Abstract
Invertebrates possess a diverse collection of pentameric Cys-loop ligand gated ion channel (LGIC) receptors whose molecular structures, evolution and relationships to mammalian counterparts have been intensely investigated in several clinically and agriculturally important species. These receptors are targets for a variety of control agents that may also harm beneficial species. However, little is known about Cys-loop receptors in spiders, which are important natural predators of insects. We assembled de novo transcriptomes from the central and peripheral nervous systems of the Central American wandering spider Cupiennius salei, a model species for neurophysiological, behavioral and developmental studies. We found 15 Cys-loop receptor subunits that are expected to form anion or cation permeable channels, plus a putative acetylcholine binding protein (AChBP) that has only previously been reported in molluscs and one annelid. We used phylogenetic and sequence analysis to compare the spider subunits to homologous receptors in other species and predicted the 3D structures of each protein using the I-Tasser server. The quality of homology models improved with increasing sequence identity to the available high-resolution templates. We found that C. salei has orthologous γ-aminobutyric acid (GABA), GluCl, pHCl, HisCl and nAChα LGIC subunits to other arthropods, but some subgroups are specific to arachnids, or only to spiders. C. salei sequences were phylogenetically closest to gene fragments from the social spider, Stegodyphus mimosarum, indicating high conservation within the Araneomorphae suborder of spiders. C. salei sequences had similar ligand binding and transmembrane regions to other invertebrate and vertebrate LGICs. They also had motifs associated with high sensitivity to insecticides and antiparasitic agents such as fipronil, dieldrin and ivermectin. Development of truly selective control agents for pest species will require information about the molecular structure and pharmacology of Cys-loop receptors in beneficial species.
Introduction
Pentameric Cys-loop receptors are ligand gated ion channels (LGIC) that mediate fast synaptic transmission in central and peripheral nervous systems. Many human diseases are caused by dysfunction of these channels, making them therapeutic targets for drugs [1]. Several invertebrate Cys-loop receptors are targets of widely used control agents, such as dieldrin, ivermectin and neonicotinoids [2–5]. Phylogenetic relationships, molecular structures and functions of these receptors have mainly been investigated in agricultural pests and parasites, rather than more beneficial arthropods. Several Cys-loop receptor genes have now been cloned from arachnids, mainly ticks and mites [6], but very little is known about these receptors in the largest arachnid order, the spiders [7]. Spiders are important natural predators of insects and may unintentionally be exposed to control agents. A recent report demonstrated that nicotinic acetylcholine (nACh) receptors of the pond wolf spider Pardosa pseudoannulata are activated by neonicotinoids [8], but Cys-loop subunit genes of other spider species have not been investigated.
Invertebrate Cys-loop receptors include anion permeable channels that are gated by γ-aminobutyric acid (GABA), glutamate (GluCl), histamine (HisCl) or protons (pHCl), and nematodes also have Cl-gated ACh and 5-HT receptors [9]. The only invertebrate cation permeable Cys-loop receptors are nicotinic acetylcholine channels (nACh) [6]. Pentameric receptors can be further divided into those containing only one conserved Cys-loop in their extracellular domain and those that have an additional disulfide bond that links the β9-β10 strands in Loop C. The glycine receptor, found only in chordates, is a prototype of receptors with two Cys-loops, and many invertebrate GluCl, HisCl and pHCl receptors are members of this group [10].
High-resolution structural information is useful for understanding how transmitter receptors function. Several Cys-loop receptor protein structures have recently become available with complete atomic resolution. These include the bacterial ligand-gated ion channels ELIC and GLIC [11,12], C. elegans GluCl channel [13,14], human GABAAβ3 receptor [15] and mouse 5-HT3 receptor [16]. The only complete nACh receptor is a medium resolution electron microscopy structure from the electric ray Torpedo marmorata [17], but structural information for the channel extracellular domain has been inferred by homology with the molluscan acetylcholine binding protein AChBP [18]. All known Cys-loop receptors show remarkable structural similarities with conserved residues that are important for receptor function, making it possible to use a combination of several templates to infer information about other receptors to create homology models [19].
The Central American wandering spider, Cupiennius salei, is a widely used experimental animal, particularly for neurophysiological [20,21], and developmental studies [22,23]. Immunohistochemical and electron microscopic investigations have found a large number of GABA and glutamate containing neurons in the spider brain; some are efferents that project to the legs where they innervate mechanosensory neurons, and some project to muscles [24,25]. GABA or glutamate effects on the spider central neurons or muscle fibers have not been investigated, but electrophysiological experiments have revealed that agonists of Cys-loop GABAA receptors (GABA and muscimol) and GluCl channels (glutamate and ibotenic acid) modulate the sensitivity of mechanosensory neurons of the lyriform VS-3 slit sensilla [26–29]. GABAA agonists depolarize the neurons and induce brief inhibition followed by long lasting excitation [26,29] while GluCl agonists induce an identical response to GABA in some, but purely inhibitory response without any change in membrane potential in most VS-3 neurons [27,28]. Hence, there are probably multiple subtypes of Cys-loop receptors in each neuron.
The recently sequenced C. salei brain and hypodermis transcriptomes [30,31] provide an opportunity to investigate the genes that form Cys-loop receptors in the spider central and peripheral nervous systems. Here, we annotate sixteen genes that encode C. salei Cys-loop subunits and analyze their phylogenetic relationships and similarity with other species. To understand the structural and functional features of these molecules and predict which neurotransmitters and insecticides bind to each of them, we used sequence analysis and homology modelling with multiple templates and evaluated our results with Ramachandran plots.
Materials and Methods
Identification of Cys-loop ligand gated ion channel subunits from spider transcriptomes
Details of the Cupiennius salei transcriptome preparation, sequencing and assembly have been described previously [30]. Briefly, hypodermis membranes containing peripheral nerves and sensory neurons, glial cells and some muscle tissue, were collected from seven adult female spider siblings and two brains from different female spiders. The Qiagen RNeasy plus mini kit was used for hypodermis RNA extraction and a Qiagen RNeasy midi kit for brain RNA extraction. The mRNA separation, cDNA library construction and Illumina sequencing were performed by McGill University and Génome Québec Innovation Centre (Montréal, Québec, Canada). Initial cDNA reads (220 million pairs of 100 nucleotide length) were groomed by requiring at least 80 contiguous nucleotides with Phred score >19 to give a final database of ~110 million pairs of reads for each library.
Cys-loop LGIC subunit sequences were identified by searching the databases at low stringency against both nucleotide and amino acid sequences of published arthropod genes. Amino acid searches were conducted using BLOSUM45, BLOSUM62 and BLOSUM80 matching matrices [32]. All putatively matching reads were compared to the non-redundant protein database using BLASTX (http://blast.ncbi.nlm.nih.gov). Candidate reads were extended by the transcriptome walking algorithm [30] using an initial minimum overlap of 60 nucleotides. Minimum overlap was increased, in steps of 10 up to a maximum of 90 nucleotides, when required to eliminate multiple possible extension pathways in a few cases of highly redundant sequence structure. Overlap was reduced in steps of 10 to a minimum of 40 nucleotides when no overlaps were found in a complete pass through the database. Walking was always continued to exhaustion in the 5′ direction and continued to include the complete protein coding sequence (mORF or CDS) and the STOP codon in the 3′ direction. In many cases, the complete sequence was continued to the poly-A tail. Relative abundance of transcribed mRNA for each gene was estimated by counting matching Illumina reads to assembled genes as described earlier [31]. Data analysis and programming was performed in the C++ language using Microsoft Visual Studio and desktop computers. Multiple threaded code was used for searching and for walking, using up to 16 parallel processing cores.
Phylogenetic analysis
Homologous sequences for the sixteen C. salei Cys-loop LGIC subunits (Table 1) found in the transcriptomes were individually searched using protein Delta-Blast. We selected representative examples of homologous invertebrate and vertebrate sequences for phylogenetic analysis. Sequence alignment was performed by the online version of the multiple sequence alignment software MAFFT Version 7 (http://mafft.cbrc.jp/alignment/server/) with default parameters (BLOSUM62, gap opening penalty 1.53) and G-Ins-I strategy with 200 SwissProt homologs and threshold E = 1e-10 [33]. We used the Guidance server (http://guidance.tau.ac.il/) to test the confidence of our alignments and removed unreliable sequences before further tests [34].
Table 1. Cupiennius salei Cys loop subunits.
| Putative Protein | Uniprot code | NCBI accession number | Number of amino acids | MW (kDa) | Clone identity to transcriptome |
|---|---|---|---|---|---|
| CsGABArdl1 | T1E1C3 | GAKT01000019.1 | 504 | 57 | F missing T /R SNP |
| CsGABArdl2 | A0A061QHV7 | GBFC01000040.1 | 478 | 54 | F yes/R yes |
| CsGABAAβ | T1DCD9 | GAKT01000067.1 | 460 | 53 | F yes/R yes |
| CsGABAgrd | T1E191 | GAKT01000104.1 | 509 | 59 | F yes |
| CsGluCl1 | T1E1T2 | GAKT01000005.1 | 399 | 46 | F yes/F SNP |
| CsGluCl2 | A0A061QFY0 | GBFC01000041.1 | 401 | 47 | F yes/R SNP |
| CsGluCl3 | N/A | KT183364 | 411 | 47 | N/A |
| CspHCl1 | T1D1W5 | GAKT01000068.1 | 468 | 54 | R SNP |
| CspHCl2 | T1D1Q6 | GAKT01000128.1 | 469 | 53 | F SNP/R SNP |
| CsHisCl1 | A0A061QLK7 | GBFC01000023.1 | 422 | 48 | N/A |
| CsHisCl2 | N/A | KT363688 | 365 | 43 | N/A |
| CsNC-LGCl | A0A061QLN6 | GBFC01000042.1 | 450 | 51 | F yes/R yes |
| CsnACh1A | A0A061QLQ4 | GBFC01000022.1 | 417 | 46 | F yes/ F SNP |
| CsnACh1B | N/A | KT183361 | 417 | 48 | R yes |
| CsnAChα | N/A | KT183362 | 539 | 62 | N/A |
| CsAChBP | N/A | KT183363 | 246 | 29 | N/A |
GluCl = glutamate-gated chloride channel; GABA = γ-amino butyric acid; rdl = resistance to dieldrin; pHCl = pH gated chloride channel; grd = GABA and glycine-like receptor; nACh = nicotinic acetylcholine receptor; HisCl = histamine gated chloride channel; NC-LGCl = Not characterized ligand gated chloride channel; AChBP = acetylcholine binding protein. N/A = not available yet, submitted to GeneBank. F indicates forward and R reverse orientation in the clone direction. Sequences indicated with “yes” were identical to the transcriptome database and SNP indicates single nucleotide polymorphism. “missing T” means that a nucleotide with the nitrogenous base thymine was deleted.
