinogenesis. Another carcinogen that is associated with p53 mutation is the food contaminant aflatoxin B1, which promotes hepatocellular carcinogenesis (28,29). Because of the clinical and preclinical studies that link benzo(a)pyrene and aflatoxin B1 with p53 mutations, regulatory agencies now recognize these compounds as human carcinogens and ensure that people are protected from or made aware of the risks associated with these environmental chemicals.
Table 1.
Key molecular and cellular mediators of growth suppression and selected environmental chemicals that potentially disrupt the functions of these growth inhibitors
| Molecular/cellular target | Potential environmental chemical disruptors | Effects that chemicals may have on the molecular targeta |
|---|---|---|
| p53 | Benzo(a)pyrene, bisphenol A, DDT, folpet, aflatoxin, chlorothalonil, mancozeb | Downregulate p53 expression, induce MDM2 expression and p53 degradation, direct mutation of p53 |
| Retinoblastoma | Benzo(a)pyrene, bisphenol A, arsenic [As(III)], DDT, radon, butadiene | Loss of heterozygosity of RB1, hyperphosphorylation of Rb via increased cdk activity, increased cyclin D1 expression or loss of INK4a |
| Transforming growth factor-beta (TGF-β) | Arsenic [sodium arsenite; NaAsO2, As(III)] | Activation of upstream growth factor signals that block TGFβ signaling |
| LKB1 | Cigarette smoke | LKB1 mutation, reduced LKB1 expression |
| Gap junctions (connexins) | Bisphenol A, DDT, polychlorinated biphenyls/ aryl hydrocarbon receptor ligands (e.g. TCDD), cigarette smoke, polycyclic aromatic hydrocarbons | Down-regulate expression of specific or multiple connexins, reduce GJIC |
| Contact inhibition | Polychlorinated biphenyls/aryl hydrocarbon receptor ligands (e.g. TCDD), DDT | Disrupt contact inhibition, prevent contact normalization |
We searched PubMed to identify potential environmental chemicals that disrupt specific growth-inhibitory mechanisms. We combined the name of the target listed in the far-left column with the term ‘environmental carcinogen’ to develop a list of proposed chemical disruptors. The specific effects of the chemicals on the molecular or cellular targets and the corresponding references are cited within the text. Chemical disruptors that appear in bold-type below were found to affect more than one molecular/cellular target in the cancer hallmark of evading growth suppression.
DDT, dichlorodiphenyltrichloroethane (organochlorine pesticide); TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin (aryl hydrocarbon receptor ligand).
aThese are potential mechanisms of evasion of growth inhibition utilized by environmental chemicals reported in the literature. The order of chemicals in the list does not necessarily align with the order of the mechanisms listed.
There are also numerous environmental chemicals that are suspected of promoting cancer but have not yet been validated as human carcinogens; many of these have been shown to affect p53 function, either on their own or in combination with other environmental chemicals. For example, the estrogenic compound bisphenol A, which is a common component of everyday plastics, has been reported to downregulate p53 expression and the expression of specific p53 targets, including p21 and Bax (30). This downregulation of p53 expression is associated with increased proliferation, which persists even when bisphenol A is removed from cell culture media (30). The pesticide component folpet, which induces gastrointestinal tumors in mice (31), has also been shown to disrupt the G1/S checkpoint (32) through multiple mechanisms. Folpet downregulates expression of the p53 target p21 and disrupts the functions of the ATM/ATR checkpoint kinases (32). Another pesticide, dichlorodiphenyltrichloroethane (DDT), induces MDM2 expression and reduces the expression and transcriptional activity of p53 (33). Other xenobiotics, such as the pesticides chlorothalonil and mancozeb, have been reported to downregulate p53 mRNA levels and to upregulate a ubiquitin ligase that triggers p53 degradation (34,35). Thus, it is important for future studies to determine if combinations of environmental pesticides and/or estrogenic compounds negatively affect p53 gene status, expression and/or checkpoint function.
Finding suitable experimental models to study the molecular effects of environmental chemicals is a challenge but is necessary for truly understanding the effects on cancer checkpoint proteins, such as p53. Intriguingly, the soft-shell clam may be a novel and relevant model system for studying the effects of environmental xenobiotics on the p53 pathway. This is because the p53 gene is highly conserved in the soft-shell clam. Mutation of p53 in the leukocytes of these clams leads to leukemia-like cancers. In particular, exposure of soft-shell clams to well-characterized carcinogens, such as benzo(a)pyrene, results in mutation of p53 (36). Thus, the use of this innovative model, and identification of additional nontransformed models will allow us to determine the potential carcinogenic and mechanistic effects of mixtures of specific environmental chemicals.
Retinoblastoma protein (pRb)
Retinoblastoma protein (pRb) is a nuclear protein encoded by the retinoblastoma susceptibility gene, RB1, which was the first tumor suppressor gene to be identified (37). Loss of RB1 induces genomic instability and the accumulation of chromosomal aberrations. Similar to p53, pRb is a critical gatekeeper of the G1/S transition, although it operates by a distinct mechanism from that of p53 (38). In addition, pRb has key roles in DNA replication, cellular senescence, differentiation and apoptosis (39). Thus, disruption of the pRb pathway affects many critical cellular processes, leading to the loss of cell cycle control and promoting cellular immortalization and transformation.
pRb blocks cell cycle progression by interacting with the E2F family of transcription factors (40). The pRb pathway includes D-type cyclins, cyclin-dependent kinases (CDKs) CDK4 and CDK6 and the CDK inhibitor 2a (p16/INK4a). When pRb is hypophosphorylated, it is active and restricts cell cycle progression by binding and repressing the E2F transcription factors. pRb must be inactivated by phosphorylation in order for the cell to progress from G1 to S phase. In response to mitogenic stimuli, CDK4/6-cyclin D and CDK2-cyclin E relieve inhibition of the pRb-E2F-containing transcription complex. pRb phosphorylation allows the E2F factors to dissociate, permitting transcription of genes that are required for DNA replication (41).
The D-type cyclins activate the G1 kinases CDK4/6 and target pRb for phosphorylation and inactivation. Cyclin D1 is a critical regulator of cellular proliferation that links extracellular signaling with cell cycle progression. In fact, the cyclin D-CDK4/6/INK4/pRb/E2F pathway integrates multiple mitogenic and antimitogenic signals, including from growth factor receptors, Ras, downstream effectors and p53. Deregulation of cyclin D1 is an important biomarker of the cancer phenotype and disease progression, and has been implicated in the development and progression of many forms of breast, esophageal, bladder and lung cancers (41). In addition to cyclin D1 deregulation, CDK4/6 overexpression is also involved in tumorigenesis. For example, overexpression of CDK4 induces uncontrolled cell growth and malignant transformation, whereas suppression of CDK4 causes terminal differentiation of erythroleukemia cells (42). Further, amplification and overexpression of CDK4 is found in multiple cancers, including sarcomas and glioblastomas (43). A somatic point mutation in CDK4 has also been identified in human cancers (44). CDK4 is inhibited by a series of inhibitory proteins (INKs). Among these, the INK4 proteins are frequently lost or inactivated by mutations in cancer and represent tumor suppressor genes; mutations in INK4-encoding genes contribute directly to the evasion of growth suppression. Loss of p16/INK4a function by gene deletion, promoter methylation and/or mutation within the reading frame leads to functional inactivation of pRb and is found in multiple types of cancers (45). Thus, although a tumor cell may not have a mutation in RB1, constitutive pRb hyperphosphorylation may represent a major mechanism of carcinogenesis through aberrant regulation of other key molecules, such as p16/INK4a, cyclin D1 and CDK4/6.
The mechanisms of pRb inactivation are often tissue-specific. For example, pRb is inactivated by loss of p16/INK4a in melanomas, whereas retinoblastomas, prostate cancers and osteosarcomas show inactivation of pRb through direct mutation or loss of the RB1 locus (46). A majority of lung cancers demonstrate pRb inactivation through functional loss of the p16/INK4A–cyclin D–CDK4/6–Rb pathway. Non-small cell lung cancers display multiple mechanisms of pRb inactivation, including mutation, excessive CDK activation, deregulated CDK4-cyclin D1 expression and loss of p16/INK4a activity by aberrant promoter methylation, homozygous deletions or point mutation (47). In addition, loss of pRb function by loss of heterozygosity has been reported in glioblastomas, breast cancer, gastric carcinoma, renal carcinoma and laryngeal cancer (48).
There is significant evidence to suggest that environmental chemicals affect the function or expression of the retinoblastoma protein (Table 1). Perhaps one of the best examples is a study demonstrating that prenatal exposure to benzene and other gasoline and diesel combustion products was significantly associated with the development of retinoblastoma (49). The major gasoline and combustion products that were associated with increased risk of retinoblastoma were toluene, 1,3-butadiene, ethyl benzene, orthoxylene and meta/paraxylene. Prenatal exposure to chloroform, chromium, paradichlorobenzene and nickel and exposure to acetaldehyde during the first year of life, were also associated with increased risk of developing retinoblastoma. In addition, butadiene, which is an industrial chemical used in the synthesis of rubber, has been shown to induce loss of heterozygosity of RB1 in mice and to promote the development of murine lung and breast tumors (50). These results implicate mixtures of gasoline combustion products or individual chemicals as potential mutagens of RB1. These reports also provide rationale for performing studies to understand if gasoline byproducts and butadiene, or mixtures of these chemicals, directly inactivate RB1.
As discussed above, a major mechanism by which the growth-inhibitory function of pRb is inactivated is via increased cyclin D1-CDK4/6 activity. Environmental contaminants, including benzo(a)pyrene (51) and radon (52), can stimulate pRb phosphorylation via cdk activation or loss of INK4a function. Benzo(a)pyrene is found in coal tar and cigarette smoke and is an established human carcinogen. Radon is a suspected carcinogen, as its inhalation is implicated in lung cancer development and suspected as the leading cause of lung cancer in nonsmokers (53). Inorganic arsenic is also routinely detected in the environment and is classified as a human carcinogen. Human embryonic lung fibroblasts are transformed by exposure to low levels of arsenite (NaAsO2) (54). One of the molecular events stimulated by low levels of arsenite is the upregulation of cyclin D1 expression with subsequent activation of CDK4/6 function and pRb hyperphosphorylation. This pRb inactivation can be rescued by blocking JNK1/c-Jun signaling, which also restores growth inhibition and suppresses transformation.
These studies collectively support additional detailed analyses and laboratory-based investigations to determine how environmental agents, including industrial pollutants, alter pRb function. In the previous section, we discussed agents, including bisphenol A and pesticides, that alter p53 expression, and discussed other agents here that disrupt pRb function. Real-life situations may involve simultaneous exposures to dozens of environmental chemicals. Thus, it will be important for future laboratory studies to model the effects of mixtures of environmental chemicals at environmentally relevant concentrations to determine the collective molecular effects of common everyday exposures.
TGF-β
Another key mediator of growth inhibition is the TGF-β signaling pathway. Escape from TGF-β-mediated growth inhibition is a critical step in tumorigenesis, because it is often coupled with the ability of cancer cells to utilize TGF-β as a pro-oncogenic factor via autocrine and paracrine mechanisms (55). Studies suggest that, in normal cells, sustained low-dose exposure to chemical mixtures in the environment may directly disrupt (i) the downstream effectors of TGF-β, transcription factors Smad2 and Smad3 (56), or their interacting partners and (ii) other signaling pathways that cross-talk with the TGF-β pathway. These two mechanisms of disruption can coexist in normal cells and favor resistance to TGF-β tumor-suppressive activities.
