Abstract
The embryonic stem cell (ESC)-enriched miR-294/302 family and the somatic cell-enriched let-7 family stabilizes the self-renewing and differentiated cell fates, respectively. The mechanisms underlying these processes remain unknown. Here we show that among many pathways regulated by miR-294/302, the combinatorial suppression of epithelial–mesenchymal transition (EMT) and apoptotic pathways is sufficient in maintaining the self-renewal of ESCs. The silencing of ESC self-renewal by let-7 was accompanied by the upregulation of several EMT regulators and the induction of apoptosis. The ectopic activation of either EMT or apoptotic program is sufficient in silencing ESC self-renewal. However, only combined but not separate suppression of the two programs inhibited the silencing of ESC self-renewal by let-7 and several other differentiation-inducing miRNAs. These findings demonstrate that combined repression of the EMT and apoptotic pathways by miR-294/302 imposes a synergistic barrier to the silencing of ESC self-renewal, supporting a model whereby miRNAs regulate complicated cellular processes through synergistic repression of multiple targets or pathways.
Embryonic stem cells (ESCs) can self-renew indefinitely and differentiate into any cell type.1 Therefore, they hold great potential for clinical applications in regenerative medicine. However, the molecular mechanisms regulating the self-renewal and differentiation of ESCs are still not fully understood. miRNAs are an important class of short non-coding RNAs that regulate ESC self-renewal and differentiation.2 miRNA-deficient ESCs proliferate at a slower rate with a slight accumulation of cells in the G1 phase, and they cannot silence the self-renewal program when induced to differentiate.3, 4, 5 Introducing individual members from an miRNA family highly expressed in ESCs partially rescues the proliferation defect and reverses the G1 accumulation.6 The family shares a seed sequence (5′-AAGUGCU-3′) and has eight members, including miR-294 and miR-302a-d. Because of their role in influencing the ESC Cell Cycle, they have been called the ESCC family of miRNAs. In addition, ESC cell cycle regulating miRNAs (ESCC miRNAs) suppress the G1 restriction point by inhibiting retinoblastoma (Rb) family proteins, preventing ESCs from exiting the cell cycle during serum starvation or contact inhibition.7 In contrast to ESCC miRNAs, the introduction of let-7 family miRNAs that are enriched in somatic cells as well as several other lineage-specific miRNAs such as miR-26a, miR-99b, miR-193, miR-199a-5p, and miR-218 silences self-renewal in Dgcr8−/− (DiGeorge syndrome critical region gene 8−/−) ESCs but not wild-type ESCs.7, 8 Interestingly, the ESCC miRNAs prevent these miRNAs from silencing ESC self-renewal. Consistent with their roles in promoting self-renewal, ESCC miRNAs dramatically enhance the de-differentiation of human and mouse fibroblasts to induced pluripotent stem cells (iPSCs).9, 10, 11, 12, 13
How ESCC miRNAs maintain self-renewal in the presence of differentiation-inducing miRNAs is not clearly understood. Genomic studies have shown that these miRNAs target hundreds of mRNAs enriched in many biological processes.8, 14, 15, 16 Functional analysis of a small number of targets chosen based on their known roles has begun to give some insights into their functions in reprogramming somatic cells to iPSCs.10, 11, 17 However, due to the inherent differences between the maintenance and establishment of pluripotency,18 what targets or pathways underlie the antagonism between the two opposing families of miRNAs in regulating ESC self-renewal remains unknown. Recent work showed that while the miR-294/302 family suppresses and let-7 induces the G1/S restriction point, this cell cycle function cannot explain their antagonistic roles in maintaining pluripotency.7 Therefore, we set out to search for additional functions of the two miRNA families that directly underlie their opposing roles in regulating pluripotency. In this study, we found that combined repression of epithelial–mesenchymal transition (EMT) and apoptotic pathways by miR-294/302 forms a synergistic barrier to block the silencing of ESC self-renewal by let-7 and other differentiation-inducing miRNAs.
Results
miRNAs suppress EMT in ESCs
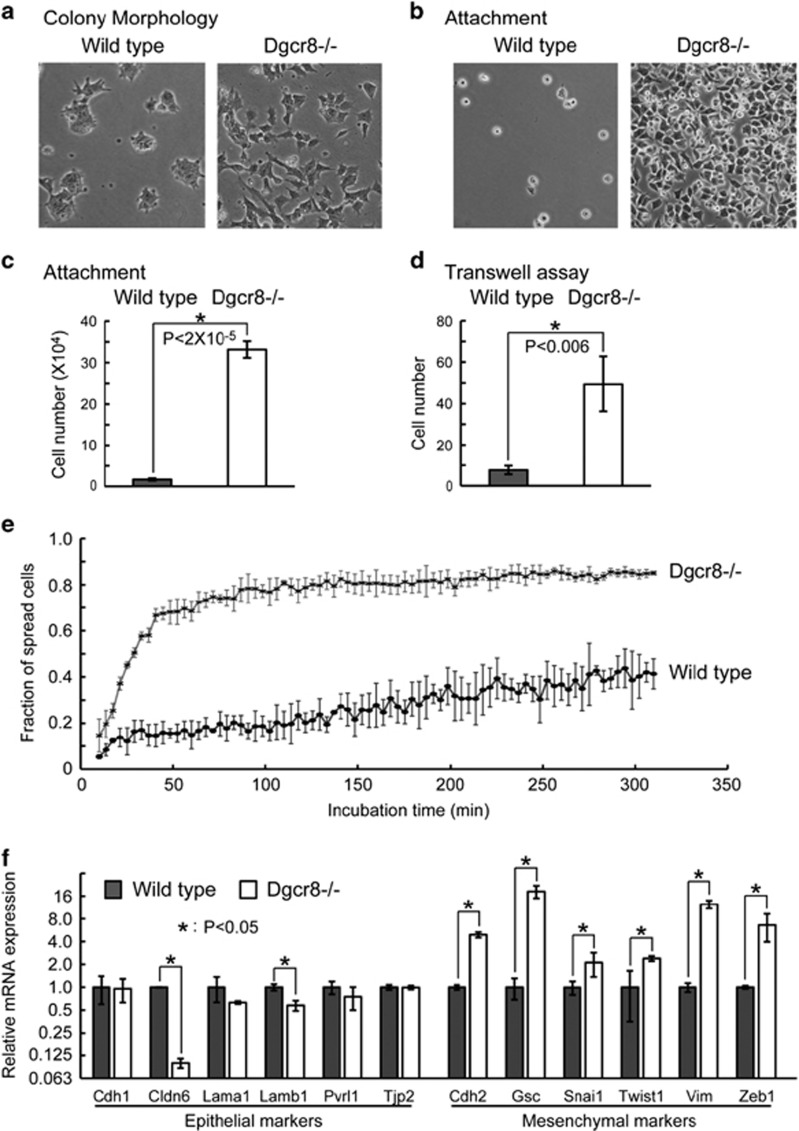
During passaging Dgcr8−/− ESCs in feeder-free conditions, we noted that the cells established a mesenchymal-like morphology (Figure 1a and Supplementary Figure S1A), suggesting that ESCs underwent EMT19, 20, 21, 22, 23 upon loss of miRNAs. Consistent with this hypothesis, Dgcr8−/− ESCs showed enhanced attachment to the culture dish during plating, higher migratory ability in transwell assays, and accelerated cell-spreading kinetics (Figures 1b–e and Supplementary Figure S1B). To determine whether the expression of epithelial and mesenchymal genes19, 20, 23, 24 changed upon Dgcr8 deletion, we analyzed the expression of representative genes using previously published mRNA microarray data25 (Supplementary Figure S1C) and by quantitative reverse transcription-PCR (qRT-PCR; Figure 1f). Although only a few epithelial genes were downregulated, most mesenchymal genes, including key EMT regulators, such as Snai1, Twist1, and Zeb1, were significantly upregulated,19, 20, 23, 24 indicating an intermediate EMT process.23 In addition, we confirmed that the protein levels of CDH1 (also known as E-Cadherin) and CDH2 (also known as N-Cadherin) were decreased and increased, respectively (Supplementary Figure S1D). These data indicate that miRNAs suppress initial EMT process in ESCs.
