Abstract
Studies of natural populations of many organisms have shown that traits are often complex, caused by contributions of mutations in multiple genes. In contrast, genetic studies in the laboratory primarily focus on studying the phenotypes caused by mutations in a single gene. However, the single mutation approach may be limited with respect to the breadth and degree of new phenotypes that can be found. We have taken the approach of isolating complex, or polygenic mutants in the lab to study the regulation of transcriptional activation distance in yeast. While most aspects of eukaryotic transcription are conserved from yeast to human, transcriptional activation distance is not. In Saccharomyces cerevisiae, the upstream activating sequence (UAS) is generally found within 450 base pairs of the transcription start site (TSS) and when the UAS is moved too far away, activation no longer occurs. In contrast, metazoan enhancers can activate from as far as several hundred kilobases from the TSS. Previously, we identified single mutations that allow transcription activation to occur at a greater-than-normal distance from the GAL1 UAS. As the single mutant phenotypes were weak, we have now isolated polygenic mutants that possess strong long-distance phenotypes. By identification of the causative mutations we have accounted for most of the heritability of the phenotype in each strain and have provided evidence that the Mediator coactivator complex plays both positive and negative roles in the regulation of transcription activation distance.
Keywords: transcription, Mediator, polygenic, Saccharomyces cerevisiae
THE correct communication between a transcriptional activator and its target gene is of crucial importance to ensure properly regulated transcription. While most fundamental aspects of transcription initiation are conserved throughout eukaryotes, the distance over which transcriptional activation can occur varies greatly between yeast and metazoans. In the compact Saccharomyces cerevisiae genome, where intergenic distances are small and upstream activation sequences (UASs) are generally found within 450 base pairs (bp) of the transcription start site (Goffeau et al. 1996; Kristiansson et al. 2009), it is important that activation occurs over only a short distance to activate the correct target gene. In contrast, in the much larger metazoan genomes, enhancers that activate transcription are often located several kilobases away, with some enhancers as far as a megabase from a target gene (Bulger and Groudine 2011; Buecker and Wysocka 2012; Erokhin et al. 2015). While many studies have focused on understanding how enhancers function over a long distance to choose the correct target (Krivega and Dean 2012), there is less understanding of the regulation of transcriptional activation distance in yeast and how it differs from that in metazoans.
Early studies of yeast UAS elements suggested that transcriptional activation distance is limited (Guarente and Hoar 1984; Struhl 1984). More recent work systematically measured the dependence of transcriptional activation on the distance between the GAL1 UAS and a core promoter (Dobi and Winston 2007) and demonstrated that transcriptional activation by Gal4 diminishes with increasing distance. This study also suggested that activation distance is repressed by particular factors and by chromatin structure, as loss-of-function mutations that allow long-distance activation of a reporter were identified in several genes, including SIN4 and SPT2, which regulate transcription, SPT10, which regulates histone levels, and the HTA1-HTB1 gene pair, which encodes histones H2A and H2B.
These results suggested that the control of activation distance in yeast involves the contributions of many factors, an idea supported by two additional results. First, although several mutants were identified that allow long-distance activation, their phenotypes were modest, with only a low level of expression of a reporter for long-distance activation over a distance of 800 bp. Second, attempts were made to isolate stronger mutants by selection for mutations that enhance the phenotype of a sin4Δ mutant. This selection successfully resulted in the isolation of a second mutation that strengthened the mutant phenotype in a sin4Δ background, but which conferred no detectable phenotype when present as a single mutant (J. Leeman, K. C. Dobi, and F. Winston, unpublished results). The isolation of such an enhancer mutation suggested the existence of other factors that regulate activation distance that might never be found by mutant selections when analysis is restricted to single mutants. Therefore, we have isolated polygenic mutants to study strains with stronger long-distance activation phenotypes.
Classical genetic studies in model organisms usually focus on single mutations to facilitate gene identification and to understand gene function. However, many traits found in nature are polygenic (or complex), caused by the combined effects of mutations in many genes, resulting in a range of phenotypes (Mackay et al. 2009; Mackay 2014). In humans, many diseases that have a genetic component are polygenic, including type 2 diabetes, schizophrenia, and hypertension. A major challenge in human genetics today is identifying the causative mutations that contribute to these diseases (Manolio et al. 2009; Womack et al. 2012).
S. cerevisiae has also been a focus for studies of natural genetic variation and polygenic traits (Liti and Louis 2012), as strains found in nature display a broad range of phenotypic variance (Ehrenreich et al. 2009; Liti and Louis 2012). Studies of yeast strain natural variance have identified the causative alleles for a number of polygenic traits including sporulation efficiency (Deutschbauer and Davis 2005; Ben-Ari et al. 2006; Gerke et al. 2006), high temperature growth (Steinmetz et al. 2002), translation efficiency (Torabi and Kruglyak 2011), and wine alcoholic fermentation (Salinas et al. 2012). A major advantage of yeast in studies of natural variation caused by multiple mutations is the ability to perform allele replacements, allowing direct tests for causative mutations.
In addition to studies of natural variation in yeast, the isolation and analysis of polygenic mutants has recently been initiated in laboratory studies to analyze specific processes, including resistance to high pH (Romano et al. 2010), the origin of multicellularity (Koschwanez et al. 2013), and the ability to grow under nutrient limitations (Gresham et al. 2008). In our current study, we have isolated polygenic mutants to study the regulation of transcriptional activation distance. Using a dual reporter system, we have isolated three polygenic mutants that allow strong long-distance activation. We have identified causative mutations in the genes MOT3, GRR1, MIT1, MSN2, and PTR3 that contribute to this activation and we show that strong long-distance activation in these mutants is not merely a result of generally stronger transcriptional activation. Furthermore, we have fully recapitulated the mutant phenotype of one of the polygenic mutants from the identified causative mutations and have partially recapitulated the phenotype of the other two. Finally, our results have suggested a role for the Mediator coactivator complex in regulating long-distance activation.
Materials and Methods
S. cerevisiae strains
All S. cerevisiae strains used in this study (Supporting Information, Table S1) are isogenic with a GAL2+ derivative of S288C (Winston et al. 1995) and were constructed by standard allele replacement methods or crosses. All PCR primers are listed in Table S2. The dual reporter strains used to select mutants that allow long-distance activation were derived from previously described strains (Dobi and Winston 2007). These strains contained two reporters constructed within large nonessential open reading frames (ORFs), bph1::kanMX-UASGAL1799-HIS3 and dug2::TRP1-UASGAL1806-URA3, and sin4Δ0::LEU2. To construct the URA3 reporter, the HIS3 ORF was replaced with the URA3 ORF. Both reporters contain the HIS3 TATA element, transcription start sites, and transcription termination sequence. Allele replacements, either to correct a mutant allele to wild type or the reverse, were done by a two-step transformation method, using strains in which the URA3 ORF was deleted from the dug2 reporter. Allele replacements to correct mutant alleles to wild type were made in strains FY3056, FY3057, and FY3058. Mutant reconstructions to replace wild-type alleles with mutations were done in sin4Δ0::LEU2 strains. The first step in each case was the integration of URA3 at the relevant site. DNA containing URA3 was made by PCR using pRS406 (Sikorski and Hieter 1989) as template DNA. For replacement of URA3 by wild-type alleles, PCR fragments ∼500 bp in length were synthesized using DNA from strain FY3046, the wild-type parental strain, as a template. PCR fragments were used to transform the URA3 transformants, followed by selection for 5-FOA resistance. For replacement of URA3 by mutant alleles, PCR fragments ∼500 bp in length were amplified from genomic DNA prepared from strains 1.2 (FY3049; for mutations in MOT3, GRR1, and SGM1), 2.3a (FY3052; for mutations in MIT1, PTR3, and YOR019W), or 2.3b (FY3053; for a mutation in MSN2). Correct alleles were all verified by DNA sequencing. Complete deletions of the MOT3, GRR1, MIT1, PTR3, and MSN2 ORFs were constructed by replacement with URA3, using pRS406 as template DNA to generate PCR fragments with URA3. Strains with the longer reporter distances of 1397 bp and 2027 bp contain the HIS3 reporter in the same location as in the 799-bp reporter, with the GAL1 UAS located at the greater distances. For construction of strains that overexpressed MED2, MED3, and CDK8, plasmids were used from a previously described collection (Hu et al. 2007). All spot tests and most Northerns contain two control strains: a strain with a wild-type HIS3 gene (strain FY76, labeled “HIS3”) and a strain with the two reporters but no mutations that contribute to long-distance activation (strain FY3045, labeled “SIN4”).
