Abstract
Infectious pancreatic necrosis virus (IPNV) is the cause of one of the most prevalent diseases in farmed Atlantic salmon (Salmo salar). A quantitative trait locus (QTL) has been found to be responsible for most of the genetic variation in resistance to the virus. Here we describe how a linkage disequilibrium-based test for deducing the QTL allele was developed, and how it was used to produce IPN-resistant salmon, leading to a 75% decrease in the number of IPN outbreaks in the salmon farming industry. Furthermore, we describe how whole-genome sequencing of individuals with deduced QTL genotypes was used to map the QTL down to a region containing an epithelial cadherin (cdh1) gene. In a coimmunoprecipitation assay, the Cdh1 protein was found to bind to IPNV virions, strongly indicating that the protein is part of the machinery used by the virus for internalization. Immunofluorescence revealed that the virus colocalizes with IPNV in the endosomes of homozygous susceptible individuals but not in the endosomes of homozygous resistant individuals. A putative causal single nucleotide polymorphism was found within the full-length cdh1 gene, in phase with the QTL in all observed haplotypes except one; the absence of a single, all-explaining DNA polymorphism indicates that an additional causative polymorphism may contribute to the observed QTL genotype patterns. Cdh1 has earlier been shown to be necessary for the internalization of certain bacteria and fungi, but this is the first time the protein is implicated in internalization of a virus.
Keywords: IPN, QTL, salmon, cadherin, disease resistance
VIRAL infections are common in farmed fish and pose a great threat to aquaculture industries and to animal welfare. In the farming of Atlantic salmon, the infectious pancreatic necrosis virus (IPNV) is a major loss factor, being prevalent in most countries where the species is farmed. IPNV was the first fish virus to be characterized (Wolf et al. 1960) and remains one of the best studied (Crane and Hyatt 2011). It belongs to the Aquabirnavirus genus of the Birnaviridae family and has a dsRNA genome enclosed in a nonenveloped, single-shelled icosahedral particle. The IPNV genome is composed of two segments, where segment A encodes a polyprotein that is cotranslationally cleaved into VP2 and VP3. VP2 is the outer capsid protein and is suggested to be the cell attachment protein (Dobos 1995). IPNV appears to enter host cells by receptor-mediated endocytosis after specific attachment to fish cells (Granzow et al. 1997), but no cellular recognition molecule has been identified. The virus can be transmitted both vertically and horizontally.
Atlantic salmon demonstrate two IPNV susceptibility windows, initially after first feeding of fry (∼0.2 g) and then soon after sea transfer of postsmolts (∼100 g) (Roberts and Pearson 2005). Experimental challenge at these specific stages can lead to >90% mortality, whereas the fish appear to be largely resistant to the virus outside the two susceptibility windows. The IPNV virion has a remarkably high physical resistance and is difficult to remove from infected premises, making recurrent outbreaks common (Murray 2006). No specific treatment is available, and although commercial vaccines against IPN have been produced, their effects are variable and they do not prevent losses completely (Sommerset et al. 2005).
There is genetic variation in resistance to IPN in commercial Atlantic salmon populations (Okamoto et al. 1993; Kjøglum et al. 2008; Guy et al. 2009), and breeding companies have selected their broodstock for increased resistance to IPN for the past 10–20 years (Storset et al. 2007). The selection has been based on survival rates of disease-challenged siblings of breeding candidates since direct phenotypic testing of the breeding candidates is not possible due to the risk of vertical transmission of the virus (Smail and Munro 2008). Consequently, genetic selection for IPN resistance has been restricted to between-family genetic variation, ignoring the equally large within-family genetic variation. Motivated by the need for selection criteria that could be employed directly on the breeding candidates, two research groups independently discovered a major QTL for IPN resistance in Atlantic salmon, explaining 80–100% of the genetic variation in both susceptibility windows (fry and postsmolt) (Houston et al. 2008; Moen et al. 2009). Here, we describe the industrial implementation of this QTL in marker-assisted selection (MAS) for IPN resistance, documenting a dramatic reduction in number of IPN outbreaks in Atlantic salmon farms. We also describe how the subsequent fine mapping of the QTL region led to the identification of epithelial cadherin (cdh1) as the causative gene. The gene product, Cdh1, has been intensively studied because of its pivotal role in tissue morphogenesis and disease (Harris 2012). This paper confirms direct binding between IPNV and Cdh1, revealing a novel mechanism in viral attachment and entry.
Materials and Methods
IPN challenge test, 2005
In 2005, a challenge test was performed on 400 full-sibling groups of fry as described earlier (Moen et al. 2009). Genetic material from this challenge test formed the basis for the development of a DNA-marker-based test for deducing QTL alleles.
Genotyping
Three microsatellites, located within the QTL region, were used for deducing QTL alleles: Alu333 (PET-TTCATAGTCCAAGAACAGTG, GCTGAGTTTACATTACACCTG, GenBank ID AY543859.1); Ssa0285BSFU (NED-CAGAACACAAACAGAGCT, CAACAGGGATCTCTCAACAT); and Ssa0374BSFU/ii (VIC-GAGTAGGCAACTGAAACAGG, CAAACTCATTCCCTCACATT). The three microsatellites were genotyped in a single PCR [94° for 5 min, 30 amplification cycles (94° for 30 sec, + 54° for 30 sec, + 72° for 1 min), 60° for 45 min], followed by fragment analysis using an ABI3730 DNA sequencer (Life Technologies, Carlsbad, CA) and GeneMapper 4.0 software (Life Technologies). SNPs were genotyped using the iPLEX platform from Agena (San Diego), following the standard iPLEX protocol provided by Agena. The sequences used for constructing iPLEX PCR and extension primers can be found in supporting information, File S1. The primers were designed using Assay Design Suite v2.0 from Agena.
Development of a marker-based test for deducing QTL alleles
Linkage phases between alleles at the three microsatellites Alu333, Ssa0285BSFU, and Ssa0374BSFU/ii were determined in a cohort of 296 individuals (mapping parents) that each had 11–44 (mean 26) genotyped offspring tested for resistance to IPN in an experimental challenge test (selective genotyping: the genotyped individuals comprised the 10% most and 10% least IPN-resistant individuals from within each challenge-tested full-sib group). This was done using a custom-made Visual Basic for Applications script, the algorithm of which was as follows: first determine origin (paternal or maternal) of alleles in offspring (obvious except when both parents were heterozygous for a biallelic polymorphism), then move from polymorphism to polymorphism according to their physical order, designating the most frequently observed linkage phase to be the true phase for the parent in question. Linkage phases between three-microsatellite haplotypes and alleles at the QTL (assuming a single QTL with two alleles) were likewise determined by tracing the cosegregation of three-microsatellite haplotypes and the phenotype (affected/resistant) from the 117 mapping parents found to be QTL heterozygous (at P < 0.01) to their offspring. All three-microsatellite haplotypes were predominantly linked to one or the other allele at the QTL. When there was ambiguity, i.e., whenever a three-microsatellite haplotype was found to be linked to one QTL allele in some individuals and to the other QTL allele in other individuals, the predominant linkage phase was chosen. For a few haplotypes, not represented among QTL-heterozygous mapping parents, linkage phases were determined using a likelihood-based approach (Moen 2010).
Deduction of QTL genotypes using marker-based test
Breeding candidates and other individuals were genotyped using three particular microsatellites, as described above. The microsatellite alleles present in each animal were phased using PHASE (Stephens and Scheet 2005), incorporating the linkage phases already found in the 296 mapping parents (File S2) as a reference. A haplotype (i.e., combination of alleles from the three microsatellites) was accepted if the likelihood of it being present in the animal in question was>0.8. QTL genotypes were assigned by replacing three-microsatellite haplotypes with the QTL allele found to be linked to that haplotype in the reference dataset (derived from the 117 QTL-heterozygous mapping parents). No QTL genotype was assigned to those individuals carrying one or two 3-microsatellite haplotypes that were not represented in the reference dataset.
Construction of a reference sequence for the QTL region
A merged SNP-microsatellite linkage map was constructed by genotyping the microsatelllite markers previously used to fine map the QTL for IPN resistance (Moen et al. 2009) in the genetic material used to create the original SNP-based linkage map (Lien et al. 2011). Using this map, and results from the initial fine mapping of the QTL (Moen et al. 2009), we defined the region flanked by markers Ssa0680BSFU and BHMS217 to be the QTL region (File S3).
