Abstract
The advance of next generation sequencing (NGS) techniques provides an unprecedented opportunity to probe the enormous diversity of the immune repertoire by deep sequencing T-cell receptors (TCRs) and B-cell receptors (BCRs). However, an efficient and accurate analytical tool is still on demand to process the huge amount of data. We have developed a high-resolution analytical pipeline, Immune Monitor (“IMonitor”) to tackle this task. This method utilizes realignment to identify V(D)J genes and alleles after common local alignment. We compare IMonitor with other published tools by simulated and public rearranged sequences, and it demonstrates its superior performance in most aspects. Together with this, a methodology is developed to correct the PCR and sequencing errors and to minimize the PCR bias among various rearranged sequences with different V and J gene families. IMonitor provides general adaptation for sequences from all receptor chains of different species and outputs useful statistics and visualizations. In the final part of this article, we demonstrate its application on minimal residual disease detection in patients with B-cell acute lymphoblastic leukemia. In summary, this package would be of widespread usage for immune repertoire analysis.
Keywords: next generation sequencing, bioinformatics, immune repertoire, TCR/BCR
THE diversity of T-cell receptors (TCRs), B-cell receptors (BCRs), and secreting form antibodies makes up the core of the complicated immune system and serves as pivotal defensive components to protect the body against invading virus, bacteria, and other pathogens. The TCR consists of a heterodimeric αβ chain (∼95%, TRA, TRB) or γδ chain (∼5%), while the BCR is assembled with two heavy chains (IGH) and two light chains (IGK or IGL). Structurally, each chain can be divided into the variable domain and the constant domain (Lefranc and Lefranc 2001a,b). The diversity of the TCR and BCR repertoire is enormous, owing to the process of V(D)J gene rearrangement, random deletion of germline nucleotides, and insertion of uncertain length of nontemplate nucleotides between V-D and D-J junctions (TRB, IGH) or V-J junctions (TRA, IGK, IGL). In humans, it has been estimated theoretically that the diversity of TCR-αβ receptors exceeds 1018 in the thymus, and the diversity of the B-cell repertoire is even larger, considering the somatic hypermutation (Janeway 2005; Benichou et al. 2012). The T- and B-cell repertoire could undergo dynamic changes under different phenotypic status. Recently, deep sequencing enabled by different platforms including Roche 454 and Illumina Hiseq (Freeman et al. 2009; Robins et al. 2009; Wang et al. 2010; Fischer 2011; Venturi et al. 2011) has been applied to unravel the dynamics of the TCR and BCR repertoire and extended to various translational applications such as vaccination, cancer, and autoimmune diseases.
Several tools and software have been developed for TCR and BCR sequence analysis, including iHMMune-align (Gaeta et al. 2007), HighV-QEUST (Li et al. 2013), IgBLAST (Ye et al. 2013), Decombinator (Thomas et al. 2013), and MiTCR (Bolotin et al. 2013). These tools are equipped with useful functions, including V(D)J gene alignment, CDR3 sequence identification, and more, yet with obvious limitations. For instance, HighV-QEUST can be adopted to analyze both TCRs and BCRs, but its online version limits maximum sequence input to 150,000 at a time for regular users. Decombinator and MiTCR can only be used to analyze the TCR sequences. Besides, most tools lack specific solutions to some common problems like systemic statistics and visualizations, PCR and sequencing errors, and amplification bias correction.
Here, we introduce a novel pipeline, Immune Monitor (IMonitor) for both TCR and BCR deep sequencing analysis. It includes four steps in its core component: basic data processing, V(D)J assignment, structural analysis, and statistics/visualization. One feature that makes IMonitor stand out is its realignment process to identify V(D)J genes and alleles with significantly enhanced precision. We simulated 15 data sets for five chains (TRA, TRB, IGH, IGK, IGL) of different sequencing error rates and hypermutation rates, together with actual rearranged sequences, to test performance of various tools. IMonitor performs quite well in accuracy and clonotype recovery. Furthermore, IMonitor incorporates a process to correct PCR and sequencing errors, utilizing the data from six plasmid mixed samples, and an in silico model was modulated to reduce the PCR bias. Finally, we validate IMonitor in detection of minimal residual disease (MRD) of B-cell acute lymphoblastic leukemia (B-ALL) to show its wide utility potential.
Materials and Methods
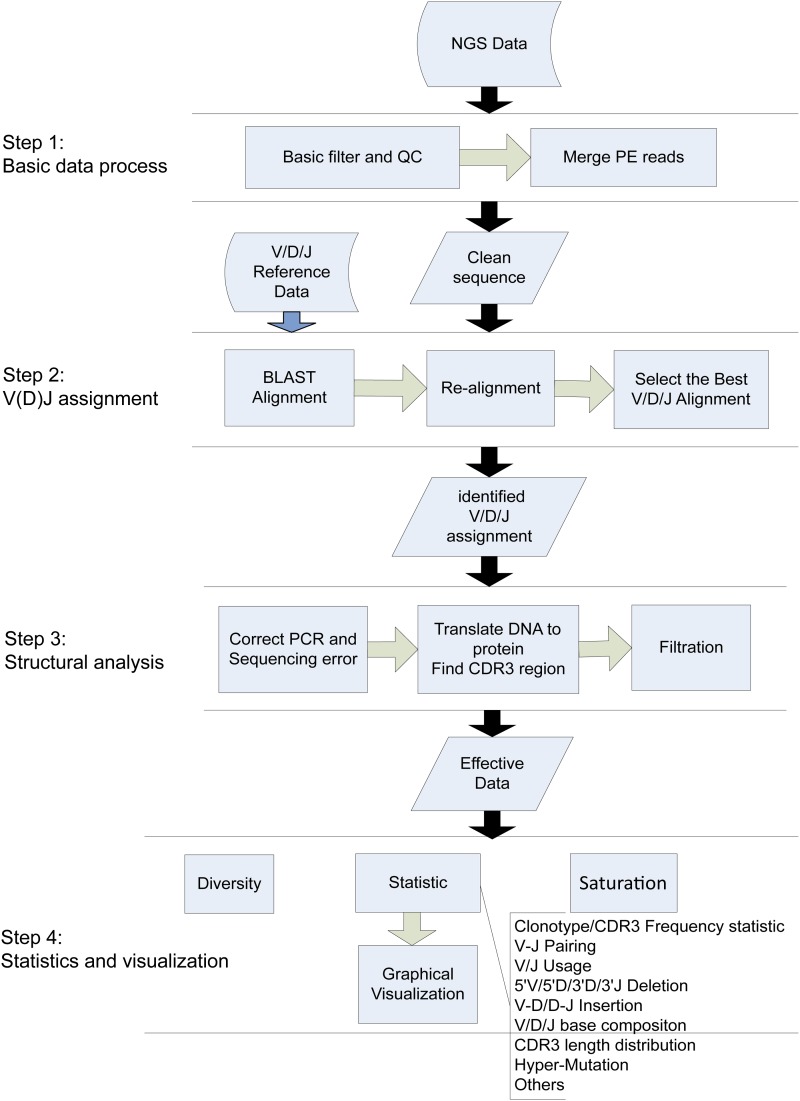
The core component of IMonitor consists of four steps: basic data processing, V(D)J assignment, structural analysis, and statistics/visualization, as shown in Figure 1. IMonitor can utilize data generated by a variety of next generation sequencing (NGS) platforms, such as Illumina, Roche 454, and Life Ion Proton, in both FASTQ and FASTA format. The final results of IMonitor include a complete map of sequences and data analysis in depth, and the latter is visualized and presented with viewer-friendly graphs and figures.
