Abstract
Cardiovirus infections inhibit nucleocytoplasmic trafficking by Leader protein-induced phosphorylation of Phe/Gly-containing nucleoporins (Nups). Recombinant Leader from encephalomyocarditis virus, Theiler’s murine encephalomyelitis virus and Saffold virus target the same subset of Nups, including Nup62 and Nup98, but not Nup50. Reporter cell lines with fluorescence mCherry markers for M9, RS and classical SV40 import pathways, as well as the Crm1-mediated export pathway, all responded to transfection with the full panel of Leader proteins, showing consequent cessation of path-specific active import/export. For this to happen, the Nups had to be presented in the context of intact nuclear pores and exposed to cytoplasmic extracts. The Leader phosphorylation cascade was not effective against recombinant Nup proteins. The findings support a model of Leader-dependent Nup phosphorylation with the purpose of disrupting Nup-transportin interactions.
Keywords: nucleocytoplasmic trafficking inhibition, cardiovirus, Leader protein, okadaic acid
Introduction
Cardioviruses, as members of the Picornaviridae family, are positive-sense, single-stranded RNA viruses. Their preferred hosts are rodents, although some will readily infect other mammals. Of the three recognized species in this genus, two are represented by encephalomyocarditis virus (EMCV) and Theiler’s murine encephalomyelitis virus (TMEV). Saffold virus (SafV), within the same Cardiovirus B species as TMEV, is one of the few members of this genus to infect humans (1). While cardioviruses have similar polyprotein organizations, each encodes a variable-length Leader (L) protein, none of which have homologs or analogs in other viruses or cells. Leader proteins are unique determinants of cardiovirus anti-host activities. Although not kinases themselves, the Leaders induce intense hyper-phosphorylation of certain Phe/Gly-containing nuclear pore proteins (Nups), including Nup62, Nup153 and Nup214 shortly after infection (2, 3). Phosphorylation of Nups within nuclear pore complexes (NPC) down-regulates active nuclear import by hindering importin association with the Nups (4, 5). This novel mechanism can be recapitulated by transfection of L-encoding cDNAs into cells or by the addition of recombinant L protein into cell extracts containing nuclei as targets (3, 6). The in vitro assays directly mimic the trafficking inhibition observed by cardiovirus infection-directed Nup phosphorylation.
The EMCV L (LE) is 67 amino acids (aa) long. The NMR solution structure for the closely related Mengo L (LM) shows an unusual N-proximal zinc-finger domain. The rest of the protein configures as random coil (7). Functionally, the LM coiled region has a C-proximal acid-rich domain and a central hinge segment which forms the primary induced-fit binding contacts with RanGTPase, a requisite partner in the anti-host activity (8-10). LE is shuttled to the nucleus after its polyprotein synthesis presumably by interactions with the viral 2A protein with which it can also interact (11). In the presence of guanine nucleotide exchange factor, RCC1, just inside the nuclear rim, LE then exchanges 2A for Ran (11). The LE interaction with this key trafficking regulator is very tight, with a measured KD of about 3 nM (12). Before, or shortly after this nuclear exchange, LE becomes phosphorylated at Thr47 and Tyr41, in steps which are obligate for the consequent LE-dependent Nup phosphorylation activities (6). The NMR orientation of LM, when bound to Ran, shows the pairing forces Ran into an allosteric conformation which mimics the RanGTP-bound active state of this transport regulator. As such, Ran (with LM) becomes competent to bind exportins and their cargos for putative shuttling to the cytoplasm (7). It has been proposed that this complex (LM:Ran:exportin), formed in the nucleus, subsequently recruits activated kinase cargos, such as p38 and/or ERK1/2 (13), and the full unit, unable to dissociate because of the bound Leader, becomes trapped in the nuclear pore, where the kinases catalyze the cell-debilitating hyper-phosphorylation of Nup62, Nup153 and Nup214 (7).
The L proteins of SafV (LS) and TMEV (LT) are similar in many respects. Cardiovirus B species Leaders are 4 (LS) to 9 (LT) aa longer than LE or LM, with the added length mostly evident as short contiguous insertions C-terminal to the Ran-contact hinge domain. Each also has an additional small relative deletion next to the N-terminal initiating Met. Like LM/LE, the TMEV and SafV proteins become dually phosphorylated in cells or in recombinant form, but at different sites (i.e. Ser57 and Thr58, respectively) and by different kinases (AMPK, not CK2) than the better studied EMCV systems (10). When recombinant LT or LS, are introduced into cells, even in the absence of infection, they can indeed induce Nup62 phosphorylation, the common assay for hyperphosphorylation (6).
There are many elements of the L-directed Nup phosphorylation model that are not well understood. It is unknown, for example, if there are other Nup proteins which are targets (or non-targets) of the activated kinase complexes. The matrix protein of vesicular stomatitis virus (VSV) causes nucleocytoplasmic trafficking inhibition (14) by complexing with Nup98 and the exportin Rae1 (15). Similar to a phenotype described for TMEV infections, where Nup98 is also reported to be phosphorylated (6, 16), VSV inhibition of Nup98-dependent trafficking stops the export of cellular mRNAs and prevents the transcription of interferon and chemokine products (3, 17-19). The full collection of Nup62, Nup98, Nup153 and Nup214 are also among the demonstrated substrates for human rhinovirus (RV) protease 2A. These cousins of the cardioviruses inactivate NPC import/export by multiple Nup cleavage reactions rather than by phosphorylation cascades (20, 21). The sequence differences in specific 2Apro which are characteristic of the multiple RV genotypes allow individual viruses to preferentially cleave selected cohorts of Nup substrate panels with different avidities and rates (21). Consequently, not all import/export pathways are equivalently disabled by every RV, allowing each 2Apro sequence to manifest as a strain-specific Nup degradation pattern (22).
With the cardioviruses, it isn’t known whether LE-directed Nup hyper-phosphorylation is aimed more generically at all transport pathways, or like the RV, is more selectively directed at only those import/export units which use particular subsets of Nups. Cardiovirus systems to test these parameters have typically linked traceable reporters (e.g. GFP) to peptide fragments (e.g. SV40) with non-specific nuclear import localization signals (NLS), or tried to follow common cellular mRNAs as the metric for nuclear egress (8). These previous experiments are less sensitive than the newer, path-specific assays recently described for the RV (22). Application of those new systems now provides clarification on the scope of LX disruption of trafficking pathways for the LE, LS and LT proteins, and as described here, show all 3 of these viruses act ubiquitously against 4 tested pathways, including a path dependent upon a nuclear export signal (NES) for protein egress. Moreover, observation of LX-dependent Nup phosphorylation becomes accelerated in cell-free systems by the presence of okadaic acid (OA), an inhibitor which prevents counterproductive phosphatase activities on both the susceptible Nups and on the required cellular kinases.