Media
Rich (YPD) and synthetic complete (SC) dropout media were prepared as previously described (Rose et al. 1990). SC Gal and SC −His Gal media contained 2% galactose as the carbon source. YP raffinose contained 2% raffinose as the carbon source and YP Gal contain 2% galactose. Specified concentrations of 3-aminotriazole (3AT) were added to SC −His Gal medium.
Selection and screen to isolate polygenic mutants that allow long-distance activation
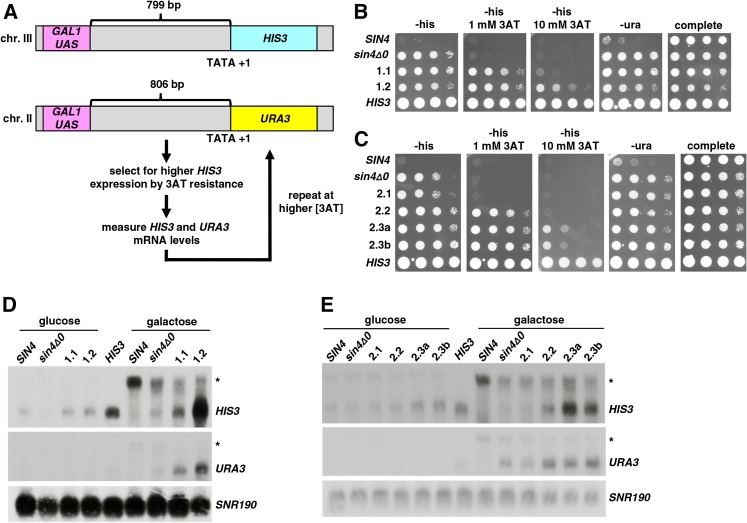
Starting with strains FY3046 and FY3047, strains of opposite mating type that each contain sin4Δ, we selected for mutants that allow long-distance activation using the selection and screening procedure diagrammed in Figure 1A. For each round of selection, 10 independent cultures of each strain were grown in liquid YPD overnight at 30°. From each culture, 200 μL were plated onto each of two SC −His Gal 3AT plates, one of which was UV irradiated (Winston 2008). The first round of mutants was selected using 1 mM 3AT, the second round using 5 mM 3AT, and the third round using 10 mM 3AT. Plates were incubated at 30° and colonies that grew were purified on SC −His Gal 3AT. Mutants 1.1, 2.1, 2.2, and 2.3a were isolated spontaneously, while strains 1.2 and 2.3b were isolated after UV mutagenesis. Strains 2.3a and 2.3b were derived from the same original colony of FY3047, but were separated for the third and final round of selection. Mutant candidates were retested by dilution spot tests for 3AT resistance and for whether the His+ phenotype was dependent on galactose. Mutants showing stronger 3AT resistance than the parent strains were then tested for HIS3 and URA3 messenger RNA (mRNA) levels by Northern analysis. Mutants that showed increased mRNA levels for both reporters relative to the parent strain were used for further selection.
Figure 1.
Isolation of polygenic mutants that allow long-distance transcriptional activation. (A) The diagram illustrates the steps for selection and screening for mutants that allow transcription of reporters with the GAL1 UAS at a distance that is normally nonpermissive for activation. (B and C) Dilution spot tests to assay expression of the long-distance reporters after each round of mutant isolation. All plates contain galactose as the carbon source and the indicated levels of 3AT. (D and E) Northern analysis was done to measure the levels of HIS3 and URA3 RNAs after each round of mutant isolation. SNR190 served as the loading control. The Northern presented in E was chosen for best overall quality, where the other two replicates showed greater increases in URA3 levels in 2.3a and 2.3b as compared to 2.2 (data not shown). The asterisks note the longer transcripts that likely initiate near the GAL1 UAS (see text and Dobi and Winston 2007). The two control strains in this and later figures are a strain with a wild-type HIS3 gene (strain FY76, labeled “HIS3”) and a strain with the two reporters but no mutations that contribute to long-distance activation (strain FY3045, labeled “SIN4”).
Bulk segregant analysis and high-throughput sequencing of yeast segregant pools to identify candidate mutations
To identify candidate mutations, we performed bulk segregant analysis (Brauer et al. 2006; Wenger et al. 2010). First, pools of strains were generated for bulk segregant analysis by backcrossing polygenic mutant 1.2 to FY3046 and backcrossing the other two polygenic mutants, 2.3a and 2.3b, to FY3047. For each cross, approximately 300 tetrads were dissected. The long-distance activation phenotypes of the progeny were tested by replica plating to SC −His Gal media with 0 mM, 1 mM, 5 mM, and 10 mM 3AT. The mutant pools for each cross were composed of segregants with the same 10-mM 3AT-resistance phenotype as in the polygenic mutant parent. The wild-type pools for each cross were composed of segregants that had the phenotype of the sin4Δ0 single mutant, which is sensitivity to 1 mM 3AT. Each pool contained at least 40 segregants, based on prior bulk segregant analysis studies (Brauer et al. 2006; Wenger et al. 2010), and the mutant and wild-type pools from each backcross contained the same number of segregants: 45 segregants (1.2), 48 segregants (2.3a), and 42 segregants (2.3b). Strain 1.2 segregants were all derived from complete tetrads while strain 2.3a and 2.3b segregants were from a mixture of complete and incomplete tetrads. To prepare genomic DNA for sequencing, a culture of each segregant was grown in YPD to saturation at 30°. For each pool, 1 ml of each culture was combined, the pooled culture was split into six fractions, and DNA was purified from each fraction as previously described (Rose et al. 1990).
Genomic libraries were prepared for DNA sequencing using the Illumina TruSeq DNA PCR-Free Sample Preparation kits. Using PCR-free sample preparation prevents additional mutations from being introduced during library amplification. The resulting libraries were sequenced on a single lane of an Illumina HighSequation 2000, producing 108-nucleotide single end reads and greater than 80-fold coverage for each library. The sequence reads, compiled in a FASTQ file, were mapped to the S. cerevisiae S288C genome using BWA (Li and Durbin 2009) producing a SAM file. Using SAMTools (Li et al. 2009), this SAM file was converted to a BAM file, which was searched for single-nucleotide polymorphisms (SNPs) using Freebayes (Garrison and Marth 2012). Freebayes generated a VCF file containing the SNPs. We then calculated, using a PERL script (File S1), the frequency of all SNPs in two matched pools: (1) segregants with the wild-type phenotype and (2) segregants with the mutant phenotype. SNPs with a higher frequency in the mutant pool were further considered as candidate SNPs that might be causative. There was no strict threshold cutoff; however, SNPs were considered candidates if they were present at a frequency >50% in pool 2. The SNPs were subsequently computationally verified by observing their frequency and location in the BAM file containing all of the aligned reads, using Integrative Genomics Viewer (IGV) (Robinson et al. 2011). Once the SNPs were confirmed by these methods, they were further confirmed to be present in the mutant parent and absent in the wild-type parent by Sanger DNA sequencing.
