Abstract
Mammalian SWI/SNF chromatin-remodeling complexes utilize either BRG1 or Brm as alternative catalytic subunits to alter the position of nucleosomes and regulate gene expression. Genetic studies have demonstrated that SWI/SNF complexes are required during cardiac development and also protect against cardiovascular disease and cancer. However, Brm constitutive null mutants do not exhibit a cardiomyocyte phenotype and inducible Brg1 conditional mutations in cardiomyocyte do not demonstrate differences until stressed with transverse aortic constriction, where they exhibit a reduction in cardiac hypertrophy. We recently demonstrated the overlapping functions of Brm and Brg1 in vascular endothelial cells and sought here to test if this overlapping function occurred in cardiomyocytes. Brg1/Brm double mutants died within 21 days of severe cardiac dysfunction associated with glycogen accumulation and mitochondrial defects based on histological and ultrastructural analyses. To determine the underlying defects, we performed nontargeted metabolomics analysis of cardiac tissue by GC/MS from a line of Brg1/Brm double-mutant mice, which lack both Brg1 and Brm in cardiomyocytes in an inducible manner, and two groups of controls. Metabolites contributing most significantly to the differences between Brg1/Brm double-mutant and control-group hearts were then determined using the variable importance in projection analysis. Increased cardiac linoleic acid and oleic acid suggest alterations in fatty acid utilization or intake are perturbed in Brg1/Brm double mutants. Conversely, decreased glucose-6-phosphate, fructose-6-phosphate, and myoinositol suggest that glycolysis and glycogen formation are impaired. These novel metabolomics findings provide insight into SWI/SNF-regulated metabolic pathways and will guide mechanistic studies evaluating the role of SWI/SNF complexes in homeostasis and cardiovascular disease prevention.
Keywords: SWI/SNF complex, BRG1, BRM, Cardiomyocyte, Metabolomics, Fatty acid, Glucose
1 Introduction
Mammalian SWI/SNF (switching defective/sucrose non-fermenting) complexes are comprised of 9–12 subunits including either BRG1 (brahma-related gene 1, also known as SMARCA4) or BRM (brahma, also known as SMARCA2) as alternative catalytic subunits with DNA-dependent ATPase activity (Wu et al. 2009). These chromatin-remodeling complexes are recruited by pioneer transcription factors to the promoters of numerous target genes, where they slide or evict nucleosomes in an ATP-dependent manner to either activate or suppress RNA Polymerase II occupancy and transcription. In addition to BRG1 or BRM, SWI/SNF complexes contain 8–11 additional subunits called BAFs (BRG1/BRM associated factors with a number referring to the protein molecular mass) that contribute to chromatin remodeling. Due to the combinatorial assembly of different BAFs, there are potentially many distinct SWI/SNF complexes that vary in subunit composition and function (Wu et al. 2009).
SWI/SNF complexes play an important role in cardiovascular development (Bevilacqua et al. 2013; Chang and Bruneau 2012). In humans, mutations in several genes encoding SWI/SNF subunits including BRG1 and BRM result in congenital syndromes that exhibit highly penetrant cardiac defects (Kosho et al. 2013a, b; Ronan et al. 2013; Santen et al. 2012; Tsurusaki et al. 2012; Van Houdt et al. 2012). Inmouse models, Baf250a, Baf180, and Baf60c constitutive null mutants exhibit a variety of cardiac defects including abnormal looping, hypoplastic ventricles, shortened outflow tracts, and septal defects that result in embryonic lethality (Lei et al. 2012; Lickert et al. 2004; Wang et al. 2004). Although Brg1 constitutive nullmutants die prior to organogenesis (Bultman et al. 2000), conditional null mutations of Brg1 in the developing endocardium, myocardium, or myocardial progenitor cells of the secondary heart field result in a variety of defects including hypoplastic ventricles, thin myocardium, shortened outflow tract, lack of septum, and hypotrebeculation that also culminate in embryonic lethality (Hang et al. 2010; Stankunas et al. 2008; Takeuchi et al. 2011). An essential role for BRG1 in cardiomyocyte development is consistent with it physically interacting with the cardiogenic transcription factors TBX5, GATA4, and NKX2.5, and the ability of BAF60c, TBX5, and GATA4 to differentiate non-cardiac mesoderm into beating cardiomyocytes (Hang et al. 2010; Kadoch et al. 2013; Takeuchi and Bruneau 2009). While Brg1 is essential for cardiomyocyte development, it is dispensable for cardiomyocyte viability in the adult animal (Hang et al. 2010). However, an inducible conditional mutation of Brg1 in adult cardiomyocytes did result in decreased hypertrophy following transverse aortic constriction to pressure overload the heart (Hang et al. 2010). In contrast, Brm constitutive null mutants are viable and do not exhibit a discernable cardiomyocyte phenotype compared to sibling wild-type controls (Han et al. 2011; Reyes et al. 1998). The combined role of Brg1 and Brm in the adult cardiomyocyte has not previously been described.
The redundancy of Brg1 and Brm in vascular endothelial cells (VECs) within the adult heart has recently been reported. Brg1 is required by VECs during embryonic development, whereas Brm is dispensable (Curtis et al. 2012; Griffin et al. 2008; Griffin et al. 2011; Nelson et al. 2005). Furthermore, the Brg1 mutant phenotype of developing VECs is not exacerbated by Brm deficiency (Griffin et al. 2008). However, an inducible conditional mutation of Brg1 in VECs from adult mice did not result in an observable phenotype (Willis et al. 2012). The lack of an adult phenotype was found to be due to the redundancy of Brg1 and Brm in the adult VECs within the heart, as double mutants died within 30 days of inducing the Brg1 deletion (Willis et al. 2012). Absence of Brg1 and Brm resulted in VEC apoptosis, vascular leakage, intra-cardiac dissection, and secondary cardiomyocyte cell death due to ischemia (Willis et al. 2012).
Considering that Brg1 and Brm are functionally redundant in adult VECs, we hypothesized that they may also have redundant functions in adult cardiomyocytes. To test this hypothesis, we analyzed Brg1/Brm double mutants where Brg1 could be mutated exclusively in cardiomyocytes in an inducible manner. Indeed, double mutants were not viable and their hearts exhibited signs of metabolic dysfunction. Therefore, we sought to characterize metabolomic changes in double mutants compared to controls. Recent advances in technology have afforded a more comprehensive analysis of a tissue’s metabolome. Both targeted and non-targeted mass spectrometry based approaches have become common, with non-targeted methods being particularly valuable to explore phenotypes that involve many metabolites in a relatively unbiased manner (Bain et al. 2009). Non-targeted technologies have been used to characterize genetic diseases that result in altered metabolism of carbohydrates, lipids, and amino acids (Frazier et al. 2006). The present studies demonstrate that BRG1 and BRM are required for metabolic homeostasis. Brg1/Brm double-mutant hearts exhibited altered metabolism of fatty acids (e.g. increased cardiac oleic and linoleic acids), glucose (e.g. decreased glucose-6-P, fructose-6-P, myoinositol), and amino acids. Both fatty acid and glucose substrate utilization has been shown to be a critical regulator of the heart’s resiliency in the face of cardiac disease, with resulting fatty acid and glucose toxicity identified when there is an imbalance matched to the stressor (Carley et al. 2014; Doenst et al. 2013; Kolwicz et al. 2013). These studies demonstrate the critical role that BRG1 and BRM play in regulating fatty acid and glucose metabolism in the intact adult cardiomyocyte, likely by supporting the activity of multiple nuclear receptors implicated in regulating fatty acid and glucose utilization in the heart (e.g. PPAR, PGC1α) (Debril et al. 2004; Gatfield et al. 2009; Li et al. 2008; Wang et al. 1996).
