Abstract
Motivation: Circadian oscillations have been observed in animals, plants, fungi and cyanobacteria and play a fundamental role in coordinating the homeostasis and behavior of biological systems. Genetically encoded molecular clocks found in nearly every cell, based on negative transcription/translation feedback loops and involving only a dozen genes, play a central role in maintaining these oscillations. However, high-throughput gene expression experiments reveal that in a typical tissue, a much larger fraction () of all transcripts oscillate with the day–night cycle and the oscillating species vary with tissue type suggesting that perhaps a much larger fraction of all transcripts, and perhaps also other molecular species, may bear the potential for circadian oscillations.
Results: To better quantify the pervasiveness and plasticity of circadian oscillations, we conduct the first large-scale analysis aggregating the results of 18 circadian transcriptomic studies and 10 circadian metabolomic studies conducted in mice using different tissues and under different conditions. We find that over half of protein coding genes in the cell can produce transcripts that are circadian in at least one set of conditions and similarly for measured metabolites. Genetic or environmental perturbations can disrupt existing oscillations by changing their amplitudes and phases, suppressing them or giving rise to novel circadian oscillations. The oscillating species and their oscillations provide a characteristic signature of the physiological state of the corresponding cell/tissue. Molecular networks comprise many oscillator loops that have been sculpted by evolution over two trillion day–night cycles to have intrinsic circadian frequency. These oscillating loops are coupled by shared nodes in a large network of coupled circadian oscillators where the clock genes form a major hub. Cells can program and re-program their circadian repertoire through epigenetic and other mechanisms.
Availability and implementation: High-resolution and tissue/condition specific circadian data and networks available at http://circadiomics.igb.uci.edu.
Contact: pfbaldi@ics.uci.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Circadian rhythms are pervasive and play a key role in ensuring homeostatic balance with the environment and coordinating many aspects of physiology including the sleep/wake cycle, eating, hormone and neurotransmitter secretion and even memory and cognitive function (Eckel-Mahan and Sassone-Corsi, 2009; Froy, 2011; Gerstner et al., 2009; Takahashi et al., 2008; Yoo et al., 2004). Disruption of circadian rhythms has been directly linked to health problems ranging from cancer, to insulin resistance, to diabetes, to obesity and to premature ageing (Antunes et al., 2010; Froy, 2010; Karlsson et al., 2001; Knutsson, 2003; Kohsaka et al., 2007; Kondratov et al., 2006; Lamia et al., 2008; Sharifian et al., 2005; Shi et al., 2013; Takahashi et al., 2008; Turek et al., 2005). Research has shown that these circadian rhythms are genetically encoded by a molecular clock found in nearly every cell, with a master clock located in the suprachiasmatic nucleus (SCN) (Moore and Eichler, 1972; Ralph et al., 1990) of the hypothalamus, coordinating and interacting with peripheral clocks throughout the body (Takahashi et al., 2008; Yoo et al., 2004). Central to the cellular clock and the rhythmicity of SCN neurons as well as other cells are transcription factors that drive the expression of their own negative regulators (Partch et al., 2014; Schibler and Sassone-Corsi, 2002). This results in a negative transcriptional and translational feedback loop, highly conserved across species, that perpetuates oscillations in gene expression that occur every 24 h. In mammals, two bHLH transcription factors, CLOCK and BMAL1 heterodimerize and bind to conserved E-box sequences in target gene promoters, thus driving the rhythmic expression of mammalian Period (Per1, Per2 and Per3) and Cryptochrome (Cry1 and Cry2) genes (Stratmann and Schibler, 2006). PER and CRY proteins form a complex that inhibits subsequent CLOCK:BMAL1-mediated gene expression (Brown et al., 2012; Dibner et al., 2010; Partch et al., 2014). In short, the core of the clock is driven by only a dozen genes (Yan et al., 2008).
In contrast, gene expression experiments (Andrews et al., 2010; Eckel-Mahan et al., 2012, 2013; Hughes et al., 2009; Masri et al., 2014; Miller et al., 2007; Panda et al., 2002; Tognini et al., unpublished data) reveal that a much larger fraction, on the order of 10%, of all transcripts in the cell are oscillating in a circadian manner and that the oscillating transcripts differ by cell or tissue type (Panda et al., 2002; Storch et al., 2002; Yan et al., 2008). Thus, the number of oscillating transcripts typically extends beyond the core clock. However, the precise extent of this phenomenon, or its applicability to other molecular species such as metabolites, has not been investigated systematically. While researchers have looked at the common denominator (the master clock genes and its interactors), little has been done to systematically understand the unique and possibly novel oscillations observed in a specific tissue or under a specific set of perturbations. In a recent study (Eckel-Mahan et al., 2013) where we contrasted the circadian profiles of both transcripts and metabolites in the liver of mice fed normal-chow and high-fat diets, we noticed considerable differences associated with a massive reprogramming occurring within the cell. By analyzing not only the transcripts and metabolites that lost their circadian oscillations as a result of the high-fat diet but also the transcripts and metabolites that gained novel circadian oscillations as a result of the perturbation, we were able to discover compensatory oscillations in important molecular species like SREBP1, a transcription factor responsible for lipid synthesis.