Dilution spot tests
Cultures were grown in liquid YPD at 30° to saturation. The next day, the cells were pelleted and resuspended in sterile water to adjust them to the same concentration based on OD600. Then 10-fold serial dilutions were made in sterile water into a microtiter dish. The dilutions were spotted on different media, and the plates were incubated at 30° for 4 days unless otherwise noted.
Northern analysis
RNA isolation and Northern hybridization experiments were performed as previously described (Ausubel et al. 1991). Strains were grown to ∼1 × 107 cells/ml in YP raffinose and then divided with one part shifted to YPD and the other shifted to YP Gal for 1 hr each. Northern hybridization analysis was conducted with probes to the coding regions of HIS3 (+400 to +651, where +1 is the ATG), URA3 (+206 to +727), and SNR190 (+1 to +190), which served as a loading control. Each Northern experiment was done at least three times with independent cultures.
Western analysis
Western blots were performed on a carboxy-terminally tagged Med3-FLAG fusion that was constructed by standard procedures, using a FLAG tag with a glycine linker (Funakoshi and Hochstrasser 2009). For Western blot analysis, protein extracts were prepared as follows. Briefly, 10-ml cultures grown to ∼1 × 107 cells/ml in YP raffinose, were shifted to YP Gal for 1 hr, and then the cells were pelleted and frozen at −70°. Cells were resuspended in 1 ml cold dH2O, 150 μl of YEX buffer (7.4 g NaOH, 7.5 ml β-mercaptoethanol, 92.5 ml dH2O) was added, and samples were incubated on ice for 10 min. A total of 150 μl of cold 50% TCA/50% dH2O was added and samples were incubated on ice for 10 min. Cells were pelleted at 12,500 rpm for 5 min at 4° and the supernatant was carefully removed. Samples were resuspended in 200 μl of 2× Laemmli buffer (2.5% SDS, 60 mM Tris base, 6.25 mM EDTA, pH 8.0, protease inhibitors), boiled for 5 min, and quantitated by Bradford assay to assure equal loading. To each sample, 5× Laemmli buffer B was added [50% glycerol 100 mM Tris base 0.1–0.05% (w/v), Bromophenol blue, 400 mM dithiothretol 10% 2-mercaptoethanol], and samples were heated to 95° for 5 min before loading on an 8% SDS-polyacrylamide gel. Primary antibodies for Western blot analysis were anti-FLAG antibody (Sigma M2) or anti-Pgk1 (Life Technologies). Secondary antibodies were LI-COR goat antimouse. Imaging was performed using the AERIUS Automated Imaging System and were quantified using ImageJ.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously (Helmlinger et al. 2008). ChIP of Med3 was performed using a Med3-FLAG fusion. The strains used for ChIP were deleted for the endogenous GAL1 UAS to allow detection of Med3 association with the GAL1 UAS at the long-distance reporter. Briefly, 50-ml cultures grown to ∼1 × 107 cells/ml in YP raffinose, were shifted to YP Gal for 1 hr and were then cross-linked (1% formaldehyde, 30 min). Next, the cells were broken open by bead beating to achieve 100% cell lysis. Wild-type cells were lysed after six rounds of bead beating and the mot3-1 grr1-1 sin4Δ0 mutant required nine rounds of bead beating to achieve the same amount of lysis. The chromatin fraction was isolated and sheared to 100- to 300-bp fragments using a Bioruptor sonicator (Diagenode). Immunoprecipitations (IPs) were performed overnight at 4° with 1 μg of anti-FLAG antisera (Sigma M2). IPs were coupled to 50 μl of protein G-Sepharose beads (GE Healthcare Life Sciences) at 4° for 4 hr. The beads were washed and eluted, and the eluate was reverse cross-linked overnight at 65° and incubated with proteinase K and glycogen for 2 hr at 37°. DNA was purified by phenol-chloroform extraction and precipitated in ethanol overnight at 80°. ChIP DNA was analyzed by qPCR on the Stratagene MX3000P cycler. Each sample was analyzed in triplicate and quantified by comparison to a standard curve (10-fold serial dilutions of input DNA). Relative percent IP (%IP) values were calculated by dividing the %IP (calculated as IP/input) at the regions of interest by the %IP at an intergenic background locus.
Microarray experiments
RNA was purified, labeled, and hybridized to Agilent expression microarrays (8× 15k custom printed yeast arrays) (AMADID 017566) as previously described (McIsaac et al. 2011). Reference RNA from strain FY3054 was labeled with Cy3 and experimental RNA was labeled with Cy5. RNA from FY3054 grown in YP raffinose was used as the reference for all arrays. After washing, arrays were scanned using Agilent Feature Extractor software version 9.5. Genes that had flagged features marked as unreliable were excluded from data analysis; this resulted in the analysis of 5610 genes. The raw signal intensity values were floored to a value of 350 (values of <350 were set to 350). The log2 ratio was calculated using the floored data values. Each microarray was performed twice for wild type (FY3054), 1.2 (FY3056), 2.3a (FY3057), 2.3b (FY3058), sin4Δ mot3-1 grr1-1 (FY3061), and sin4Δ mot3-1 mit1-1 (FY3067). Analysis of the log2 ratios revealed that one of the 1.2 replicates and one of the 2.3a replicates were unusable, so these were discarded from analysis. For the remaining replicates, the log2 ratios were averaged and these averages were used for subsequent analysis. We used a two-fold cutoff for calling RNA levels as increased or decreased (log2 value of 1 or −1). Hierarchical clustering was performed using Cluster 3.0 with average linkage using the Pearson correlation distance as the metric of similarity between genes (de Hoon et al. 2004). K-means clustering was performed with MultiExperiment Viewer using a setting of 10 clusters and using Euclidean distance as the metric of similarity between genes (Saeed et al. 2003, 2006).
Data availability
File S1 describes the code used to analyze the sequencing results. Yeast strains are listed in Table S1 and are available upon request. Table S2 lists all oligonucleotide sequences used. Table S3 lists crosses used for bulk segregant analysis. Table S4 lists sequencing coverage. Table S5 lists causative mutant candidates. Table S6 lists microarray results. Table S7 describes enrichment for transcriptional effects on adjacent gene pairs. Figure S1 shows evidence for disomy in two mutants. Figure S2 presents Western analysis of Med3 levels. The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) (Edgar et al. 2002) and are accessible through GEO series accession no. GSE72093.
Results
Selection of polygenic mutants that allow increased transcription activation distance
To study the regulation of transcriptional activation distance, we decided to isolate polygenic mutants, based on two sets of previous results. First, the single mutants that we previously identified that allow long-distance activation (Dobi and Winston 2007) have modest phenotypes, with weak expression of the HIS3 reporter. Second, additional pilot mutant selections demonstrated that we could obtain double mutants with stronger phenotypes, but when the mutations were separated genetically, one of the mutations often caused no detectable phenotype. Therefore, we rationalized that the isolation of polygenic mutants would allow the isolation of mutants with stronger phenotypes as well as the identification of additional genes involved in the regulation of activation distance.
We selected for polygenic mutants that allow long-distance activation using the dual reporter strains described in Materials and Methods (Figure 1A). Each strain also contained a sin4Δ0 mutation, as sin4 mutants were the strongest and most frequent class of single mutants previously isolated (Dobi and Winston 2007). Two reporters were used, as selection using only a single reporter often identified cis-acting mutations and chromosomal rearrangements in addition to the desired class of trans-acting mutations (C. T. Reavey and F. Winston, unpublished results).