Sequences of the markers (SNPs and microsatellites) identified within the IPN-resistance QTL region were screened for suspected and known repetitive elements in salmonids using RepeatMasker and the salmon v2.0 repeat library (http://lucy.ceh.uvic.ca/repeatmasker/cbr_repeatmasker.py). Bacterial artificial chromosome clones (BACs) containing these markers were identified from the Atlantic salmon CHORI-214 BAC library (Thorsen et al. 2005) using oligonucleotide probes designed from the masked sequences flanking the markers. The probes and PCR primers to amplify genomic DNA in the IPN-resistance QTL region were designed using Primer3 v0.4.0 software (Koressar and Remm 2007; Untergrasser et al. 2012) (File S4). The Atlantic salmon BAC library was screened as described by Phillips et al. (2009). Briefly, 5′-end 32P-labeled probes were added to prehybridized BAC filters in 5× saline–sodium citrate buffer (SSC), 0.5% sodium dodecyl sulfate (SDS), and 5× Denhardt’s solution and incubated overnight at 65°. To remove nonhybridized probe, three 1-hr washes were performed at 50° in 1× SSC and 0.1% SDS. The filters were exposed overnight to phosphor screens, which were subsequently imaged using a Typhoon Trio + Variable Mode Imager (GE Healthcare). Hybridization positive BACs were picked from the library and incubated with shaking (at 250 rpm) in 5 ml of LB medium at 37° overnight in presence of 20 µg/ml chloramphenicol. Grown clones were stored as 15% glycerol stocks at −80°. The presence of IPN-resistance QTL markers in the BACs was verified by PCR using 1/40 dilution of the glycerol stock as a template. BAC clones containing the IPN-resistance QTL markers were associated with fingerprint scaffolds fps378, fps787, fps2100, fps334, and fps591 in the Atlantic salmon physical map (Ng et al. 2005) as shown in the Atlantic Salmon database: ASalBase (www.asalbase.org). To construct minimum tiling paths (MTPs) across each fingerprint scaffold, a subset of clones with available SP6 and T7 end sequence were chosen to design sequence tagged sites (STSs) for PCR amplification. The relative orientation of the BACs was determined on the basis of which STS they had in common. The MTP for each fingerprint scaffold represents the minimum overlapping BACs required to cover the entire fingerprint scaffold. BAC end sequences of the outermost clones in the scaffolds were used to design oligonucleotide probes to check for possible joins between fingerprint scaffolds. Procedures to identify and check STS positive BACs were the same as described above. It was possible to join four scaffolds: fps378, fps787, fps2100, and fps334 (File S5). DNA was extracted from 29 BACs covering the IPN-resistance QTL region using the Qiagen Large-Construct kit and following manufacturer’s protocol.
The reference sequence was annotated by first running nucleotide BLAST (Altschul et al. 1990) against a set of full-length cDNA sequence contigs (assembled from EST sequences and kindly provided by Ben Koop, University of Victoria), and then mapping exons of each positive nucleotide BLAST hit (i.e., cDNA sequence) onto the QTL reference sequence using EST2GENOME (Mott 1997).
The 29 BACs were individually sequenced using an Illumina HiSequation 2000 (Illumina, San Diego; paired-end sequencing, 350-bp fragment length). Residual adapter and poor quality sequences (quality score threshold of 10) were trimmed from the raw data using the FASTX toolkit (Pearson et al. 1997), and reads shorter than 20 bp were discarded. De novo assembly of each BAC sequence was performed with clc_novo_assemble from the CLC Assembly Cell Software suite (version 4 beta, http://www.clcbio.com/) using the default parameters and a minimum contig size of 200. Reads from each BAC were realigned to the BAC contigs using clc_ref_assemble_long (discarding multiple hit alignments), and subsequently contigs with lower than 100× coverage were removed. Individual BAC sequences were then combined with Phrap software (http://www.phrap.org/phredphrapconsed.html#block_phrap) using the parameters -new_ace -bypasslevel 0 -forcelevel 0 -minmatch 50 -node_seg 4 -node_space 2 -repeat_stringency 0.97 -minscore 50 -maxgap 30 -revise_greedy. Later, as they became available, reads provided (and made public) by the International Consortium to Sequence the Atlantic Salmon Genome (ICSASG; http://www.icisb.org/salmon-sequencing-project) were used in order to make a de novo assembly of the Atlantic salmon genome; this was done using the Celera assembler (Myers et al. 2000). Scaffolds from the BAC sequencing and the whole genome sequencing (ICSASG) were merged together using CAP3 software (Huang and Madan 1999) and positioned to the BAC-based physical map. To bridge gaps in the map and produce a reference sequence spanning the whole QTL region, we developed a pipeline that utilized scaffolds from the QTL region on Atlantic salmon chromosome 26 (Ssa26) together with scaffolds in the corresponding homeologous region on Ssa11 to build super scaffolds in both regions. The pipeline used the LASTZ software (Harris 2007) to identify the homeologous regions and custom Python scripts (H. Grove et al., unpublished results) were used in order to construct the super scaffolds. Scaffolds were repeat masked prior to the alignment using RepeatMasker (http://lucy.ceh.uvic.ca/repeatmasker/cbr_repeatmasker.py) and a repeat library was generated for Atlantic salmon (kindly provided by Ben Koop, University of Victoria).
Fine mapping using next-generation sequencing
Twenty-two individuals deduced by the marker-based test to have QTL genotype QQ (i.e., two copies of the high-resistance allele), and 23 individuals likewise deduced to have QTL genotype qq, were selected for whole genome resequencing. In addition to having a particular QTL genotype, the individuals all met the following two criteria. First, the survival rate of their (IPN-challenge tested) offspring was within the upper or lower end, respectively, of the distribution of survival rates typical for genotype group QQ or qq. Second, within each pool, as many three-microsatellite haplotypes as possible were to be represented, so that each pool would cover as much of the genetic variation in the population as possible. The 45 individuals were individually sequenced on HiSequation 2000 (paired-end 2 × 100 bp reads), producing an average of 5.9 Gb (range 0.8–11.6, standard deviation 1.9) of sequence per individual that represented ∼2× genome coverage on average. The raw sequence reads were submitted to the NCBI Sequence Read Archive (File S8). The fragments (reads + interread fragments) were 336 bp long on average. The reads were aligned using Bowtie2 (Langmead and Salzberg 2012) in end-to-end mode with very-sensitive preset options, with the exception of the -N option (maximum number of mismatches in the seed) that was set at 1, and the -X option (maximum fragment length) that was set at 800. Prior to alignment, the reference was repeat masked. SNPs (and other polymorphisms) were detected using freebayes (http://arxiv.org/abs/1207.3907) using the parameter string “-use-best-n-alleles 2 –min-alternate-total 3 –pooled-discrete-ploidy 90.” Based on the output from freebayes, a Pearson’s chi-squared test was used to test for independence between the SNP allele and QTL allele at each individual SNP. The most significant SNPs from this test were genotyped in the 117 QTL-heterozygous mapping parents and in their IPN-challenge-tested offspring (20–40 offspring of each parent), using Sequenom iPlex assays (the SNPs and their flanking sequences can be found in File S1, and the (phased) genotypes can be found in File S6). From the SNP data, haplotypes comprising a QTL allele and a SNP allele were derived by following the segregation of SNP alleles into the groups of most- and least-IPN-resistant offspring (using a custom-made Python script). A Pearson’s chi-squared test was used to test for independence between QTL allele and SNP allele within each haplotype.
Search for putative secondary polymorphisms
A SNP-chip using Axiom technology from Affymetrix (San Diego) containing 930,000 putative SNPs from Atlantic salmon (or more precisely, two SNP chips, each with 465,000 SNPs) had been developed on the basis of resequencing data from 29 diverse AquaGen individuals and three double haploid individuals (T. Moen et al., unpublished results). Approximately 650,000 of the SNPs were found to be polymorphic in the AquaGen population. The arrays were used to genotype 200 mapping parents, including 16 individuals that were heterozygous for the QTL (Qq) and homozygous (qq) for an already identified (putative) primary causative mutation termed SNP4 (i.e., these individuals were QqQTL/qqSNP4) and 26 individuals that were homozygous for the susceptibility allele of the QTL (qq) and also homozygous (qq) for SNP4 (i.e., these individuals were qqQTL/qqSNP4). This dataset was used to identify putative “secondary” functional SNPs that could explain the QTL-heterozygous nature of QqQTL/qqSNP4 individuals. For each SNP, the (pseudo) likelihood of the genotypes observed at the 16 QqQTL/qqSNP4 individuals and 26 qqQTL/qqSNP4 individuals was calculated, using a custom-made Python script, in two ways: (H1) by assuming that the SNP was truly QqSNP for all 16 QqQTL/qqSNP4 individuals and qqSNP for all 26 qqQTL/qqSNP4 individuals, and assuming a particular error rate (more precisely, the likelihood was multiplied with 0.985, 0.01, or 0.005 depending on whether the 0, 1, or 2 allele substitutions had to be made to move from the observed genotype to the “requested” genotype); and (H0) by assuming that the genotype was random, i.e., determined by the allele frequencies observed in the population (allele frequencies were calculated on the basis of all 200 genotyped mapping parents, and genotype probabilities were calculated from allele frequencies by assuming Hardy–Weinberg disequilibrium). The ratio of H1 to H0 was taken as a measure of how likely H1 was relative to H0.
A subset of the most likely secondary polymorphisms identified in this manner was genotyped in all mapping parents. All possible pairwise combinations of a putative primary mutation and a putative secondary mutation were tested (using a custom-made Python script) for their association with the QTL: First, two-polymorphism haplotypes were constructed in the 117 QTL-heterozygous mapping parents. Next, the resulting four-allele haplotypes were transformed into a two-allele system (QHAP and qHAP) by naming the haplotype alleles according to the most prevalent linkage phase between the two-polymorphism haplotypes and QTL. Finally, a Pearson’s chi-squared test for independence between haplotype and QTL alleles was performed.