After several rounds of pruning, re-aligning and testing, the final phylogenetic trees for the putative anion and cation channel subunits were created separately with MEGA 6 software using the Maximum likelihood method [35]. Based on the results from MEGA 6 “Models” the best protein model for the MAFFT aligned sequences using all sites was the Gamma distributed Le-Gascuel (LG + G) model for both groups [36]. Analysis of the putative cation channel group required addition of invariant sites (LG + G + I). The maximum likelihood analysis was performed with 1000 bootstraps using subtree pruning and extensive regrafting option with otherwise default settings. Accession codes and nomenclature for C. salei sequences are shown in Table 1 and sequences from other species that were used in the final analyses are listed in the S1 Table. The final trees were exported from MEGA6 as emf files and images created in Adobe illustrator CS5 (Adobe systems inc, San Jose, CA).
Sequence analysis
The MAFFT aligned protein sequences were viewed and analyzed using Jalview 2.9. software (http://www.jalview.org/) [37]. For functional analysis, prediction of signal-peptide cleavage sites and membrane spanning regions we used the InterProScan 5 server (http://www.ebi.ac.uk/Tools/pfa/iprscan5/), which scans the sequence against the protein signatures of several databases [38]. Further sequence analysis was done with the MemPype server (http://mu2py.biocomp.unibo.it/mempype/default/index) [39].
Pairwise identity matrices were created using a Sequence demarcation tool with the MAFFT alignment and clustering the sequences with Neighbour joining tree [40]. The identity was also tested in Jalview by pairwise comparison and results from these two tests were closely similar.
Protein structure prediction
We used the I-Tasser server to generate three-dimensional structure predictions of the C. salei Cys-loop subunits [41]. I-Tasser is a free user-friendly program that has been repeatedly ranked as the No 1 server in the Community Wide Experiments on the Critical Assessment of Techniques for Protein Structure Prediction (CASP). The sequences were uploaded in FASTA format and ran without restraints allowing the software to select the templates. Prior to the homology modelling, we removed the short N-terminal segments in front of the signal peptide cleavage sites that were found by InterPro scan for all other sequences except CsGABAgrd and CsGABAHisCl1 and 2 (S1 Fig and S2 Fig). These signal peptides are eliminated from mature proteins. We used the PyMOL Molecular Graphics System, Version 1.7.4 (Schrödinger, LLC.) software to generate images of the homology models. The stereochemical quality of predicted homology models were investigated by Ramachandran plot analysis using the RAMPAGE server [42].
PCR analysis and cDNA cloning
Total RNA was extracted from the leg hypodermal tissues using a Qiagen RNeasy plus mini kit as described previously [31]. Reverse transcription (RT) was performed using 200 ng total RNA and oligo d(T)23VN primer with ProtoScript II reverse transcriptase (New England BioLabs, Whitby, ON, Canada). The synthesized cDNA was used as a template to amplify the Open Reading Frame (ORF) regions of the genes using gene specific primers with designs based on the assembled sequences using Q5 High-Fidelity DNA Polymerase (New England BioLabs). The PCR product size was verified by running with a DNA marker on an agarose gel and then gel-purified using GenElute Gel Extraction Kit (Sigma Aldrich, Oakville, ON, Canada). A-tailing reaction was then performed by incubating the purified PCR product with dATP and Taq DNA polymerase (New England BioLabs). After the tailing reaction, the DNA was used for ligation with pGEM®-T Easy Vector and then transformed into JM109 competent cells (Promega, Madison, WI). After verifying by colony PCR, the positive clones were selected for plasmid purification using GenElute Plasmid Miniprep kit (Sigma Aldrich) and sequenced by the McGill University and Génome Québec Innovation Centre.
Results and Discussion
1. Cupiennius salei brain and hypodermis transcriptomes have sixteen putative Cys-loop ligand gated ion channel subunits
All Cys-loop LGICs are composed from a ring of five subunits that have an extracellular ligand binding domain made mainly from β-sheets and an α-helical transmembrane domain [13,15,16]. Deduced proteins from sixteen mRNA sequences in C. salei brain and hypodermis transcriptomes (Table 1, S1 Fig and S2 Fig) had conserved features of Cys-loop LGIC subunits. They had extracellular N-terminal regions with the characteristic Loops A to G that form the ligand-binding site and a cysteine loop in the extracellular domain. Five subunits also had a disulphide bond in the Loop C. This feature is similar to the vertebrate glycine receptor and invertebrate GluCl channels [10]. Most sequences had the typical four membrane spanning regions (TM1-TM4) and a highly variable loop between TM3 and TM4 that has been shown to associate with cytoskeletal proteins such as gephyrin or rapsyn, and is believed to contribute to channel kinetics [43,44]. However, one subunit, the CsAChBP lacked the transmembrane regions completely.
We analyzed twelve C. salei Cys-loop gene sequences by PCR and cloned them. In most cases cloning produced identical sequences to the transcriptome database (Table 1). Some cloned sequences had single nucleotide polymorphisms (SNP) that encoded the same amino acids and were present, but less well represented in the transcriptomes than the canonical sequence. The only gene that could not be amplified by PCR was CsHisCl1.
InterPro scan identified all 16 proteins from the transcriptomes as subunits of neurotransmitter gated ion channels. Based on conserved motifs in transmembrane regions, twelve subunits were predicted to form pentameric anion permeable channels. The remaining four subunits had typical ligand binding motifs for cation channels in the nicotinic acetylcholine receptor (nACh) family. CsnAChBP did not have TM regions, but the other three subunits had TM motifs that were also typical for nACh channels. Various InterPro databases provided evidence for further classification of some subunits. We have combined this information with the evidence from phylogenetic and sequence analysis (below) to classify the sequences as shown in Table 1. Complete C. salei Cys-loop amino acid sequences are aligned with homologous sequences in S1 Fig and S2 Fig.
2. All sixteen Cys-loop genes are present in the C. salei brain and leg hypodermis
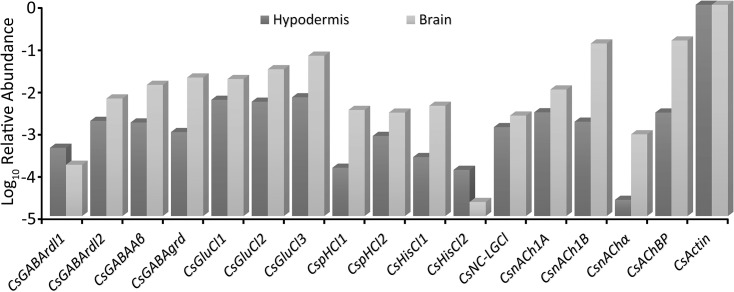
We estimated the relative abundance of transcribed mRNA for each Cys-loop sequence by searching the groomed transcriptome data for matches to the main open reading frame (mORF) of each gene [31]. Depending on their abundances, we tested 40–230 million reads for each gene requiring a minimum of 90/100 identical nucleotide matches to score each read as derived from the gene. We normalized the total counts by mORF length, and then expressed them as abundance relative to the putative gene that encodes actin. Actin RNA accounted for about 0.4% of all reads [31]. Fig 1 shows the relative abundances of each C. salei Cys-loop gene in the brain and hypodermis transcriptomes. We found all 16 sequences in both tissues, but in most cases, the relative abundance was higher in brain than in hypodermis. Exceptions were the CsGABArdl1 and the CsHisCl2 whose relative abundances were higher in the hypodermis than in the brain. The nACh sequences were generally much sparser in hypodermis than in the brain, and the CsnAChα had a significantly lower abundance in the hypodermis than any other gene.
Fig 1. Cys-loop subunit abundances.
Relative abundances of Cys-loop receptor subunits in C. salei brain and hypodermis transcriptomes compared to the abundance of actin. The data were obtained by counting total reads in the transcriptome libraries with at least 90 consecutive identical nucleotides to the reading frame of each gene, then normalizing by reading frame length. The scale is logarithmic. Accession numbers and sequence nomenclature are in Table 1.
Both the brain and hypodermis tissue that we used for the transcriptome preparation contain mainly neurons and glial cells, and most of the Cys-loop subunits are likely to originate from neurons. However, some muscle tissue was attached to the hypodermis and it is possible that some of the Cys-loop subunits originate from leg muscle. Arthropod skeletal muscles are innervated by glutamatergic motor neurons [45] and excitatory glutamate receptors that are not Cys-loop receptors are likely to mediate muscle contraction. However, nerve fibers containing GABA also innervate C. salei and other arthropod muscle fibers (45, 25). Thus, Cys-loop receptor subunits are also likely to be present in muscle.
3. Molecular phylogenetic analysis found members of C. salei Cys-loop superfamily in eight major clades
To clarify the evolutionary relationships of the C. salei Cys-loop subunits to homologous sequences in other species we performed molecular phylogenetic analysis based on the Maximum likelihood algorithm. C. salei Cys-loop sequences clustered with high bootstrap values into eight orthologous clades found in several invertebrate phyla with the closest homologs in other arachnid species. Some are also orthologous to mammalian and other vertebrate Cys-loop receptors. One radiation tree (Fig 2) includes representative examples of the seven clades in the anion selective channel group, while another (Fig 3) shows the nACh receptors. Alternative phylogenetic analyses based on the Neighbour-joining method did not reveal significant differences in the topology of either tree. Figs 4 and 5 show pairwise identity matrices for most of the same sequences as the phylogenetic trees. These figures also include a small number of sequences that were not included into the final trees, but whose 3-dimensional structures and/or physiological functions have been characterized and are discussed below.
Fig 2. Molecular phylogenetic analysis of putative anion permeable channel subunits.
Evolutionary history inferred by the Maximum Likelihood method. The unrooted radiation tree is drawn to scale, with branch lengths measured in numbers of substitutions per site. The analysis involved 72 MAFFT aligned amino acid sequences and was run with 1000 bootstraps. Numbers indicate bootstrap values. There were a total of 1385 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [35]. Accession numbers and nomenclature for C. salei sequences are in Table 1 and for other sequences in S1 Table.
Fig 3. Molecular phylogenetic analysis of putative nACh channel subunits.
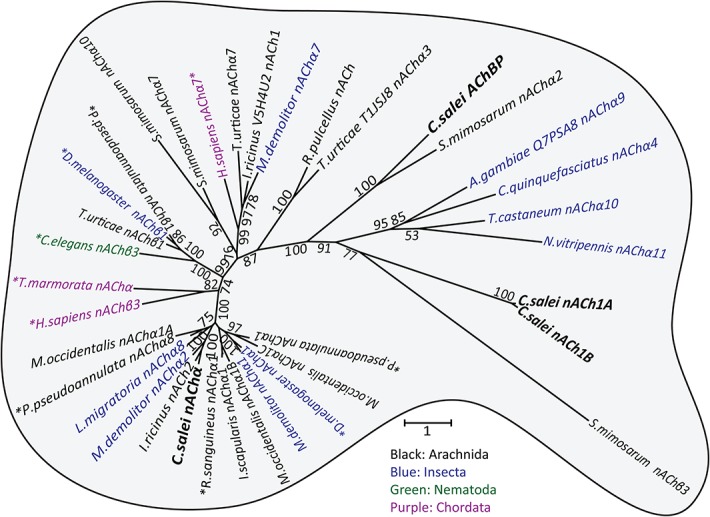
Evolutionary history inferred by the Maximum Likelihood method. The unrooted radiation tree is drawn to scale, with branch lengths measured in numbers of substitutions per site. The analysis involved 36 amino acid sequences and was run with 1000 bootstraps. Numbers indicate bootstrap values. There were a total of 903 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [35]. Accession numbers for C. salei sequences are in Table 1 and for other sequences in S1 Table.