Beginning with single colonies, one from strain FY3046 and the other from strain FY3047, multiple rounds of selection and screening for increased long-distance activation were performed (Materials and Methods), resulting in the isolation of three strains that allowed strong long-distance activation, named 1.2 (FY3049), 2.3a (FY3052), and 2.3b (FY3053). The latter two strains derived from the same original colony of FY3046, but were separated for the third and final round of selection. Additional rounds of selection were attempted for each, but did not yield any mutants with increased long-distance activation. We will refer to these strains as 1.2, 2.3a, and 2.3b to denote their lineage relationship.
To analyze the strength of long-distance activation at each round of mutant isolation, we assayed both 3AT resistance (a phenotypic readout of HIS3 expression levels) and the levels of the HIS3 and URA3 reporter mRNAs by Northern analysis. Our results show that the mutants generally show an increased level of 3AT resistance after each round of selection (Figure 1, B and C), as well as increased HIS3 and URA3 mRNA levels (Figure 1, D and E). In addition, all the mutants require galactose for their phenotypes, showing that, as expected, the activation of the reporter is galactose dependent. In addition to the HIS3 and URA3 transcripts, we also see a longer transcript for each reporter. Previous 5′-RACE experiments showed that the HIS3 long transcript initiates proximal to the UAS (Dobi and Winston 2007). Although the level of this long transcript is variable and we do not understand its regulation, we note that transcripts initiating near enhancers have been observed in metazoans (Ashe et al. 1997; Kong et al. 1997; Carninci et al. 2005; Manak et al. 2006). Overall, our results demonstrate the successful isolation of mutants with strong long-distance activation phenotypes.
Identification of candidate causative mutations by bulk segregant analysis and whole-genome sequencing
To identify candidate causative mutations, we used bulk segregant analysis followed by whole-genome sequencing (Materials and Methods). Briefly, the three polygenic mutant strains, 1.2, 2.3a, and 2.3b, were crossed to sin4Δ0, the resulting diploids were sporulated, and ∼300 tetrads were dissected and analyzed for each cross (Table S3). Consistent with multiple mutations causing the strong 3AT-resistance phenotype, the progeny displayed a continuum of 3AT-resistance phenotypes ranging from sensitivity to 1 mM 3AT (the phenotype of the sin4Δ0 single mutant parent) to resistance to 10 mM 3AT (the phenotype of the polygenic mutant parent).
To identify candidate mutations based on DNA sequence, progeny with each parental phenotype were pooled, each pool was sequenced (Table S4), and candidate mutations were identified as those present in the mutant (3AT-resistant) pool at a level >50%, while present in the 3AT-sensitive pool at ∼50%. We set the threshold below 100% for the mutant pool as we considered the possibility that different combinations of causative mutations might be sufficient to confer the strong mutant phenotype. By this reasoning, every mutation might not be required in every segregant to produce the strong mutant phenotype. To qualify as a candidate, we also required that the putative mutant allele be present in the original polygenic mutant parent and the wild-type allele to be present in the sin4Δ single mutant parent. Using these requirements, we established a list of genes to be tested for causality (Table S5).
Identification of causative mutations in lineage 1
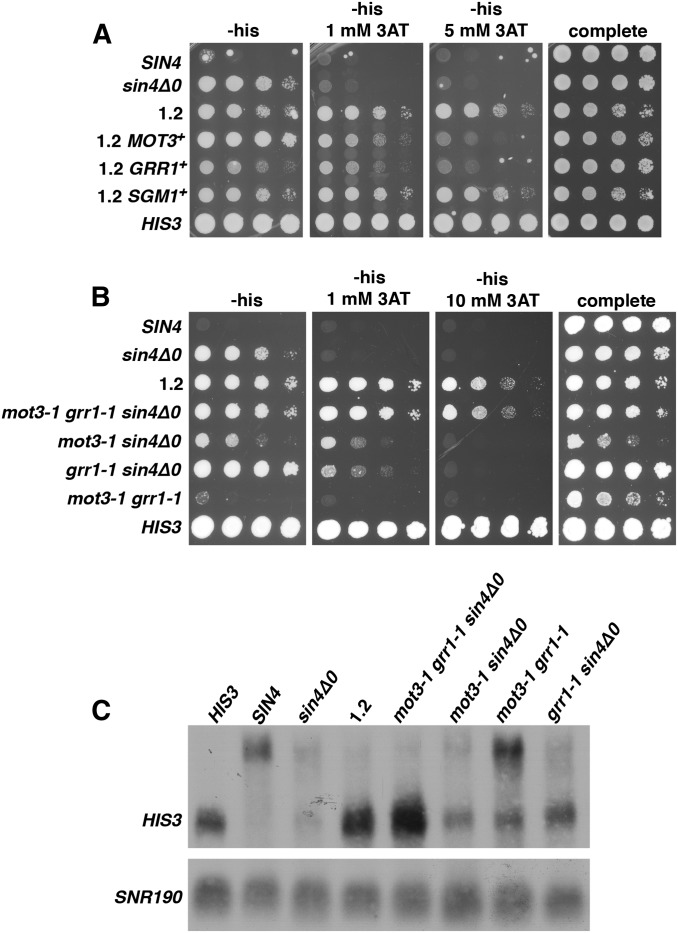
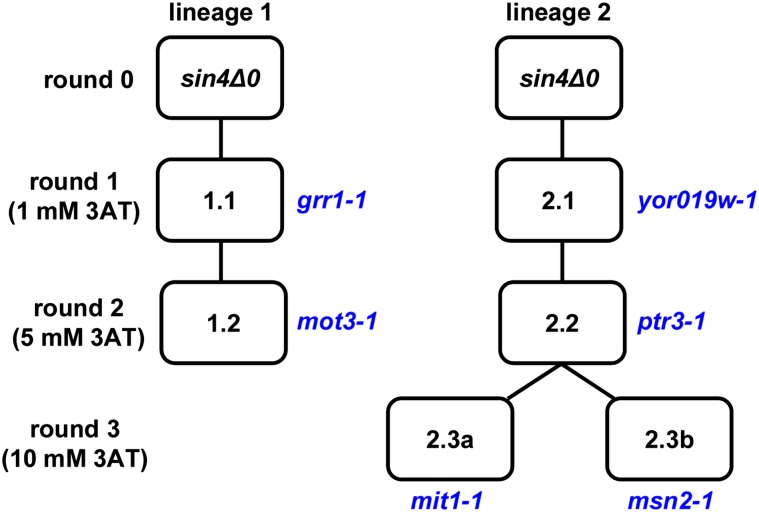
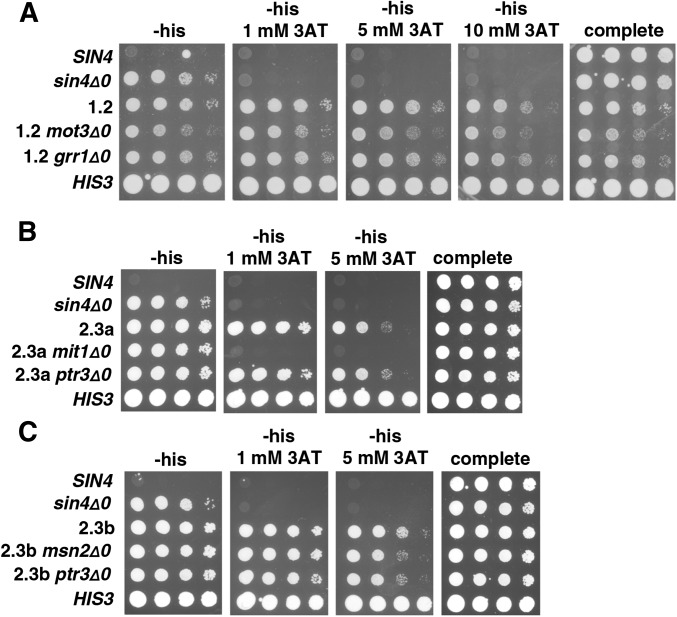
To test the candidate mutations present in strain 1.2 for causality, we replaced them with the wild-type allele and then tested the mutant phenotypes. In these experiments, strains contained only the HIS3 reporter as the allele replacement method required that URA3 not be expressed. Our results showed that replacement of either grr1-1 or mot3-1 causes a reduction in 3AT resistance (Figure 2A). In contrast, replacement of another mutation, sgm1-1, with the wild-type allele does not alter 3AT resistance. The sgm1-1 mutation was likely present at a high frequency in the mutant pool because SGM1 is linked to GRR1. Sanger sequencing of GRR1 and MOT3 showed that the grr1-1 arose in the first round of selection while mot3-1 arose in the second round (Figure 3). These results demonstrate that grr1-1 and mot3-1 are causative.
