Abstract
Across the diversity of life, organisms have evolved different strategies to thrive in hypoxic environments, and microbial eukaryotes (protists) are no exception. Protists that experience hypoxia often possess metabolically distinct mitochondria called mitochondrion-related organelles (MROs). While there are some common metabolic features shared between the MROs of distantly related protists, these organelles have evolved independently multiple times across the breadth of eukaryotic diversity. Until recently, much of our knowledge regarding the metabolic potential of different MROs was limited to studies in parasitic lineages. Over the past decade, deep-sequencing studies of free-living anaerobic protists have revealed novel configurations of metabolic pathways that have been co-opted for life in low oxygen environments. Here, we provide recent examples of anaerobic metabolism in the MROs of free-living protists and their parasitic relatives. Additionally, we outline evolutionary scenarios to explain the origins of these anaerobic pathways in eukaryotes.
Keywords: mitochondrion-related organelles, anaerobic metabolism, eukaryotic evolution
1. Introduction
Mitochondria and related organelles are ubiquitous among eukaryotes. The last half-century of cellular and molecular evolutionary research has conclusively shown that mitochondria are descended from an α-proteobacterial endosymbiont that took up residence within a host cell prior to the last eukaryotic common ancestor (LECA) [1]. However, the precise phylogenetic position of the mitochondrial lineage among α-proteobacteria [2,3], as well as the nature of the host lineage that engulfed it [4], remain active areas of debate and investigation.
Mitochondria are best known for their role in ATP synthesis by oxidative phosphorylation. In this pathway, pyruvate from glycolysis is imported into mitochondria where it is oxidatively decarboxylated to acetyl-CoA by pyruvate dehydrogenase (PDH) and fed into the Krebs cycle to produce NADH and FADH2. These reduced cofactors are oxidized by the electron transport chain (ETC) to generate a proton gradient across the inner mitochondrial membrane and ultimately reduce O2 to H2O. The proton motive force drives ATP synthesis by an F1Fo-ATP synthase. However, mitochondria are known to carry out many other metabolic and biosynthetic functions. In addition to possessing genomes that are replicated, transcribed and translated, they function in iron–sulfur (Fe–S) cluster generation (via the iron–sulfur cluster (ISC) system), haem biosynthesis and amino and fatty acid, phospholipid, vitamin and steroid metabolism. Indeed, although the proteomes of mitochondria of model system organisms vary in composition, they typically comprise over 1000 proteins [5,6], which is significantly more than the coding potential of any known mitochondrial genome [7]. Most mitochondrial proteins are therefore nucleus-encoded, and, via targeting signals (either N-terminal or internal), are imported into mitochondria post-translationally via a complex translocation and refolding apparatus. A fraction of the proteins that function in mitochondria were originally encoded by the α-proteobacterial endosymbiont genome (approx. 15%) [8], and most were later transferred to the host nuclear genome via endosymbiotic gene transfer, and subsequently lost from the mitochondrial genome [9]. A substantial fraction (approx. 40%) of mitochondrial proteins are found only in eukaryotes, or have non-α-proteobacterial prokaryotic affinities (approx. 45%) [8].
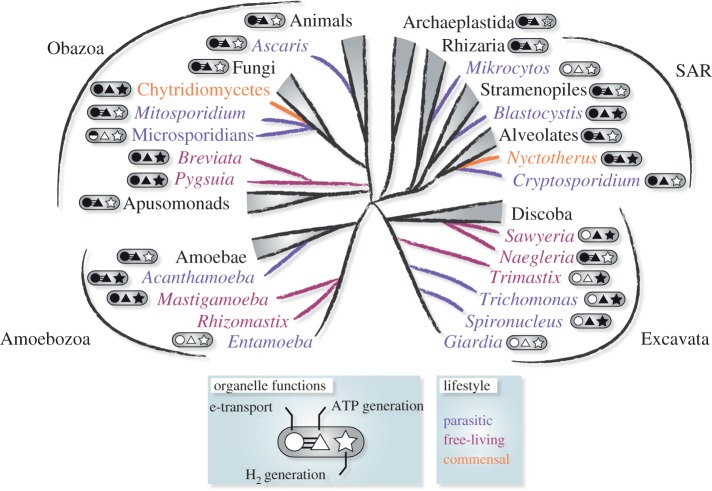
A number of eukaryotic microbes (protists) from low oxygen environments, including the parasitic microsporidia, diplomonads and parabasalids, lack typical cristate mitochondria. These organisms possess small double-membrane-bound organelles of mitochondriate ancestry to which canonical mitochondrial proteins are targeted [10–17]. These organelles were variously named hydrogenosomes, mitosomes and ‘mitochondrion-like organelles'. Here, we refer to them collectively as ‘mitochondrion-related organelles' (MROs), to indicate their evolutionary relationship with mitochondria. The number of eukaryotes discovered to lack canonical mitochondria has grown over the last few decades and most such lineages have been placed on the latest consensus phylogeny of eukaryotes (figure 1). Recently, Müller et al. [18] proposed a novel five-class scheme for mitochondria and MROs based on the different types of energy metabolism they possess. Although this system has practical value, it should be noted that these classes are provisional. As we characterize MROs from various novel protist lineages, their metabolisms increasingly appear as a spectrum of phenotypes that defy strict classification.
Figure 1.
Distribution of MROs across the major supergroups of eukaryotes. Organisms with parasitic (purple), commensal (orange) or free-living (red) lifestyles are indicated. Metabolic functions of each organism's MRO are indicated: shaded shapes represent the presence of electron transporting complexes (circle), ATP synthesis (triangle) and hydrogen production (star). Where at least one proton-pumping complex (CI, CIII, CIV) and ATP synthase (CV) was identified, the circle and triangle are joined by three lines. ‘*’ represents algal lineages where hydrogen production is located in the plastid.
Here, we review recent progress in characterizing this diversity of function in MROs of eukaryotes, focusing especially on anaerobic ATP generation. First, we summarize what is known about the MROs of each of the eukaryote lineages (figures 2 and 3) that underwent modifications of their aerobic mitochondria to function in low oxygen conditions. Then, we provide plausible scenarios by which the anaerobic energy metabolic pathways of these MROs could have originated.
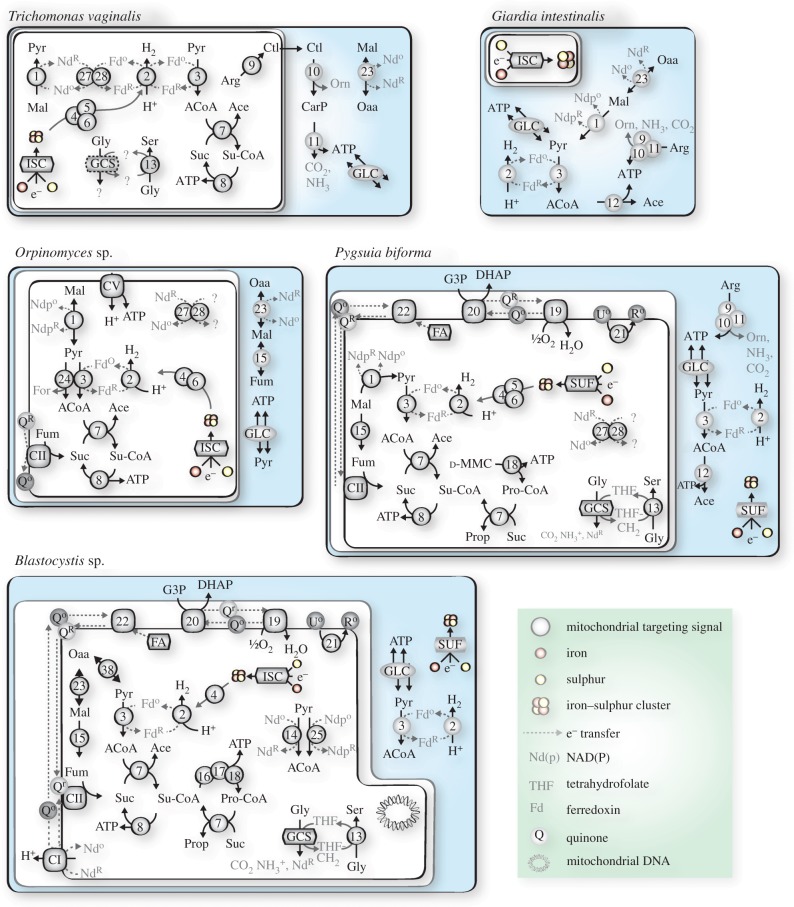
Figure 2.
Major metabolic pathways in the MROs of selected anaerobic protists. Oaa, oxaloacetate; Mal, malate; Pyr, pyruvate; Fum, fumarate; ACoA, acetyl-CoA; Ace, acetate; Suc, succinate; Su-CoA, succinyl-CoA; d-MMC, d-methylmalonyl-CoA; Prop, propionate; Pro-CoA, propionyl-CoA; Arg, arginine; Ctl, citrulline; Orn, ornithine; CarP, carbamoyl phosphate; Gly, glycine; Ser, serine. GLC, glycolysis; GCS, glycine cleavage system; ISC, iron–sulfur cluster system of ISC assembly; SUF, sulfur assimilation (SUF) system of ISC assembly; NIF, nitrogen fixation system of ISC assembly. THF, tetrahydrofolic acid. For the following, O: oxidized, R: reduced: Fd, ferredoxin; Nd, nicotinamide adenine dinucleotide (NAD+); Ndp, nicotinamide adenine dinucleotide phosphate (NADP+); Q, quinone; U, ubiquinone; R, rhodoquinone. 1, malic enzyme; 2, [FeFe]-hydrogenase (HYDA); 3, pyruvate : ferredoxin oxidoreductase (PFO); 4–6, hydrogenase maturases HydE, F G; 7, acetate : succinate CoA transferase (ASCT); 8, succinyl-CoA synthetase (SCS); 9, arginine deiminase (ADI); 10, ornithine transcarbamylase (OTC); 11, carbamate kinase (CK); 12, acetyl-CoA synthetase (ACS); 13, serine hydroxymethyltransferase (SHMT); 14, pyruvate dehydrogenase (PDH); 15, fumarase (FUM); 16, methylmalonyl-CoA epimerase (MCE); 17, methylmalonyl-CoA mutase (MCM); 18, propionyl-CoA carboxylase (PCC); 19, alternative oxidase (AOX); 20, glycerol-3-phosphate dehydrogenase (G3PDH); 21, rhodoquinone biosynthesis protein RQUA; 22, electron transport flavoprotein (ETF); 23, malate dehydrogenase (MDH); 24, pyruvate formate lyase (PFL); 25, pyruvate : NADP+ oxidoreductase (PNO); 27, NuoE/24 kDa subunit of ETC CI; 28, NuoF/51 kDa subunit of ETC CI; CI, complex I; CII, complex II (at least A and B subunits); CV, complex V.