Results
Nup98 and Nup50
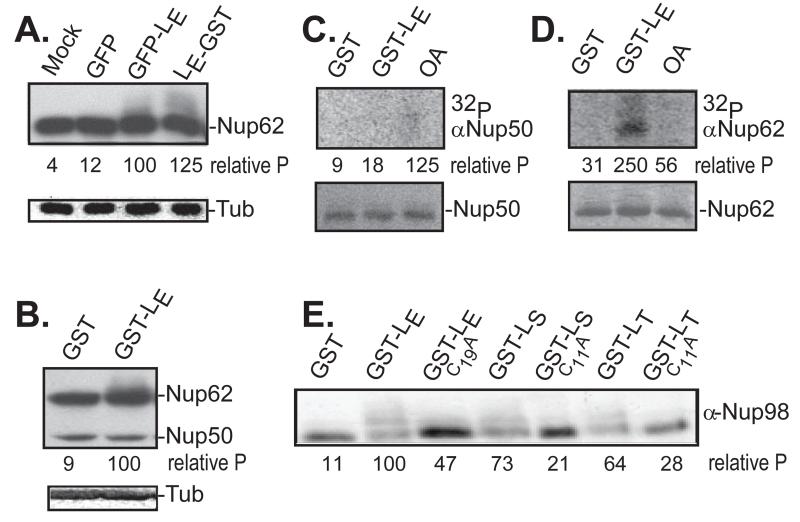
Transfection of cells with LX-encoding cDNAs, followed by Western analyses is the standard assay for LX-dependent Nup phosphorylation (8). Some prior experiments also evaluated Westerns or γ32P incorporation after incubation of recombinant LX proteins with fractionated cell nuclei and cytosol (9). The size (67-76 aa) and charge (pI ~3) of LX proteins presents technical issues unless these segments are linked to fusion tags, such as GST or GFP (13). For LE, the observed Nup phosphorylation after cDNA transfection was comparably strong (6) whether the tag was N-terminal (e.g. GFP-LE), or C-terminal (e.g. LE-GST), as illustrated in Fig 1A. The common detection antibody (mAb414) recognizes multiple Phe/Gly-containing Nups, albeit with differing affinities. Typically, Nup62 modifications manifest on gels as a “smear” towards higher mobility when multiple phosphates are added sequentially. The change is distinctive whether LE is assayed after transfection of cDNA (e.g. Fig 1A), or as recombinant protein in cell-free extracts (e.g. Fig 1B). The same is true for Nup153 and Nup214 (3). Nup50 and Nup98, however, are only intermittently detectable with this mAb (e.g. Fig 1B). For these, evaluation of the LX-dependent changes required different reagents. Cytosol/nuclei mixtures similar to Fig 1B, were labeled with γ32P-ATP, and then extracted with mAbs specific to Nup50 or Nup62. While Nup50 was observable by Western analysis (Fig 1B), the presence of GST-LE, did not direct detectable label incorporation (Fig 1C), nor did it shift in molecular weight. Nup62 on the other hand, was demonstrably labeled with 32P by the inclusion of GST-L (Fig 1D). Equivalent Nup98 reagents are not effective in similar immunoprecipitation experiments. For this evaluation, HeLa cells were transfected with cDNAs encoding LE-GST, LS-GST, LT-GST, and also with corresponding cognates encoding Cys-to-Ala LX-inactivating mutations (6). In every case when there was active LX protein, the Nup98 mAb detected the upward “smear” of LX-dependent phosphorylation. The activities ranked as LE>LS>LT for these particular conditions. Therefore, Nup98 but not Nup50, is a target of the LE-dependent phosphorylation cascades (Fig. 1E).
Figure 1.
Nup phosphorylation assays. (A) HeLa cells were transfected with cDNAs encoding the indicated proteins. After 16 hrs, harvested lysates were probed in Western assays using mAb414 (Nup62) or α-tubulin (Tub). (B) Recombinant LE-GST or GST-LE proteins (5 μg) were incubated with HeLa cytosol supplemented with isolated nuclei. After 45 min, the samples were fractionated, then probed by Western analyses as in A. (C) HeLa nuclei, cytosol and recombinant GST or GST-LE were incubated with γ32P-ATP in the presence or absence of okadaic acid (OA). After incubation at 37° for 45 minutes, proteins reactive with α-Nup50 were extracted and fractionated. Upper panel is an autoradiogram. Lower panel is a silver stain of the same materials. (D) Same as C, except immunoprecipitation was with α-Nup62. (E) Similar to A, unlabeled transfected lysates were fractionated, then probed in Western assays with α-Nup98. In panels A, B, E, the “relative P” is pixel count (TotalLab software) in the phosphorylated product, normalized to GFP-LE or GST-LE controls. For C, D, these values are the relative pixels in the labeled bands, normalized to the silver stain signals.
Import/export Pathway Imaging
During EMCV infections, LX cDNA transfections, or cell-free reactions with recombinant proteins, NPC active import/export is abrogated. Small proteins (<40 kD) and metabolites then diffuse across the NPC to equilibrium (3). Visualization of this process requires addition of fluorescent reporters linked to NLS sequences to record relative changes in nuclear/cytoplasmic cellular distribution. Recombinant GST-GFPNLS, for example, was previously tracked in digitonin-treated HeLa cells to document GST-LE concentration and rate effects (3). HTNLS (Halotag), transfected as cDNA into cells, showed similar relocalization (2, 3, 18). In all these previous experiments however, the tested reporter-NLS was from SV40, which traffics via the importin α/β pathways and is responsive only to particular segments of the Nup cohort (Table 1) specific to that karyopherin passage through the NPC (23-25).
