Abstract
Long noncoding RNA (lncRNA) is a pivotal factor regulating various aspects of genome activity. Genome regulation via DNA methylation and posttranslational histone modifications is a well-documented function of lncRNA in plants, fungi, and animals. Here, we summarize evidence showing that lncRNA also controls chromatin structure including nucleosome positioning and chromosome looping. We focus on data from plant experimental systems, discussed in the context of other eukaryotes. We explain the mechanisms of lncRNA-controlled chromatin remodeling and the implications of the functional interplay between noncoding transcription and several different chromatin remodelers. We propose that the unique properties of RNA make it suitable for controlling chromatin modifications and structure.
Keywords: chromatin remodeling, nucleosome, chromosome looping, RNA Polymerase V
Noncoding RNA and chromatin
Regulation of genome activity is one of the most fundamental processes in living organisms. Many different genomic functions are tightly controlled; the most notable being gene expression and genome integrity. One class of prominent factors controlling genomes are noncoding RNAs, which can be further grouped into small RNAs such as miRNAs, siRNAs, or piRNAs (reviewed in [1]), or categorized as long noncoding RNAs (lncRNAs) [2]. LncRNAs are typically defined as long, functional ribonucleic acids that do not encode proteins or function independent of potentially encoded polypeptides. However, the definition of lncRNAs remains controversial. Indeed, early reports of up to 90% of the eukaryotic genomes that give rise to RNA [3] did not provide much evidence of lncRNAs being functional [4], suggesting that a significant fraction of RNAs produced outside of coding regions originate from transcriptional noise or artefacts of sensitive detection methods. Therefore, an RNA should only be categorized as a lncRNA if there is evidence of functionality meeting at least the “causal role” criteria [5]. The definition of lncRNA also requires it to work independently of its coding potential. This is important because RNAs assumed to be noncoding may encode polypeptides [6] and messenger RNAs may have functions independent of encoded proteins [7]. Moreover, various lncRNAs often share no common evolutionary origins, biological functions, or molecular mechanisms. Therefore, the term “lncRNA” should be used cautiously to avoid implying mechanistic, functional or evolutionary conservation.
There are numerous, characterized lncRNAs with various well-documented functions and this number is quickly growing. lncRNAs have been shown to control genome activity on the chromatin level across the eukaryotic kingdom [2,8]. For instance, the mammalian Xist RNA controls chromatin-mediated inactivation of the X chromosome [9], while the lncRNA HOTAIR recruits chromatin-modifying enzymes and mediates histone modifications on specific target loci in mammals [10]. Moreover, a class of lncRNAs produced by RNA Polymerase V in plants is essential for the process of RNA-mediated transcriptional gene silencing (TGS, see Glossary) [11–13]. These and other well-studied regulatory lncRNAs are not evolutionarily conserved but share a common mode of action by controlling DNA methylation and posttranslational histone modifications [2,8].
In this review, we discuss evidence that lncRNAs control genome activity by affecting chromatin structure, which includes nucleosome positioning and chromosome looping. We focus on data obtained in plant model systems, where a unique genetic toolset allows for gaining deep mechanistic insights into functions of specific classes of lncRNAs. We present recent advances made in plants in the context of related processes across other eukaryotes to provide a broader overview of lncRNA effects on chromatin structure. We discuss how functional interplay between noncoding transcription and several chromatin remodelers allows the establishment of a repressed chromatin status. Finally, we propose that unique properties of RNA make it a versatile factor controlling structure and function of chromatin.
RNA-directed DNA methylation
RNA-mediated Transcriptional Gene Silencing (TGS) is a conserved process where small RNA (sRNA) and lncRNA direct repressive chromatin modifications to specific regions in the genomes (reviewed in [14]). This occurs by the recruitment of DNA methyltransferases and histone modifying enzymes, which mediate DNA methylation and repressive histone modifications, respectively [14]. However, accumulating evidence suggests that TGS also works by controlling chromatin structure, including chromatin remodeling and chromosome looping. TGS has been studied extensively in S. pombe and Arabidopsis thaliana. Both model systems provide comprehensive toolsets, which make TGS highly traceable and allow for deep mechanistic insights to be gained.
