Abstract
Protein docking protocols are used for the prediction of both small molecule binding to DNA and protein macromolecules and of complexes between macromolecules. These protocols are becoming increasingly automated and powerful tools for computer-aided drug design. We review the basic methodologies and strategies used for analyzing molecular recognition by computer docking algorithms and discuss recent experiments in which (i) substrate and substrate analogues are docked to the active site of isocitrate dehydrogenase and (ii) maltose binding protein is docked to the extracellular domain of the receptor, which signals maltose chemotaxis.
Full text
PDF




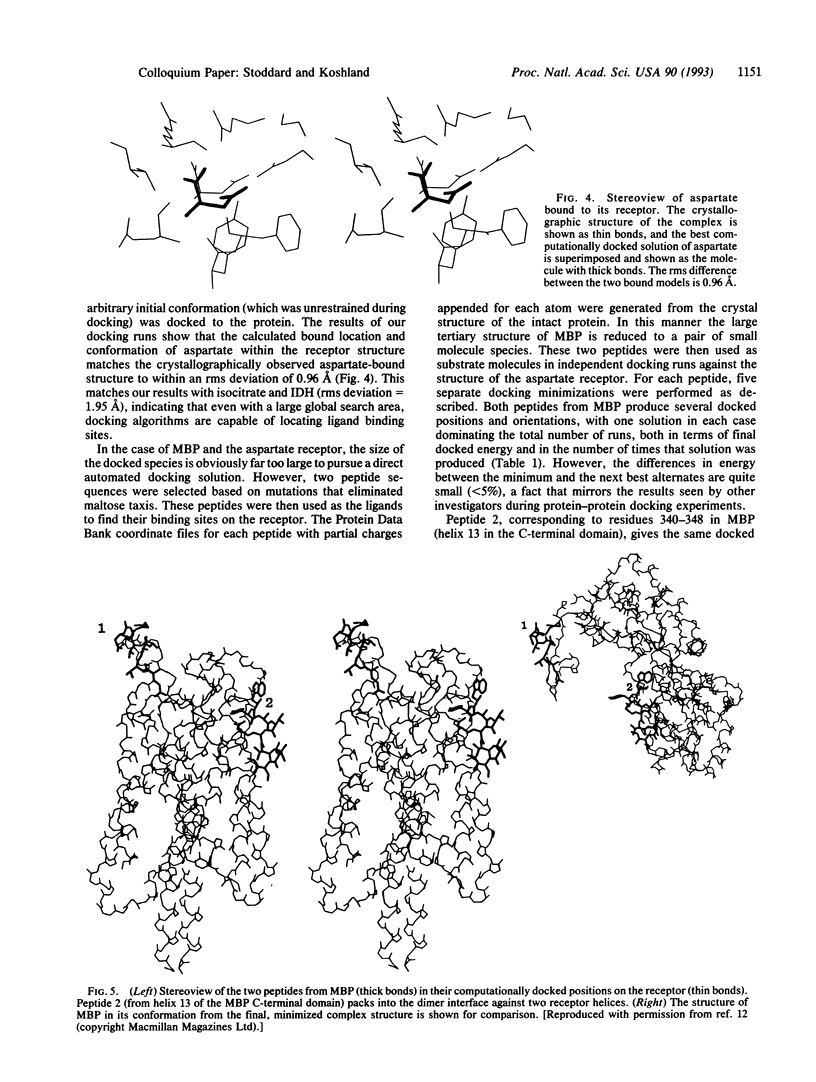
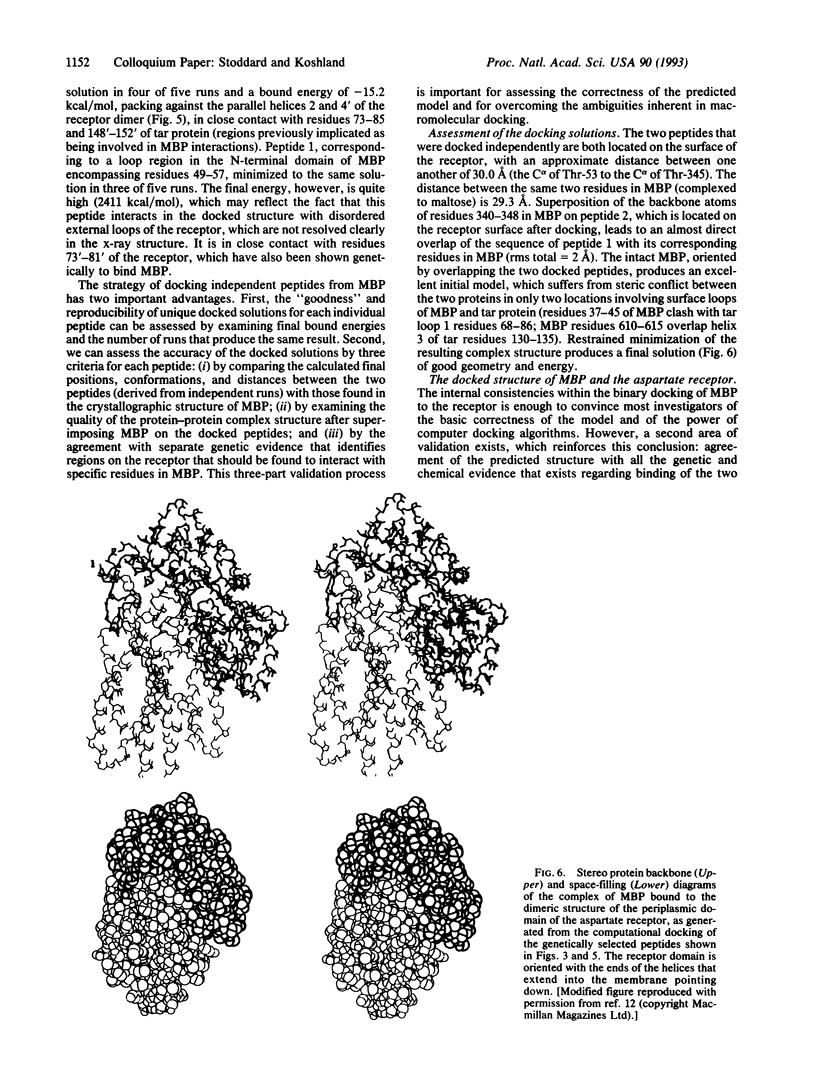

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bone R., Silen J. L., Agard D. A. Structural plasticity broadens the specificity of an engineered protease. Nature. 1989 May 18;339(6221):191–195. doi: 10.1038/339191a0. [DOI] [PubMed] [Google Scholar]
- Cherfils J., Duquerroy S., Janin J. Protein-protein recognition analyzed by docking simulation. Proteins. 1991;11(4):271–280. doi: 10.1002/prot.340110406. [DOI] [PubMed] [Google Scholar]
- DesJarlais R. L., Sheridan R. P., Seibel G. L., Dixon J. S., Kuntz I. D., Venkataraghavan R. Using shape complementarity as an initial screen in designing ligands for a receptor binding site of known three-dimensional structure. J Med Chem. 1988 Apr;31(4):722–729. doi: 10.1021/jm00399a006. [DOI] [PubMed] [Google Scholar]
- Gardina P., Conway C., Kossman M., Manson M. Aspartate and maltose-binding protein interact with adjacent sites in the Tar chemotactic signal transducer of Escherichia coli. J Bacteriol. 1992 Mar;174(5):1528–1536. doi: 10.1128/jb.174.5.1528-1536.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodford P. J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem. 1985 Jul;28(7):849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- Goodsell D. S., Olson A. J. Automated docking of substrates to proteins by simulated annealing. Proteins. 1990;8(3):195–202. doi: 10.1002/prot.340080302. [DOI] [PubMed] [Google Scholar]