Figure 2.
Tests to identify causative mutations in lineage 1. (A) Allele replacements were done to test each candidate mutation. Starting with polygenic mutant 1.2, three mutations mot3-1, grr1-1, and sgm1-1 were each replaced with the wild-type allele and were then tested for long-distance activation by expression of the HIS3 reporter on the plates shown. All plates contain galactose as the carbon source. Plates were incubated at 30° for 4 days. (B) To test the sufficiency of the identified causative mutations, beginning with the sin4Δ strain, FY3055, strains were constructed that contained each mutation singly (data not shown), as different combinations of double mutants, and as the mot3-1 grr1-1 sin4Δ0 triple mutant. All plates contain galactose as the carbon source. Plates were incubated at 30° for 4 days. (C) Northern analysis was done to measure HIS3 RNA levels in the reconstructed mutants and to compare the levels to that in the original polygenic mutant 1.2.
Figure 3.
Lineages of the polygenic mutants. Diagrammed are the steps in the isolation of the polygenic mutants 1.2, 2.3a, and 2.3b. Shown in blue are the causative mutations isolated after each round. The strains 2.3a and 2.3b were derived independently from 2.2 as described in the text.
As a complementary approach, we tested whether we could recapitulate the original mutant phenotypes of strain 1.2 by integrating the causative mutations into a wild-type background. To do this, we constructed a sin4Δ0 mot3-1 grr1-1 triple mutant, as well as all possible combinations of double mutants, and analyzed the strains for their phenotypes by 3AT resistance and Northern analysis. Significantly, the sin4Δ0 mot3-1 grr1-1 triple mutant is resistant to 10 mM 3AT and shows a similar level of HIS3 mRNA expression as strain 1.2 (Figure 2, B and C), strongly suggesting that these three mutations account for the strain 1.2 mutant phenotypes. In contrast, all of the double mutant combinations have much weaker phenotypes, especially the mot3-1 grr1-1 mutant. From these results, we conclude that the sin4Δ0, mot3-1, and grr1-1 mutations together can account for the strain 1.2 mutant phenotype.
Identification of causative mutations in lineage 2
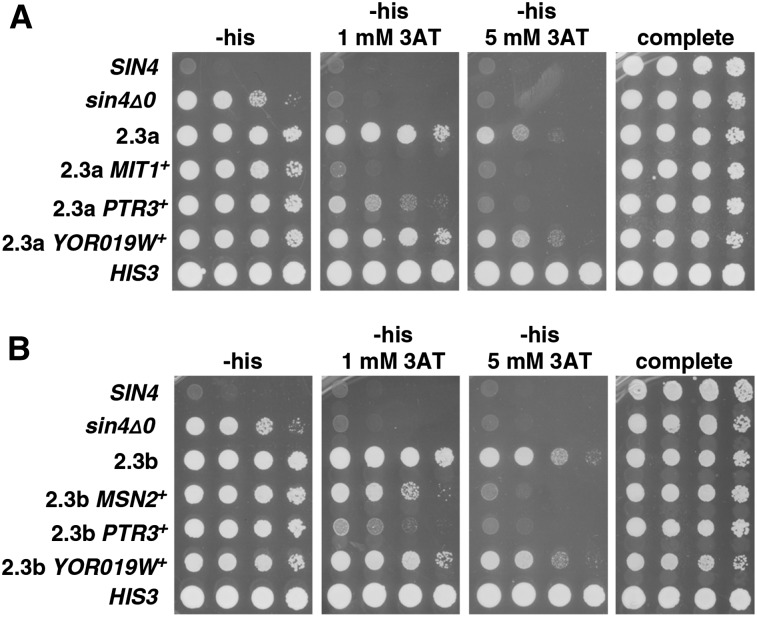
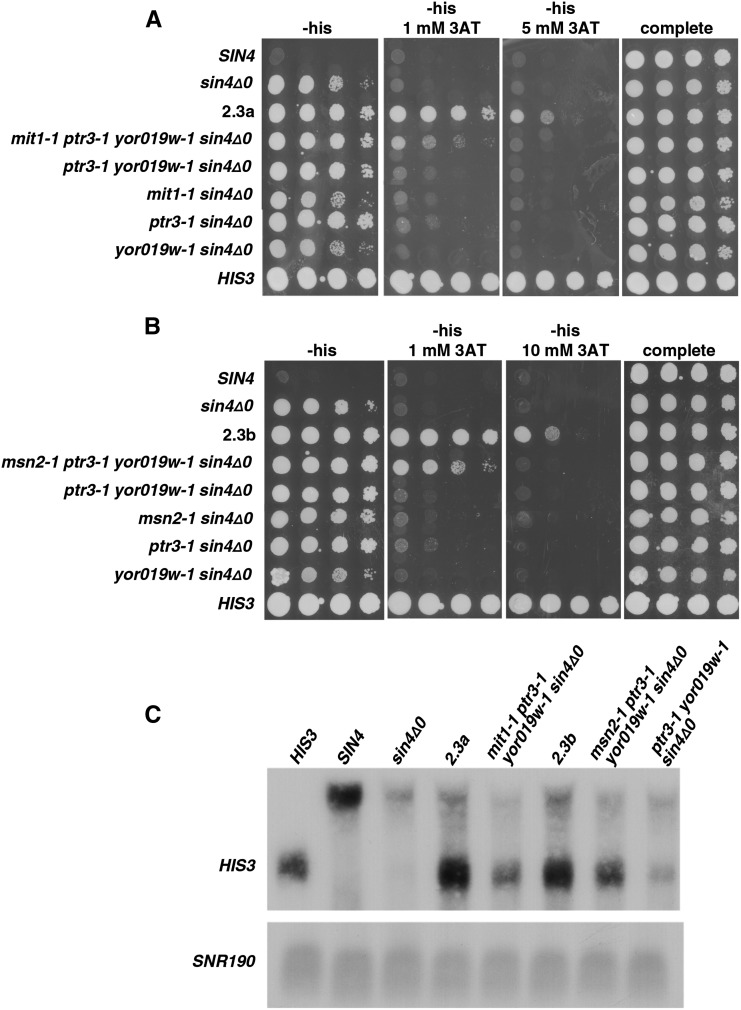
Similar steps were taken with strains 2.3a and 2.3b to determine the causative mutations. By replacement with wild-type alleles, our results showed that ptr3-1 is causative in both strains 2.3a and 2.3b, while mit1-1 is causative in 2.3a and msn2 in 2.3b (Figure 4). Our results indicate that yor019w-1 contributes very weakly to the mutant phenotype, as replacement with the wild-type allele causes a very modest reduction in growth on 3AT media in the 2.3b strain background. Sanger sequencing of each relevant gene in strains 2.1, 2.2, 2.3a, and 2.3b showed that yor019w-1 arose in the first round of selection, ptr3-1 in the second round, and mit1-1 and msn2-1 independently in the third round (Figure 3).