Table 1. Nups Associated with NPC Pathways.
| NLS/NES Motif sequence | Required Nups1 | Karyopherin1 | |
|---|---|---|---|
| Classical import pathway. NLS is from the SV40 large T- antigen protein |
Nup62 | Nup214 | importin α/β |
| Nup98 | Nup358 | ||
| Nup153 | |||
|
| |||
| M9-mediated import pathway. NLS is from the M9 domain of mRNA binding protein hnRNPA1 |
Nup62 | transportin 1 | |
| Nup98 | |||
| Nup153 | |||
|
| |||
| RS-mediated import pathway. NLS is from the RS domain of splicing factor 2, an SR (Arg/Ser-rich) protein. |
Nup98 | transportin 3 | |
| Nup153 | |||
|
| |||
| Leucine-rich NES-mediated export pathway. NES is from the protein kinase A inhibitor (PKI) |
Nup62 | Nup214 | Crm1 |
| Nup98 | Nup358 | ||
| Nup153 | |||
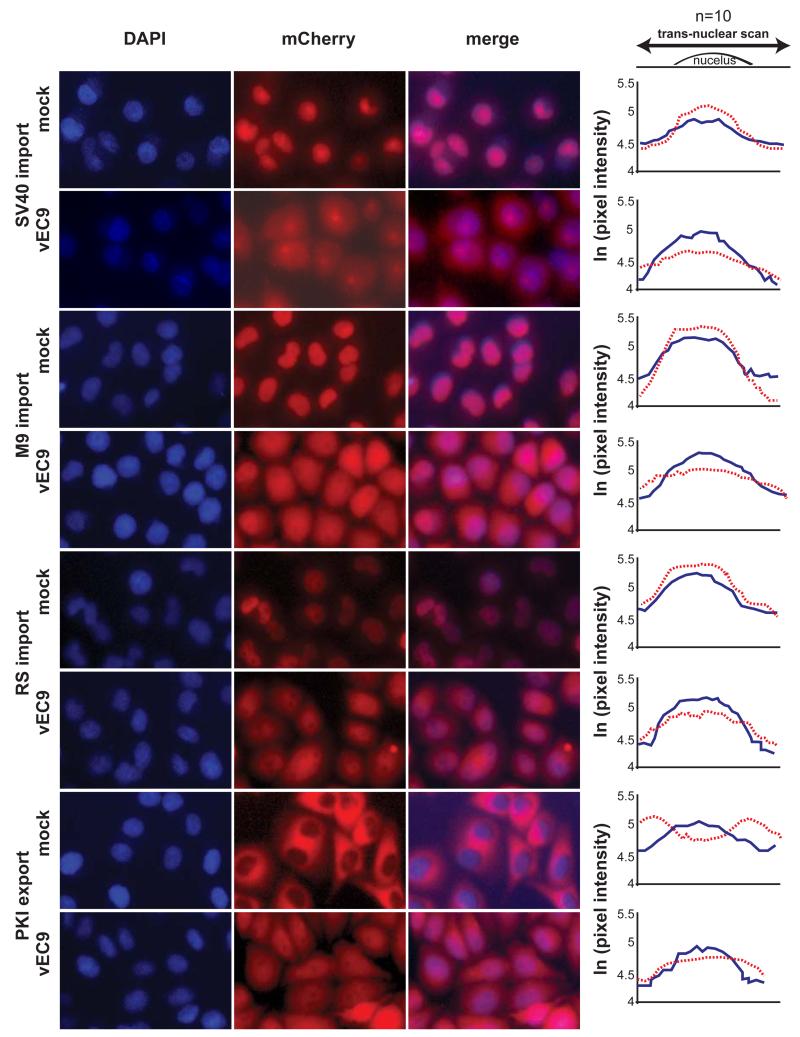
The impact of LX on other characterized transport pathways was assessed with HeLa cell lines transduced with mCherry reporter genes (~30 kD) linked to additional NLS/NES segments (~15-45 aa; (22)). After infection with vEC9 (3 hr), the cells and controls were fixed, stained with DAPI and imaged (Fig 2). Averaged pixel scans centered over the width of individual nuclei showed that the mCherry signals, compared to steady-state DAPI, diminished measurably after infection of cells, if the reporter was linked to the SV40 NLS, the M9 NLS (26), or the RS domain NLS from an SR protein, splicing factor 2 (27). Previous characterization of these cells with rhinovirus reagents confirmed the cell-wide stability of the total mCherry signal (22). The reporter signal redistributes out of nuclei during infection, but is not degraded. The fourth tested cell line expressed mCherry linked to the leucine-rich NES from PKI (28). This segment is sensitive to Crm1-mediated active nuclear egress. Here, the initial nuclear exclusion of the reporter was reversed after infection, allowing a stronger mCherry signal to accumulate in the nuclei relative to control cells. For each of these 4 lines, infection with vEC9 impacted the respective mCherry-labeled NPC transport pathway. Virus disruption of active transport into or out of the nuclei resulted in reporter redistribution by diffusion relative to the steady-state DAPI signals.
Figure 2.
Trafficking disruption by virus. Transduced HeLa cells expressing mCherry fusion proteins linked to NLS (SV40, M9 or RS) or NES (PKI) import/export sequences, were infected with vEC9 (MOI=15). At 3 hrs, the cells were fixed, stained with DAPI and imaged. For each condition, randomly selected individual cells (n=10) were scanned for pixel intensity (linear stretch of 80 pixels), centered on the DAPI signal. The values were averaged and plotted as the natural log of each signal for DAPI (blue solid line), and mCherry (dashed red line). The average standard deviation was 0.210 ln(x) (range: 0.124 to 0.274).
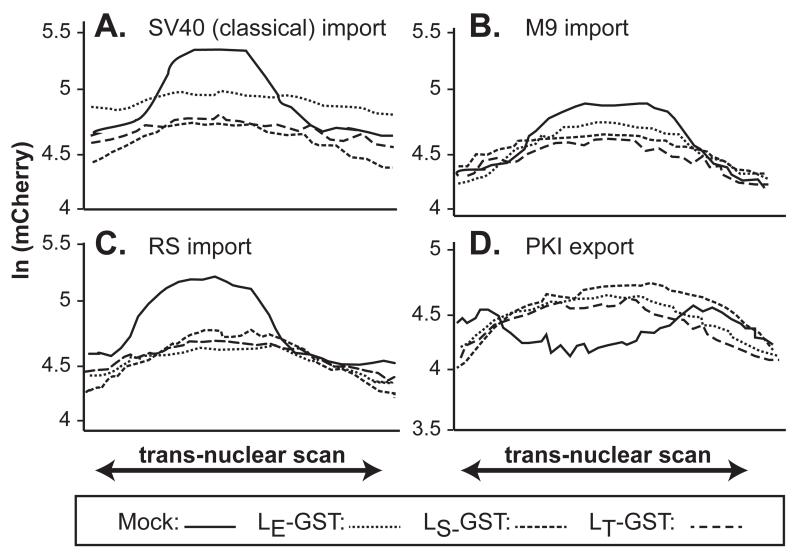
The LE protein of intact vEC9 is the effector for the experimental set depicted in Fig 2. Cell visualization assays with the same mCherry cell lines were repeated after transfection with cDNAs encoding LE-GST, LT-GST or LS-GST. Again, averaged pixel scans centering on the nuclei illustrated the impact of LX on mCherry relocalization (Fig 3), this time in live, unfixed cells. Within these graphs, the solid lines now represent the mCherry profiles observed in control cells (i.e. equivalent to Fig 2, mock). Relative to this, expression of all three LX proteins mediated measurable reporter diffusion out of (NLS lines), or into (NES line) nuclei. Unlike the rhinovirus 2Apro which can discriminate these respective import/export systems according to virus genotype (22), the 3 cardiovirus LX proteins seemed equivalently adept at disrupting all 4 tested transport pathways (Fig 3).