2 Materials and Methods
2.1 Mouse Lines
All mouse experiments were approved by the Institutional Animal Care and Use Committees (IACUC) review board at the University of North Carolina at Chapel Hill and were performed in accordance with federal guidelines. The αMHC-Cre-ERT mice [also known as B6.Cg-Tg(Myh6-cre/Esr1)1JmkJ or αMHC-MerCreMer] were obtained from The Jackson Laboratory (#005657, Bar Harbor, ME) and genotyped as previously described (Sohal et al. 2001). The Brg1 conditional mutant mouse line and Brm constitutive mutant mouse line have been described previously (Bultman et al. 2000; Reyes et al. 1998; Sumi-Ichinose et al. 1997). Genotyping of the Brg1 floxed and Δfloxed alleles and the Brm mutation were genotyped by PCR as previously described (Bultman et al. 2000; Reyes et al. 1998; Sumi-Ichinose et al. 1997).
2.2 Tamoxifen Induction of Brg1 Mutation
To induce the Brg1 conditional mutation in adult cardiomyocytes, 3–6 month old male and female mice were provided rodent chow containing tamoxifen (Sigma-Aldrich #T5648, St. Louis, MO) over a 7-day period. The route of delivery and dose were selected to minimize a previously described artifact caused by high doses of tamoxifen in the presence of the αMHC-Cre-ERT transgene (Koitabashi et al. 2009). Briefly, 500 mg of tamoxifen was mixed with 1 kg of ground-up rodent chow and then mixed with water, kneaded into pellets, and dried in a hood. Provided to mice ad libitum, the dose was estimated to be 80 mg/kg/day. After the 7-day treatment period, the tamoxifen-fortified chow was removed and replaced with the same chow lacking tamoxifen.
2.3 Echocardiography
Conscious cardiac transthoracic echocardiography was performed on mice at the indicated time points using a VisualSonics Vevo 2100 ultrasound biomicroscopy system (VisualSonics, Inc., Toronto, Ontario, Canada) as previously described (Files et al. 2012; Oakley et al. 2013; Willis et al. 2009a,b). Two-dimensional M-mode echocardiography was performed in the parasternal long-axis view at the level of the papillary muscle on loosely restrained mice. Anterior and posterior wall thickness was measured as distance from epicardial to endocardial leading edges. Left ventricular internal diameters were also measured. Left ventricular systolic function was assessed by ejection fraction (LV EF% = [(LV Vol; d-LV Vol; s/LV Vol; d) × 100] and fractional shortening (%FS = [(LVEDD − LVESD)/LVEDD] × 100). Investigators were blinded to mouse genotype from collection through waveform measurements. Each measurement represents the average of three cardiac cycles from each mouse.
2.4 Histopathological and Ultrastructural Analyses
Histology was performed by fixing heart tissues in 4 % paraformaldehyde, embedding in paraffin, cutting 5-μm sections, and staining sections with hematoxylin and eosin (H&E) according to standard procedures. For transmission electron microscopy (TEM), heart tissues were fixed in 2 % paraformaldehyde and 2.5 % glutaraldehyde in 0.15 M sodium phosphate buffer (pH 7.4) overnight and then post-fixed with 1 % osmium tetroxide in 0.15 sodium phosphate buffer. Samples were dehydrated with increasing concentrations of ethanol, infiltrated and embedded in Polybed 812 epoxy resin (Polysciences, Warrington, PA), and 70-nm ultrathin sections were cut with a diamond knife. Sections were mounted on 200-mesh copper grids and staining with 4 % aqueous uranyl acetate and Reynold’s lead citrate. Sections were observed with a LEO EM910 transmission electron microscope operating at 80 kV (LEO Electron Microscopy, Thornwood, NY) and photographed with a Gatan-Orius SC1000 CCD Digital Camera and Digital Micrograph 3.11.0 (Gatan, Pleasanton, CA).
2.5 RT-qPCR Analysis
RNA from double-mutant and control hearts was prepared using Trizol reagent (Life Technologies) and reverse transcribed using random hexamers and SuperScript II RT (Life Technologies) according to standard procedures. Pfk-fb1 was amplified using the following primers (5′GAGTG CAAGACCACGTTCAA3′ and 5′GGAGCTGATGCTTT GAGACC3′ at 300 nM final concentration for each) and Power SYBR Green Master Mix (Life Technologies) under the following cycling parameters (95C 45 s, 60C 30 s, 72C 45 s). Dissociation curves and agarose gels demonstrated a single PCR product in each case without primer dimmers. Gapdh was amplified using a TaqMan assay (Life Technologies) and default cycling parameters. Negative control reactions lacking RT yielded little or no Pfk or Gapdh signal, and relative expression levels were determined using the ΔΔCt method.
2.6 Metabolomics Determination by GC–MS Instrumentation
Cardiac tissue was flash frozen in a liquid nitrogen cooled biopress, a fraction of it weighed (~25–50 mg wet weight), then the finely cut up tissue quickly added to fresh pre-made buffer (50 % acetyl-nitrile, 50 % water, 0.3 % formic acid) at a standard concentration of 25 mg/475 mcl buffer then fully homogenized on ice for 10–25 s and placed on dry ice/stored at −80C. The samples were “crash” deprotonized by methanol precipitation and spiked with D27-deuterated myristic acid (D27-C14:0) as an internal standard for retention-time locking and dried. The trimethylsilyl (TMS)-D27-C14:0 standard retention time (RT) was set at ~16.727 min. Reactive carbonyls were stabilized at 50 °C with methoxyamine hydrochloride in dry pyridine. Metabolites were made volatile with TMS groups using N-methyl-N-(trimethylsilyl) trifluoroacetamide or MSTFA with catalytic trimethylchlorosilane at 50 °C. GC/MS methods generally follow those of Roessner et al. (2000), Fiehn et al. (2008), and Kind et al. (2009), and used a 6890 N GC connected to a 5975 Inert single-quadrupole MS (Agilent Technologies, Santa Clara, CA). The two wall-coated, open-tubular (WCOT) GC columns connected in series are both from J&W/Agilent (part 122–5512), DB5-MS, 15 meters in length, 0.25 mm in diameter, with an 0.25-μm luminal film. Positive ions generated with conventional electron-ionization (EI) at 70 eV are scanned broadly from 600 to 50 m/z in the detector throughout the 45 min cycle time.
Data were acquired using an MSD ChemStation (Agilent Technologies) by identifying metabolites on their mass fragmentation patterns and RT. Rawdata formatted files were exported for further analysis in Automatic Mass Spectral Deconvolution and Identification Software or AMDIS (freeware developed by Drs. Steve Stein, W. Gary Mallard, and their coworkers at National Institute of Standards and Technology or NIST (Mallard and Reed 1997; Halket et al. 1999; Stein 1999). Deconvoluted spectra are annotated as metabolites, to the extent possible, using an orthogonal approach that incorporates both RT from GC and the fragmentation pattern observed in EI-MS. Peak annotation is based primarily on our own RT-locked spectral library of metabolites. The library is built upon the Fiehn GC/MS Metabolomics RTL Library (a gift from Agilent, part number G1676-90000; Kind et al. (2009), Golm Metabolome Library (courtesy of Dr. Joachim Kopka and coworkers at the Max Planck Institute of Molecular Plant Physiology, Golm, Germany (Kopka 2005) the Wiley 9th-NIST 2011 commercial library (Agilent G1730-64000), and other spectral libraries. Once annotation was complete, a cross-tabulated spreadsheet was created, listing the integrated peak area for each metabolite versus sample identity. This was accomplished using a custom Visual Basic program in Microsoft Excel that grouped peaks across samples based on identical metabolite annotation and RT proximity. Peak alignment across samples was further confirmed using SpectConnect (Styczynski et al. 2007) to assess similarity in spectral fragmentation patterns, and by manual curation. The raw, transformed, and sorted data used for each of the three comparisons in the metabolomic analyses (next) can be found in Supplemental Table 1.