Fig 4. Pairwise identity matrix for anion permeable Cys-loop sequences.
Each cell represents a percentage identity score between two sequences, one indicated horizontally to the left and the other vertically at the top. The 11 C. salei sequences are in bold and their scores are shaded in grey. The sequence demarcation tool for MAFFT aligned sequences [40] was used to create the matrix.
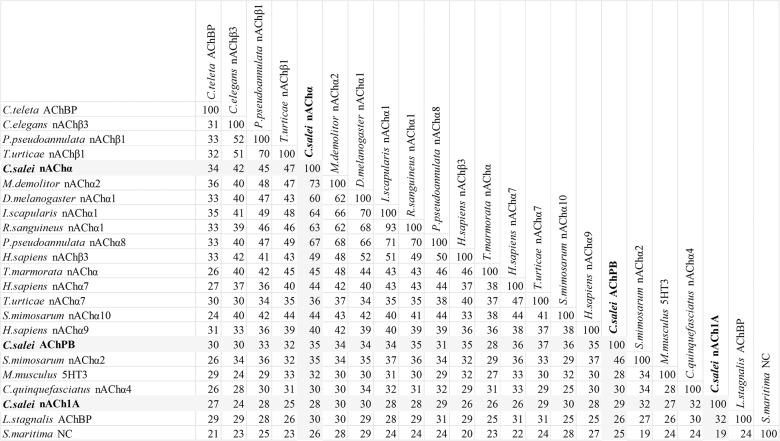
Fig 5. Pairwise percentage identity score matrix for cation selective Cys-loop sequences.
Each cell shows the score between two sequences, one indicated horizontally to the left and the other vertically on the top. Three C. salei sequences are in bold and their scores are shaded in grey. The sequence demarcation tool for MAFFT aligned sequences [40] was used to create the matrix.
The phylogenetic analyses indicated that C. salei transcriptomes contained RNA that encode most of the same types of Cys-loop receptors that have been described in other invertebrate species, although some sequences appear to be more specific to arachnids or spiders. In addition, we found a putative ACh binding protein (AChBP) that has only previously been described in molluscs and in one annelid [46,47]. Inclusion of C. elegans Cys-loop sequences into analysis produced trees with low bootstrap values, so we excluded them from the final analyses. However, we tested the phylogenetic relationships of spider and C. elegans sequences separately and found orthologous groups, except for the nematode Cl-gated ACh or 5-HT receptors [9].
Most invertebrate sequences included in our analyses have previously been annotated based on homology, not actual function. Sequences that have been functionally tested by expression in heterologous systems and/or by agonist binding studies are indicated by asterisks in Figs 2 and 3 and noted in the S1 Table. Some invertebrate sequences have been annotated as glycine receptors by homology, but we do not know of any evidence that these sequences are glycine receptors, which are otherwise found exclusively in chordates [10].
3.1. GABAA receptor subunits
Of the four putative GABAA receptor subunits (Fig 2) two are in the invertebrate “resistance to dieldrin’ (rdl) clade that has no vertebrate orthologues, one is in the “GABA and glycine-like receptor” (grd) clade that is orthologous to vertebrate GABAAα subunits, and one is in the GABAAβ clade that is a highly conserved clade in vertebrates and invertebrates. This clade is named “ligand gated chloride ion channel homolog 3”, lcch3 in insects [48]. Mammalian pentameric GABAA receptors typically have two α, two β and one γ subunit, and GABA binding sites are located between the α-β interface [49]. Invertebrate GABAA receptor subunit composition is not known, but expression of insect rdl genes in oocytes produced functional homopentameric receptors that had similar properties to their native GABA receptors; they were Cl-channels that could be blocked by picrotoxin, but not by bicuculline [50–52]. Conversely, co-expression of Drosophila rdl and lcch3 genes formed channels that had different properties to native receptors [53]. Expression of neither Drosophila lcch3 nor grd gene on their own produced functional channels, but when co-expressed in Xenopus oocytes, they formed GABA-gated channels that were selective to cations [54]. However, receptors with similar properties have not been found in native insect tissue. The mechanosensory VS-3 neurons in C. salei leg patella respond to GABA with depolarization and this response is blocked by picrotoxin but not by bicuculline [26], suggesting that these cells have GABArdl receptors. Whether the GABAAβ and grd are also subunits of these receptors, or have other roles in spider tissue remains to be investigated.
Insects have one GABArdl gene that encodes several alternatively spliced subunits with different agonist sensitivities [55]. They also have one lcch3 and one grd gene [56–59]. The spider mite, T. urticae, has three rdl genes but no β or grd genes [6]. Multiple rdl genes are also common in other arachnid species, and some appear to lack genes that encode other GABAA subunits [6]. However, we found arachnid GABA subunits in all three clades (Fig 2). For example, the African social velvet spider Stegodyphus mimosarum has a homologous gene for each of the C. salei GABAA sequences, but only one rdl gene. Therefore, the numbers of different GABAA sequences vary even in closely related arachnid species.
3.2. Glutamate-Gated Chloride Channel (GluCl) subunits
We found three putative GluCl genes (CsGluCl1-3) in the C. salei transcriptomes. Each had a homologous counterpart in the S. mimosarum genome (Fig 2). Most insects have only one GluCl subunit [57,58] and these are phylogenetically closest to the CsGluCl1. The spider mite T. urticae has six orthologous GluCl subunits [6] and although only one is shown in Fig 2, they were all in the same sub-clade with the CsGluCl1, more distant to the other two CsGluCl subunits. We also tested six C. elegans GluCl sequences separately [9] and found that they are also more closely related to the CsGluCl1 than to the other two C. salei GluCl subunits. The above-mentioned S. mimoasarum sequences where the only closely homologous sequences to CsGluCl2 and CsGluCl3, suggesting that these sub-clades may be specific to spiders. Similar to the insect and T. urticae 12344 Cl-channel clade, the CsGluCl2 and CsGluCl3 sequences lack the second Cys-loop that is usually found in GluCl channels [6]. However, the C. salei transcriptomes did not have any sequences in the 12344 clade and it was not included in our phylogenetic tree.
Insect and Aplysia GluClα subunits formed homomultimeric glutamate gated Cl-channels that were activated irreversibly by ivermectin in expression systems [10,52,60]. However, a functional C. elegans GluCl receptor required both α and β subunits [13,61]. Drosophila and Musca domestica GluCl subunits were shown to co-assemble with GABArdl subunits and the latter formed heteromultimeric channels with higher GABA sensitivity than the homomultimeric GABArdl channel [52,62]. Arachnid GluCl or GABA channels have not been tested in expression systems. Spider VS-3 neurons had variable responses to glutamate in electrophysiological experiments [27,28]: Some cells responded to both glutamate and GABA with a large depolarization and brief inhibition followed by excitation. However, glutamate did not depolarize most neurons at all or only slightly, but inhibited responses to mechanical and electrical stimulation. These differences suggest a heterogeneous population of GluCl channels in different VS-3 neurons with varying agonist sensitivities and ionic selectivities.
3.3. pH gated chloride channels (pHCl)
Two C. salei subunits are in the pHCl clade that includes representatives from many insect and arachnid species [6,58]. The pH sensitivity of these channels was demonstrated in Drosophila [63] and itch mite Sarcoptes scabiei [64] pHCl subunits expressed in oocytes. Both subunits formed homopentameric Cl-permeable channels that were insensitive to Cys-loop receptor agonists including GABA, glutamate and ACh, but alkaline pH induced inward currents. Both channels were also sensitive to ivermectin, which produced a slowly deactivating or irreversible inward current that was larger at alkaline pH [63,64]. The Drosophila pHCl sequence shares 54% identity with both C. salei pHCl subunits, but the mite pHCl sequence is less similar to C. salei pHCl, or any insect pHCl sequences (Fig 4). Furthermore, we were not able to obtain reliable results with phylogenetic analysis when we added the S. scabiei subunit to our database. The structural features that provides pH sensitivity to the Drosophila or S. scabiei channels are unknown.
3.4. Histamine-gated Cl-channels (HisCl)
Two C. salei subunits are in the HisCl-channel clade. The CsHisCl1 subunit is in a distant branch to most insect, arachnid and crustacean HisCl subunits. It forms a separate branch with arachnid subunits that have been annotated as glycine or glutamate gated Cl-channels (Fig 2). Since none of these channels has been functionally tested, we do not know which transmitter(s) activate them, or their functional roles. The CsHisCl2 is orthologous to the insect HisCl channel that has been shown to mediate neurotransmission in Drosophila eye where histamine activates HisCl2 (also called hclA or ort) channels in the second order lamina neurons [65,66]. Drosophila HisCl1 (also called hclB) subunits also formed histamine gated Cl-channels in expression systems, but had different properties than the native receptors and were only found in laminar glial cells [66,67]. Our phylogenetic analysis found orthologous sequences to CsHisCl2 subunits in several arachnid species, including the spider S. mimosarum. Our transcriptomes did not originate from spider eyes, indicating that CsHisCl2 is not exclusively expressed in the eye. However, CsHisCl2 RNA abundance was very low in the brain and higher in the leg hypodermis (Fig 1), which has a large number of sensory neurons.
3.5. Non-Characterized Ligand Gated Chloride Channels (NC-LGCl)
One of the C. salei sequences is in a distinct group of putative ligand gated Cl-channels that has only arachnid members. Arachnid orthologues to this sequence have been named either GABA, glycine or GluCl subunits, but there is no experimental evidence that any of these transmitters gate these channels. Therefore, we named the C. salei sequence NC-LGCl. The most closely related functionally expressed sequences are the American dog tick, Dermacentor variabilis, GluCl10-2, GluCl1 and GluCl11 (US patents 7267964 and EP2009021). Homomultimeric GluCl10-2 expressed in Xenopus oocytes were not sensitive to glutamate or ivermectin, while homomultimeric GluCl1 and 11 formed Cl-channels that were activated by both glutamate and ivermectin.
3.6. Nicotinic Acetylcholine Receptor Subunits (nACh)
Compared to other invertebrate nACh receptor subunits our C. salei transcriptome searches revealed relatively few nACh subunits. Most insect genomes have 10–16 nACh subunits, while the T. urticae genome had 10 [6]. It is likely that spider has more nACh subunits in tissues other than the brain and hypodermis. Classification of nACh receptor subunits is complicated by differences in nomenclature among different species. Generally, α-subunit Loop C has two vicinal cysteines that are necessary for agonist binding, while non-α subunits lack the vicinal cysteines and are sometimes classified as β subunits [68,69]. One of the C. salei nACh proteins was orthologous to nAChα subunits (Fig 3). Closely related proteins include sequences from the wolf spider Pardosa pseudoannulata and the tick Rhipicephalus sanguineus that share 67% and 63% similarity with CsnAChα (Fig 5). Both of these sequences have been expressed in Xenopus oocytes, but similarly to most invertebrate nACh subunits, they failed to function as homomultimers [8,70]. When co-expressed with nAChβ subunits, they formed functional ACh and nicotine sensitive receptors.