Figure 3.
Trafficking disruption by cDNAs. Similar to Fig 2, transduced HeLa cells expressing mCherry fusion proteins linked to the SV40 NLS (A), M9 NLS (B), RS NLS (C) or PKI NES (D) were transfected with cDNAs for LE-GST, LS-GST or LT-GST. After 16 hrs, individual live cells (n=8) were imaged for mCherry pixel intensity (linear stretch of 80 pixels) centered on the nucleus. Mock cells (solid lines) were transfected with cDNA for GST alone. The average standard deviation was 0.1274 ln(x) (range: 0.64 to 0.244).
Context for Nup Phosphorylation
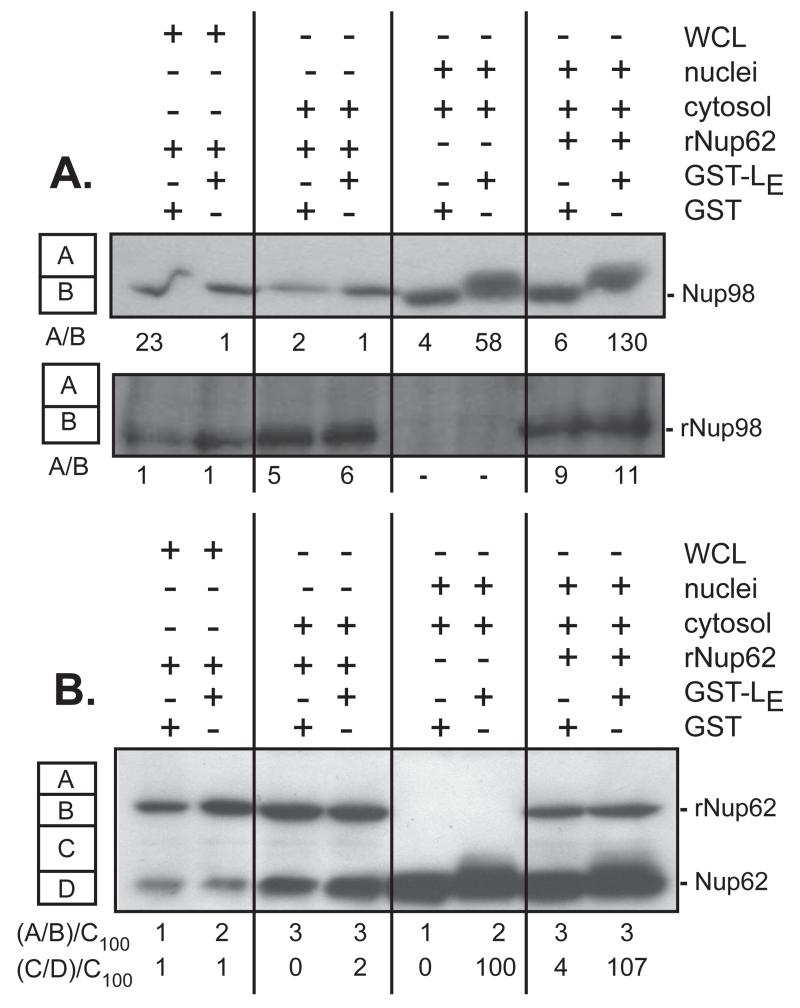
LX-dependent Nup phosphorylation assays typically present the substrates in the context of intact NPC, either by testing within whole cells (transfection, infection), or by reconstituting isolated cytoplasm and nuclei in the presence of recombinant protein (3, 13). The current hypothesis predicts that Ran-bound LX, complexed with an exportin and activated kinase cargo, becomes trapped within an NPC, leading to Nup hyperphosphorylation (7). Recombinant GST-Nup62 (21) and His-Nup98 (personal communications, K.E. Watters) have been demonstrated as native-like substrates for RV 2Apro. But when either protein was added to dounce-homogenized HeLa cell whole-cell extracts, or to subcellular fractionated HeLa cytosol, GST-LE failed to induce detectable phosphorylation (Fig 4A,B). The whole-cell lysates and HeLa cytosol each contain endogenous Nups, either as precursors to nascent NPC assembly (cytosol), or from the cell disruption (whole cell lysates). For Nup62 the native and recombinant forms are easy to distinguish by size (Fig 4B). For Nup98, the proteins co-migrate, but rNup98 phosphorylation would still be evident by measuring the A/B area ratios on the Western blot as detected with the His-tag mAb (Fig 4A). Although all these samples also contain Ran, Crm1, and kinases, in the absence of intact nuclei, neither the recombinant nor the endogenous Nups became phosphorylated in an LE-dependent manner. It required the addition of intact nuclei back to these mixtures to see evidence of Nup phosphorylation. Even then, however, recombinant proteins were still not viable substrates. The Nup phosphorylation mechanism was capable of discriminating context and modified only the native Nups, presumably those presented by the nuclei NPC.
Figure 4.
Nup format. Cell-free systems containing HeLa whole-cell lysates (WCL), cytosol, or cytosol plus isolated nuclei, were reacted with recombinant GST or GST-LE, in the presence or absence of recombinant His-Nup98 (A), or recombinant GST-Nup62 (B). After 45 min of incubation, the samples were fractionated, probed by Western analyses using α-Nup98 and α-His (A), or α-Nup62 (B). Pixel counts in the A, B, C, D boxes for each lane show the areas recorded and normalized to the control samples, as relative measures of observed phosphorylation.
