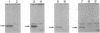Abstract
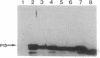
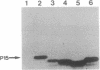
We cloned in Escherichia coli truncated versions of the protein p15 encoded by open reading frame III of cauliflower mosaic virus. We then compared the ability of the wild-type p15 (129 amino acids) and the deleted p15 to bind viral double-stranded DNA genome. Deletions of > 11 amino acids in the C-terminal proline-rich region resulted in loss of DNA binding activity of wild-type p15. Moreover, a point mutation of the proline at position 118 sharply reduced the interaction between the viral protein and DNA. These results suggest that cauliflower mosaic virus p15 belongs to the family of DNA binding proteins having a proline-rich motif involved in interaction with double-stranded DNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Al Ani R., Pfeiffer P., Lebeurier G. The structure of cauliflower mosaic virus. II. Identity and location of the viral polypeptides. Virology. 1979 Feb;93(1):188–197. doi: 10.1016/0042-6822(79)90286-1. [DOI] [PubMed] [Google Scholar]
- Cheng R. H., Olson N. H., Baker T. S. Cauliflower mosaic virus: a 420 subunit (T = 7), multilayer structure. Virology. 1992 Feb;186(2):655–668. doi: 10.1016/0042-6822(92)90032-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Franck A., Guilley H., Jonard G., Richards K., Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980 Aug;21(1):285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- Giband M., Mesnard J. M., Lebeurier G. The gene III product (P15) of cauliflower mosaic virus is a DNA-binding protein while an immunologically related P11 polypeptide is associated with virions. EMBO J. 1986 Oct;5(10):2433–2438. doi: 10.1002/j.1460-2075.1986.tb04518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givord L., Xiong C., Giband M., Koenig I., Hohn T., Lebeurier G., Hirth L. A second cauliflower mosaic virus gene product influences the structure of the viral inclusion body. EMBO J. 1984 Jun;3(6):1423–1427. doi: 10.1002/j.1460-2075.1984.tb01987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn T., Hohn B., Lesot A., Lebeurier G. Restriction map of native and cloned cauliflower mosaic virus DNA. Gene. 1980 Oct;11(1-2):21–31. doi: 10.1016/0378-1119(80)90083-9. [DOI] [PubMed] [Google Scholar]
- Katouzian-Safadi M., Laine B., Chartier F., Cremet J. Y., Belaiche D., Sautiere P., Charlier M. Determination of the DNA-interacting region of the archaebacterial chromosomal protein MC1. Photocrosslinks with 5-bromouracil-substituted DNA. Nucleic Acids Res. 1991 Sep 25;19(18):4937–4941. doi: 10.1093/nar/19.18.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Linderoth N. A., Ziermann R., Haggård-Ljungquist E., Christie G. E., Calendar R. Nucleotide sequence of the DNA packaging and capsid synthesis genes of bacteriophage P2. Nucleic Acids Res. 1991 Dec;19(25):7207–7214. doi: 10.1093/nar/19.25.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Fulton A. M., Dobson M. J., Roberts N. A., Wilson W., Kingsman A. J., Kingsman S. M. The Ty transposon of Saccharomyces cerevisiae determines the synthesis of at least three proteins. Nucleic Acids Res. 1985 Sep 11;13(17):6249–6263. doi: 10.1093/nar/13.17.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnard J. M., Geldreich A., Xiong C., Lebeurier G., Hirth L. Expression of a putative plant viral gene in Escherichia coli. Gene. 1984 Nov;31(1-3):39–47. doi: 10.1016/0378-1119(84)90193-8. [DOI] [PubMed] [Google Scholar]
- Mesnard J. M., Kirchherr D., Wurch T., Lebeurier G. The cauliflower mosaic virus gene III product is a non-sequence-specific DNA binding protein. Virology. 1990 Feb;174(2):622–624. doi: 10.1016/0042-6822(90)90118-b. [DOI] [PubMed] [Google Scholar]
- Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992 Jul;66(7):4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C., Lebeurier G., Hirth L. Detection in vivo of a new gene product (gene III) of cauliflower mosaic virus. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6608–6612. doi: 10.1073/pnas.81.21.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]