Figure 1.
miRNAs suppress the EMT in ESCs. (a) Morphology of wild-type and Dgcr8−/− ESC colonies. (b) Morphology and (c) the number of attached wild-type and Dgcr8−/− ESCs 40 min after plating. Shown are mean±S.D., n=3. (d) The number of migrated wild-type and Dgcr8−/− ESCs by transwell assays. Shown are mean±S.D., n=3. (e) The fraction of spread cells at different time points during 5 h of incubation. (f) qRT-PCR analysis of epithelial and mesenchymal markers. β-Actin gene was used as a control. For each gene, data were normalized to the mRNA level of wild-type ESCs. Shown are mean±S.D., n=3
The miR-294/302 and let-7 families antagonistically regulate EMT in ESCs
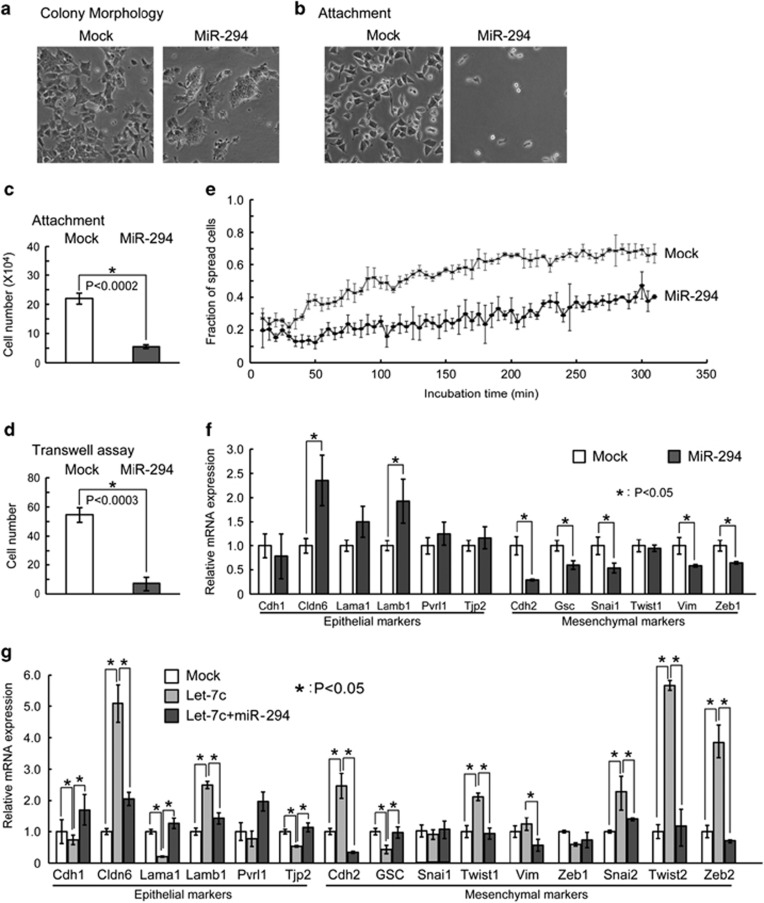
The miR-294/302 family includes the most highly expressed miRNAs in mouse ESCs.26, 27, 28 Therefore, we asked whether this family normally suppresses EMT in ESCs. Introducing members of this family, including miR-294, miR-295, and miR-302a, but not a control mimic or miR-293 (an miRNA from the same cluster as miR-294 and miR-295 but with a different seed sequence), reversed morphological changes of Dgcr8−/− ESCs, leading to more compact cell colonies (Figure 2a and Supplementary Figure S2A) and decreased cell attachment (Figures 2b and c and Supplementary Figure S2B). Because these miRNAs share a common seed sequence and have similar functions in the cell cycle,6 blocking differentiation7, 8 and repressing EMT, we then focused on miR-294 as a representative member of the miR-294/302 family. Consistent with this miRNA being an inhibitor of EMT, miR-294 decreased the migratory ability (Figure 2d) and slowed the cell-spreading kinetics (Figure 2e) of Dgcr8−/− ESCs. Moreover, qRT-PCR analysis showed that mesenchymal genes, including key EMT regulators19, 20, 23, 24 such as Snai1 and Zeb1, were suppressed by miR-294 (Figure 2f). In addition, western blotting showed that mesenchymal marker CDH2 protein was significantly downregulated by miR-294 (Supplementary Figure S2C). These data show that the miR-294/302 family suppresses EMT in ESCs.
Figure 2.
MiR-294/302 suppresses and let-7 promotes the EMT in ESCs. (a) Colony morphology of mock- and miR-294-transfected Dgcr8−/− ESCs. (b) Morphology and (c) the number of attached mock- and miR-294-transfected Dgcr8−/− ESCs 40 min after plating. Shown are mean±S.D., n=3. (d) The number of migrated mock- and miR-294-transfected Dgcr8−/− ESCs by transwell assays. Shown are mean±S.D., n=3. (e) The fraction of spread mock- and miR-294-transfected Dgcr8−/− ESCs at different time points during 5 h of incubation. (f) qRT-PCR analysis of epithelial and mesenchymal markers. (g) qRT-PCR analysis of epithelial and mesenchymal markers in mock, Let7-c, and Let7-c plus miR-294 co-transfected Dgcr8−/− ESCs. For RT-PCR, β-actin gene was used as a control. Data were normalized to the mRNA level of mock-transfected Dgcr8−/− ESCs. Shown are mean±S.D., n=3
Given that the miR-294 and let-7 miRNA families have antagonistic effects on ESC differentiation, we next asked whether let-7 exerts the opposite effect by promoting EMT. Indeed, qRT-PCR analysis revealed that let-7c further upregulated the expression of Cdh2 as well as key EMT regulators19, 20, 23, 24 Snai2, Twist1, Twist2, and Zeb2 starting from 1 day after transfection and continuing for at least 2 more days (Supplementary Figure S2D). Western blotting confirmed that CDH1 and CDH2 proteins were downregulated and upregulated by let-7c, respectively (Supplementary Figure S2E). Furthermore, co-transfection with miR-294 blocked the upregulation of Cdh2, Snai2, Twist1, Twist2, and Zeb2 (Figure 2g). These data demonstrate an antagonistic relationship between miR-294/302 and let-7 in regulating EMT in ESCs.