The spider transcriptomes had two non-α subunit isomers, CsnACh1A and B that lacked the vicinal Loop C cysteines. Phylogenetically these sequences formed a different branch to other nACh sequences that was closest to a branch containing only a S. mimosarum subunit named nAChβ3. Exhaustive GeneBank search did not find any closer homologues to these subunits than those shown in Fig 3. It is not clear what the functions of these subunits are, whether they form heteromultimeric channels with the CsnAChα or some other subunits. Since the mRNA level of these non-α subunits are higher in the brain and hypodermis than the level of α subunits (Fig 1) they are likely to have an important role in regulating the functions of both central and sensory neurons.
3.7. Acetylcholine Binding Protein (AChBP)
CsAChBP shares a branch of the phylogenetic tree with the S. mimosarum fragment that has been named nAChα2, but may be an ACh binding protein. Since AChBPs have previously only been found in molluscan and annelid species, we ran additional phylogenetic analysis by adding AChBP sequences to the nACh group and a separate analysis that only included AChBPs, but these tests produced trees with low bootstrap values. This is probably because AChBP sequence conservation is low, even within molluscan species [46]. Although several AChBPs have been shown to bind ACh and nicotine, their physiological roles remain unclear [18,47,71]. Molluscan glial cells secrete AChBPs that are believed to regulate synaptic transmission by buffering ACh levels in the synapses instead of, or in addition to, acetylcholine esterase (AChE) [46]. AChE is widely found in metazoans and is important in regulating synaptic transmission by reducing ACh levels [47]. We also found four AChE sequences in the C. salei transcriptomes, making it unlikely that AChBP would be needed to regulate ACh levels in spider tissue.
4. Sequence analysis
Functional Cys-loop receptors consist of five subunits. Agonists bind to a specific binding pocket region on the large extracellular N-terminal domains of two neighbouring subunits. It is formed by three loops A–C of the principal face that span β strands and contain key aromatic residues. The adjacent subunit, which forms the complementary face, contributes three to four β strands with residues clustered in segments called loops D–F [18,44,72,73]. The binding sites of all Cys-loop receptors are dominated by aromatic residues but the critical residues are not necessarily equivalent even for closely related receptors [74–76]. Each subunit has four transmembrane helices TM1-TM4. The TM2 helix is located centrally and forms the channel pore; its amino acid sequence determines ion selectivity [43]. Portions of the C. salei sequences aligned with representative homologous sequences from other species containing the agonist binding loops and the TM2 are shown in Figs 6 and 7, respectively. The complete sequence alignments are in S1 Fig and S2 Fig.
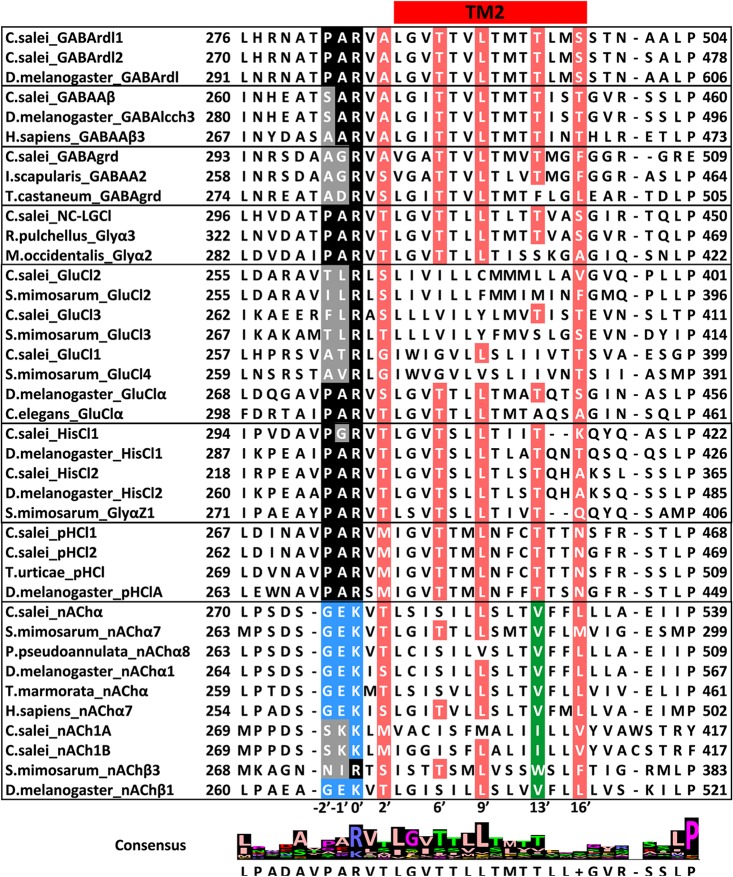
Fig 6. Alignment of the amino acid sequences in the N-terminal agonist binding Loops.
Conserved residues involved in agonist binding to the principal face are highlighted in blue and those to the complementary face are shown in red. Cysteine residues of the second Cys-loop in GluCl, HisCl and pHCl subunits are indicated in brown and the vicinal cysteine residues in the Loop C of the nAChα subunits are also shown in brown. Numbering is shown for the C. salei GABArdl1 subunit. Sequences were aligned using MAFFT.
Fig 7. Alignment of the amino acid sequences in transmembrane domain 2 (TM2).
Residues that line the pore in most Cys-loop receptors are shown in red. Arginine in position 0’ is critical for Cl-selective channels. The equivalent position for cation selective channels (-1’) generally has negatively charged residue (E or D). Note that the two isomers of C. salei nACh1 have a positively charged amino acid in this location. Position 13’, which is occupied by hydrophobic residues in cation permeable channels, is shown in green. Sequences were aligned with MAFFT.
4.1. Extracellular ligand binding domain in C. salei Cys-loop subunits
All twelve subunits in the anion permeable channel family and the CsnAChα subunit have well-conserved Cys-loops with 13 amino acids between the cysteine residues (S1 Fig and S2 Fig) [1]. The CsnACh1A and 1B subunits have 14 residues in their Cys-loops, which is unusual. CsAChBP has the most atypical Cys-loop with 16 amino acids. AChBPs have variable numbers of residues in their Cys-loops, even in different molluscan species [18,77]. Since AChBPs do not have the channel region, the Cys-loop must have a different role than in “normal” receptors where it is involved in communication between the neurotransmitter binding site and the pore [73]. The second Cys-loop, which is commonly found in glycine and GluCl receptors, occurs only in one of the C. salei GluCl subunits (CsGluCl1), but it is present in CsHisCl1 and 2 as well as CspHCl1 and 2 subunits (Fig 6, S1 Fig).
GABA binds to a range of vertebrate and invertebrate Cys-loop receptors, but the actual binding site varies [75]. The CsGABArdl1 and 2, the CsNS-LGIC and CsGluCl3 are the only C. salei anion channel sequences that have all the residues believed to be involved in agonist binding in both the principal face (blue residues in Fig 6) and the complementary face (red residues in Fig 6). The CsGABAAβ and CsGluCl1 subunits have all of these residues only on the principal face and lack some of them on the complementary face while the CsGABAgrd subunit has all the same complementary face amino acids involved in agonist binding, but lacks one on the principal face. All other putative anion selective subunits lack some or all of these residues. This arrangement suggests that, at least in some of the pentameric channels, different subunits form the principal and complementary faces for agonist binding. Since activation of Cys-loop receptors generally requires binding of at least two agonist molecules [78], additional subunits may have different roles.
In addition to the aromatic ring required for agonist binding in all Cys-loop receptors, arthropod and nematode GluCl channel glutamate recognition was recently shown to depend on an arginine residue in Loop G [79]. In C. salei, this arginine is only present in the CsGluCl1 subunit (Fig 6), but not in the other two GluCl subunits. Interestingly, along with all other complementary face agonist binding residues, this arginine is also present in the CsGABAgrd sequence. The Loop F lysine that was shown to interact with glutamate in C. elegans GluCl channel [13] is missing from all C. salei GluCl subunits and was recently shown to be unnecessary for glutamate binding [79].
The nAChα subunit and the CsnAChBP share all the agonist binding residues with both invertebrate and vertebrate nAChα receptors (Fig 6). However, these residues are not present in the two isomers of CsnACh1 where the Loop A tyrosine is replaced with another aromatic, histidine. The nicotine binding tryptophan in Loop D is occupied with a tyrosine that is commonly found in anion selective channels [80]. Based on the lack of Loop C vicinal cysteines that are essential for agonist binding [68], these sequences can be classified as non-α subunits.
4.2. Transmembrane helix 2 (TM2)
Fig 7 uses the standard numbering system for LGIC TM2 domain [43]. Starting from highly conserved residue 0’ close to the intracellular C-terminal end, moving inward through the channel, residues in positions 2’, 6’, 13’ and 16’ line the Cys-loop channel pore and have important roles in controlling channel selectivity and conductance [81]. These residues in CsGABArdl are identical to the Drosophila rdl sequence (red residues in Fig 7). Alanine in position 2’ is the residue that, when mutated to serine in Drosophila and many other insect subunits, renders the channel resistant to the insecticide dieldrin [82,83]. The same residue is also implicated in binding other drugs, such as the Cl-channel blocker picrotoxin and the antiparasitic ivermectin [4,13,84]. Additional positions for picrotoxin and dieldrin block are threonine in position 6’ [84–86] and leucine in position 9’ [87]. Alanine in position 2’ is present in all four C. salei GABA subunits, the threonine 6’ and leucine 9’ can be found in most anion permeable subunits, except the CsGluCl2 and 3, while the GluCl1 has only one of these residues (Fig 7). Picrotoxin binds between 2’ alanine and -2’ proline in C. elegans GluCl channel [13]. The -2’ proline is present in both CsGABArdl and CspHCl subunits as well as in CsNC-LGCl. In electrophysiological experiments, the C. salei VS-3 neuron responses to both GABAA and GluCl receptor agonists were blocked by picrotoxin, but the latter required a significantly higher concentration [26,27]. Since CsGluCl subunits lack all or most of the above mentioned picrotoxin binding residues, it is likely that the VS-3 neuron glutamate response is mediated by heteromultimeric channels.
Anion selective Cys-loop channel α-subunits typically have a PAR motif (proline-alanine-arginine) in the intracellular region between TM1 and TM2 (locations -2’,-1’,0’ in Fig 7), while other subunits have either the proline or alanine, or both, replaced with a different amino acid [10]. All putative C. salei anion permeable subunits have positively charged arginine in position 0’. This residue is absolutely conserved in Cl-channels [81]. The complete PAR motif is present in both CsGABArdl and CspHCl subunits, as well as the CsHisCl2 and the CsNC-LGCl subunits. CsGABAAβ, CsGABAgrd and their orthologues in other species do not have proline in the -2’ position. Proline in this position is typical for anion selective channels [10,43,88]. In all grd subunits, alanine in the -1’ position is also replaced. It is likely that the CsGABAAβ and grd are subunits of heteropentameric channels, rather than either of them forming homopentameric channels in native invertebrate tissues.