2.7 Metabolomic Analyses
Metaboanalyst (v2.0) run on the statistical package R (v2.14.0) used metabolite peaks areas (as representative of concentration) (Xia et al. 2009, 2012). These data were first analyzed by an unsupervised principal component analysis (PCA), which identified the presence of the Brg1/Brm double mutant as the principal source of variance. To sharpen the separation between our three groups, data were next analyzed using a partial least squares discriminant analysis (PLS-DA) to further determine which metabolites were responsible for separating these two groups. The specific metabolites contributing most significantly to the differences identified by PLS-DA between Brg1/Brm double mutant and control group hearts were determined using the variable importance in projection (VIP) analysis in the metaboanalyst environment. The metabolites that best differentiated the groups were then individually tested using the Student’s t test (Microsoft Excel 2011, Seattle, WA). The t test significant metabolites were matched to metabolomics pathways using the Pathway Analysis feature in Metaboanalyst 2.0. Heat maps of the metabolite data (individual and grouped) were generated using the GENE-E software (http://www.broadinstitute.org/cancer/software/GENE-E/index.html).
3 Results and Discussion
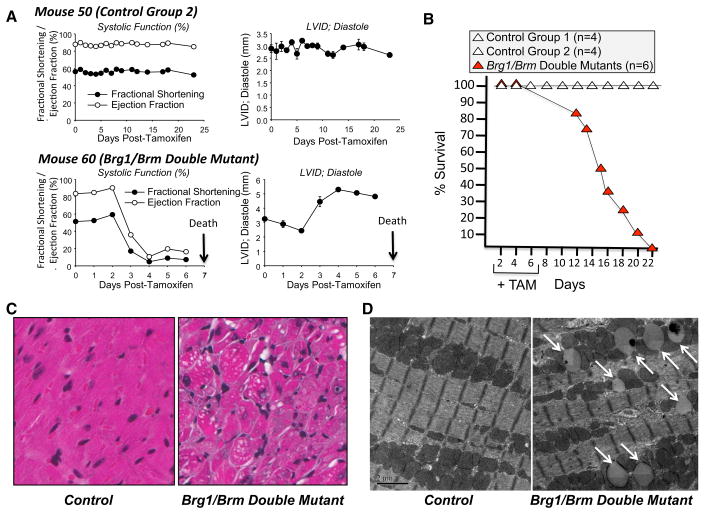
To determine whether there is functional compensation of Brg1 and Brm in adult cardiomyocytes, we generated Brg1 floxed/floxed mice carrying the αMHC-Cre-ERT transgene on a Brm-deficient background. To induce the Brg1 conditional mutation in cardiomyocytes, these mice were treated with oral tamoxifen by providing it in their chow for 7 days (Brg1/Brm double mutant). As a control, the same Brg1floxed/floxed; αMHC-Cre-ERT+/0; Brm−/− mice did not receive any tamoxifen treatment (Control Group 1). This control group addressed the potential for transgene leakiness (i.e., tamoxifen-independent induction of the Brg1 mutation). As a second control, Brg1 floxed/floxed mice lacking the αMHC-Cre-ERT transgene on a Brm-deficient background (Brg1floxed/floxed; αMHC-Cre-ERT0/0; Brm−/−) were treated with tamoxifen (Control Group 2). This control group addressed the potential for tamoxifen having off-target effects unrelated to the Cre-ERT transgene. Rapid onset of cardiac dysfunction (Fig. 1a; Table 1), evidenced by significant decreases in Brg1-Brm DM mice ejection fraction % (42.0 vs. 88.6 % in Control Groups 1 & 2) and factional shortening % (21.3 vs. 57.3 % in Control Groups 1 & 2), led to the rapid onset of death (Fig. 1b) in Brg1/Brm double mutants, but not in either control group. Kaplan–Meier analysis illustrates the first deaths occurring at day 13, with all other Brg1/Brm double mutants dying by day 22 relative to the first day of tamoxifen-fortified chow (which was provided on days 1–7). No histological changes were detected in the Brg1/Brm double mutant hearts, but increases in LV Mass/BW were identified (6.2 ± 0.3 mg/g vs. 4.2 ± 0.1 mg/g Brm−/− flx/flx Brg1 Tg+ given 7 days of Tamoxifen at 1 day premortem, average day 11.6 ± 1.5 days, N = 27 and 30, respectively, *p<0.05 by Student’s t test). The cardiac phenotype of double mutants included changes in mitochondrial dynamics and accumulation storage vacuoles based on our analysis of H&E-stained sections (Fig. 1c) and TEM (Fig. 1d). Therefore, we considered the possible role of SWI/SNF complexes in regulating cardiomyocyte metabolism. To detect proximal events leading to heart failure and death, we investigated changes in the metabolomic profiles of Brg1/Brm double-mutant mice on day 9, which was 2 days after tamoxifen treatment ended. Day 9 was also a point prior to any signs of adverse health and 4 days before any of the mice were found to die (Fig. 1b).
Fig. 1.
The phenotype of Brg1/Brm double-mutant mice. a Kaplan–Meier survival curve of mice following the administration of tamoxifen (+TAM) on days 1 through 7. b Longitudinal echo time course of an example control group 2 (Brg1floxed/floxed; αMHC-Cre-ERT0/0(no transgene); Brm−/− mice plus tamoxifen treatment) and a Brg1/Brm double mutant (Brg1floxed/floxed; αMHC-Cre-ERT+/0; Brm−/− mice plus tamoxifen treatment) illustrating the rapid decline in systolic function and dilation upon deletion of Brg1 with tamoxifen induction (via chow). c Representative H&E-stained heart sections from control (left) and double mutant (right) mice at ×200 magnification. d Representative transmission electron micrographs of cardiomyocytes from control (left) and double mutant (right) mice at ×5000 magnification. Illustrating accumulation of vacuoles in the mitochondria interspersed in the sarcomere (white arrows) that were found adjacent to fragmented mitochondria
Table 1.
High-resolution transthoracic echocardiography performed on conscious mice in Groups 1–5 at baseline and after initiating tamoxifen/chow or chow diet for 7 days
| Group 1 Brg1floxed/floxed; αMHC-Cre-ERT+/0; Brm−/− No TAM N = 3 |
Group 2 Brg1floxed/floxed; αMHC-Cre-ERT0/0; Brm−/−) +TAM Baseline N = 5 |
Group 3 Brg1floxed/floxed; αMHC-Cre-ERT+/0; Brm−/− +TAM (Brg1/Brm DM) Baseline N = 5 |
Groups 1,2 (Controls) Day matched pre-mortem Group 1 N = 5 Group 2 N = 10 Total N = 15 |
GBrg1/Brmroup 3 DM Brg1floxed/floxed; αMHC-Cre-ERT+/0; Brg1/Brm−/− +TAM 1 Day pre-mortem (day 11.6 ± 1.5) Total N = 10 |
|
|---|---|---|---|---|---|
| AWTD (mm) | 1.10 ± 0.02 | 1.06 ± 0.03 | 1.06 ± 0.03 | 1.11 ± 0.03 | 1.03 ± 0.08 |
| AWTS (mm) | 1.79 ± 0.06 | 1.82 ± 0.05 | 1.76 ± 0.11 | 1.87 ± 0.05 | 1.42 ± 0.15# |