Kinase Activation
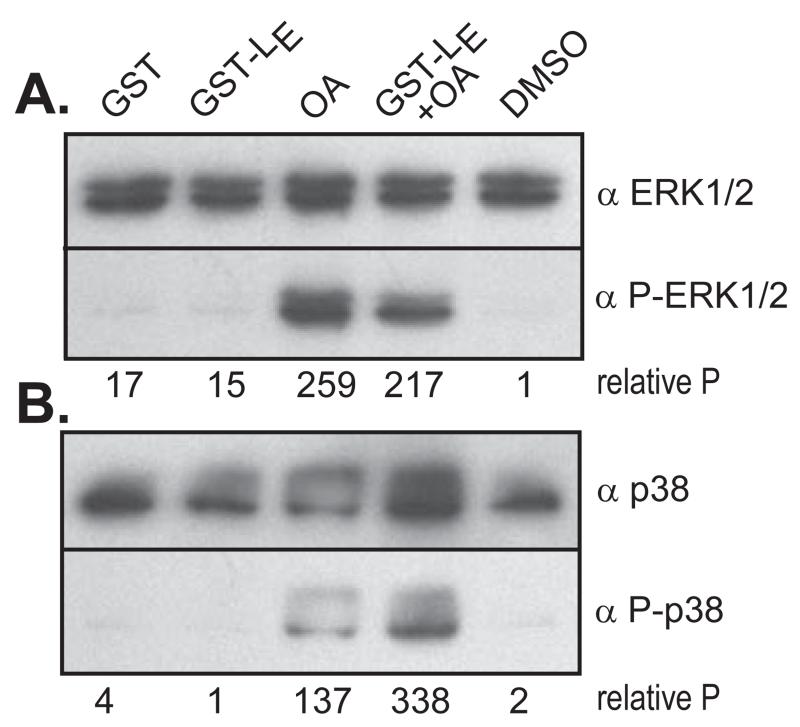
Inhibitor experiments have implicated mitogen-activated protein kinases (MAPK), particularly ERK1/2 and p38, as the probable pathways involved in Nup phosphorylation during EMCV infections or LE-dependent cDNA transfections (13). In intact cells, activation of both enzymes can be observed in the presence of LE, independent of the simultaneous activation of their upstream signaling cascades, including MEK1/2, MKK3, MKK6 and cRaf. The pathway activation points must then be at or near the effector enzymes themselves (13), potentially involving phosphatases rather than kinases as the regulatory mediators. This idea was tested with the reconstituted cytosol and nuclei mixes, supplemented with GST-LE (Fig 5). As described above, this experimental combination gives effective Nup hyperphosphorylation. Surprisingly though, only very weak signals were detected with mAbs specific to the phosphorylated effector kinases (Fig 5AB, GST-LE lanes). Reasoning that the activated kinases could be cycling, the protein phosphatase 1/2a inhibitor, okadaic acid (OA) was added at a concentration that inhibits such enzymes (29). The OA increased the phosphorylated ERK1/2 and p38 signals by at least 10-20 fold, presumably by preventing dephosphorylation within the MAPK pathways and allowing the intermediates to accumulate (30). In the absence of GST-LE, OA had only a modest effect on Nup62 phosphorylation in these reactions, as measured by autoradiography (Fig 1D, OA lane). It is known that some Nups, including p150 in Drosophila cells, can become phosphorylated by CDK1 in the presence of OA (31), as are other non-specific Nups in mammalian cells (4). Nup50 contains a phosphatase-sensitive phosphorylation site (5). Indeed, in the cell-free reconstitution assays this observation was confirmed by low-level incorporation of γ32P into Nup50-specific material (Fig 1C) in the presence of OA.
Figure 5.
Kinase activation. HeLa cytosol was reacted with recombinant GST or GST-LE in the presence or absence of okadaic acid (OA) and/or DMSO. After 45 min of incubation, the samples were fractionated then probed by Western analyses using (A) α-ERK/12, α-P-ERK1/2, or (B), α-p38, α-P-p38. Pixel normalization for each lane was relative to the respective, unactivated forms (unphosphorylated) of the kinases.
Discussion
ERK1/2 and p38 have been implicated as effector kinases in LE-dependent Nup hyper-phosphorylation (13). In phosphor-proteomics studies (5) Nup50 has been suggested as a potential substrate for ERK1/2, and it was therefore of interest to assay this protein’s putative alterations in the presence of LE. Although in the presence of the phosphatase inhibitor OA (without GST-LE), Nup50 did become labeled with γ32P, LE did not direct this event (Fig 1C). Under similar circumstances, in addition to the previously described phosphorylation of Nup62, Nup153 and Nup214, the phosphorylation of Nup98 was readily observed and dependent upon the presence of active LE, LT and/or LS sequences. The Nup phosphorylation by LE required the substrates to be presented in the context of intact nuclei, because when tested with parallel recombinant versions of Nup62 or Nup98 that do not become incorporated into nuclear pores, the LE-dependent mechanism only altered the native proteins (Fig 5). Though some substrate-altering aberration of the recombinant Nups cannot be entirely ruled out, proximity (possibly through trapping the LX:Ran complexes within the NPC) is the most likely explanation for this observed mechanistic preference. If captured within the NPC, limited catalytic amounts of the LX complexes might then direct massive hyperphosphorylation of the preferred substrates.
When rhinoviruses infect cells, the virus-encoded 2Apro cleave many of the same subset of NPC Nups affected by LX-dependent hyperphosphorylation (20, 21). As it is with the cardioviruses, the effect is down regulation of active nuclear import/export and redistribution of diffusible proteins throughout the cell. A special characteristic of the rhinovirus system, though, is that not all import/export pathways are affected equivalently. The genotype-specific sequences of individual 2Apro have different Nup cleavage preferences (21), and therefore, the order, rate and location of each NPC cleavage can be used by these viruses to regulate the specific activities of nucleocytoplasmic cargo exchange. This has been demonstrated for the SV40, M9 and RS NLS-dependent import pathways and the Crm1-dependent export pathway (22). The classical NLS (SV40) pathway uses karyopherins importin α/β to transport a broad range of cargos (32, 33). After the cargo:karyopherin complex is formed, it traverses the NPC via transient interactions with preferred Phe/Gly sites on multiple Nups (see Table 1), including Nup62, Nup98, Nup153, Nup214 and Nup358 (34). The M9 nuclear localization signal is from the mRNA binding protein hnRNPA1. It is recognized by transportin1 (karyopherin B2 in yeast, reviewed in (35), which is also responsible for import of HIV-1 Rev into the nucleus. The RS sequence from splicing factor 2 is recognized by transportin 3, a member of the karyopherin β family, and mediates the transport of SR proteins containing Arg-Ser-rich domains (RS) involved in the regulation of pre-mRNA splicing (36, 37). The PKI nuclear export pathway uses Crm1 (XPO1), a transportin which interacts with cargos or adaptors bearing leucine-rich motifs (38) such as the HIV-1 Rev protein, the protein kinase A inhibitor (PKI), 5S rRNAs, and U snRNAs (28, 39).