In combination, these results raise several fundamental questions (Patel et al., 2014). Exactly how pervasive are circadian oscillations at the molecular level, i.e. how far do they extend beyond the core clock? What is the overlap in circadian oscillations across different tissues and conditions? How flexible and programmable are these oscillations and what are the underlying mechanisms controlling rhythmicity? To begin to address these questions, we conduct a large-scale aggregated analysis of multiple circadian transcriptome and metabolome datasets.
2 Methods
To ensure quality and consistency, we use circadian data from experiments all carried in mice, with at least six time points and, in most cases, at least three replicates.
2.1 Transcriptome analysis
Time-resolved gene expression microarrays from 16 published (Andrews et al., 2010; Eckel-Mahan et al., 2013; Hughes et al., 2009; Masri et al., 2014; Miller et al., 2007; Panda et al., 2002) and 2 unpublished experiments (Tognini et al., unpublished data) were gathered for analysis. The datasets were downloaded from Circa (Hughes et al., 2009) or GEO (Edgar et al., 2002) or provided by the authors. The transcriptomes along with tissue (e.g. liver, muscle) and condition (e.g. wild-type, knockout, high-fat diet) are listed in Supplementary Table S1. To compare gene lists across different microarray platforms/experiments, we used DAVID (Huang et al., 2009, 2007) for all gene/transcript ID conversions.
2.2 Metabolome analysis
Time-resolved metabolite levels measured using LC/GC chromatography were obtained from eight published (Dyar et al., 2014; Eckel-Mahan et al., 2013; Masri et al., 2014) and two unpublished experiments (Abbondante et al., unpublished data). The metabolomes along with corresponding tissues and conditions are listed in Supplementary Table S1. The unique compound identifier reported by the Metabolon (Durham, NC) system was used to compare the list of metabolites across the different experiments.
2.3 Circadian analysis
Gene expression and metabolite levels from all experimental datasets were analyzed using JTK_CYCLE with typical default parameters. JTK_CYCLE (Hughes et al., 2010) implements a nonparametric statistical test which can be used to determine cycling events in gene expression and other time series. A gene was considered circadian, if at least one of its transcripts was found to be circadian by JTK_CYCLE. To correct for multiple testing, we use the Bonferroni-corrected P values produced by JTK_CYCLE (Supplementary). Furthermore, all analyses are conducted at three different cutoffs.
2.4 Creation and analysis of circadian networks
We constructed comprehensive networks maps using CircadiOmics (Eckel-Mahan et al., 2012). The methods to build these networks are described in detail in Patel et al. (2012). Briefly, CircadiOmics combines information about molecular species and their interactions from several databases to provide information-rich and tissue-specific views of the underlying networks in a circadian context. These networks include metabolic and enzymatic reactions, protein–protein interactions and regulatory edges from MotifMap (Daily et al., 2011; Xie et al., 2009) and published ChIP experiments, along with the time series profiles available from the corresponding experimental data. Thus, for instance, through CircadiOmics, we can investigate any particular experiment to find out whether a transcription factor or enzyme has a circadian expression profile or not and which of the genes or metabolites it may control has a circadian profile or not.
3 Results
3.1 Comparison of transcriptomes
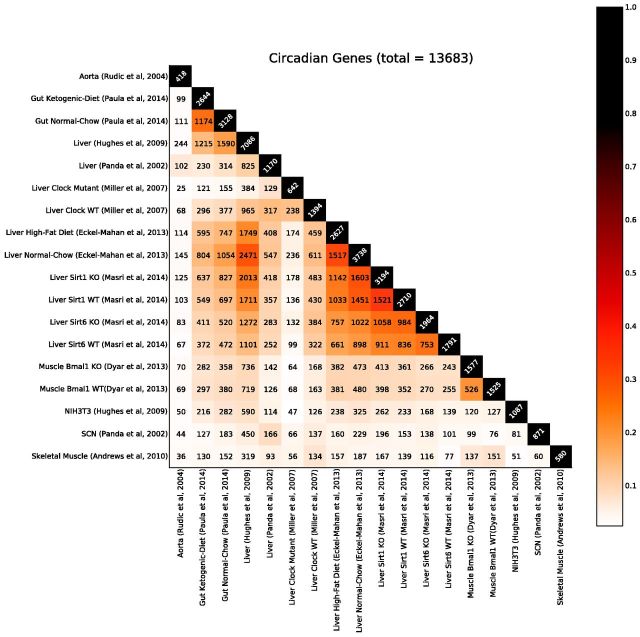
Analysis across different tissue types and conditions reveals surprisingly little overlap between the molecular species that oscillate in one tissue/condition versus another (Fig. 1) beyond the core clock genes. The figure displays the matrix of the sizes of all pairwise intersections of circadian genes across all experiments, color coded so that darker colors correspond to larger intersections. When the results of circadian experiments conducted over different tissues and conditions (listed in Supplementary Table S1) are aggregated with a stringent oscillatory cutoff (P < 0.05), over 13 600 genes are found to oscillate in at least one tissue or condition in mouse. Thus, a significant fraction (∼68%) of the protein coding genes in a mammalian genome is capable of generating mRNAs which oscillate in a circadian fashion in at least one tissue or condition. At a more stringent P-value cutoff of 0.01, we still find ∼8650 genes (Supplementary Fig. S2) exhibiting circadian oscillations in at least one experiment. And at the even more stringent P-value cutoff of 0.005, we still find ∼7877 genes (Supplementary Fig. S4) exhibiting circadian oscillations in at least one experiment. These numbers are likely to increase in the future as more tissues/conditions are studied and aggregated. Even comparing transcriptomes from perturbation experiments performed on the same tissue (liver, see Supplementary Fig. S1) show that a major fraction of genes can produce circadian transcripts under some condition in that tissue alone. Thus, tissue-type, environmental, genetic and even diet perturbations all lead to significant differences in the list of oscillating genes.