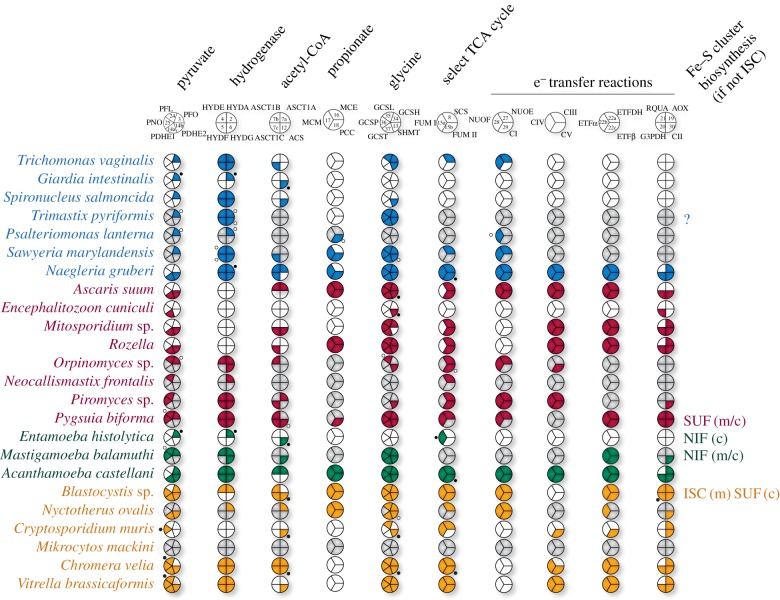
Figure 3.
Coulson plots showing the presence and absence of various proteins and/or protein subunits in anaerobic protists. Blue, Excavata; red, Obazoa; green, Amoebozoa; yellow, SAR clade (Stramenopiles, Alveolata and Rhizaria). Coloured sections indicate proteins found to be present in genome or transcriptome data; white sections, proteins absent from complete genome data; grey sections, proteins absent from transcriptome or incomplete genome data [19]. Black and white circles indicate proteins with a predicted cytosolic location based on Mitoprot predictions or incomplete coding sequences. Unless indicated otherwise, all lineages use the ISC system for the biosynthesis of Fe–S clusters. Enzyme abbreviations are as indicated in figures 2. Accession numbers for each protein can be found in the electronic supplementary material, S1.
2. Excavates
(a). Metamonada
The ‘hydrogenosomes’ of parasitic trichomonads were first described in the 1970s [20,21] and those of the human urogenital parasite Trichomonas vaginalis have become the best-studied MROs to date. They function in several canonical mitochondrial processes including amino acid metabolism, oxygen detoxification and Fe-S cluster biosynthesis, and possess two components of complex I (CI; the 51 and 24 kDa subunits) [22–26]. Unlike typical aerobic mitochondria, the hydrogenosomes of Trichomonas metabolize pyruvate using an ‘extended glycolysis’ pathway, whereby pyruvate is oxidized by pyruvate : ferredoxin oxidoreductase (PFO) to acetyl-CoA, with the concomitant reduction of ferredoxin (figures 2 and 3). Electrons from ferredoxin (or NADH via the two soluble CI subunits) are transferred to protons via an [FeFe]-hydrogenase (HYD), ultimately generating H2. Three maturase proteins (HYDE, HYDF and HYDG) are typically required for correct assembly of the mature HYD protein. The CoA moiety of acetyl-CoA is transferred to succinate via acetate : succinate CoA transferase subtype 1C (ASCT1C). The resulting succinyl-CoA can be used by the Krebs cycle succinyl-CoA synthetase (SCS) to generate ATP by substrate-level phosphorylation. Trichomonas also uses the arginine dihydrolase pathway to synthesize ATP. One of the three enzymes involved in this pathway (arginine deiminase) is localized in the MRO, while the other two enzymes (ornithine transcarbamylase and carbamate kinase) are cytoplasmic [27].
The diplomonad Giardia intestinalis is a parasite of humans that thrives in the low oxygen environment of the gastrointestinal tract [28]. Giardia's MROs do not synthesize ATP and are thus called ‘mitosomes’. Currently, their only major known function is Fe–S cluster biogenesis, via the ISC system [29]. Giardia also has a cytosolic extended glycolysis pathway (figures 2 and 3). Like Trichomonas, they use a PFO/ferredoxin and HYD pathway to oxidatively decarboxylate pyruvate, but in this case the acetyl-CoA is converted to acetate by an acetyl-CoA synthetase (ACS), that generates ATP directly [29–31]. Like Trichomonas, Giardia also synthesizes ATP by the arginine dihydrolase pathway; however, this pathway is entirely cytosolic in this organism [32]. The MROs of Spironucleus salmonicida, a related diplomonad parasite of salmonid fish [33,34], resemble Trichomonas hydrogenosomes in possessing a hydrogen-producing extended glycolysis pathway, Fe–S cluster generation, and possibly glycine metabolism. ATP is produced in these organelles by the Giardia-like ACS system, and not the ASCT/SCS system seen in Trichomonas [33,34].
Putative MROs have been described in many different species of the flagellated metamonad genus Trimastix [35–38]. At least two HYD maturases are predicted to localize to the MROs of Trimastix pyriformis. However, it is still unclear whether other proteins linked to hydrogen production, including HYD, and PFO, are organellar or cytosolic [39]. The predicted proteome of the T. pyriformis MROs (based on the presence of predicted mitochondrial targeting peptides) also includes amino acid metabolism enzymes, and even the Krebs cycle enzyme aconitase. To date, the exact mechanism of ATP generation in this organism, beyond glycolysis, is unknown, as ASCT, SCS and ACS have not been identified in T. pyriformis.
(b). Heterolobosea
In recent years, many transcriptomic or genomic sequencing initiatives have allowed the prediction of the MRO proteomes of heteroloboseid excavates, such as Psalteriomonas lanterna, Sawyeria marylandensis and Naegleria gruberi. The Psalteriomonas expressed sequence tag (EST) survey reported homologues of HSP70, HYD, PFO and the 51 kDa subunit of CI [40]. The larger EST survey of Sawyeria revealed homologues of many of the proteins found in T. vaginalis hydrogenosomes, including HYD proteins, PFO, SCS, ASCT, chaperones, CI subunits and Fe–S cluster biosynthesis proteins [41]. Interestingly, Sawyeria appears to encode enzymes for a variety of canonical mitochondrial pathways linked to amino acid metabolism that are not present in Trichomonas, such as a full glycine cleavage system, proline, serine and ornithine metabolism, and branched chain amino acid degradation, although lack of complete sequences hinders predictions of their localization [41].
Naegleria gruberi is a free-living aerobic amoeboflagellate that can tolerate low oxygen conditions and possesses classical mitochondria [42]. HYD, HYD maturases and ASCT1B and 1C were identified in the genome, and were predicted to function in mitochondria (possibly in hypoxic conditions) based on the presence of predicted N-terminal targeting sequences [42], although a recent report argues that hydrogen production occurs exclusively in the cytoplasm [43].
3. Obazoa (opisthokonts, breviates and apusomonads)
(a). Animals
Some platyhelminths and nematodes transiently experience hypoxic conditions [18,44–47]. Through regulated expression of various proteins, the metabolism of the anaerobic mitochondria of these organisms is reconfigured to generate ATP by substrate-level phosphorylation, F1Fo-ATP synthase and a modified respiratory chain [18,48–50]. As in aerobic mitochondria, CI oxidizes NADH to pump protons into the intermembrane space, fuelling ATP synthesis via complex V (CV). However, electrons are transferred from CI to a specialized quinone, rhodoquinone (RQ), that has a lower redox potential than the usual mitochondrial electron carrier ubiquinone. A process called ‘malate dismutation’ helps to replenish the oxidized RQ pool using fumarase and complex II (CII), which preferentially catalyse the reverse reactions to their aerobic mitochondrial counterparts (i.e. malate dehydration and rhodoquinol-dependent fumarate reduction) [51,52]. These organisms also generate ATP via propionyl-CoA-mediated substrate-level phosphorylation, generating propionate.
(b). Microsporidians
Microsporidia are deep-branching fungi [53–55] that are obligate intracellular parasites of animals [56]. The MROs (mitosomes) of these organisms are not known to produce ATP, but participate in ISC-mediated Fe–S cluster biosynthesis [57–61]. Microsporidia typically have reduced nuclear genomes, often lacking amino acid and nucleotide metabolism; some even lack core sugar metabolism [57,59,62,63]. For example, Encephalitozoon cuniculi has a significantly reduced ATP generation machinery and must instead rely on ATP import from the host cell into the parasite cytoplasm, and eventually into the mitosomes, using bacteria-derived ADP/ATP translocators [64,65]. The mitosomes of Antonospora locustae and Trachypleistophora hominis encode an MRO-targeted alternative oxidase (AOX) and glycerol-3-phosphate dehydrogenase (G3PDH) indicating the likely presence of an electron-carrying quinone [64,66,67]; these may contribute to cytosolic NAD+ regeneration, rather than generating energy in the mitosomes. Two newly discovered deep-branching relatives of microsporidians, Rozella and Mitosporidium, have recently been shown to have more complex MROs compared with previously studied microsporidia (e.g. they each have mitochondrial DNA, a Krebs cycle, complexes II–IV of the ETC and an ATP synthase [55,68]).
(c). Chytrid fungi
Anaerobic fungi such as Neocallimastix, Piromyces and Orpinomyces thrive in low oxygen animal rumen environments, and possess MROs that ultrastructurally resemble those found in Trichomonas [69–73]. Previous reports have detected activity of pyruvate formate lyase (PFL)—an oxygen-sensitive enzyme that catalyses the non-oxidative conversion of pyruvate to acetyl-CoA—in the MROs of Neocallimastix and Piromyces, and HYD and PFO activity in Neocallimastix species [69,74–76]. The Orpinomyces MRO is predicted to function in hydrogen production, pyruvate metabolism using PFL, and ATP generation using the ASCT/SCS system [77]. However, it also appears to have an F1Fo-ATP synthase, suggesting Orpinomyces may rely on an organellar proton gradient for ATP synthesis. We have identified previously unreported MRO-targeted homologues of PFO, HYDE, HYDF, HYDG, as well as enzymes of amino acid metabolism and other pathways summarized in figure 2 in the Orpinomyces genome.