The TM2 regions of all three C. salei and S. mimosarum GluCl subunits are different to typical anion selective channels. However, even though the -2’and -1’positions do not have proline or alanine, they are not occupied by negatively charged residues as would be expected for cation selective channels [10,43,54,88]. The GluCl sequences in the two spider species also have different residues occupying the locations that line the channel pore [6’,9’,13’] than most anion selective channels. Anion selective channels typically have the polar threonine in position 13’ while this residue is hydrophobic in cation selective channels [43,54,88]. All C. salei nACh subunits have a hydrophobic residue in this location (either isoleucine or valine), but it is also hydrophobic in CsGluC1 and 2 (isoleucine and leucine, respectively). Therefore, the spider GluCl subunits are quite different to other arthropod sequences and this may contribute to a variability in ion selectivity and channel kinetics.
The CsnAChα subunit shares all of the amino acids in TM2 helix that line the pore and determine the cation selectivity of nACh channels (Fig 7), but both CsnACh1 isomers have significant differences in this helix. Importantly, negatively charged glutamic acids in positions -1’ and 20’ that serve as selectivity filters in cation selective channels [43,73] are occupied by a positively charged lysine and neutral serine, respectively. Therefore, the roles of these subunits are probably different to typical nACh receptors.
4.3. Binding sites for insecticides and antiparasitic agents
Since spiders are ecologically important natural predators for many insects, it is important to determine if widely used insecticides and antiparasitic agents that target Cys-loop receptors also affect spider receptors. As discussed earlier, all four C. salei GABA receptors have the three TM2 residues (A2’, T6’ and L9’, Fig 7 and S1 Fig) that have been proposed to be involved in binding the cyclodiene insecticide dieldrin, which is now generally banned due to non-specific toxicity [89]. These same residues are also suggested to be involved in binding phenylpyrazoles such as the widely used slow acting insect poison fipronil [6,87]. An additional residue for fipronil binding is a threonine in the TM3 motif (S1 Fig), which is found in the two C. salei GABArdl subunits. Based on these similarities, all C. salei GABA subunits are likely to bind dieldrin while CsGABArdl1 and 2 would also be affected by fipronil. These same residues are also present in S. mimosarum GABAAβ1 and β2 subunits, indicating that the same control agents would affect these behaviorally different spiders.
Avermectins such as ivermectin and abamectin are antiparasitics that are used against worms causing river blindness in humans, as well as worms and mites in livestock and pets. Abamectins are also used in ant baits and to protect fruit, vegetable and other crops from mites and insects [81]. Avermectins are allosteric modulators directed against GluCl channels; they bind to the receptor transmembrane domain and stabilize the receptor in an open conformation [13]. When tested in electrophysiological experiments, ivermectin depolarized C. salei VS-3 neurons irreversibly [27], indicating that some receptors in these neurons have binding sites for this drug. No generic amino acid motif has been shown to bind ivermectin in Cys-loop receptors of all species [5]. However, Hibbs and Gouaux [13] predicted three residues to be involved in hydrogen-bonding and 12 additional residues with van der Waals interactions with ivermectin in the C. elegans GluClα subunit. Of these, the TM3 glycine was later shown to be essential for high ivermectin [90] and abamectin sensitivity [6]. This location is also important in binding new types of insecticides called meta-diamides that are non-competitive antagonists of GABArdl receptors [91]. Fig 8 shows the M1-M3 region of C. elegans GluClα sequence aligned with the C. salei anion channel subunits. Many of the residues associated with ivermectin binding are present in several C. salei subunits and the GABArdl1 and 2, NC-LGCl, and GluCl1 and 3 also have the TM3 glycine, suggesting that these subunits may be involved in ivermectin binding. Although the main targets of ivermectin are GluCl receptors, many other receptors, including GABA, glycine, HisCl and pHCl receptors are modulated by this drug, usually at higher concentrations than GluCl channels [4,81]. If ivermectin binding sites in C. salei were similar to the C. elegans GluClα subunit, the two CsGABArdl subunits would be most strongly affected by this drug.
Fig 8. Residues potentially involved in ivermectin binding.
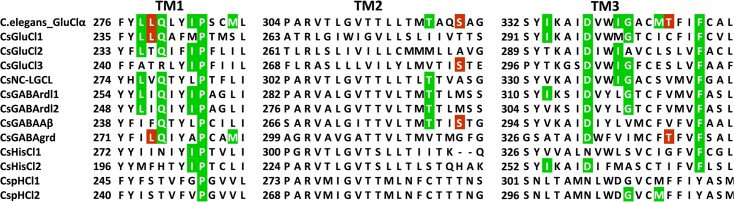
Partial alignment of putative C. salei anion channel subunits with the C. elegans GluClα sequence. Residues in the C. elegans sequence that were predicted to be involved in hydrogen-bonding and van der Waals interactions with ivermectin are shown in brown and green respectively [13]. Alignment was done by MAFFT.
Neonicotinoids, such as the currently most widely used agricultural insecticide Imidacloprid, bind to insect nACh receptors and, since they are not broken down by AChE, cause overstimulation, paralysis and death. Differently to nicotinoids, neonicotinoids cannot cross vertebrate blood brain barriers and they have a negatively charged nitro or cyano group, which is proposed to interact with positively charged amino acids present on insect, but not mammalian nAChRs [2,3,11]. They have not been shown to be toxic for spider mites T. urticae [6] or wolf spiders P. pseudoannulata, although the functionally expressed nACh receptors of the latter were sensitive to several neonicotinoids [8]. Although the aromatic residues in the agonist binding region (Fig 7) are highly conserved in different species, there are differences in other residues in this region even within species that are insensitive to neonicotioids [2,6]. The high affinity for neonicotinoids at the insect nAChR has been attributed to their specific binding orientation and different positioning compared to nicotinoids at the vertebrate nAChR [2,3]. Although we do not know the functions or subunit compositions of spider nACh receptors, they are expressed in both the central and peripheral nervous systems (Fig 1), and it is possible that some of them bind neonicotinoids.
5. C. salei Cys-loop receptor homology modelling
Related proteins usually have similar three-dimensional structures and these structures are often evolutionally even more conserved than would be expected from the sequence similarity [92]. We investigated the structural similarity of C. salei Cys-loop sequences compared to the templates currently available in the Protein Data Bank (PDB) using the I-TASSER server [41]. Each run created five models and the best model based on I-TASSER scores (Table 2) was selected for further analysis. The C-score is in the range of -5 to 2 and indicates the quality of predicted models; higher C-score value signifies a model with more confidence. The TM score indicates the structural similarity between predicted model and the native structure. A TM-score > 0.5 indicates a model of correct topology and a TM-score < 0.17 means random similarity. The results show that the models were of good to excellent quality, with the exception of CsGABAgrd, which had a C-score of -2.27 and TM score of 0.45. Table 2 also shows the main templates that I-TASSER server used to create the models. The templates with highest identity to each C. salei sequence are indicated in bold.
Table 2. The C- and TM scores of Cupiennius Cys-loop subunit 3D structures predicted by I-TASSER server, the Protein data bank (PDB) templates used by I-TASSER to create the structures and percentages of residues in favored, allowed and outlier regions as evaluated by Ramachandran plot analysis.
| Subunit | C-score | TM score | PDB Templates | #of residues in favored region | #of residues in allowed region | #of residues in outlier region |
|---|---|---|---|---|---|---|
| CsGABArdl1 | -1.45 | 0.54±0.15 | 4cof,4pir,4aq5 | 81.4% | 11.6% | 7.0% |
| CsGABArdl2 | -1.12 | 0.57±0.14 | 4cof | 84.4% | 9.1% | 6.5% |
| CsGABAAβ | -1.33 | 0.55±0.15 | 4cof,4aq5,4pir | 88.3% | 7.7% | 4.0% |
| CsGABAgrd | -2.27 | 0.45±0.14 | 4cof,3eam,4aq5,4pir | 82.6% | 12.5% | 4.9% |
| CsGluCl1 | -0.37 | 0.67±0.13 | 4tnw,3rhw,3eam,4cof | 88.4% | 9.7% | 1.9% |
| CsGluCl2 | -0.39 | 0.66±0.13 | 4tnw,3rhw,4cof,4aq5 | 86% | 9.8% | 4.3% |
| CsGluCl3 | -1.10 | 0.58±0.14 | 4tnw,3rhw,4cof,4hfi | 84.3% | 10.7% | 5% |
| CspHCl1 | -0.53 | 0.65±0.13 | 3rhw,4tnw,4cof,4pir | 81.5% | 11.4% | 7.1% |
| CspHCl2 | -0.59 | 0.64±0.13 | 3rhw,4tnw,4pir,4cof | 79.9% | 12.9% | 7.2% |
| CsHisCl1 | -1.04 | 0.58±0.14 | 3rhw,4tnw,4cof | 80.4% | 10.6% | 9% |
| CsHisCl2 | -0.28 | 0.68±0.12 | 3rhw, 4cof, 3eam | 79.9% | 13.8% | 6.3% |
| CsNC-LGCl | -0.80 | 0.61±0.14 | 4tnw,3rhw,4cof | 85.8% | 9.1% | 5.1% |
| CsnACh1A | -0.91 | 0.60±0.14 | 4pir,4aq5,4cof | 84.1% | 11.4% | 4.7% |
| CsnACh1B | -0.64 | 0.63±0.13 | 4pir,4aq5,2qc1 | 84.1% | 11.1% | 4.8% |
| CsnAChα | -1.48 | 0.53±0.15 | 2bg9,4pir,4aq5 | 75.5% | 15.3% | 9.2% |
| CsAChBP | -0.26 | 0.68±0.12 | 3sq6,4d01,2qc1,4aq5,2bg9 | 81% | 13.7% | 5.3% |
4cof H. sapiens GABAAβ3, 4pir M. musculus 5-HT3; 4aq5 and 2bg9 T. marmorata nACh receptor, 3eam and 4hfi G. violaceus GLIC, 3rhw C. elegans GluClα open channel, 4tnw C. elegans GluClα closed channel, 2qc1 M. musculus extracellular domain of nAChRα1, 3sq6 H. sapiens and L. stagnalis ligand binding domain nAChα7 Chimera, 4d01 H. sapiens nAChα9.
A predicted cartoon model for CsGABAAβ (Fig 9) is aligned with a ribbon representation of the human GABAAβ subunit (4cof). Similar images from all C. salei sequences aligned to the templates with highest identities are shown in S3 Fig. All models have the large, β-sheet-rich, extracellular domain with ligand-binding loops and an α-helical pore-forming domain. The intracellular domain, which is believed to interact with cellular scaffolding proteins in most Cys-loop receptors, is present in all but the three CsGluCl and the CsNC-LGCL models. This intracellular domain is also missing in the C. elegans GluClα crystal structure [13].