| LVEDD (mm) | 2.93 ± 0.17 | 3.13 ± 0.11 | 2.76 ± 0.13 | 2.83 ± 0.08 | 4.19 ± 0.22* |
| LVESD (mm) | 1.25 ± 0.01 | 1.31 ± 0.04 | 1.16 ± 0.07 | 1.21 ± 0.04 | 3.35 ± 0.30* |
| PWTD (mm) | 1.11 ± 0.01 | 1.07 ± 0.04 | 1.02 ± 0.02 | 1.12 ± 0.02 | 1.02 ± 0.08 |
| PWTS (mm) | 1.81 ± 0.07 | 1.82 ± 0.03 | 1.60 ± 0.05 | 1.81 ± 0.04 | 1.23 ± 0.10* |
| EF % | 88.6 ± 0.6 | 89.0 ± 0.2 | 88.4 ± 0.5 | 88.6 ± 0.5 | 42.0 ± 6.4* |
| FS % | 56.3 ± 1.0 | 58.0 ± 0.4 | 58.2 ± 0.7 | 57.3 ± 0.7 | 21.3 ± 3.6* |
| LV mass (mg) | 116.9 ± 7.0 | 120.6 ± 4.9 | 98.1 ± 9.5 | 113.0 ± 4.8 | 177.9 ± 15.4* |
| LV vol; d (μl) | 33.2 ± 2.8 | 34.1 ± 3.3 | 78.5 ± 7.6 | 30.8 ± 2.0 | 81.0 ± 9.2* |
| LV vol; s (μl) | 3.7 ± 0.1 | 4.3 ± 0.3 | 3.1 ± 0.5 | 3.6 ± 0.3 | 51.0 ± 9.4* |
| LV mass/BW (mg/g) | 3.7 ± 0.1 | 3.8 ± 0.2 | 3.9 ± 0.3 | 3.6 ± 0.1 | 6.9 ± 0.5* |
| BW | 31.5 ± 0.9 | 32.0 ± 0.8 | 25.6 ± 2.3 | 31.2 ± 1.0 | 26.1 ± 1.7 |
| HR (bpm) | 707 ± 45 | 707 ± 13 | 690 ± 9 | 744 ± 11 | 484 ± 40* |
Data represent mean ± SEM. A one way analysis of variance was performed, followed by an all pairwise multiple comparison procedure (Holm-Sidak method), when overall significance <0.05
HR heart rate, ExLVD external left ventricular diameter, bpm heart beats per minute, AWTD anterior wall thickness in diastole, AWTS anterior wall thickness in systole, PWTD posterior wall thickness in diastole, PWTS posterior wall thickness in systole, LVEDD left ventricular end-diastolic dimension, LVESD left ventricular end-systolic dimension, FS fractional shortening calculated as (LVEDD − LVESD)/LVEDD × 100, EF% ejection fraction calculated as (end Simpson’s diastolic volume − end Simpson’s systolic volume)/end Simpson’s diastolic volume * 100, ND not determined
p<0.001 versus all other groups;
versus Groups 1, 2 Controls day matched pre-mortem
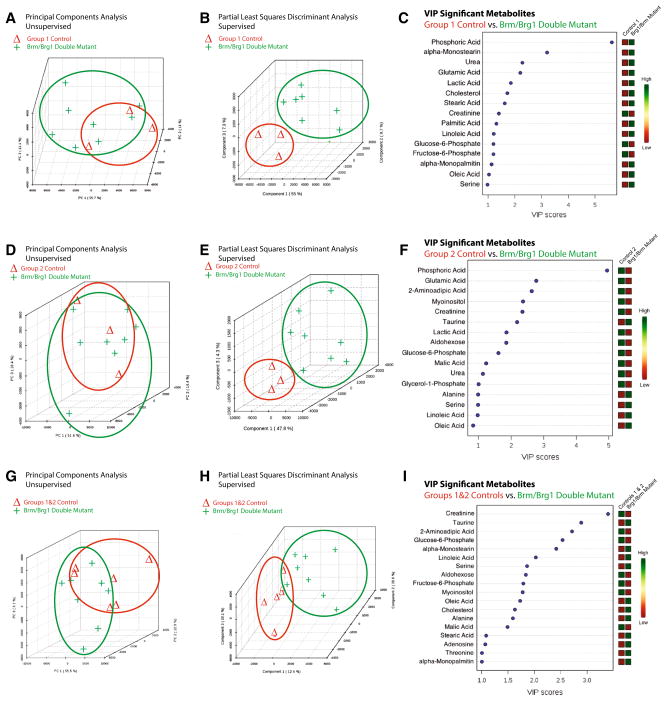
Compared to the group 1 sibling-matched controls, quantitative non-targeted metabolomics profiling identified distinct differences in the Brm/Brg1 double mutant hearts by PCA and PLS-DA analysis (Fig. 2a, b, respectively). In both analyses, the principal component 1 accounted for 55 % of the differences between the two groups. Further analysis of the top 15 metabolites using a VIP that differentiated the Brg1/Brm double-mutant hearts from the group 1 controls include phosphoric acid, α-monostearin, urea, glutamic acid, and lactic acid (Fig. 2c).
Fig. 2.
The metabolomic profiles of Brg1/Brm double-mutant hearts are significantly different from control hearts prepared and analyzed in parallel. Principal components analysis (PCA) illustrating the unsupervised relationship between Brg1/Brm double-mutant hearts (Brg1floxed/floxed; αMHC-Cre-ERT+/0; Brm−/− mice plus tamoxifen treatment) compared to a Control Group 1 (Brg1floxed/floxed; αMHC-Cre-ERT+/0; Brm−/− minus tamoxifen treatment) d Control Group 2 (Brg1floxed/floxed; αMHC-Cre-ERT0/0(no transgene); Brm−/− mice plus tamoxifen treatment), and g Combined Control Groups 1 & 2 hearts. Partial least squares discriminant analysis (PLS-DA) score visualizes supervised (assigned group) clustering of Brg1/Brm double-mutant heart metabolomics compared to b Control Group 1 e Control Group 2, and h Control Groups 1 & 2. Data points that are closer together indicate higher degree of similarity than data points that are further apart. PLS-DA score plots for Brg1/Brm double-mutant heart metabolites (red) compared to metabolites identified in control hearts (green). Each point represents the combined metabolite profile in an individual mouse. Variable importance in projection (VIP) plot of VIP statistical analysis identifying the top 15 metabolites contributing to the differences between the Brg1/Brm double mutant and c Control Group 1 f Control Group 2, and i Control Groups 1 & 2 hearts (relative concentration to the right of the figure). N = 8 (Brg1/Brm double-mutant hearts), N = 3 (Control Group 1), N = 3 (Control Group 2), N = 6 (Control Groups 1 & 2) hearts per group analyzed. Each point represents an individual mouse heart run in parallel with the other samples (Color figure online)
Compared to the group 2 sibling-matched controls, metabolomics non-targeted profiling identified distinct differences in Brg1/Brm double-mutant mice by PCA and PLS-DA analysis (Fig. 2d, e). Between 47.8 and 51.6 % of the differences between these groups accounted for the variability described by principal component 1. Like the analysis of the Brg1/Brm double-mutant mice compared to the group 1 controls, the top 15 metabolites using a VIP analysis to differentiate Brg1/Brm double-mutant mice from group 2 controls included phosphoric acid, glutamic acid, 2-aminoadipic acid, myoinositol, creatinine, and taurine (Fig. 2f). Since these two analyses differed in ways that may have been due to the presence of tamoxifen treatment, we chose next to combine the two control groups (Groups 1 and 2) and compared them to the Brg1/Brm double-mutant mice (Fig. 2g, h, i). By PCA analysis, principal component 1 accounted for 55.5 % of the variability between the two groups and by PLS-DA, principal components 1–3 accounted for 12.4, 39.6, and 20.1 %, respectively and separated out the two groups distinctly (Fig. 2h). VIP analysis identified creatinine, taurine, 2-aminoadipic acid, glucose-6-phosphate, α-monostearin, and linoleic acid as the top significant hits (Fig. 2i). Comparison of the top 15 VIP hits between the three analyses performed, creatinine, linoleic acid, and glucose-6-phosphate were consistently found and represent differences not dependent upon the types of controls used (Table 2, in bold).
Table 2.