It has been proposed that rhinoviruses, by subtly balancing the trafficking unique to these pathways, can tailor the cell’s cytokine responses to needs of each genotype (22). The cardiovirus LX activities, however, when tested with these same pathways (Fig 2, Fig 3) seemed to show an indiscriminant, brute force NPC attack mode. Although activated kinases frequently transit the NPC in their roles as transcription regulators (40), there are few reports of coincidental (or accidental?) Nup phosphorylation. In uninfected cells, some Nups do become partially phosphorylated during mitosis, contributing to the transient dissociation of the NPC, but the cardiovirus-fostered events are unique biological phenomena in the strength and extent of Nup targeting. Not only were all 4 tested import/export pathways compromised within 3 hr of vEC9 infection, but all 3 LX-GST proteins, LE, LT and LS, were equivalently effective against all of the pathways. Apparently, phosphorylation is a much less discerning method of NPC disruption than rhinoviruses’ incisive proteolytic cleavage. The monitored processes also included measurable disruption of karyopherin-mediated NPC cargo export (i.e. Crm1). Although not directly tested here, one might expect the β3 importin 5 pathway, responsible for the transport of various ribosomal proteins, to be additionally hindered by LX, as it requires the same subsets of altered Nups, including Nup62, Nup98, Nup153, Nup214 and Nup358 (41, 42). Still, despite the obvious broad-based NPC changes brought about by hyper-phosphorylation, it was somewhat surprising to find a level of LE-dependent substrate selection within these pores. Nup50, which is not phosphorylated by LE, is also not implicated in any of the above transport pathways. This is despite observations that Nup50 can indeed, during normal cell cycling, be phosphorylated by ERK1/2 (5), among the enzymes implicated in the LE mechanism (13). The potent phosphatase 1 and phosphatase 2a inhibitor OA, increased the pools of activated kinases, including ERK1/2 in the cell extracts, and allowed Nup50 phosphorylation. Phosphatases generally serve custodial roles during certain normal cellular events. In particular, protein phosphatases 1 and 2a interfere with MAPK pathways and are responsible for reversing any cyclin dependent kinase conferred- Nup phosphorylations acquired during mitosis (31). Full reconstitution of NPC during cell cycling actually requires this type of phosphate curating, and these processes are therefore sensitive to OA (31).
Conclusions
Here, we present data that Nup hyper-phosphorylation pathways by LE, LT and LS, all target Nup98, in addition to Nup62 (3). Nup50, on the other hand was not phosphorylated by recombinant LE protein, and by inference, during infection. Furthermore, LE-dependent Nup phosphorylation in cell-free assays requires the substrates to be presented in the context of full nuclear pores (i.e. intact nuclei), as supplemented with cytosol (8). When this happens, at least 3 independent cellular importin pathways (importin α/β, transportin-1, transportin-3), and an exportin pathway (Crm1) become compromised, as monitored with cell lines transduced to express path-specific mCherry NLS/NES reporters.
Materials and Methods
HeLa Cell Lines
Suspension cultures were maintained in modified Eagle’s medium (37°C, 10% calf serum, 2% FBS, under 5% CO2). In addition to standard cells (ATCC CRL 1958), stable transduced cell lines carrying mCherry reporter genes linked to defined NLS/NES sequences have been described (22). Briefly, the SV40 large T antigen NLS, M9 NLS (26), the RS NLS from splicing factor 2 (27), or the leucine-rich NES from PKI (28) were engineered in-frame, C-terminal to an mCherry gene in the context of a retroviral vector plasmid encoding neomycin resistance (43). Moloney murine leukemia virus vectors were used in combination with these plasmids to create virus stocks harboring the respective genes. Infection of HeLa cells, genome integration and antibiotic selection gave cell lines with highly visible, constitutive expression of the mCherry derivatives. Before visualization, plated cells were rinsed with PBS (2×, 137 mM NaCl, 2.7 mM KCl, 10mM Na2HPO4, pH 7.4), treated overnight at 4 C with 4% paraformaldehyde, and then rinsed with PBS (3×) before the addition of DAPI (1ug/ml).
Cell Fractionation
HeLa cytosol was isolated after swelling suspension cells in hypotonic buffer (0.75 Mg(OAc)2, 0.15 mM EDTA, 1 mM PMSF, 0.01 mg/ml leupeptin, 20 mM pepstatin A, 3 mM DTT), followed by dounce homogenization and clarification (16,000×g, 20 min, 4°C). TB (1/10 volume, 10× at 20 mM hepes pH 7.3, 2 mM Mg(OAc)2, 110 mM K(OAc), 1 mM EGTA) was added before storage at −80°C. Whole cell lysates were prepared similarly, by the addition of WCLB (50mM Tris pH7.4, 50 mM NaF, 5mM Na4O7P2, 1 mM EDTA, 1mM EGTA, 250mM mannitol, 1% triton ×100; 1mM DTT, 1mM PMSF, 20 mM pepstatin A, and 1× protein phosphatase inhibitor cocktail 3, Sigma Aldrich) to cells, followed by sonication (2×, 30 sec, 4°C) and clarification (16,000×g, 10 min). The materials were aliquoted and snap-frozen before storage at −80°C. Nuclei were isolated from suspension cells following treatment with digitonin and clarification to remove cytosol (44). Briefly, the cells were collected, washed (2×, PBS), then incubated (10 min, 4°C) in RSB (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 2.5 mM MgCl) with digitonin (40ug/ml, Sigma Aldrich). The samples were clarified (2000×g, 8 min), and the pellets were rinsed with RSB, then TB (plus 1 mM DTT, 3×). Quantitation was with a hemocytometer after staining with tryphan blue.
Transfection and Infections
Eukaryotic expression plasmids for GST, GFP, GFP-LE, LE-GST, LT-GST (BeAn), and LS-GST (SafV-2) were as described (10, 13). The GST-LX panel also had matched cognates encoding corresponding, inactive LX sequences (C19A, or C11A). Cells were transfected (3×105 cells per well, 1 ug cDNA) using lipofectamine (1 ul, Invitrogen) in Opti-MEM media (Invitrogen), and then incubated (37°C under 5% CO2). Infections with vEC9 (45) used an MOI=15, in PBS. At harvest, the wells were rinsed (2×, PBS) before 2× SDS buffer was added. Cell materials were collected and boiled (15 min) before fractionation by SDS-PAGE and protein detection in Western assays.