Fig. 1.
Pairwise comparison matrix across 18 transcriptomic experiments. The numbers correspond to the number of protein coding genes capable of producing a circadian oscillatory transcript () that are common to both tissues/conditions (i.e. ). The color intensity corresponds to the Tanimoto–Jaccard index (). In total, there are 13 683 (∼67%) protein coding genes that can produce a circadian transcript in at least one tissue or condition
3.2 Comparison of metabolomes
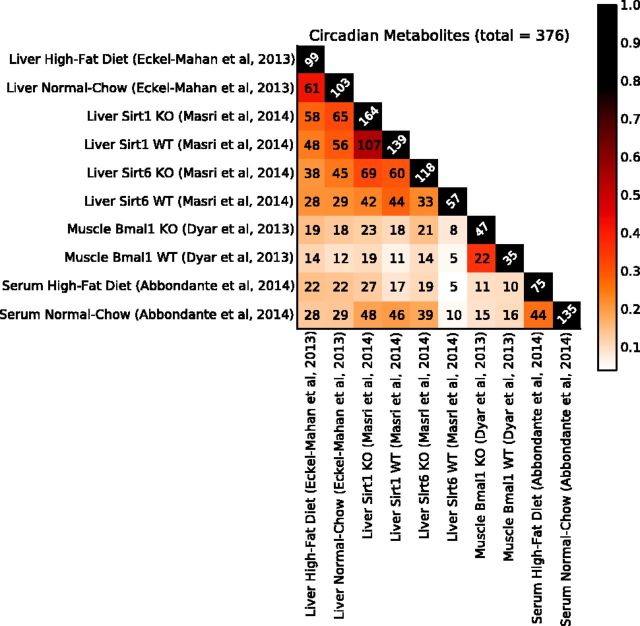
Similar results are seen when time-dependent metabolite levels from different tissues and conditions are aggregated (Fig. 2). The figure displays the matrix of the sizes of all pairwise intersections of circadian metabolites. At a P-value cutoff of 0.05, out of the 554 measured metabolites, we find 376 metabolites oscillating in a circadian manner in at least one of the conditions. Hence, ∼67% of the measured metabolites oscillate even when using a relatively small set of tissue/conditions. At a more stringent P-value cutoff of 0.01, we still find ∼300 metabolites (Supplementary Fig. S3) exhibiting circadian oscillations in at least one experiment. And at the even more stringent P-value cutoff of 0.005, we still find ∼270 metabolites (Supplementary Fig. S5) exhibiting circadian oscillations in at least one experiment.
Fig. 2.
Pairwise comparison matrix across 10 metabolomic experiments. The numbers correspond to the number of oscillating genes/metabolites () that are common to both tissues/conditions (i.e. ). The color intensity corresponds to the Tanimoto–Jaccard index (). In total, there are 376 (∼68%) measured metabolites that oscillate in at least one tissue or condition
Taken together, these results from across tissue and within tissue comparisons establish that a significant fraction of transcripts and metabolites is capable of circadian oscillations in at least one type of tissue or condition. We hypothesize that similar results hold also for protein levels, although systematic high-throughput circadian proteomic measurements are not yet available (see Section 4).
3.3 Circadian oscillations are plastic: effects of perturbations
We observe that genetic or environmental perturbations tend to disrupt circadian oscillations in given system in several ways. As expected, such perturbations can:
Change the amplitude of pre-existing circadian oscillations for some of the molecular species;
Change the phase of pre-existing circadian oscillations for some of the molecular species;
Disrupt or even suppress the pre-existing oscillations of some of the molecular species.