Figure 1.
Overview of workflow of IMonitor. Although the program includes four steps, we have several parameters to control whether the module runs or not. The program takes raw NGS (FASTA or FASTQ) as input and outputs the V(D)J assignment of the sequence, some statistics, and corresponding figures.
IMonitor for basic data processing
In the first step, the reads were checked for inclusion of adaptor sequences. If any adaptor sequence was detected and located within 50 bp of the 3′end of the read, it was deleted from the read. Reads bearing adaptor sequence at the 5′ end or >5% “N” bases were discarded. The average base quality of each read was calculated after removing the low-quality bases (base quality <10) at the 3′ end. Further filtration left out reads with average quality <15. For Illumina paired-end (PE) sequencing, the PE reads were merged at their overlapping region. For PE reads with insertion length longer than the length of a single read, the COPE (Liu et al. 2012) tool was used; otherwise reads were assembled by an in-house program. The main parameters for both tools included the maximum overlapping length (read length), minimum overlapping length (10 bp), mismatch rate (10%) at the overlapping region, and ratio (best overlap length/second-best overlap length, 0.7).
IMonitor for V(D)J assignment
The V/D/J reference sequences were downloaded from the IMGT database, the international ImMunoGeneTics information system (http://www.imgt.org/). Processed sequences were aligned to the V, (D), J references, respectively, by BLAST (Altschul et al. 1990; Zhang et al. 2000; Ye et al. 2006) and specific parameters were applied to accommodate the differences in lengths of V, (D), J segments (BLAST parameters: V, -W 15 -K 3 -v 1 -b 3; D, -W 4 -K 3 -v 3 -b 5; and J, -W 10 -K 3 -v 1 -b 3).
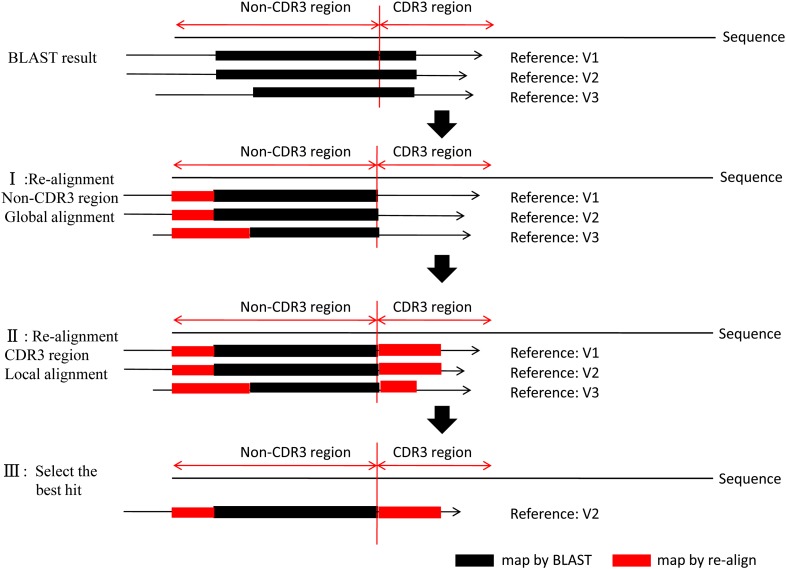
The high similarity among the genes and alleles of the germline sequences, along with the diversity of V/D/J gene rearrangement, gave rise to difficulties for accurate alignment. This might eventually lead to an incorrect structural analysis (CDR3 identification, deletion, or insertion). To improve the accuracy, a second alignment procedure was developed to identify exactly the V/D/J genes (Figure 2). First, a global alignment strategy, which attempted to align every base in every sequence, was used for the non-CDR3 region of the sequence. The mapped region generated from BLAST became a new seed and served as starting points for bootstrapping (base-by-base) extension to both directions, until the entire non-CDR3 region in the query was mapped to the target (reference) sequence. The mapping score was calculated according to these rules: reward for a nucleotide match was 5 and penalty for a nucleotide mismatch was −4. Second, the M-mismatch extension model of local alignment strategy was applied to locate the exact end positions of V and J genes during CDR3 region realignment. The procedure began at the CDR3 start position in the V gene or the CDR3 end position in the J gene and continuously extended in one direction until the preset mismatch limit was reached, generating the longest possible interval with the highest score. The mismatch numbers allowed for V/D/J genes were determined based on the analysis result of publicly available rearrangement sequences (http://www.imgt.org/ligmdb/) (Supporting Information, Figure S2A) and adjusted accordingly for different TCR and BCR chains (mismatches allowed: TRBV/J, TRAV/J, 0; IGHV/J, 2; IGKV/J, IGLV/J, 7). As shown in Figure S2A, these mismatch limits took mutations into consideration and covered >99.5% of all defined rearrangement sequences. Because the entire D gene was located within the CDR3 region, only the M-mismatch extension model was used for its realignment (mismatches allowed: TRBD, 0; IGHD, 4). Finally, all data including alignment score, identity, mismatch number, and alignment length were processed, and the alignment with highest score and identity larger than the threshold (>80%) was selected as the best hit. However, there might be several best hits with the same score due to the homology among the germline genes and alleles. In this case, the reference with the fewest deletions was selected, as shorter deletions are more likely to happen according to previous reported results (Warren et al. 2009) and our analysis from actual public rearrangement data (Figure S2B).
Figure 2.
The workflow of realignment. The program takes the BLAST alignment results as input, realigns the sequence to reference for both the non-CDR3 region and the CDR3 region, calculating the score and identity, and then selects the maximal score as the best hit. The reference with shortest length of deletion is preferred if it finds multiple references with the same maximal scores. It outputs the optimal alignment result.