Microscopy
Cells (live or fixed with paraformaldehyde) were visualized with a Ti-E ECLIPSE inverted wide-field microscope (Nikon Corporation). The images were collected using a CoolSnapHQ camera (Photometrics). Excitation/emission filter sets detected 460 nm (DAPI) and 632 nm (mCherry). Nikon NIS Elements software (version 4.30.01) was used to tabulate individual pixel intensities across an 80 pixel-window, centered on the nucleus, per cell. DAPI and mCherry data for a minimum of 8 live cells or 10 fixed cells per experimental condition were collected. The values were compiled in Excel and averaged across the 80 pixel windows. To compare conditions with fixed cells, the averaged DAPI levels were normalized. The natural log of each point in the intensity profile was plotted.
Recombinant Proteins
Recombinant engineering, bacterial expression and protein isolation for GST and GST-LE have been described (8). The isolation of recombinant GST-Nup62 (human) is also described (21). Purified recombinant His-Nup98 (human) was a gift from Dr. Kelly Watters.
Westerns and Antibodies
Samples were boiled with SDS buffer before the proteins were resolved by SDS-PAGE, and transferred to polyvinylidene difluororide membranes (Immobilon-P, Millipore). The membranes were blocked (30 min) in TBST (20 mM Tris, pH 7.6, 150 mM NaCl, 0.5% Tween 20) with 10% dry milk as described (3). For Western assays, the membranes were incubated with a primary antibody (in TBST, 1% dry milk, overnight, 4°C), before rinsing (3×, TBST), and addition of a secondary antibody (1 hr, 20°C). After additional rinses (3×, TBST), the membranes were developed according to manufacturers’ specifications for enhanced chemiluminescence (Pierce, GE Lifesciences). Antibodies included: α-Nup98 (goat mAb, IgG, Sigma, 1:10000), α–FG-repeat Nups (murine mAb414, IgG, Covance, 1:2000), α-Nup62 (murine Ab, IgG, BD Transduction Laboratories, 1:2000), α-tubulin (murine Ab, IgG, Sigma, 1:10000), α-Nup50 (goat Ab, Abcam, 1:2000), α-GST (murine mAb, IgG, Novagen, 1:10000), α-His (6× His, murine Ab, Abcam, 1:2000), α-P-p38 (activated, phospho-Thr180/Tyr182; rabbit Ab, IgG, Cell Signaling Technology, 1:3000), α–p38 (unactivated, rabbit Ab, IgG, Cell Signaling Technology, 1:3000), α-P-ERK1/2 (activated, phospho-Thr202/Tyr204 ERK 1/2; rabbit Ab, IgG, Cell Signaling Technology, 1:2000), α-ERK1/2 (unactivated, ERK1/2, rabbit Ab, IgG, Upstate, Millipore, 1:5000), α-mouse (secondary Ab, IgG, Sigma Aldrich, 1:8000), α-rabbit (secondary Ab, IgG, Promega, 1:8000) α-goat (secondary Ab, IgG, Sigma, 1:8000).
Nup50 and Nup62 Immunoprecipitation
HeLa nuclei (106 cell equivalents) and fractionated cytosol (3×105 cell equivalents) were combined with γ-32P-ATP (10 μCi). Recombinant GST or GST-LE (2 μg), were added. Some samples were supplemented with okadaic acid (250 nM). Reactions were incubated at 37°C for 45 min before the addition of RIPA (300 μl/sample, 50 mM Tris, pH 7.4, 150 mM NaCl, 0.1 % SDS, 1% Triton ×100, 0.5% Na deoxycholate, 1mM PMSF, 20 mM pepstatin A, and protein phosphatase inhibitor cocktail 3, Sigma Aldrich) and sonication. Protein G-conjugated beads (10 μl, G.E. Lifesciences) were added. Incubation was for 1 hr before the beads were removed by centrifugation. Fresh protein-G beads conjugated to α-Nup50 or α-Nup62 (saturated) were incubated with agitation (4°C, 3 hr) and then collected. After extensive washing (6×, PBS with 0.02% triton), samples were denatured with SDS buffer, boiled and fractionated by SDS-PAGE. Protein bands were detected by silver-staining and autoradiography, with densitometry performed using a Typhoon imager (GE Healthcare).
Highlights.
Nup98, but not Nup50 becomes phosphorylated by cardiovirus Leader protein-dependent mechanisms.
At least four independent nucleocytoplasmic trafficking pathways are inhibited by this process.
Nups must be presented in a nuclear pore context for Leader-directed phosphorylation.
Leader, by itself, does not cause activation of cellular kinases.
Acknowledgements
The authors thank Drs. Nathan Sherer and Kelly Watters for microscopy assistance and the generation of mCherry cell lines, Jaye Gardiner for assistance with NIS Elements software, and Nathan Ciomperlik for assistance with data analysis. We thank Dr. Howard Lipton for the generous gift of SafV-2 and TMEV (BeAn) cDNAs, and Kelly Watters for the purified recombinant His-Nup98. This work was supported by NIH grant AI017331 to ACP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones MS, Lukashov VV, Ganac RD, Schnurr DP. Discovery of a novel human picornavirus in a stool sample from a pediatric patient presenting with fever of unknown origin. J. Clin. Microbiol. 2007;45:2144–2150. doi: 10.1128/JCM.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardina MV, Lidsky P, Sheval EV, Fominykh KV, van Kuppeveld FJ, Polyakov VY, Agol V. Mengovirus-induced rearrangements of the nuclear pore complex: Hijacking cellular phosphorylation machinery. J.Virol. 2009;83:3150–3161. doi: 10.1128/JVI.01456-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porter FW, Palmenberg AC. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition of cardioviruses. J.Virol. 2009;83:1941–1951. doi: 10.1128/JVI.01752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kehlenbach RH, Gerace L. Phosphorylation of the nuclear transport machinery down-regulates nuclear protein import in vitro. J. Biol. Chem. 2000;275:17848–17856. doi: 10.1074/jbc.M001455200. [DOI] [PubMed] [Google Scholar]
- 5.Kosako H, Yamaguchi N, Aranami C, Ushiyama M, Kose S, Imamoto N, Taniguchi H, Nishida E, Hattori S. Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat Struct Mol Biol. 2009;16:1026–1035. doi: 10.1038/nsmb.1656. [DOI] [PubMed] [Google Scholar]