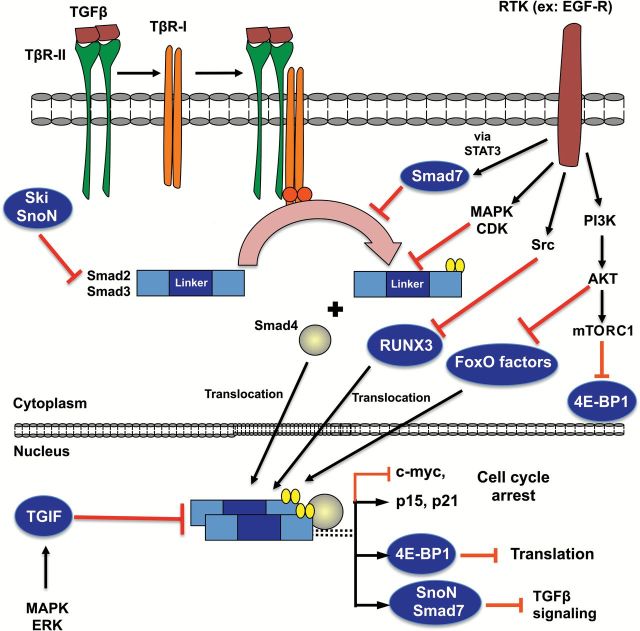
One major upstream regulator of TGF-β signaling is the epidermal growth factor receptor (EGFR) (Figure 1), which activates MAPK/ERK, JNK, p38, CDKs and glycogen synthase kinase 3β (GSK3β), inducing phosphorylation of critical linker regions in the TGF-β effectors Smad2 and Smad3 (reviewed in [57]). Smad2 and Smad3 linker phosphorylation directly inhibits the transcription factor functions of these Smad proteins, resulting in reduced transcription of TGF-β-target genes, including the p15 INK4B and p21 WAF1/CIP1 growth inhibitors (57,58). In addition, signals that activate EGFR-RAS-MEK-ERK increase the stability and levels of the Smad2 competitor, TGF beta-induced factor homeobox 1 (TGIF), which inhibits TGF-β (59,60). Similarly, hepatocyte growth factor stabilizes TGIF (61) and upregulates the TGF-β negative regulators c-Ski and SnoN (62). Inhibition of TGF-β signaling can also occur by overexpression of the TGF-β negative regulator, Smad7, which has been shown to induce premalignant pancreatic ductal lesions in mice (63). One mechanism by which Smad7 levels are upregulated is by activation of EGFR and downstream STAT3 signaling, which induces expression of Smad7 and causes loss of TGF-β-mediated growth inhibition (64).
Figure 1.
Targets for disruption of TGFβ tumor suppression (dark blue background), initiated by TGFβ binding and TβR-I receptor-mediated C-terminal phosphorylation of Smad2/3 (yellow ovals) and followed by translocation and transcriptional activation/repression.
Another regulator of TGF-β-mediated growth inhibition is the tumor suppressor RUNX3. RUNX3 promotes growth arrest and apoptosis in stomach epithelial cells by cooperating with Smads to induce TGF-β-dependent p21WAF1/CIP1 expression (65) and to transcriptionally upregulate Bim (66). The gastric epithelia of Runx3−/− adult mice are hyperplastic. In contrast to wild-type animals, these mice develop adenocarcinomas in response to the alkylating agent N-methyl-N-nitrosourea. These data suggest that loss of RUNX3 promotes the development of chemically induced cancer in gastric epithelial cells (67). One mechanism through which RUNX3 cytoplasmic mislocalization/inactivation occurs is by direct tyrosine phosphorylation of RUNX3 by Src (68). Thus, similar to the other mechanisms of TGFβ regulation discussed above, chemical stimulation of receptor tyrosine kinases may also lead to RUNX3 phosphorylation and inactivation, resulting in the inhibition of TGFβ tumor suppressive activities.
TGFβ signaling mediates growth inhibition in part by stimulating the Smad transcription factors to complex with FoxO factors, which then bind and activate transcription of p21 (69) and p15 (70). This process is negatively regulated by AKT, which phosphorylates FoxO factors, leading to their cytoplasmic retention (71). A hyperactive PI3K/AKT pathway excludes FoxO factors from the nucleus, preventing p21 expression and conferring resistance to TGF-β-induced cytostasis (69). Hyperactive AKT may also prevent BIM-mediated apoptosis induced by TGF-β in cell types where FoxO-induced Bim plays a role in that process (reviewed in [57]). The antiproliferative effects of TGF-β also require expression of the PI3K/AKT/mammalian target of rapamycin (mTOR) downstream target 4E-BP1 (72). TGF-β activates 4E-BP1 promoter activity through Smad4; silencing 4E-BP1 in normal and pancreatic cancer cells prevents the growth-inhibitory effects of TGF-β. Thus, sustained growth factor stimulation or direct activation of PI3K/AKT/mTORC1 by chronic exposure to environmental chemicals would favor 4E-BP1 phosphorylation/inactivation and resistance to TGF-β-mediated growth inhibition.
As described above, the antiproliferative and proapoptotic activities of TGF-β are tightly controlled by Smads, Smad cofactors (FoxO, RUNX3), TGF-β negative regulators (Smad linker phosphorylation, c-ski, SnoN, TGIF, Smad7) and 4E-BP1. Importantly, stimulation of one receptor tyrosine kinase, the epidermal growth factor receptor (EGFR), simultaneously activates AKT and MAPK, increases Smad7 expression, stabilizes TGIF, excludes FoxO factors and RUNX3 from the nucleus, and/or frees eIF4E from 4E-BP1; all of these events block TGF-β-stimulated growth inhibition. Therefore, TGF-β growth-inhibitory signaling could be disrupted in normal cells through chronic, sustained exposures to chemicals that stimulate a major upstream regulator of TGF-β, such as EGFR, or chemical mixtures that directly activate multiple downstream signals, such as MAPK and STAT3. An example of one such environmental contaminant is arsenic [sodium arsenite; NaAsO2, As(III)] (Table 1), which contaminates many drinking water reservoirs (73). Arsenic and its metabolites activate MAPK signaling (73) and increase expression of EGFR ligands (74). In addition, As(III) has been shown to abrogate TGFβ signaling and reduce phosphorylation of Smads (75); these effects were observed with low, environmentally relevant levels of arsenic. As discussed above, a loss of TGFβ-Smad function prevents expression of cell cycle arrest mediators, such as p21 and p15. Thus, sustained exposures to chemicals or mixtures of chemicals that increase cell surface or cytoplasmic growth factor signaling may ultimately increase the risk of carcinogenesis in part by disrupting TGFβ-mediated growth inhibition.
Gap junctions
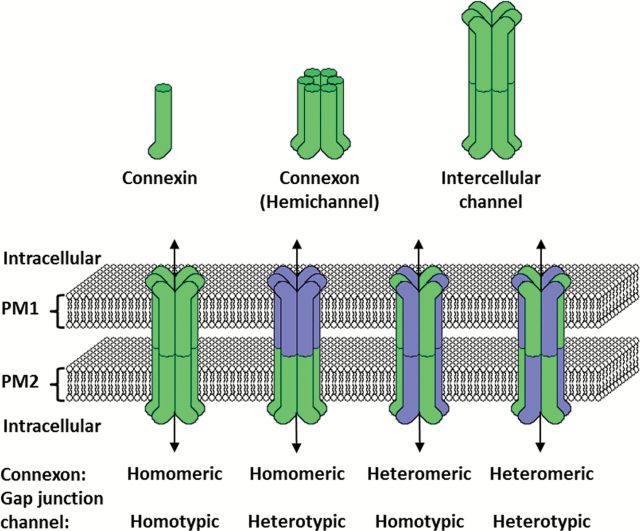
Gap junctions are clusters of tightly packed intercellular channels assembled from connexin (Cx) family members. These junctions are important for the intercellular exchange of metabolites, ions and small molecules (e.g. cAMP, IP3, ATP, Ca2+) through a process known as gap junctional intercellular communication (GJIC) (76). The connexin family includes 21 members. Cells can simultaneously express different connexins. These connexin molecules selectively intermix to form homomeric or heteromeric channels (Figure 2). Thus, there are many different subtypes of gap junctions that can form. After cotranslational insertion into the endoplasmic reticulum, connexins oligomerize to form ‘connexons’, which traffic to the plasma membrane. At the cell surface, connexons become functional ‘hemichannels’ and allow molecular exchanges to occur between the cytoplasm and extracellular environment. However, these hemichannels appear to quickly seek out and dock with other hemichannels on a contacting cell to form a gap junction intercellular channel; the other hemichannel may be of the same (homotypic) or different (heterotypic) type. Individual gap junction channels form tight arrays called gap junction plaques (77).
Figure 2.
Schematic diagram depicting connexins, connexons (hemichannels) and gap junction (GJ) intercellular channels. PM1 and PM2 represent plasma membranes from two adjacent cells. Blue and green represent two different connexin family members.
Early studies indicate that gap junctions are impacted by a variety of chemical carcinogens and oncogenes, with reduced numbers or function of gap junction channels being associated with tumor formation (78,79). Loewenstein suggested that GJIC plays a role in the dispersion and dilution of growth-promoting signals, suppressing cell proliferation (80). The loss of gap junctions was predicted to increase intracellular signaling, enhancing proliferation and tumor formation (81). Although this model remains viable, the significance of gap junctions in tumor biology has expanded to also include nonchannel functions. Connexins are now recognized as tumor suppressors that reduce tumor cell growth in vitro and in vivo and partially redifferentiate transformed cells (79,82–86). Genetically modified mice that lack a connexin family member (e.g., Cx32) have increased susceptibility for chemical- or radiation-induced liver and lung tumors (87–89). Similarly, genetically modified mice with a Cx43 mutation have increased numbers of lung metastases (90). There is ample evidence to suggest that the tumor-suppressive role of gap junctions is mediated in part by the molecules that pass through these channels (78,79,87–89,91). However, GJIC-independent mechanisms may also mediate tumor suppression in some contexts, including molecular exchanges between the extracellular environment and cytoplasm via hemichannels (92–95) and Cx-binding partners that may mediate tumor-suppressive activities.
The tumor suppressor classification of connexins has been challenging to validate, as large-scale, retrospective studies examining cancer susceptibility in patients with loss-of-function connexin mutations are difficult to perform due to the small cohort of patients with connexin-linked diseases. However, some studies support roles for connexins in late-stage disease progression, metastasis and development of life-threatening secondary tumors in a variety of tumor types (96,97). Thus, connexins are viewed as ‘conditional tumor suppressors’ (79), consistent with the biphasic functions of connexins depending on the stage of disease progression.
Paradoxically, connexins demonstrate cancer-promoting effects, such as invasion and metastasis, in the advanced stages of some tumor types (79,97,98). This may be a reflection of the inter-dependence of connexins and other types of junctions, i.e. tight and adherent junctions (77,99). Cell adhesion mediated by cadherin family members is necessary for gap junction formation and maintenance (77). Thus, reduced expression of cadherins can lead to the subsequent destabilization and loss of gap junctions. Interestingly, however, the down-regulation of gap junctions can also lead to reduced cell adhesion (100,101) and cell migration (102). This bidirectional crosstalk could partially explain how gap junctions serve as tumor suppressors in early-onset disease but serve to promote extravasation in late-stage disease.
Down-regulation of GJIC by tumor-promoting compounds is a proposed mechanism by which cancer cells evade growth suppressive signals (103,104). Further, GJIC inhibition is a proposed mechanism for the cancer-promoting activities of chemical compounds (105). Indeed, numerous carcinogenic or tumor-promoting chemicals downregulate GJIC and/or connexin expression in vitro or in vivo in various experimental models. Polychlorinated biphenyls (PCBs) downregulate GJIC and/or Cx expression in a wide range of tissue and cellular models, including rodent hepatocytes, rodent liver epithelial cells, human keratinocytes and human breast epithelial cells (106–110). A thorough overview of chemical compounds that affect liver gap junctions has been provided in a recent review by Vinken et al. (111). Toxic compounds that have been proposed to negatively affect GJIC include: (i) incomplete combustion products and industrial contaminants, such as polycyclic aromatic hydrocarbons (PAHs), halogenated aromatic hydrocarbons, including PCBs and polychlorinated dibenzo-p-dioxins (e.g. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)), organic solvents (such as carbon tetrachloride and trichloroethylene), or phthalates; (ii) organochlorine pesticides and herbicides (DDT, endosulfane, chlordane, heptachlor, dieldrin, lindane, hexachlorobenzene or pentachlorophenol); (iii) heavy metals, such as mercury and cadmium; and (iv) some biological toxins, such as lipopolysaccharide, mycotoxins, cyanotoxins and especially phorbol esters, which are prototypical tumor promoters (reviewed in [111]). Downregulation of GJIC has been observed also for some complex mixtures of chemical compounds, such as cigarette smoke, cigarette smoke condensates, extracts of airborne particulate matter or commercial PCB mixtures (112–115).