(d). Breviates
The breviates are a recently described lineage of microaerophilic/anaerobic amoeboid flagellated protists that branch sister to Opisthokonta (animals and fungi) and Apusomonada [78]. To date, a detailed prediction of MRO metabolism has only been performed for Pygsuia biforma [79]. The Pygsuia genome encodes MRO-targeted proteins involved in protein import and folding, amino acid, fatty acid and lipid metabolism. Pygsuia has an extended glycolysis pathway that localizes to the MRO and the cytoplasm; however, the HYD maturases are only predicted to function in the MRO (figures 2 and 3). Within the organelle, Pygsuia uses ASCT1C and SCS to generate ATP by substrate-level phosphorylation like T. vaginalis, although it may also generate ATP in the cytosol using ACS (a putative cytosolic ACS homologue is present in the Pygsuia transcriptome [78]). Interestingly, it also encodes an MRO-targeted ASCT1B and some components of the propionyl-CoA carboxylase pathway, suggesting it might use this pathway to synthesize ATP. Unlike many other MROs, Pygsuia encodes a large selection of MRO-targeted quinone-using enzymes including CII, electron-transferring flavoprotein (ETF) dehydrogenase, G3PDH and AOX but no other components of the ETC, suggesting that Pygsuia has an ETC that does not appear to function in proton transport or energy generation. The ISC system for the biosynthesis of Fe–S clusters of Pygsuia has been replaced by a methanoarchaeal SUF biosynthesis system that was acquired by lateral gene transfer (LGT) [79].
4. Amoebozoa
(a). Archamoebae
The mitosomes of the human parasite Entamoeba histolytica are the best-studied MROs in the supergroup Amoebozoa [80,81]. As in Giardia, the extended glycolysis and ACS-catalysed ATP synthesis pathway of Entamoeba is localized exclusively in the cytoplasm. However, the ISC system for the biosynthesis of Fe–S clusters has been lost in this organism, and replaced with a homologous system called the nitrogen fixation (NIF) system that was acquired by LGT from ε-proteobacteria [82–84]. While initial reports suggested that the NIF system is dually localized to the cytoplasm and mitosomes [84], more recent proteomic studies have been unable to detect NIF system components in the organelle, calling into question the widely held notion that the main reason for maintaining MROs is the biosynthesis of Fe–S clusters [80]. Currently, the only known function of the MROs of Entamoeba is ATP import via mitochondrial carrier proteins [85] and sulfate activation for the generation of sulfated compounds [80,86].
Mastigamoeba balamuthi is a free-living relative of Entamoeba that inhabits low oxygen freshwater environments [87]. Mastigamoeba balamuthi was the first non-parasitic organism in which MROs were characterized [88]; these participate in pyruvate oxidation, hydrogen evolution, sulfate activation and amino acid metabolism [89]. Like the Spironucleus organelles, the MROs of Mastigamoeba generate ATP using extended glycolysis and an ACS. Mastigamoeba encodes some proteins linked to the respiratory chain and electron transport (CII and ETF) but none of the canonical proton-pumping components; therefore, it is unclear if the M. balamuthi MROs have a proton gradient across their inner membranes. Mastigamoeba uses the ε-proteobacterial-derived NIF system for the biosynthesis of Fe–S clusters; two copies are present, one of which is localized in the MROs and the other in the cytoplasm [90].
Recent studies have identified other anaerobic/microaerophilic protists that are members of the archamoebae, including Iodamoeba and Rhizomastix, that lack typical cristate mitochondria [91,92], but have MRO-like organelles in electron micrographs.
(b). Discosea
Acanthamoeba castellanii inhabits a range of environments, including soil bacterial biofilms or human contact lenses [93]; it shows a preference for low oxygen conditions [94]. The ultrastructure and general metabolism of Acanthamoeba mitochondria resemble those of classical aerobic organelles [95]. However, like Naegleria, Acanthamoeba possesses enzymes involved in the extended glycolysis pathway and ATP generation via SCS and ASCT1B. All of these proteins have predicted N-terminal mitochondrial targeting sequences. PFO, a HYD maturase and ASCT were shown experimentally to localize to the mitochondria of Acanthamoeba [96].
5. SAR clade
(a). Stramenopiles
The Blastocystis genus comprises strictly anaerobic unicellular stramenopile parasites that inhabit animal intestines [97]. Genomic and transcriptomic data available for different subtypes of Blastocystis show that these organisms express multiple acetyl-CoA generating proteins including PDH, PFO and pyruvate : NADP+ oxidoreductase (PNO; a specialized PFO with a C-terminal P450 reductase domain) [98–100]. However, to date, only PNO activity has been detected biochemically within the MROs of Blastocystis sp. subtype 7 [98]. The acetyl-CoA generated by this reaction is predicted to be used by ASCT1B and 1C to generate propionyl-CoA or succinyl-CoA, which is a substrate for SCS or the propionyl-CoA pathway to generate ATP, much like the anaerobic mitochondria from facultative anaerobes (Acanthamoeba or Ascaris). While hydrogen production has not actually been detected in the MROs of Blastocystis subtypes 1 and 7, genomic and transcriptomic evidence suggests that there are multiple MRO-targeted HYDs that are fused to flavodoxin domains [99]. The unique domain composition of the Blastocystis HYDs might explain the inability to detect activity biochemically as the enzymatic assays used are designed for canonical HYD proteins [98]; it is even possible these enzymes do not produce H2 but instead shuttle electrons to some other electron carrier. Analysis of the genome sequence of Blastocystis subtype 7 indicates that its MRO participates in a variety of pathways including amino acid metabolism, Fe–S cluster biogenesis, reactive oxygen species (ROS) defence and fatty acid biosynthesis [100] (figures 2 and 3).
(b). Alveolates
The MRO of the ciliate Nyctotherus uses PDH to generate acetyl-CoA and malate dismutation to generate succinate by oxidizing RQH2 [101]. The organelles use ASCT (subtype 1A) and SCS to generate ATP from acetyl-CoA [101,102]. These MROs use a specialized HYD that uniquely possesses C-terminal modules that are most closely related to domains of bacterial [NiFe] HYDs, and are distant homologues of the 51 and 24 kDa subunits of CI [103]. This enzyme presumably oxidizes NADH by reducing protons to generate H2, thus replenishing the NAD+ pool under anaerobic conditions. Interestingly, rumen ciliates including Nyctotherus are often observed in symbiotic relationships with methanogenic endosymbiotic archaea that specifically associate with the MROs, consuming H2 produced by the organelle [104,105]. Additional functions of these MROs include amino acid metabolism and ROS defence. H2-producing MROs of anaerobic ciliates appear to have evolved multiple independent times from canonical mitochondria in ciliate evolution [106], although most of these organelles have not been investigated in detail.
The apicomplexan coccidians Cryptosporidium parvum and Cryptosporidium muris are intracellular parasites of different animals including (but not limited to) humans and rodents. While the MROs of both species are predicted to synthesize ATP by substrate-level phosphorylation, only the MROs of C. muris are predicted to synthesize ATP via oxidative phosphorylation [107–111]. Both C. muris and C. parvum have relict ETCs composed of AOX, malate : quinone oxidoreductase and an alternative NADH dehydrogenase (NDH2). Cryptosporidium muris encodes ATP synthase, SDH and a Krebs cycle, whereas C. parvum encodes a partial F1Fo-ATP synthase. Reports suggest that the C. parvum organelle imports ATP (generated in the cytoplasm by PNO and ACS) using an ADP/ATP carrier [111,112].
(c). Rhizaria
The rhizarian Mikrocytos is an intracellular parasite of oysters [113,114]. In a recent transcriptomic survey, the authors were only able to identify ISC system components and not genes that encode typical mitochondrial proteins (e.g. protein import and folding) or proteins of an extended glycolytic pathway. This suggests the mitosomal metabolism of Mikrocytos is highly reduced and streamlined for Fe–S cluster biogenesis. There are no obvious non-glycolytic candidates for ATP-generating enzymes in the transcriptome of Mikrocytos, suggesting it relies exclusively on fermentative metabolism or another ATP generation mechanism and/or the parasite imports ATP from host cytoplasms like other intracellular parasites [65,115].
6. Anaerobic metabolism: ancestral or acquired?
The evolutionary origin(s) of the aforementioned pathways of anaerobic energy generation have been debated in recent years [116–118]. Two general kinds of evolutionary accounts have been articulated. The first suggests that the ancestor of all extant eukaryotes was facultatively anaerobic and that the mitochondrial endosymbiont was ancestrally capable of respiring both aerobically and anaerobically, depending on the availability of oxygen [116]. The second suggests that various genes related to anaerobic metabolism were acquired multiple times in different eukaryote lineages via LGT [34,41,89,90,99,118–121].
(a). Hydrogen hypothesis
In 1998, Martin & Müller [116] proposed the ‘Hydrogen hypothesis’ in which they argued that the ancestor of mitochondria was a H2-producing, facultatively anaerobic α-proteobacterium that formed a syntrophic relationship with a H2-dependent methanogenic archaeon. In an anaerobic environment, the α-proteobacterium produced ATP by the anaerobic extended glycolysis pathway discussed above, producing H2, CO2 and acetate as waste products that were consumed by the methanogen. Over time, the host archaeon maximized surface area contact with the symbiont (without phagocytosis) to acquire these waste products. At that point, the host–symbiont system could exist in anaerobic and aerobic environments. This proto-eukaryote had an archaeal cytoplasm and a H2-producing ‘organelle’ also capable of oxygen-dependent respiration. Later, after the major lineages of extant eukaryotes diverged from the LECA, aerobic and anaerobic metabolisms were differentially lost in anaerobic and aerobic lineages, respectively, generating the diversity of energy metabolism and the MROs that we see today.
Although this hypothesis has received much attention, several of its predictions are not straightforwardly compatible with current data. Most importantly, there is no direct evidence supporting an α-proteobacterial symbiotic origin of genes encoding anaerobic enzymes of energy metabolism within eukaryotes [34,41,89,90,99,118,119]. Although all recent phylogenies of the pyruvate metabolizing enzymes associated with anaerobes (PFO, PNO and PFL) show eukaryotes as monophyletic, the closest related prokaryotic homologues to eukaryotes are never α-proteobacterial; instead, they are sequences from δ-, ɛ-proteobacteria or firmicutes, although the relationships among bacterial groups in these trees are often mixed up [79,89,96,119]. Phylogenies of HYD resolve at least two phylogenetically distinct eukaryotic clades affiliated with sequences from separate bacterial groups. To date, α-proteobacteria homologues do not branch sister to either of these eukaryotic groups in these analyses. Furthermore, inter-eukaryotic relationships observed in the phylogeny of HYD are incongruent with known organismal relationships and some of these inconsistencies are clearly the result of eukaryote-to-eukaryote LGT [79,96,118]. Finally, it is important to note that the vast majority of modern-day α-proteobacterial genomes do not encode these enzymes for anaerobic metabolism, and those that do appear to have recently acquired them from other bacterial lineages by LGT [79,96,118].