VIP significant metabolites in Brm/Brg1 double mutant mice (Flx/Flx, Cre Tg+, +tamoxifen) 12 days after feeding initiated (7 days total, Groups 1 and 3 only). Brm/Brg1 double mutant
| Control Group 1 (Flx/Flx, No Brg1 Tg, Chow+tamoxifen) versus Brm/Brg1 double mutant (VIP Rank) | Control Group 2 (Flx/Flx, Brg1 Tg+, Chow diet − no tamoxifen) versus Brm/Brg1 double mutant (VIP Rank) | Control Groups 1 & 2 versus Brm/Brg1 double mutant (Chow+tamoxifen) (VIP Rank) |
|---|---|---|
| Phosphoric Acid (1) | Phosphoric acid (1) | * |
| Alpha-monostearin (2) | * | Alpha-monostearin (5) |
| Urea (3) | Urea (11) | * |
| Glutamic acid (4) | Glutamic acid (2) | * |
| Lactic acid (5) | Lactic acid (7) | * |
| Cholesterol (6) | * | Cholesterol (12) |
| Stearic acid (7) | * | Stearic acid (15) |
| Creatinine (8) | Creatinine (5) | Creatinine (1) |
| Palmitic acid (9) | * | * |
| Linoleic acid (10) | Linoleic acid (15) | Linoleic acid (6) |
| Glucose-6-phosphate (11) | Glucose-6-phosphate (9) | Glucose-6-phosphate (4) |
| Fructose-6-phosphate (12) | * | Fructose-6-phosphate (9) |
| Alpha-monopalmitin (13) | * | Alpha-Monopalmitin (18) |
| Oleic acid (14) | Oleic acid (16) | Oleic acid (11) |
| Serine (15) | Serine (14) | Serine (7) |
| * | 2-Aminoadipic acid (3) | 2-Aminoadipic acid (3) |
| * | Myoinositol (4) | Myoinositol (10) |
| * | Taurine (6) | Taurine (2) |
| * | Aldohexose (8) | Aldohexose (8) |
| * | Malic acid (10) | Malic acid (14) |
| * | Glycerol-1-phosphate (12) | * |
| * | Alanine (13) | Alanine (13) |
| * | * | Adenosine (16) |
| * | * | Threonine (17) |
The ranking (1–15) indicates the ranking given to the metabolites in their respective groups (Control Group 1, Control Group 2, Brg/Brm Double mutant Group 3)
Bold present in all three analyses
Indicates this metabolite was not found to be a VIP significant metabolite
The comparisons of Brg1/Brm double-mutant mice to Group 1, Group 2, and Groups 1 & 2 were performed to demonstrate the need for multiple controls (i.e. how tamoxifen and genetic backgrounds influence cardiac metabolites individually). The consistent differences Brg1/Brm double-mutant mice have independent of their genotype and strain (Group 1) and Tamoxifen (Group 2) was most dependably determined when Groups 1 and 2 were combined (Fig. 2g–i). Differences in Groups 1 and Group 2 were identified, as expected due to the tamoxifen treatment. For example, differences were detected by PCA, PLS-DA, and t test analysis (Supplemental Fig. 1). The differences between Groups 1 and 2 are limited, due to the low number (N = 3) of biological replicates run in these subgroups. The comparison of Brg1/Brm double-mutant samples to the combined Groups 1 and 2 allowed identification of metabolic signatures that are independent of the Brg1/Brm-null phenotype.
The studies presented here have limitations that should be acknowledged. While the GC–MS method applied here allows global definitions of metabolism, it is not expected that all of the possible metabolites present would be identified. Identification of metabolites is also limited to molecules with unambiguous chemical annotation in established metabolite libraries; the exploratory nature of non-targeted metabolomics methodologies has the limitation of being semi-quantitative based on the their RTs (vs. an internal standard) and mass fragmentation patterns. The use of non-targeted metabolomics in the present study was to determine the underlying metabolic changes, to allow insight into the systems level changes. However, specific mechanisms identified need further validation to make specific conclusions. Additionally, a second limitation is the limited number of experimental replicates per group (6–8/group, Fig. 2g–i). Previous studies have noted an increasing chance of false-positive findings with reduced statistical power when replicates are limited, a common problem also associated with high throughput technologies in general; it is also possible that relevant markers have been similarly dismissed (Drogan et al. 2014; Floegel et al. 2013). For these reasons, the use of an internal control for each chemical class of specific metabolites in question should be used to validate these results. Based on this context, we focus our discussion here on the metabolomics signatures and our limited validation of specific enzyme effects.
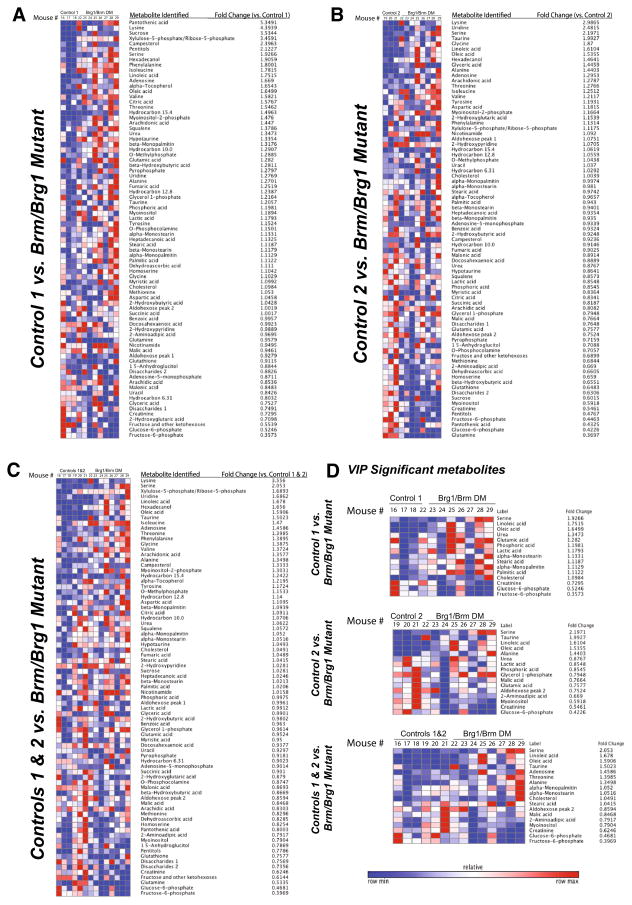
The unsupervised heat maps illustrating the differences in metabolite quantities for each of the three individual analyses, identified metabolites that were different from the controls by 5.3 to 0.35 fold (Fig. 3a, b, c). When individual VIP significant metabolites were identified in the three analyses, serine, linoleic acid, oleic acid, and taurine were >2.0 fold and fructose-6-phosphate, glucose-6-phosphate, creatinine, myoinositol were decreased <0.8 fold and compared to controls (Fig. 3d). We next took the VIP significant metabolites that were also t test significant in the three comparisons (control groupings) and performed a pathway enrichment analysis (Table 3). All three comparisons consistently identified oleic acid and linoleic acid as significant, with the combined controls additionally identifying fructose-6-phosphate, creatinine, and alanine as significant.
Fig. 3.
Unsupervised heat maps of all identified metabolites and their associated fold changes in Brg1/Brm double-mutant hearts compared to a Control Group 1 (Brg1floxed/floxed; αMHC-Cre-ERT+/0; Brm−/− minus tamoxifen treatment), b Control Group 2 (Brg1floxed/floxed; αMHC-Cre-ERT0/0(no transgene); Brm−/− mice plus tamoxifen treatment), and c Combined Groups 1 & 2 controls. d Identification of the VIP significant metabolites (p<0.05). N = 8 (Brg1/Brm double mutant hearts), N = 3 (Control Group 1), N = 3 (Control Group 2), N = 6 (Control Groups 1 & 2) hearts per group analyzed
Table 3.