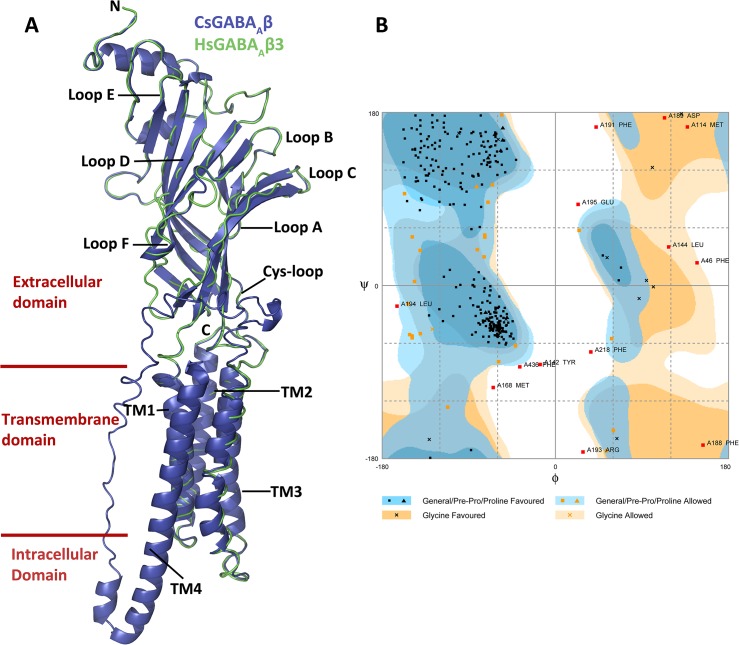
Fig 9. Structural model of the C. salei GABAAβ subunit and its evaluation by Ramachandran plot analysis.
(A) Homology model of CsGABAAβ created by I-TASSER server is shown in blue as cartoon representation aligned with the human HsGABAAβ3 crystal structure in green ribbon representation. The structures are shown as viewed from the outside of the pentameric ring. The agonist binding Loops A-B in the principal face and D-F in the complementary face and the transmembrane helices TM1-M4 are indicated. Image was created with PYMOL software. (B) Ramachandran plot analysis of CsGABAAβ protein structure calculated by the Rampage server.
All three C. salei GABA subunit models shared highest identity (43–59%) with the human GABAAβ3 crystal structure [15]. C. salei models were well aligned with this structure, except in the N-terminal ligand binding region of CsGABAgrd. All other putative anion channel subunits shared highest identity (31–44%) with the C. elegans GluCl crystal structure [14,15]. The CsGluCl1-3 and CsNC-LGCl models were well aligned with this model, but CspHCl1 and 2 as well as CsHisCl1 and 2 N-terminal extracellular domains did not match as well. Interestingly, the two isomers of C. salei nACh1 subunits were best aligned with the mouse 5-HT3 crystal structure [16] rather than any of the available nACh structures. Even though the sequence identity of these proteins was only 23%, their ligand binding extracellular regions and TM regions were well aligned. Extracellular regions of nAChα and AChBP sequences have all the basic structures predicted to be required for ligand binding, but they were poorly aligned with any available models. They share relatively high sequence identity with several nACh receptor crystal structures, but most of these are rather low resolution models.
We evaluated the overall stereochemical qualities of each homology model with Ramachandran plots using the RAMPAGE server [42]. The highly variable segment between TM3 and TM4 is not included in several of the Protein data bank templates that I-TASSER used as models. These segments are clearly visible in many of the C. salei models and not aligned with their templates (Fig 9, S3 Fig). Therefore, these segments were removed before the models were submitted to the Rampage server. An example of a Ramachandran plot is shown in Fig 9 and the overall results are listed in Table 2. The results indicated that the predicted three-dimensional models are of good quality. All models had some residues in the outlier region, indicating that the combination of phi and psi torsion angles were unusual, but these were mainly in regions distant from the binding site or channel pore, close to the C- or N-terminal regions or in the region between the extracellular ligand-binding and transmembrane segments. Only two outlier residues were in transmembrane helices; in models CsGABArdl2 and CspHCl1, in both cases a highly conserved proline in TM1 was indicated to be an outlier. Most residues in the extracellular agonist binding loops were in the favoured or allowed regions, but 1–3 outliers were present in loop C and also in the coil region close to loop C. The only sequences without these outliers were CsGluCl1 and CspHCl1 and 2. It is not clear if these outliers are truly unusual angles in the protein folding in these functionally important regions, or model errors. Only a small number of high-resolution Cys-loop protein models are available in PDB and even fewer have been crystallized in more than one state (open and closed). From the agonist binding loops, Loop C undergoes most pronounced movement when the agonist or antagonist binds to the receptor [1], making it more difficult to predict homology models from this area.
Conclusions
C. salei transcriptome searches revealed a relatively small number of Cys-loop receptor subunits compared to available insect or arachnid genomes that have more than 20 such subunits [6] or C. elegans with 102 Cys-loop subunits [9]. It is likely that additional subunits are expressed in C. salei organs and tissues that were not included in the transcriptomes. It is also possible that we missed very low abundance genes in the brain and hypodermis. However, we found representatives of all major groups of invertebrate Cys-loop receptors, in some cases more than one orthologous subunit, indicating that these proteins have important roles in the spider central and peripheral nervous systems.
Phylogenetic analysis showed that although the same Cys-loop receptor clades are found in most arthropods, some arachnid clades are distant to insects and some subclades had only spider genes. Unfortunately, there is little molecular, and even less physiological, information currently available about Cys-loop receptors in other spider species. Several S. mimosarum Cys-loop gene fragments have been submitted to the GeneBank [93] and annotated based on homology, but their functions are unknown. Our analysis suggests that some annotated functions of S. mimosarum, and other arachnid subunits may not be accurate. It is interesting that even though C. salei and S. mimosarum are members of different Araneomorhae families (Lycosoidea and Eresidae, respectively) their Cys-loop receptor genes are closely related. This includes the putative AChBP that in our analysis had both the start and stop codons, indicating that it encodes a complete protein, while S. mimosarum has a gene fragment in the same clade but we do not know if it also encodes AChBP rather than the complete nACh receptor subunit as annotated.
Although subunit compositions of native invertebrate Cys-loop receptors are unknown, expression of some genes form homomultimeric receptors. Examples are the GABArdl, GluClα and pHCl subunits of several insects [50,52,60,63,64]. However, evidence from several arthropod species indicate that multiple pharmacologically different receptors exist, suggesting that many native Cys-loop receptors are heteromultimeric [59,62]. Examples of these are the invertebrate and most mammalian nACh subunits that alone do not produce functional nACh receptors, but require at least two different subunits [94,95]. When two wolf spider (P. pseudoannulata) nAChα subunits were each co-expressed with the nAChβ subunit, both formed functional ACh receptors, but not when they were expressed alone [8]. We did not find any C. salei subunits that would be orthologous to the P. pseudoannulata nAChβ, but we found two isomers of non-α nACh subunits that may be needed to form functional receptors.
Our sequence analysis and homology models are helpful in functional annotation of the C. salei Cys-loop receptors. They indicated that many features are similar to currently available templates and suggest that they have similar abilities to bind ligands and drugs, some likely as harmful to spiders as to insect pests and parasites. However, we also found differences that probably lead to different binding properties as well as differences in ion channel kinetics and selectivity. It would be important to express the genes functionally to determine properties of each receptor. Control agent development generally considers safety for humans and other mammals, but little effort has been placed on potential harm to beneficial species such as spiders.
There is significant physiological information about the anion selective Cys-loop receptors that modulate the sensitivity of VS-3 mechanoreceptor neurons innervating slit sensilla in the spider patella [26–29]. We now have molecular tools to identify the proteins that form each of these receptors and determine the roles they play in regulating the flow of information from the spider’s environment to its central nervous system.
Supporting Information
Signal peptides for C. salei sequences are indicated in red. Secondary structure based on I-Tasser prediction with high confidence (score over 8) is presented for the CsGABArdl1 protein. β-sheets are indicated in green and α-helices in red. Blue lines indicate amino acid sequences that form agonist-binding loops A to G. Cys-loops are identified in brown. TM1-TM4 are the transmembrane segments. Residues potentially involved in dieldrin and fipronil binding in TM2 and TM3 are circled in green. Gray color indicates conserved amino acids based on Jalview consensus threshold of 50% where darker gray indicates higher degree of conservation. Accession numbers for C. salei sequences are in Table 1 and for other species in the S1 Table.
(PDF)
Signal peptides for all C. salei sequences are shown in red. Secondary structure as predicted by I-Tasser for CsnAChα protein shows β-sheets in green and α-helices in red. I-Tasser confidence score was over 8 for these predictions. Amino acid sequences that form agonist-binding loops A to F are indicated as blue lines. Cys-loop and the double cysteine are shown in brown. TM1-TM4 are the transmembrane segments. Gray color indicates conserved amino acids based on Jalview consensus threshold of 50% where darker gray means higher conservation. Accession numbers for C. salei sequences are in Table 1 and for the other species in S1 Table.
(PDF)
C. salei structural models are shown in blue cartoon representations aligned with their PDB templates in ribbon representations. Human GABAAβ3 (HsGABAAβ3, green 4cof), C. elegans GluClα (CeGluCl, red 3rhw), mouse 5-HT3 (Mm5-HT3, orange 4pir), Torpedo marmorata nAChα (TmnAChα, cyan 4aq5) and Lymnae stagnalis/Human nAChα7 ligand binding domain chimera (LsHsnAChα7, cyan 3sq6). Images were created with PYMOL software.
(PDF)
(PDF)
Data Availability
All relevant data are within the paper and its supporting information files.
Funding Statement
Funding provided by Canadian Institutes of Health Research, Grant number MOP-106602, http://www.cihr.ca/e/193.html (PHT and ASF); Natural Sciences and Engineering Research Council of Canada RGPIN 05565, http://www.nserc-crsng.gc.ca/index_eng.asp (PHT).