To reconcile these observations with an endosymbiotic mitochondrial origin of these enzymes, it has been argued that their phylogenetic affinities have been obscured by rampant LGTs among prokaryotes since the endosymbiosis; the genomes of original endosymbionts that gave rise to the mitochondrion were therefore very different from the modern-day α-proteobacteria [18]. This is, of course, possible. However, many typical endosymbiont-derived genes associated with aerobic metabolism that are encoded either on mitochondrial genomes (e.g. cytochrome oxidase 1) or in the nucleus (e.g. PDH) have not had their origins obscured by LGT in this way; α-proteobacterial homologues are, phylogenetically, their closest relatives [122]. It would seem a strange coincidence, then, that all of the eukaryotic enzymes that function exclusively in anaerobic metabolism discussed above lack the characteristic α-proteobacterial affinity of mitochondrial proteins. Even if LGT has completely obscured the phylogenetic origins of these enzymes in eukaryotes, given that prokaryote-to-eukaryote LGT is now a well-established phenomenon [123–126], there is no strong reason to suspect they originated from the mitochondrial symbiont genome rather than some other bacterial source.
(b). Laterally acquired anaerobic metabolism
Here, we propose an LGT model whereby genes encoding enzymes that exclusively function in anaerobic ATP metabolism in eukaryotes were initially acquired from anaerobic bacteria (possibly food or transient endosymbionts) by one or more eukaryotic lineages, and subsequently transferred between eukaryotes after the diversification of extant eukaryotes. The selective advantage was the ability to continue to produce acetyl-CoA and ultimately ATP from pyruvate (and/or malate) under hypoxic conditions commonly encountered by free-living and anaerobic protists. The model rests on the general assumption that, in adapting to new environments, protists can acquire and express genes from prokaryotic or eukaryotic donors that allow them to better thrive. LGT in eukaryotes has been extensively documented for more than a decade and often these gene acquisitions are adaptive [127–129]. Furthermore, the transfer patterns observed are consistent with known ecological co-occurrences of the protist recipients and the bacterial donors [130–132], as observed in cases of prokaryotic LGT [133]. Various specific mechanisms by which LGT can occur in eukaryotes have also been proposed and discussed [134–136], but will not be elaborated in detail here.
Support for an LGT-based model comes primarily from the patchy distribution of the enzymes across the eukaryotic tree, heterogeneous bacterial affinities of the eukaryotic homologues and the unusual inter-eukaryote relationships sometimes observed in phylogenies of the HYD, ASCT, ACS, PFO, HYD and PFL enzymes [34,79,96,118,119,137]. Curiously, eukaryotes are recovered as monophyletic in the most recent phylogenies of PFO, PFL and the HYD maturases. This can be explained in two ways: (i) these enzymes were acquired prior to LECA, and then vertically inherited [118,120] and lost many times in distinct eukaryote lineages, or (ii) they were acquired by one eukaryote lineage (post-LECA) and then passed among eukaryotes via LGT. The latter is plausible if genes acquired from other eukaryotes were more frequently expressed and retained than those acquired from prokaryotes because their cis-acting signals and regulatory elements (e.g. promoters, polyadenylation signals, etc.) were more ‘compatible’ with the recipient genome. In the case that multiple proteins are required to perform a function, LGT-based models are more plausible if these genes were transferred in a single event. This can be accomplished by transfers of: (i) operons in the case of bacterial donors, and (ii) adjacent or fused genes for eukaryote donors (e.g. [79,119]).
(c). A new scenario
The previous sections aimed to highlight both the remarkable diversity and the extensive similarities in the metabolisms of MROs across the tree of eukaryotes. Although MROs have evolved independently in dozens of lineages, they show functional similarities because several different ‘anaerobic’ enzymes of energy metabolism co-occur in MROs of different lineages. Using modules made up of laterally acquired and ancestral proteins, the MROs of anaerobic protists have convergently acquired similar mechanisms to metabolize pyruvate, generate hydrogen and synthesize ATP. Below, we elaborate rationales for the ordering of events that occur during these evolutionary transitions.
(i). Reduction of the electron transport chain in response to hypoxia
During transient hypoxia, the subunits of CIII and CIV of the ETC are typically downregulated in the mitochondria of many organisms [48,138] including mammalian cells [139–141]. Therefore, in adapting to hypoxia as a permanent condition, it seems likely that an organism would no longer express CIII and CIV subunits, and, over time, the lack of purifying selection would ultimately lead to their loss from the nuclear and mitochondrial genomes. In this scenario, the first four enzymes of the Krebs cycle (citrate synthase to SCS) would still function to convert citrate to succinate and NADH. However, the remaining enzymes of the Krebs cycle were probably functioning in reverse in the malate dismutation pathway, transforming malate to fumarate, as they do in anaerobic mitochondria and some cancer cells [18,48]. In this situation, CI continues to oxidize NADH and pump protons fuelling ATP synthesis via ATP synthase. However, the lack of CIII and CIV results in a build-up of ubiquinol generated by CI. This build-up of ubiquinol and fumarate would have favoured the functioning of CII in reverse as a fumarate reductase, to regenerate ubiquinone. This scenario is plausible, as some CIIs of aerobes perform fumarate reduction with sufficient concentrations of substrate [142]. Metabolically, this organelle would resemble the mitochondria of some animals (e.g. Ascaris) when functioning in hypoxic conditions [18]. In some lineages of eukaryotes, the ability to synthesize and use RQ by the acquisition of an enzyme like RQUA [143] or an analogous protein by LGT would have greatly increased the efficiency of the fumarate reductase activity of CII.
(ii). Hydrogenase and complex I
Curiously, organisms that encode PFO always possess a HYD homologue. However, the converse is not true: the genomes of some organisms such as Naegleria, Neocallimastix, Piromyces and Nyctotherus (figure 3) encode only HYD, suggesting that this enzyme need not rely on reduced ferredoxin produced by PFO activity. Studies have shown that the Thermotoga HYD forms a trimeric complex with the NuoE/24 kDa and NuoF/51 kDa subunits of CI to simultaneously oxidize both ferredoxin and NADH using a unique electron bifurcation mechanism [144]. This is consistent with studies of T. vaginalis hydrogenosomes that showed that homologues of these CI subunits (responsible for oxidation of NADH) can reduce ferredoxin that is then oxidized by HYD [23] or possibly even shuttle electrons directly to HYD [24]. Interestingly, other anaerobic protists with MRO-targeted HYDs have retained the same two subunits of CI (figure 3), and in Nyctotherus bacterial homologues of these proteins are fused to HYD [103]. This suggests that coupling of NADH oxidation with H2 production (ferredoxin) could also occur in these organisms [18].
In this scenario, once HYD was acquired by a protist, the NuoE and F subunits could have had dual roles. Under aerobic conditions, they would be predominantly bound to the membrane-embedded subunits functioning as part of CI. However, under low oxygen conditions when the ETC is functioning less efficiently, NuoE and F could instead mediate electron transfer from NADH to HYD, ultimately generating hydrogen gas and regenerating NAD+ required by multiple mitochondrial pathways. In some lineages, most CI subunits appear to have been completely lost, along with ATP synthase, and only the NuoE and F subunits were retained to carry out this putative HYD-related function. It is unclear why a full CI persists in organisms that lack ATP synthase, such as Blastocystis and Nyctotherus; it may be related to maintaining a proton gradient required for protein import [145], small molecule transport (e.g. SLC25 carrier proteins [146]) and NADH/NADPH exchange (for review, see [147]).
(iii). Changes to pyruvate metabolism and ATP generation
When aerobic organisms experience hypoxia, the ETC becomes inefficient causing NADH levels to rise, inhibiting PDH via negative allosteric regulation and phosphorylation by PDH kinase [148,149]. Under these conditions, the energy output of mitochondria decreases [150] and limits the efficiency of pathways that rely on acetyl-CoA as a substrate (e.g. long-chain fatty acid biosynthesis [151]). For organisms adapting to hypoxia, acquiring an NADH-insensitive pyruvate-oxidizing enzyme such as PFO would be selectively advantageous. In this scenario, electrons from PFO-mediated pyruvate oxidation could be ferried directly to the HYD via ferredoxin. The MRO would then possess both PDH and a PFO/HYD system as found in Blastocystis sp. [99] and Acanthamoeba [96]. Once PFO and HYD were acquired, as long as the organism was living under predominantly hypoxic conditions (as PFO is an oxygen-sensitive enzyme [152]), PDH would become unnecessary and eventually be lost.
At this point, the hypothetical MRO had a partially functioning ETC (CI and CII), PFO, HYD and a partial Krebs cycle and, possibly, mtDNA; it synthesized ATP via CV and ultimately excreted succinate (propionate and/or acetate) as the end product of metabolism. It is possible that, at this stage in the evolutionary transition of the MRO, succinate could be directly converted to succinyl-CoA by an ASCT. The timing of the acquisition of one of the subtypes of these enzymes is unclear as ASCT homologues are present in MROs of a number of anaerobically adapted organisms including those with or without the PFO and/or HYD enzymes [18]. In any case, ASCT allows the synthesis of succinyl-CoA as substrate for SCS to generate ATP independent of the Krebs cycle.
A reduction in NADH production (because of loss of PDH and/or reduced output from the partial Krebs cycle) and lowered proton motive force (because of loss of CIII and CIV) probably led to a reduced efficiency, and eventual loss, of the F1Fo-ATP synthase, as observed in Blastocystis [99] and Nyctotherus [102].
(iv). Loss of complex I, mtDNA and malate dismutation
The loss of the majority of CI subunits and the Krebs cycle would yield an MRO resembling that of P. biforma (figure 2)—an organelle lacking a mitochondrial genome, and retaining only CII, two CI subunits, PFO, HYD, ASCT and SCS. Eventually, the CII (and thus malate dismutation) could have been lost if succinate (the substrate for ASCT) was imported (or generated by an alternative mechanism), as presumably occurs in T. vaginalis hydrogenosomes. Instead of acquiring ASCT, some lineages acquired ACS and generated ATP directly from acetyl-CoA in their MROs (e.g. M. balamuthi [89] or S. salmonicida [34]). Interestingly, Mastigamoeba has a mysterious combination of enzymes; its genome encodes only the four subunits of CII and not ASCT, SCS or a MRO-targeted fumarase.