A wide range of mechanisms contributing to downregulation of specific Cx species and/or GJIC by xenobiotics have been proposed. Most chemical contaminants that inhibit Cx32- and/or Cx26-mediated GJIC in liver tissue do so through downregulation of the Cx mRNA and/or protein. TCDD and closely related ligands of the aryl hydrocarbon receptor (AhR), such as dioxin-like PCBs, have been reported to decrease hepatic Cx32 protein and/or mRNA levels in association with reduced levels of gap junction plaques (106,116–118). Similar effects have been reported for a wider spectrum of chemical contaminants, including heavy metals, lipopolysaccharide, carbon tetrachloride and hexachlorobenzene (119–122). Some compounds, such as ochratoxin A, have more prominent effects, such as simultaneously downregulating Cx26, Cx32 and Cx43 in rat liver (123). Nevertheless, it should be mentioned that downregulation of Cx32 is not always accompanied with reduced GJIC (121). Transient or permanent inhibition of Cx43-mediated GJIC has been observed for many contaminants, such as low-molecular weight PAHs (including parent PAH compounds, their metabolites, ozonation products or methylated PAH derivatives), perfluorinated fatty acids, nondioxin-like PCBs (or their metabolites), mycotoxins and pesticides (111). Most of these compounds induce rapid closure of gap junctions formed by Cx43. The mechanisms responsible for such effects are compound- and cell-specific and are associated with Cx43 phosphorylation through activated extracellular signal-regulated kinases 1/2 (ERK1/2), phosphatidylcholine-specific phospholipase C-dependent pathway, p38 MAP kinase or other signaling pathways (124–128). Phorbol esters employ ERK1/2 and protein kinase C (PKC) to induce Cx43 hyperphosphorylation, ubiquitination, internalization and degradation (129–131). Chemical contaminants may also reduce Cx43 levels through enhanced endocytosis and degradation via lysosomal or proteasomal pathways, leading to long-term GJIC inhibition (132–134). Several compounds, such as pentachlorophenol or ochratoxin A, have been found to downregulate Cx43 mRNA levels (123,135).
Contact inhibition
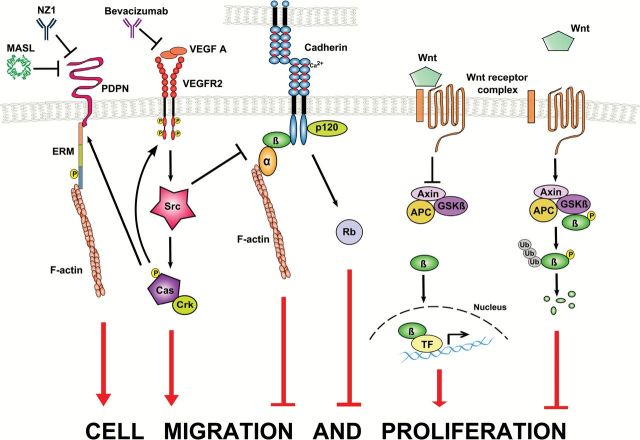
Nontransformed adherent cells generally undergo a density-dependent decrease in cell division and/or G1 arrest when they become confluent (136). In contrast, many tumor cells do not undergo contact growth inhibition and continue to proliferate at confluence. Thus, tumor cells often display higher cell-saturation densities in culture than their nontransformed precursors. Thus, loss of contact inhibition is recognized as a hallmark of tumor cell growth (137). Contact growth inhibition results from signal transduction events initiated by intercellular junctions, in which cadherins play important roles (Figure 3) (138). Cadherins are type 1 transmembrane proteins that mediate calcium-dependent adhesive interactions between adjacent cells. Although there are at least 80 members in the cadherin family, ‘classical’ cadherins share properties that underlie their effects on cell adhesion and growth control. These include an extracellular region of several modular domains that associate in a zipper-like fashion with corresponding domains presented by neighboring cells, followed by a transmembrane domain, and ending in a carboxyl-terminal region that interacts with the actin cytoskeleton via associations with β-catenin (139).
Figure 3.
Contact growth inhibition and the control of cell proliferation and motility. Cadherins interact with other cadherins on adjacent cells and intracellular proteins including p120 and β-catenin (β), which associates with α-catenin (α) to control the actin cytoskeleton and inhibit cell motility and proliferation. Normally, free β-catenin is phosphorylated by GSKβ and associates with the Axin-APC complex to undergo ubiquitin-mediated proteasomal degradation. However, Wnt signaling can prevent β-catenin degradation and allow it to augment transcription of genes that promote cell migration and proliferation (TF). Contact growth inhibition also promotes the expression of tumor suppressors including Rb to prevent cell cycle progression. Transforming agents exemplified by the Src tyrosine kinase can disrupt intercellular junctions and augment the activity of tumor promoters such as VEGFR2, Crk, Cas and PDPN to promote cancer invasion and metastasis. For example, PDPN associates with ezrin proteins (ERM) to modify the actin cytoskeleton and promote cell migration. Receptors that allow tumor cells to overcome contact growth inhibition may serve as cancer biomarkers and chemotherapeutic targets. For example, bevacizumab inhibits VEGF signaling, whereas monoclonal antibodies (NZ1) or lectins (MASL) target PDPN on malignant cells.
E-cadherin is commonly expressed by non-transformed epithelial cells and maintains intercellular contact between cells to form ‘epithelial sheets’ that are essentially contact-inhibited monolayers in cell culture. This process involves active recruitment of cdk inhibitors, including p16, p21 and p27, which block cdk4-cyclin D and cdk2-cyclin E catalytic activities to arrest cells at the G1 phase of the cell cycle (136,140–142). Contact growth inhibition also induces expression of tumor suppressors, including pRb, p53 and p27. These tumor suppressors are frequently mutated in cancers, allowing escape from contact inhibition and cell cycle progression (143–145).
The intracellular domain of cadherins interacts with cytoplasmic proteins, such as β-catenin and p120. These molecules act as a nexus between cadherins and the actin cytoskeleton to control cell growth and motility. For example, vascular endothelial cadherin (VE-cadherin) can prevent VEGF signaling in contact-inhibited endothelial cells (146). In addition, alterations in cadherin junctions that reduce E-cadherin expression and increase N-cadherin promote epithelial-mesenchymal transition and are strongly correlated with increased invasion and metastasis in a variety of cancer cells (137). In addition to changes in cadherin expression, oncogenes and tumor promoters can disrupt cadherin junctions to allow tumor cells to escape contact growth inhibition. For example, the Src tyrosine kinase can phosphorylate connexins and cadherins to disrupt junctional communication between tumor cells. Other agents, including phorbol esters, can also disrupt intercellular junctions and enable cells to overcome contact growth inhibition (147,148).
In addition to homotypic interactions between transformed cells, heterotypic communication between transformed cells and their nontransformed neighbors can inhibit tumor cell growth and migration. This ‘contact normalization’ can control the morphology and phenotype of transformed cells within the tumor microenvironment (149–152). Importantly, cells that are transformed by chemicals (153) can be normalized by communication with adjacent normal cells. As shown in Figure 3, elucidating the effectors and pathways that regulate contact growth inhibition and contact normalization will provide key information regarding potential mechanisms by which environmental chemicals promote cancer. For example, specific tumor suppressor genes that are induced in transformed cells undergoing contact normalization (154) include miR126 (which targets the Crk oncogene), Fhl1 (four and a half LIM domains) and Sdpr (serum deprivation response protein). FHL1 and miR126 inhibit anchorage independence and motility, and SDPR plays a role in serum independence. In addition to inducing tumor suppressors, contact normalization inhibits the expression of powerful tumor promoters (154). These proteins tend to promote migration of malignant cells out of their microenvironment to become invasive and metastatic. These findings are particularly exciting, because they may identify functionally relevant targets that may be compromised by environmental chemicals. For example, Tmem163 (transmembrane protein 163), Vegfr2 (vascular endothelial growth factor receptor 2) and Pdpn (podoplanin) are all extracellular receptors that promote the motility of tumor cells that escape contact normalization.
In contrast to GJIC, contact inhibition has not been extensively studied in the context of environmental carcinogenesis. Early work showed that phorbol esters may induce loss of contact inhibition in human fibroblasts (155), and there has been recent interest in the effects of environmental contaminants on the deregulation of cell-to-cell communication. Particular attention has been paid to compounds that activate the aryl hydrocarbon receptor. Deregulated AhR activity contributes to altered cell-to-cell communication, including at adherens junctions, and also affects cell adhesion (156–158). Activation of AhR by various ligands disrupts contact inhibition and induces cell proliferation in some cell types (156,159,160). In some cases, there is a link between disruption of growth suppression via deregulation of contact inhibition and loss of response to GJIC-mediated growth-inhibitory signals from neighboring cells. Recently, TCDD and other AhR ligands were shown to simultaneously alter cell proliferation, leading to the disruption of contact inhibition and downregulation of GJIC via enhanced Cx43 degradation in rat liver epithelial cells (161). Thus, environmental chemicals that affect connexins and gap junctions may negatively impact contact inhibition and further promote evasion of growth suppression.
Expert perspective
Many of the environmental chemicals that we have discussed in the individual sections above affect multiple growth-inhibitory mediators, as shown in Table 1. Thus, sustained exposure to one environmental chemical can result in major effects on a single cancer hallmark. Two examples of such chemicals are bisphenol A and DDT. Bisphenol A promotes cell cycle progression by disrupting multiple targets that have been discussed here. In addition to reducing functional p53, low nanomolar concentrations of bisphenol A reduce expression of Cx43, compromising gap junction communication (162). Bisphenol A-mediated effects on GJIC are connexin-selective, as reduced expression of Cx43 has been observed after bisphenol A exposure, but Cx26 is unaffected (163). DDT also disrupts GJIC in a dose-dependent manner (164,165) and increases expression of the p53-degrading protein Mdm2 (33). In addition, DDT increases transcription of Ccnd1 (cyclin D1) and E2f1 and induces phosphorylation of pRb (33). Thus, bisphenol A and DDT represent examples of environmental chemicals that affect multiple targets within the hallmark of evading growth suppression.
In addition to evading growth suppression, cancer cells coordinate deregulation of multiple mechanisms that constitute other hallmarks of tumorigenesis. Each of these mechanisms is a potential target for therapeutic intervention; similarly, each mechanism can be targeted or activated by environmental chemicals. In fact, it is likely that the carcinogenic effects of an environmental chemical depend on the simultaneous activation of multiple cancer hallmark mechanisms. Thus, we performed literature searches to determine the roles of the molecular targets discussed in this manuscript in the context of other cancer hallmarks, as defined by Hanahan and Weinberg (137) (Tables 2 and 3). We found that individual growth-inhibitory mediators discussed in this review have variable roles in other cancer hallmarks. Based on our search results presented in Tables 2 and 3, it is likely that a chemical or chemical mixture that disrupts any one of the selected molecular targets will disrupt multiple cellular functions, promoting the establishment of multiple cancer hallmarks. For example, our literature search indicated that p53 has roles in each of the cellular processes involved in the various cancer hallmarks. Thus, a chemical that disrupts p53 function is likely to promote each of the established or suspected hallmarks of cancer, which may explain why p53 loss or mutation on its own is considered a major procarcinogenic event.
Table 2.
Roles of selected mediators of growth suppression in the cancer hallmarks of metabolism, angiogenesis, genetic instability, immune evasion and cell death
| Target | Metabolism | Angiogenesis | Genetic instability | Immune evasion | Cell death |
|---|---|---|---|---|---|
| p53 | p53 inactivation is associated with increased cancer cell metabolism (166) | p53 inactivation is associated with angiogenesis (167) | P53 inactivation is associated with genetic instability (168) | Wild-type p53 activates immune function; mutant p53 may promote immune evasion (169–171) | p53 inactivation promotes resistance to cell death (172) |
| pRB | Inactivation of pRb is associated with increased cancer cell metabolism (173) | Inactivation of pRb is associated with angiogenesis (174) | Inactivation of pRb is associated with genetic instability (175) | Unestablisheda | pRb induces cell death (176) |
| TGF-beta | TGF-beta signaling promotes cancer cell metabolism (177) | TGF-beta promotes angiogenesis (178) | TGF-beta suppresses genetic instability (179,180) | TGF-beta promotes cancer cell immune evasion (181) | TGF-beta induces cell death (182) |
| LKB1 | Loss of LKB1 promotes cancer cell metabolism (183). | LKB1 promotes angiogenesis (184) | LKB1 suppresses genetic instability (185) | Unestablished | LKB1 can inhibit or activate cell death (186,187) |
| Connexins | Loss of gap junctions may promote cancer cell metabolism (188) | Unestablished | Connexins suppress genetic instability (189) | Unestablished | Unestablished |
| Contact inhibition | Contact inhibition can inhibit or activate cancer cell metabolism (190,191) | Unestablished | Contact inhibition suppresses genetic instability (165) | Contact inhibition promotes cancer cell immune evasion (192) | Unestablished |
We searched PubMed to determine if the listed growth-inhibitory molecular target had roles in other cancer hallmarks. We combined the name of the target listed in the far-left column with the word ‘cancer’ and the name of each of the specific cancer hallmarks that appear in the top row (e.g. ‘p53 cancer metabolism’, ‘p53 cancer angiogenesis’, etc.).
aIf no literature support was found to document the role of a specific molecular target in a particular hallmark, we stated that the target has an ‘unestablished’ role in that hallmark.
Table 3.