References
- 1. Smart TG, Paoletti P. Synaptic neurotransmitter-gated receptors. Cold Spring Harb Perspect Biol. 2012; 4: a009662 10.1101/cshperspect.a009662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tomizawa M, Casida JE. Molecular recognition of neonicotinoid insecticides: the determinants of life or death. Acc Chem Res. 2009; 42: 260–269. 10.1021/ar800131p [DOI] [PubMed] [Google Scholar]
- 3. Tomizawa M, Casida JE. Unique neonicotinoid binding conformations conferring selective receptor interactions. J Agric Food Chem. 2011; 59: 2825–2828. 10.1021/jf1019455 [DOI] [PubMed] [Google Scholar]
- 4. Wolstenholme AJ. Recent progress in understanding the interaction between avermectins and ligand-gated ion channels: putting the pests to sleep. 2010; Invert Neurosci. 10: 5–10. 10.1007/s10158-010-0105-y [DOI] [PubMed] [Google Scholar]
- 5. Zemkova H, Tvrdonova V, Bhattacharya A, Jindrichova M. Allosteric modulation of ligand gated ion channels by ivermectin. Physiol Res. 2014; 63 Suppl 1: S215–S224. [DOI] [PubMed] [Google Scholar]
- 6. Dermauw W, Ilias A, Riga M, Tsagkarakou A, Grbic M, Tirry L, et al. The cys-loop ligand-gated ion channel gene family of Tetranychus urticae: implications for acaricide toxicology and a novel mutation associated with abamectin resistance. Insect Biochem Mol Biol. 2012; 42: 455–465. 10.1016/j.ibmb.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 7. Foelix RF. Biology of spiders New York, Oxford: Oxford University Press, Georg Thieme Verlag; 2011. [Google Scholar]
- 8. Meng X, Zhang Y, Guo B, Sun H, Liu C, Liu Z. Identification of key amino acid differences contributing to neonicotinoid sensitivity between two nAChRα subunits from Pardosa pseudoannulata . Neurosci Lett. 2014; 584C: 123–128. [DOI] [PubMed] [Google Scholar]
- 9. Jones AK, Sattelle DB. The cys-loop ligand-gated ion channel gene superfamily of the nematode, Caenorhabditis elegans . Invert Neurosci. 2008; 8: 41–47. 10.1007/s10158-008-0068-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kehoe J, Buldakova S, Acher F, Dent J, Bregestovski P, Bradley J. Aplysia cys-loop glutamate-gated chloride channels reveal convergent evolution of ligand specificity. J Mol Evol. 2009; 69: 125–141. 10.1007/s00239-009-9256-z [DOI] [PubMed] [Google Scholar]
- 11. Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009; 457: 115–118. 10.1038/nature07461 [DOI] [PubMed] [Google Scholar]
- 12. Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009; 457: 111–114. 10.1038/nature07462 [DOI] [PubMed] [Google Scholar]
- 13. Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011; 474: 54–60. 10.1038/nature10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Althoff T, Hibbs RE, Banerjee S, Gouaux E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature. 2014; 512: 333–337. 10.1038/nature13669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller PS, Aricescu AR. Crystal structure of a human GABA receptor. Nature 2014; 512: 270–275. 10.1038/nature13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hassaine G, Deluz C, Grasso L, Wyss R, Tol MB, Hovius R, et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 2014; 512: 276–281. 10.1038/nature13552 [DOI] [PubMed] [Google Scholar]
- 17. Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4Å resolution. J Mol Biol. 2005; 346: 967–989. [DOI] [PubMed] [Google Scholar]
- 18. Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001; 411: 269–276. [DOI] [PubMed] [Google Scholar]
- 19. Comitani F, Cohen N, Ashby J, Botten D, Lummis SC, Molteni C. Insights into the binding of GABA to the insect RDL receptor from atomistic simulations: a comparison of models. J Comput Aided Mol Des. 2014; 28: 35–48. 10.1007/s10822-013-9704-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barth FG. A spider's world: Senses and behavior Heidelberg: Springer Verlag; 2002. [Google Scholar]
- 21. French AS, Torkkeli PH, Seyfarth E-A. From stress and strain to spikes: mechanotransduction in spider slit sensilla. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2002; 188: 739–752. [DOI] [PubMed] [Google Scholar]
- 22. McGregor AP, Hilbrant M, Pechmann M, Schwager EE, Prpic NM, Damen WG. Cupiennius salei and Achaearanea tepidariorum: Spider models for investigating evolution and development. Bioessays. 2008; 30: 487–498. 10.1002/bies.20744 [DOI] [PubMed] [Google Scholar]
- 23. Wolff C, Hilbrant M. The embryonic development of the Central American wandering spider Cupiennius salei . Front Zool. 2011; 8: 15 10.1186/1742-9994-8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fabian-Fine R, Seyfarth E-A, Meinertzhagen IA. Peripheral synaptic contacts at mechanoreceptors in arachnids and crustaceans: Morphological and immunocytochemical characteristics. Microsc Res Tech. 2002; 58: 283–298. [DOI] [PubMed] [Google Scholar]
- 25. Fabian-Fine R, Meisner S, Torkkeli PH, Meinertzhagen IA. Co-localization of gamma-aminobutyric acid and glutamate in neurons of the spider central nervous system. Cell Tissue Res. 2015; Epub ahead of print. . [DOI] [PubMed] [Google Scholar]
- 26. Panek I, French AS, Seyfarth E-A, Sekizawa S-i, Torkkeli PH. Peripheral GABAergic inhibition of spider mechanosensory afferents. Eur J Neurosci. 2002; 16: 96–104. [DOI] [PubMed] [Google Scholar]
- 27. Panek I, Torkkeli PH. Inhibitory glutamate receptors in spider peripheral mechanosensory neurons. Eur J Neurosci. 2005; 22: 636–646. [DOI] [PubMed] [Google Scholar]
- 28. Torkkeli PH, Meisner S, Pfeiffer K, French AS. GABA and glutamate receptors have different effects on excitability and are differentially regulated by calcium in spider mechanosensory neurons. Eur J Neurosci. 2012; 36: 3602–3614. 10.1111/j.1460-9568.2012.08275.x [DOI] [PubMed] [Google Scholar]
- 29. Pfeiffer K, Panek I, Höger U, French AS, Torkkeli PH. Random stimulation of spider mechanosensory neurons reveals long-lasting excitation by GABA and muscimol. J Neurophysiol. 2009; 101: 54–66. 10.1152/jn.91020.2008 [DOI] [PubMed] [Google Scholar]
- 30. French AS. Transcriptome walking: a laboratory-oriented GUI-based approach to mRNA identification from deep-sequenced data. BMC Res Notes. 2012; 5: 673 10.1186/1756-0500-5-673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. French AS, Li AW, Meisner S, Torkkeli PH. Upstream open reading frames and Kozak regions of assembled transcriptome sequences from the spider Cupiennius salei. Selection or chance? Gene. 2014; 539: 203–208. 10.1016/j.gene.2014.01.079 [DOI] [PubMed] [Google Scholar]
- 32. Henikoff S, Henikoff JG. Performance evaluation of amino acid substitution matrices. Proteins. 1993; 17: 49–61. [DOI] [PubMed] [Google Scholar]
- 33. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013, 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 2010; 38: W23–W28. 10.1093/nar/gkq443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008; 25: 1307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- 37. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009; 25: 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001; 17: 847–848. [DOI] [PubMed] [Google Scholar]
- 39. Pierleoni A, Indio V, Savojardo C, Fariselli P, Martelli PL, Casadio R. MemPype: a pipeline for the annotation of eukaryotic membrane proteins. Nucleic Acids Res. 2011; 39: W375–W380. 10.1093/nar/gkr282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One. 2014; 9: e108277 10.1371/journal.pone.0108277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010; 5: 725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, et al. Structure validation by Cα geometry Φ, ψ and Cβ deviation. Proteins. 2003; 50: 437–450. [DOI] [PubMed] [Google Scholar]
- 43. Keramidas A, Moorhouse AJ, Schofield PR, Barry PH. Ligand-gated ion channels: mechanisms underlying ion selectivity. Prog Biophys Mol Biol. 2004; 86: 161–204. [DOI] [PubMed] [Google Scholar]
- 44. Bartos M, Corradi J, Bouzat C. Structural basis of activation of cys-loop receptors: the extracellular-transmembrane interface as a coupling region. Mol Neurobiol. 2009; 40: 236–252. 10.1007/s12035-009-8084-x [DOI] [PubMed] [Google Scholar]
- 45. Burrows M. The neurobiology of an insect brain Oxford, New York, Tokyo: Oxford University Press; 1996. [Google Scholar]
- 46. Sixma TK, Smit AB. Acetylcholine binding protein (AChBP): a secreted glial protein that provides a high-resolution model for the extracellular domain of pentameric ligand-gated ion channels. Annu Rev Biophys Biomol Struct. 2003; 32: 311–334. [DOI] [PubMed] [Google Scholar]
- 47. McCormack T, Petrovich RM, Mercier KA, DeRose EF, Cuneo MJ, Williams J. Identification and functional characterization of a novel acetylcholine-binding protein from the marine annelid Capitella teleta . Biochemistry. 2010; 49: 2279–2287. 10.1021/bi902023y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. ffrench-Constant RH, Anthony N, Aronstein K, Rocheleau T, Stilwell G. Cyclodiene insecticide resistance: from molecular to population genetics. Annu Rev Entomol. 2000; 45: 449–466. [DOI] [PubMed] [Google Scholar]
- 49. Sigel E, Steinmann ME. Structure, function, and modulation of GABAA receptors. J Biol Chem. 2012; 287: 40224–40231. 10.1074/jbc.R112.386664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen R, Belelli D, Lambert JJ, Peters JA, Reyes A, Lan NC. Cloning and functional expression of a Drosophila γ-aminobutyric acid receptor. Proc Natl Acad Sci USA. 1994; 91: 6069–6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shotkoski F, Lee HJ, Zhang HG, Jackson MB, ffrench-Constant RH. Functional expression of insecticide-resistant GABA receptors from the mosquito Aedes aegypti . Insect Mol Biol. 1994; 3: 283–287. [DOI] [PubMed] [Google Scholar]
- 52. Eguchi Y, Ihara M, Ochi E, Shibata Y, Matsuda K, Fushiki S, et al. Functional characterization of Musca glutamate- and GABA-gated chloride channels expressed independently and coexpressed in Xenopus oocytes. Insect Mol Biol. 2006; 15: 773–783. [DOI] [PubMed] [Google Scholar]
- 53. Zhang HG, Lee HJ, Rocheleau T, Ffrench-Constant RH, Jackson MB. Subunit composition determines picrotoxin and bicuculline sensitivity of Drosophila γ-aminobutyric acid receptors. Mol Pharmacol. 1995; 48: 835–840. [PubMed] [Google Scholar]
- 54. Gisselmann G, Plonka J, Pusch H, Hatt H. Drosophila melanogaster GRD and LCCH3 subunits form heteromultimeric GABA-gated cation channels. Br J Pharmacol. 2004; 142: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hosie AM, Buckingham SD, Presnail JK, Sattelle DB. Alternative splicing of a Drosophila GABA receptor subunit gene identifies determinants of agonist potency. Neuroscience. 2001; 102: 709–714. [DOI] [PubMed] [Google Scholar]
- 56. Jones AK, Sattelle DB. The cys-loop ligand-gated ion channel superfamily of the honeybee, Apis mellifera . Invert Neurosci. 2006, 6: 123–132. [DOI] [PubMed] [Google Scholar]