A final step in organelle reduction has occurred in those lineages that have MROs that do not function in ATP synthesis (i.e. mitosomes). In such lineages, ATP is either synthesized in the cytoplasm (by PFO, HYD and ACS in Giardia and Entamoeba) (figure 2) or imported from host cells (in Microsporidia) [64,65].
7. Final remarks
Above, we have discussed a set of evolutionary transitions that could lead to the metabolic diversity seen in anaerobic protists today. It is likely that the true evolutionary pathways that led to MRO diversity in extant anaerobes were more complex than the foregoing scenarios might suggest. We advance these hypotheses only to stimulate further investigation into, and discussion of, the evolutionary events and selective forces leading to adaptation of mitochondria to hypoxia.
Although we have focused predominantly on ATP and carbohydrate metabolism, we observe the general pattern that MROs of free-living organisms tend to maintain more anabolic pathways than those of parasites, which presumably rely on the host to fulfil these needs. For instance, the organelles of Pygsuia have retained a variety of lipid and amino acid metabolic pathways not found in those of parasites like Trichomonas, Giardia or Entamoeba. Furthermore, the MROs of free-living organisms (e.g. Pygsuia, chytrids, Mastigamoeba) often express several different enzymes with redundant functions; for example, many encode several enzymes that convert pyruvate to acetyl-CoA (PFL, PFO and/or PNO). This contrasts with the more streamlined metabolism found in parasites (e.g. Trichomonas, Spironucleus, Entamoeba and Giardia, in which PFO is the only acetyl-CoA-producing enzyme). This observation is harder to reconcile with respect to reliance on a host. It is possible that since free-living organisms tend to occupy dynamic environments (compared with parasites), several enzymes with different cofactor dependencies could provide metabolic versatility. More information on the regulation of these various enzymes under different conditions is needed to determine how they might allow these organisms to adapt to changing environments.
Supplementary Material
Acknowledgements
The authors thank Dr Alastair Simpson, Dr Laura Eme, and Dr Bernard Lemire and Dr Martin Kolisko for helpful discussion.
Authors' contributions
C.W.S. and M.M.L. collected data summarized in the electronic supplementary material. C.W.S., M.M.L. and A.J.R. wrote the manuscript. C.W.S. designed and constructed the figures.
Competing interests
We declare we have no competing interests.
Funding
Work by C.W.S. was funded in part by Killam Trusts, National Science and Engineering Research Council of Canada Canadian Graduate Scholarship. M.M.L. was funded in part by the Nova Scotia Health Research Foundation and the National Research Fund of Luxembourg. This work was supported by a Regional Partnerships Program grant (FRN#62809) from the Canadian Institutes of Health Research (CIHR) and the NSHRF awarded to A.J.R.
References
- 1.Andersson SG, et al. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–140. ( 10.1038/24094) [DOI] [PubMed] [Google Scholar]
- 2.Thrash JC, et al. 2011. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci. Rep. 1, 13 ( 10.1038/srep00013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodríguez-Ezpeleta N, Embley TM. 2012. The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS ONE 7, e30520 ( 10.1371/journal.pone.0030520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poole AM, Gribaldo S. 2014. Eukaryotic origins: how and when was the mitochondrion acquired? Cold Spring Harb. Perspect. Biol. 6, a015990 ( 10.1101/cshperspect.a015990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvo SE, Mootha VK. 2010. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 11, 25–44. ( 10.1146/annurev-genom-082509-141720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith AC, Blackshaw JA, Robinson AJ. 2012. MitoMiner: a data warehouse for mitochondrial proteomics data. Nucleic Acids Res. 40, D1160–D1167. ( 10.1093/nar/gkr1101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burger G, Gray MW, Forget L, Lang BF. 2013. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol. Evol. 5, 418–438. ( 10.1093/gbe/evt008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szklarczyk R, Huynen MA. 2010. Mosaic origin of the mitochondrial proteome. Proteomics 10, 4012–4024. ( 10.1002/pmic.201000329) [DOI] [PubMed] [Google Scholar]
- 9.Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. ( 10.1038/nrg1271) [DOI] [PubMed] [Google Scholar]
- 10.Clark CG, Roger AJ. 1995. Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proc. Natl Acad. Sci. USA 92, 6518–6521. ( 10.1073/pnas.92.14.6518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bozner P. 1996. The heat shock response and major heat shock proteins of Tritrichomonas mobilensis and Tritrichomonas augusta. J. Parasitol. 82, 103–111. ( 10.2307/3284124) [DOI] [PubMed] [Google Scholar]
- 12.Germot A, Philippe H, Le Guyader H. 1996. Presence of a mitochondrial-type 70-kDa heat shock protein in Trichomonas vaginalis suggests a very early mitochondrial endosymbiosis in eukaryotes. Proc. Natl Acad. Sci. USA 93, 14 614–14 617. ( 10.1073/pnas.93.25.14614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horner DS, Hirt RP, Kilvington S, Lloyd D, Embley TM. 1996. Molecular data suggest an early acquisition of the mitochondrion endosymbiont. Proc. R. Soc. Lond. B 263, 1053–1059. ( 10.1098/rspb.1996.0155) [DOI] [PubMed] [Google Scholar]
- 14.Roger AJ, Clark CG, Doolittle WF. 1996. A possible mitochondrial gene in the early-branching amitochondriate protist Trichomonas vaginalis. Proc. Natl Acad. Sci. USA 93, 14 618–14 622. ( 10.1073/pnas.93.25.14618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AGB, Roger AJ. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic ‘supergroups’. Proc. Natl Acad. Sci. USA 106, 3859–3864. ( 10.1073/pnas.0807880106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roger AJ, Svard SG, Tovar J, Clark CG, Smith MW, Gillin FD, Sogin ML. 1998. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc. Natl Acad. Sci. USA 95, 229–234. ( 10.1073/pnas.95.1.229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horner DS, Embley TM. 2001. Chaperonin 60 phylogeny provides further evidence for secondary loss of mitochondria among putative early-branching eukaryotes. Mol. Biol. Evol. 18, 1970–1975. ( 10.1093/oxfordjournals.molbev.a003737) [DOI] [PubMed] [Google Scholar]
- 18.Müller M, et al. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495. ( 10.1128/MMBR.05024-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Field HI, Coulson RMR, Field MC. 2013. An automated graphics tool for comparative genomics: the Coulson plot generator. BMC Bioinform. 14, 141 ( 10.1186/1471-2105-14-141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindmark DG, Muller M. 1973. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J. Biol. Chem. 248, 7724–7728. [PubMed] [Google Scholar]
- 21.Lindmark DG, Müller M, Shio H. 1975. Hydrogenosomes in Trichomonas vaginalis. J. Parasitol. 61, 552–554. ( 10.2307/3279345) [DOI] [Google Scholar]
- 22.Tachezy J, Sanchez LB, Muller M. 2001. Mitochondrial type iron-sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol. Biol. Evol. 18, 1919–1928. ( 10.1093/oxfordjournals.molbev.a003732) [DOI] [PubMed] [Google Scholar]
- 23.Hrdy I, Hirt RP, Dolezal P, Bardonová L, Foster PG, Tachezy J, Embley TM. 2004. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432, 618–622. ( 10.1038/nature03149) [DOI] [PubMed] [Google Scholar]
- 24.Dyall SD, Yan W, Delgadillo-correa MG, Lunceford A, Loo JA, Clarke CF, Johnson PJ. 2004. Non-mitochondrial complex I proteins in a hydrogenosomal oxidoreductase complex. Nature 28, 1103–1107. ( 10.1038/nature02990) [DOI] [PubMed] [Google Scholar]
- 25.Schneider RE, Brown MT, Shiflett AM, Dyall SD, Hayes RD, Xie Y, Loo JA, Johnson PJ. 2011. The Trichomonas vaginalis hydrogenosome proteome is highly reduced relative to mitochondria, yet complex compared with mitosomes. Int. J. Parasitol. 41, 1421–1434. ( 10.1016/j.ijpara.2011.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlton JM, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315, 207–212. ( 10.1126/science.1132894) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morada M, Smid O, Hampl V, Sutak R. 2011. Hydrogenosome-localization of arginine deiminase in Trichomonas vaginalis. Mol. Biochem. Parasitol. 176, 51–54. ( 10.1016/j.molbiopara.2010.10.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ankarklev J, Jerlström-Hultqvist J, Ringqvist E, Troell K, Svärd SG. 2010. Behind the smile: cell biology and disease mechanisms of Giardia species. Nat. Rev. Microbiol. 8, 413–422. ( 10.1038/nrmicro2317) [DOI] [PubMed] [Google Scholar]
- 29.Tovar J, León-Avila G, Sánchez LB, Sutak R, Tachezy J, van der Giezen M, Hernández M, Müller M, Lucocq JM. 2003. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature 426, 172–176. ( 10.1038/nature01945) [DOI] [PubMed] [Google Scholar]
- 30.Sanchez LB, Galperin MY, Muller M. 2000. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming). J. Biol. Chem. 275, 5794–5803. ( 10.1074/jbc.275.8.5794) [DOI] [PubMed] [Google Scholar]
- 31.Emelyanov VV, Goldberg AV. 2011. Fermentation enzymes of Giardia intestinalis, pyruvate : ferredoxin oxidoreductase and hydrogenase, do not localize to its mitosomes. Microbiology 157, 1602–1611. ( 10.1099/mic.0.044784-0) [DOI] [PubMed] [Google Scholar]