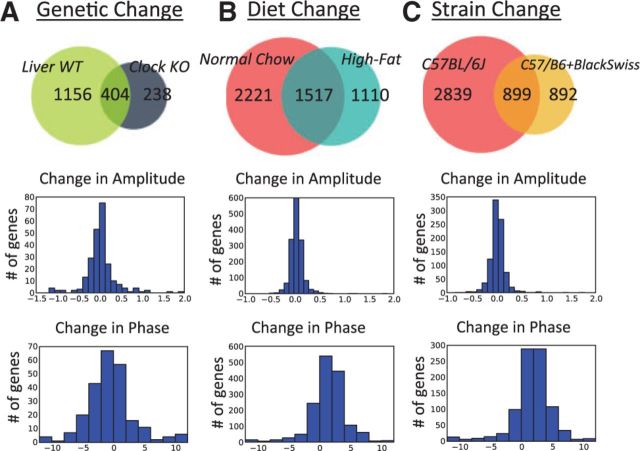
Indeed, experiments involving genetic knockouts, diet changes or even simply different mice strains, show these effects (Fig. 3). For instance, when comparing gene expression in liver tissue from Clock mutant and wild-type mice (Miller et al., 2007), ∼1160 genes show a loss of circadian rhythmicity. However, ∼400 genes oscillate in both conditions but with a difference in amplitude or phase (Fig. 3A). Similarly, when comparing gene expression and metabolite levels in liver tissue from 10-week high-fat-fed versus normal-chow-fed mice (Eckel-Mahan et al., 2013), ∼2200 genes and ∼40 measured metabolites show a loss of circadian rhythmicity, whereas ∼1520 genes and ∼60 measured metabolites oscillate in both conditions, but with a difference in amplitude or phase.
Fig. 3.
First row: Venn diagrams comparing (A) wild-type and Clock mutant liver gene expression, as an example of genetic perturbation; (B) normal-chow fed and high-fat fed liver gene expression, as an example of environmental perturbation and (C) C57BL/6J and C57/B6 + Black Swiss liver gene expression, as an example of ‘strain perturbation’. In all cases, there is massive reprogramming leading to a large number of new oscillations. Second row: histogram showing the changes in amplitudes. Third row: histogram showing the changes in phases (measured in hours)
More importantly perhaps, perturbations can also:
Create new circadian oscillations in many molecular species that were not oscillating in the control case.
Unlike the changes above, the massive creation of new oscillations, all at the same frequency, in such complex systems is puzzling and requires further analysis and explanations (see also Section 4).
3.4 Emergence of new oscillations
When comparing liver samples from Clock mutant and wild-type mice (Miller et al., 2007), ∼240 genes oscillate in the mutants but not in the controls (Fig. 3A). Interestingly, these new oscillations arise in spite of mutating the Clock gene. Similarly, when comparing liver samples from mice fed with high-fat chow versus normal-chow (Eckel-Mahan et al., 2013), ∼1110 genes and ∼40 measured metabolites oscillate in the high-fat but not in the normal-chow condition (Fig. 3B). Important transcription factors, enzymes and metabolites are found among the new oscillating species. For example, SREBF1 a key transcription factor regulating enzymes involved in lipid synthesis shows a new, robust (), circadian oscillation in the high-fat condition.
Remarkably, novel oscillations are also seen in data collected from the same tissue of wild-type mice but corresponding to different genetic strains. Specifically, a comparison of liver tissue from C57BL/6J (Eckel-Mahan et al., 2013) and C57/B6 + Black Swiss (Masri et al., 2014) mice strains uncovers ∼2840 genes that oscillate only in C57BL/6J, ∼890 genes that oscillate only in C57/B6 + Black Swiss and ∼900 genes that oscillate in both strains (Fig. 3C). This is also true when comparing liver samples from two animals with a slightly different mixture of C57/B6 and Black Swiss strains (see Liver Sirt1 WT and Liver Sirt6 WT in Fig. 1). Note that this may in part also explain why differences in oscillatory behavior are seen in assays that are presumed to be identical (e.g. ‘wild-type liver tissue’) but in reality are not, due to significant differences between wild-type strains (Yalcin et al., 2012) and the environments in which the mice are raised. Even the concept of ‘normal chow’ is vague and can differ substantially from one laboratory to another.
In aggregate, these results suggest that the physiological state of a cell or a tissue is strongly characterized by its circadian profile, essentially the list of molecular species that oscillate in a circadian fashion and the characteristics of their oscillations (e.g. phases and amplitudes). Furthermore, one can predict that any significant perturbation (genetic, epigenetic or environmental) will significantly change this profile, by changing amplitudes and phases of existing oscillations and by introducing novel oscillations.
3.5 Web server and visualization
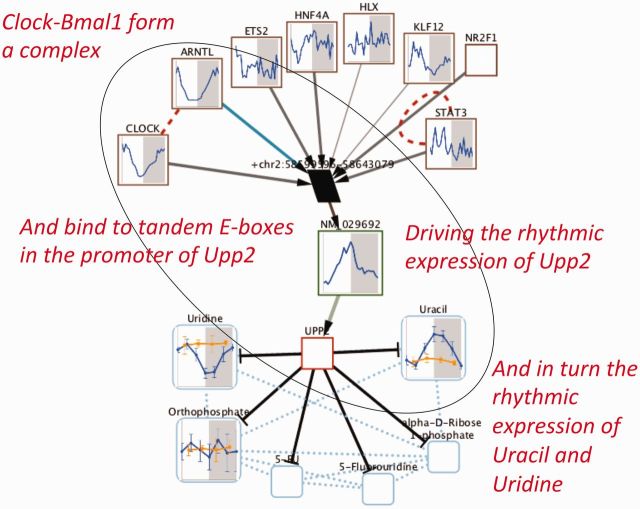
All the transcriptome and metabolome data used in the analyses presented here have been imported into the CircadiOmics database and web server (Patel et al., 2012) and can be analyzed and visualized online at: http://circadiomics.igb.uci.edu. CircadiOmics produces high-resolution biological networks displaying, for instance, metabolites, enzymes, transcription factors and their interactions and concentration changes over time throughout the circadian cycle (Fig. 4).