The miR-294/302 and let-7 families antagonistically regulate apoptosis in ESCs
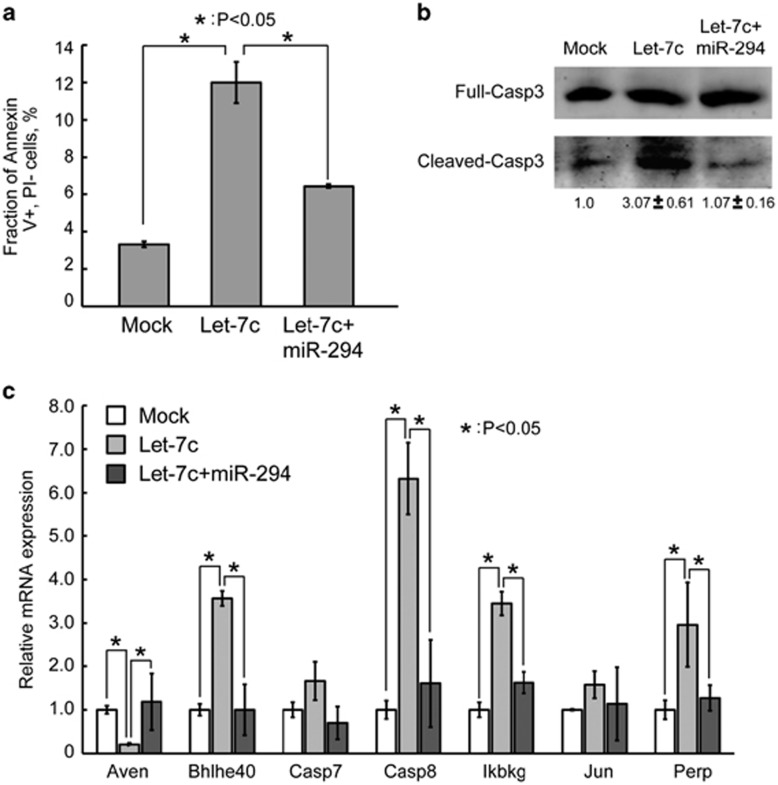
Previous work has shown that deleting miR-290 or Dgcr8 in ESCs results in a hypersensitive apoptotic phenotype in association with either doxorubicin treatment or serum starvation.7, 29 Interestingly, we noted that transfecting let-7c similarly resulted in increased apoptosis in Dgcr8−/− ESCs (Figure 3a). Co-transfecting miR-294 suppressed the let-7c-induced apoptosis. In addition, quantification of cleaved/full-length CASP3 showed that miR-294 and let-7c antagonistically regulated the activation of CASP3 (Figure 3b). Furthermore, the apoptosis-promoting factors Bhlhe40, Casp8, Ikbkg, and Perp were upregulated, and the apoptosis-inhibiting factor Aven was downregulated upon let-7c transfection. Consistent with miR-294 suppressing apoptosis, the changes in the expression levels of these genes were all rescued by miR-294 (Figure 3c). These data show that the let-7 and miR-294/302 families have opposing roles in regulating apoptosis in ESCs.
Figure 3.
MiR-294/302 suppresses and let-7 promotes apoptosis in ESCs. (a) Apoptosis analysis by Annexin V and propidium iodide staining in Dgcr8−/− ESCs mock transfected, transfected with let-7c, or co-transfected with let-7c and miR-294. Shown are mean±S.D., n=3. (b) Western blotting analysis of activated CASP3. Data were normalized to mock-transfected Dgcr8−/− ESCs. Shown are mean±range. n=2. (c) qRT-PCR analysis of regulators of apoptosis. β-Actin gene was used as a control. Data were normalized to the mRNA level of mock-transfected Dgcr8−/− ESCs. Shown are mean±S.D., n=3
Activation of either EMT or apoptotic pathways is sufficient to silence ESC self-renewal in Dgcr8−/− ESCs
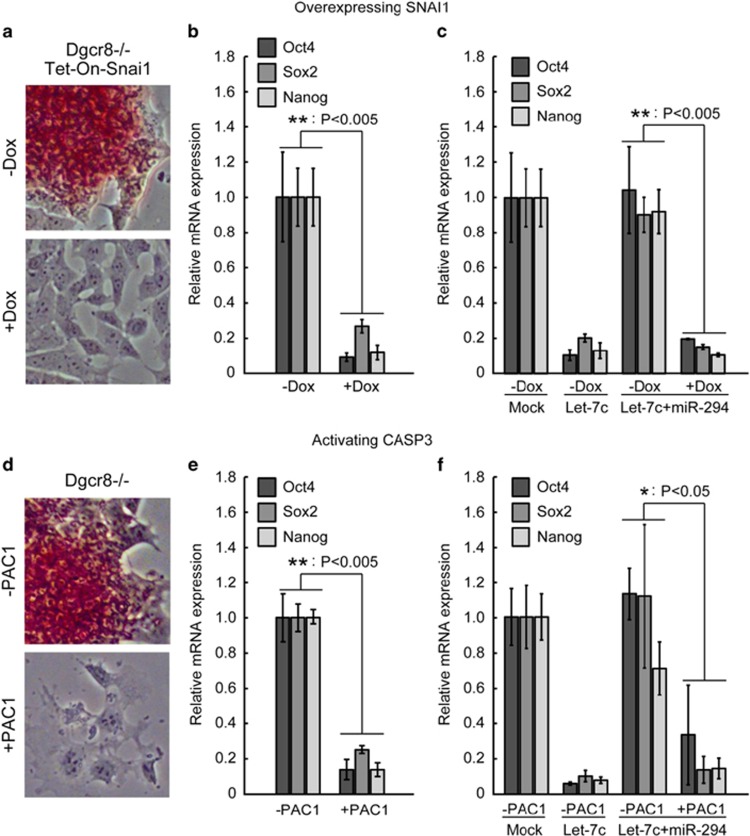
Next we asked whether promotion of either EMT or apoptosis serves as the driving force to silence the self-renewal in Dgcr8−/− ESCs, which normally are refractory to the silencing of pluripotency programs even under differentiation conditions.5 To evaluate the effect of EMT, we generated cell lines overexpressing Snai1, an important transcription factor promoting EMT,19, 20, 23, 24 in a doxycycline (Dox)-inducible manner. Snai1 overexpression successfully induced EMT as revealed by the downregulation of CDH1 protein and the upregulation of CDH2 protein (Supplementary Figure S3A). More interestingly, alkaline phosphatase (AP) staining, a marker of the pluripotent state, was lost upon Snai1 overexpression even in culture conditions that normally promote self-renewal (Figure 4a). Furthermore, the expression of Oct4, Sox2, and Nanog was dramatically downregulated upon overexpressing Snai1 (Figure 4b). These data suggest that let-7 silences ESC self-renewal partially through upregulating the expression of EMT regulators. Snai1 overexpression also significantly reduced the expression of pluripotency genes in wild-type ESCs (Supplementary Figure S3B) and in Dgcr8−/− ESCs co-transfected with let-7 and miR-294 (Figure 4c), suggesting that Snai1 functions downstream of the point at which miR-294 suppresses differentiation.
Figure 4.
Activation of either the EMT or apoptotic pathways silences self-renewal in Dgcr8−/− ESCs. (a) AP staining and (b) qRT-PCR analysis of pluripotency markers for Dgcr8−/− ESCs without (−Dox) or with (+Dox) Snai1 overexpression. Data were normalized to Dgcr8−/− ESCs without (−Dox) Snai1 overexpression. (c) qRT-PCR analysis of pluripotency markers for Dgcr8−/− ESCs co-transfected with let-7c and miR-294 and without (−Dox) or with (+Dox) Snai1 overexpression. Data were normalized to mock-transfected Dgcr8−/− ESCs without (−Dox) Snai1 overexpression. (d) AP staining and (e) qRT-PCR analysis of pluripotency markers forDgcr8−/− ESCs without or with PAC1 treatment. Data were normalized to Dgcr8−/− ESCs without PAC1 treatment. (f) qRT-PCR analysis of pluripotency markers for Dgcr8−/− ESCs co-transfected with let-7c and miR-294 and without or with PAC1 treatment. Data were normalized to mock-transfected Dgcr8−/− ESCs without PAC1 treatment. For all RT-PCR results in this figure, β-actin gene was used as a control. Shown are mean±S.D., n=3
To evaluate the effect of apoptosis on silencing ESC self-renewal, we used a small-molecule procaspase-activating compound 1 (PAC1), an activator of caspase-3.30 PAC1 treatment downregulated the pluripotency program in Dgcr8−/− ESCs (Figures 4d and e). Similar, albeit reduced effects were seen in PAC1-treated wild-type ESCs (Supplementary Figure S3C) and Dgcr8−/− ESCs co-transfected with miR-294 and let-7c (Figure 4f), suggesting that the activation of caspase-3 is downstream of the differentiation-suppressing effect of the miR-294/302 family. These findings show that the failure of Dgcr8−/− ESCs to silence self-renewal upon differentiation can be overcome by promoting either EMT or apoptotic programs and suggest that let-7 silences ESC self-renewal at least partially by activating these programs.