- 32.Schofield PJ, Costello M, Edwards MR, O'sullivan WJ. 1990. The arginine dihydrolase pathway is present in Giardia intestinalis. Int. J. Parasitol. 20, 697–699. ( 10.1016/0020-7519(90)90133-8) [DOI] [PubMed] [Google Scholar]
- 33.Xu F, Jerlström-Hultqvist J, Einarsson E, Astvaldsson A, Svärd SG, Andersson JO. 2014. The genome of Spironucleus salmonicida highlights a fish pathogen adapted to fluctuating environments. PLoS Genet. 10, e1004053 ( 10.1371/journal.pgen.1004053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jerlström-Hultqvist J, et al. 2013. Hydrogenosomes in the diplomonad Spironucleus salmonicida. Nat. Commun. 4, 2493 ( 10.1038/ncomms3493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brugerolle G, Patterson D. 1997. Ultrastructure of Trimastix convexa hollande, an amitochondriate anaerobic flagellate with a previously undescribed organization. Eur. J. Protistol. 33, 121–130. ( 10.1016/S0932-4739(97)80029-6) [DOI] [Google Scholar]
- 36.O'Kelly CJ, Farmer MA, Nerad TA. 1999. Ultrastructure of Trimastix pyriformis (Klebs) Bernard et al.: similarities of Trimastix species with retortamonad and jakobid flagellates. Protist 150, 149–162. ( 10.1016/S1434-4610(99)70018-0) [DOI] [PubMed] [Google Scholar]
- 37.Simpson AGB, Bernard C, Patterson DJ. 2000. The ultrastructure of Trimastix marina Kent 1880 (Eukaryota), an excavate flagellate. Eur. J. Protistol. 36, 229–251. ( 10.1016/S0932-4739(00)80001-2) [DOI] [Google Scholar]
- 38.Stechmann A, Baumgartner M, Silberman JD, Roger AJ. 2006. The glycolytic pathway of Trimastix pyriformis is an evolutionary mosaic. BMC Evol. Biol. 6, 101 ( 10.1186/1471-2148-6-101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zubáčová Z, et al. 2013. The mitochondrion-like organelle of Trimastix pyriformis contains the complete glycine cleavage system. PLoS ONE 8, e55417 ( 10.1371/journal.pone.0055417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Graaf RM, Duarte I, van Alen TA, Kuiper JWP, Schotanus K, Rosenberg J, Huynen MA, Hackstein JHP. 2009. The hydrogenosomes of Psalteriomonas lanterna. BMC Evol. Biol. 9, 287 ( 10.1186/1471-2148-9-287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barberà MJ, Ruiz-Trillo I, Tufts JYA, Bery A, Silberman JD, Roger AJ. 2010. Sawyeria marylandensis (Heterolobosea) has a hydrogenosome with novel metabolic properties. Eukaryot. Cell 9, 1913–1924. ( 10.1128/EC.00122-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fritz-Laylin LK, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140, 631–642. ( 10.1016/j.cell.2010.01.032) [DOI] [PubMed] [Google Scholar]
- 43.Tsaousis AD, Nyvltová E, Sutak R, Hrdy I, Tachezy J. 2014. A nonmitochondrial hydrogen production in Naegleria gruberi. Genome Biol. Evol. 6, 792–799. ( 10.1093/gbe/evu065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rew R. 1974. Enzyme localization in the anaerobic mitochondria of Ascaris lumbricoides. J. Cell Biol. 63, 125–135. ( 10.1083/jcb.63.1.125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Föll RL, Pleyers A, Lewandovski GJ, Wermter C, Hegemann V, Paul RJ. 1999. Anaerobiosis in the nematode Caenorhabditis elegans. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 124, 269–280. ( 10.1016/S0305-0491(99)00130-3) [DOI] [PubMed] [Google Scholar]
- 46.Klockiewicz M. 1995. Rhodoquinone and complex II of the electron transport chain in anaerobically functioning eukaryotes. J. Biol. Chem. 270, 31 065–31 070. ( 10.1074/jbc.270.52.31065) [DOI] [PubMed] [Google Scholar]
- 47.Tielens AG, Rotte C, van Hellemond JJ, Martin W. 2002. Mitochondria as we don't know them. Trends Biochem. Sci. 27, 564–572. ( 10.1016/S0968-0004(02)02193-X) [DOI] [PubMed] [Google Scholar]
- 48.Sakai C, Tomitsuka E, Esumi H, Harada S, Kita K. 2012. Mitochondrial fumarate reductase as a target of chemotherapy: from parasites to cancer cells. Biochim. Biophys. Acta Gen. Subj. 1820, 643–651. ( 10.1016/j.bbagen.2011.12.013) [DOI] [PubMed] [Google Scholar]
- 49.Amino H, et al. 2003. Isolation and characterization of the stage-specific cytochrome b small subunit (CybS) of Ascaris suum complex II from the aerobic respiratory chain of larval mitochondria. Mol. Biochem. Parasitol. 128, 175–186. ( 10.1016/S0166-6851(03)00074-4) [DOI] [PubMed] [Google Scholar]
- 50.Iwata F, Shinjyo N, Amino H, Sakamoto K, Islam MK, Tsuji N, Kita K. 2008. Change of subunit composition of mitochondrial complex II (succinate-ubiquinone reductase/quinol-fumarate reductase) in Ascaris suum during the migration in the experimental host. Parasitol. Int. 57, 54–61. ( 10.1016/j.parint.2007.08.002) [DOI] [PubMed] [Google Scholar]
- 51.Tielens AGM. 1994. Energy generation in parasitic helminths. Parasitol. Today 10, 346–352. ( 10.1016/0169-4758(94)90245-3) [DOI] [PubMed] [Google Scholar]
- 52.Tielens AGM, Van Hellemond JJ, Van Hellemond JJ. 1998. The electron transport chain in anaerobically functioning eukaryotes. Biochim. Biophys. Acta 1365, 71–78. ( 10.1016/S0005-2728(98)00045-0) [DOI] [PubMed] [Google Scholar]
- 53.Hirt RP, Logsdon JM, Healy B, Dorey MW, Doolittle WF, Embley TM. 1999. Microsporidia are related to fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl. Acad. Sci. USA 96, 580–585. ( 10.1073/pnas.96.2.580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lara E, Moreira D, López-García P. 2010. The environmental clade LKM11 and Rozella form the deepest branching clade of fungi. Protist 161, 116–121. ( 10.1016/j.protis.2009.06.005) [DOI] [PubMed] [Google Scholar]
- 55.Jones MDM, Forn I, Gadelha C, Egan MJ, Bass D, Massana R, Richards TA. 2011. Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474, 200–203. ( 10.1038/nature09984) [DOI] [PubMed] [Google Scholar]
- 56.Goodgame RW. 1996. Understanding intestinal spore-forming protozoa: Cryptosporidia, Microsporidia, Cyclospora. Ann. Intern. Med. 124, 429 ( 10.7326/0003-4819-124-4-199602150-00008) [DOI] [PubMed] [Google Scholar]
- 57.Katinka MD, et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414, 450–453. ( 10.1038/35106579) [DOI] [PubMed] [Google Scholar]
- 58.Williams BAP, Hirt RP, Lucocq JM, Embley TM. 2002. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature 418, 865–869. ( 10.1038/nature00949) [DOI] [PubMed] [Google Scholar]
- 59.Corradi N, Akiyoshi DE, Morrison HG, Feng X, Weiss LM, Tzipori S, Keeling PJ. 2007. Patterns of genome evolution among the microsporidian parasites Encephalitozoon cuniculi, Antonospora locustae and Enterocytozoon bieneusi. PLoS ONE 2, e1277 ( 10.1371/journal.pone.0001277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldberg AV, et al. 2008. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature 452, 624–628. ( 10.1038/nature06606) [DOI] [PubMed] [Google Scholar]
- 61.Cornman RS, et al. 2009. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 5, e1000466 ( 10.1371/journal.ppat.1000466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keeling PJ, Fast NM. 2002. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol. 56, 93–116. ( 10.1146/annurev.micro.56.012302.160854) [DOI] [PubMed] [Google Scholar]
- 63.Keeling PJ, Corradi N, Morrison HG, Haag KL, Ebert D, Weiss LM, Akiyoshi DE, Tzipori S. 2010. The reduced genome of the parasitic microsporidian Enterocytozoon bieneusi lacks genes for core carbon metabolism. Genome Biol. Evol. 2, 304–309. ( 10.1093/gbe/evq022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heinz E, et al. 2012. The genome of the obligate intracellular parasite Trachipleistophora hominis: new insights into microsporidian genome dynamics and reductive evolution. PLoS Pathog. 8, e1002979 ( 10.1371/journal.ppat.1002979) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsaousis AD, Kunji ER, Goldberg AV, Lucocq JM, Hirt RP, Embley TM. 2008. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature 453, 553–556. ( 10.1038/nature06903) [DOI] [PubMed] [Google Scholar]
- 66.Williams BAP, Elliot C, Burri L, Kido Y, Kita K, Moore AL, Keeling PJ. 2010. A broad distribution of the alternative oxidase in microsporidian parasites. PLoS Pathog. 6, e1000761 ( 10.1371/journal.ppat.1000761) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dolgikh VV, Senderskiy IV, Pavlova OA, Naumov AM, Beznoussenko GV. 2011. Immunolocalization of an alternative respiratory chain in Antonospora (Paranosema) locustae spores: mitosomes retain their role in microsporidial energy metabolism. Eukaryot. Cell 10, 588–593. ( 10.1128/EC.00283-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haag KL, James TY, Pombert J-F, Larsson R, Schaer TMM, Refardt D, Ebert D. 2014. Evolution of a morphological novelty occurred before genome compaction in a lineage of extreme parasites. Proc. Natl Acad. Sci. USA 111, 15 480–15 485. ( 10.1073/pnas.1410442111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yarlett N, Orpin CG, Munn EA, Yarlett NC, Greenwood CA. 1986. Hydrogenosomes in the rumen fungus Neocallimastix patriciarum. Biochem. J. 236, 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marvin-Sikkema FD, Gomes TMP, Grivet JP, Gottschal JC, Prins RA. 1993. Characterization of hydrogenosomes and their role in glucose metabolism of Neocallimastix sp. L2. Arch. Microbiol. 160, 388–396. ( 10.1007/BF00252226) [DOI] [PubMed] [Google Scholar]
- 71.Van der Giezen M, Rechinger KB, Svendsen I, Durand R, Hirt RP, Fevre M, Embley TM, Prins RA. 1997. A mitochondrial-like targeting signal on the hydrogenosomal malic enzyme from the anaerobic fungus Neocallimastix frontalis: support for the hypothesis that hydrogenosomes are modified mitochondria. Mol. Microbiol. 23, 11–21. ( 10.1046/j.1365-2958.1997.1891553.x) [DOI] [PubMed] [Google Scholar]