Fig. 4.
Network and concentration time series visualization showing how the transcription factors in the clock can drive the oscillations of enzymes and corresponding metabolites. In CircadiOmics, users can select an experiment and a specific gene or metabolite and visualize the molecular network in the vicinity of the corresponding node, as well as the time series of the corresponding concentrations. Each box represents a molecular species with, when available, a plot of the time series of its concentration throughout the day–night cycle in the control (blue) and perturbed condition (red)
4 Discussion
Previous gene expression studies conducted with single conditions have already revealed that a significant fraction of the transcriptome can oscillated in a circadian fashion in simple organisms: a third of the transcriptome in the plant Arabidopsis thaliana (Covington et al., 2008; Harmer et al., 2000), 64% of the transcriptome in the cyanobacterium Synechococcus elongatus (Vijayan et al., 2009) and almost the entire transcriptome in the marine unicellular alga Ostreococcus tauri (Monnier et al., 2010). In a recent independent study conducted in 12 mouse organs, 43% of all protein coding genes showed circadian rhythms in transcription somewhere in the body (Zhang et al., 2014). Here, we have extended these studies by aggregating and analyzing both high-throughput transcriptomic and metabolomic data in a mammalian system over multiple tissues and conditions, revealing that a large fraction of the molecular species in a mouse cell/tissue is capable of circadian oscillations under at least some set of conditions. Furthermore, while the experiments used here were conducted using tissue preparations, it is reasonable to infer that even more diversity in oscillations would be observed if one were to aggregate experiments done at the level of single cells both within tissues and across tissues, as well as many other conditions. Thus, our results provide only lower bounds on the total number of transcripts or metabolites that could be found to oscillate in a circadian manner under at least one set of conditions by the same methods. Additional non-transcriptional circadian oscillations, for instance, in the levels of post-translationally modified proteins, are also known to exist (O’Neill et al., 2011; van Ooijen and Millar, 2012) leading us to conjecture that almost any molecular species in the cell is potentially capable of circadian oscillations.
To better interpret these puzzling results, we briefly develop a coupled circadian-oscillators framework logically articulated around four questions: (i) what are the oscillators? (ii) what are their periods? (iii) how are they coupled? and (iv) what is the role of the clock?
4.1 Coupled circadian-oscillators framework
4.1.1 Molecular loops as oscillators
The first observation is that a transcript (or any other molecular species) cannot oscillate in isolation. What really oscillate are entire loops of interacting molecular species comprising different kinds of interactions such as regulatory (transcriptional), protein–protein and enzymatic interactions. Oscillatory loops typically contain an odd number of negative interactions (Fig. 5). Biological network contain a large number of such directed loops and thus many potential oscillators. For instance, in a network consisting of 21 826 genes/proteins with 120 988 edges (114 493 regulatory edges and 6495 physical protein–protein interactions), we found over 3600 directed loops of size 3 and over 71 100 directed loops of size 4 (see also Supplementary Table S3). These numbers are not meant to be precise, as it is well known that there are several sources of noise in reconstructed biological networks, but they are indicative of the general trends and it is reasonable to estimate that the number of potential oscillators in the cell is in the 105 range.
Fig. 5.
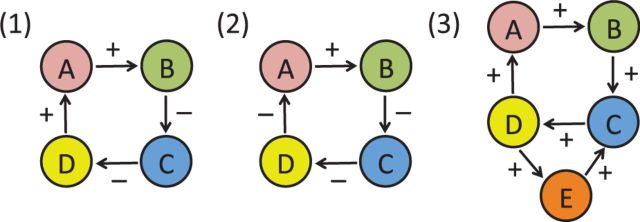
(1) A cycle between four molecular species with an even number of negative interactions. Increasing the concentration of A, increases the concentration of B, which decreases the concentration of C, which increases the concentration of D, which further increases the concentration of A (and vice versa if the concentration of A is decreased). Thus in general, such a system does not oscillate and will tend to converge to one of several fixed-point attractors. (2) A cycle between four molecular species with an odd number of negative interactions. Increasing the concentration of A, increases the concentration of B, which decreases the concentration of C, which decreases the concentration of D, which then decreases the concentration of A. Thus such a system will tend to oscillate. (3) Example of two interlocked cycles one of length three and another of length four sharing one edge (between C and D) with fixed-point attractors. Changing the sign of the shared interaction creates two oscillatory loops, exemplifying how a small change can have a large effect
4.1.2 Circadian periodicity
In the complex molecular circuitry of a cell, having many loops--and thus many potential oscillators-- does not explain why a large fraction of them would oscillate with a circadian frequency. When a complex physical system with many components is perturbed in many different ways, one does not expect to see each time a different subset of its component oscillating at the same constant frequency, unless this frequency is deeply built-in into the system as a resonant frequency. Indeed, high time resolution circadian data (Hughes et al., 2009) show that most oscillating genes have a period of ∼24 h, with some genes oscillating at harmonic periods of about 12 h and 8 h. Very short periods (e.g. periodicity of one hour or less) and periods not commensurate with the day–night cycle (e.g. periodicity of 7 h) are not observed as they are probably not physiological. The key question then is why so many loops exhibit the same 24-h periodicity? We believe evolution provides the answer to this question as the world is drastically different during the day and the night, for instance in terms of temperature, light, winds and predators. Thus paying attention to these differences is likely to have conferred major survival benefits to the corresponding organisms in the course of evolution. It is important to note that some of the earliest unicellular precursors of current living systems were highly circadian. For example, Cyanobacteria which were present 3.4 billion years ago are highly circadian (Vijayan et al., 2009) since they use photosynthesis. Thus circadian oscillations at the molecular level were discovered very early by evolution and subsequently refined and propagated throughout the tree of life over two trillion day–night cycles (Supplementary). Thus evolution has deeply sculpted the relentless circadian rhythm into many of the molecular oscillators present in each cell, so that the circadian frequency is the main resonant frequency of these networks.