Roles of selected mediators of growth suppression in the cancer hallmarks of replicative immortality, sustained proliferation, invasion and metastasis, inflammation and tumor microenvironment
| Target | Replicative immortality | Sustained proliferation | Invasion and metastasis | Inflammation | Microenvironment |
|---|---|---|---|---|---|
| p53 | p53 inhibits replicative immortality (193–195) | p53 inactivation is associated with sustained proliferative signaling (196) | p53 inactivation is associated with invasion and metastasis (197) | p53 promotes inflammatory processes (198) | p53 maintains the tumor microenvironment (199) |
| pRB | pRb inhibits replicative immortality (200,201) | pRb inhibits sustained proliferation (202) | Inactivation of pRb is associated with invasion and metastasis (203) | pRb promotes inflammatory processes (204) | pRb maintains the tumor microenvironment (205). |
| TGF beta | TGF-beta inhibits replicative immortality (206) | TGF-beta promotes sustained proliferation (202) | TGF-beta promotes invasion and metastasis (207,208) | TGF-beta promotes inflammatory processes (209) | TGF-beta maintains the tumor microenvironment (210) |
| LKB1 | Unestablisheda | LKB1 promotes sustained proliferation (211,212) | LKB1 inhibits invasion and metastasis (213,214) | LKB1 promotes inflammatory processes (215) | LKB1 maintains the tumor microenvironment (216) |
| Connexins | Unestablished | Connexins inhibit sustained proliferation (217) | Connexins have been shown to inhibit and activate invasion and metastasis depending on the cell model (218–220) | Connexins promote inflammatory processes (221) | Connexins maintain the tumor microenvironment (222) |
| Contact inhibition | Unestablished | Contact inhibition suppresses sustained proliferation (223) | Contact inhibition inhibits invasion and metastasis (224) | Contact inhibition promotes inflammatory processes (225) | Contact inhibition has an inhibitory role in maintenance of the tumor microenvironment (222) |
We searched PubMed to determine if the listed growth-inhibitory molecular target had roles in other cancer hallmarks. We combined the name of the target listed in the far-left column with the word ‘cancer’ and the name of each of the specific cancer hallmarks that appear in the top row (e.g. ‘p53 cancer replicative immortality’, ‘p53 cancer sustained proliferation’, etc.).
aIf no literature support was found to document the role of a specific molecular target in a particular hallmark, we stated that the target has an ‘unestablished’ role in that hallmark.
In addition to the fact that one molecular target affects multiple hallmarks, our literature search also indicated that each single hallmark is regulated by most of the molecular targets that we reviewed. For example, the newly proposed cancer hallmark of cellular metabolism is regulated by LKB1, p53 and pRb, among other molecular mediators. One of the central regulators of cellular metabolism is the LKB1 substrate AMPK. LKB1-AMPK signaling tightly controls signaling from mTOR, such that loss of LKB1 activates mTOR, promoting extensive protein synthesis and expression of glycolytic enzymes. Thus, LKB1 loss induces glycolysis and metabolic changes through the so-called Warburg effect (183). This describes the process by which highly proliferating cells rely on glycolysis to convert glucose to lactic acid in order to generate ATP (226). Tumors lacking LKB1 have underlying mechanisms that drive the Warburg effect. These changes are evident from the high levels of metabolites and the expression of glycolytic enzymes (227,228). Wild-type p53 also inhibits mTOR signaling by activating AMPK-mediated phosphorylation of TSC2 and increasing expression of PTEN (229). Importantly, glycolysis in the absence of wild-type p53 is not only due to loss of mTOR regulation, but can also be caused by a gain-of-function mutant p53 that causes translocation of the glucose transporter 1 (Glut 1) to the cell membrane and stimulation of the Warburg effect (230). AMPK also phosphorylates pRb on serine 804, which is important for maintaining brain development and neural stem and progenitor cell growth (231). However, hypophosphorylation of pRb has also been documented in the context of increased AMPK signaling secondary to inhibition of Glut 1, which is important for mediating glycolysis and the Warburg effect (232). Thus, pRb appears to have a complex role in the AMPK-mediated signaling cascade that regulates cellular metabolism. These examples demonstrate that the major growth-inhibitory molecules reviewed here also play roles in other cancer hallmarks. Thus, loss of function of any one molecular target can establish multiple cancer hallmarks, not just evasion of growth suppression. This is important from the perspective of understanding that a complex mixture of environmental chemicals should be studied for effects on multiple molecular targets and multiple cancer hallmarks to truly understand the mechanisms by which any given mixture promotes carcinogenesis.
Similar to the fact that most of the proposed molecules affect multiple hallmarks, many of the chemicals that we selected affect not only the hallmark of evading growth suppression, but also other cancer hallmarks (Tables 4 and 5). The majority of data regarding the effects of these compounds on cancer hallmarks has been collected in animal models; thus, the effects on human carcinogenesis remain extremely controversial. However, the animal data provide substantial rationale for performing rigorous investigations to understand if single environmental chemicals or mixtures of chemicals simultaneously promote the development of numerous cancer hallmarks. One of the best examples of a potential environmental agent that affects almost all of the established cancer hallmarks is bisphenol A. Bisphenol A may be a prevalent disruptor of multiple hallmarks due to its abilities to inactivate p53, activate mTOR and promote estrogenic effects. The presence of a p53 mutation, particularly in combination with high TERT activity [319], promotes sustained proliferative signaling, resistance to cell death, angiogenesis, tissue invasion and metastasis and a proinflammatory environment. Similarly, mTOR activation promotes multiple cellular processes, such as sustained proliferation, cell survival, glycolysis and motility and invasion (233). Increased proliferation has been documented in various cancer models of bisphenol A exposure. For example, staining for the proliferation marker Ki-67 was increased in DMBA-induced breast tumor tissues collected from 50-day-old female rats exposed to bisphenol A-contaminated breast milk (234). In addition, prenatal exposure to bisphenol A promoted the development of preneoplastic lesions in the mammary glands of rats (234). Further, bisphenol A altered the expression levels of multiple proteins involved in angiogenesis, including VEGF and annexin A2, and proliferation and cell survival, such as PI3K and MAPK signaling (162,234–237). Activation of mTOR and activated estrogen receptor function by bisphenol A also increased proliferation and blocked apoptosis in animal models of mammary cancer (238). Cellular metabolism may also be affected by bisphenol A due to mTOR activation. Additional mechanisms by which bisphenol A can promote the development or progression of cancer include inhibition of DNA repair (239,240), increased replicative immortality due to the estrogenic effect of inducing expression of the telomerase catalytic subunit hTERT (241), increased expression of factors, such as matrix metalloproteases, which drive motility and invasion (242), and potential activation of inflammatory processes due to the accumulation of reactive oxygen species (243).
Table 4.
Roles of selected chemicals on the cancer hallmarks of metabolism, angiogenesis, genetic instability, immune evasion and cell death
| Chemical | Metabolism | Angiogenesis | Genetic instability | Immune evasion | Cell death |
|---|---|---|---|---|---|
| Bisphenol A | Bisphenol A promotes cancer metabolism (233,234,238) | Bisphenol A promotes angiogenesis (162,234,235) | Bisphenol A promotes genetic instability (239,240) | Unestablisheda | Bisphenol A can promote or block cell death (30,238) |
| DDT | Unestablished | DDT may promote angiogenesis (244) | DDT promotes genetic instability (245) | DDT can block the function of natural killer cells, causing immune evasion (246) | DDT inhibits cell death (33) |
| Folpet | Unestablished | Unestablished | Folpet may promote genetic instability (31,247) | Unestablished | Folpet shows toxic effects inititally, but prolonged exposure may block apoptotic signals (31,247,248) |
| Triazine herbicides (atrazine) | Unestablished | Unestablished | Triazine herbicides promote genetic instability (249) | Atrazine promotes immune system evasion (250) | Unestablished |
We searched PubMed to determine if the selected chemicals had roles in other cancer hallmarks. We combined the name of the chemical listed in the far-left column with the name of the specific cancer hallmark that appears in the top row (e.g. ‘bisphenol metabolism’, ‘bisphenol angiogenesis’, etc.). DDT, Dichlorodiphenyltrichloroethane (organophosphate pesticide).
aIf no literature support was found to document the role of a specific molecular target in a particular hallmark, we stated that the target has an ‘unestablished’ role in that hallmark.
Table 5.
Roles of selected chemicals on the cancer hallmarks of replicative immortality, sustained proliferation, invasion and metastasis, inflammation and tumor microenvironment
| Chemical | Replicative immortality | Sustained proliferation | Invasion and metastasis | Inflammation | Microenvironment |
|---|---|---|---|---|---|
| Bisphenol A | Bisphenol A promotes replicative immortality (241) | Bisphenol A activates sustained proliferative signaling (30,234,236,237) | Unestablisheda | Bisphenol A promotes inflammatory processes (243) | Unestablished |
| DDT | DDT promotes replicative immortality (251) | DDT promotes sustained proliferative signaling (244,252) | Unestablished | DDT promotes inflammatory processes (253) | Unestablished |
| Folpet | Unestablished | Folpet activates proliferative signaling (31,247,248,254) | Unestablished | Folpet promotes inflammatory processes (31) | Unestablished |
| Triazine herbicides (atrazine) | Unestablished | Triazine herbicides promote proliferative signaling (255–257) | Unestablished | Triazine herbicides promote inflammatory processes (257,258) | Unestablished |
We searched PubMed to determine if the selected chemicals had roles in other cancer hallmarks. We combined the name of the chemical listed in the far-left column with the name of the specific cancer hallmark that appears in the top row (e.g. ‘bisphenol replicative immortality’, ‘bisphenol sustained proliferation’, etc.). DDT, dichlorodiphenyltrichloroethane (organophosphate pesticide).
aIf no literature support was found to document the role of a specific molecular target in a particular hallmark, we stated that the target has an ‘unestablished’ role in that hallmark.
Similar to bisphenol A, the organochlorine pesticide component DDT appears to promote multiple cancer-related processes or hallmarks, including sustained proliferation through pathways, such as Wnt/beta-catenin (252) and MAPK (244), and increased cell survival (33,244). In addition, DDT increases expression levels of VEGF (244), cyclooxygenase-2 (253) and reactive oxygen species (252), thus potentially promoting the cancer hallmarks of angiogenesis and inflammation. In vitro data also suggest that DDT causes DNA damage and genetic instability (245) and may cause telomere shortening (251). Interestingly, DDT also suppresses the function of immune natural killer cells in part by blocking interactions between natural killer cells and target proteins on cancer cells (246). The immune effects of most environmental chemicals remain poorly understood. However, the reported effects of DDT on immune cell function warrant further investigations into the immune-suppressive abilities of environmental chemicals or mixtures of chemicals. Because we are most likely exposed to numerous chemicals simultaneously, either through polluted air or drinking water reservoirs, improved attempts should be made to model and study chemical mixtures, rather than individual chemicals, at environmentally relevant concentrations. Ultimately, our analysis indicates that a greater level of research is required to not only look at the disruptive effects of environmental chemicals in the evasion of growth arrest, but also synergy between mixtures of chemicals that simultaneously enable multiple hallmarks. Current research should thus be aimed at understanding the molecular and cellular mechanisms through which environmentally relevant doses of chemical mixtures disrupt multiple cancer hallmarks.
Funding
Winship Cancer Institute (R.N.); FONDECYT (#1120006), MINEDUC (G.M.C.); NIH/NCI (R01CA172392 to R.C.C.); American Cancer Society (116683-RSG-09-087-01-TBE to K.C.-S.); Canadian Breast Cancer Foundation and the Canadian Institute of Health Research (D.L., C.N.); Dalhousie Medical Research Foundation, Beatrice Hunter Cancer Research Institute (P.M.); Czech Science Foundation (13-07711S to J.V.); Fondazione Cariplo (2011-0370 to C.M.); Kuwait Institute for the Advancement of Sciences (2011-1302-06 to F. al-M.); Grant University Scheme (RUGs) Ministry of Education Malaysia (04-02-12-2099RU to R.A.H.); Italian Ministry of University and Research (2009FZZ4XM_002 to A.A); the University of Florence (ex60%2012 to A.A.); US Public Health Service Grants (RO1 CA92306, RO1 CA92306-S1, RO1 CA113447 to R.R.); Department of Science and Technology, Government of India (SR/FT/LS-063/2008 to N.S.); Osteopathic Heritage Foundation to G.S.G..