- 6.Basta HA, Bacot-Davis VR, Ciomperlik JJ, Palmenberg AC. Encephalomyocarditis virus Leader is phosphorylated by CK2 and syk as a requirement for subsequent phosphorylation of cellular nucleoporins. J.Virol. 2014;88:2219–2226. doi: 10.1128/JVI.03150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacot-Davis VR, Ciomperlik JJ, Cornilescu CC, Basta HA, Palmenberg AC. Solution structures of Mengovirus leader protein, its phosphorylated derivatives, and in complex with RanGTPase. Proc Natl Acad Sci USA. 2014;111:15792–15797. doi: 10.1073/pnas.1411098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter FW, Bochkov YA, Albee AJ, Wiese C, Palmenberg AC. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc Natl Acad Sci USA. 2006;103:12417–12422. doi: 10.1073/pnas.0605375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacot-Davis VR, Palmenberg AC. Encephalomyocarditis virus Leader protein hinge domain is responsible for interactions with Ran GTPase. Virology. 2013;443:177–185. doi: 10.1016/j.virol.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basta HA, Palmenberg AC. AMP-activated protein kinase phosphorylates EMCV, TMEV, and SafV Leader proteins at different sites. Virology. 2014;262:236–240. doi: 10.1016/j.virol.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petty RV, Basta HA, Bacot-Davis VR, Brown BA, Palmenberg AC. Binding interactions between the encephalomyocarditis virus leader and protein 2A. J.Virol. 2014;88:13503–13509. doi: 10.1128/JVI.02148-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petty RV, Palmenberg AC. Guanine-nucleotide exchange factor RCC1 facilitates a tight binding between EMCV Leader and cellular Ran GTPase. J. Virol. 2013;87:6517–6520. doi: 10.1128/JVI.02493-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter FW, Brown B, Palmenberg A. Nucleoporin phosphorylation triggered by the encephalomyocarditis virus leader protein is mediated by mitogen-activated protein kinases. J.Virol. 2010;84:12538–12548. doi: 10.1128/JVI.01484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Her L-S, Lund E, Dahlberg JE. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 15.Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17:93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 16.Ricour C, Borghese F, Sorgeloos F, Hato SV, van Kuppeveld FJ, Michiels T. Random mutagenesis defines a domain of Theiler’s virus leader protein that is essential for antagonism of nucleocytoplasmic trafficking and cytokine gene expression. J.Virol. 2009;83:11223–11232. doi: 10.1128/JVI.00829-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Sage V, Mouland AJ. Viral subversion of the nuclear pore complex. Viruses. 2013;5:2019–2042. doi: 10.3390/v5082019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lidsky PL, Hato S, Bardina MV, Aminev AG, Palmenberg AC, Sheval EV, Polyakov VY, van Kuppeveld FJ, Agol V. Nucleo-cytoplasmic traffic disorder induced by cardioviruses. J.Virol. 2006;80:2705–2717. doi: 10.1128/JVI.80.6.2705-2717.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricour C, Delhaye S, Hato SV, Olenyik TD, Michel B, van Kuppeveld FJ, Gustin KE, Michiels T. Inhibition of mRNA export and dimerization of interferon regulatory factor 3 by Theiler’s virus leader protein. J.Gen.Virol. 2009;90:177–186. doi: 10.1099/vir.0.005678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustin KE, Sarnow P. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 2002;76:8787–8796. doi: 10.1128/JVI.76.17.8787-8796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watters K, Palmenberg A. Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J.Virol. 2011;85:10874–10883. doi: 10.1128/JVI.00718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watters K, Inankur B, Warrick J, Sherer NM, Yin J, Palmenberg A. Differential disruption of nucleocytoplasmic trafficking by rhinovirus 2A proteases. Mol.Biol. of Cell. 2015 doi: 10.1128/JVI.02472-16. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fagerlund R, Kinnuen L, Kohler M, Julkunen I, Melen K. NF-kappa B is transported into the nucleus by importin alpha3 and importin alpha4. J. Biol. Chem. 2005;280:15942–15951. doi: 10.1074/jbc.M500814200. [DOI] [PubMed] [Google Scholar]
- 24.Kumar KP, McBride KM, Weaver BK, Dingwall C, Reich NC. Regulated nuclear-cytoplasmic localization of interferon Regulatory Factor 3, a subunit of double stranded RNA-Activaed Factor 1. Mol and Cell Biol. 2000;20:4159–4168. doi: 10.1128/mcb.20.11.4159-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride KM, Banninger G, Reich NC. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO. 2002;21:1754–1763. doi: 10.1093/emboj/21.7.1754. M. C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kataoka N, Bachorik JL, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 28.Wen W, Meinkotht JL, Tsein RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 29.Cohen P, Holmes CFB, Tsukitani Y. Okadaic acid: a new probe for the study of cellular regulation. Trends Biochem Sci. 1990;15:92–102. doi: 10.1016/0968-0004(90)90192-e. [DOI] [PubMed] [Google Scholar]
- 30.Ho DT, Shayan H, Murphy TH. Okadaic acid induces hyperphosphorylation of tau independently of mitogen-activated protein kinase activation. J. Neurochem. 1997;68:106–111. doi: 10.1046/j.1471-4159.1997.68010106.x. [DOI] [PubMed] [Google Scholar]
- 31.Onischenko EA, Gubanova NV, Kiseleva EV, Hallberg E. CDK1 and okadaic acid-sensitive phosphatases control assembly of nuclear pore complexes in drosophila embryos. Mol. Biol. Cell. 2005;16:5152–5162. doi: 10.1091/mbc.E05-07-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adam EJH, Adam SA. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J. Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 34.Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers; alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci U S A. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 36.Hedley ML, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc Natl Acad Sci U S A. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci U S A. 2001;98:10154–10159. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 39.Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 40.Zehorai E, Yao Z, Plotnikov A, Seger R. The subcellular localiation of MEK and ERK--a novel translocation signal (NTS) paves a way to the nucleus. Mol Cell Endocrinol. 2010;314:213–220. doi: 10.1016/j.mce.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Yaseen NR, Blobel G. Cloning and characterization of human karyopherin β3. Proc Natl Acad Sci U S A. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jakel S, Gurlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection as is supressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 44.Mili S, Shu HJ, Zhao Y, Pinol-Roma S. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: Candidate intermediates in formation and export of mRNA. Mol. Cell Biol. 2001;21:7307–7319. doi: 10.1128/MCB.21.21.7307-7319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hahn H, Palmenberg AC. Encephalomyocarditis viruses with short poly(C) tracts are more virulent than their Mengo virus counterparts. J.Virol. 1995;69:2697–2699. doi: 10.1128/jvi.69.4.2697-2699.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryotic Cell. 2009;8:1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryan KJ, Wente SR. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr Opin Cell Biol. 2000;12:361–371. doi: 10.1016/s0955-0674(00)00101-0. [DOI] [PubMed] [Google Scholar]