miR-294/302 miRNAs repress multiple genes involved in EMT
To identify mRNA targets of miR-294/302 that are responsible for the EMT phenotypes (Supplementary Figure S4A), we analyzed previously published microarray data that compared gene expression in miR-294- versus mock-transfected Dgcr8−/− ESCs and in wild-type versus Dgcr8−/− ESCs.8, 25 Approximately, 1500 genes were downregulated at least 20% in wild-type versus Dgcr8−/− ESCs and miR-294- versus mock-transfected ESCs. Out of these genes, 741 genes were predicted targets of miR-294/302 miRNAs by Targetscan.31 We then picked 19 genes that have been indicated to have a role in EMT process (Supplementary Table S1). These include genes encoding proteins of the Akt pathway, RhoGEF pathway, transforming growth factor (Tgf)-beta pathway, extracelluar matrix proteins, and glycogen synthase kinase 3 (Gsk3)-beta. Among them, RhoC, Smad2, and Tgfbr2 have been shown to be regulated by miR-294/302 during reprogramming of fibroblasts to iPSCs.10, 11, 17, 32 To confirm whether these genes are direct targets of miR-294/302 miRNAs, we cloned their 3′ untranslated regions (UTRs) downstream of a Renilla luciferase gene. Luciferase reporter assays showed that 3′UTRs of nine genes were significantly inhibited in wild-type ESCs and by miR-294 (Supplementary Figure S4B). We then used qRT-PCR analysis to confirm that the transcript levels of these targets were reduced by miRNAs (Supplementary Figures S4C and D). Six of the nine genes, Arhgef3, Fndc3a, Gsk3b, Mmp23, Tgfbr2, and Vim (Figures 1f and 2f), were down in both wild-type and miR-294-transfected Dgcr8−/− ESCs versus Dgcr8−/− ESCs. We also confirmed that the protein levels of GSK3B and VIM were downregulated in wild-type and miR-294-transfected Dgcr8−/− ESCs (Supplementary Figure S4E). These data show that multiple genes involved in EMT are the direct targets of miR-294/302 miRNAs.
miR-294/302 miRNAs suppress EMT through repressing Tgfbr1 and 2 and Gsk3-beta
To identify the functional targets of miR-294/302 miRNAs in suppressing EMT, we ordered siRNAs for each of the six above-mentioned genes. Successful knockdown of target genes by these siRNAs was confirmed by qRT-PCR (Supplementary Figure S5A). All except one siRNA to Mmp23 failed to cause morphological changes in Dgcr8−/− ESCs (Supplementary Figure S5B and data not shown). We reasoned that monitoring morphological changes may not be sensitive enough to identify functional targets; so we decided to use the cell attachment assay for a more quantitative assessment. In this assay, all siRNAs against the six miRNA targets led to decreased cell attachment when introduced into Dgcr8−/− ESCs, while a control siRNA and siRNAs to RhoC did not (Supplementary Figure S5C). These data suggest that miR-294/302 miRNAs suppress EMT through combinatorial effects on multiple targets.
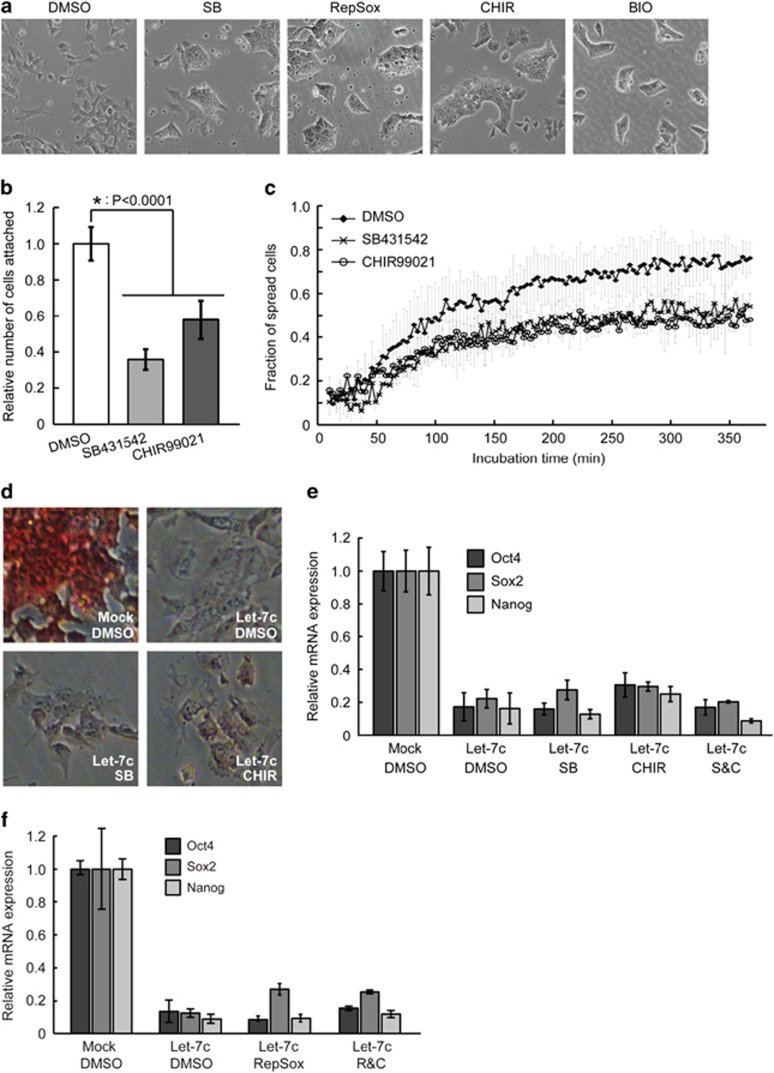
Regulation of Tgfbr2 and Gsk3-beta by miR-294/302 suggests that miR-294/302 could suppress EMT through inhibiting TGF-beta and GSK3 pathways. Consistent with this, Tgfbr1 was also indirectly inhibited by miR-294/302 miRNAs (Supplementary Figures S4B and D) and knocking down Tgfbr1 led to decreased cell attachment (Supplementary Figure S5C). To test the functional role for these targets in promoting a mesenchymal fate in Dgcr8−/− ESCs, cultures were treated with small-molecule inhibitors of TGF-beta receptors (SB431542) and GSK3 kinases (CHIR99021). Both inhibitors promoted morphological changes in Dgcr8−/− ESCs similar to those caused by miR-294/302 miRNAs (Figure 5a). Furthermore, similar to miR-294/302, SB431542 (SB) and CHIR99021 (CHIR) also caused decreased cell attachment and slower spreading kinetics (Figures 5b and c). Treating Dgcr8−/− ESCs with another TGF-beta receptor inhibitor RepSox33 or another GSK3 inhibitor BIO induced similar phenotypes (Figure 5a); therefore, the observed phenotypes were unlikely to be secondary to off-target effects. These data show that miR-294/302 suppresses EMT in ESCs in part through repressing Tgf-beta receptors and Gsk3-beta kinase.
Figure 5.