- 72.Li J, Heath IB, Bauchop T. 1990. Piromyces mae and Piromyces dumbonica, two new species of uniflagellate anaerobic chytridiomycete fungi from the hindgut of the horse and elephant. Can. J. Bot. 68, 1021–1033. ( 10.1139/b90-129) [DOI] [Google Scholar]
- 73.Li J, Heath IB, Cheng K-J. 1991. The development and zoospore ultrastructure of a polycentric chytridiomycete gut fungus, Orpinomyces joyonii comb.nov. Can. J. Bot. 69, 580–589. ( 10.1139/b91-079) [DOI] [Google Scholar]
- 74.Akhmanova A, Voncken FG, Hosea KM, Harhangi H, Keltjens JT, op den Camp HJ, Vogels GD, Hackstein JH. 1999. A hydrogenosome with pyruvate formate-lyase: anaerobic chytrid fungi use an alternative route for pyruvate catabolism. Mol. Microbiol. 32, 1103–1114. ( 10.1046/j.1365-2958.1999.01434.x) [DOI] [PubMed] [Google Scholar]
- 75.Gelius-Dietrich G, Henze K. 2004. Pyruvate formate lyase (PFL) and PFL activating enzyme in the chytrid fungus Neocallimastix frontalis: a free-radical enzyme system conserved across divergent eukaryotic lineages. J. Eukaryot. Microbiol. 51, 456–463. ( 10.1111/j.1550-7408.2004.tb00394.x) [DOI] [PubMed] [Google Scholar]
- 76.Boxma B, et al. 2004. The anaerobic chytridiomycete fungus Piromyces sp. E2 produces ethanol via pyruvate:formate lyase and an alcohol dehydrogenase E. Mol. Microbiol. 51, 1389–1399. ( 10.1046/j.1365-2958.2003.03912.x) [DOI] [PubMed] [Google Scholar]
- 77.Youssef NH, Couger MB, Struchtemeyer CG, Liggenstoffer AS, Prade RA, Najar FZ, Atiyeh HK, Wilkins MR, Elshahed MS. 2013. The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol. 79, 4620–4634. ( 10.1128/AEM.00821-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brown MW, Sharpe SC, Silberman JD, Heiss AA, Lang BF, Simpson AGB, Roger AJ. 2013. Phylogenomics demonstrates that breviate flagellates are related to opisthokonts and apusomonads. Proc. R. Soc. B 280, 20131755 ( 10.1098/rspb.2013.1755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stairs CW, Eme L, Brown MW, Mutsaers C, Susko E, Dellaire G, Soanes DM, Van Der Giezen M, Roger AJ. 2014. A SUF Fe-S cluster biogenesis system in the mitochondrion-related organelles of the anaerobic protist Pygsuia. Curr. Biol. 24, 1176–1186. ( 10.1016/j.cub.2014.04.033) [DOI] [PubMed] [Google Scholar]
- 80.Mi-ichi F, Abu Yousuf M, Nakada-Tsukui K, Nozaki T. 2009. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc. Natl Acad. Sci. USA 106, 21 731–21 736. ( 10.1073/pnas.0907106106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dolezal P, et al. 2010. The essentials of protein import in the degenerate mitochondrion of Entamoeba histolytica. PLoS Pathog. 6, e1000812 ( 10.1371/journal.ppat.1000812) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ali V, Shigeta Y, Tokumoto U, Takahashi Y, Nozaki T. 2004. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron-sulfur cluster assembly under anaerobic conditions. J. Biol. Chem. 279, 16 863–16 874. ( 10.1074/jbc.M313314200) [DOI] [PubMed] [Google Scholar]
- 83.Van der Giezen M, Cox S, Tovar J. 2004. The iron-sulfur cluster assembly genes iscS and iscU of Entamoeba histolytica were acquired by horizontal gene transfer. BMC Evol. Biol. 4, 7 ( 10.1186/1471-2148-4-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maralikova B, Ali V, Nakada-Tsukui K, Nozaki T, van der Giezen M, Henze K, Tovar J. 2010. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell. Microbiol. 12, 331–342. ( 10.1111/j.1462-5822.2009.01397.x) [DOI] [PubMed] [Google Scholar]
- 85.Chan KW, et al. 2005. A novel ADP/ATP transporter in the mitosome of the microaerophilic human parasite Entamoeba histolytica. Curr. Biol. 15, 737–742. ( 10.1016/j.cub.2005.02.068) [DOI] [PubMed] [Google Scholar]
- 86.Mi-Ichi F, Miyamoto T, Takao S, Jeelani G, Hashimoto T, Hara H, Nozaki T, Yoshida H. 2015. Entamoeba mitosomes play an important role in encystation by association with cholesteryl sulfate synthesis. Proc. Natl Acad. Sci. USA 112, E2884–E2890. ( 10.1073/pnas.1423718112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chavez LA, Balamuth W, Gong T. 1986. A light and electron microscopical study of a new, polymorphic free-living amoeba, Phreatamoeba balamuthi n. g., n. sp. J. Eukaryot. Microbiol. 33, 397–404. [DOI] [PubMed] [Google Scholar]
- 88.Gill EE, et al. 2007. Novel mitochondrion-related organelles in the anaerobic amoeba Mastigamoeba balamuthi. Mol. Microbiol. 66, 1306–1320. ( 10.1111/j.1365-2958.2007.05979.x) [DOI] [PubMed] [Google Scholar]
- 89.Nývltová E, Stairs CW, Hrdý I, Rídl J, Mach J, Pačes J, Roger J, Tachezy J. 2015. Lateral gene transfer and gene duplication played a key role in the evolution of Mastigamoeba balamuthi hydrogenosomes. Mol. Biol. Evol. 32, 1039–1055. ( 10.1093/molbev/msu408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nyvltová E, Suták R, Harant K, Sedinová M, Hrdy I, Paces J, Vlcek C, Tachezy J. 2013. NIF-type iron-sulfur cluster assembly system is duplicated and distributed in the mitochondria and cytosol of Mastigamoeba balamuthi. Proc. Natl Acad. Sci. USA 110, 7371–7376. ( 10.1073/pnas.1219590110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zaman V, Howe J, Ng M. 1998. Ultrastructure of the nucleus of the Iodamoeba bütschlii cyst. Parasitol. Res. 84, 421–422. ( 10.1007/s004360050421) [DOI] [PubMed] [Google Scholar]
- 92.Ptáčková E, Kostygov AY, Chistyakova LV, Falteisek L, Frolov AO, Patterson DJ, Walker G, Cepicka I. 2013. Evolution of Archamoebae: morphological and molecular evidence for pelobionts including Rhizomastix, Entamoeba, Iodamoeba, and Endolimax. Protist 164, 380–410. ( 10.1016/j.protis.2012.11.005) [DOI] [PubMed] [Google Scholar]
- 93.Van Klink F, Alizadeh H, Stewart GL, Pidherney MS, Silvany RE, He Y, McCulley JP, Niederkorn JY. 1992. Characterization and pathogenic potential of a soil isolate and an ocular isolate of Acanthamoeba castellanii in relation to Acanthamoeba keratitis. Curr. Eye Res. 11, 1207–1220. ( 10.3109/02713689208999546) [DOI] [PubMed] [Google Scholar]
- 94.Cometa I, Schatz S, Trzyna W, Rogerson A. 2011. Tolerance of naked amoebae to low oxygen levels with an emphasis on the genus Acanthamoeba. Acta Protozool. 50, 33–40. [Google Scholar]
- 95.Gawryluk RMR, Chisholm KA, Pinto DM, Gray MW. 2014. Compositional complexity of the mitochondrial proteome of a unicellular eukaryote (Acanthamoeba castellanii, supergroup Amoebozoa) rivals that of animals, fungi, and plants. J. Proteomics 109C, 400–416. ( 10.1016/j.jprot.2014.07.005) [DOI] [PubMed] [Google Scholar]
- 96.Leger MM, Gawryluk RMR, Gray MW, Roger AJ. 2013. Evidence for a hydrogenosomal-type anaerobic ATP generation pathway in Acanthamoeba castellanii. PLoS ONE 8, e69532 ( 10.1371/journal.pone.0069532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zierdt CH. 1983. Blastocystis hominis, a protozoan parasite and intestinal pathogen of human beings. Clin. Microbiol. Newsl. 5, 57–59. ( 10.1016/S0196-4399(83)80079-8) [DOI] [Google Scholar]
- 98.Lantsman Y, Tan KS, Morada M, Yarlett N. 2008. Biochemical characterization of a mitochondrial-like organelle from Blastocystis sp. subtype 7. Microbiology 154, 2757–2766. ( 10.1099/mic.0.2008/017897-0) [DOI] [PubMed] [Google Scholar]
- 99.Stechmann A, Hamblin K, Pérez-Brocal V, Gaston D, Richmond GS, van de Giezen M, Clark CG, Roger AJ. 2008. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr. Biol. 18, 580–585. ( 10.1016/j.cub.2008.03.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Denoeud F, et al. 2011. Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol. 12, R29 ( 10.1186/gb-2011-12-3-r29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boxma B, et al. 2005. An anaerobic mitochondrion that produces hydrogen. Nature 434, 74–79. ( 10.1038/nature03343) [DOI] [PubMed] [Google Scholar]
- 102.De Graaf RM, et al. 2011. The organellar genome and metabolic potential of the hydrogen-producing mitochondrion of Nyctotherus ovalis. Mol. Biol. Evol. 28, 2379–2391. ( 10.1093/molbev/msr059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Boxma B, et al. 2007. The [FeFe] hydrogenase of Nyctotherus ovalis has a chimeric origin. BMC Evol. Biol. 7, 230 ( 10.1186/1471-2148-7-230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gijzen HJ, Broers CA, Barughare M, Stumm CK. 1991. Methanogenic bacteria as endosymbionts of the ciliate Nyctotherus ovalis in the cockroach hindgut. Appl. Environ. Microbiol. 57, 1630–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Hoek AHAM, van Alen TA, Sprakel VSI, Leunissen JAM, Brigge T, Vogels GD, Hackstein JHP. 2000. Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates. Mol. Biol. Evol. 17, 251–258. ( 10.1093/oxfordjournals.molbev.a026304) [DOI] [PubMed] [Google Scholar]
- 106.Embley TM, Finlay BJ, Dyal PL, Hirt RP, Wilkinson M, Williams AG. 1995. Multiple origins of anaerobic ciliates with hydrogenosomes within the radiation of aerobic ciliates. Proc. R. Soc. Lond. B 262, 87–93. ( 10.1098/rspb.1995.0180) [DOI] [PubMed] [Google Scholar]
- 107.Keithly JS. 2007. The mitochondrion-related organelle of Cryptosporidium parvum. Microbiol. Monogr. 9, 231–253. ( 10.1007/7171_2007_115) [DOI] [Google Scholar]
- 108.Xu P, et al. 2004. The genome of Cryptosporidium hominis. Nature 431, 1107–1112. ( 10.1038/nature02977) [DOI] [PubMed] [Google Scholar]
- 109.Abrahamsen MS, et al. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304, 441–445. ( 10.1126/science.1094786) [DOI] [PubMed] [Google Scholar]