Acknowledgments
We acknowledge the efforts of the cofounders of Getting to Know Cancer, Leroy Lowe and Dr Michael Gilbertson and the NIEHS for support of this project. The authors would like to thank Dr John Kelly for providing the schematic image used in Figure 2.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AhR
aryl hydrocarbon receptor
- AMPK
5′ adenosine monophosphate-activated protein kinase
- ATM
Ataxia-Telangiectasia-mutated
- ATR
Ataxia-Telangiectasia-related
- CDKs
cyclin-dependent kinases
- EGFR
epidermal growth factor receptor
- Chk2
checkpoint kinase 2
- DDT
dichlorodiphenyltrichloroethane
- pRb
retinoblastoma protein
- GJIC
gap junctional intercellular communication
- INKs
inhibitory proteins
- mTOR
mammalian target of rapamycin
- PAHs
polycyclic aromatic hydrocarbons
- PCB
polychlorinated biphenyls
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- TGF-β
transforming growth factor-beta
References
- 1. Lee W., et al. (1994) Solution structure of the tetrameric minimum transforming domain of p53. Nat. Struct. Biol., 1, 877–890. [DOI] [PubMed] [Google Scholar]
- 2. Bieging K.T., et al. (2012) Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol., 22, 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goh A.M., et al. (2011) The role of mutant p53 in human cancer. J. Pathol., 223, 116–126. [DOI] [PubMed] [Google Scholar]
- 4. Kussie P.H., et al. (1996) Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science, 274, 948–953. [DOI] [PubMed] [Google Scholar]
- 5. Popowicz G.M., et al. (2008) Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle, 7, 2441–2443. [DOI] [PubMed] [Google Scholar]
- 6. Pant V., et al. (2011) Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc. Natl. Acad. Sci. USA, 108, 11995–12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang L., et al. (2011) The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo . Proc. Natl. Acad. Sci. USA, 108, 12001–12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gu J., et al. (2002) Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem., 277, 19251–19254. [DOI] [PubMed] [Google Scholar]
- 9. Ray-Coquard I., et al. (2012) Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol., 13, 1133–1140. [DOI] [PubMed] [Google Scholar]
- 10. Wade M., et al. (2013) MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer, 13, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith J., et al. (2010) The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res., 108, 73–112. [DOI] [PubMed] [Google Scholar]
- 12. Bartek J., et al. (2003) Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell, 3, 421–429. [DOI] [PubMed] [Google Scholar]
- 13. Jin J., et al. (2008) Differential roles for checkpoint kinases in DNA damage-dependent degradation of the Cdc25A protein phosphatase. J. Biol. Chem., 283, 19322–19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou B.B., et al. (2003) Targeting DNA checkpoint kinases in cancer therapy. Cancer Biol. Ther., 2(4 Suppl 1), S16–S22. [PubMed] [Google Scholar]
- 15. Yadavilli S., et al. (2009) Mechanism of diepoxybutane-induced p53 regulation in human cells. J. Biochem. Mol. Toxicol., 23, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giardiello F.M., et al. (1987) Increased risk of cancer in the Peutz-Jeghers syndrome. N. Engl. J. Med., 316, 1511–1514. [DOI] [PubMed] [Google Scholar]
- 17. Avizienyte E., et al. (1999) LKB1 somatic mutations in sporadic tumors. Am. J. Pathol., 154, 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen R.W., et al. (1999) PTEN and LKB1 genes in laryngeal tumours. J. Med. Genet., 36, 943–944. [PMC free article] [PubMed] [Google Scholar]
- 19. Guldberg P., et al. (1999) Somatic mutation of the Peutz-Jeghers syndrome gene, LKB1/STK11, in malignant melanoma. Oncogene, 18, 1777–1780. [DOI] [PubMed] [Google Scholar]
- 20. Zeng P.Y., et al. (2006) LKB1 is recruited to the p21/WAF1 promoter by p53 to mediate transcriptional activation. Cancer Res., 66, 10701–10708. [DOI] [PubMed] [Google Scholar]
- 21. Scott K.D., et al. (2007) LKB1 catalytically deficient mutants enhance cyclin D1 expression. Cancer Res., 67, 5622–5627. [DOI] [PubMed] [Google Scholar]
- 22. Nath-Sain S., et al. (2009) LKB1 catalytic activity contributes to estrogen receptor alpha signaling. Mol. Biol. Cell, 20, 2785–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gurumurthy S., et al. (2008) LKB1 deficiency sensitizes mice to carcinogen-induced tumorigenesis. Cancer Res., 68, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ratovitski E.A. (2010) LKB1/PEA3/ΔNp63 pathway regulates PTGS-2 (COX-2) transcription in lung cancer cells upon cigarette smoke exposure. Oxid. Med. Cell. Longev., 3, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pfeifer G.P., et al. (2002) Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene, 21, 7435–7451. [DOI] [PubMed] [Google Scholar]
- 26. Hussain S.P., et al. (2001) Mutability of p53 hotspot codons to benzo(a)pyrene diol epoxide (BPDE) and the frequency of p53 mutations in nontumorous human lung. Cancer Res., 61, 6350–6355. [PubMed] [Google Scholar]
- 27. Jiang Y., et al. (2007) Ataxia-telangiectasia mutated expression is associated with tobacco smoke exposure in esophageal cancer tissues and benzo[a]pyrene diol epoxide in cell lines. Int. J. Cancer, 120, 91–95. [DOI] [PubMed] [Google Scholar]
- 28. Ozturk M. (1991) p53 mutation in hepatocellular carcinoma after aflatoxin exposure. Lancet, 338, 1356–1359. [DOI] [PubMed] [Google Scholar]
- 29. Hamid A.S., et al. (2013) Aflatoxin B1-induced hepatocellular carcinoma in developing countries: Geographical distribution, mechanism of action and prevention. Oncol. Lett., 5, 1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dairkee S.H., et al. (2013) Bisphenol-A-induced inactivation of the p53 axis underlying deregulation of proliferation kinetics, and cell death in non-malignant human breast epithelial cells. Carcinogenesis, 34, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cohen S.M., et al. (2010) Carcinogenic mode of action of folpet in mice and evaluation of its relevance to humans. Crit. Rev. Toxicol., 40, 531–545. [DOI] [PubMed] [Google Scholar]
- 32. Santucci M.A., et al. (2003) Cell-cycle deregulation in BALB/c 3T3 cells transformed by 1,2-dibromoethane and folpet pesticides. Environ. Mol. Mutagen., 41, 315–321. [DOI] [PubMed] [Google Scholar]
- 33. Kazantseva Y.A., et al. (2013) Dichlorodiphenyltrichloroethane technical mixture regulates cell cycle and apoptosis genes through the activation of CAR and ERα in mouse livers. Toxicol. Appl. Pharmacol., 271, 137–143. [DOI] [PubMed] [Google Scholar]
- 34. Pariseau J., et al. (2009) Potential link between exposure to fungicides chlorothalonil and mancozeb and haemic neoplasia development in the soft-shell clam Mya arenaria: a laboratory experiment. Mar. Pollut. Bull., 58, 503–514. [DOI] [PubMed] [Google Scholar]
- 35. Pariseau J., et al. (2011) Effects of pesticide compounds (chlorothalonil and mancozeb) and benzo[a]pyrene mixture on aryl hydrocarbon receptor, p53 and ubiquitin gene expression levels in haemocytes of soft-shell clams (Mya arenaria). Ecotoxicology, 20, 1765–1772. [DOI] [PubMed] [Google Scholar]
- 36. Liu Z., et al. (2005) p53 mutations in benzo(a)pyrene-exposed human p53 knock-in murine fibroblasts correlate with p53 mutations in human lung tumors. Cancer Res., 65, 2583–2587. [DOI] [PubMed] [Google Scholar]
- 37. Wang J.Y., et al. (1994) The retinoblastoma tumor suppressor protein. Adv. Cancer Res., 64, 25–85. [DOI] [PubMed] [Google Scholar]
- 38. Brown M., et al. (1991) Fidelity of mitotic chromosome transmission. Cold Spring Harb. Symp. Quant. Biol., 56, 359–365. [DOI] [PubMed] [Google Scholar]
- 39. Weinberg R.A. (1995) The retinoblastoma protein and cell cycle control. Cell, 81, 323–330. [DOI] [PubMed] [Google Scholar]
- 40. Weintraub S.J., et al. (1995) Mechanism of active transcriptional repression by the retinoblastoma protein. Nature, 375, 812–815. [DOI] [PubMed] [Google Scholar]
- 41. Lundberg A.S., et al. (1998) Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol. Cell. Biol., 18, 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kiyokawa H., et al. (1994) Suppression of cyclin-dependent kinase 4 during induced differentiation of erythroleukemia cells. Mol. Cell. Biol., 14, 7195–7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. An H.X., et al. (1999) Gene amplification and overexpression of CDK4 in sporadic breast carcinomas is associated with high tumor cell proliferation. Am. J. Pathol., 154, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Papp T., et al. (1999) Mutational analysis of the N-ras, p53, p16INK4a, CDK4, and MC1R genes in human congenital melanocytic naevi. J. Med. Genet., 36, 610–614. [PMC free article] [PubMed] [Google Scholar]
- 45. Konishi N., et al. (2002) Heterogeneous methylation and deletion patterns of the INK4a/ARF locus within prostate carcinomas. Am. J. Pathol., 160, 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nielsen G.P., et al. (1998) CDKN2A gene deletions and loss of p16 expression occur in osteosarcomas that lack RB alterations. Am. J. Pathol., 153, 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Myong N.H. (2008) Cyclin D1 overexpression, p16 loss, and pRb inactivation play a key role in pulmonary carcinogenesis and have a prognostic implication for the long-term survival in non-small cell lung carcinoma patients. Cancer Res. Treat., 40, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pietruszewska W., et al. (2008) Loss of heterozygosity for Rb locus and pRb immunostaining in laryngeal cancer: a clinicopathologic, molecular and immunohistochemical study. Folia Histochem. Cytobiol., 46, 479–485. [DOI] [PubMed] [Google Scholar]
- 49. Heck J.E., et al. (2013) Retinoblastoma and ambient exposure to air toxics in the perinatal period. J. Expo. Sci. Environ. Epidemiol, 25, 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wiseman R.W., et al. (1994) Allelotyping of butadiene-induced lung and mammary adenocarcinomas of B6C3F1 mice: frequent losses of heterozygosity in regions homologous to human tumor-suppressor genes. Proc. Natl. Acad. Sci. USA, 91, 3759–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jeong J.B., et al. (2010) 2-Methoxy-4-vinylphenol can induce cell cycle arrest by blocking the hyper-phosphorylation of retinoblastoma protein in benzo[a]pyrene-treated NIH3T3 cells. Biochem. Biophys. Res. Commun., 400, 752–757. [DOI] [PubMed] [Google Scholar]
- 52. Bastide K., et al. (2009) Molecular analysis of the Ink4a/Rb1-Arf/Tp53 pathways in radon-induced rat lung tumors. Lung Cancer, 63, 348–353. [DOI] [PubMed] [Google Scholar]
- 53. Torres-Durán M., et al. (2014) Residential radon and lung cancer in never smokers. A systematic review. Cancer Lett., 345, 21–26. [DOI] [PubMed] [Google Scholar]
- 54. Li Y., et al. (2011) Up-regulation of cyclin D1 by JNK1/c-Jun is involved in tumorigenesis of human embryo lung fibroblast cells induced by a low concentration of arsenite. Toxicol. Lett., 206, 113–120. [DOI] [PubMed] [Google Scholar]
- 55. Ikushima H., et al. (2010) TGFbeta signalling: a complex web in cancer progression. Nat. Rev. Cancer, 10, 415–424. [DOI] [PubMed] [Google Scholar]
- 56. Massagué J. (2012) TGFβ signalling in context. Nat. Rev. Mol. Cell Biol., 13, 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lasfar A., et al. (2010) Resistance to transforming growth factor β-mediated tumor suppression in melanoma: are multiple mechanisms in place? Carcinogenesis, 31, 1710–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Matsuzaki K. (2011) Smad phosphoisoform signaling specificity: the right place at the right time. Carcinogenesis, 32, 1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang S., et al. (2008) EGF antagonizes TGF-beta-induced tropoelastin expression in lung fibroblasts via stabilization of Smad corepressor TGIF. Am. J. Physiol. Lung Cell. Mol. Physiol., 295, L143–L151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lo R.S., et al. (2001) Epidermal growth factor signaling via Ras controls the Smad transcriptional co-repressor TGIF. EMBO J., 20, 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dai C., et al. (2004) Hepatocyte growth factor antagonizes the profibrotic action of TGF-beta1 in mesangial cells by stabilizing Smad transcriptional corepressor TGIF. J. Am. Soc. Nephrol., 15, 1402–1412. [DOI] [PubMed] [Google Scholar]
- 62. Tan R., et al. (2007) Molecular basis for the cell type specific induction of SnoN expression by hepatocyte growth factor. J. Am. Soc. Nephrol., 18, 2340–2349. [DOI] [PubMed] [Google Scholar]