Suppression of TGF-beta receptors and GSK3 kinases blocks the EMT in Dgcr8−/−ESCs but not the silencing of self-renewal by let-7c. (a) Morphology of Dgcr8−/− ESCs treated with dimethyl sulfoxide (DMSO), SB, CHIR, RepSox, or BIO. (b) Cell-attachment analysis of Dgcr8−/− ESCs treated with DMSO, SB, or CHIR. Shown are mean±S.D., n=3. (c) Fraction of spread Dgcr8−/− ESCs treated with DMSO, SB, or CHIR at different time points during 6 h of incubation. (d) AP staining for let-7c-transfected Dgcr8−/− ESCs treated with DMSO, SB, or CHIR. (e) qRT-PCR analysis of pluripotency markers in let-7c-transfected Dgcr8−/− ESCs treated with DMSO, SB, CHIR, or SB plus CHIR (S&C). β-Actin gene was used as a control. Data were normalized to the mRNA level of mock-transfected Dgcr8−/− ESCs. Shown are mean±S.D., n=6. (f) qRT-PCR analysis of pluripotency markers in let-7c-transfected Dgcr8−/− ESCs treated with DMSO, RepSox, CHIR, or RepSox plus CHIR (R&C). β-Actin gene was used as a control. Data were normalized to the mRNA level of mock-transfected Dgcr8−/− ESCs. Shown are mean±S.D., n=3
Suppression of TGF-beta and GSK3 pathways is not sufficient to block let-7-induced silencing of ESC self-renewal
We then asked whether miR-294/302 blocks let-7-induced silencing of ESC self-renewal by suppressing the TGF-beta34, 35, 36 and GSK3 pathways.37, 38 AP staining (Figure 5d) and qRT-PCR analysis of pluripotency genes Oct4, Sox2, and Nanog showed that SB and CHIR individually or in combination (S&C) failed to block let-7c-induced differentiation (Figure 5e). Similarly, treating cells with RepSox or RepSox plus CHIR (R&C) was also unsuccessful in rescuing the ESC self-renewal in the presence of let-7c (Figure 5f), even though R&C treatment downregulated the expression of CDH2 protein (Supplementary Figure S6A). These data suggest that the suppression of TGF-beta and GSK3 pathways is not sufficient to maintain ESC self-renewal when challenged by let-7c.
Suppression of apoptosis is not sufficient to block let-7-induced silencing of ESC self-renewal
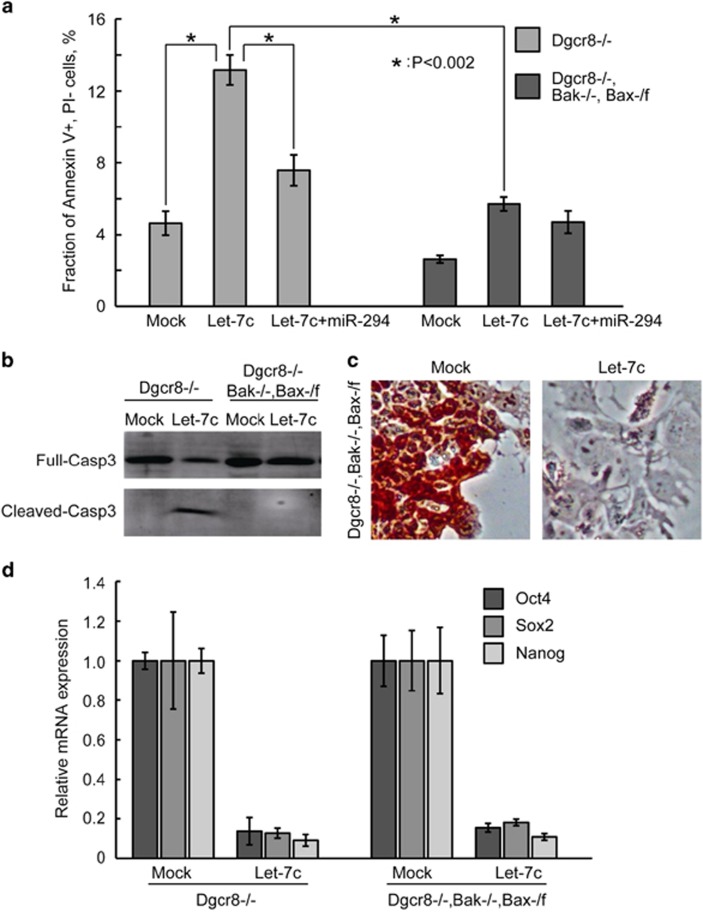
We next asked whether the suppression of apoptosis blocks the silencing of ESC self-renewal by let-7. Previously, we showed that deletion of two alleles of BCL2-antagonist/killer 1 (Bak) and one allele of BCL2-associated X protein (Bax) blocks the apoptosis induced by serum starvation in Dgcr8−/− ESCs.7 We therefore tested whether Bak−/−, Bax-/flox rescue the apoptosis and silencing of ESC self-renewal induced by let-7c. The Bak−/−, Bax-/flox deletions successfully blocked the apoptosis induced by let-7c to an extent similar to the co-introduction of miR-294 (Figure 6a). In addition, the Bak−/−, Bax-/flox deletions prevented the let-7c-induced cleavage of caspase-3 (Figure 6b). These data suggest that let-7c is upstream of Bax/Bak in inducing apoptosis. We then checked whether the Bak−/−, Bax-/flox deletions suppress let-7c from silencing the ESC self-renewal. However, AP staining and qRT-PCR analysis showed that blocking apoptosis had no effect on inhibiting let-7c-induced differentiation (Figures 6c and d). Together, these data suggest that suppression of neither apoptotic nor EMT pathways alone is sufficient to block the let-7c-induced silencing of ESC self-renewal.
Figure 6.
Suppression of apoptosis is not sufficient to block the silencing of self-renewal by let-7c in Dgcr8−/− ESCs. (a) Apoptosis analysis by Annexin V and propidium iodide staining in mock-, let-7c-, and let-7c plus miR-294-transfected Dgcr8−/− or Bak−/−, Bax-/flox, Dgcr8−/− ESCs. Shown are mean±S.D., n=3. (b) Western blotting analysis of activated CASP3 in mock- and let-7c-transfected Dgcr8−/− or Bak−/−, Bax-/flox, Dgcr8−/− ESCs. (c) AP staining of mock- and let-7c-transfected Bak−/−, Bax-/flox, Dgcr8−/− ESCs. (d) qRT-PCR analysis of pluripotency markers in mock- and let-7c-transfected Dgcr8−/− or Bak−/−, Bax-/flox, Dgcr8−/− ESCs. β-Actin gene was used as a control. For each gene, data were normalized to the mRNA level of mock-transfected ESCs with the same genotype. Shown are mean±S.D., n=3
Combined suppression of EMT and apoptotic pathways promotes ESC self-renewal
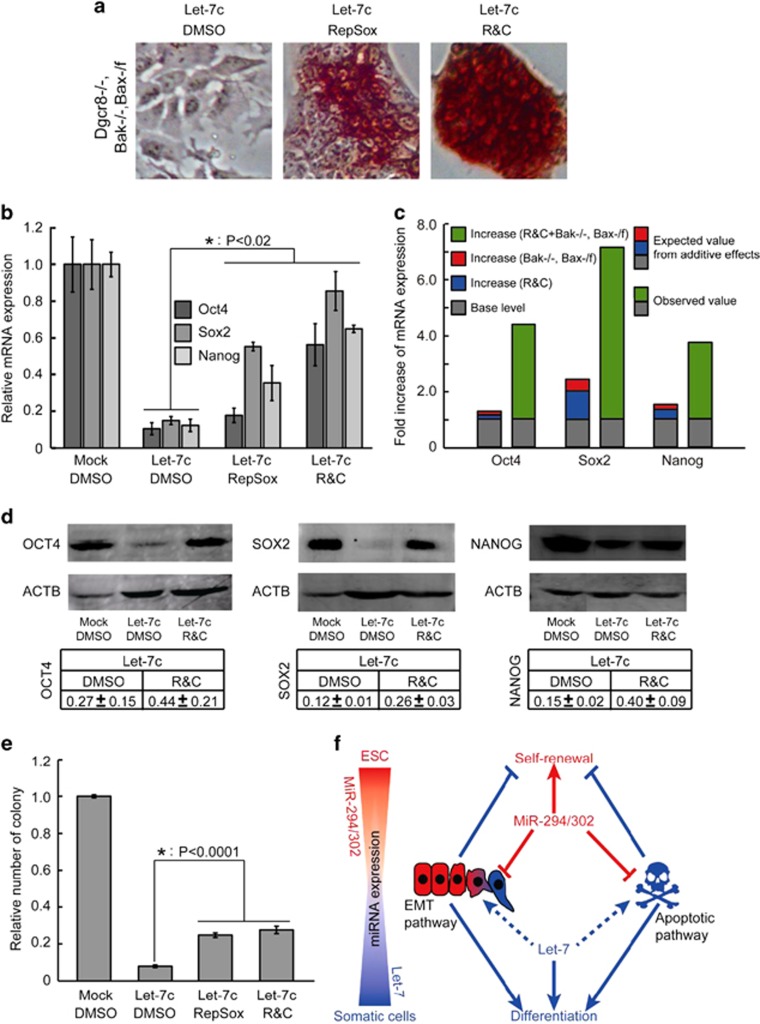
We then asked whether suppressing EMT and apoptotic pathways together inhibits the let-7-induced silencing of ESC self-renewal. To answer this question, we cultured Bak−/−, Bax-/flox, Dgcr8−/− ESCs in the presence of the TGF-beta receptor and/or GSK3 small-molecule inhibitors. The RepSox effectively suppressed let-7c silencing AP activity in these cells. Adding CHIR further inhibited the loss of AP activity (Figure 7a), consistent with a block in the silencing of self-renewal. This conclusion was further supported by qRT-PCR analysis of a panel of pluripotency genes (Figures 7b and c and Supplementary Figure S6B) and western blotting analysis of OCT4, SOX2, and NANOG (Figure 7d), as well as colony-formation assays (Figure 7e). Expression of the pluripotency factors and colony formation were not fully reversed, but the inhibition was highly significant. These data show that, while neither pathway alone blocks the let-7-induced silencing of the pluripotency program, inhibition of both apoptosis and EMT act synergistically to do so.