- 110.Henriquez FL, Richards TA, Roberts F, McLeod R, Roberts CW. 2005. The unusual mitochondrial compartment of Cryptosporidium parvum. Trends Parasitol. 21, 68–74. ( 10.1016/j.pt.2004.11.010) [DOI] [PubMed] [Google Scholar]
- 111.Mogi T, Kita K. 2010. Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol. Int. 59, 305–312. ( 10.1016/j.parint.2010.04.005) [DOI] [PubMed] [Google Scholar]
- 112.Ctrnacta V, Ault JG, Stejskal F, Keithly JS. 2006. Localization of pyruvate : NADP+ oxidoreductase in sporozoites of Cryptosporidium parvum. J. Eukaryot. Microbiol. 53, 225–231. ( 10.1111/j.1550-7408.2006.00099.x) [DOI] [PubMed] [Google Scholar]
- 113.Hine PM, Bower SM, Meyer GR, Cochennec-Laureau N, Berthe FCJ. 2001. Ultrastructure of Mikrocytos mackini, the cause of Denman Island disease in oysters Crassostrea spp. and Ostrea spp. in British Columbia, Canada. Dis. Aquat. Organ. 45, 215–227. ( 10.3354/dao045215) [DOI] [PubMed] [Google Scholar]
- 114.Burki F, Corradi N, Sierra R, Pawlowski J, Meyer GR, Abbott CL, Keeling PJ. 2013. Phylogenomics of the intracellular parasite Mikrocytos mackini reveals evidence for a mitosome in Rhizaria. Curr. Biol. 23, 1541–1547. ( 10.1016/j.cub.2013.06.033) [DOI] [PubMed] [Google Scholar]
- 115.Schmitz-Esser S, Linka N, Collingro A, Beier CL, Neuhaus HE, Wagner M, Horn M. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J. Bacteriol. 186, 683–691. ( 10.1128/JB.186.3.683-691.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Martin W, Müller M. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41. ( 10.1038/32096) [DOI] [PubMed] [Google Scholar]
- 117.López-García P, Moreira D. 1999. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem. Sci. 24, 88–93. ( 10.1016/S0968-0004(98)01342-5) [DOI] [PubMed] [Google Scholar]
- 118.Hug LA, Stechmann A, Roger AJ. 2010. Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes. Mol. Biol. Evol. 27, 311–324. ( 10.1093/molbev/msp237) [DOI] [PubMed] [Google Scholar]
- 119.Stairs CW, Roger AJ, Hampl V. 2011. Eukaryotic pyruvate formate lyase and its activating enzyme were acquired laterally from a Firmicute. Mol. Biol. Evol. 28, 2087–2099. ( 10.1093/molbev/msr032) [DOI] [PubMed] [Google Scholar]
- 120.Horner DS, Hirt RP, Embley TM. 1999. A single eubacterial origin of eukaryotic pyruvate : ferredoxin oxidoreductase genes: implications for the evolution of anaerobic eukaryotes. Mol. Biol. Evol. 16, 1280–1291. ( 10.1093/oxfordjournals.molbev.a026218) [DOI] [PubMed] [Google Scholar]
- 121.Horner DS, Heil B, Happe T, Embley TM. 2002. Iron hydrogenases—ancient enzymes in modern eukaryotes. Trends Biochem. Sci. 27, 148–153. ( 10.1016/S0968-0004(01)02053-9) [DOI] [PubMed] [Google Scholar]
- 122.Emelyanov VV. 2003. Common evolutionary origin of mitochondrial and rickettsial respiratory chains. Arch. Biochem. Biophys. 420, 130–141. ( 10.1016/j.abb.2003.09.031) [DOI] [PubMed] [Google Scholar]
- 123.Berriman M. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422. ( 10.1126/science.1112642) [DOI] [PubMed] [Google Scholar]
- 124.Kondrashov FA, Koonin EV, Morgunov IG, Finogenova TV, Kondrashova MN. 2006. Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biol. Direct 1, 31 ( 10.1186/1745-6150-1-31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nakashima K, Yamada L, Satou Y, Azuma JI, Satoh N. 2004. The evolutionary origin of animal cellulose synthase. Dev. Genes Evol. 214, 81–88. ( 10.1007/s00427-003-0379-8) [DOI] [PubMed] [Google Scholar]
- 126.Ricard G, et al. 2006. Horizontal gene transfer from Bacteria to rumen Ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics 7, 22 ( 10.1186/1471-2164-7-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. 2006. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr. Biol. 16, 1857–1864. ( 10.1016/j.cub.2006.07.052) [DOI] [PubMed] [Google Scholar]
- 128.Andersson JO, Roger AJ. 2003. Evolution of glutamate dehydrogenase genes: evidence for lateral gene transfer within and between prokaryotes and eukaryotes. BMC Evol. Biol. 3, 14 ( 10.1186/1471-2148-3-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Andersson JO. 2009. Gene transfer and diversification of microbial eukaryotes. Annu. Rev. Microbiol. 63, 177–193. ( 10.1146/annurev.micro.091208.073203) [DOI] [PubMed] [Google Scholar]
- 130.Strese A, Backlund A, Alsmark C. 2014. A recently transferred cluster of bacterial genes in Trichomonas vaginalis--lateral gene transfer and the fate of acquired genes. BMC Evol. Biol. 14, 119 ( 10.1186/1471-2148-14-119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Degnan SM. 2014. Think laterally: horizontal gene transfer from symbiotic microbes may extend the phenotype of marine sessile hosts. Front. Microbiol. 5, 638 ( 10.3389/fmicb.2014.00638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alsmark C, Foster PG, Sicheritz-Ponten T, Nakjang S, Martin Embley T, Hirt RP. 2013. Patterns of prokaryotic lateral gene transfers affecting parasitic microbial eukaryotes. Genome Biol. 14, R19 ( 10.1186/gb-2013-14-2-r19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wiedenbeck J, Cohan FM. 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35, 957–976. ( 10.1111/j.1574-6976.2011.00292.x) [DOI] [PubMed] [Google Scholar]
- 134.Doolittle WF. 1998. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14, 307–311. ( 10.1016/S0168-9525(98)01494-2) [DOI] [PubMed] [Google Scholar]
- 135.Andersson JO. 2005. Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 62, 1182–1197. ( 10.1007/s00018-005-4539-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Richards TA, Soanes DM, Jones MDM, Vasieva O, Leonard G, Paszkiewicz K, Foster PG, Hall N, Talbot NJ. 2011. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl Acad. Sci. USA 108, 15 258–15 263. ( 10.1073/pnas.1105100108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hampl V, Stairs CW, Roger AJ. 2011. The tangled past of eukaryotic enzymes involved in anaerobic metabolism. Mob. Genet. Elements 1, 71–74. ( 10.4161/mge.1.1.15588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Millar AH, Trend AE, Heazlewood JL. 2004. Changes in the mitochondrial proteome during the anoxia to air transition in rice focus around cytochrome-containing respiratory complexes. J. Biol. Chem. 279, 39 471–39 478. ( 10.1074/jbc.M406015200) [DOI] [PubMed] [Google Scholar]
- 139.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. 2006. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 3, 187–197. ( 10.1016/j.cmet.2006.01.012) [DOI] [PubMed] [Google Scholar]
- 140.Fukuda R, Zhang H, Kim J, Shimoda L, Dang CV, Semenza GL. 2007. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122. ( 10.1016/j.cell.2007.01.047) [DOI] [PubMed] [Google Scholar]
- 141.Vijayasarathy C, Damle S, Prabu SK, Otto CM, Avadhani NG. 2003. Adaptive changes in the expression of nuclear and mitochondrial encoded subunits of cytochrome c oxidase and the catalytic activity during hypoxia. Eur. J. Biochem. 270, 871–879. ( 10.1046/j.1432-1033.2003.03447.x) [DOI] [PubMed] [Google Scholar]
- 142.Ackrell BAC, Armstrong FA, Cochran B, Sucheta A, Yu T. 1993. Classification of fumarate reductases and succinate dehydrogenases based upon their contrasting behaviour in the reduced benzylviologen/fumarate assay. FEBS Lett. 326, 92–94. ( 10.1016/0014-5793(93)81768-U) [DOI] [PubMed] [Google Scholar]
- 143.Lonjers ZT, Dickson EL, Chu T-PT, Kreutz JE, Neacsu FA, Anders KR, Shepherd JN. 2012. Identification of a new gene required for the biosynthesis of rhodoquinone in Rhodospirillum rubrum. J. Bacteriol. 194, 965–971. ( 10.1128/JB.06319-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schut GJ, Adams MWW. 2009. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 191, 4451–4457. ( 10.1128/JB.01582-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Martin J, Mahlke K, Pfanner N. 1991. Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J. Biol. Chem. 266, 18 051–18 057. [PubMed] [Google Scholar]
- 146.Palmieri F. 2004. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 447, 689–709. ( 10.1007/s00424-003-1099-7) [DOI] [PubMed] [Google Scholar]
- 147.Jackson JB. 2012. A review of the binding-change mechanism for proton-translocating transhydrogenase. Biochim. Biophys. Acta 1817, 1839–1846. ( 10.1016/j.bbabio.2012.04.006) [DOI] [PubMed] [Google Scholar]
- 148.Bremmer J. 1969. Pyruvate dehydrogenase, substrate specificity and product inhibition. Eur. J. Biochem. 8, 636–640. ( 10.1111/j.1432-1033.1969.tb00559.x) [DOI] [PubMed] [Google Scholar]
- 149.Kerbey BAL, Randle PJ, Cooper RH, Whitehouse S, Pask HT, Denton RM. 1976. Regulation of pyruvate dehydrogenase in rat heart. Biochem J. 154, 327–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brown GK, Otero LJ, LeGris M, Brown RM. 1994. Pyruvate dehydrogenase deficiency. J. Med. Genet. 31, 875–879. ( 10.1136/jmg.31.11.875) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hiltunen JK, Autio KJ, Schonauer MS, Kursu VAS, Dieckmann CL, Kastaniotis AJ. 2010. Mitochondrial fatty acid synthesis and respiration. Biochim. Biophys. Acta 1797, 1195–1202. ( 10.1016/j.bbabio.2010.03.006) [DOI] [PubMed] [Google Scholar]
- 152.Chabrière E, Charon MH, Volbeda A, Pieulle L, Hatchikian EC, Fontecilla-Camps JC. 1999. Crystal structures of the key anaerobic enzyme pyruvate : ferredoxin oxidoreductase, free and in complex with pyruvate. Nat. Struct. Biol. 6, 182–190. ( 10.1038/5870) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.