In plants, RNA-mediated TGS is also known as RNA-directed DNA Methylation (RdDM; reviewed in detail in [15]). One of its most striking features is the involvement of two specialized RNA polymerases Pol IV and Pol V. Pol IV is responsible for siRNA biogenesis and is believed to produce precursor transcripts [16,17]. These single stranded RNAs are converted to a double-stranded form by an RNA-dependent RNA polymerase and then processed into siRNAs by Dicer [18,19]. By contrast, Pol V is believed to produce scaffold transcripts, which provide binding sites for several RNA-binding proteins that recruit chromatin-modifying enzymes [11,20–26]. RNA produced by Pol V or the process of transcription itself is likely to be functionally relevant since mutations in Pol V subunits cause loss of transcriptional gene silencing [20,21]. Pol V transcripts are also believed to not encode proteins [11]. Even if they do, they seem to work independent of any coding potential as shown by a strong overlap between Pol V binding to chromatin and Pol V-dependent repressive chromatin modifications [24,27,28]. Given these characteristics, scaffold transcripts produced by Pol V are considered lncRNA.
Despite the lack of sequence conservation, lncRNAs produced by Pol V share several similarities with scaffold transcripts produced by Pol II during TGS in S. pombe and arguably also by the piRNA pathway in metazoans [14,29]. The most important similarity is the ability to provide binding sites for sRNA-directed silencing complexes [30]. Moreover, these lncRNAs display comparable functional associations with key proteins involved in TGS including Dicer, Argonaute, and chromatin-modifying enzymes, as well as the ability to transcribe heterochromatic sequences, which are generally presumed to be refractory to transcription [14,29]. RdDM also has several unique features specific to plants that have allowed several pioneering discoveries. Indeed, because Pol IV and Pol V are not essential for viability in Arabidopsis thaliana [16,17,20,21], specific classes of lncRNAs can be eliminated by mutating unique subunits of these RNA polymerases [11,19]. This approach can be used to distinguish lncRNAs from mRNAs, to determine which chromatin modifiers are recruited by lncRNAs and to identify chromatin modifications, which are directed by lncRNAs.
Chromatin modifications established by RdDM in plants and TGS pathways in other organisms include methylation of lysine 9 of histone H3 and/or DNA methylation [14]. There is, however, a category of chromatin-related mechanisms that affect transcription in a fundamentally different way from DNA methylation or posttranslational histone modifications. These processes affect structural aspects of chromatin, which have the potential to directly affect transcription factor binding and RNA polymerase activity without the involvement of reader proteins. They include nucleosome positioning and chromosome looping. We refer to these levels of chromatin organization as chromatin structure.
lncRNA controls nucleosome positioning
Recruitment of chromatin remodelers by lncRNA in Arabidopsis
One of the documented functions of TGS in controlling chromatin structure is nucleosome positioning. Nucleosomes are the fundamental unit of chromatin whereby DNA is wrapped around histone octamers. Tight interaction with histone cores can strongly affect DNA accessibility. Moreover, nucleosome positioning is a critical factor in controlling gene expression, and it is determined by a combination of local DNA features and active remodeling [31,32]. Modulation of nucleosome positioning by lncRNA employs various molecular mechanisms from the recruitment of ATP-dependent remodelers through the negative regulation of chromatin remodeling to transcription-mediated nucleosome stabilization.
In Arabidopsis, lncRNA produced by Pol V serves as a binding scaffold for several RNA-binding proteins including INVOLVED IN DE NOVO 2 (IDN2). This protein was discovered in forward genetic screens and was shown to be required for RdDM [33,34]. IDN2 physically interacts with SWI3B, a core subunit of the SWI/SNF complex [25], which is a putative ATP-dependent chromatin remodeler [35]. This interaction guides the SWI/SNF complex to loci transcribed by Pol V, where, in turn, specific nucleosomes are stabilized [25] (Fig. 1, top). This way, lncRNA produced by Pol V in Arabidopsis is involved in active nucleosome positioning.