4.1.3 Coupling of the oscillators and cellular programming/reprogramming
Armed with an understanding or what the oscillators are and why they may have a built-in resonant period of 24 h, we can now consider how these oscillators are coupled to each other and how biological systems can manipulate the oscillatory landscape and its couplings to adapt to internal or external perturbations.
Many different biological mechanisms couple these oscillators together, but at the root of the coupling, there is always the sharing of vertices (or even edges or paths) between oscillating loops forming an intricate network of coupled circadian oscillators. The couplings in such a network are likely to be non-linear, heterogeneous and condition specific. Reprogramming at the cellular level (Obviously reprogramming occurs at many levels and may involve, for instance hormonal signals triggered by the SCN and cell-to-cell communication within a tissue) occurs by (i) suppressing existing molecular interactions thereby breaking loops; (ii) enabling new molecular interactions thereby creating new loops or (iii) changing the sign of existing molecular interactions thereby modifying the oscillatory behavior of existing loops.
There are several possible non-exclusive mechanisms by which the cell can create, suppress or modify interactions between the different species to rapidly reprogram its oscillatory repertoire. For instance, dynamic changes in the epigenome, like methylation, acetylation and chromatin remodeling can play a central role in selecting the fraction of oscillating species. An epigenetic modification in the promoter of a gene can prevent the expression of a gene permanently, thus suppressing the oscillatory behavior of all the loops containing the corresponding transcript or protein. Removing the modification has the opposite effect. Recent studies have also identified circadian long-range interactions (Aguilar-Arnal et al., 2013) and the role of CLOCK protein as a histone acetyltransferase (Doi et al., 2006). Similarly, a post-translational modification may enable the interaction of two proteins and thus the creation of corresponding loops. Furthermore, nodes or edges associated with many loops act as hubs that can couple and simultaneously influence many other oscillators.
4.1.4 Role of molecular hubs and the clock
Molecular hubs associated with highly connected species usually affect many oscillating loops and are capable of setting up cascades of changes in amplitude, phase and oscillatory behavior. An example is provided by nicotinamide adenine dinucleotide+, a metabolite that participates in many reactions and plays a central role in regulating circadian rhythms (Nakahata et al., 2008; Peek et al., 2013; Ramsey et al., 2009). Not surprisingly, transcription factors also tend to behave like hubs, and the clock itself behaves as a central hub intersecting many loops and helping cellular reprogramming and the selection of a significant fraction of which loops actually oscillate under a given set of internal and external conditions (Fig. 4).
In particular, the main transcription factors in the clock, Clock and Bmal1, are densely connected (see Supplementary Tables S2 and S3). They are known to bind to a single or pair of E-box sites. E-box sites are short (canonical sequence CACGTG) and frequent in the genome (Fig. 6). With a stringent Bayesian Branch Length Score (Xie et al., 2009) greater than 1, we found over 23 800 conserved E-box sites in the mouse genome using MotifMap– several of which are in the promoters of transcription factors. Using time-resolved ChIP-seq data for BMAL1, Rey et al. (2011) identified 2049 E-box binding sites in mouse liver. Among these, ∼60% (1319) showed a rhythmic binding of BMAL1 and 13% of all BMAL1 sites had a pair of E-box elements with spacers of 6–7 base pairs. Thus, in a given environment, cells can reveal or hide a fraction of E-box sites thereby controlling which loops are directly, or indirectly, affected and possibly entrained by Clock and Bmal1.
Fig. 6.