IMonitor for structural analysis
The IMGT collaboration (Yousfi Monod et al. 2004) outlined the CDR3 region of all chains, starting from the second conserved cysteine encoded by the V segment and ending with the conserved phenylalanine or tryptophane encoded by the J segment. Combining this information with our selected reference from the previous step, the CDR3 region of target sequence could be readily identified. For unmapped sequences, the CDR3 region was determined by searching through for a conservative amino acids module within both ends of the CDR3 region (“YXC” for start and “[FW]GXG” for end, where “X” stands for any amino acid). The rearrangement frame was tagged as “in-frame” if the length of CDR3 was a multiple of three and no stop codon was found in whole sequence; otherwise it was tagged “out-of-frame.” The structure of the sequence was clearly described, including V, (D), J segments used, the CDR3 region, and the deletions and insertions at rearrangement sites. Then the nucleotide sequences were translated into peptides. However, some sequences must be filtered out to ensure the accuracy of the immune repertoire, which include (1) sequences without CDR3 region and (2) sequences with V and J alignment orientation conflict. The sequences that were aligned to pseudogenes, were out-of-frame, and included a stop codon were marked.
IMonitor for statistics and visualization
The basic statistics of IMonitor include CDR3 frequency distribution, V-J paring, V/J usage, 5′V/5′D/3′D/5′J deletion length distribution, V-D/D-J insertion length distribution, V/J base composition, CDR3 length distribution, CDR3 segmental frequency statistics, Top10 CDR3 frequency, hypermutation of BCRs, etc. Figures were plotted to visually demonstrate each result. For V-J pairing, a three-dimensional figure was generated. R script was used to draw most of the figures while the V/J base composition was plotted with weblogo 2.8 (Crooks et al. 2004). To completely eliminate sequencing error effect, a sequence detected <10 times was excluded to calculate hypermutation. The mutation rate consisted of base mutation and sequence mutation. The former was the content of mutational bases in total bases, and the latter was rate of the sequence containing mutation in total sequences.
The Shannon–Weiner index (Shannon 1997), as shown below, used as an immunological diversity value in several previous works (Sherwood et al. 2013), provided a good estimate of diversity in a large-scale study and was suitable for immune repertoire. We used it to calculate the diversity of CDR3, the V gene, the J gene, and V-J pairing,
| (1) |
where CDR3 is an example, S denotes the total number of unique CDR3, and denotes the frequency of CDR3.
IMonitor also provided the saturation analysis with the Chao1 algorithm, which was used to estimate the target richness for individual-based data in previous ecological studies. Estimated values generated by the Chao1 bias-corrected algorithm were used to predict the maximum number of clones in the sample, while observed values were drawn separately with the rarefaction curves (Chao 1984, 1987):
| (2) |
In Equation 2, stands for total number of observed clonotypes in a sample; F denotes the number of clonotypes (, the number of clonotypes detected one time; , the number of clonotypes detected two times).
PCR and sequencing error correction
PCR and sequencing error of NGS is one of the toughest problems in immune repertoire analysis. The method we developed to correct this error could be utilized on either the whole sequence or just the CDR3 region. The procedure consisted of three steps. First, sequences were divided into three groups: (1) high-quality sequences whose base qualities all were >Q20 (Q20 were the best cutoff according to Figure S5, A and B; (2) sequences with more than five (three for only CDR3 region correction) low-quality bases were unwanted and discarded; and (3) the rest were defined as low-quality sequences. Second, the low-quality sequences were mapped to the high-quality ones. When the mismatches were no more than five (three for only CDR3 region correction) and all located at low-quality positions, the mismatches were corrected; otherwise the sequence was discarded. Finally, to eliminate PCR errors, sequences with low abundance were compared to ones with high abundance (at least fivefold difference). If fewer than three mismatches were found in the low-abundance sequences, they were corrected to the corresponding high-abundance sequences. To test the effectiveness of this method, samples made from mixtures of six plasmids were used for error characteristics analysis and further evaluation.
Multiplex PCR bias minimization
We established a new bioinformatic approach to minimize PCR amplification bias. The approach is built on the hypothesis that there are two factors affecting a clone’s frequency during multiplex PCR (MPCR): the template’s concentration and the multiple primers’ efficiency. Using six plasmid mixture samples (Table S4), we could compare the observed with the expected frequency and simulate an effective formula.
The streamlined procedure of this method is demonstrated by the flowchart in Figure 6. Two factors, templates’ concentration and primers’ efficiency, were considered to be affecting the bias and examined here. The samples were mixed properly, and clones were grouped to explore PCR bias rules. First, we analyzed the bias’ correlation with templates’ concentration (concentration analysis without primer effect). Each clone had three different concentrations in all samples. To eliminate potential effects caused by the multiple primers efficiencies, the clones that had the same concentration ratio among all samples were grouped together, generating five groups in total [groups were named 10_2E4_1E5, 1000_2E4, 100_1000_2E4, 100_1E4_2E4, and 10_1E4_2E4; for instance, 10_2E4_1E5 denoted the three concentrations (10, 2E4, 1E5) in respective samples]. Then, within each group, the clone frequencies were normalized by multiplying the same coefficient k, which could generate a minimal sum of absolute deviation Dmin(i). The 10_2E4_1E5 group sets an example as follows:
| (3) |
| (4) |
| (5) |
where n is clone number in a group, i is a clone, and f is clone frequency. k was set consecutively from 0 and a series of D(i) was calculated, after which the k that generated the smallest D(i) was selected.
Figure 6.
Minimize MPCR bias flowchart. Six samples are mixed together and search the bias rules under two independent pathways, concentration analysis and primer analysis. For concentration analysis, six groups are created to eliminate primer effect, and each group is normalized; then five groups are combined together and a curve is fitted. For primer analysis, six groups (10, 100, 1000, 1E4, 2E4, 1E5) are created to eliminate the concentration effect. Each group is normalized by multiplying a constant term and combined together, and then the primer efficiencies are obtained. A formula for reducing PCR bias is generated by integrating these two factors. Finally, cross-validation is used to evaluate this method’s robustness. Each time, five samples are used as training data, and the remaining one is used as testing data. The residual sum of squares, , where is the observed value, and is the expected value.
After normalization, five groups were combined on the basis of 2E4 copies, which existed in all groups. We used a regression module to fit a curve (Equation 6) that reflected the relationship between concentration and PCR bias,
| (6) |
where y is the observed sequence’s frequency and x is the expected sequence’s frequency.
Second, we analyzed the bias caused only by primer efficiencies (primer analysis without concentration effect). To remove the effect of clone concentration, clones with the same concentration in all samples were collected into one group; thus six groups were generated (10, 100, 1E3, 1E4, 2E4, and 1E5, where group 10 contained all clones that had the concentration 10 in any of the samples) (Figure 6). After calculation, each group was normalized by multiplying the same coefficient l, with the following details,
| (7) |
| (8) |
| (9) |
where, n is clone number in a group, i is a clone, and f is clone frequency. l was set consecutively from 0 and a series of R(i) was calculated, after which the l that generated the minimal R(i) was selected.
Each primer’s efficiency was calculated after normalization. Then, analysis of the two factors was integrated into a formula that minimized the PCR bias (Equation 10),
| (10) |
where fcorrect is the corrected frequency, Si is the clone’s observed abundance, S is the sum abundance of the sample, p(v) is the primer efficiency value for the V gene, and p(j) is the primer efficiency value for the J gene.