IPN challenge test, 2007
A challenge test was performed to compare IPN-induced mortalities of salmon originating from eggs selected using MAS with IPN-induced mortalities of standard (not selected using MAS) eggs. In this challenge test, the MAS fry (offspring of random females and males that were homozygous for the resistance allele at the QTL) were compared to standard fry (offspring of random males and random females) and IPN susceptible fry (offspring of random females and males that were homozygous for the susceptibility allele at the QTL). Fry at an average weight of 0.2 g at start feeding were transferred to a research station (Havbruksstasjonen, Tromsø, Norway) and allowed 1 week of acclimation before challenge. The trial was conducted by Nofima AS (Tromsø, Norway). A total of 1,600 fry were included in the study, distributed across eight tanks each containing 200 fry. Three tanks contained MAS fry, three tanks contained standard fry, and two tanks contained IPN-sensitive fry. One tank held standard fry that were kept as uninfected controls, whereas the fry in the remaining tanks were bath challenged by adding IPN virus at a concentration of 105 TCID50/ml water. Normal water flow was paused for 3 hr while the water was aerated. Two tanks holding MAS fry, two tanks holding standard fry, and one tank holding IPN-sensitive fry were reserved for mortality registrations. One tank with IPN-sensitive and one tank with MAS fry were reserved for sampling of alive but preferably diseased fish; 10 fish were sampled from each tank 2, 4, and 12 days after infection, with an additional 20 fish sampled from the same two tanks 6, 7, 8, and 10 days after infection. The livers of the sampled fish were dissected and placed in RNAlater (Qiagen) for later determination of viral load and gene expression. The trial was terminated 34 days after challenge. At termination, 10 surviving fry from each of the eight tanks were sampled for determination of carrier status. Quantification of viral loads of infected fish was determined by a TaqMan real-time PCR assay provided by an accredited commercial lab (PatoGen Analyze AS, Ålesund, Norway) and compared to the expression of a reference gene (elongation factor 2-α). The tails of all sampled fish were frozen before DNA extraction and subsequent deduction of QTL alleles. The frequencies of the high-resistance allele in the test groups were determined by genotyping 30 random fry sampled prechallenge.
Tissue culture and immunofluorescence
Juvenile genotyped Atlantic salmon at the freshwater stage were anesthetized according to Norwegian regulations. A liver biopsy was taken immediately after sacrification and embedded in ultralow melting temperature agarose (Sigma-Aldrich). Slices of 300 μm were prepared in ice cold Hank’s buffer using a VF300 tissue slicer (Precisionary Instruments) and transferred to Netwells (Corning, NY) with L15 Glutamax with 5% FBS and 1% Penstrep (Invitrogen) and cultured at 15° for 3 days. The liver slices were then challenged with IPNV (TCID 50 ml−1) for 2 days, before being thorough washing in ice-cold Hank’s buffer and fixation by freeze substitution. Localization of Cdh1-1 and route of IPNV entry was visualized by immunofluorescence using an IPNV polyclonal rabbit antibody (kindly provided by Øystein Evensen, the Norwegian Veterinary Institute, Oslo), a Cdh1-1-specific antibody (Dako), and a Clathrin light chain antibody (Abcam). Image stacks were captured on a Zeiss Axio Observer Z1 microscope and deconvolved in Zen Blue software (Carl Zeiss Microimaging).
Coimmunoprecipitation
Coimmunoprecipitation was carried out using 80 mg/ml of frozen liver in NP40 buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol). The tissues were lysed using a tissue disruptor (Qiagen) and centrifuged at 4 °C for 10 min at 6000 rpm before 1 ml of lysate was mixed with 5 μl of IPNV. After a 30-min incubation at 4 °C with agitation, the sample was mixed with Protein A-coated Dynabeads (Invitrogen) coupled to IPNV antibodies. After a second 30-min incubation and washing, the bound proteins were eluted and subjected to Western blotting using a Cdh1 antibody.
Estimation of viral load
An IPN challenge test was performed in 2007, as described above. Fish were sampled at 34 days postchallenge (i.e., at termination of the test). The livers of the sampled fish were carefully dissected and placed in RNAlater (Qiagen) for later determination of viral load. Ten surviving fry from each of the eight tanks were sampled for determination of carrier status. Quantification of viral loads of infected fish was determined by a TaqMan real-time PCR assay provided by an accredited commercial lab (PatoGen Analyze AS, Ålesund, Norway) and compared to the expression of a reference gene (elongation factor 2-α).
Results
Development of a marker-based test for deducing the QTL allele
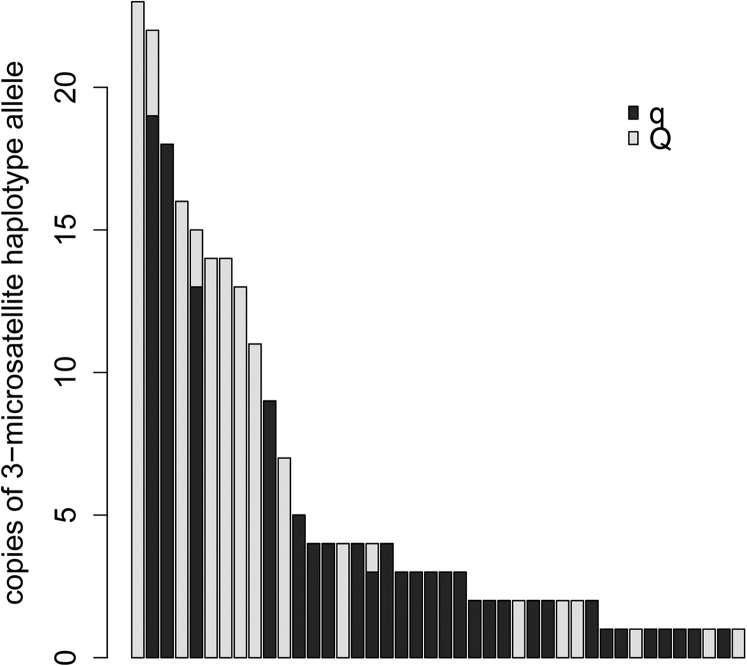
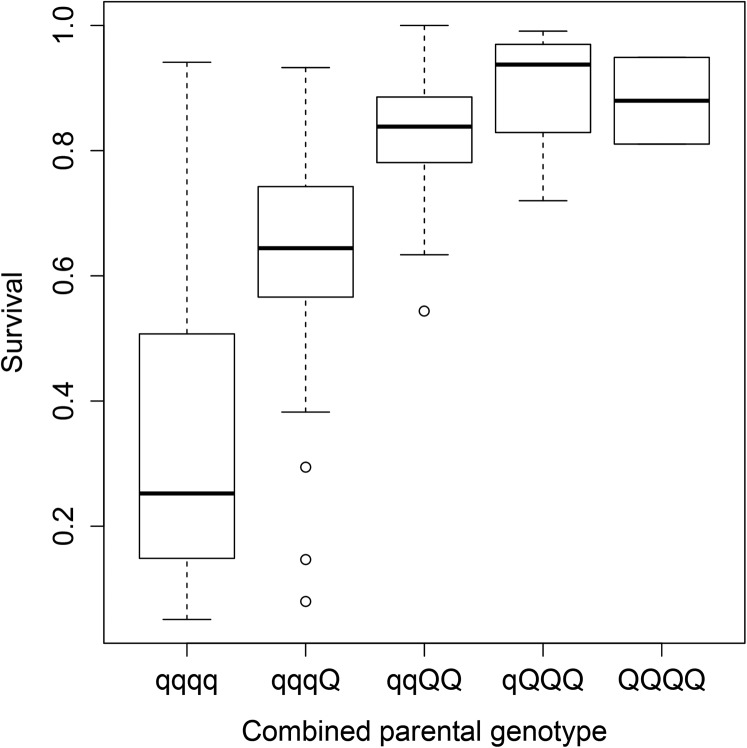
Moen et al. (2009) earlier found population-level linkage disequilibrium between the QTL for IPN resistance and haplotypes constituted by three microsatellites located within the QTL region. The three microsatellites were now genotyped in the first 10 mortalities and the last 10 survivors from each of 207 full-sibling groups tested for IPN resistance in a controlled challenge test performed at the fry stage (100 individuals challenge tested per full-sib group), as well as in the 146 mothers and 150 fathers of these groups, facilitating the identification of 117 QTL-heterozygous mapping parents (Pearson’s chi-squared test for independence between affected/survivor status and allele inherited from common parent within half-sibling groups, α = 0.01). Haplotypes found in QTL-heterozygous parents were designated as Q (high resistance) or q (low resistance) according to the coinheritance of haplotype and QTL allele within QTL-heterozygous parents. Additionally, some haplotypes found only in homozygous mapping parents were designated as Q or q using maximum likelihood, as described elsewhere (Moen 2010). Haplotypes were found to be predominantly linked to either Q or q (Figure 1). When the most likely linkage phases between haplotype and QTL alleles were extrapolated to all 296 parents of the genotyped sibling groups, in addition to 140 parents of not-genotyped sibling groups coming from the same IPN challenge test, a strong correlation was found between the challenge-test survival rates within full-sibling groups and the number of Q alleles shared by the parents of the full-sibling groups (Figure 2). Thus, the three microsatellites formed the basis for a test that could predict QTL genotypes of individual animals at the population level. In this test, individuals were genotyped for the three microsatellites, haplotypes carried by the individuals were inferred using PHASE (Stephens and Scheet 2005), and QTL alleles were assigned according to the key provided in the reference dataset (i.e., by the predominant haplotype–QTL linkage relationships found in QTL-heterozygous mapping parents). The test was applicable on all individuals originating from the population that the reference dataset was derived from (the breeding nucleus of the Atlantic salmon egg provider AquaGen), as long as the haplotypes carried by the individual were represented in the reference dataset.
Figure 1.
Number of copies of each microsatellite haplotype found among QTL-heterozygous mapping parents. Each haplotype copy has been labeled according to the QTL allele (Q or q) it is linked to.
Figure 2.
Effect of QTL genotype on IPN resistance. Survival rates, in a challenge test for IPN resistance performed on Atlantic salmon fry, of offspring of parental pairs sharing 0, 1, 2, 3, or 4 copies of the high-resistance allele (Q) of the QTL. Thick line, median; box, quartiles; whiskers, 1.5 times distance from median to quartile; circles, data points outside the “whiskers” region.