Figure 1.
Mechanisms preventing negative feedback between non-coding transcription and lncRNA-mediated nucleosome positioning. Pol V produces lncRNA, which serves as a binding scaffold for RNA-binding proteins including IDN2 (top). IDN2 recruits the SWI/SNF chromatin remodeling complex, which positions nucleosomes. Positioned nucleosomes contribute to repression of Pol II transcription. In a subsequent round of transcription (bottom) the previously positioned nucleosome could prevent Pol V from transcribing and reinforcing silencing. The RDD complex, which includes the putative chromatin remodeler DRD1, is proposed to remove the nucleosome and facilitate Pol V transcription, thereby preventing negative feedback between lncRNA and lncRNA-mediated nucleosome positioning.
Recruitment of the SWI/SNF complex to loci targeted by RdDM is also likely to involve additional lncRNA-binding proteins. IDN2 binding to lncRNA has been shown to require the prior presence of ARGONAUTE4 (AGO4) [26], which is the primary Argonaute involved in RdDM in Arabidopsis [36]. AGO4 incorporates siRNAs, which provide it with sequence specificity towards particular genomic regions, possibly by base pairing between siRNA and lncRNA [24,37]. Because SWI3B is recruited by IDN2 [25], SWI/SNF binding to RdDM targets may likely require AGO4 and siRNA. Another lncRNA-binding protein involved in RdDM is SUPPRESSOR OF TY INSERTION 5 - LIKE (SPT5L) [38,39], which binds silenced loci in parallel to AGO4 [23]. Although the function of SPT5L and its effects on IDN2 binding to lncRNA remain unknown, it is required for transcriptional silencing at least at a subset of RdDM targets [23,38,39]. This suggests that SPT5L may also be involved in SWI/SNF recruitment to chromatin. Similarly, a maize homolog of the RNA-dependent RNA polymerase, which is required for siRNA production, has been shown to affect nucleosome positioning on specific loci [40]. Although there is no evidence of RdDM-mediated recruitment of SWI/SNF in maize, this further suggests that additional RdDM components are involved in nucleosome positioning.
Recruitment of chromatin remodelers by lncRNA in S. pombe
A similar mechanism exists in S. pombe, where pericentromeric and other heterochromatic regions are transcribed and these transcripts are bound by Seb1, a homolog of the conserved RNA-binding protein Nrd1 [41]. Seb1 in turn recruits the SHREC complex, which contains the putative Snf2 chromatin remodeler Mit1 and is required for proper nucleosome positioning [41– 43]. SHREC eliminates nucleosome-free regions and establishes dimethylation of lysine 9 of histone H3 (H3K9me2) [41,44,45]. Thereby, sites of transcription initiation may become inaccessible and Pol II association may be inhibited [44]. Together, these results indicate that in fission yeast lncRNA controls nucleosome positioning by recruiting ATP-dependent chromatin remodeling factors. This mechanism is similar to nucleosome positioning in plant RdDM, where a chromatin remodeler is recruited by heterochromatic lncRNA. An important difference is that SHREC recruitment does not involve siRNAs or Argonaute and appears to work in parallel to RNAi [41].
Although several lines of evidence show that lncRNAs control nucleosome positioning in various organisms, it remains unknown if recruitment of chromatin remodelers is the primary mechanism responsible for this phenomenon. lncRNA-mediated transcriptional silencing is known to direct DNA methylation and H3K9me2 [14]. These repressive chromatin modifications may then be recognized by reader proteins, which in turn could recruit chromatin remodelers. Such an alternative scenario has been shown in fission yeast where the SHREC complex is recruited not only by lncRNAs but also by HP1 [42]. Similarly, in Arabidopsis, a lncRNA-directed DNA methyltransferase has been shown to affect at least a subset of nucleosomes [25]. This finding suggests that nucleosome positioning may be controlled not only by proteins directly recruited by lncRNAs but also by lncRNA-mediated chromatin modifications.