Multiplex PCR amplification
To amplify rearranged CDR3 regions, multiple forward primers in the V region and reverse primers in the J region were designed. For the RNA sample, the first-strand cDNAs were synthesized using SuperScript II Enzyme according to the manufacturer’s instructions. Then two individual equimolar pools of the forward primers and the reverse primers were used for a multiplex PCR (QIAGEN, Valencia, CA) of 30 cycles according to the provided protocol. The fractions between 110 and 180 bp of the PCR products were excised and purified.
Simulation of in silico sequences
A total of 105 sequences were generated in silico for each data set with a length of 200–300 bp by simulating the relevant biological processes that occur during B-cell and T-cell development. The sequences of V(D)J genes of TRA/TRB/IGH/IGK/IGL were downloaded from IMGT (http://www.imgt.org/). First, to simulate recombination, a V allele and a J allele (an extra D allele for TRB and IGH) were selected at random to generate a V-D-J (V-J for TRA and light chain) combination. Second, to simulate deletion and insertion, some bases were deleted at the 3′ end of V and the 5′ end of J (both ends of D for TRB and IGH) according to the deletion length distribution from previous research (Figure S1); meanwhile some random bases were inserted at V-D and D-J junctions (V-J junction for TRA and light chain), using the established insertion length distribution (Figure S1). Third, both somatic hypermutation during BCR maturation and sequencing error were also taken into consideration. Although the typical error rate of Illumina HTS technology is ∼1%, it can be reduced after merging paired-end reads (Figure 5A). Therefore, for each chain, the error rate was set to 0.1%, 0.5%, and 2% evenly at every position of the sequence. The hypermutation rate was set to 1% for IGH and 4% for IGK/IGL.
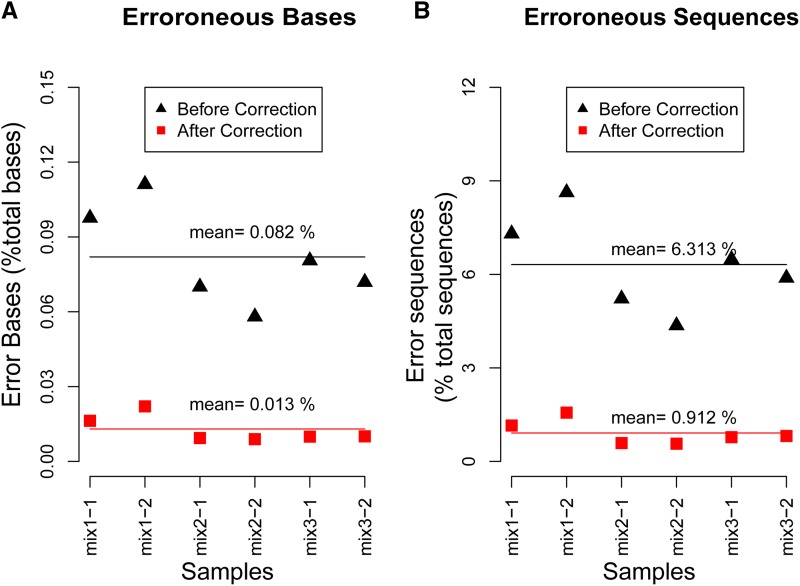
Figure 5.
Error rate before and after correction. (A) The percentage of erroneous nucleotide bases divided by the total bases sequenced. (B) The percentage of sequences containing erroneous bases divided by total sequences. The primers at each sequence are excluded for the calculation of error rate.
Public rearrangement data sets
The data sets were obtained from the IMGT/LIGM-DB database(http://www.imgt.org/ligmdb/); searched by “Homo sapiens,” “rearranged,” “TRB,” or “IGH”; and then the selected sequences were annotated manually (Annot. level==“manual”) and annotated by V, D, J genes. Finally, 24 TRB and 1763 IGH sequences met these requirements. The sequence was in FASTA format, without sequencing quality. The length of sequences downstream of the J gene was limited to 50 bp.
Samples
Plasmid mix samples:
Thirty-three different functional TCR β-chain sequences, which included all the TCR β, V, and J genes, were integrated into plasmid vectors. Three mixing pattern pools were used: one with an equal mole number of each plasmid and the other two pools with different pooling gradients. The mix patterns are listed in Table S4, and each pattern was replicated to produce six plasmid mix samples.
Spiked-in DNA samples:
Spiked-in samples were generously donated by Karen Cerosaletti (Benaroya Research Institute, Seattle, WA) (Robins et al. 2012). Five CD4+ T-cell clones were spiked in a background of sorted CD4+CD45RA+ naive T cells and each unit had 1 million cells. The five clones in three different units had different numbers of unique TCRβ CDR3 sequences and are shown in Table S7. The background cells for these doped samples were sorted from a fresh peripheral blood mononuclear cell (PBMC) sample obtained from a control donor with informed consent. DNA was extracted from the cells with a commercial kit.
Healthy donor samples:
Samples of peripheral blood from two healthy human donors (H-H-1 and H-B-1) were obtained by venipuncture with informed consent. PBMCs were isolated immediately and RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA). DNA was extracted with a QIAamp DNA Blood Mini Kit and stored at −20°. The CDR3 region was amplified by multiplex PCR (Table S9) and sequenced by an Illumina platform (Table S6).
MRD samples:
Bone marrow samples from two patients (M001 and M002) with B-ALL were provided with informed consent. The samples of pretreatment, day 15, and day 33 post-treatment were assayed. A total of 1.2 μg DNA of each sample was used for multiplex PCR amplification (Table S9). The library of ∼150–270 bp insert-size length was extracted and sequenced using the 2×100 PE Illumina platform (Table S6).
The research was prospectively reviewed and approved by a duly constituted ethics committee.
Data availability
The source code of IMonitor is freely available for download at https://github.com/zhangwei2015.
Table S1: Simulated TRB with 0.1%, 0.5% and 2% sequencing error.
Table S2: Simulated IGH with 0.1%, 0.5% and 2% sequencing error and 0.1% hayper-mutation.
Table S3: Simulated Data with 0.5% sequencing error (TRA/IGK/IGL) and 4% hyper-mutation.
Table S4: Plasmid mixing pattern.
Table S5: Data process for PCR and sequencing error statistics.
Table S6: Samples information.
Table S7: Experimental design for five CD4+ T cell clones in the three spiked in mix.
Table S8: Performance of IMonitor and other tools on the simulated dataset.
Table S9: TRB and IGH V/J primers.
Figure S1: Insertion and deletion length distribution for simulated data.
Figure S2: IGH-VDJ Mutation and deletion/insertion analysis on the public sequences.
Figure S3: Outputs of IMonitor, H-B-01 as an example.
Figure S4: H-B-01 sample output figure of IMonitor.