Dominance of the favorable allele
To test whether the QTL displayed a dominance effect, the frequencies of the different QTL genotypes were estimated within each full-sib family, using the deduced parental QTL genotypes and assuming Mendelian segregation ratios (the frequencies could not be calculated directly, since only 20% of the individuals within each full-sib group had been sampled and genotyped). Using a weighted (by family size) regression model, estimated mortalities (±SE) within the challenge-tested offspring were 0.66 ± 0.02, 0.09 ± 0.02, and 0.05 ± 0.06 for individuals deduced to have genotypes qq, Qq, and QQ, respectively. The majority of the between-family variation (R2 = 0.56) was explained by simple presence/absence of the Q allele (P < 0.0001), with no significant difference between Qq and QQ genotypes (P = 0.61). Hence, the favorable allele appears to be largely dominant, at least under conditions similar to those of the challenge test in question. However, based on these data, it cannot be ruled out that the observed dominance may be due to interactions between genetic resistance of the host and a potential nonlinear dose–response relationship for observed mortality (Bishop and Wooliams 2010) (i.e., even a moderate increase in resistance may dramatically increase survival at low environmental pathogen doses).
Construction of a reference sequence for the QTL region
The microsatellite-based test for deducing QTL genotypes provided a foundation for further fine mapping of the QTL. However, since a reference sequence for the Atlantic salmon genome was not available at the time the project was initiated, fine mapping also required the construction of a reference DNA sequence of the QTL region. First, the microsatellite markers used for the initial interval mapping of the QTL (Moen et al. 2009) were mapped onto a SNP-based linkage map (Lien et al. 2011). The QTL region was defined as the region flanked by markers Ssa0680BSFU and BHMS217 on the merged microsatellite-SNP map, covering 22.2 cM (40.1–62.3) on the integrated female linkage map of Atlantic salmon chromosome 26 (www.asalbase.org; see File S3). From an Atlantic salmon physical map based on BACs (Ng et al. 2005), 29 BAC clones were identified that formed a partial minimal tiling path of this region. Based on next-generation sequencing of these BACs, a 3.3-Mbp partial reference sequence of the QTL region, distributed on 254 contigs, was generated. As they became available, contigs and raw reads produced by ICSASG (Davidson et al. 2010) were used to bridge and correct the BAC-based contigs and for ordering them into scaffolds. We also used sequence from the corresponding (homeologous) region on Atlantic salmon chromosome 11, to construct a super-scaffold covering 10.19 Mb of the QTL region, including 2.5 Mb of undefined sequence (gaps). Following the public release of the second version of the Atlantic salmon genome by ICSASG (downloadable from http://www.icisb.org/atlantic-salmon-genome-sequence/), all data and results were mapped against a 7.8-Mb long ICSASG scaffold (ccf1000000016_0_0) centered on the QTL region. This scaffold displayed very good concordance with the reference sequence for the QTL region.
Fine mapping and identification of a putative functional polymorphism
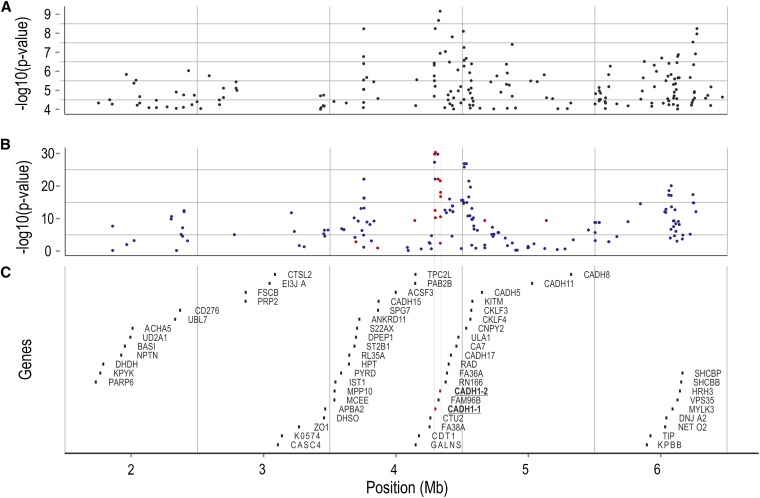
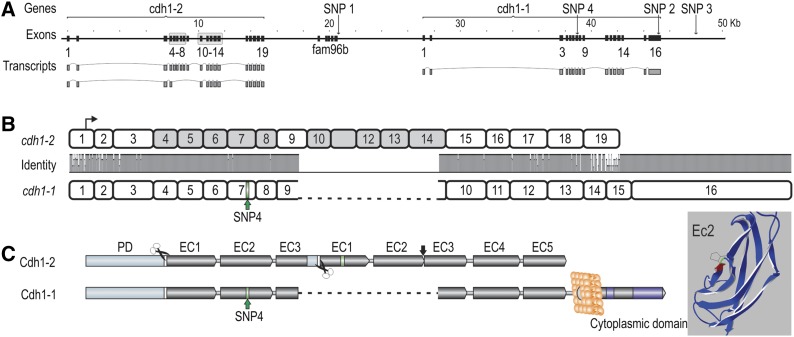
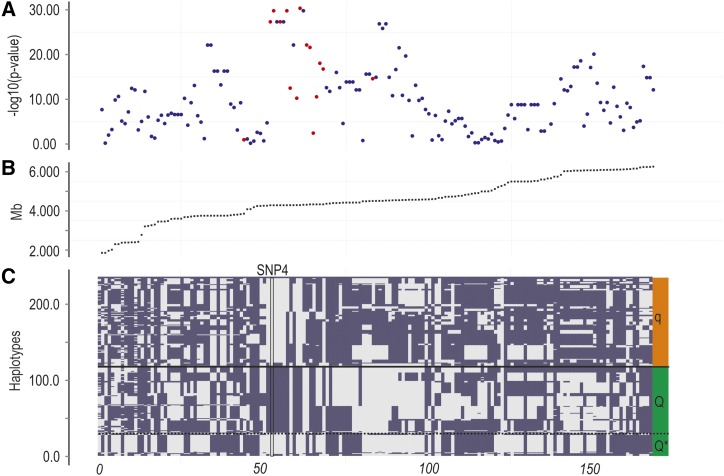
Utilizing the reference sequence of the QTL region, and individuals with deduced QTL genotype, next-generation sequencing was used to identify DNA polymorphisms more strongly associated to the QTL. Twenty-two individuals deduced to have QTL genotype QQ (QQQTL) and 23 individuals deduced to have QTL genotype qq (qqQTL) were individually sequenced to ∼2× genome coverage each, using Illumina technology (Illumina, San Diego). The resulting reads were aligned to the reference sequences of the QTL region, to identify DNA polymorphisms exhibiting large allele frequency differences between QQQTL and qqQTL individuals. Several highly significant DNA polymorphisms were found across the entire region (Pearson’s chi-squared test; Figure 3A). The most significant polymorphisms were genotyped in the 117 QTL-heterozygous mapping parents and in their (IPN challenged) offspring, and a Pearson’s test was applied on haplotyped data to confirm the results (Figure 3B; File S7). The three DNA polymorphisms most strongly associated with the QTL were located within a region harboring two epithelial cadherin genes (which we termed cdh1-1 and cdh1-2), flanking a fam96b gene-encoding family with sequence similarity 96, member B [Fam96b, also known as MSS19-Interacting Protein of 18 kDa (MIP18)]. These three DNA polymorphisms were in complete linkage disequilibrium (LD) with each other, and in strong LD (r2 = 0.64) with the QTL. One of the three IPN-associated polymorphisms (SNP1) was located 26 bp upstream of the first exon of the fam96b gene, another (SNP2) was located within the 3′ untranslated region of the cdh1-1 gene, while the third (SNP3) was located further downstream of the cdh1-1 gene (Figure 3C, Figure 4). Following further Illumina- and Sanger-based resequencing of genomic DNA and cDNA from individuals with deduced QTL genotypes (using genotyping of QTL-heterozygous mapping parents to confirm the results), we later discovered a fourth polymorphism (SNP4) in perfect LD with the first three, causing a serine-to-proline amino acid shift within the second extracellular cadherin domain of Cdh1-1 (Figure 4), in addition to other DNA polymorphisms in almost as strong LD with the QTL (Figure 3B). Phasing of genotypes from the 117 QTL-heterozygous mapping parents confirmed the presence of a single, high-frequency haplotype carrying the alternate linkage phase between the most significant SNPs and the QTL (Figure 5), indicating that the lack of a complete correspondence between SNP1–SNP4 and the QTL was due to a “deviating” haplotype segregating within the population, rather than being the result of genotyping errors or false negatives or positives in the test for segregation of the QTL.
Figure 3.
Fine mapping of QTL for IPN resistance. Positions on the x-axis are relative to the ICSASG scaffold, ccf1000000016_0_0. (A) Chi-square test for independence between SNP alleles and QTL alleles, performed on Illumina reads coming from QTL-heterozygous mapping parents, aligned against ccf1000000016_0_0. (B) Chi-square test for independence between SNP alleles and QTL alleles performed on phased genotype data coming from 117 QTL-heterozygous mapping parents. Blue dots, SNPs identified in the “scan,” based on Illumina sequence data; orange dots, SNPs identified by targeted Sanger-based resequencing of the “core” QTL region. (C) Genes located within the QTL region, according to an annotation based on BLASTN + EST2GENOME, performed against a set of 34,794 Atlantic salmon cDNA/EST sequences.
Figure 4.