Figure 7.
Combined suppression of the EMT and apoptotic pathways synergistically promotes ESC self-renewal. (a) AP staining of let-7c-transfected Bak−/−, Bax-/flox, Dgcr8−/− ESCs treated with dimethyl sulfoxide (DMSO), RepSox, or RepSox plus CHIR (R&C). (b) qRT-PCR analysis of pluripotency markers in let-7c-transfected Bak−/−, Bax-/flox, Dgcr8−/− ESCs treated with DMSO, RepSox, or R&C. β-Actin gene was used as a control. For each gene, data were normalized to mock-transfected Bak−/−, Bax-/flox, Dgcr8−/− ESCs treated with DMSO. Shown are mean±S.D., n=3. (c) Evaluation of synergy between suppression of the EMT pathway and suppression of the apoptotic pathway in promoting the expression of Oct4, Sox2, and Nanog. In all cases, the measured value was at least twofold greater than the value expected from additive effects. (d) Western blotting analysis of OCT4, SOX2, and NANOG. Representative gels are shown. Data were first normalized to actin and then to mock-transfected Bak−/−, Bax-/flox, Dgcr8−/− ESCs treated with DMSO. Shown are mean±range. n=2. (e) Colony-formation assays. Data were normalized to mock-transfected Bak−/−, Bax-/flox, Dgcr8−/− ESCs treated with DMSO. Shown are mean±S.D., n=3. (f) Graphic model of the opposing functions of miR-294/302 and let-7 in regulating ESC self-renewal through antagonistically regulating the EMT and apoptotic pathways
To check whether the synergistic effects of inhibiting the EMT and apoptotic pathways impose a general barrier to the silencing of ESC self-renewal, we tested other differentiation models. In addition to let-7c, five other miRNAs (miR-26a, miR-99b, miR-193, miR-199a-5p, and miR-218) have been shown to silence the self-renewal of Dgcr8−/− ESCs, but not wild-type ESCs.7 Similar to let-7c, the silencing of self-renewal by these miRNAs was suppressed by the co-introduction of miR-294/302. We transfected these other differentiation-inducing miRNAs into Dgcr8−/− or Bak−/−, Bax-/flox, Dgcr8−/− ESCs in the presence or absence of RepSox and CHIR. Consistent with the let-7 findings, blocking the EMT or apoptosis alone had little to no effect in blocking differentiation triggered by these miRNAs (Supplementary Figures S6C and D). However, the combination of Bak−/−, Bax-/flox with RepSox plus CHIR significantly inhibited miR-26a, miR-99b, and miR-218 from silencing the self-renewal of Dgcr8−/− ESCs (Supplementary Figures S6C–E). miR-193 and miR-199a-5p remained unaffected. Interestingly, similar to let-7c, miR-26a, miR-99b, and miR-218 significantly induced the expression of multiple EMT regulators and apoptosis, while miR-193 and miR-199a-5p were not as potent as these miRNAs (Supplementary Figures S6F–G), suggesting that these miRNAs silence ESC self-renewal through different mechanisms. Together, these data showed that the combined inhibition of EMT and apopototic pathways reduces the ability of multiple, but not all, differentiation-inducing miRNAs to silence the self-renewal of ESCs.
Discussion
Here we showed that the let-7 family promoted and the miR-294/302 family inhibited the induction of mesenchymal programs and apoptosis. Although the causal link between let-7 and the induction of two programs requires further investigation to be elucidated, this antagonism between two miRNA families appears to have a critical role in regulating the switch between ESC self-renewal and differentiation (Figure 7f). miR-294/302 is a remarkable miRNA family that has pleiotropic functions in ESCs and during reprogramming.2 Studies from Filipowicz's and Blasco's laboratories showed that these miRNAs regulate telomere length and the methylation of Oct4 promoter through Rbl2/Dnmt3 pathway in ESCs.14, 39 Studies by us and other researchers showed that Tgfbr2 is targeted by miR-294/302 and the repression of Tgfbr2 promoted mesenchymal–epithelial transition (MET) and the efficiency of iPSC generation during reprogramming.10, 11, 17, 32 Sharp's laboratory showed that these miRNAs have a latent role in protecting ESCs from apoptosis induced by DNA damage reagents.29 Recently, we also observed the similar anti-apoptotic function of these miRNAs in a serum-starvation model.7 In addition, these miRNAs promote proliferation in ESCs in part by targeting regulators of the G1/S restriction point, including Cdkn1a, Lats2, and Rb family proteins.6, 7 However, although let-7 induces and miR-294 suppresses the G1 restriction point, removal of the restriction point fails to inhibit differentiation.7 Whether any other above-mentioned targets or pathways are responsible for miR-294/302 in maintaining pluripotency was not clear. Here we show that the repression of EMT and apoptotic pathways in combination, but not individually, is sufficient to maintain the pluripotency program of ESCs in the presence of differentiation-inducing miRNAs. To our knowledge, these findings provide the first example of multiple pathways targeted by a single miRNA that not only collaborate but also act synergistically to maintain a specific cell fate. That is, blocking either pathway alone had little to no effect while the combined suppression of the two dramatically inhibited miRNA-induced differentiation. This synergism among targets is likely to be a common paradigm for miRNA function.
The functions of EMT and the reverse process MET have been well documented in the reprogramming of differentiated cells to iPSCs.24, 40, 41, 42, 43 The EMT has recently been described in various ESC differentiation models.44, 45, 46 Calcineurin-NFAT signaling is necessary and sufficient to cause ESC differentiation and functions through promoting EMT.47 Furthermore, overexpressing Snail induces EMT and accelerates ESC differentiation in the DKK-induced ESC differentiation model.48 In contrast, overexpressing the EMT inhibitor miR-200 family of miRNAs stalls differentiating ESCs at the pluripotent epiblast stem cell stage.48 We found that overexpression of Snail silenced the ESC pluripotency program in 48 h, even in culture conditions that normally support ESC self-renewal. It will be important to determine whether EMT regulators other than Snail induce the silencing of ESC self-renewal and how these factors mechanistically silence the pluripotency program.