Figure S5: Error characteristics of 6 plasmid mix samples.
Figure S6: V-J pairing dynamics for M002.
Figure S7: MiTCR and IMonitor performance in 3 spiked-in samples.
Figure S8: Nucleotide composition of V/J genes.
Results
System design of IMonitor
Four steps are described in Figure 1:
Basic data process: Sequence containing adapter sequence was processed and low-quality bases at the 3′ end of the sequence were removed. PE reads were merged to one sequence by an in-house program and COPE (Liu et al. 2012).
V(D)J assignment: The reference germline sequences were downloaded directly from IMGT (http://www.imgt.org/). Processed data were aligned to the references by BLAST (Altschul et al. 1990; Zhang et al. 2000; Ye et al. 2006) and realigned to improve the map accuracy, after which the optimal alignment was selected for every sequence (Figure 2).
Structural analysis: A novel method was established to correct PCR and sequencing errors. The CDR3 region was identified with the help of both V/J references and conservative amino acids and then translated into amino acids.
Statistics and visualization. Characteristic data of the immune repertoire of the samples, such as repertoire diversity, clonotype frequency, CDR3 length distribution, V/J usage, V-J pairing, hypermutation, deletion, and insertion, were collected (Figure S3) and presented with corresponding graphs. More specifically, the V-J pairing was visualized by a three-dimensional graph (Figure 4, Figure S4, and Figure S8).
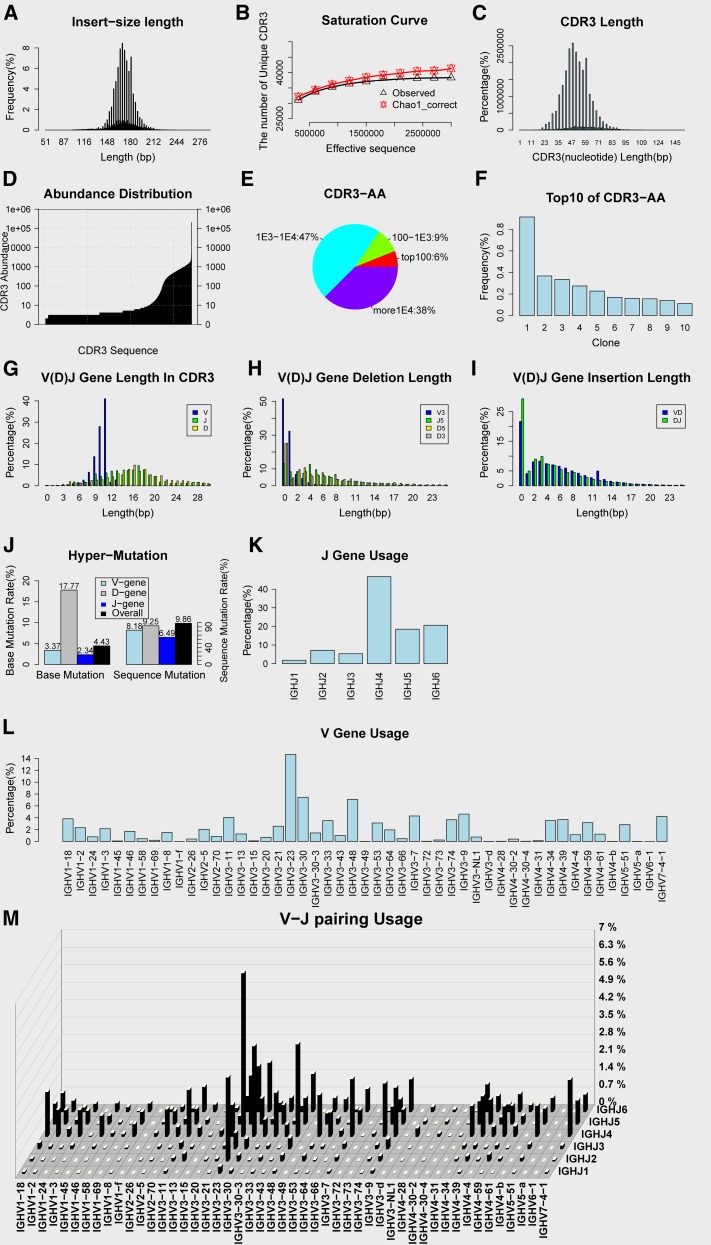
Figure 4.
Output figure of IMonitor. H-H-01 sample is shown as an example. (A) Sequence length distribution. (B) Saturation curve, rarefaction studies of sequences. Subsequences are randomly selected and the observed unique CDR3 number and predicted CDR3 number (Chao1-corrected algorithm) are calculated. (C) CDR3 nucleotide length distribution. (D) CDR3 abundance distribution. (E) CDR3 amino acid frequencies sectional content. (F) Top 10 frequency of CDR3 amino acid. (G) The length distribution of the V/D/J gene in the CDR3 region. (H) Deletion length distribution of the V/D/J gene. (I) Insertion length distribution between V and D genes, D and J genes. (J) Hypermutation, only for Ig. (K) J-gene usage. (L) V-gene usage. (M) Three-dimensional graph of V-J pairing.
IMonitor outperforms other analytical tools in various aspects
To evaluate the performance of IMonitor, we designed a head-to-head comparison between IMonitor and other publicly available tools with both simulated data and public rearrangement sequences. The TRA/TRB data were analyzed by HighV-QEUST (Li et al. 2013), Decombinator (Thomas et al. 2013), IgBLAST (Ye et al. 2013), and IMonitor, while the IGH/IGK/IGL data were analyzed by HighV-QEUST, IgBLAST (Ye et al. 2013), and IMonitor. Thomas et al. (2013) reported that iHMMune-align (Gaeta et al. 2007) generated a similar result to IgBLAST so it was excluded from the comparison. MiTCR (Bolotin et al. 2013) performance is strongly related to sequencing quality, so neither simulated data nor public sequences were suitable for it. Three spiked-in samples sequenced by Illumina were used to compare MiTCR and IMonitor, the result of which is shown in Figure S7. Although MiTCR finished the run much faster than IMonitor, clone G was missed because of erroneous J-gene assignment and an incorrect CDR3 region. All tools employed their default parameters.