- 63. Kuang C., et al. (2006) In vivo disruption of TGF-beta signaling by Smad7 leads to premalignant ductal lesions in the pancreas. Proc. Natl. Acad. Sci. U. S. A., 103, 1858–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luwor R.B., et al. (2013) Targeting Stat3 and Smad7 to restore TGF-β cytostatic regulation of tumor cells in vitro and in vivo . Oncogene, 32, 2433–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chi X.Z., et al. (2005) RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor {beta}-activated SMAD. Mol. Cell. Biol., 25, 8097–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yano T., et al. (2006) The RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells undergoing transforming growth factor beta-induced apoptosis. Mol. Cell. Biol., 26, 4474–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ito K., et al. (2011) Loss of Runx3 is a key event in inducing precancerous state of the stomach. Gastroenterology, 140, 1536–46.e8. [DOI] [PubMed] [Google Scholar]
- 68. Goh Y.M., et al. (2010) Src kinase phosphorylates RUNX3 at tyrosine residues and localizes the protein in the cytoplasm. J. Biol. Chem., 285, 10122–10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Seoane J., et al. (2004) Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell, 117, 211–223. [DOI] [PubMed] [Google Scholar]
- 70. Gomis R.R., et al. (2006) C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell, 10, 203–214. [DOI] [PubMed] [Google Scholar]
- 71. Brunet A., et al. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell, 96, 857–868. [DOI] [PubMed] [Google Scholar]
- 72. Azar R., et al. (2009) 4E-BP1 is a target of Smad4 essential for TGFbeta-mediated inhibition of cell proliferation. EMBO J., 28, 3514–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bailey K.A., et al. (2012) Transcriptional Modulation of the ERK1/2 MAPK and NF-κB Pathways in Human Urothelial Cells After Trivalent Arsenical Exposure: Implications for Urinary Bladder Cancer. J. Can. Res. Updates, 1, 57–68. [PMC free article] [PubMed] [Google Scholar]
- 74. Germolec D.R., et al. (1996) Arsenic induces overexpression of growth factors in human keratinocytes. Toxicol. Appl. Pharmacol., 141, 308–318. [DOI] [PubMed] [Google Scholar]
- 75. Allison P., et al. (2013) Disruption of canonical TGFβ-signaling in murine coronary progenitor cells by low level arsenic. Toxicol. Appl. Pharmacol., 272, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Goodenough D.A., et al. (1996) Connexins, connexons, and intercellular communication. Annu. Rev. Biochem., 65, 475–502. [DOI] [PubMed] [Google Scholar]
- 77. Laird D.W. (2006) Life cycle of connexins in health and disease. Biochem. J., 394, 527–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cronier L., et al. (2009) Gap junctions and cancer: new functions for an old story. Antioxid. Redox Signal., 11, 323–338. [DOI] [PubMed] [Google Scholar]
- 79. Naus C.C., et al. (2010) Implications and challenges of connexin connections to cancer. Nat. Rev. Cancer, 10, 435–441. [DOI] [PubMed] [Google Scholar]
- 80. Loewenstein W.R., et al. (1967) Intercellular communication and tissue growth. I. Cancerous growth. J. Cell Biol., 33, 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rose B., et al. (1993) Gap-junction protein gene suppresses tumorigenicity. Carcinogenesis, 14, 1073–1075. [DOI] [PubMed] [Google Scholar]
- 82. Naus C.C., et al. (1992) In vivo growth of C6 glioma cells transfected with connexin43 cDNA. Cancer Res., 52, 4208–4213. [PubMed] [Google Scholar]
- 83. Zhu D., et al. (1991) Transfection of C6 glioma cells with connexin 43 cDNA: analysis of expression, intercellular coupling, and cell proliferation. Proc. Natl. Acad. Sci. USA, 88, 1883–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Leithe E., et al. (2006) Downregulation of gap junctions in cancer cells. Crit. Rev. Oncog., 12, 225–256. [DOI] [PubMed] [Google Scholar]
- 85. Mesnil M. (2002) Connexins and cancer. Biol. Cell, 94, 493–500. [DOI] [PubMed] [Google Scholar]
- 86. Yamasaki H., et al. (1999) Connexins in tumour suppression and cancer therapy. Novartis Found. Symp., 219, 241–254. [DOI] [PubMed] [Google Scholar]
- 87. Moennikes O., et al. (1999) The effect of connexin32 null mutation on hepatocarcinogenesis in different mouse strains. Carcinogenesis, 20, 1379–1382. [DOI] [PubMed] [Google Scholar]
- 88. King T.J., et al. (2004) The gap junction protein connexin32 is a mouse lung tumor suppressor. Cancer Res., 64, 7191–7196. [DOI] [PubMed] [Google Scholar]
- 89. King T.J., et al. (2004) Mice deficient for the gap junction protein Connexin32 exhibit increased radiation-induced tumorigenesis associated with elevated mitogen-activated protein kinase (p44/Erk1, p42/Erk2) activation. Carcinogenesis, 25, 669–680. [DOI] [PubMed] [Google Scholar]
- 90. Plante I., et al. (2011) Cx43 suppresses mammary tumor metastasis to the lung in a Cx43 mutant mouse model of human disease. Oncogene, 30, 1681–1692. [DOI] [PubMed] [Google Scholar]
- 91. McLachlan E., et al. (2006) Connexins act as tumor suppressors in three-dimensional mammary cell organoids by regulating differentiation and angiogenesis. Cancer Res., 66, 9886–9894. [DOI] [PubMed] [Google Scholar]
- 92. Evans W.H., et al. (2006) The gap junction cellular internet: connexin hemichannels enter the signalling limelight. Biochem. J., 397, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kandouz M., et al. (2010) Gap junctions and connexins as therapeutic targets in cancer. Expert Opin. Ther. Targets, 14, 681–692. [DOI] [PubMed] [Google Scholar]
- 94. Decrock E., et al. (2009) Connexin 43 hemichannels contribute to the propagation of apoptotic cell death in a rat C6 glioma cell model. Cell Death Differ., 16, 151–163. [DOI] [PubMed] [Google Scholar]
- 95. Vinken M., et al. (2006) Connexins and their channels in cell growth and cell death. Cell. Signal., 18, 592–600. [DOI] [PubMed] [Google Scholar]
- 96. Ito A., et al. (2000) A role for heterologous gap junctions between melanoma and endothelial cells in metastasis. J. Clin. Invest., 105, 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Stoletov K., et al. (2013) Role of connexins in metastatic breast cancer and melanoma brain colonization. J. Cell Sci., 126(Pt 4), 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sin W.C., et al. (2012) Opposing roles of connexin43 in glioma progression. Biochim. Biophys. Acta, 1818, 2058–2067. [DOI] [PubMed] [Google Scholar]
- 99. Duffy H.S., et al. (2006) Cardiac connexins: genes to nexus. Adv. Cardiol., 42, 1–17. [DOI] [PubMed] [Google Scholar]
- 100. Meyer R.A., et al. (1992) Inhibition of gap junction and adherens junction assembly by connexin and A-CAM antibodies. J. Cell Biol., 119, 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wei C.J., et al. (2005) Connexin43 associated with an N-cadherin-containing multiprotein complex is required for gap junction formation in NIH3T3 cells. J. Biol. Chem., 280, 19925–19936. [DOI] [PubMed] [Google Scholar]
- 102. Matsuuchi L., et al. (2012) Gap junction proteins on the move: connexins, the cytoskeleton and migration. Biochim. Biophys. Acta, 1828, 94–108. [DOI] [PubMed] [Google Scholar]
- 103. Ruch R.J., et al. (2001) Gap-junction communication in chemical carcinogenesis. Drug Metab. Rev., 33, 117–124. [DOI] [PubMed] [Google Scholar]
- 104. Yamasaki H. (1996) Role of disrupted gap junctional intercellular communication in detection and characterization of carcinogens. Mutat. Res., 365, 91–105. [DOI] [PubMed] [Google Scholar]
- 105. Rosenkranz H.S., et al. (2000) Exploring the relationship between the inhibition of gap junctional intercellular communication and other biological phenomena. Carcinogenesis, 21, 1007–1011. [DOI] [PubMed] [Google Scholar]
- 106. Bager Y., et al. (1997) Altered function, localization and phosphorylation of gap junctions in rat liver epithelial, IAR 20, cells after treatment with PCBs or TCDD. Environ. Toxicol. Pharmacol., 3, 257–266. [DOI] [PubMed] [Google Scholar]
- 107. Hemming H., et al. (1991) Inhibition of dye transfer in rat liver WB cell culture by polychlorinated biphenyls. Pharmacol. Toxicol., 69, 416–420. [DOI] [PubMed] [Google Scholar]
- 108. Kang K.S., et al. (1996) Inhibition of gap junctional intercellular communication in normal human breast epithelial cells after treatment with pesticides, PCBs, and PBBs, alone or in mixtures. Environ. Health Perspect., 104, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ruch R.J., et al. (1986) Effects of tumor promoters, genotoxic carcinogens and hepatocytotoxins on mouse hepatocyte intercellular communication. Cell Biol. Toxicol., 2, 469–483. [DOI] [PubMed] [Google Scholar]
- 110. Swierenga S.H., et al. (1990) Effects on intercellular communication in human keratinocytes and liver-derived cells of polychlorinated biphenyl congeners with differing in vivo promotion activities. Carcinogenesis, 11, 921–926. [DOI] [PubMed] [Google Scholar]
- 111. Vinken M., et al. (2009) Gap junctional intercellular communication as a target for liver toxicity and carcinogenicity. Crit. Rev. Biochem. Mol. Biol., 44, 201–222. [DOI] [PubMed] [Google Scholar]
- 112. Rivedal E., et al. (2003) Supplemental role of the Ames mutation assay and gap junction intercellular communication in studies of possible carcinogenic compounds from diesel exhaust particles. Arch. Toxicol., 77, 533–542. [DOI] [PubMed] [Google Scholar]
- 113. Krutovskikh V.A., et al. (1995) Inhibition of rat liver gap junction intercellular communication by tumor-promoting agents in vivo. Association with aberrant localization of connexin proteins. Lab. Invest., 72, 571–577. [PubMed] [Google Scholar]
- 114. Rutten A.A., et al. (1988) Effect of retinol and cigarette-smoke condensate on dye-coupled intercellular communication between hamster tracheal epithelial cells. Carcinogenesis, 9, 315–320. [DOI] [PubMed] [Google Scholar]
- 115. Roemer E., et al. (2013) Characterization of a gap-junctional intercellular communication (GJIC) assay using cigarette smoke. Toxicol. Lett. [DOI] [PubMed] [Google Scholar]
- 116. Baker T.K., et al. (1995) Inhibition of gap junctional intercellular communication by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in rat hepatocytes. Carcinogenesis, 16, 2321–2326. [DOI] [PubMed] [Google Scholar]
- 117. Herrmann S., et al. (2002) Indolo[3,2-b]carbazole inhibits gap junctional intercellular communication in rat primary hepatocytes and acts as a potential tumor promoter. Carcinogenesis, 23, 1861–1868. [DOI] [PubMed] [Google Scholar]
- 118. Mally A., et al. (2002) Non-genotoxic carcinogens: early effects on gap junctions, cell proliferation and apoptosis in the rat. Toxicology, 180, 233–248. [DOI] [PubMed] [Google Scholar]
- 119. Plante I., et al. (2002) Decreased gap junctional intercellular communication in hexachlorobenzene-induced gender-specific hepatic tumor formation in the rat. Carcinogenesis, 23, 1243–1249. [DOI] [PubMed] [Google Scholar]
- 120. De Maio A., et al. (2000) Interruption of hepatic gap junctional communication in the rat during inflammation induced by bacterial lipopolysaccharide. Shock, 14, 53–59. [DOI] [PubMed] [Google Scholar]
- 121. Cowles C., et al. (2007) Different mechanisms of modulation of gap junction communication by non-genotoxic carcinogens in rat liver in vivo . Toxicology, 238, 49–59. [DOI] [PubMed] [Google Scholar]
- 122. Jeong S.H., et al. (2000) Cadmium decreases gap junctional intercellular communication in mouse liver. Toxicol. Sci., 57, 156–166. [DOI] [PubMed] [Google Scholar]
- 123. Gagliano N., et al. (2006) Early cytotoxic effects of ochratoxin A in rat liver: a morphological, biochemical and molecular study. Toxicology, 225, 214–224. [DOI] [PubMed] [Google Scholar]
- 124. Ren P., et al. (1998) Inhibition of gap junctional intercellular communication by tumor promoters in connexin43 and connexin32-expressing liver cells: cell specificity and role of protein kinase C. Carcinogenesis, 19, 169–175. [DOI] [PubMed] [Google Scholar]
- 125. Horvath A., et al. (2002) Determination of the epigenetic effects of ochratoxin in a human kidney and a rat liver epithelial cell line. Toxicon, 40, 273–282. [DOI] [PubMed] [Google Scholar]
- 126. Upham B.L., et al. (2008) Tumor promoting properties of a cigarette smoke prevalent polycyclic aromatic hydrocarbon as indicated by the inhibition of gap junctional intercellular communication via phosphatidylcholine-specific phospholipase C. Cancer Sci., 99, 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Upham B.L., et al. (2009) Structure-activity-dependent regulation of cell communication by perfluorinated fatty acids using in vivo and in vitro model systems. Environ. Health Perspect., 117, 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Machala M., et al. (2003) Inhibition of gap junctional intercellular communication by noncoplanar polychlorinated biphenyls: inhibitory potencies and screening for potential mode(s) of action. Toxicol. Sci., 76, 102–111. [DOI] [PubMed] [Google Scholar]