The core QTL region, containing cdh1-1, cdh1-2, and fam96b. (A) Organization of the cdh1 and fam96b genes in the core QTL region, with positions relative to ICSASG scaffold, ccf1000000016_0_0. Main transcripts of the cdh1 genes are shown. Gray boxes correspond to the duplicated exon blocks in cdh1-2. (B) Homology between full-length cdh1-1 and cdh1-2 transcripts. The former is encoded by 16 exons, whereas cdh1-2 consists of 19 exons. Transcriptional start is denoted by arrow and SNP4 by green bar on cdh1-1. Note the duplicated blocks of exons in cdh1-2 not present in cdh1-1. (C) The left figure shows the domain architecture of Cdh1-1 and Cdh1-2, where the former contains all epithelial cadherin domains of a fully functional protein, including a prodomain (orange), proteolytic cleavage signals (white bar/scissor), extracellular (EC) domains 1–5 (gray), transmembrane domain (light gray circle), and cytoplasmic tail with catenin binding motifs (purple). The protein residue in EC2 corresponding to SNP4 has been labeled in green. The full-length Cdh1-2 protein contains several anomalies when compared to a classical Cdh1. The duplicated exon blocks leads to duplications of EC1–EC3, though EC3 is truncated and fused to EC1, interspersed by a proteolytic cleavage site. EC2 and EC3 of the second exon block are fused and the residues C-terminal to EC5 are absent; hence, the protein is most likely not anchored to the cell membrane. The 3D structure of Cdh1-1 EC2 with the SNP4 S167P mutation is highlighted in green. The amino acid colored in red corresponds to the side of the hydrophobic pocket involved in cis-dimerization (Harrison et al. 2011).
Figure 5.
Haplotypes in QTL-heterozygous mapping parents. (A) Chi-square test for independence between SNP alleles and QTL alleles, performed on phased genotype data coming from 117 QTL-heterozygous mapping parents. Blue dots, SNPs identified in the “scan,” based on Illumina sequence data; orange dots, SNPs identified by targeted Sanger-based resequencing of the “core” QTL region. (B) Position of polymorphisms within the reference sequence, the ICSASG scaffold, ccf1000000016_0_0. (C) Alleles at haplotypes. Blue, one allele; gray, the other allele; green, haplotypes containing QQTL; red, haplotypes containing qQTL; Q*, copies of the haplotype having the alternate linkage phase between SNP4 and the QTL.
Among the SNPs strongly associated with the QTL, SNP4 was the only one having an easily decipherable function. The sequence flanking SNP4 could be found in four different locations within the Atlantic salmon genome: in cdh1-1, in both copies of a duplicated exon block within chd1-2, and in a cdh1 gene found on chromosome 11. SNP4 was polymorphic both within cdh1-1 and within the second copy of the duplicated exon block of cdh1-2; however, the genotype patterns of the latter copy of SNP4 did not agree with the segregation pattern of the QTL in any way (data not shown). A comparison of Cdh1 sequences from Atlantic salmon and related species (Figure 6) reveals that the DNA variant corresponding to the susceptibility variant of the Atlantic salmon protein (serine) can be found in the most closely related species (brown trout), but not in the more distantly related species (rainbow trout), indicating that the proline residue (at the amino acid encoded by the SNP4 codon) represents the ancestral allele, i.e., the IPN-resistance variant of the protein is the ancestral variant.
Figure 6.

Amino acid sequence corresponding to exon 7 of the cdh1-1 gene from Atlantic salmon (Ssa_Cdh-1-1), aligned against corresponding sequences. The other sequences are the amino acid sequences of the corresponding exon within each of the two exon blocks of cdh1-2 (Ssa_Cdh-1-2.1 and Ssa_Cdh-1-2.2), as well as seven corresponding sequences found within the rainbow trout (Oncorhyncys mykiss) genome (Berthelot et al. 2014) and one sequence found within the brown trout (Salmo trutta) genome (Str_Cdh1). The rainbow trout sequences are named according to the GenBank references of the genome contigs/scaffolds they are part of. Some of the rainbow trout sequences may represent the same genome locus. The underlined residues are polymorphic [S/P]. The S. trutta sequence was taken from a rough draft of the brown trout genome sequence, developed by Lien et al. (M. Kent, S. Lien, and M. Sodeland unpublished results), and was the only corresponding sequence found within that genome draft.
Functional studies of IPNV and E-cadherin interaction
We targeted Cdh1-1 for functional studies for three reasons. First, the protein product of cdh1-1 is a full-length epithelial cadherin, containing all necessary functional domains. In contrast, cdh1-2 lacks the exons encoding the transmembrane- and C- terminal cytoplasmic domains, presumably preventing Cdh1-2 from embedding itself in the cell membrane and from communicating with the cell interior. Second, cdh1-2 displays some exotic features, such as a near-perfect duplication of a block containing five exons (Figure 4), and third cdh1-1 harbors SNP4, the best candidate for being a functional polymorphism.
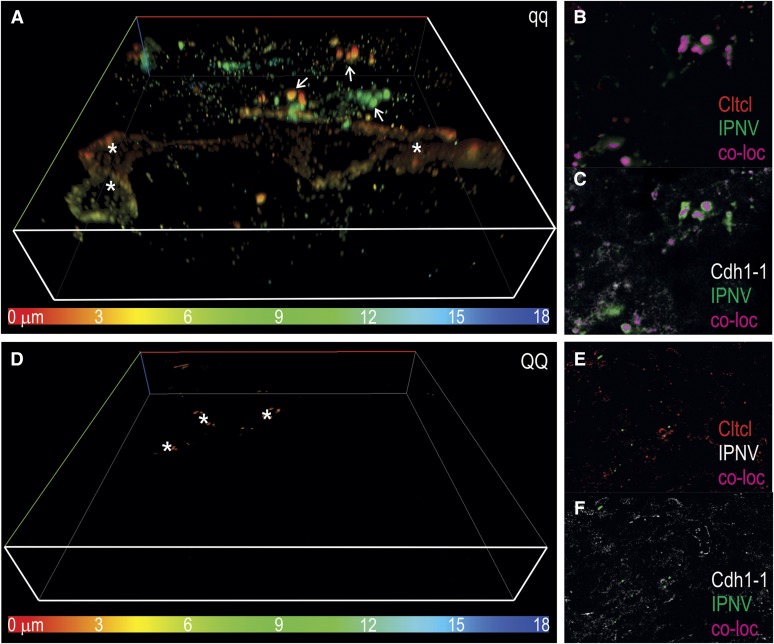
Investigation of IPN virus entry and binding to Cdh1-1 was carried out in vitro using IPNV-infected organotypic liver slices (LeCluyse et al. 2012). Immunofluorescence of the slices using antibodies specific for IPNV revealed a widespread distribution of virions in hepatocytes from qqQTL individuals (Figure 7A). On the cell membrane of hepatocytes, IPNV colocalized with Cdh1-1 and clathrin light chains in coated pits, forming early endosomes (Figure 7B). These findings imply that IPNV enters cells through binding to Cdh1-1, and that subsequent entry depends upon clathrin-mediated endocytosis. Analysis of liver sections from QQQTL fish revealed the presence of only a very few viruses, trapped by cellular debris on the surface of the tissue slices only (Figure 7C). Consequently no colocalization was observed between IPNV and Cdh1-1 or clathrin in fish with the resistant genotype, implying that the IPN virus is unable to bind to its receptor and enter hepatocytes.
Figure 7.
Immunofluorescence analysis of Cdh1-1 and IPNV. IPNV infection of in vitro cultured liver slices was carried out to visualize virus uptake and localization of Cdh1-1. (A) Depth-coded 3D visualization of IPNV in a liver section from a susceptible fish (qq) shows heavily infected hepatocytes (asterisks), several virus particles in early endosomes together with Cdh1-1 and Cltcl1 (arrows, see B and C for colocalization images) and presence of virus throughout the depth of the image stack. (B) Colocalization between IPNV and Cltcl1 (pseudo colored pink) in early endosomes. (C) Colocalization between IPNV and Cdh1-1 in the same early endosomes as shown in B. (D) Depth coded 3D visualization of IPNV in a liver slice from IPNV-resistant salmon (QQ) describe only a few virions on the surface of the tissue slice. (E) In the resistant fish there is no colocalization (pink color) with Cltcl1 (E) or Cdh1-1 (F).
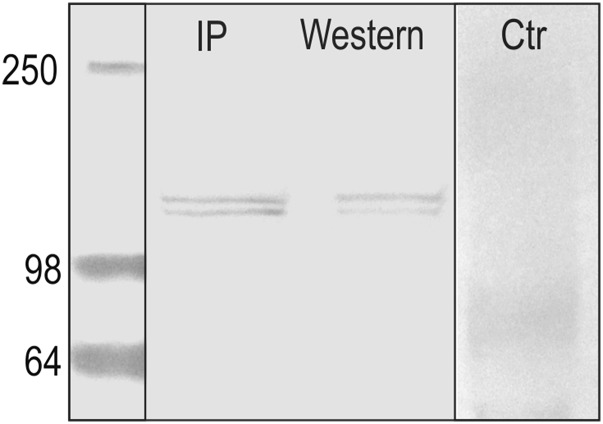
Colocalization of IPNV and Cdh1-1 suggested that Cdh1-1 could act as a receptor or a coreceptor for the virus. To prove direct protein–protein interaction between the IPN virus and Cdh1-1, we carried out a coimmunoprecipitation analysis using IPNV as bait, followed by a Western blot using a Cdh1-1 C-terminal-specific antibody (Figure 8). Cdh1-1 was detected as two fragments of ∼100 kDa, similar to what was observed in a liver protein lysate.
Figure 8.
Direct binding of IPNV to Cdh1-1. Direct protein interaction between IPNV and Cdh1-1 as verified by coimmunoprecipitation (IP). An antibody specificity control of the Cdh1-1 polyclonal antibody using a protein lysate (W) and negative control (exclusion of IPNV) are also shown.