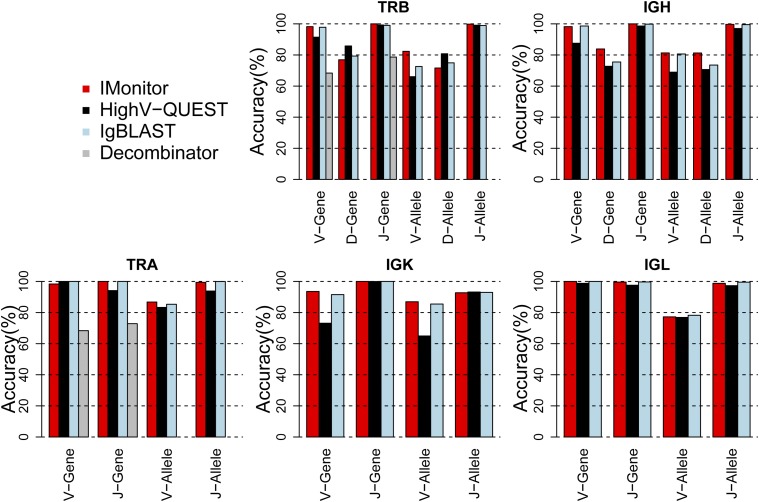
By processing the in silico data with different tools, we calculated the accuracy of V/D/J genes and V/D/J alleles of each tool analyzed (Figure 3 and Table S1, Table S2, and Table S3). For all 24 V/D/J genes and alleles in all TCR/BCR chains, IMonitor showed superior performance in 14 of them (58.33%), with 7 of 24 slightly lower in performance than the best tool (no more than 1% difference) and the remaining 3 from 1.6 to 3.25% difference (TRAV gene, TRBD gene, and TRBD allele). For the highly homologous V-gene family (at least 40 for each chain), the accuracy of IMonitor was >95% for almost all chains and exceeded 99% for all J genes. For D genes, which were short and embedded with deletions and insertions at both ends, IMonitor performed significantly better for IGH (>80% accuracy), while slightly worse for TRB. Decombinator was not designed to identify alleles and D genes.
Figure 3.
Performance of different types of software on 105 in silico sequences. Shown is accuracy of TRA, TRB, IGH, IGK, and IGL sequences with 0.5% sequencing error. The accuracy is calculated for both gene and allele, but Decombinator has only V and J genes identified.
In addition to the simulating data, public rearranged sequences that are annotated manually with clear V(D)J genes and extracted from the IMGT/LIGM-DB database (http://www.imgt.org/ligmdb/) were utilized to test IMonitor. Twenty-four TRB sequences and 1763 IGH sequences were analyzed by different tools (Table 1). For TRB, IMonitor and IgBLAST performed better than Decombinator in general, whereas IMonitor outperformed IgBLAST in D genes. For IGH, IMonitor performed similarly to IgBLAST in V and D alleles, but was superior in all other genes and alleles. The good performance in D genes by IMonitor demonstrated the effectiveness of the M-mismatch extension model during D gene realignment. The accuracies of IGH-J genes and alleles were both slightly lower for these two tools, because some public IGH sequences have only a partial J segment (<30 bp) and they are difficult to distinguish from other homologous genes and alleles.
Table 1. Number of sequences with correctly identified V, D, and J genes or alleles in public sequence data sets.
| Datasets/Tools | V_gene | V_allele | D_gene | D_allele | J_gene | J_allele |
|---|---|---|---|---|---|---|
| Data set of TRB (24 sequences) | ||||||
| IMonitor | 24 (100%) | 23 (96%) | 17 (71%) | 12 (50%) | 24 (100%) | 24 (100%) |
| IgBLAST | 23 (96%) | 22 (92%) | 13 (54%) | 11 (46%) | 24 (100%) | 24 (100%) |
| Decombinatora | 21 (87%) | — | — | — | 23 (96%) | — |
| Data set of IGH (1763 sequences) | ||||||
| IMonitor | 1735 (98%) | 1509 (86%) | 1037 (59%) | 952 (54%) | 1619 (92%) | 1533 (87%) |
| IgBLAST | 1716 (97%) | 1518 (86%) | 986 (56%) | 956 (54%) | 1563 (89%) | 1498 (85%) |
The data sets were obtained from the IMGT/LIGM-DB database (http://www.imgt.org/ligmdb/); searched by “Homo sapiens,” “rearranged,” “TRB,” or “IGH”; and then the selected sequences were annotated manually (Annot. level==“manual”) and annotated by V, D, J genes. So these sequences have a fairly high level of annotation confidence. The data sets and HighV-QEUST came from the same website, so HighV-QEUST was not used here. The references used for tools were the same and were from the IMGT database (http://www.imgt.org).
Decombinator analyzed just the gene level of V and J.
To assess running time and memory needed, 105 simulated TRB and IGH data sets were analyzed by IMonitor, IgBLAST, and Decombinator separately; the results are shown in Table S8. When only one CPU was used, IMonitor took 12 min 52 sec and 21 min 96 sec to analyze the two data sets separately, with peak memory of 226 ∼ 325 Mb. Of all tools tested, Decombinator was the fastest and IMonitor ranked second.
Overall, IMonitor produced satisfactory results for both simulated and published sequences. It generated similar results to IgBLAST in some genes, while it outperformed other tools in most occasions. It is also direct proof that the realignment strategy for V(D)J identification is useful.
The output of IMonitor
One of the features that distinguish IMonitor from others is its ability to export comprehensive statistics for characteristics of TCR/BCR repertoire and accessible graphs. The statistics include not only basic statistics but also in-depth statistics (Figure S3). The former elucidates the process from raw data to effective sequences, such as clean data rate and V(D)J gene mapped rate, which all provide sequence number, rate of input, and rate of raw data. The latter consists of multiple statistics based on effective sequence, such as functional classification, V/J/V-J gene usage rate, clone number, diversity calculated by Shannon index, and hypermutation. IMonitor is also able to translate obscure data into self-explanatory graphs. Important statistics like V/J usage, top 10 clone frequencies, CDR3 segmental frequency statistics (split into four segments after frequency sorted: top100, 100-1E3, 1E3-1E4, >1E4), insertion and deletion length distribution, V/J nucleotide composition, and V-J pairing can all be presented as figures (Figure 4, Figure S4, and Figure S8) . V-J pairing diversity is visualized (Figure 4M, Figure S4M), which can be applied to track the changes of the immune system over time and reveal immunological conditions.
PCR and sequencing errors correction
IMonitor also integrates a process to correct PCR and sequencing errors. The six plasmid mixture samples with three different pooling gradients (Materials and Methods and Table S4) were used to analyze the error characteristics and evaluate the effectiveness of error correction. The processed sequences were mapped to V/D/J references, and the final effective data were used to summarize the error characteristics shown in Figure S5. The known template sequences were used as references to calculate the error rate, and the results are shown in Figure 5 and Table S5. The average percentage of high-quality sequences was 74.88%. A total of 12.86% of sequences with low quality were corrected according to high-quality sequences, while the remaining 12.26% were discarded. Thereafter, an average 6.33% of low-abundance sequences were also corrected (Table S5). In consideration of the influence posed on error statistics by impurity of the plasmids during the cell culture before we mixed the plasmids, if the “erroneous” sequence was found in on less than four samples and was detected >100 times in each sample, it was excluded from error rate calculation. After the correction process, the mean error rate of all sequences was decreased from 0.082 to 0.013%, and the percentage of error-bearing sequences was decreased from 6.313 to 0.912% (Figure 5).