Until recently apoptosis, or programmed cell death, has mainly been associated with the elimination of unwanted or overproduced cells. However, several recent studies show that apoptosis-regulating factors are potentially involved in ESC self-renewal and the induction of pluripotency. Overexpression of anti-apoptotic Bcl2 cooperates with leukemia-inhibitory factor to maintain mouse ESC self-renewal in the absence of serum and feeder cells49 and obviates the requirement of serum factors for maintaining human ESCs.50 Furthermore, the apoptosis-inducer p53 is directly involved in controlling the differentiation of ESCs.51 More direct evidence for the involvement of apoptosis regulators in ESC self-renewal come from a functional study of Casp3.52 Knocking out Casp3, an executor of apoptosis, delays ESC differentiation in the presence of retinoic acid and in a teratoma model. Interestingly, CASP3 and another pro-apoptotic factor CASP8 are activated during the reprogramming of iPSCs.53 The activation of these caspases is critical for iPSC induction, as knocking down Casp3 and Casp8 significantly reduces iPSC formation. These results were unexpected as activation of CASP3 promotes the silencing of self-renewal in ESCs, suggesting very complicated roles of apoptotic pathway or factors in differentiation and de-differentiation processes. Consistent with pro-differentiation roles of apoptosis, we showed that a small-molecule activator of CASP3, PAC1, markedly downregulated the pluripotency program in both wild-type and Dgcr8−/− ESCs. Furthermore, inhibition of apoptosis by deleting Bax and Bak promoted ESC self-renewal in cooperation with EMT inhibitors, when challenged with differentiation-inducing miRNAs. As Bax and Bak are responsible for processing a variety of apoptotic signals and trigger many downstream factors,54, 55 it will be interesting to study their regulatory function in self-renewal in other contexts and the exact molecular mechanisms.
These previous studies identified the potential function of EMT and apoptotic pathways in regulating ESC self-renewal. However, by focusing on a single pathway these studies missed the interaction between the two pathways in this regulation. By identifying the functional miRNA targets, our study demonstrated that suppression of the EMT pathway and suppression of the apoptotic pathway combine to achieve a synergistic effect in blocking ESC differentiation. These results trigger many interesting questions, such as whether and how these two pathways crosstalk with each other and whether suppression of both pathways has synergistic effects in promoting iPSC induction. Answering these questions can further our understanding of the regulation of the pluripotency program in ESCs and during iPSC induction. As most miRNAs are predicted to target multiple genes and pathways to carry out their functions, we propose that functional analysis of miRNA targets is a powerful tool for discovering novel interactions between miRNA-regulated targets and/or pathways.
Materials and Methods
Cell culture, transfection, and AP staining
Mouse ESCs were grown on gelatin-coated plates or irradiated mouse embryonic fibroblast feeder cells (MEFs) in 15% FBS as previously described.5, 6 Dox-inducible cell lines were constructed as previously described.56 For ESC differentiation assays, 20 000 Dgcr8−/− or Bak−/−, Bax-/flox, Dgcr8−/− ESCs were plated in gelatinized 12-well plates on day 0 in LIF medium. On day 1, miRNA mimics (Dharmacon, ThermoFisher, Lafayette, CO, USA) were transfected at 25 nM or 50 nM using the DharmaFECT1 transfection reagent (Dharmacon, ThermoFisher) following the manufacturer's protocol. Sequences of siRNAs are listed in Supplementary Table S2. On day 3 after transfection, cells were processed for qRT-PCR analysis, AP staining, apoptosis analysis, or western blotting analysis. Colony-reformation assays were performed as previously described.5, 8 In brief, cells were exposed to miRNA mimics for 3 days and then trypsinized and counted. One thousand cells were re-plated on MEFs to form colonies for 7 days.
Cell attachment, migration, and spreading assays
For cell attachment experiments, ESCs grown for 48 h were trypsinized to obtain a single-cell suspension. Three hundred thousand cells per well of a gelatinized or uncoated 24-well plate were plated and incubated for 40 min at 37 °C. Unattached cells were washed away with PBS, and attached cells were trypsinized, re-suspended in ESC medium, and counted with an automated cell counter (Counter Star, Shanghai, China). For migration assays, 0.5 million cells were seeded into the upper chamber of a transwell insert (pore size, 8 μm; Costar, Washington, DC, USA) in 100 μl serum-free medium per well for 24 h at 37 °C. Medium (600 μl) containing 15% serum was placed in the lower chamber to function as a chemoattractant. Non-migratory cells were removed from the upper chamber by scraping with a cotton bud. The cells remaining on the lower surface of the insert were fixed with 4% paraformaldehyde and stained with crystal violet (Sigma, St. Louis, MO, USA). For cell-spreading assays, 50 000 cells were plated on gelatinized 6-well or 12-well plates and analyzed in Cell-IQ MLF (CM Technologies, Tampere, Finland). Briefly, three locations in each well were imaged every 4 min continuously for ~6 h. Images were viewed and analyzed in the Cell-IQ Analyzer program. Examples of spread and un-spread cells are shown in Supplementary Figure S1B.
Quantitative RT–PCR analysis
RNA for qRT-PCR analyses was prepared using Trizol (Invitrogen, Grand Island, NY, USA) and quantified on a BiodropsisBD2000 (OSTC, Beijing, China). Five hundred nanograms of RNA were treated with DNase I and reverse-transcribed using the Superscript III first complementary DNA (cDNA) synthesis kit (Invitrogen). Quantitative PCR was performed on AB Step One Plus (Applied Biosystems, Foster City, CA, USA). Primers are listed in Supplementary Table S3.
Western blotting analysis
Wild-type (0.15 million) and Dgcr8−/− (0.3 million) ESCs were plated in six-well plates and grown for ~24 h prior to transfection. Proteins were extracted using lysis buffer (Kangweishiji, Beijing, China) containing 1 × protease inhibitor usually 48 h after transfection or plating. For western blotting of OCT4, SOX2, and NANOG, proteins were extracted on day 3 after transfection of miRNAs. Antibodies against GSK3-beta and full-length or cleaved CASP3 were from Bioworld (St. Louis Park, MN, USA), against OCT4 and SOX2 from Santa Cruz Biotechnology (Dallas, TX, USA), and against NANOG from Calbiochem (Darmstadt, Germany).
Apoptosis analysis
Cells were prepared as for ESC differentiation assays, trypsinized 48 h after transfection, labeled with propidium iodide and FITC-Annexin V (Annexin), and analyzed by flow cytometry. The fraction of PI-negative and Annexin-V-positive cells in the total population was calculated to quantify the early stage of apoptosis.
Luciferase reporter assay
Luciferase constructs were produced as follows. 3′UTRs for candidate genes were amplified from ESC cDNA or genomic DNA and cloned into the NotI and XhoI sites in psiCheck-2 vector (Promega, Madison, WI, USA) as described previously.8 Sequences of cloning primers are listed in Supplementary Table S1. Approximately 24 h prior to transfection of miRNAs, 8000 Dgcr8−/− or 4000 wild-type ESCs were plated in ESC media in a 96-well plate pretreated with 0.2% gelatin. For the co-transfection of miRNAs and reporter plasmids, miRNA mimics were first transfected using DharmaFECT1 (Dharmacon, Thermo Fisher), and approximately 10 h later 200 ng luciferase reporter plasmids were transfected. Cells were lysed 36 h later and processed for luciferase assay using the Dual-Luciferase Reporter Assay System (Promega).
Statistical analysis
The data were presented as mean±S.D. except where indicated. We performed two-tailed unpaired Student's T-test to determine statistical significance. P-value <0.05 was considered as statistically significant.
Small molecules
The TGF-beta and GSK3 pathways were inhibited with SB431542 (15 μM), RepSox (25 μM), CHIR99021 (3 μM), and BIO (3 μM). The caspase-3 activator PAC1 was used at a final concentration of 5 μM. In the ESC differentiation assays, all the inhibitors were added along with the miRNAs and incubated for 3 days. In the other assays (spreading kinetics and cell attachment), cells were treated with small molecules for 2 days before processing.
Acknowledgments
We thank the members of Wang laboratory for critical reading and discussion of the manuscript. YW was supported by funds from the Chinese Ministry of Science and Technology (2011CBA01100 and 2012CB966700) and the NSFC (Project 31221002); CM by a National Science Foundation predoctoral training award; AS by a CIRM predoctoral training award (TG2-01153); SAO by the National Institutes of Health (RO1CA136577 and R01DK095306), an American Cancer Society Research Scholar Award, and the Juvenile Diabetes Research Foundation; and RB by the National Institutes of Health (R01 NS057221, GM101180) and CIRM (RN2-00906).