Figure 4.
Tests to identify causative mutations in lineage 2. (A and B) Allele replacements were done to test each candidate mutation. Starting with polygenic mutants 2.3a and 2.3b, the mutations mit1-1, ptr3-1, yor019w1-1, and msn2-1 were each replaced with the wild-type allele and were then tested for long-distance activation by expression of the HIS3 reporter on the plates shown. All plates contain galactose as the carbon source.
As we did for lineage 1, we tested whether combining the identified causative mutations can recapitulate the phenotypes of strains 2.3a and 2.3b. Our results show that, while each reconstructed strain can activate the long-distance reporter, the phenotypes are more modest than those of the original strains 2.3a and 2.3b (Figure 5). Therefore, we are able to partially recapitulate the phenotype of 2.3a and 2.3b from the known causative mutations, although there is some missing heritability.
Figure 5.
Attempts to reconstruct the phenotypes of strains 2.3a and 2.3b from causative mutations. (A and B) To test the sufficiency of the identified causative mutations for strains 2.3a and 2.3b, beginning with the sin4Δ strain, FY3055, strains were constructed that contained each mutation singly (data not shown), as different combinations of double and triple mutants, and as the quadruple mutants. All plates contain galactose as the carbon source. Pictured is the highest 3AT concentration with strong 2.3a or 2.3b growth on day 4. (C) Northern analysis was done to measure HIS3 RNA levels in the reconstructed mutants and to compare the levels to that in the original polygenic mutants 2.3a and 2.3b.
Some of the missing heritability can be accounted for by aneuploidy. Analysis of the DNA sequencing results for strains 2.3a and 2.3b suggests that they are both disomic for chromosome III, the chromosome that contains the HIS3 reporter, and that only 2.3a is also disomic for chromosome XIV (Figure S1). Additional genetic analysis showed that the chromosome III disomy contributes to the strength of the phenotype in these strains, based on increased 3AT resistance, although it does not completely account for the missing heritability (data not shown).
The mit1-1 and mot3-1 mutations do not cause loss of function
To test whether the causative mutations confer their phenotypes due to loss of function, we deleted each causative gene in the polygenic mutants, replacing the original mutant allele, and tested the mutant phenotypes. From these results, the deletion alleles grr1Δ0, ptr3Δ0, and msn2Δ0 each confer the same level of 3AT resistance as the original alleles (Figure 6), suggesting that the original alleles cause a loss of function. As MSN2 has a paralogue, MSN4, we also tested the effect of msn4Δ0 and of msn2Δ0 msn4Δ0 in a sin4Δ0, ptr3-1, and yor019w-1 background. We found that msn4Δ0 confers a level of 3AT resistance similar to msn2Δ0, while the msn2Δ0 msn4Δ0 combination allows greater 3AT resistance (data now shown), consistent with distinct roles for the two proteins (Berry and Gasch 2008). Therefore, loss of the factors Grr1, Ptr3, Msn2, and Msn4 contributes to long-distance activation.
Figure 6.
Tests of of loss-of-function mutations. To test whether the mutations isolated in polygenic mutants 1.2, 2.3a, and 2.3b cause a loss of function, each causative allele was replaced by a complete deletion of the gene and the strains were compared to the original polygenic mutants. All plates contain galactose as the carbon source.
In contrast, mot3Δ0 and mit1Δ0 do not confer the same mutant phenotype as the mot3-1 and mit1-1 alleles, respectively, as each deletion mutation weakens the original mutant phenotype (Figure 6). In the case of the mot3Δ0 mutation, this is a partial effect, as we see three strengths of mutant phenotypes (mot3-1 > mot3Δ0 > MOT3+), while in the case of the mit1 alleles, the difference is clearer as mit1Δ0 confers the same phenotype as MIT1+. Both the mot3-1 and mit1-1 mutations encode changes in the DNA binding domains of these transcription factors (Madison et al. 1998; Cain et al. 2012), as mot3-1 causes an N388H change in a position that directly contacts DNA (Grishin et al. 1998), while mit1-1 causes an H187R change at a position that likely causes a conformational change of the DNA binding domain (Lohse et al. 2014) (M. B. Lohse and A. D. Johnson, personal communication). These results demonstrate that mit1-1 and mot3-1 are not loss-of-function alleles.
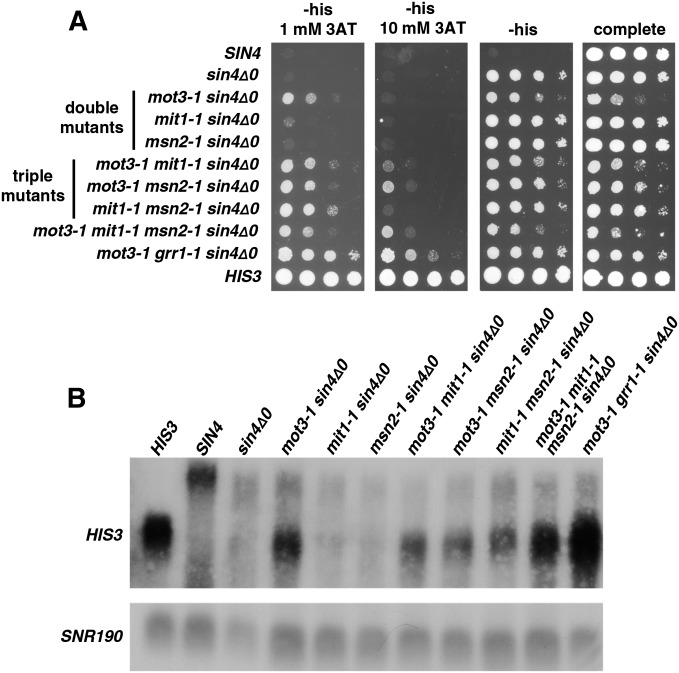
Combinatorial analysis of transcription factor mutations
We were interested in testing for genetic interactions between the mutations isolated in the different lineages. To do this, we constructed a new series of mutant strains, testing combinations of the mot3, mit1, and msn2 mutations. Our results (Figure 7) show that combining the mutations in a sin4Δ background both increases 3AT resistance and causes corresponding increases in HIS3 mRNA levels. These results show that interactions between the mutations are not confined to the lineages in which they arose.
Figure 7.
Combinations of mutations from different lineages allow long-distance activation. (A) To test whether mutations isolated in different lineages can function together to allow long-distance activation, we constructed the strains shown in the figures and tested them for 3AT resistance by spot tests. All plates contain galactose as the carbon source. (B) The same strains were also assayed for HIS3 RNA levels by Northern analysis. Strains are compared to the original sin4Δ0 parent and the reconstructed lineage 1.2 strain, mot3-1 grr1-1 sin4Δ0.