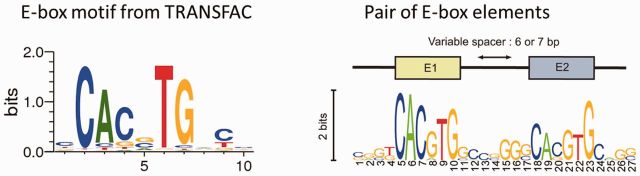
Left: E-box motif logo from TRANSFAC representing the corresponding DNA sequence preferences. Right: Tandem E-box motif logo from ChIP-seq studies (Rey et al., 2011)
To further understand the factors that confer to the cell its circadian reprogramming capabilities, we analyzed the role of the core clock genes in the context of the underlying global molecular network. Using a network with regulatory and protein–protein interaction edges, we calculated the distance of all nodes from Clock or Bmal1 and also the total number of directed loops that contain Clock or Bmal1. We found that ∼10% of genes are one hop away and ∼60–70% genes are two hops away from Clock or Bmal1 (see Supplementary Table S2). In addition, ∼10% of genes are connected to Clock or Bmal1 through a directed loop (see Supplementary Table S3) of size 6 or less. In short, in this network of coupled oscillators, Clock and Bmal1 form a central hub coupling and modulating many other circadian oscillators.
4.2 Formal models of coupled oscillators networks
Formal models of circadian oscillators and coupled oscillators (Baldi and Meir, 1990; Brandt et al., 2006; Goel and Ermentrout, 2002; Goldbeter, 1997; Strogatz, 2000) are briefly discussed in Supplementary Material.
4.3 Differential gene expression analysis
A simple consequence of having so many transcripts oscillating, or with the potential for oscillating, is that some caution should be exercised when drawing differential conclusions from gene expression experiments, especially when these are conducted at a single time point, which is the majority of the cases. While one cannot reasonably expect that all expression experiments be carried at multiple time points along the circadian cycle, it is clear that circadian oscillations of transcripts can impact the lists of genes that appear to be differentially expressed between two conditions and their interpretation.
4.4 Conclusion
At the behavioral level, circadian rhythms are paradoxically both relentless and flexible. Relentless because they turn us into robots executing the same routine every 24 hours. Flexible because this routine is elastic and let us accommodate, for instance, an occasional early meal or a late bedtime without suffering major consequences. Remarkably, these paradoxical features of pervasiveness and plasticity are found at multiple levels of biological organization, including tissues, cells and molecular networks as shown in this work.
Two trillion night-and-day cycles during the course of evolution have deeply sculpted the molecular networks of the cell and made 24-h oscillations pervasive. Aggregation of high-throughput transcriptomic and metabolomic experiments across tissue types and conditions indeed reveal that a large fraction of the molecular network of a cell is primed for and potentially capable of oscillating in a circadian manner. In a given environment taken in its broadest sense (to include for instance genetic modifications or inter-cell/inter-organ communications), through epigenetic and other modifications, a cell or tissue selects which fraction of molecular species out of its entire repertoire exhibit circadian oscillations, including entirely novel oscillations with respect to the corresponding control case.
Ongoing and future work should provide the data to better model and understand networks of coupled-circadian oscillators, predict how they respond to perturbations and use these responses to explain biology and direct therapeutic intervention.
Supplementary Material
Acknowledgements
We thank Paola Tognini and Mari Murakami for providing the unpublished gut ketogenic diet dataset and Yu Liu for doing the circadian analysis. We thank Serena Abbondante for providing the unpublished serum high-fat diet dataset and Nicholas Ceglia for doing the circadian analysis. We thank Yuzo Kanomata for helping develop and maintain the CircadiOmics systems and web site.
Funding
This work was supported by the National Science Foundation [IIS-1321053 to P.B.] and the National Institutes of Health [NIH LM010235 and NIH NLM T15 LM07443 to P.B.].
Conflict of Interest: none declared.
References
- Aguilar-Arnal L., et al. (2013) Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat. Struct. Mol. Biol., 20, 1206–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews J.L., et al. (2010) Clock and bmal1 regulate myod and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Natl. Acad. Sci. U S A, 107, 19090–19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes L.C., et al. (2010) Obesity and shift work: chronobiological aspects. Nutr. Res. Rev., 23, 155–168. [DOI] [PubMed] [Google Scholar]
- Baldi P., Meir R. (1990) Computing with arrays of coupled oscillators: an application to preattentive texture discrimination. Neural Comput., 2, 458–471. [Google Scholar]
- Brandt S., et al. (2006) Synchronization from disordered driving forces in arrays of coupled oscillators. Phys. Rev. Lett., 96, 34104. [DOI] [PubMed] [Google Scholar]
- Brown S.A., et al. (2012) (Re)inventing the circadian feedback loop. Dev. Cell, 22, 477–87. [DOI] [PubMed] [Google Scholar]
- Covington M.F., et al. (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol., 9, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily K., et al. (2011) Motifmap: integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinformatics, 12, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C., et al. (2010) The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol., 72, 517–549. [DOI] [PubMed] [Google Scholar]
- Doi M., et al. (2006) Circadian regulator clock is a histone acetyltransferase. Cell, 125, 497–508. [DOI] [PubMed] [Google Scholar]
- Dyar K.A., et al. (2014) Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Molecular metabolism. 3(1), 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan K., Sassone-Corsi P. (2009) Metabolism control by the circadian clock and vice versa. Nat. Struct. Mol. Biol., 16, 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan K.L., et al. (2012) Coordination of the transcriptome and metabolome by the circadian clock. Proc. Natl. Acad. Sci. U S A, 109, 5541–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan K.L., et al. (2013) Reprogramming of the circadian clock by nutritional challenge. Cell, 155, 1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., et al. (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O. (2010) Metabolism and circadian rhythms—implications for obesity. Endocr. Rev., 31, 1–24. [DOI] [PubMed] [Google Scholar]