Author Contributions
W-TG performed all the experiments with the help from other authors. All authors were involved in the interpretation of data. YW and RB conceived the project and wrote the manuscript with help from W-TG.
Glossary
- AP
alkaline phosphatase
- Bak
BCL2-antagonist/killer 1
- Bax
BCL2-associated X protein
- cDNA
complementary DNA
- Dgcr8
DiGeorge syndrome critical region gene 8
- Dox
doxycycline
- EMT
epithelial–mesenchymal transition
- ESC
embryonic stem cell
- ESCC miRNA
ESC cell cycle regulating miRNA
- Gsk3
glycogen synthase kinase 3
- iPSC
induced pluripotent stem cell
- MET
mesenchymal–epithelial transition
- PAC1
procaspase-activating compound 1
- Rb
retinoblastoma
- qRT-PCR
quantitative reverse transcription-PCR
- Tgf
transforming growth factor
- UTR
untranslated region
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- 1NIH, Stem Cell BasicsIn In Stem Cell Information[http://stemcells.nih.gov/info/basics/pages/basics3.aspx]Bethesda, MD: National Institutes of Health, U.S. Department of Health and Human Services, 2009. [Google Scholar]
- 2Guo WT, Wang XW, Wang Y. Micro-management of pluripotent stem cells. Protein Cell 2014; 5: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA 2005; 102: 12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 2005; 19: 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet 2007; 39: 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 2008; 40: 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Wang Y, Melton C, Li YP, Shenoy A, Zhang XX, Subramanyam D et al. miR-294/miR-302 promotes proliferation, suppresses G1-S restriction point, and inhibits ESC differentiation through separable mechanisms. Cell Rep 2013; 4: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010; 463: 621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 2009; 27: 459–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 2011; 29: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Liao B, Bao X, Liu L, Feng S, Zovoilis A, Liu W et al. MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem 2011; 286: 17359–17364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011; 8: 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011; 8: 633–638. [DOI] [PubMed] [Google Scholar]
- 14Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 2008; 15: 259–267. [DOI] [PubMed] [Google Scholar]
- 15Hanina SA, Mifsud W, Down TA, Hayashi K, O'Carroll D, Lao K et al. Genome-wide identification of targets and function of individual MicroRNAs in mouse embryonic stem cells. PLoS Genet 2010; 6: e1001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol 2011; 18: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Judson RL, Greve TS, Parchem RJ, Blelloch R. MicroRNA-based discovery of barriers to dedifferentiation of fibroblasts to pluripotent stem cells. Nat Struct Mol Biol 2013; 20: 1227–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet 2013; 14: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139: 871–890. [DOI] [PubMed] [Google Scholar]
- 20Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119: 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Kalluri R. EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest 2009; 119: 1417–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest 2009; 119: 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 2009; 119: 1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol 2013; 25: 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Shenoy A, Blelloch R. Genomic analysis suggests that mRNA destabilization by the microprocessor is specialized for the auto-regulation of Dgcr8. PLoS One 2009; 4: e6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 2008; 22: 2773–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008; 134: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Calabrese JM, Seila AC, Yeo GW. & Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA 2007; 104: 18097–18102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Zheng GX, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S et al. A latent pro-survival function for the mir-290-295 cluster in mouse embryonic stem cells. PLoS Genet 2011; 7: e1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Putt KS, Chen GW, Pearson JM, Sandhorst JS, Hoagland MS, Kwon JT et al. Small-molecule activation of procaspase-3 to caspase-3 as a personalized anticancer strategy. Nat Chem Biol 2006; 2: 543–550. [DOI] [PubMed] [Google Scholar]
- 31Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Faherty N, Curran SP, O'Donovan H, Martin F, Godson C, Brazil DP et al. CCN2/CTGF increases expression of miR-302 microRNAs, which target the TGFβ type II receptor with implications for nephropathic cell phenotypes. J Cell Sci 2012; 125: 5621–5629. [DOI] [PubMed] [Google Scholar]
- 33Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D et al. A small-molecule inhibitor of Tgf-Beta signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell 2009; 5: 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Zavadil J, BÖttinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene 2005; 24: 5764–5774. [DOI] [PubMed] [Google Scholar]
- 35Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res 2009; 19: 156–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol 2012; 13: 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem J 2001; 359: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol 2001; 2: 769–776. [DOI] [PubMed] [Google Scholar]
- 39Benetti R, Gonzalo S, Jaco I, Muñoz P, Gonzalez S, Schoeftner S et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 2008; 15: 268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Liu X, Sun H, Qi J, Wang L, He S, Liu J et al. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat Cell Biol 2013; 15: 829–838. [DOI] [PubMed] [Google Scholar]
- 41Esteban MA, Bao X, Zhuang Q, Zhou T, Qin B, Pei D et al. The mesenchymal-to-epithelial transition in somatic cell reprogramming. Curr Opin Genet Dev 2012; 22: 423–428. [DOI] [PubMed] [Google Scholar]
- 42Li R, Liang J, Ni S, Zhou T, Qing X, Li H et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010; 7: 51–63. [DOI] [PubMed] [Google Scholar]
- 43Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010; 7: 64–77. [DOI] [PubMed] [Google Scholar]
- 44Eastham AM, Spencer H, Soncin F, Ritson S, Merry CL, Stern PL et al. Epithelial-mesenchymal transition events during human embryonic stem cell differentiation. Cancer Res 2007; 67: 11254–11262. [DOI] [PubMed] [Google Scholar]
- 45Spencer HL, Eastham AM, Merry CL, Southgate TD, Perez-Campo F, Soncin F et al. E-cadherin inhibits cell surface localization of the pro-migratory 5T4 oncofetal antigen in mouse embryonic stem cells. Mol Biol Cell 2007; 18: 2838–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Martínez-Estrada OM, Lettice LA, Essafi A, Guadix JA, Slight J, Velecela V et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet 2010; 42: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Li X, Zhu L, Yang A, Lin J, Tang F, Jin S et al. Calcineurin-NFAT signaling critically regulates early lineage specification in mouse embryonic stem cells and embryos. Cell Stem Cell 2011; 8: 46–58. [DOI] [PubMed] [Google Scholar]
- 48Gill JG, Langer EM, Lindsley RC, Cai M, Murphy TL, Kyba M et al. Snail and the microRNA-200 family act in opposition to regulate epithelial-to-mesenchymal transition and germ layer fate restriction in differentiating ESCs. Stem Cells 2011; 29: 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Yamane T, Dylla SJ, Muijtjens M, Weissman IL. Enforced Bcl-2 expression overrides serum and feeder cell requirements for mouse embryonic stem cell self-renewal. Proc Natl Acad Sci USA 2005; 102: 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Ardehali R, Inlay MA, Ali SR, Tang C, Drukker M, Weissman IL et al. Overexpression of BCL2 enhances survival of human embryonic stem cells during stress and obviates the requirement for serum factors. Proc Natl Acad Sci USA 2011; 108: 3282–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 2005; 7: 165–171. [DOI] [PubMed] [Google Scholar]
- 52Fujita J, Crane AM, Souza MK, Dejosez M, Kyba M, Flavell RA et al. Caspase activity mediates the differentiation of embryonic stem cells. Cell Stem Cell 2008; 2: 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Li F, He Z, Shen J, Huang Q, Li W, Liu X et al. Apoptotic caspases regulate induction of iPSCs from human fibroblasts. Cell Stem Cell 2010; 7: 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Westphal D, Dewson G, Czabotar PE, Kluck RM. Molecular biology of Bax and Bak activation and action. Biochim Biophys Acta 2011; 1813: 521–531. [DOI] [PubMed] [Google Scholar]
- 55Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 2010; 11: 621–632. [DOI] [PubMed] [Google Scholar]
- 56Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 2005; 121: 465–477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.