Analysis of shorter and longer activation distances
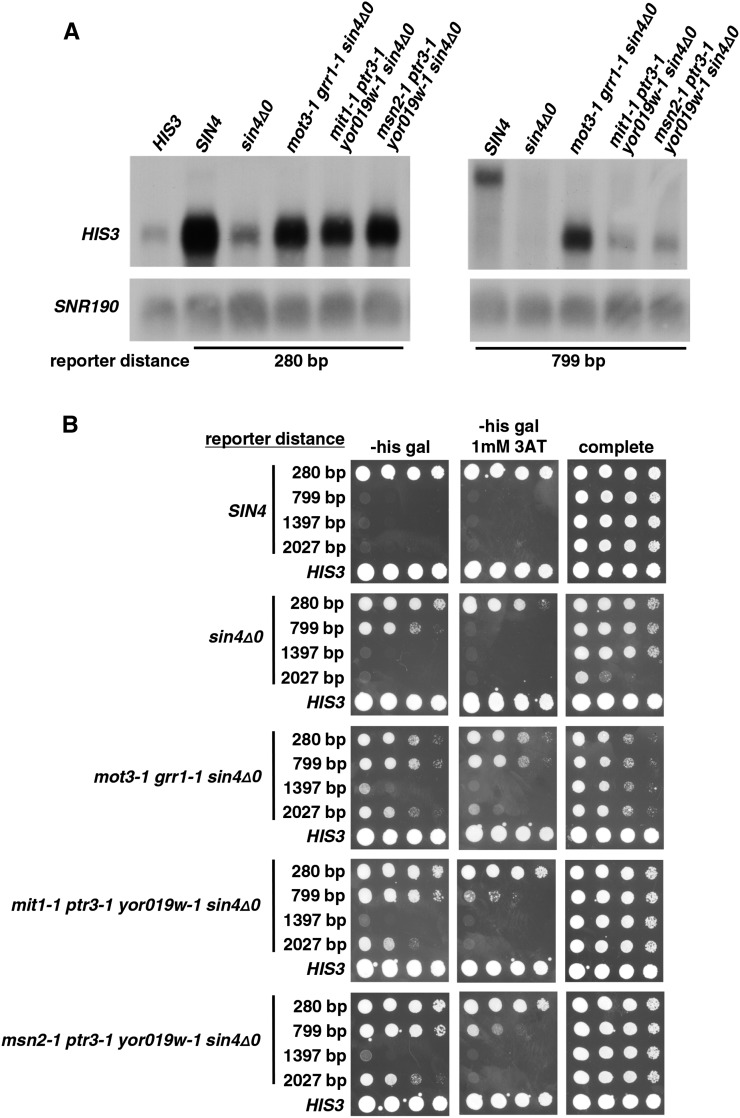
Previous studies have suggested a correlation between activation strength and activation distance (Carey et al. 1990), suggesting that our mutants might allow long-distance activation by a general increase in Gal4 activation strength. By this model, we expect to observe increased activation in our mutant backgrounds even over a short distance. However, when we tested this idea by analyzing expression of a HIS3 reporter where activation occurs over a distance of only 280 bp, we found that the highest level of activation occurs in the wild-type strain, with a lower level in the polygenic mutant and an even lower level in a sin4Δ mutant (Figure 8A). These results show that the mutants do not act through an overall increase in activation strength and may specifically have acquired the ability to activate over long distances.
Figure 8.
Tests of activation at shorter and longer distances. (A) Northern analysis was performed to test the polygenic mutants for activation of a reporter with a shorter distance of 280 bp and to compare the relative levels of activation to the long-distance reporter with a distance of 799 bp. (B) Spot tests were done to measure the level of 3AT resistance in strains that carry reporters with four different distances between the GAL1 UAS and the HIS3 TATA: 280 bp, 799 bp, 1397 bp, and 2027 bp. For reasons we do not understand, we reproducibly observed poor growth of the sin4Δ mutant with the 2027-bp reporter on complete medium. All plates contain galactose as the carbon source. Plates were incubated at 30° for 3 days.
To test whether the mutant strains are able to activate transcription at distances greater than the 799 bp in our reporter, we constructed and tested two longer reporters of 1397 bp and 2027 bp. All three reconstructed polygenic mutants strains show the ability to activate, albeit modestly, at these longer distances (Figure 8B). Surprisingly, there appears to be stronger activation with the 2027-bp reporter strain than with the 1397-bp reporter strain. Attempts to detect transcription at these long distances by Northern analysis were not successful, presumably due to the low level of the transcripts. We additionally tested the polygenic mutants at each distance for growth using glucose as the carbon source and did not see any growth on SC −His selective medium (data not shown), suggesting that there are not other activator sites within the reporter contributing to the phenotype in the 1397- and 2027-bp reporter strains. Our results strongly suggest that the polygenic mutants are able to activate transcription at distances up to 2 kb.
A role for the Mediator coactivator complex in regulation of activation distance
Several lines of evidence suggest a role for the Mediator coactivator complex in the regulation of transcription activation distance. Our initial studies of long-distance activation showed that Sin4 and Rgr1 of Mediator repress long-distance activation (Dobi and Winston 2007; De Santa et al. 2010; Kim et al. 2010; K. C. Dobi, C. T. Reavey, and F. Winston, unpublished results). While our current study did not isolate additional mutations in genes encoding Mediator components, a recent study (Gonzalez et al. 2014) has connected Grr1, a ubiquitin ligase, to Mediator function, as it suggested a role for Grr1 in regulating the stability of Med3 of Mediator. That study demonstrated that Grr1 normally destabilizes Med3, dependent upon Med3 phosphorylation by Cdk8, another Mediator component. In addition, previous studies have provided biochemical and genetic evidence that Mediator is required for activation by Gal4 (Park et al. 2000; Jeong et al. 2001; Bryant and Ptashne 2003; Kuras et al. 2003; Larschan and Winston 2005; Reeves and Hahn 2005).
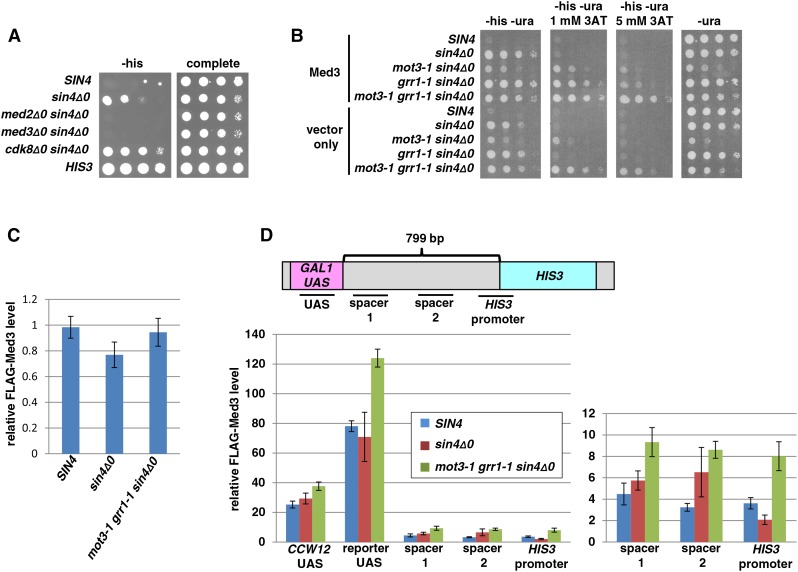
Based on these previous studies, we tested for roles for Med3, Cdk8, and other Mediator components in long-distance activation by double mutant analysis with sin4Δ. Both the sin4Δ0 med2Δ0 and sin4Δ0 med3Δ0 double mutants show complete loss of long-distance activation (Figure 9A). In contrast, the sin4Δ0 cdk8Δ0 double mutant shows a modest increase in long-distance activation relative to the sin4Δ0 single mutant. These results suggest that Med2 and Med3 are required for long-distance activation and Cdk8 plays a role in repressing it.
Figure 9.