- Goodsell D., Dickerson R. E. Isohelical analysis of DNA groove-binding drugs. J Med Chem. 1986 May;29(5):727–733. doi: 10.1021/jm00155a023. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Dean A. M., Koshland D. E., Jr, Stroud R. M. Catalytic mechanism of NADP(+)-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes. Biochemistry. 1991 Sep 3;30(35):8671–8678. doi: 10.1021/bi00099a026. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Dean A. M., Sohl J. L., Koshland D. E., Jr, Stroud R. M. Regulation of an enzyme by phosphorylation at the active site. Science. 1990 Aug 31;249(4972):1012–1016. doi: 10.1126/science.2204109. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Dean A. M., Thorsness P. E., Koshland D. E., Jr, Stroud R. M. Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the free enzyme. J Biol Chem. 1990 Mar 5;265(7):3599–3602. doi: 10.2210/pdb4icd/pdb. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Thorsness P. E., Ramalingam V., Helmers N. H., Koshland D. E., Jr, Stroud R. M. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8635–8639. doi: 10.1073/pnas.86.22.8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick S., Gelatt C. D., Jr, Vecchi M. P. Optimization by simulated annealing. Science. 1983 May 13;220(4598):671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- Kossmann M., Wolff C., Manson M. D. Maltose chemoreceptor of Escherichia coli: interaction of maltose-binding protein and the tar signal transducer. J Bacteriol. 1988 Oct;170(10):4516–4521. doi: 10.1128/jb.170.10.4516-4521.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz I. D., Blaney J. M., Oatley S. J., Langridge R., Ferrin T. E. A geometric approach to macromolecule-ligand interactions. J Mol Biol. 1982 Oct 25;161(2):269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- Kuntz I. D. Structure-based strategies for drug design and discovery. Science. 1992 Aug 21;257(5073):1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- Manson M. D., Kossmann M. Mutations in tar suppress defects in maltose chemotaxis caused by specific malE mutations. J Bacteriol. 1986 Jan;165(1):34–40. doi: 10.1128/jb.165.1.34-40.