Other ways lncRNA may affect nucleosome positioning
While nucleosome positioning in TGS requires lncRNA to recruit ATP-dependent chromatin remodelers, additional mechanisms have emerged that point to lncRNAs that are evolutionarily and functionally unrelated to those involved in TGS. lncRNAs not involved in TGS can direct nucleosome positioning through negative regulation of ATP-dependent chromatin remodelers and transcriptional interference. Indeed, the human SChLAP1 lncRNA, which is overexpressed in a subset of cancer cells, physically interacts with a subunit of SWI/SNF and prevents or weakens SWI/SNF binding to chromatin, causing widespread changes in gene expression [46]. lncRNA may also mediate co-transcriptional nucleosome positioning. The SER3 gene in yeast is controlled in this manner, and transcription of the intergenic noncoding transcript SRG1, which overlaps the SER3 promoter [47], causes the deposition of nucleosomes along its sequence and represses SER3 [48].
Together, these data indicate that lncRNA is capable of controlling more than DNA methylation and histone modifications, as it can also use a variety of mechanisms to regulate nucleosome positioning. lncRNA-mediated control of nucleosome positioning has been reported in plants, fungi, and animals, which indicates that it may be a broad function of various lncRNAs.
Conflict between silencing and silencers
Nucleosomes and transcription feedback
lncRNAs may position nucleosomes, which in turn, might repress mRNA production. However, this lncRNA-mediated nucleosome positioning may repress noncoding transcription itself. The same logic applies to all repressive events on chromatin and constitutes a well recognized conflict between silencing and cis-acting silencers [49]. It implies a negative feedback between silencing and silencers, which is not compatible with maintenance of transcriptional silencing. In the case of DNA methylation and histone modifications, repressive marks are recognized by reader proteins, which affect the ability of Pol II to transcribe mRNAs. The conflict between silencing and silencer lncRNA is resolved if the RNA polymerase, which produces lncRNAs, is not functionally connected to repressive reader proteins. This scenario is predicted for Pol IV and Pol V in plants, which are able to transcribe methylated DNA [19,27]. However, much less is known about Pol II-mediated noncoding transcription on silenced regions in S. pombe.
Nonetheless an alternative explanation has been proposed where variation of chromatin modifications during the cell cycle provides a window of opportunity for Pol II to transcribe silenced loci [50,51].
Nucleosome positioning is fundamentally different than DNA methylation or repressive histone modifications because it is more transient and provides a direct steric hindrance to transcription over well-positioned nucleosomes. Therefore, transcription of cis-acting lncRNAs, which mediate nucleosome positioning, is expected to require active removal of nucleosomes prior to every round of transcription. Evidence of such mechanisms exists in A. thaliana, where two putative ATP-dependent chromatin remodelers have been genetically identified to work upstream of Pol IV and Pol V.
Nucleosome positioning upstream of lncRNA production
CLASSY1 is a SNF2 protein that is required for Pol IV-dependent production of siRNA [52]. It physically interacts with Pol IV and is believed to be necessary for Pol IV transcription [19,53,54]. DRD1 is a related protein [55] and is required for Pol V binding to chromatin and transcription [11]. Existence of these putative chromatin remodelers suggests that production of lncRNAs by Pol IV and Pol V requires active remodeling of nucleosomes, which is essential for preventing negative feedback between silencing and cis-acting silencers (Fig. 1, bottom). How CLASSY1 and DRD1 exactly work, remains unknown. Their chromatin remodeling activities were predicted based on their amino acid sequences without biochemical corroboration. Both CLASSY1 and DRD1 belong to the Rad54-like group of Snf2 ATPases, which has been implicated in homologous recombination [35], suggesting that RdDM may involve single-stranded DNA [55,56]. It is also unknown if CLASSY1 and DRD1 are required for initiation or elongation of transcription.