VIP significant metabolites that were also t test significant (p < 0.05) in the three comparisons made in this study, along with the related matched pathways from pathway enrichment analysis
| Comparison | Metabolite (p value) | Pathway (hits) | p value |
|---|---|---|---|
| Control 1 versus Brg1/Brm double mutant | Oleic acid (0.005) | Biosynthesis of unsaturated fatty acids (2) | 0.0009 |
| Linoleic acid (0.008) | Linoleic acid metabolism (1) | 0.0080 | |
| Fatty acid biosynthesis (1) | 0.0598a | ||
| Control 2 versus Brg1/Brm double mutant | Alanine (0.005) | Biosynthesis of unsaturated fatty acids (2) | 0.0049 |
| Oleic acid (0.010) | Linoleic acid metabolism (1) | 0.0168 | |
| Linoleic acid (0.0014) | Ascorbate and aldarate metabolism (1) | 0.0252 | |
| Myoinositol (0.0021) | Galactose metabolism (1) | 0.0715a | |
| Inositol phosphate metabolism (1) | 0.0768a | ||
| Fatty acid biosynthesis (1) | 0.1161a | ||
| Control 1 & 2 versus Brg1/Brm double mutant | Oleic acid (0.007) | Biosynthesis of unsaturated fatty acids (2) | 0.0081 |
| Linoleic acid (0.010) | Linoleic acid metabolism (1) | 0.0210 | |
| Fructose-6-phosphate (0.016) | Amino sugar and nucleotide sugar metabolism (1) | 0.1241a | |
| Creatinine (0.017) | Fatty acid biosynthesis (1) | 0.1429a | |
| Alanine (0.033) |
VIP significance was based on the analysis of Control group 1 (Flx/Flx, No Brg1 Tg, Chow+tamoxifen, N = 3), Control Group 2 (Flx/Flx, Brg1 Tg+, Chow diet − no tamoxifen, N = 3), Control Groups 1 & 2 (Flx/Flx, No Brg1 Tg, Chow+tamoxifen AND Flx/Flx, Brg1 Tg+, Chow diet − no tamoxifen, N = 6), and Brg1/Brm Double Mutant (Chow+tamoxifen, N = 5) hearts
Not significant
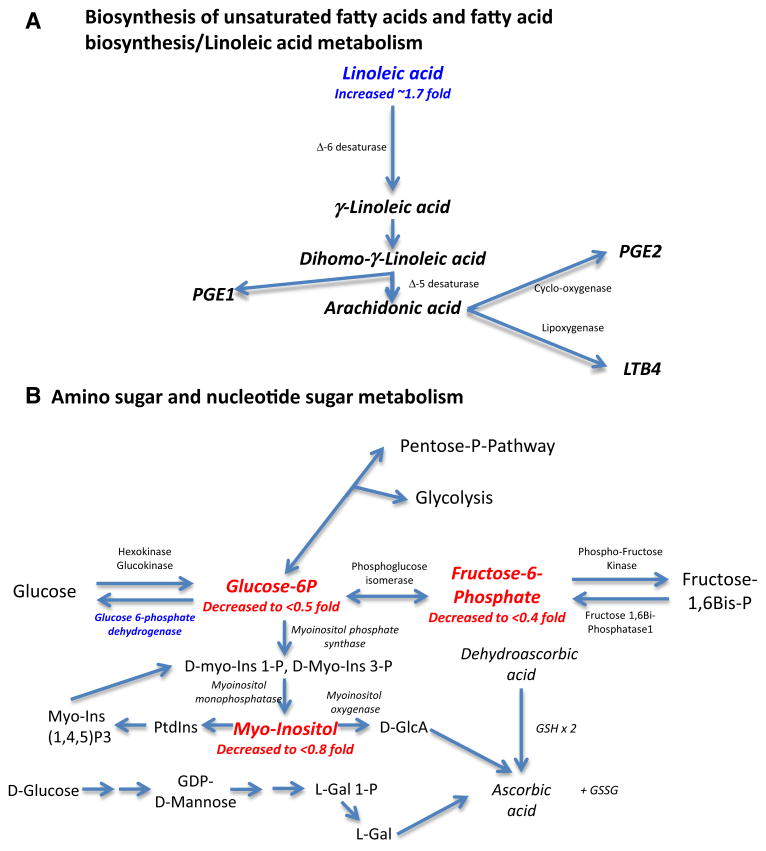
The significant metabolic pathways that were identified included (1) biosynthesis of unsaturated fatty acids and (2) linoleic acid metabolism; (Table 3, right column). In the context of VIP significant metabolites (Fig. 3d), increased linoleic acid may broadly involve arachidonic acid pathways (Fig. 4a). Decreased glucose-6-phophate and fructose-6-phosphate (<0.5 fold) and downstream myoinositol (decreased <0.8 fold) (Fig. 3d), illustrate multiple points in which glucose metabolism is affected in amino sugar and nucleotide sugar metabolism (Fig. 4b). Similarly, evidence of alterations in alanine metabolism (increases in threonine, alanine, and serine metabolites, Fig. 3d) represents potential increases in the ability to create pyruvate through these pathways (Supplemental Fig. 2a). Finally, decreased creatinine and increases in urea (Fig. 3d) illustrate where reducing cardiomyocyte Brg1/Brm expression affects creatine and creatinine metabolism in vivo (Supplemental Fig. 2b). The mechanistic relevance of these findings in this mouse model is not clear, but the importance of each of these metabolites in cardiac (patho)physiology may offer insights for future investigations.
Fig. 4.
Metabolic pathways related to cardiac Brg1/Brm double-mutant activity in vivo determined by non-targeted metabolomics analysis of ventricular cardiac tissue (see Table 3 for analytic details). Metabolites identified by VIP and t test significance are shown in red in the context of generalized pathways analysis (identified by metaboanalyst). a Biosynthesis of unsaturated fatty acids and fatty acid biosynthesis/linoleic acid metabolism; b Amino sugar and nucleotide sugar metabolism (adapted from http://www.genome.jp/kegg-bin/show_pathway?map00520+C00031) (Color figure online)
Both linoleic acid and oleic acid are polyunsaturated fatty acids involved in critical cellular processes in the heart. Recent metabolomics studies of angiotensin II (Ang II)-induced cardiac hypertrophy in Sprague–Dawley and double transgenic rats harboring human renin and angiotensinogen genes challenged with Ang II blockade found that the double transgenic rats had the greatest differences in the use of fatty acids, having decreased oleic and linoleic acids (Mervaala et al. 2010). Morphological changes suggesting an increase in mitochondrial fusion and altered cardiac expression of the redox-sensitive and cardioprotective metabolic sensor sirtuin 1 was increased in the double transgenic rats (Mervaala et al. 2010). The metabolomics changes and mitochondrial biogenesis induced by Ang II was inhibited by valsartan, illustrating the specificity of the Ang II in the regulation of mitochondrial biogenesis and alterations in metabolomics (Mervaala et al. 2010).
Increased oleic and linoleic acid in the Brg1/Brm double- mutant hearts may be linked directly or indirectly to the role of the SWI/SNF complex in the regulation of fatty acid utilization. The SWI/SNF complex has been implicated in regulating fatty acid oxidation by its interactions with peroxisome proliferator activated receptors (PPARs). The PPAR nuclear receptors, in coordination with RXRα and co-activators such as PGC-1α, act as transcription factors for genes encoding proteins responsible for fatty acid uptake and oxidation. BRG1 and BRM-catalyzed SWI/SNF complexes functionally interact with a variety of nuclear hormone receptors (transcription factors) including the PPAR isoforms (Trotter and Archer 2007; Viswakarma et al. 2010). For example, BAF180, which binds the PPARγ-RXRα complex, contains six bromodomains that bind selectively to acetylated histone tails required for targeting the co-regulator complex to chromatin (Lemon et al. 2001). BRG1 and BRM themselves have been identified in the PPAR-γ promoter, and the SWI/SNF complex itself is critical to the transcriptional regulation of PPAR-γ during the formation of preinitiation complexes (Salma et al. 2004). Other BAFs are critical for PPAR activity, with BAF60c interacting with the ligand-binding domain of PPAR-γ and BAF60c reported to bind the C-terminal PPAR-γ domain in a ligand-dependent manner (Debril et al. 2004). BAF60a is a partner of the PPAR co-activator PGC-1α (Li et al. 2008) and is involved in regulating lipid metabolism by its crosstalk with PPARβ/δ (Gatfield et al. 2009; Wang et al. 1996). The changes in fatty acids used as energy substrates, including oleic acid in the present study, may reflect alterations in fatty acid utilization in the absence of SWI/SNF support of PPAR activity. It is also possible that SWI/SNF alters linoleic acid by its alterations in the mitochondria. Specifically, alterations in the physicochemical properties of membranes, including cardiolipins, could contribute to the cardiac phenotypes reported here (Sparagna and Lesnefsky 2009). Recent studies have identified that linoneic acid preserves mitochondrial cardiolipin found in the inner membrane and attenuates mitochondrial dysfunction in heart failure (He and Han 2014; Mulligan et al. 2012). In the present study, the increases in linoleic acid may reflect this response to protect against heart failure. Direct evidence of altered PPAR activity or mitochondrial cardiolipin is beyond the scope of the present study but merits consideration in the underlying mechanisms that SWI/SNF plays in preserving adult cardiomyocyte function.