- 129. Matesic D.F., et al. (1994) Changes in gap-junction permeability, phosphorylation, and number mediated by phorbol ester and non-phorbol-ester tumor promoters in rat liver epithelial cells. Mol. Carcinog., 10, 226–236. [DOI] [PubMed] [Google Scholar]
- 130. Leithe E., et al. (2004) Ubiquitination and down-regulation of gap junction protein connexin-43 in response to 12-O-tetradecanoylphorbol 13-acetate treatment. J. Biol. Chem., 279, 50089–50096. [DOI] [PubMed] [Google Scholar]
- 131. Rivedal E., et al. (2005) Connexin43 synthesis, phosphorylation, and degradation in regulation of transient inhibition of gap junction intercellular communication by the phorbol ester TPA in rat liver epithelial cells. Exp. Cell Res., 302, 143–152. [DOI] [PubMed] [Google Scholar]
- 132. Guan X., et al. (1996) Gap junction endocytosis and lysosomal degradation of connexin43-P2 in WB-F344 rat liver epithelial cells treated with DDT and lindane. Carcinogenesis, 17, 1791–1798. [DOI] [PubMed] [Google Scholar]
- 133. Šimečková P, Vondráček J, Andrysík Z, et al. The 2,2’,4,4’,5,5’-hexachlorobiphenyl-enhanced degradation of connexin 43 involves both proteasomal and lysosomal activities. Toxicol Sci 2009; 107: 9–18. [DOI] [PubMed] [Google Scholar]
- 134. Gakhar G., et al. (2009) Regulation of gap junctional intercellular communication by TCDD in HMEC and MCF-7 breast cancer cells. Toxicol. Appl. Pharmacol., 235, 171–181. [DOI] [PubMed] [Google Scholar]
- 135. Sai K., et al. (1998) Inhibitory effect of pentachlorophenol on gap junctional intercellular communication in rat liver epithelial cells in vitro . Cancer Lett., 130, 9–17. [DOI] [PubMed] [Google Scholar]
- 136. Polyak K., et al. (1994) p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev., 8, 9–22. [DOI] [PubMed] [Google Scholar]
- 137. Hanahan D., et al. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 138. Lieberman M.A., et al. (1981) Density-dependent regulation of cell growth: an example of a cell-cell recognition phenomenon. J. Membr. Biol., 63, 1–11. [DOI] [PubMed] [Google Scholar]
- 139. Halbleib J.M., et al. (2006) Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev., 20, 3199–3214. [DOI] [PubMed] [Google Scholar]
- 140. Dietrich C., et al. (1997) Differences in the mechanisms of growth control in contact-inhibited and serum-deprived human fibroblasts. Oncogene, 15, 2743–2747. [DOI] [PubMed] [Google Scholar]
- 141. Kato A., et al. (1997) Inactivation of the cyclin D-dependent kinase in the rat fibroblast cell line, 3Y1, induced by contact inhibition. J. Biol. Chem., 272, 8065–8070. [DOI] [PubMed] [Google Scholar]
- 142. Levenberg S., et al. (1999) p27 is involved in N-cadherin-mediated contact inhibition of cell growth and S-phase entry. Oncogene, 18, 869–876. [DOI] [PubMed] [Google Scholar]
- 143. Deffie A., et al. (1995) Cyclin E restores p53 activity in contact-inhibited cells. Mol. Cell. Biol., 15, 3926–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Swat A., et al. (2009) Cell density-dependent inhibition of epidermal growth factor receptor signaling by p38alpha mitogen-activated protein kinase via Sprouty2 downregulation. Mol. Cell. Biol., 29, 3332–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Ewen M.E., et al. (1993) TGF beta inhibition of Cdk4 synthesis is linked to cell cycle arrest. Cell, 74, 1009–1020. [DOI] [PubMed] [Google Scholar]
- 146. Grazia Lampugnani M., et al. (2003) Contact inhibition of VEGF-induced proliferation requires vascular endothelial cadherin, beta-catenin, and the phosphatase DEP-1/CD148. J. Cell Biol., 161, 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Wang C.Z., et al. (2009) DDR1/E-cadherin complex regulates the activation of DDR1 and cell spreading. Am. J. Physiol. Cell Physiol., 297, C419–C429. [DOI] [PubMed] [Google Scholar]
- 148. Shen Y., et al. (2007) SRC utilizes Cas to block gap junctional communication mediated by connexin43. J. Biol. Chem., 282, 18914–18921. [DOI] [PubMed] [Google Scholar]
- 149. Alexander D.B., et al. (2004) Normal cells control the growth of neighboring transformed cells independent of gap junctional communication and SRC activity. Cancer Res., 64, 1347–1358. [DOI] [PubMed] [Google Scholar]
- 150. Rubin H. (2008) Contact interactions between cells that suppress neoplastic development: can they also explain metastatic dormancy? Adv. Cancer Res., 100, 159–202. [DOI] [PubMed] [Google Scholar]
- 151. Li X., et al. (2009) Regulation of miRNA expression by Src and contact normalization: effects on nonanchored cell growth and migration. Oncogene, 28, 4272–4283. [DOI] [PubMed] [Google Scholar]
- 152. Jhon Alberto Ochoa-Alvarez CG, et al. (2011) Goldberg contact normalization: mechanisms and pathways to biomarkers and chemotherapeutic targets. In Extracellular and intracellular signaling. RSC Publishing, pp. 105–115. [Google Scholar]
- 153. Panse J., et al. (1997) Fibroblasts transformed by chemical carcinogens are sensitive to intercellular induction of apoptosis: implications for the control of oncogenesis. Carcinogenesis, 18, 259–264. [DOI] [PubMed] [Google Scholar]
- 154. Krishnan H., et al. (2012) SRC points the way to biomarkers and chemotherapeutic targets. Genes Cancer, 3, 426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Oesch F., et al. (1988) 12-O-tetradecanoylphorbol-13-acetate releases human diploid fibroblasts from contact-dependent inhibition of growth. Carcinogenesis, 9, 1319–1322. [DOI] [PubMed] [Google Scholar]
- 156. Dietrich C., et al. (2010) The aryl hydrocarbon receptor (AhR) in the regulation of cell-cell contact and tumor growth. Carcinogenesis, 31, 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Barouki R., et al. (2007) The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett., 581, 3608–3615. [DOI] [PubMed] [Google Scholar]
- 158. Kung T., et al. (2009) The aryl hydrocarbon receptor (AhR) pathway as a regulatory pathway for cell adhesion and matrix metabolism. Biochem. Pharmacol., 77, 536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Andrysík Z., et al. (2007) The aryl hydrocarbon receptor-dependent deregulation of cell cycle control induced by polycyclic aromatic hydrocarbons in rat liver epithelial cells. Mutat. Res., 615, 87–97. [DOI] [PubMed] [Google Scholar]
- 160. Weiss C., et al. (2008) TCDD deregulates contact inhibition in rat liver oval cells via Ah receptor, JunD and cyclin A. Oncogene, 27, 2198–2207. [DOI] [PubMed] [Google Scholar]
- 161. Andrysík Z., et al. (2013) Aryl hydrocarbon receptor-mediated disruption of contact inhibition is associated with connexin43 downregulation and inhibition of gap junctional intercellular communication. Arch. Toxicol., 87, 491–503. [DOI] [PubMed] [Google Scholar]
- 162. Andersson H., et al. (2012) Proangiogenic effects of environmentally relevant levels of bisphenol A in human primary endothelial cells. Arch. Toxicol., 86, 465–474. [DOI] [PubMed] [Google Scholar]
- 163. Li M.W., et al. (2010) Connexin 43 is critical to maintain the homeostasis of the blood-testis barrier via its effects on tight junction reassembly. Proc. Natl. Acad. Sci. U. S. A., 107, 17998–18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Lin Z.X., et al. (1986) Inhibition of gap junctional intercellular communication in human teratocarcinoma cells by organochlorine pesticides. Toxicol. Appl. Pharmacol., 83, 10–19. [DOI] [PubMed] [Google Scholar]
- 165. Ruch R.J., et al. (1987) Inhibition of intercellular communication between mouse hepatocytes by tumor promoters. Toxicol. Appl. Pharmacol., 87, 111–120. [DOI] [PubMed] [Google Scholar]
- 166. Liu J., et al. (2015) Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett., 356, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Ravi R., et al. (2000) Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev., 14, 34–44. [PMC free article] [PubMed] [Google Scholar]
- 168. Hanel W., et al. (2012) Links between mutant p53 and genomic instability. J. Cell. Biochem., 113, 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Hacke K., et al. (2010) Regulation of MCP-1 chemokine transcription by p53. Mol. Cancer, 9, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Tang X., et al. (2012) p53 is an important regulator of CCL2 gene expression. Curr. Mol. Med., 12, 929–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Wang L., et al. (2011) PI3K pathway activation results in low efficacy of both trastuzumab and lapatinib. BMC Cancer, 11, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Chipuk J.E., et al. (2006) Dissecting p53-dependent apoptosis. Cell Death Differ., 13, 994–1002. [DOI] [PubMed] [Google Scholar]
- 173. Nicolay B.N., et al. (2013) Loss of RBF1 changes glutamine catabolism. Genes Dev., 27, 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Gabellini C., et al. (2006) Involvement of RB gene family in tumor angiogenesis. Oncogene, 25, 5326–5332. [DOI] [PubMed] [Google Scholar]
- 175. Amato A., et al. (2009) CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer, 8, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Attardi L.D., et al. (2013) RB goes mitochondrial. Genes Dev., 27, 975–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Guido C., et al. (2012) Metabolic reprogramming of cancer-associated fibroblasts by TGF-β drives tumor growth: connecting TGF-β signaling with “Warburg-like” cancer metabolism and L-lactate production. Cell Cycle, 11, 3019–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Geng L., et al. (2013) TGF-beta suppresses VEGFA-mediated angiogenesis in colon cancer metastasis. PLoS One, 8, e59918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Glick A.B., et al. (1996) Transforming growth factor beta 1 suppresses genomic instability independent of a G1 arrest, p53, and Rb. Cancer Res., 56, 3645–3650. [PubMed] [Google Scholar]
- 180. Rao C.V., et al. (2013) Genomic instability and colon carcinogenesis: from the perspective of genes. Front. Oncol., 3, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Taylor M.A., et al. (2011) Role of TGF-β and the tumor microenvironment during mammary tumorigenesis. Gene Expr., 15, 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Mishra S., et al. (2013) Androgen receptor and microRNA-21 axis downregulates transforming growth factor beta receptor II (TGFBR2) expression in prostate cancer. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Shackelford D.B. (2013) Unravelling the connection between metabolism and tumorigenesis through studies of the liver kinase B1 tumour suppressor. J. Carcinog., 12, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Londesborough A., et al. (2008) LKB1 in endothelial cells is required for angiogenesis and TGFbeta-mediated vascular smooth muscle cell recruitment. Development, 135, 2331–2338. [DOI] [PubMed] [Google Scholar]
- 185. Forcet C., et al. (2007) Dialogue between LKB1 and AMPK: a hot topic at the cellular pole. Sci. STKE, 2007, pe51. [DOI] [PubMed] [Google Scholar]
- 186. Gurumurthy S., et al. (2010) The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature, 468, 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Takeda S., et al. (2007) LKB1 is crucial for TRAIL-mediated apoptosis induction in osteosarcoma. Anticancer Res., 27, 761–768. [PubMed] [Google Scholar]
- 188. Gatenby R.A., et al. (2003) The glycolytic phenotype in carcinogenesis and tumor invasion: insights through mathematical models. Cancer Res., 63, 3847–3854. [PubMed] [Google Scholar]
- 189. Zhu W., et al. (1997) Increased genetic stability of HeLa cells after connexin 43 gene transfection. Cancer Res., 57, 2148–2150. [PubMed] [Google Scholar]
- 190. Fiaschi T., et al. (2012) Reciprocal metabolic reprogramming through lactate shuttle coordinately influences tumor-stroma interplay. Cancer Res., 72, 5130–5140. [DOI] [PubMed] [Google Scholar]
- 191. Gregory S.H., et al. (1979) Glycolytic enzyme activities in malignant cells grown in vitro and in vivo . Cancer Lett., 7, 319–324. [DOI] [PubMed] [Google Scholar]
- 192. Feyler S., et al. (2012) Tumour cell generation of inducible regulatory T-cells in multiple myeloma is contact-dependent and antigen-presenting cell-independent. PLoS One, 7, e35981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. González-Suárez E., et al. (2002) Cooperation between p53 mutation and high telomerase transgenic expression in spontaneous cancer development. Mol. Cell. Biol., 22, 7291–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Li H., et al. (1999) Molecular interactions between telomerase and the tumor suppressor protein p53 in vitro . Oncogene, 18, 6785–6794. [DOI] [PubMed] [Google Scholar]