Both CLASSY1 and DRD1 have been shown to associate with other proteins required for noncoding transcription. CLASSY1 associates with SHH1, which binds histone H3 tails containing repressive posttranslational modifications (methylated H3K9 and unmethylated H3K4) [57]. DRD1 is a subunit of the DDR complex, which also includes the hinge domain protein DMS3 [58], and another protein, RDM1 [59,60]. The DDR complex has been shown to associate with two catalytically inactive SET-domain proteins (SUVH2 and SUVH9), which bind methylated DNA [61–64]. Identification of these protein-protein interactions and subsequent ChIP studies [28,57,64] suggest that noncoding transcription by Pol IV and Pol V requires recognition of pre-existing repressive chromatin modifications followed by nucleosome remodeling.
It is intriguing that production of lncRNA requires both the recognition of repressive chromatin marks and nucleosome remodeling. Control of Pol IV and Pol V transcription by pre-existing chromatin modifications allows RdDM to work in a positive feedback loop, where RNA-mediated establishment of H3K9me2 and DNA methylation further enhance lncRNA production and silencing [57,64]. In contrast, nucleosome remodeling upstream of Pol IV and Pol V is likely to prevent a negative feedback loop between lncRNA-mediated nucleosome positioning by SWI/SNF and noncoding transcription (Fig. 2).
Figure 2.
Feedback between non-coding transcription and chromatin in TGS. lncRNA mediates repressive chromatin modifications (DNA methylation and H3K9me2), which facilitate non-coding transcription. Therefore, lncRNA-mediated repressive chromatin modifications are maintained by a positive feedback loop. lncRNA-mediated nucleosome positioning creates a physical obstacle to subsequent rounds of transcription. This way, there is negative feedback between non-coding transcription and nucleosome positioning, which could prevent efficient silencing. Additional chromatin remodelers are presumed to prevent this negative feedback and facilitate efficient silencing.
A similar positive feedback loop mechanism exists in S. pombe TGS, where binding of the RITS complex to chromatin requires not only an interaction with scaffold RNA [30] but also recognition of methylated H3K9 by the chromodomain of Chp1 [65,66]. It is, however, unclear if there are any mechanisms preventing a negative feedback between RNA-directed nucleosome positioning and scaffold RNA production in S. pombe, comparable to the events in Arabidopsis.
Together, the literature discussed above indicates that the presence of a lncRNA-controlled nucleosome positioning mechanism in plant RdDM is accompanied by nucleosome remodeling upstream of lncRNA production. It is likely that this is necessary to prevent a negative feedback loop between silencing and silencers. Although there is no evidence yet of similar processes in other organisms, it could be a more general feature of RNA-mediated silencing pathways.
lncRNA controls chromosome looping
lncRNA may also control other important aspects of chromatin structure such as chromosome looping. A compelling example of such looping in plants has been observed for the APOLO lncRNA, which controls the expression of PID, a transporter of the plant hormone auxin [67]. The APOLO locus is transcribed by Pol V and targeted by RdDM. When the APOLO locus is repressed (transcribed by Pol V but not by Pol II), it loops to PID and causes its transcriptional repression. However, when Pol V transcription is lost, APOLO is transcribed by Pol II and its looping to PID is inhibited, causing the activation of PID [67]. This indicates that lncRNA produced by Pol V is involved in establishing a chromosome loop. Although the exact mechanism of this looping event remains unknown, one could speculate that nascent lncRNA produced from APOLO physically interacts with PID and brings APOLO and PID together. This is supported by lncRNA interacting with chromatin at both loci. Alternatively, RNA-mediated chromatin modifications could directly affect chromosome looping.