The inability of IPNV to infect QQQTL individuals was confirmed by qPCR-based quantification of viral load in QQQTL, QqQTL, and qqQTL individuals, sampled during a IPN challenge test performed at the fry stage. Only qq fish displayed the high virus levels characteristic of diseased/moribund fish. QQ and Qq fish were predominantly negative, except for a few positive fish with very low virus levels.
Investigations into a two-locus model
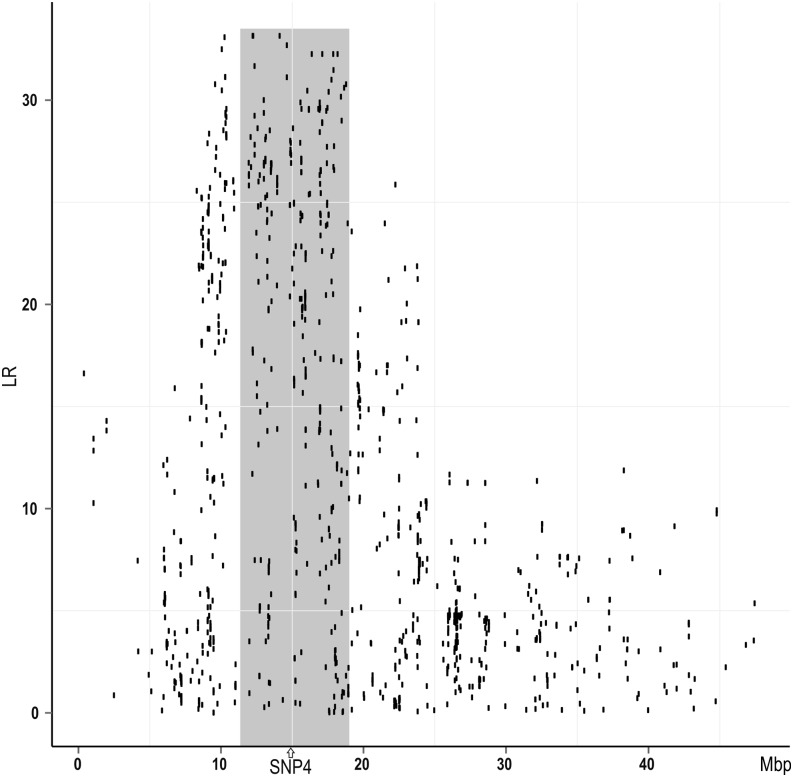
Since we had not been able to find a single “all-explaining” polymorphism, we hypothesized that the observed (in the 435 mapping parents) QTL genotypes might be determined by SNP4 and an additional, causative polymorphism. Using a custom-made Affymetrix SNP chip for the Atlantic salmon, containing 650,000 SNP polymorphisms within the AquaGen population, we scanned the genome for SNPs that might, in conjunction with SNP4, be entirely compatible with the QTL. More precisely, we searched for SNPs that were heterozygous in 16 genotyped QqQTL/qqSNP4 individuals and homozygous for one and the same allele in 26 genotyped qqQTL/qqSNP4 individuals; this was done using a (pseudo) maximum likelihood approach, to take the overall allele frequency in the population into account. Of the 300 highest-scoring SNPs, 89% were located on chromosome 26. Within chromosome 26, the highest-scoring SNPs were distributed across a genome region centered on SNP4 (Figure 9); the most promising of these SNPs were genotyped in all QTL-heterozygous mapping parents. After testing all possible pairs from among all genotyped polymorphisms, several pairs of polymorphisms were found whose haplotypes were exclusively linked to either QQTL or qQTL in QTL-heterozygous mapping parents. The pairs that were in perfect LD with the QTL all contained SNP1, SNP2, SNP3, or SNP4, in addition to a “second” polymorphism whose position varied within a ∼1-Mb region. None of the most likely second polymorphisms had an easily decipherable function such as causing an amino acid shift within a protein. Thus, the two-locus model was in agreement with the genetic data, although no definite candidate for a second functional mutation was found. The identified pairs of polymorphisms, being in perfect LD with the QTL, were later utilized in marker-assisted selection.
Figure 9.
A genome scan searching for a “second” causative mutation. The scan was based on genotypes obtained using a SNP chip with 650,000 polymorphic SNPs and assumes that SNP4 (or a polymorphism in approximately perfect LD with SNP4) is but one of two polymorphisms underlying the observed QTL genotypes. The figure displays only chromosome 26, as the majority of significant SNPs were found within this chromosome. The region corresponding to ICSASG scaffold ccf1000000016_0_0 is highlighted, as is the position of SNP4.
Industrial implementation of marker-assisted selection in salmon aquaculture
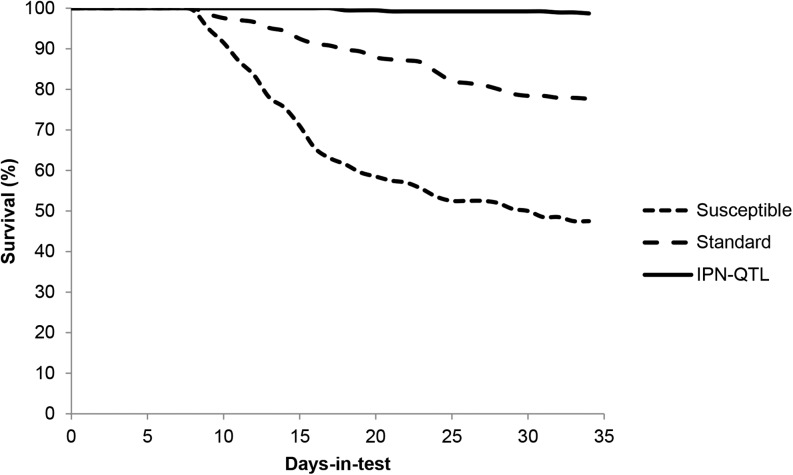
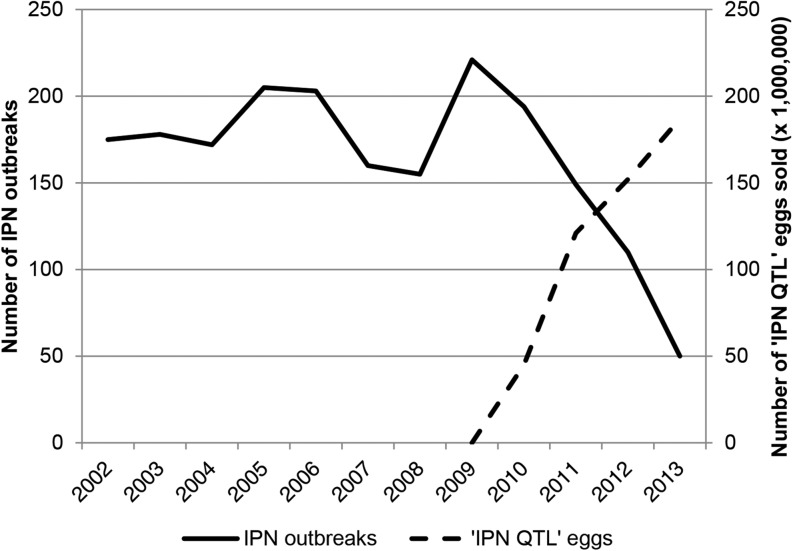
AquaGen, supplying ∼55% of the salmon eggs used by the Norwegian Atlantic salmon farming industry, implemented marker-assisted selection for IPN resistance in egg production from 2009. Haplotypes constructed from the above-mentioned microsatellites were used from the outset, but were later substituted for haplotypes constructed from SNP4 and a second SNP. MAS was generally implemented by the testing of candidate fathers for egg production, fathers deduced to be homozygous QQQTL being mated to random mothers. In some years, when the population of mothers had a low frequency of QQTL, selection was performed on the female side as well by culling all qq females. A controlled challenge test showed that salmon fry whose fathers were homozygous QQQTL (the mother being random individuals from the population) were almost entirely resistant to IPN (99% survival), whereas offspring of random fathers and random mothers had a survival rate of 78%. In offspring of sensitive fathers (homozygous qqQTL) the survival rate was only 48% (Figure 10). A field study initiated in 2010, monitoring the performance of salmon selected using MAS (29 million) vs. standard salmon (22 million) from the same egg provider, demonstrated improved results for the producers both in the freshwater and the seawater phase of the production. In freshwater, the incidence of IPN outbreaks was 0% in the 15 hatcheries receiving MAS eggs, an improvement from an incidence of 47% (seven outbreaks) the year before. In the 14 hatcheries receiving standard eggs there was no improvement, as the incidence of outbreaks was comparable to the year before, six vs. a previous five outbreaks. As IPN often reappears after seawater transfer of smolts, IPN-related mortality is normally registered within the first 90 days in sea. The accumulated mortality 90 days after sea transfer for salmon transferred to sea in the autumn 2010 was 1.1% for the MAS-selected fish, as compared to 6.4% for the standard fish. For fish transferred in the spring of 2011, the accumulated mortality was 4.6 vs. 9% in the standard group. This improvement in fish health is also evident in the Norwegian national statistics of fish diseases. In 2013, the number of IPN outbreaks in Norway had dropped 75% relative to the 2009 level (Figure 11), attributable in large part, according to Norwegian fish health authorities, to MAS for IPN resistance (Hjeltnes 2014).
Figure 10.
Mortalities in Atlantic salmon selected using MAS, compared to standard and negatively selected Atlantic salmon. The test was performed at the fry stage. “IPN-QTL” individuals were offspring of QQQTL fathers and random mothers, “standard” individuals were offspring of random fathers and random mothers, and “susceptible” individuals were offspring of qqQTL fathers and random mothers. x-axis: days postinfection.