- Froy O. (2011) Circadian rhythms, aging, and life span in mammals. Physiology (Bethesda), 26, 225–235. [DOI] [PubMed] [Google Scholar]
- Gerstner J.R., et al. (2009) Cycling behavior and memory formation. J. Neurosci., 29, 12824–12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel P., Ermentrout B. (2002) Synchrony, stability, and firing patterns in pulse-coupled oscillators. Phys. D Nonlinear Phenomena, 163, 191–216. [Google Scholar]
- Goldbeter A. (1997) Biochemical oscillations and cellular rhythms: the molecular bases of periodic and chaotic behaviour. Cambridge university press. [Google Scholar]
- Harmer S.L., et al. (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science, 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Huang D.W., et al. (2007) David bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res., 35(Web Server issue), W169–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., et al. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Hughes M.E., et al. (2009) Harmonics of circadian gene transcription in mammals. PLoS Genet., 5, e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M.E., et al. (2010) JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms, 25, 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson B., et al. (2001) Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27, 485 people. Occup. Environ. Med., 58, 747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsson A. (2003) Health disorders of shift workers. Occup. Med. (Lond), 53, 103–108. [DOI] [PubMed] [Google Scholar]
- Kohsaka A., et al. (2007) High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab., 6, 414–421. [DOI] [PubMed] [Google Scholar]
- Kondratov R.V., et al. (2006) Early aging and age-related pathologies in mice deficient in bmal1, the core component of the circadian clock. Genes Dev., 20, 1868–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia K.A., et al. (2008) Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U S A, 105, 15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S., et al. (2014) Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell, 158, 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B.H., et al. (2007) Circadian and clock-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. USA, 104, 3342–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier A., et al. (2010) Orchestrated transcription of biological processes in the marine picoeukaryote ostreococcus exposed to light/dark cycles. BMC Genomics, 11, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.Y., Eichler V.B. (1972) Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res., 42, 201–206. [DOI] [PubMed] [Google Scholar]
- Nakahata Y., et al. (2008) The nad+-dependent deacetylase sirt1 modulates clock-mediated chromatin remodeling and circadian control. Cell, 134, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J.S., et al. (2011) Circadian rhythms persist without transcription in a eukaryote. Nature, 469, 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S., et al. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell, 109, 307–320. [DOI] [PubMed] [Google Scholar]
- Partch C.L., et al. (2014) Molecular architecture of the mammalian circadian clock. Trends Cell Biol., 24, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V., et al. (2014) How pervasive are circadian oscillations?. Trends Cell Biol., 24, 329–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V.R., et al. (2012) Circadiomics: integrating circadian genomics, transcriptomics, proteomics, and metabolomics. Nat. Methods, 9, 772–773. [DOI] [PubMed] [Google Scholar]
- Peek C.B., et al. (2013) Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science, 342, 1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph M.R., et al. (1990) Transplanted suprachiasmatic nucleus determines circadian period. Science, 247, 975–978. [DOI] [PubMed] [Google Scholar]
- Ramsey K.M., et al. (2009) Circadian clock feedback cycle through nampt-mediated nad+ biosynthesis. Science, 324, 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G., et al. (2011) Genome-wide and phase-specific DNA-binding rhythms of bmal1 control circadian output functions in mouse liver. PLoS Biol., 9, e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Sassone-Corsi P. (2002) A web of circadian pacemakers. Cell, 111, 919–922. [DOI] [PubMed] [Google Scholar]
- Sharifian A., et al. (2005) Shift work as an oxidative stressor. J. Circadian Rhythms, 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.-Q., et al. (2013) Circadian disruption leads to insulin resistance and obesity. Curr. Biol., 23, 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch K.F., et al. (2002) Extensive and divergent circadian gene expression in liver and heart. Nature, 417, 78–83. [DOI] [PubMed] [Google Scholar]
- Stratmann M., Schibler U. (2006) Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J. Biol. Rhythms, 21, 494–506. [DOI] [PubMed] [Google Scholar]
- Strogatz S. (2000) From kuramoto to crawford: exploring the onset of synchronization in populations of coupled oscillators. Phys. D Nonlinear Phenomena, 143, 1–20. [Google Scholar]
- Takahashi J.S., et al. (2008) The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet., 9, 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek F.W., et al. (2005) Obesity and metabolic syndrome in circadian clock mutant mice. Science, 308, 1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G., Millar A.J. (2012) Non-transcriptional oscillators in circadian timekeeping. Trends Biochem. Sci., 37, 484–492. [DOI] [PubMed] [Google Scholar]
- Vijayan V., et al. (2009) Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. USA, 106, 22564–22568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., et al. (2009) Motifmap: a human genome-wide map of candidate regulatory motif sites. Bioinformatics, 25, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., et al. (2012) Next-generation sequencing of experimental mouse strains. Mamm. Genome, 23, 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., et al. (2008) Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput. Biol., 4, e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.H., et al. (2004) Period2::luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U S A, 101, 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., et al. (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. USA, 111, 16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.