- 57. Jones AK, Sattelle DB. The cys-loop ligand-gated ion channel gene superfamily of the red flour beetle, Tribolium castaneum . BMC Genomics. 2007; 8: 327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jones AK, Bera AN, Lees K, Sattelle DB. The cys-loop ligand-gated ion channel gene superfamily of the parasitoid wasp, Nasonia vitripennis . Heredity (Edinb). 2010; 104: 247–259. [DOI] [PubMed] [Google Scholar]
- 59. Knipple DC, Soderlund DM. The ligand-gated chloride channel gene family of Drosophila melanogaster . Pestic Biochem Physiol. 2010; 97: 140–148. [Google Scholar]
- 60. Cully DF, Paress PS, Liu KK, Schaeffer JM, Arena JP. Identification of a Drosophila melanogaster glutamate-gated chloride channel sensitive to the antiparasitic agent avermectin. J Biol Chem. 1996; 271: 20187–20191. [DOI] [PubMed] [Google Scholar]
- 61. Cully DF, Vassilatis DK, Liu KK, Paress PS, Van der Ploeg LH, Schaeffer JM, et al. Cloning of an avermectin-sensitive glutamate-gated chloride channel from Caenorhabditis elegans . Nature. 1994; 371: 707–711. [DOI] [PubMed] [Google Scholar]
- 62. Ludmerer SW, Warren VA, Williams BS, Zheng Y, Hunt DC, Ayer MB, et al. Ivermectin and nodulisporic acid receptors in Drosophila melanogaster contain both γ-aminobutyric acid-gated Rdl and glutamate-gated GluClα chloride channel subunits. Biochemistry. 2002; 41: 6548–6560. [DOI] [PubMed] [Google Scholar]
- 63. Schnizler K, Saeger B, Pfeffer C, Gerbaulet A, Ebbinghaus-Kintscher U, Methfessel C, et al. A novel chloride channel in Drosophila melanogaster is inhibited by protons. J Biol Chem. 2005; 280: 16254–16262. [DOI] [PubMed] [Google Scholar]
- 64. Mounsey KE, Dent JA, Holt DC, McCarthy J, Currie BJ, Walton SF. Molecular characterisation of a pH-gated chloride channel from Sarcoptes scabiei . Invert Neurosci. 2007; 7: 149–156. [DOI] [PubMed] [Google Scholar]
- 65. Hardie RC. A histamine-activated chloride channel involved in neurotransmission at a photoreceptor synapse. Nature. 1989; 339: 704–706. [DOI] [PubMed] [Google Scholar]
- 66. Pantazis A, Segaran A, Liu CH, Nikolaev A, Rister J, Thum AS, et al. Distinct roles for two histamine receptors (hclA and hclB) at the Drosophila photoreceptor synapse. J Neurosci. 2008; 28: 7250–7259. 10.1523/JNEUROSCI.1654-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gisselmann G, Pusch H, Hovemann BT, Hatt H. Two cDNAs coding for histamine-gated ion channels in D. melanogaster . Nat Neurosci. 2002; 5: 11–12. [DOI] [PubMed] [Google Scholar]
- 68. Dent JA. Evidence for a diverse Cys-loop ligand-gated ion channel superfamily in early bilateria. J Mol Evol. 2006; 62: 523–535. [DOI] [PubMed] [Google Scholar]
- 69. Thany SH, Tricoire-Leignel H. Emerging pharmacological properties of cholinergic synaptic transmission: Comparison between mammalian and insect synaptic and extrasynaptic nicotinic receptors. Curr Neuropharmacol. 2011; 9: 706–714. 10.2174/157015911798376343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lees K, Jones AK, Matsuda K, Akamatsu M, Sattelle DB, Woods DJ, et al. Functional characterisation of a nicotinic acetylcholine receptor α subunit from the brown dog tick, Rhipicephalus sanguineus . Int J Parasitol. 2014; 44: 75–81. 10.1016/j.ijpara.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Saur M, Moeller V, Kapetanopoulos K, Braukmann S, Gebauer W, Tenzer S, et al. Acetylcholine-binding protein in the hemolymph of the planorbid snail Biomphalaria glabrata is a pentagonal dodecahedron (60 subunits). PLoS One. 2012; 7: e43685 10.1371/journal.pone.0043685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Corringer PJ, Le Novere N, Changeux JP. Nicotinic receptors at the amino acid level. Annu Rev Pharmacol Toxicol. 2000; 40: 431–458. [DOI] [PubMed] [Google Scholar]
- 73. Miller PS, Smart TG. Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol Sci. 2010; 31: 161–174. 10.1016/j.tips.2009.12.005 [DOI] [PubMed] [Google Scholar]
- 74. McGonigle I, Lummis SC. Molecular characterization of agonists that bind to an insect GABA receptor. Biochemistry. 2010; 49: 2897–2902. 10.1021/bi901698c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lummis SC, McGonigle I, Ashby JA, Dougherty DA. Two amino acid residues contribute to a cation-π binding interaction in the binding site of an insect GABA receptor. J Neurosci. 2011; 31: 12371–12376. 10.1523/JNEUROSCI.1610-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lummis SC, Harrison NJ, Wang J, Ashby JA, Millen KS, Beene DL, et al. Multiple tyrosine residues contribute to GABA binding in the GABAC receptor binding pocket. ACS Chem Neurosci. 2012; 3: 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Karlin A. The acetylcholine-binding protein: “What's in a name?” Pharmacogenomics J. 2001; 1: 221–223. [DOI] [PubMed] [Google Scholar]
- 78. Rayes D, De Rosa MJ, Sine SM, Bouzat C. Number and locations of agonist binding sites required to activate homomeric Cys-loop receptors. J Neurosci. 2009; 29: 6022–6032. 10.1523/JNEUROSCI.0627-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lynagh T, Beech RN, Lalande MJ, Keller K, Cromer BA, Wolstenholme AJ, et al. Molecular basis for convergent evolution of glutamate recognition by pentameric ligand-gated ion channels. Sci Rep. 2015; 5: 8558 10.1038/srep08558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Olsen RW, Li GD, Wallner M, Trudell JR, Bertaccini EJ, Lindahl E, et al. Structural models of ligand-gated ion channels: Sites of action for anesthetics and ethanol. Alcohol Clin Exp Res. 2014; 38: 595–603. 10.1111/acer.12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lynagh T, Lynch JW. Molecular mechanisms of Cys-loop ion channel receptor modulation by ivermectin. Front Mol Neurosci. 2012; 5: 60 10.3389/fnmol.2012.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. ffrench-Constant RH, Rocheleau TA, Steichen JC, Chalmers AE. A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature. 1993; 363: 449–451. [DOI] [PubMed] [Google Scholar]
- 83. Ozoe Y. γ-aminobutyrate- and glutamate-gated chloride channels as targets of insecticides. Adv Insect Physiol. 2013; 44: 211–286. [Google Scholar]
- 84. Shan Q, Haddrill JL, Lynch JW. A single β subunit M2 domain residue controls the picrotoxin sensitivity of αβ heteromeric glycine receptor chloride channels. J Neurochem. 2001; 76: 1109–1120. [DOI] [PubMed] [Google Scholar]
- 85. Erkkilä BE, Sedelnikova AV, Weiss DS. Stoichiometric pore mutations of the GABAAR reveal a pattern of hydrogen bonding with picrotoxin. Biophys J. 2008; 94: 4299–4306. 10.1529/biophysj.107.118455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hope M, Menzies M, Kemp D. Identification of a dieldrin resistance-associated mutation in Rhipicephalus (Boophilus) microplus (Acari: Ixodidae). J Econ Entomol. 2010; 103: 1355–1359. [DOI] [PubMed] [Google Scholar]
- 87. Casida JE, Durkin KA. Novel GABA receptor pesticide targets. Pestic Biochem Physiol. 2015; 121: 22–30. 10.1016/j.pestbp.2014.11.006 [DOI] [PubMed] [Google Scholar]
- 88. Wotring VE, Miller TS, Weiss DS. Mutations at the GABA receptor selectivity filter: a possible role for effective charges. J Physiol. 2003; 548: 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ozoe Y, Asahi M, Ozoe F, Nakahira K, Mita T. The antiparasitic isoxazoline A1443 is a potent blocker of insect ligand-gated chloride channels. Biochem Biophys Res Commun. 2010; 391: 744–749. 10.1016/j.bbrc.2009.11.131 [DOI] [PubMed] [Google Scholar]
- 90. Lynagh T, Lynch JW. A glycine residue essential for high ivermectin sensitivity in Cys-loop ion channel receptors. Int J Parasitol 2010; 40: 1477–1481. 10.1016/j.ijpara.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 91. Nakao T, Banba S. Minireview: Mode of action of meta-diamide insecticides. Pestic Biochem Physiol. 2015; 121: 39–46. 10.1016/j.pestbp.2014.09.010 [DOI] [PubMed] [Google Scholar]
- 92. Kaczanowski S, Zielenkiewicz P. Why similar protein sequences encode similar three-dimensional structures? Theor Chem Acc. 2015; 125: 643–650. [Google Scholar]
- 93. Sanggaard KW, Bechsgaard JS, Fang X, Duan J, Dyrlund TF, Gupta V, et al. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun. 2014; 5: 3765 10.1038/ncomms4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lansdell SJ, Millar NS. Cloning and heterologous expression of Dα4, a Drosophila neuronal nicotinic acetylcholine receptor subunit: identification of an alternative exon influencing the efficiency of subunit assembly. Neuropharmacology. 2000; 39: 2604–2614. [DOI] [PubMed] [Google Scholar]
- 95. Elliott KJ, Ellis SB, Berckhan KJ, Urrutia A, Chavez-Noriega LE, Johnson EC, et al. Comparative structure of human neuronal α2-α7 and β2-β4 nicotinic acetylcholine receptor subunits and functional expression of the α2, α3, α4, α7, β2, and β4 subunits. J Mol Neurosci. 1996; 7: 217–228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Signal peptides for C. salei sequences are indicated in red. Secondary structure based on I-Tasser prediction with high confidence (score over 8) is presented for the CsGABArdl1 protein. β-sheets are indicated in green and α-helices in red. Blue lines indicate amino acid sequences that form agonist-binding loops A to G. Cys-loops are identified in brown. TM1-TM4 are the transmembrane segments. Residues potentially involved in dieldrin and fipronil binding in TM2 and TM3 are circled in green. Gray color indicates conserved amino acids based on Jalview consensus threshold of 50% where darker gray indicates higher degree of conservation. Accession numbers for C. salei sequences are in Table 1 and for other species in the S1 Table.
(PDF)
Signal peptides for all C. salei sequences are shown in red. Secondary structure as predicted by I-Tasser for CsnAChα protein shows β-sheets in green and α-helices in red. I-Tasser confidence score was over 8 for these predictions. Amino acid sequences that form agonist-binding loops A to F are indicated as blue lines. Cys-loop and the double cysteine are shown in brown. TM1-TM4 are the transmembrane segments. Gray color indicates conserved amino acids based on Jalview consensus threshold of 50% where darker gray means higher conservation. Accession numbers for C. salei sequences are in Table 1 and for the other species in S1 Table.
(PDF)
C. salei structural models are shown in blue cartoon representations aligned with their PDB templates in ribbon representations. Human GABAAβ3 (HsGABAAβ3, green 4cof), C. elegans GluClα (CeGluCl, red 3rhw), mouse 5-HT3 (Mm5-HT3, orange 4pir), Torpedo marmorata nAChα (TmnAChα, cyan 4aq5) and Lymnae stagnalis/Human nAChα7 ligand binding domain chimera (LsHsnAChα7, cyan 3sq6). Images were created with PYMOL software.
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its supporting information files.