Minimization of multiplex PCR bias
We successfully developed a novel method to minimize the MPCR bias under a given set of multiplex primers. Details of this method are shown in Figure 6 and Materials and Methods. Cross-validation was used to evaluate this method, as shown in Figure 6. Residual sum of squares (RSS) (Draper and Smith 1998) was calculated for each test, where , is the observed frequency, and is the expected frequency. Except for the mix 1-2 sample, the other five samples reduced the RSS value, which demonstrated that this method is evidently effective.
Moreover, the method was tested using three spiked-in samples, in which five known clones were spiked in 106 cells. Compared to the expected frequency, clones B, C, D, and G had obvious bias. After modifying the frequency with this method, the bias was relieved to a certain extent (Figure 7, A–C). The bias ratio was defined as the observed frequency bias divided by the corrected frequency bias. If the ratio is >1, it means the frequency is corrected positively. A total of 86.7% of the clones (except clone A in Index-R/T) generated a ratio >1, particularly clone D in Index-R. These results conclude that the method is indeed capable of minimizing MPCR bias (Figure 7, D and E).
Figure 7.
Evaluation of bias reduction in three spiked-in samples. Both observed frequencies and PCR bias-corrected frequencies were calculated for the five clones in three samples, and each clone provides a bias ratio to show bias change after correction. The bias ratio is defined as the observed frequency bias (observed frequency minus expected frequency) divided by corrected frequency bias (corrected frequency minus expected frequency). (A) Sample: Index-K, five clone frequencies. (B) Sample: Index-R, five clone frequencies. (C) Sample: Index-T, five clone frequencies. (D) Sample: Index-K, bias ratio. (E) Sample: Index-R, bias ratio. (F) Sample: Index-T, bias ratio.
IMonitor to monitor MRD
To test the feasibility of IMonitor in translational research, we applied it to analyze the data from two patients with B-ALL. Samples were collected upon diagnosis and on day 15 and day 33 post-treatment (details in Materials and Methods). We first tried to identify the cancer clones in the B-cell receptor repertoire. Patients with B-ALL showed a clear pattern of deficient clonal diversity (Figure 8A, Figure S6). By the cutoff of 10% for the cancer clone frequency, the cancer clone was identified in patient M001 with clonal frequency of 87.02% upon diagnosis, whereas it substantially decreased to 14.65% on day 15 and further to 0.28% on day 33, suggesting the effect of treatment (Figure 8B). However, with the increased sensitivity of our method, MRD was detected on day 33. In contrast, flow cytometry was not able to detect any MRD in this time slot. Interestingly, two cancer clones (48.39% and 42.13%) were identified in patient M002, and the data of day 33 post-treatment showed an MRD level of 2.54% (1.46% and 1.08% corresponding to each clone), compared with the negative result in detection with flow cytometry. Although follow-up of the patients’ condition was required to demonstrate its prognostic value, BCR sequencing plus IMonitor analysis showed its superiority in convenience and sensitivity for MRD detection. More importantly, the three-dimensional V-J pairing figure outputted by IMonitor revealed the remodeling of clonal diversity in the BCR repertoire following treatment, demonstrating its application in monitoring immune reconstruction (Figure 8A and Figure S6).
Figure 8.
Detection of MRD in B-ALL using IMonitor. (A) Repertoire (V-J pairing) of IGH is shown for pretreatment (day 0) and post-treatment (day 15 and day 33). (B) Cancer clone frequency is shown for each clone in the two patients (M001 and M002) before treatment (day 0) and post-treatment (day 15 and day 33).
Discussion
We have developed a comprehensive methodology for analysis of the T-cell receptor repertoire and B-cell receptor repertoire made available by next generation sequencing technology. IMonitor provides an arsenal of solutions for four steps: basic data processing, V(D)J assignment, structural analysis, and statistics visualization. IMonitor distinguishes itself from other analysis tools with several features. The first important feature is its realignment process. The high homology among genes and alleles together with random base deletion and insertion at gene junctions have been affecting the accuracy of alignment. Therefore, global or local alignment by itself is not sufficient to complete the whole picture. During the realignment process, CDR3 regions are scrutinized with the M-mismatch extension model of local alignment while non-CDR3 regions are covered by global alignment. The test using simulated data and published rearrangement sequences demonstrated IMonitor’s unquestionably better performance than other tools. The second feature of IMonitor is its ability to correct PCR and sequencing error and minimize MPCR bias, whose usage can be extended to other fields of research. IMonitor can be used to analyze any chain of T- and B-cell receptors and multiple species such as humans, monkeys, and rabbits. Furthermore, IMonitor results are presented with intuitive graphs. For example, the overall diversity of the immune system can be interpreted easily from a three-dimensional V-J pairing graph.
PCR and sequencing error of NGS is one of the problems that remain untackled for immune repertoire analysis. Preliminary results from previous studies show that a significant number of errors accumulate, and these errors can potentially lead to overestimating the actual TCR clonotypes. Besides, sequencing error also results in artificially increased diversity of the TCR repertoire (Nguyen et al. 2011; Warren et al. 2011). Simply filtering out all low-quality sequences not only removes the sequencing error bases, but also leaves out a lot of genuine sequences. Our method, however, manages to decrease the error rate while rescuing most of the sequences. Actually, the efficiency of the approach would be improved if more factors are considered when correcting the errors. For example, erroneous bases have some bias for certain sequencing platforms, and the occurrence probability of sequencing error at each position of sequence is different. Using the six plasmid mixture samples, the characteristics of PCR and sequencing errors can be analyzed, as shown in Figure S5. The base error rate declines as the base quality in the sequence improves, whereas the percentage of discarded sequences increases sharply (Figure S5, A and B). The quality indexes of incorrect bases mainly fall into two categories: Q ≤ 10 and Q ≥ 35. Apparently, the former mostly results from sequencing error, while the cause of the latter is mostly PCR error (Figure S5D) . More than 85% of sequences have only one error base, and the rate rises after removing the sequences with minimal base quality of Q20 (Figure S5C).
Here we have introduced a new bioinformatics methodology to reduce the PCR bias of MPCR samples. We found that the bias originated from two factors: template concentration and inconsistent primer efficiencies. Using six plasmid mixture samples, we designed a formula to reduce the bias. By applying it to the spiked-in samples, we validated its effectiveness. However, due to the limited size of training data, some bias persisted in spiked-in samples. We believe when the training data contain ≥100 templates, the effect of the approach would be more significant. Besides, different primer sets should be trained to generate a suitable formula to reduce bias, so this article mainly introduces a bioinformatics approach showing how to create a suitable formula to adjust the bias. Previous literature reports that it reduce the bias mainly through an experimental method to optimize the primers and primer concentration (Carlson et al. 2013). It is a scientific and systemic experimental method to adjust primers. It would be ideal to use this method for optimizing primers in the first step and then to use our bioinformatics method for further reducing PCR bias. Stephen R. Quake and colleagues developed a consensus read sequencing approach that incorporated unique barcode labels (UIDs) on each starting RNA molecule (Vollmers et al. 2013). It could eliminate PCR bias completely in theory if the synthetic UIDs were random enough.