Figure 11.
Number of recorded IPN outbreaks at Norwegian salmon farms. The data were collected by the Norwegian Veterinary Institute (Hjeltnes 2014).
Discussion
Aquaculture is a fast-growing food-producing sector, supplying nearly 50% of the seafood consumed worldwide (FAO 2014). Salmonid farming is an important contributor to global aquaculture, being the most industrialized and technologically advanced branch of aquaculture. Atlantic salmon production is steadily increasing and reached 2.8 million tons round bled weight in 2013 (Salmon World 2014, Kontali analyze, Norway), but important issues within fish health and fish welfare must be solved to ensure sustainable development of the industry. Progress in vaccinology has resulted in effective protection against several bacterial diseases, but less so against intracellular bacteria and viruses. This paper describes identification of cdh1-1 as the major determinant of resistance to one of the most important viral diseases in Atlantic salmon, IPN. We further document the successful implementation of MAS for IPNV resistance in farmed Atlantic salmon and the resulting dramatic reduction of losses due to this disease across the industry.
Fine mapping based on next-generation sequencing implicated a genome region containing cdh1-1, fam96b, and cdh1-2 as responsible for resistance to IPN. Within this region, the SNPs most strongly associated with the QTL were located. The very low virus levels observed in challenge-tested QQQTL individuals, and the absence of intercellular IPNV in QQQTL liver slices, suggest that the gene product underlying resistance is acting as a receptor or a coreceptor for the virus, i.e., the protein is probably a membrane protein. Fam96b, being involved in mitotic spindle formation, prelaminA accumulation, and assembly of the cytoplasmic Fe–S (CIA) machinery (Gari et al. 2012; Stehling et al. 2012; van Wietmarschen et al. 2012; Xiong et al. 2013), is unlikely to have such a function. Also, no putatively functional polymorphisms (strongly associated with the QTL) were detected within fam96b, and no differences in expression between QQQTL and qqQTL individuals were detected (data not shown). The protein encoded by cdh1-1, on the other hand, was a strong candidate for being involved in virus uptake due to its location on the cell membrane. The cdh1-1 gene encodes a classical epithelial cadherin, containing all functional domains necessary for dimerization with neighboring cells (trans-dimers) and with other epithelial cadherins on the same cell (cis-dimers). Immunofluorescence analysis of Cdh1-1 and IPNV in qqQTL individuals confirmed colocalization, and a coimmunoprecipitation assay showed that IPNV binds to Cdh1-1, strongly indicating that Cdh1-1 is part of the cell machinery that the virus uses for infection. The cdh1-1 gene also contains a putatively functional polymorphism (SNP4), which is one of the polymorphisms most strongly associated with the QTL. SNP4 is located within exon 7 of cdh1-1 (S167P), corresponding to the second extracellular domain (EC2) of the protein. EC2 interacts with EC1 to form cis-dimers between E-cadherins located on the same cell, and the formation of these cis-dimers are a prerequisite for the formation of (the more stable) trans-dimers; in human, for example, a hydrophobic-to-charged amino acid shift (V175D) in EC2 has been shown to abolish cis-dimerization and stability of adherence (Harrison et al. 2011). The resistance allele of SNP4 corresponds to a proline residue, likely to increase the hydrophobicity of the part of the EC2 domain that binds to EC1. The stability of adherence junctions is affected by constant Cdh1 turnover via endocytosis and exocytosis where both cis- and trans-dimers are directly endocytosed from mature adherence junctions (de Beco et al. 2009). Thus the increased hydrophobicity of the EC2 domain of the “resistant” Cdh1 variants (Cdh1Pro) in salmon may lead to more stable cis-dimers, and consequently to more stable Cdh1 trans-complexes and a lower rate of Cdh1 (and thus, IPNV) endocytosis. This hypothesis could also explain the apparent dominance of the QTL; if one assumes that both Cdh1Pro/Cdh1Pro and Cdh1Pro/Cdh1Ser cis-dimers are significantly more stable than Cdh1Ser/Cdh1Ser cis-dimers, only Cdh1Ser/Cdh1Ser cis-dimers would correspond to low resistance. Cdh1 dimerization dynamics could also explain the “windows” of IPN susceptibility in salmon (after start feeding and after smoltification), since these are time points where major physiological changes take place and adherence junctions are remodeled.
Although we regard SNP4 as a putatively causative polymorphism, SNP4 does not explain all observed genotypes at the QTL. We therefore cannot rule out the possibility that we have missed the true (single) underlying causative polymorphism, although this would mean that we have also missed all other polymorphisms in near-perfect LD with this putative polymorphism. Assuming that a yet undiscovered single causative polymorphism exists, the mapping data suggest it is most likely to be located in the vicinity of the epithelial cadherin genes. It could be located outside coding regions, facilitating (for example) differences in expression between the variants of cdh1-1. However, expression analysis of Cdh1-1 and 1-2 from QQQTL, QqQTL, and qqQTL animals, using qPCR, oligonucleotide microarrays, and RNA-seq, did not reveal consistent differences between the genotypes in terms of expression levels of splicing patterns (data not shown). Alternatively, the QTL genotypes may be the product of genotypes at two causative mutations. Such an additional mutation could be located within cdh1-1 itself, facilitating a change in the protein product similar to that of SNP4. Alternatively, it could be located within cdh1-2, assuming that the protein products of cdh1-1 and cdh1-2 interact, for example by forming heterodimers. It could also be located in any other gene within the QTL region, if that gene was also involved in the internalization of IPNV. Our results indicated that under such a two-locus model, SNP4 is likely to be one of the two loci, since no other combination of two polymorphisms was more strongly associated with IPN than those that included SNP4. Assuming that SNP4 is in fact one of two causative mutations, the number of potential secondary polymorphisms is large, since the discrepancy between SNP4 and the QTL is caused by one single haplotype. In addition to the SNP-based approach described here, we also sequenced (on Illumina HiSequation 2000) QqQTL individuals carrying the haplotype with the alternate linkage phase between SNP4 and the QTL, contrasting these sequences with sequences from qqQTL and QQQTL individuals to identify putative secondary causative mutations (data not shown). This attempt also did not lead to conclusive results and was complicated by the highly complex nature of the QTL region. Thus, further research is necessary to explain the existence of a particular haplotype in which QQTL is linked to qSNP4. The function and role of Cdh1-2 also calls for further research; cdh1-2 encodes a protein that lacks important functional domains, but it was nevertheless found to be expressed in the tissues investigated (data not shown). Interestingly, an ortholog of cdh1-2 is found within the genome of rainbow trout (Berthelot et al. 2014) (GenBank ID FR904752.1), displaying the same duplication of exon blocks, the same truncated C terminus, and having a juxtaposed neighboring full-length cdh-1, thus indicating that the segmental duplication predates the split between the Salmo and Oncorhyncus genera, ∼27–29 million years ago (Crete-Lafreniere et al. 2012). The cdh1-2 gene also displays two hypervariable regions, one located within each copy of the duplicated exon block.
Since a locus with a large positive effect on a trait is expected to be fixed relatively rapidly by natural or artificial selection, the continued segregation of a locus with such a large effect as the IPN QTN appears as a paradox. However, the Atlantic salmon has a very short history of artificial selection, only 11 generations having passed since the foundations of the first aquaculture strains (and selection for IPN resistance has been carried out for only the latest of these generations). Also, IPN has not been found to cause mortalities among Atlantic salmon living in the wild, disease pressures being very different under high-density aquaculture conditions than in the wild (Bebak and McAllister 2008). It is therefore plausible that the IPN QTL is segregating in Atlantic salmon populations because the locus is neutral in wild-living populations, while being under ongoing selection, though not yet fixed, in farmed populations. The alternative explanation is that the locus has a detrimental effect on another trait under selection. Our own data, however, do not show evidence of such negative correlations; all genetic correlations to other traits are neutral or positive (data not shown).
In conclusion, we have identified epithelial cadherin as the major determinant of the resistance of Atlantic salmon individuals to IPNV. Epithelial cadherin has earlier been shown to mediate internalization of the bacterium Listeria monocytogenes (Mengaud et al. 1996) and the fungus Candida albicans (Phan et al. 2007), but this is the first report of interaction between an epithelial cadherin and a virus. Selection of individuals carrying the resistance-related allele at the cdh1-1 gene has led to a rapid decline in the number of IPN outbreaks in Atlantic salmon aquaculture, leading to significant improvement with regard to animal welfare and the economy of Atlantic salmon production.
Table 1. Polymorphisms strongly associated with IPN resistance and/or putative causative mutations.
| Short name | Position within CCF1000000016_0_0 | Location | Polymorphism | Amino acids | r2 |
|---|---|---|---|---|---|
| SNP1 | 4313673 | 30 bp upstream of fam96b exon 1 | C/T | NA | 0.57 |
| SNP2 | 4289405 | 3′ UTR of cdh1-1 | -/A | NA | 0.57 |
| SNP3 | 4288035 | Downstream of cdh1-1 | -/AT | NA | 0.57 |
| SNP4 | 4295389 | Exon 7 of cdh1-1 | C/T | Pro/Ser | 0.58 |
r2 = the square of the correlation coefficient between the polymorphism and the QTL, based on genotypes in 117 QTL-heterozygous mapping parents. The alleles are relative to the DNA sequences found in File S1.