Similar effects of lncRNA on chromosome looping have been documented in mammals, where enhancers are transcribed and give rise to enhancer RNAs (eRNAs) [68,69]. These eRNAs control gene expression [70], possibly by affecting looping between enhancers and promoters [70,71]. Such a mechanism has been shown for genes controlled by the mammalian transcription factor ER-α [72]. Upon activation, ER-α binds enhancers and causes an increase in eRNA production. eRNAs mediate chromosome looping between enhancers and target promoters, thereby increasing the transcription of target genes [72]. eRNA may directly contribute to looping by interacting with proteins bound to target promoters. This was shown for the eRNA ncRNA-a7, which physically interacts with the Mediator complex. This interaction mediates enhancer-promoter looping and causes the activation of target genes [73].
The functional relationship between lncRNA and chromosome looping is probably much more complex than the effects described above. lncRNAs unrelated to eRNAs have also been shown to mediate long range chromosomal interactions [74,75] and to negatively control loop formation [76]. Moreover, the expression of lncRNA itself may be controlled by chromosome looping [77], and chromosome looping may control the spreading of lncRNA throughout the chromosomes, as has been proposed for Xist [78].
Together, these data show that lncRNA may affect chromatin structure beyond nucleosome positioning – by mediating three-dimensional organization of chromosomes. Different classes of lncRNAs in various organisms control chromosome looping using different mechanisms, suggesting a broader role of lncRNA in controlling chromatin structure.
Concluding remarks
The sheer variety of different mechanisms used by lncRNAs to control chromatin structure raises an important question. Are any aspects of this process mechanistically conserved? These mechanisms appear to be conserved only when lncRNAs share a common evolutionary origin. Informative examples are scaffold transcripts in plant RdDM and S. pombe TGS, which both originate from an ancestral RNAi pathway.
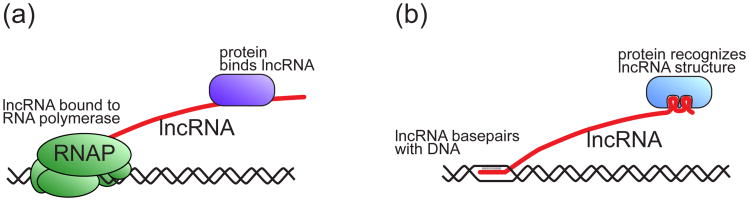
Although processes involving unrelated lncRNAs usually share little mechanistic conservation, they tend to follow some general rules, possibly indicating convergent evolution. These rules likely reflect the fundamental properties of RNA, which make lncRNAs such versatile molecules. One of those features is that, while still being transcribed, RNA may be used to recognize and bind specific genomic loci [2] (Figure 3a). Nascent RNA may serve as a more universal binding platform than DNA on regions where repressive chromatin modifications are actively established. Once lncRNA is produced, protein binding to this RNA should not be affected by chromatin modifications or structure. Thereby, efficient noncoding transcription may be sufficient to avoid a negative feedback between silencing and silencers. Examples of lncRNA following this general rule include scaffold transcripts in TGS and enhancer RNAs.
Figure 3.
Proposed unique features that make lncRNA a versatile factor in controlling chromatin structure.
(a) lncRNA may work in cis by staying attached to the transcribing RNA polymerase. Nascent lncRNA provides a binding site for RNA binding proteins. RNA-binding proteins interact with lncRNA by a sequence- or structure-specific mechanism.
(b) lncRNA may work in trans by combining nucleotide sequence and complex three-dimensional structure. Sequence complementarity may facilitate lncRNA binding to specific loci in the genome. A complex three-dimensional structure may allow interaction with proteins that are recruited to chromatin or mediate chromosome looping.
RNA has another unique property – it combines nucleotide sequence with the ability to acquire complex structures. Nucleotide sequence may allow binding to RNA or DNA by base pairing. Such a binding mechanism has a clear advantage over complex sequence-specific DNA-or RNA-binding protein domains. The ability of RNA to form complex structures may also allow structure-based interactions with proteins. Combining sequence- and structure-based interactions may be a versatile mechanism for guiding proteins to DNA in trans, targeting proteins to RNA or bringing two genomic regions together (Fig. 3b). Such a mechanism has been recently suggested to be involved in class switch recombination [79].