Effects on glucose storage as glycogen may also be seen in the Brg1/Brm double-mutant hearts. Alterations in cardiac glucose-6-phosphate (G-6-P) and fructose-6-phosphate (F-6-P) has been reported in previous metabolomics studies. Specifically, increased levels of G-6-P and F-6-P have been seen when phosphofructokinase (PFK) activity was experimentally inhibited (McDonald et al. 2014). By mediating the flow of substrate from F-6-P to Fructose-1,6 BisP, PFK initiates start of glycolysis and downstream utilization of glucose ultimately for generating energy. Mice lacking muscle PFK (PFKM−/−) exhibit cardiac hypertrophy with age, impaired oxidative metabolism, muscle fiber necrosis, and impaired exercise tolerance (Garcia et al. 2009). Similarly, mutations in muscle PFK are found in patients resulting in glycogen storage diseases (Raben and Sherman 1995), characterized by muscle weakness, exercise intolerance, and the inability to utilize carbohydrates for energy production well (Nakajima et al. 2002). Increases in threonine, serine, and alanine in alanine metabolism and decreases in cardiac creatinine (Supplemental Fig. 2a, b, respectively) have not previously been reported in metabolomics studies or linked to the SWI/SNF complex previously and may represent mechanistic links unique to the cardiomyocyte or the unique redundant role of Brg1/Brm in the maintenance of the adult cardiomyocyte in vivo.
Mice lacking liver SNF5/BAF47 die due to post-natal hypoglycemia linked to impaired glycogen storage due to transcriptional down regulation of phosphofructose kinase fructose biphosphatase (PFKFB1), illustrating the critical role of the SWI/SNF complex in glucose metabolism(Gresh et al. 2005). By specifically inhibiting PFK activity, increases in G-6-P and F-6-P levels may have been identified as in the present study, where we impaired the SWI/SNF complex (by inducing cardiomyocyte-specific loss of Brg1/Brm). In contrast to the liver SNF5/BAF47 study, we did not observe a significant difference in PFKFB1 mRNA levels (Supplemental Fig. 3). This finding is not necessarily surprising because the SNF5/BAF47 and BRG1 subunits are known to regulate different target genes and to have different effects in tumor suppression.
4 Concluding Remarks
Previously, the BRG1 and BRM catalytic subunits of SWI/SNF complexes were shown to have a functionally redundant role in maintaining the viability in VECs, resulting in primary defects in the heart, including cardiac dissection, cardiac necrosis, and death (Willis et al. 2012). In the present study, we took a non-targeted approach to metabolomically analyze the effects of inducing the simultaneous loss of Brg1 and Brm in adult cardiomyocytes at an early time point before the lethal effects were seen in vivo. Since the Brg1/Brm double-mutant model had both a genetic component and a tamoxifen feeding component, two types of controls were used and analyzed separately and together to identify significant differences in creatinine, linoleic acid, glucose-6-phosphate, oleic acid, and serine (Table 2). The increased oleic and linoleic acid in the Brg1/Brm double-mutant hearts may be linked to the role of the SWI/SNF complex in the regulation of PPARs, instrumental to the mainly fatty acid oxidation the heart uses as a primary energy source (Neubauer 2007). While the link between BRG1 and BRM regulation of PPARs is limited, impairment of the SWI/SNF complex leads to altered PPAR activity in non-cardiomyocyte cells has been previously identified (Debril et al. 2004; Gatfield et al. 2009; Lemon et al. 2001; Salma et al. 2004; Wang et al. 1996) and may offer clues to novel ways in which BRG1 and BRM redundantly regulate cardiomyocyte fatty acid oxidation for future studies. Similarly, the decreases in cardiac G-6-P and F-6-P (Fig. 2) illustrate a dysregulation in downstream glycolysis and glycogen formation, and may reflect more complex alterations of enzymes both up and downstream (Garcia et al. 2009; Nakajima et al. 2002; Raben and Sherman 1995).
Supplementary Material
Acknowledgments
The authors would like to thank Kumar Pandya for providing the αMHC-Cre-ERT transgenic mice and advice on adding tamoxifen to the rodent chow. The authors would like to acknowledge Janice Weaver (University of North Carolina Animal Histopathology Laboratory) and Victoria Madden (University of North Carolina Microscopy Services Laboratory) for assistance in preparing histological specimens and performing the TEM, respectively. Finally, the authors would also like to thank Tim Koves for his guidance and valuable discussion and suggestions for harvesting and preparing heart samples for metabolomics analysis. This work was supported by the National Institutes of Health (R01HL104129 to M.W. and RO1 CA125237 to S.J.B.), a Jefferson-Pilot Corporation Fellowship (to M.W.), and the Fondation Leducq (to M.W.).
Abbreviation
- Ang II
Angiotensin II
- BAFs
BRG1- or BRM-associated factors
- Brg1
Brahma-related gene 1
- Brm
Brahma
- Flx
LoxP-flanked DNA polymerase gene
- F-6-P
Fructose-6-phosphate
- G-6-P
Glucose-6-phosphate
- α-MHC-Cre-ERT
Cre recombinase fused to a mutated ligand-binding domain of human estrogen receptor
- RXRα
Retinoid X receptor alpha
- PPAR
Peroxisome proliferator activated receptor
- PGC-1α
PPAR-gamma coactivator 1-alpha
- VECs
Vascular endothelial cells
- VIP
Variable importance in projection
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11306-015-0786-7) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflict of interest.
Compliance with Ethical Standards All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Contributor Information
Ranjan Banerjee, University of North Carolina School of Medicine, Chapel Hill, NC, USA.
Scott J. Bultman, Department of Genetics, University of North Carolina, Chapel Hill, NC, USA
Darcy Holley, Department of Genetics, University of North Carolina, Chapel Hill, NC, USA.
Carolyn Hillhouse, Department of Pathology & Laboratory Medicine, University of North Carolina, Chapel Hill, NC, USA.