IMonitor for analyzing the TCRs and BCRs repertoire in human and other animal models has the widest applications among the available tools in basic and translational research. We have demonstrated its utility in identifying the cancer clonotypes and monitoring MRD in B-ALL, while at the same time evaluating the clonal diversity for immune remodeling following treatment by its graphic visualization. We believe that IMonitor can also be applied in many other areas, such as tracing emerging clonotypes upon vaccination and following their frequencies during the process, selecting monoclonal antibodies based on sequencing the immune repertoire. With the importance of immune repertoire research becoming more recognized, we believe IMonitor will play a role in advancing our understanding of the immune system.
Supplementary Material
Acknowledgments
We thank the Science and Technology Planning Project of Guangdong Province (no. 2012A031100010) for support and the Shenzhen Municipal Government of China (no. GJHZ20130417140835564) for support. This study is supported by the International Science and Technology Cooperation Program of Shenzhen, China (GJHZ20130417140835564).
Footnotes
Communicating editor: G. D. Stormo
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.176735/-/DC1.
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Benichou J., Ben-Hamo R., Louzoun Y., Efroni S., 2012. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology 135: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D. A., Shugay M., Mamedov I. Z., Putintseva E. V., Turchaninova M. A., et al. , 2013. MiTCR: software for T-cell receptor sequencing data analysis. Nat. Methods 10: 813–814. [DOI] [PubMed] [Google Scholar]
- Carlson C. S., Emerson R. O., Sherwood A. M., Desmarais C., Chung M. W., et al. , 2013. Using synthetic templates to design an unbiased multiplex PCR assay. Nat. Commun. 4: 2680. [DOI] [PubMed] [Google Scholar]
- Chao A., 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11: 265–270. [Google Scholar]
- Chao A., 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43: 783–791. [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper N. R., Smith H., 1998. Applied Regression Analysis. Wiley, New York. [Google Scholar]
- Fischer N., 2011. Sequencing antibody repertoires: the next generation. MAbs 3: 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J. D., Warren R. L., Webb J. R., Nelson B. H., Holt R. A., 2009. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 19: 1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta B. A., Malming H. R., Jackson K. J., Bain M. E., Wilson P., et al. , 2007. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics 23: 1580–1587. [DOI] [PubMed] [Google Scholar]
- Janeway C., 2005. Immunobiology: The Immune System in Health and Disease. Garland Science, New York. [Google Scholar]
- Lefranc M.-P., Lefranc G., 2001a The Immunoglobulin Factsbook. Academic Press, San Diego. [Google Scholar]
- Lefranc M.-P., Lefranc G., 2001b The T Cell Receptor Factsbook. Academic Press, San Diego. [Google Scholar]
- Li S., Lefranc M. P., Miles J. J., Alamyar E., Giudicelli V., et al. , 2013. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat. Commun. 4: 2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Yuan J., Yiu S. M., Li Z., Xie Y., et al. , 2012. COPE: an accurate k-mer-based pair-end reads connection tool to facilitate genome assembly. Bioinformatics 28: 2870–2874. [DOI] [PubMed] [Google Scholar]
- Nguyen P., Ma J., Pei D., Obert C., Cheng C., et al. , 2011. Identification of errors introduced during high throughput sequencing of the T cell receptor repertoire. BMC Genomics 12: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H., Desmarais C., Matthis J., Livingston R., Andriesen J., et al. , 2012. Ultra-sensitive detection of rare T cell clones. J. Immunol. Methods 375: 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H. S., Campregher P. V., Srivastava S. K., Wacher A., Turtle C. J., et al. , 2009. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 114: 4099–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C. E., 1997. The mathematical theory of communication. 1963. MD Comput. 14: 306–317. [PubMed] [Google Scholar]
- Sherwood A. M., Emerson R. O., Scherer D., Habermann N., Buck K., et al. , 2013. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol. Immunother. 62: 1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N., Heather J., Ndifon W., Shawe-Taylor J., Chain B., 2013. Decombinator: a tool for fast, efficient gene assignment in T-cell receptor sequences using a finite state machine. Bioinformatics 29: 542–550. [DOI] [PubMed] [Google Scholar]
- Venturi V., Quigley M. F., Greenaway H. Y., Ng P. C., Ende Z. S., et al. , 2011. A mechanism for TCR sharing between T cell subsets and individuals revealed by pyrosequencing. J. Immunol. 186: 4285–4294. [DOI] [PubMed] [Google Scholar]
- Vollmers C., Sit R. V., Weinstein J. A., Dekker C. L., Quake S. R., 2013. Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proc. Natl. Acad. Sci. USA 110: 13463–13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Sanders C. M., Yang Q., Schroeder H. W., Jr, Wang E., et al. , 2010. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc. Natl. Acad. Sci. USA 107: 1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. L., Nelson B. H., Holt R. A., 2009. Profiling model T-cell metagenomes with short reads. Bioinformatics 25: 458–464. [DOI] [PubMed] [Google Scholar]
- Warren R. L., Freeman J. D., Zeng T., Choe G., Munro S., et al. , 2011. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 21: 790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., McGinnis S., Madden T. L., 2006. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 34: W6–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Ma N., Madden T. L., Ostell J. M., 2013. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41: W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousfi Monod M., Giudicelli V., Chaume D., Lefranc M. P., 2004. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics 20(Suppl 1): i379–i385. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W., 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203–214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source code of IMonitor is freely available for download at https://github.com/zhangwei2015.
Table S1: Simulated TRB with 0.1%, 0.5% and 2% sequencing error.
Table S2: Simulated IGH with 0.1%, 0.5% and 2% sequencing error and 0.1% hayper-mutation.
Table S3: Simulated Data with 0.5% sequencing error (TRA/IGK/IGL) and 4% hyper-mutation.
Table S4: Plasmid mixing pattern.
Table S5: Data process for PCR and sequencing error statistics.
Table S6: Samples information.
Table S7: Experimental design for five CD4+ T cell clones in the three spiked in mix.
Table S8: Performance of IMonitor and other tools on the simulated dataset.
Table S9: TRB and IGH V/J primers.
Figure S1: Insertion and deletion length distribution for simulated data.
Figure S2: IGH-VDJ Mutation and deletion/insertion analysis on the public sequences.
Figure S3: Outputs of IMonitor, H-B-01 as an example.
Figure S4: H-B-01 sample output figure of IMonitor.
Figure S5: Error characteristics of 6 plasmid mix samples.
Figure S6: V-J pairing dynamics for M002.
Figure S7: MiTCR and IMonitor performance in 3 spiked-in samples.
Figure S8: Nucleotide composition of V/J genes.