Supplementary Material
Acknowledgments
We thank Olai Einen for important contributions in the initiation of the project; Ben Koop (University of Victoria, BC, Canada) for his kind sharing of DNA sequence libraries; the Sven Arild Korsvoll and the rest of the AquaGen team for their support in the implementation of marker-assisted selection in salmon egg production; the fish farmers reporting performance data of both QTL-innOva and standard salmon; Heidi E. Mikalsen and Lill-Heidi Johansen (Nofima, Tromsø, Norway) for carrying out the challenge test for IPN resistance; Hege Munck, Katrine Hånes Kirste, (Nofima, Ås, Norway), and Helene Meaas Svendsen, Ida Johansson Schneider, Kristil Sundsaasen, Kristina Vagonyte-Hallen, Mariann Arnyasi, and Silje Karoliussen (CIGENE, Ås, Norway) for technical laboratory support; Simon Taylor, Teshome Dagne Mulugeta, and Jon Olav Vik for informatics support; and Fabian Grammes, Harald Grove, and Torfinn Nome for their contributions to analysis of RNAseq data, genome scaffolding, and sequence annotation. This study was supported by grants from the Research Council of Norway, division Havbruk (project 192322/S40).
Footnotes
Communicating editor: D. J. de Koning
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.175406/-/DC1.
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bebak J., McAllister P., 2008. Continuous exposure to infectious pancreatic necrosis virus during early life stages of rainbow trout, Oncorhynchus mykiss (Walbaum). J. Fish Dis. 32: 173–181. [DOI] [PubMed] [Google Scholar]
- Berthelot C., Brunet F., Chalopin D., Juanchich A., Bernard M., et al. , 2014. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 5: 3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, S. C., and J. A. Wooliams, 2010 Understanding field disease data. In: Contribution 0401 from Proceedings from the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany. [Google Scholar]
- Crane M., Hyatt A., 2011. Viruses of fish: an overview of significant pathogens. Viruses 3: 2025–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crete-Lafreniere A., Weir L. K., Bernatchez L., 2012. Framing the Salmonidae family phylogenetic portrait: a more complete picture from increased taxon sampling. PLoS ONE 7: e46662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson W. S., Koop B. F., Jones S. J. M., Iturra P., Vidal R., et al. , 2010. Sequencing the genome of the Atlantic salmon (Salmo salar). Genome Biol. 11: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beco S., Gueudry C., Amblard F., Coscoy S., 2009. Endocytosis is required for E-cadherin redistribution at mature adherens junctions. Proc. Natl. Acad. Sci. USA 106: 7010–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobos P., 1995. The molecular biology of infectious pancreatic necrosis virus (IPNV). Annu. Rev. Fish Dis. 5: 25–54. [Google Scholar]
- FAO, 2014 FAO Global Aquaculture Production Volume and Value Statistics Database Updated to 2012. Available at: http://www.fao.org/3/a-i3720e.pdf.
- Gari K., Ortiz A. M. L., Borel V., Flynn H., Skehel M. J., et al. , 2012. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 337: 243–245. [DOI] [PubMed] [Google Scholar]
- Granzow H., Weiland F., Fichtner D., Enzmann P. J., 1997. Studies of the ultrastructure and morphogenesis of fish pathogenic viruses grown in cell culture. J. Fish Dis. 20: 1–10. [Google Scholar]
- Guy D. R., Bishop S. C., Woolliams J. A., Brotherstone S., 2009. Genetic parameters for resistance to infectious pancreatic necrosis in pedigreed Atlantic salmon (Salmo salar) post-smolts using a reduced animal model. Aquaculture 290: 229–235. [Google Scholar]
- Harris, R., 2007 Improved pairwise alignment of genomic DNA. Ph.D. Thesis, The Pennsylvania State University, Graduate School College of Engineering, University Park, PA. [Google Scholar]
- Harris T., 2012. An introduction to adherens junctions: from molecular mechanisms to tissue development and disease, pp. 1–5 in Adherens Junctions: from Molecular Mechanisms to Tissue Development and Disease, edited by Harris T. Springer-Verlag, New York. [DOI] [PubMed] [Google Scholar]
- Harrison O. J., Jin X., Hong S., Bahna F., Ahlsen G., et al. , 2011. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure 19: 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjeltnes B. (Editor), 2014. Fish Health Report 2013, Norwegian Veterinary Institute, Oslo. [Google Scholar]
- Houston R. D., Haley C. S., Hamilton A., Guy D. R., Tinch A. E., et al. , 2008. Major quantitative trait loci affect resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar). Genetics 178: 1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Madan A., 1999. CAP3: a DNA sequence assembly program. Genome Res. 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjøglum S., Henryon M., Aasmundstad T., Korsgaard I., 2008. Selective breeding can increase resistance of Atlantic salmon to furunculosis, infectious salmon anaemia and infectious pancreatic necrosis. Aquacult. Res. 39: 498–505. [Google Scholar]
- Koressar T., Remm M., 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 15: 1289–1291. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCluyse E. L., Witek R. P., Andersen M. E., Powers M. J., 2012. Organotypic liver culture models: meeting current challenges in toxicity testing. Crit. Rev. Toxicol. 42: 501–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S., Gidskehaug L., Moen T., Hayes B. J., Berg P. R., et al. , 2011. A dense SNP-based linkage map for Atlantic salmon (Salmo salar) reveals extended chromosome homeologies and striking differences in sex-specific recombination patterns. BMC Genomics 12: 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud J., Ohayon H., Gounin P., Mege R.-M., Cossart P., 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84: 923–932. [DOI] [PubMed] [Google Scholar]
- Moen T., Baranski M., Sonesson A. K., Kjøglum S., 2009. Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): population-level associations between markers and trait. BMC Genomics 10: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen, T., 2010 Response to selection at a single locus: large change in allele frequency between two consecutive generations at a major QTL for IPN resistance in Atlantic salmon (Salmo Salar). In: Proceedings from the 9th World Congress on Genetics Applied to Livestock Production, Leipzig. [Google Scholar]
- Mott R., 1997. EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. Comput. Appl. Biosci. 13: 477–478. [DOI] [PubMed] [Google Scholar]
- Murray A. G., 2006. Persistence of infectious pancreatic necrosis virus (IPNV) in Scottish salmon (Salmo salar L.) farms. Prev. Vet. Med. 76: 97–108. [DOI] [PubMed] [Google Scholar]
- Myers E. W., Sutton G. G., Delcher A. L., Dew I. M., Fasulo D. P., et al. , 2000. A whole-genome assembly of Drosophila. Science 287: 2196–2204. [DOI] [PubMed] [Google Scholar]
- Ng S. H. S., Artieri C. G., Bosdet I. E., Chiu R., Danzmann R. G., et al. , 2005. A physical map of the genome of Atlantic salmon, Salmo salar. Genomics 86: 396–404. [DOI] [PubMed] [Google Scholar]
- Okamoto N., Tayama T., Kawanobe M., Fujiki N., Yasuda Y., et al. , 1993. Resistance of a rainbow trout strain to infectious pancreatic necrosis. Aquaculture 117: 71–76. [Google Scholar]
- Pearson W., Wood T., Zhang Z., Miller W., 1997. Comparison of DNA sequences with protein sequences. Genomics 46: 24–36. [DOI] [PubMed] [Google Scholar]
- Phan Q. T., Myers C. L., Fu Y., Sheppard D. C., Yeaman M. R., et al. , 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R. B., Keatley K. A. , M. R. Morasch, A. B. Ventura, K. P. Lubieniecki et al, 2009. Assignment of Atlantic salmon (Salmo salar) linkage groups to specific chromosomes: conservation of large syntenic blocks corresponding to whole chromosome arms in rainbow trout (Oncorhynchus mykiss). BMC Genet. 10: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R., Pearson M., 2005. Infectious pancreatic necrosis in Atlantic salmon, Salmo salar L. J. Fish Dis. 28: 383–390. [DOI] [PubMed] [Google Scholar]
- Smail D. A., Munro E. S., 2008. Isolation and quantification of infectious pancreatic necrosis virus from ovarian and seminal fluids of Atlantic salmon, Salmo salar L. J. Fish Dis. 31: 49–58. [DOI] [PubMed] [Google Scholar]
- Sommerset I., Krossøy B., Biering E., Frost P., 2005. Vaccines for fish in aquaculture. Expert Rev. Vaccines 4: 89–101. [DOI] [PubMed] [Google Scholar]
- Stehling O., Vashisht A. A., Mascarenhas J., Jonsson Z. O., Sharma T., et al. , 2012. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337: 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Scheet P., 2005. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 76: 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storset A., Strand C., Wetten M., Kjøglum S., Ramstad A., 2007. Response to selection for resistance against infectious pancreatic necrosis in Atlantic salmon (Salmo salar L.). Aquaculture 272: S62–S68. [Google Scholar]
- Thorsen J., Zhu B., Frengen E., Osoegawa K., de Jong P. J. et al, 2005. A highly redundant BAC library of Atlantic salmon (Salmo salar): an important tool for salmon projects. BMC Genomics 6: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergrasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., et al. , 2012. Primer3: new capabilities and interfaces. Nucleic Acids Res. 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wietmarschen N., Moradian A., Morin G. B., Lansdorp P. M., Uringa E.-J., 2012. The mammalian proteins MMS19, MIP18, and ANT2 are involved in cytoplasmic iron-sulfur cluster protein assembly. J. Biol. Chem. 287: 43351–43358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Snieszko S. F., Dunbar C. E., Pyle E., 1960. Virus nature of infectious pancreatic necrosis in trout. Exp. Biol. Med. 104: 104–108. [DOI] [PubMed] [Google Scholar]
- Xiong X.-D., Wang J., Zheng H., Jing X., Liu Z., et al. , 2013. Identification of FAM96B as a novel prelamin A binding partner. Biochem. Biophys. Res. Commun. 440: 20–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.