- 195. Maniwa Y., et al. (2001) Association of p53 gene mutation and telomerase activity in resectable non-small cell lung cancer. Chest, 120, 589–594. [DOI] [PubMed] [Google Scholar]
- 196. Lee J.S., et al. (2014) A novel tumor-promoting role for nuclear factor IA in glioblastomas is mediated through negative regulation of p53, p21, and PAI1. Neuro. Oncol., 16, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Goh A.M., et al. (2011) The role of mutant p53 in human cancer. J. Pathol., 223, 116–126. [DOI] [PubMed] [Google Scholar]
- 198. Cooks T., et al. (2013) Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell, 23, 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Lorin S., et al. (2013) Autophagy regulation and its role in cancer. Semin. Cancer Biol., 23, 361–379. [DOI] [PubMed] [Google Scholar]
- 200. Rayess H., et al. (2012) Cellular senescence and tumor suppressor gene p16. Int. J. Cancer, 130, 1715–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Sperka T., et al. (2012) DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol., 13, 579–590. [DOI] [PubMed] [Google Scholar]
- 202. Gore A.J., et al. (2014) Pancreatic cancer-associated retinoblastoma 1 dysfunction enables TGF-β to promote proliferation. J. Clin. Invest., 124, 338–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Chou N.H., et al. (2006) Expression of altered retinoblastoma protein inversely correlates with tumor invasion in gastric carcinoma. World J. Gastroenterol., 12, 7188–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Ying L., et al. (2005) Chronic inflammation promotes retinoblastoma protein hyperphosphorylation and E2F1 activation. Cancer Res., 65, 9132–9136. [DOI] [PubMed] [Google Scholar]
- 205. Mankame T.P., et al. (2012) The RB tumor suppressor positively regulates transcription of the anti-angiogenic protein NOL7. Neoplasia, 14, 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Li H., et al. (2006) Transforming growth factor beta suppresses human telomerase reverse transcriptase (hTERT) by Smad3 interactions with c-Myc and the hTERT gene. J. Biol. Chem., 281, 25588–25600. [DOI] [PubMed] [Google Scholar]
- 207. Fabregat I., et al. (2013) TGF-beta signaling in cancer treatment. Curr. Pharm. Des, 20, 2934–2947. [DOI] [PubMed] [Google Scholar]
- 208. Hasegawa T., et al. (2013) Cancer-associated fibroblasts might sustain the stemness of scirrhous gastric cancer cells via transforming growth factor-β signaling. Int. J. Cancer. [DOI] [PubMed] [Google Scholar]
- 209. Bierie B., et al. (2010) Transforming growth factor beta (TGF-beta) and inflammation in cancer. Cytokine Growth Factor Rev., 21, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Bailey JM, Leach SD. (2012) Signaling pathways mediating epithelial- mesenchymal crosstalk in pancreatic cancer: Hedgehog, Notch and TGFbeta. In Grippo PJ, Munshi HG. (eds) Pancreatic cancer and tumor microenvironment. Trivandrum, India. [PubMed] [Google Scholar]
- 211. Sanchez-Cespedes M. (2011) The role of LKB1 in lung cancer. Fam. Cancer, 10, 447–453. [DOI] [PubMed] [Google Scholar]
- 212. Shackelford D.B., et al. (2009) The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer, 9, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Kline E.R., et al. (2013) LKB1 represses focal adhesion kinase (FAK) signaling via a FAK-LKB1 complex to regulate FAK site maturation and directional persistence. J. Biol. Chem., 288, 17663–17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Xu X., et al. (2013) LKB1 controls human bronchial epithelial morphogenesis through p114RhoGEF-dependent RhoA activation. Mol. Cell. Biol., 33, 2671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Wingo S.N., et al. (2009) Somatic LKB1 mutations promote cervical cancer progression. PLoS One, 4, e5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Salem A.F., et al. (2012) Two-compartment tumor metabolism: autophagy in the tumor microenvironment and oxidative mitochondrial metabolism (OXPHOS) in cancer cells. Cell Cycle, 11, 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Spath C., et al. (2013) Inverse relationship between tumor proliferation markers and connexin expression in a malignant cardiac tumor originating from mesenchymal stem cell engineered tissue in a rat in vivo model. Front. Pharmacol., 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Ezumi K., et al. (2008) Aberrant expression of connexin 26 is associated with lung metastasis of colorectal cancer. Clin. Cancer Res., 14, 677–684. [DOI] [PubMed] [Google Scholar]
- 219. Ogawa K., et al. (2012) Silencing of connexin 43 suppresses invasion, migration and lung metastasis of rat hepatocellular carcinoma cells. Cancer Sci., 103, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Zhang D., et al. (2012) Cx31.1 acts as a tumour suppressor in non-small cell lung cancer (NSCLC) cell lines through inhibition of cell proliferation and metastasis. J. Cell. Mol. Med., 16, 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Czyz J. (2008) The stage-specific function of gap junctions during tumourigenesis. Cell. Mol. Biol. Lett., 13, 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Corn P.G. (2012) The tumor microenvironment in prostate cancer: elucidating molecular pathways for therapy development. Cancer Manag. Res., 4, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Li J.F., et al. (2014) The effects of cell compressibility, motility and contact inhibition on the growth of tumor cell clusters using the Cellular Potts Model. J. Theor. Biol., 343, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Dixon J.S. (1987) Short-term clinical trials of anti-rheumatoid drugs–an opinion. Agents Actions, 21, 89–92. [DOI] [PubMed] [Google Scholar]
- 225. Schrader J., et al. (2009) Restoration of contact inhibition in human glioblastoma cell lines after MIF knockdown. BMC Cancer, 9, 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. WARBURG O. (1956) On the origin of cancer cells. Science, 123, 309–314. [DOI] [PubMed] [Google Scholar]
- 227. Shackelford D.B., et al. (2009) The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer, 9, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Andrade-Vieira R., et al. (2013) Loss of LKB1 expression reduces the latency of ErbB2-mediated mammary gland tumorigenesis, promoting changes in metabolic pathways. PLoS One, 8, e56567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Sen N., et al. (2012) p53 and metabolism: old player in a new game. Transcription, 3, 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Zhang C., et al. (2013) Tumour-associated mutant p53 drives the Warburg effect. Nat. Commun., 4, 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Dasgupta B., et al. (2009) AMP-activated protein kinase phosphorylates retinoblastoma protein to control mammalian brain development. Dev. Cell, 16, 256–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Liu Y., et al. (2012) A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo . Mol. Cancer Ther., 11, 1672–1682. [DOI] [PubMed] [Google Scholar]
- 233. Fillon M. (2012) Getting it right: BPA and the difficulty proving environmental cancer risks. J. Natl. Cancer Inst., 104, 652–655. [DOI] [PubMed] [Google Scholar]
- 234. Betancourt A.M., et al. (2012) Altered carcinogenesis and proteome in mammary glands of rats after prepubertal exposures to the hormonally active chemicals bisphenol a and genistein. J. Nutr., 142, 1382S–1388S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Durando M., et al. (2011) Prenatal exposure to bisphenol A promotes angiogenesis and alters steroid-mediated responses in the mammary glands of cycling rats. J. Steroid Biochem. Mol. Biol., 127, 35–43. [DOI] [PubMed] [Google Scholar]
- 236. Ptak A., et al. (2012) Bisphenol A induces leptin receptor expression, creating more binding sites for leptin, and activates the JAK/Stat, MAPK/ERK and PI3K/Akt signalling pathways in human ovarian cancer cell. Toxicol. Lett., 210, 332–337. [DOI] [PubMed] [Google Scholar]
- 237. Pupo M., et al. (2012) Bisphenol A induces gene expression changes and proliferative effects through GPER in breast cancer cells and cancer-associated fibroblasts. Environ. Health Perspect., 120, 1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Goodson W.H.3rd et al. (2011) Activation of the mTOR pathway by low levels of xenoestrogens in breast epithelial cells from high-risk women. Carcinogenesis, 32, 1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Allard P., et al. (2010) Bisphenol A impairs the double-strand break repair machinery in the germline and causes chromosome abnormalities. Proc. Natl. Acad. Sci. USA, 107, 20405–20410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Ito Y., et al. (2012) Identification of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) as a novel target of bisphenol A. PLoS One, 7, e50481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Takahashi A., et al. (2004) Bisphenol A from dental polycarbonate crown upregulates the expression of hTERT. J. Biomed. Mater. Res. B. Appl. Biomater., 71, 214–221. [DOI] [PubMed] [Google Scholar]
- 242. Zhu H., et al. (2010) Environmental endocrine disruptors promote invasion and metastasis of SK-N-SH human neuroblastoma cells. Oncol. Rep., 23, 129–139. [PubMed] [Google Scholar]
- 243. Hassan Z.K., et al. (2012) Bisphenol A induces hepatotoxicity through oxidative stress in rat model. Oxid. Med. Cell. Longev., 2012, 194829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Bratton M.R., et al. (2012) The organochlorine o,p’-DDT plays a role in coactivator-mediated MAPK crosstalk in MCF-7 breast cancer cells. Environ. Health Perspect., 120, 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Yáñez L., et al. (2004) DDT induces DNA damage in blood cells. Studies in vitro and in women chronically exposed to this insecticide. Environ. Res., 94, 18–24. [DOI] [PubMed] [Google Scholar]
- 246. Hurd-Brown T., et al. (2013) Effects of DDT and triclosan on tumor-cell binding capacity and cell-surface protein expression of human natural killer cells. J. Appl. Toxicol., 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Arce G.T., et al. (2010) Genetic toxicology of folpet and captan. Crit. Rev. Toxicol., 40, 546–574. [DOI] [PubMed] [Google Scholar]
- 248. Canal-Raffin M., et al. (2008) Cytotoxicity of folpet fungicide on human bronchial epithelial cells. Toxicology, 249, 160–166. [DOI] [PubMed] [Google Scholar]
- 249. Campos-Pereira F.D., et al. (2012) Early cytotoxic and genotoxic effects of atrazine on Wistar rat liver: a morphological, immunohistochemical, biochemical, and molecular study. Ecotoxicol. Environ. Saf., 78, 170–177. [DOI] [PubMed] [Google Scholar]
- 250. Pinchuk L.M., et al. (2007) In vitro atrazine exposure affects the phenotypic and functional maturation of dendritic cells. Toxicol. Appl. Pharmacol., 223, 206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Hou L., et al. (2013) Lifetime pesticide use and telomere shortening among male pesticide applicators in the Agricultural Health Study. Environ. Health Perspect., 121, 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Jin X.T., et al. (2014) Dichlorodiphenyltrichloroethane exposure induces the growth of hepatocellular carcinoma via Wnt/β-catenin pathway. Toxicol. Lett., 225, 158–166. [DOI] [PubMed] [Google Scholar]
- 253. Han E.H., et al. (2008) o,p’-DDT induces cyclooxygenase-2 gene expression in murine macrophages: Role of AP-1 and CRE promoter elements and PI3-kinase/Akt/MAPK signaling pathways. Toxicol. Appl. Pharmacol., 233, 333–342. [DOI] [PubMed] [Google Scholar]
- 254. Gordon E., et al. (2012) Folpet-induced short term cytotoxic and proliferative changes in the mouse duodenum. Toxicol. Mech. Methods, 22, 54–59. [DOI] [PubMed] [Google Scholar]
- 255. Albanito L., et al. (2008) G-protein-coupled receptor 30 and estrogen receptor-alpha are involved in the proliferative effects induced by atrazine in ovarian cancer cells. Environ. Health Perspect., 116, 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 256. Tsuda H., et al. (2005) High susceptibility of human c-Ha-ras proto-oncogene transgenic rats to carcinogenesis: a cancer-prone animal model. Cancer Sci., 96, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Ueda M., et al. (2005) Possible enhancing effects of atrazine on growth of 7,12-dimethylbenz(a) anthracene-induced mammary tumors in ovariectomized Sprague-Dawley rats. Cancer Sci., 96, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Wetzel L.T., et al. (1994) Chronic effects of atrazine on estrus and mammary tumor formation in female Sprague-Dawley and Fischer 344 rats. J. Toxicol. Environ. Health, 43, 169–182. [DOI] [PubMed] [Google Scholar]