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn M. V., Privé G. G., Milligan D. L., Scott W. G., Yeh J., Jancarik J., Koshland D. E., Jr, Kim S. H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991 Nov 29;254(5036):1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Milligan D. L., Koshland D. E., Jr Intrasubunit signal transduction by the aspartate chemoreceptor. Science. 1991 Dec 13;254(5038):1651–1654. doi: 10.1126/science.1661030. [DOI] [PubMed] [Google Scholar]
- Mowbray S. L., Koshland D. E., Jr Additive and independent responses in a single receptor: aspartate and maltose stimuli on the tar protein. Cell. 1987 Jul 17;50(2):171–180. doi: 10.1016/0092-8674(87)90213-3. [DOI] [PubMed] [Google Scholar]
- Salemme F. R. An hypothetical structure for an intermolecular electron transfer complex of cytochromes c and b5. J Mol Biol. 1976 Apr 15;102(3):563–568. doi: 10.1016/0022-2836(76)90334-x. [DOI] [PubMed] [Google Scholar]
- Shoichet B. K., Kuntz I. D. Protein docking and complementarity. J Mol Biol. 1991 Sep 5;221(1):327–346. doi: 10.1016/0022-2836(91)80222-g. [DOI] [PubMed] [Google Scholar]
- Spurlino J. C., Lu G. Y., Quiocho F. A. The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem. 1991 Mar 15;266(8):5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- Stoddard B. L., Koshland D. E., Jr Prediction of the structure of a receptor-protein complex using a binary docking method. Nature. 1992 Aug 27;358(6389):774–776. doi: 10.1038/358774a0. [DOI] [PubMed] [Google Scholar]
- Wang E. A., Koshland D. E., Jr Receptor structure in the bacterial sensing system. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7157–7161. doi: 10.1073/pnas.77.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwicker J. Investigating protein-protein interaction surfaces using a reduced stereochemical and electrostatic model. J Mol Biol. 1989 Mar 20;206(2):381–395. doi: 10.1016/0022-2836(89)90487-7. [DOI] [PubMed] [Google Scholar]
- Wilson C., Mace J. E., Agard D. A. Computational method for the design of enzymes with altered substrate specificity. J Mol Biol. 1991 Jul 20;220(2):495–506. doi: 10.1016/0022-2836(91)90026-3. [DOI] [PubMed] [Google Scholar]
- Wodak S. J., Janin J. Computer analysis of protein-protein interaction. J Mol Biol. 1978 Sep 15;124(2):323–342. doi: 10.1016/0022-2836(78)90302-9. [DOI] [PubMed] [Google Scholar]