In summary, accumulating evidence identifies lncRNA as an important factor controlling chromatin structure. Therefore, elucidating mechanisms used by lncRNA in controlling chromatin structure has become an exciting and important goal for future research (Outstanding questions box). Considering that lncRNA is potentially mechanistically versatile, it will be especially interesting to discover if lncRNA may use its unique properties to affect chromatin in ways that would be much harder for proteins to achieve.
Outstanding Questions.
Do chromatin modifications such as DNA methylation and posttranslational histone modifications affect nucleosome positioning?
Does ATP-dependent chromatin remodeling affect DNA methylation and posttranslational histone modifications?
Is the ability to recruit ATP-dependent chromatin remodelers or to modulate their activity a general feature of lncRNA?
Is lncRNA-mediated nucleosome positioning usually accompanied by the removal of nucleosomes prior to non-coding transcription?
Is the control of chromosome looping a general mechanism contributing to gene regulation by several classes of lncRNA?
Are any other structural features of chromatin controlled by lncRNA?
Trends box.
Long noncoding RNA (lncRNA) modulates genome activity not only by affecting DNA\ methylation and postranslational histone modifications but also by controlling chromatin structure.
Plant model systems offer a unique genetic toolset for studying lncRNA and chromatin structure. These tools include a multitude of viable mutants and a specialized RNA polymerase producing scaffold lncRNA.
Various lncRNAs have been shown to control nucleosome positioning and chromosome looping in plants and other eukaryotes.
Noncoding transcription involved in transcriptional gene silencing in plants cooperates with at least two putative ATP-dependent chromatin remodelers.
Unique features of RNA make it especially suited for controlling chromatin structure.
Acknowledgments
Our work on the topic of this review is supported by a grant from the National Institutes of Health R01GM108722 to A.T.W. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. G.B. was supported by the Austrian Science Fund (FWF) fellowship J3199-B09. We thank Gyorgyi Csankovszki for critical reading of the manuscript.
Glossary
- Nucleosome
a unit of chromatin organization, where ∼150 bp of DNA is wrapped around a core made of histone proteins
- DNA methylation
a covalent modification of DNA, in eukaryotes predominantly involved in controlling gene expression
- Histone modifications
covalent modifications of histone proteins, which provide binding sites for reader proteins and affect the activity of specific genomic regions
- RNA-mediated Transcriptional Gene Silencing (TGS)
a broad group of RNAi-related mechanisms, where small RNAs silence specific regions of the genome by mediating the establishment of repressive chromatin modifications
- RNA-directed DNA Methylation (RdDM)
a plant TGS pathway characterized by the involvement of two specialized RNA polymerases, Pol IV and Pol V
- Chromatin remodeler
an enzyme, which uses energy from ATP to modify positioning of nucleosomes on DNA
- Enhancer RNA (eRNA)
a class of long non-coding RNA implicated in enhancer function
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ENCODE Project Consortium et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doolittle WF. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci U S A. 2013;110:5294–5300. doi: 10.1073/pnas.1221376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graur D, et al. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 2013;5:578–590. doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz-Orera J, et al. Long non-coding RNAs as a source of new peptides. eLife. 2014;3:e03523. doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabin LR, et al. Dogma derailed: the many influences of RNA on the genome. Mol Cell. 2013;49:783–794. doi: 10.1016/j.molcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Froberg JE, et al. Guided by RNAs: X-inactivation as a model for lncRNA function. J Mol Biol. 2013;425:3698–3706. doi: 10.1016/j.jmb.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wierzbicki AT, et al. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wierzbicki AT. The role of long non-coding RNA in transcriptional gene silencing. Curr Opin Plant Biol. 2012;15:517–522. doi: 10.1016/j.pbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 14.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matzke MA, et al. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu Rev Plant Biol. 2014 doi: 10.1146/annurev-arplant-043014-114633. [DOI] [PubMed] [Google Scholar]
- 16.Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Herr AJ, et al. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 18.Haag JR, et al. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol Cell. 2012;48:811–818. doi: 10.1016/j.molcel.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, et al. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 2014 doi: 10.1101/gr.182238.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanno T, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 21.Pontier D, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wierzbicki AT, et al. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowley MJ, et al. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet. 2011;7:e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Q, et al. RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J Cell Mol Biol. 2012 doi: 10.1111/tpj.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y, et al. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Böhmdorfer G, et al. RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J Cell Mol Biol. 2014 doi: 10.1111/tpj.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wierzbicki AT, et al. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012;26:1825–1836. doi: 10.1101/gad.197772.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong X, et al. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 2012;19:870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bühler M, et al. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 31.Narlikar GJ, et al. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ausin I, et al. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol. 2009;16:1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Z, et al. An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis. Plant J Cell Mol Biol. 2010;62:92–99. doi: 10.1111/j.1365-313X.2010.04130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knizewski L, et al. Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 2008;13:557–565. doi: 10.1016/j.tplants.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Havecker ER, et al. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zilberman D, et al. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 38.He XJ, et al. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bies-Etheve N, et al. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labonne JDJ, et al. Changes in nucleosome position at transcriptional start sites of specific genes in Zea mays mediator of paramutation1 mutants. Epigenetics Off J DNA Methylation Soc. 2013;8 doi: 10.4161/epi.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marina DB, et al. A conserved ncRNA-binding protein recruits silencing factors to heterochromatin through an RNAi-independent mechanism. Genes Dev. 2013;27:1851–1856. doi: 10.1101/gad.226019.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugiyama T, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 43.Yamane K, et al. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell. 2011;41:56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia JF, et al. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 2010;24:1758–1771. doi: 10.1101/gad.1946410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Creamer KM, et al. The Mi-2 homolog Mit1 actively positions nucleosomes within heterochromatin to suppress transcription. Mol Cell Biol. 2014;34:2046–2061. doi: 10.1128/MCB.01609-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prensner JR, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martens JA, et al. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 48.Hainer SJ, et al. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 2011;25:29–40. doi: 10.1101/gad.1975011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grewal SIS, Elgin SCR. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kloc A, et al. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol CB. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 52.Smith LM, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Law JA, et al. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci U S A. 2013;110:8290–8295. doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanno T, et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol CB. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 56.Pikaard CS, et al. A transcription fork model for Pol IV and Pol V-dependent RNA-directed DNA methylation. Cold Spring Harb Symp Quant Biol. 2012;77:205–212. doi: 10.1101/sqb.2013.77.014803. [DOI] [PubMed] [Google Scholar]
- 57.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanno T, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 59.Law JA, et al. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol CB. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao Z, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson LM, et al. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 2008;4:e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuhlmann M, Mette MF. Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Mol Biol. 2012;79:623–633. doi: 10.1007/s11103-012-9934-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu ZW, et al. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 2014;10:e1003948. doi: 10.1371/journal.pgen.1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson LM, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507:124–128. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Debeauchamp JL, et al. Chp1-Tas3 interaction is required to recruit RITS to fission yeast centromeres and for maintenance of centromeric heterochromatin. Mol Cell Biol. 2008;28:2154–2166. doi: 10.1128/MCB.01637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schalch T, et al. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34:36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ariel F, et al. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell. 2014;55:383–396. doi: 10.1016/j.molcel.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 68.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ørom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Melo CA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao H, et al. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crevillén P, et al. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013;32:140–148. doi: 10.1038/emboj.2012.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan-Wong SM, et al. Gene loops enhance transcriptional directionality. Science. 2012;338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng S, et al. Non-coding RNA Generated following Lariat Debranching Mediates Targeting of AID to DNA. Cell. 2015;161:762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]