James R. Bain, Sarah W. Stedman Nutrition and Metabolism Center, Duke Molecular Physiology Institute, Duke University Medical Center, Durham, NC, USA. Division of Endocrinology, Metabolism, and Nutrition, Department of Medicine, Duke University Medical Center, Durham, NC, USA
Christopher B. Newgard, Sarah W. Stedman Nutrition and Metabolism Center, Duke Molecular Physiology Institute, Duke University Medical Center, Durham, NC, USA. Division of Endocrinology, Metabolism, and Nutrition, Department of Medicine, Duke University Medical Center, Durham, NC, USA
Michael J. Muehlbauer, Sarah W. Stedman Nutrition and Metabolism Center, Duke Molecular Physiology Institute, Duke University Medical Center, Durham, NC, USA
Monte S. Willis, Email: monte_willis@med.unc.edu, Department of Pathology & Laboratory Medicine, University of North Carolina, Chapel Hill, NC, USA. McAllister Heart Institute, University of North Carolina, Chapel Hill, NC, USA
References
- Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Metabolomics applied to diabetes research: Moving from information to knowledge. Diabetes. 2009;58:2429–2443. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua A, Willis MS, Bultman SJ. SWI/SNF chromatin-remodeling complexes in cardiovascular development and disease. Cardiovascular Pathology. 2013 doi: 10.1016/j.carpath.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Molecular Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Carley AN, Taegtmeyer H, Lewandowski ED. Matrix revisited: Mechanisms linking energy substrate metabolism to the function of the heart. Circulation Research. 2014;114:717–729. doi: 10.1161/CIRCRESAHA.114.301863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annual Review of Physiology. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- Curtis CD, Davis RB, Ingram KG, Griffin CT. Chromatin-remodeling complex specificity and embryonic vascular development. Cellular and Molecular Life Sciences. 2012 doi: 10.1007/s00018-012-1023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. Journal of Biological Chemistry. 2004;279:16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- Doenst T, Nguyen TD, Abel ED. Cardiac metabolism in heart failure: Implications beyond ATP production. Circulation Research. 2013;113:709–724. doi: 10.1161/CIRCRESAHA.113.300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogan D, et al. Untargeted metabolic profiling identifies altered serum metabolites of type 2 diabetes mellitus in a prospective, nested case-control study. Clinical Chemistry. 2014 doi: 10.1373/clinchem.2014.228965. [DOI] [PubMed] [Google Scholar]
- Fiehn O, et al. Quality control for plant metabolomics: Reporting MSI-compliant studies. Plant Journal. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- Files DC, et al. A critical role for muscle ring finger-1 in acute lung injury-associated skeletal muscle wasting. American Journal of Respiratory and Critical Care Medicine. 2012;185:825–834. doi: 10.1164/rccm.201106-1150OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floegel A, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62:639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier DM, et al. The tandem mass spectrometry newborn screening experience in North Carolina: 1997–2005. Journal of Inherited Metabolic Disease. 2006;29:76–85. doi: 10.1007/s10545-006-0228-9. [DOI] [PubMed] [Google Scholar]
- Garcia M, et al. Phosphofructo-1-kinase deficiency leads to a severe cardiac and hematological disorder in addition to skeletal muscle glycogenosis. PLOS Genetics. 2009;5:e1000615. doi: 10.1371/journal.pgen.1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes and Development. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresh L, et al. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO Journal. 2005;24:3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135:493–500. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proceedings of the National Academy of Sciences USA. 2011;108:2282–2287. doi: 10.1073/pnas.1013751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halket JM, Przyborowska A, Stein SE, Mallard WG, Down S, Chalmers RA. Deconvolution gas chromatography/mass spectrometry of urinary organic acids–potential for pattern recognition and automated identification of metabolic disorders. Rapid Communications in Mass Spectrometry. 1999;13:279–284. doi: 10.1002/(SICI)1097-0231(19990228). [DOI] [PubMed] [Google Scholar]
- Han P, Hang CT, Yang J, Chang CP. Chromatin remodeling in cardiovascular development and physiology. Circulation Research. 2011;108:378–396. doi: 10.1161/CIRCRESAHA.110.224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang CT, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Han X. Cardiolipin remodeling in diabetic heart. Chemistry and Physics of Lipids. 2014;179:75–81. doi: 10.1016/j.chemphyslip.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature Genetics. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, et al. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Analytical Chemistry. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koitabashi N, et al. Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circulation Research. 2009;105:12–15. doi: 10.1161/CIRCRESAHA.109.198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolwicz SC, Jr, Purohit S, Tian R. Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circulation Research. 2013;113:603–616. doi: 10.1161/CIRCRESAHA.113.302095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmueller E, Doermann P, Weckwerth W, Gibon Y, Stitt M, Willmitzer L, Fernie AR, Steinhauser D. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics. 2005;21(8):1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- Kosho T, Kuniba H, Tanikawa Y, Hashimoto Y, Sakurai H. Natural history and parental experience of children with trisomy 18 based on a questionnaire given to a Japanese trisomy 18 parental support group. American Journal of Medical Genetics A. 2013a;161A:1531–1542. doi: 10.1002/ajmg.a.35990. [DOI] [PubMed] [Google Scholar]
- Kosho T, et al. Clinical correlations of mutations affecting six components of the SWI/SNF complex: Detailed description of 21 patients and a review of the literature. American Journal of Medical Genetics A. 2013b;161:1221–1237. doi: 10.1002/ajmg.a.35933. [DOI] [PubMed] [Google Scholar]
- Lei I, Gao X, Sham MH, Wang Z. SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. Journal of Biological Chemistry. 2012;287:24255–24262. doi: 10.1074/jbc.M112.365080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- Li S, et al. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metabolism. 2008;8:105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Mallard WG, Reed J. Automated mass spectral deconvolution and identification system: AMDIS user guide. Gaithersburg: National Institute of Standards and Technology, US Department of Commerce; 1997. p. iv.p. 58. [Google Scholar]
- McDonald TS, Tan KN, Hodson MP, Borges K. Alterations of hippocampal glucose metabolism by even versus uneven medium chain triglycerides. Journal of Cerebral Blood Flow and Metabolism. 2014;34:153–160. doi: 10.1038/jcbfm.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mervaala E, et al. Metabolomics in angiotensin II-induced cardiac hypertrophy. Hypertension. 2010;55:508–515. doi: 10.1161/HYPERTENSIONAHA.109.145490. [DOI] [PubMed] [Google Scholar]
- Mulligan CM, et al. Dietary linoleate preserves cardiolipin and attenuates mitochondrial dysfunction in the failing rat heart. Cardiovascular Research. 2012;94:460–468. doi: 10.1093/cvr/cvs118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H, Raben N, Hamaguchi T, Yamasaki T. Phosphofructokinase deficiency; past, present and future. Current Molecular Medicine. 2002;2:197–212. doi: 10.2174/1566524024605734. [DOI] [PubMed] [Google Scholar]
- Nelson TJ, Balza R, Jr, Xiao Q, Misra RP. SRF-dependent gene expression in isolated cardiomyocytes: Regulation of genes involved in cardiac hypertrophy. Journal of Molecular and Cellular Cardiology. 2005;39:479–489. doi: 10.1016/j.yjmcc.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Neubauer S. The failing heart–an engine out of fuel. New England Journal of Medicine. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- Oakley RH, et al. Essential role of stress hormone signaling in cardiomyocytes for the prevention of heart disease. Proceedings of the National Academy of Sciences USA. 2013;110:17035–17040. doi: 10.1073/pnas.1302546110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N, Sherman JB. Mutations in muscle phosphofructokinase gene. Human Mutation. 1995;6:1–6. doi: 10.1002/humu.1380060102. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO Journal. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Technical advance: Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: Unexpected roles for chromatin. Nature Reviews Genetics. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salma N, Xiao H, Mueller E, Imbalzano AN. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Molecular and Cellular Biology. 2004;24:4651–4663. doi: 10.1128/MCB.24.11.4651-4663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santen GW, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nature Genetics. 2012;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- Sohal DS, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circulation Research. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- Sparagna GC, Lesnefsky EJ. Cardiolipin remodeling in the heart. Journal of Cardiovascular Pharmacology. 2009;53:290–301. doi: 10.1097/FJC.0b013e31819b5461. [DOI] [PubMed] [Google Scholar]
- Stankunas K, et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Developmental Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein SE. An integrated method for spectrum extraction and compound identification from GC/MS data. Journal of the American Society for Mass Spectrometry. 1999;10(8):770–781. [Google Scholar]
- Styczynski MP, Moxley JF, Tong LV, Walther JL, Jensen KL, Stephanopoulos GN. Systematic identification of conserved metabolites in GC/MS data for metabolomics and biomarker discovery. Analytical Chemistry. 2007;79:966–973. doi: 10.1021/ac0614846. [DOI] [PubMed] [Google Scholar]
- Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Molecular and Cellular Biology. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nature Communications. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter KW, Archer TK. Nuclear receptors and chromatin remodeling machinery. Molecular and Cellular Endocrinology. 2007;265–266:162–167. doi: 10.1016/j.mce.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurusaki Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nature Genetics. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- Van Houdt JK, et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nature Genetics. 2012;44(445–9):S1. doi: 10.1038/ng.1105. [DOI] [PubMed] [Google Scholar]
- Viswakarma N, et al. Coactivators in PPAR-regulated gene expression. PPAR Research. 2010 doi: 10.1155/2010/250126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes and Development. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes and Development. 2004;18:3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, et al. Muscle ring finger 1 mediates cardiac atrophy in vivo. American Journal of Physiology Heart and Circulatory Physiology. 2009a;296:H997–H1006. doi: 10.1152/ajpheart.00660.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, et al. Cardiac muscle ring finger-1 increases susceptibility to heart failure in vivo. Circulation Research. 2009b;105:80–88. doi: 10.1161/CIRCRESAHA.109.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, et al. Functional redundancy of SWI/SNF catalytic subunits in maintaining vascular endothelial cells in the adult heart. Circulation Research. 2012;111:e111–e122. doi: 10.1161/CIRCRESAHA.112.265587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0–a comprehensive server for metabolomic data analysis. Nucleic Acids Research. 2012;40:W127–W133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Research. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.