Tests for the requirements of Mediator components. (A) Spot tests to determine the requirement for Mediator components Med2, Med3, and Cdk8. All plates contain galactose as the carbon source. Plates were incubated at 30° for 3 days. (B) Spot tests to determine the phenotype of overexpression of MED3 from a high-copy-number plasmid. All plates contain galactose as the carbon source. (C) To determine the level of Med3 protein in our strains, Western analysis was performed. Shown is a bar graph showing the mean and standard error of three experiments normalized to the levels of Pgk1. T-tests indicated no significant difference between Med3 levels in WT, sin4Δ0, and mot3-1 grr1-1 sin4Δ0 for P < 0.05. (D) At the top is a diagram of the HIS3 reporter showing the regions assayed for Med3 association by ChIP. At the bottom left is a bar graph showing the ChIP results. The CCW12 promoter is a previously characterized site where Mediator is known to associate. Shown are the mean and standard error for six experiments (WT and mot3-1 grr1-1 sin4Δ0) or five experiments (sin4Δ0). T-tests indicated a significant difference between WT and mot3-1 grr1-1 sin4Δ0 at all primer sets for P < 0.05. Shown at the bottom right are the same data for three of the regions, plotted on a different scale to make the results easier to view.
Given the proposed role of Grr1 in controlling Med3 levels, we tested whether the levels of Mediator proteins may play a role in long-distance activation. To do this, we overexpressed MED2, MED3, and CDK8. Our results show that overexpression of MED3 in some of our polygenic mutants led to an increase in long-distance activation, based on the level of 3AT resistance (Figure 9B), while overexpression of MED2 and CDK8 had no detectable effect (data not shown). The overexpression of MED3 in wild-type and sin4Δ0 strains had no effect, suggesting that overexpression of MED3 enhances long-distance activation only in a grr1-1 mutant background.
If Med3 levels affect long-distance activation, we might also expect to see an increase in either Med3 levels or Med3 recruitment to the long-distance reporter in our mutants that are permissive for long-distance activation. To test these possibilities, we measured Med3 levels and performed ChIP of FLAG-Med3. By Western analysis, we did not detect any change in Med3 levels in a mutant that allows strong long-distance activation (Figure 9C, Figure S2). However, by ChIP, we measured a modest 1.5- to 2-fold increase in the recruitment of Med3 in the mot3-1 grr1-1 sin4Δ0 mutant (Figure 9D). This increase is not seen in the sin4Δ single mutant, suggesting that all three mutations are required for elevated Med3 recruitment. Additionally, we see a 1.5-fold increase of Med3 at the CCW12 UAS, a known Med3 binding site, suggesting Mediator recruitment may also be increased at other sites in the genome. These results suggest that the combination of mutations in the polygenic mutant results in an increased level of Mediator recruited to the UAS, which may be important in long-distance activation.
Discussion
In this study, we have isolated and analyzed yeast mutants that are capable of strong transcriptional activation over distances much greater than can occur in wild type. To identify these strains, we used a dual reporter system that allowed multiple rounds of selection and screening for an increased level of long-distance activation, resulting in the isolation of polygenic mutants. Importantly, these mutants do not generally increase all transcriptional activation; over the short distance that is typical of most yeast promoters, the mutants actually activate more poorly than wild type, suggesting they have become specifically permissive for long-distance activation. Although the different polygenic mutants acquired distinct sets of mutations, combining mutations from different lineages also allows strong long-distance activation, showing that specific mutant interactions are not necessary for this phenotype. Given the approximately additive contribution of each mutation, long-distance activation can be considered to be a quantitative trait. Our results suggest that many factors contribute to regulate long-distance activation in yeast where it is normally repressed, and possibly, in larger eukaryotes where long-distance activation normally occurs.
Our results have implicated the Mediator coactivator complex in the regulation of transcriptional activation distance and suggest that some of the mutations that we have isolated strengthen Mediator with respect to long-distance activation. Interestingly, human Mediator has been shown to be required for long-distance activation by enhancers in human embryonic stem cells (Kagey et al. 2010). Our current results, as well as our previous studies (Dobi and Winston 2007; K. C. Dobi, C. T. Reavey, F. Winston, unpublished results) show that the ability of Mediator to activate transcription over a long distance is normally repressed by the Mediator components Sin4, Rgr1, and Cdk8 and is dependent upon the Mediator components Med2 and Med3. Interestingly, previous results demonstrated a positive role for Cdk8 at the GAL1 UAS (Larschan and Winston 2005), suggesting that the role of Cdk8 may depend upon the distance between the GAL1 UAS and the target gene. Our results are also consistent with earlier results that showed positive roles for Med2, Med3, and Gal11/Med15 in activation by Gal4 (Piruat et al. 1997; Lee et al. 1999; Myers et al. 1999; Bryant and Ptashne 2003; Kuras et al. 2003; Reeves and Hahn 2005; Ansari and Morse 2012, 2013). Finally, previous results showed that the tail domain of Mediator (Med2, Med3, and Med15) can be recruited independently of the rest of Mediator in a sin4Δ mutant (Zhang et al. 2004; He et al. 2008; Ansari et al. 2009, 2012). Our results, showing a modestly increased level of Med3 recruitment have not distinguished between recruitment of the tail domain or the entire complex. Tests of additional Mediator components will shed light on the role of the rest of the complex in the regulation of activation distance.
Our isolation of a grr1 mutation also implicates Mediator based on recent evidence that Grr1 destabilizes Med3 in a Cdk8-dependent fashion (Gonzalez et al. 2014). However, in contrast to that study, we did not detect elevated levels of Med3 in a grr1Δ background. This difference may be because we measured Med3 levels with the MED3 gene in single copy, while Gonzalez et al. (2014) made their observations when MED3 was overexpressed from a plasmid. Since we failed to detect a change in Med3 levels, Grr1 may normally repress long-distance activation by controlling the level of other Mediator components or by a mechanism independent of Mediator.
The requirement for Mediator for long-distance activation in yeast suggests a possible mechanism for activation, as Mediator is required for the enhancer to loop to the core promoter in human embryonic stem cells (Kagey et al. 2010). To date, we have little evidence either for or against looping as a mechanism in yeast long-distance activation. Our previous results show that looping does occur at our long-distance reporter; however, it was detected in both wild-type and mutant strains (Dobi and Winston 2007). Possibly, looping is necessary but not sufficient for long-distance activation. Tracking, another proposed mechanism for long-distance activation by enhancers, is unlikely, as we previously showed that long-distance activation still occurs in the presence of a terminator between the UAS and core promoter (Dobi and Winston 2007). Clearly, additional experiments are required to elucidate the mechanism for the long-distance activation that occurs in our mutants, including tests for a role of cohesin, which interacts with Mediator to generate looping between enhancers and core promoters in human cells (Kagey et al. 2010).
The roles of the other factors identified by our mutations remain to be determined. Three of the factors, Mot3, Mit1, and Msn2, are site-specific DNA-binding transcriptional regulators. While the msn2 mutation clearly causes loss of Msn2 function, the mot3 and mit1 mutations are not loss-of-function mutations. Both the mot3-1 and the mit1-1 alleles are predicted to cause changes in their DNA binding domains, which could result in a novel function for either transcription factor, causing direct or indirect effects on transcriptional activation at the long-distance reporters. The isolation of the ptr3-1 mutation was surprising, as Ptr3 is known to be important for nutrient sensing and transport (Forsberg et al. 2001; Forsberg and Ljungdahl 2001). Our results suggest the possibility of additional functions for Ptr3. Conceivably, the ptr3-1 mutation or other mutations we identified could affect RNA stability instead of synthesis.
An important question is whether our polygenic mutants allow long-distance activation from endogenous UAS elements in the yeast genome. To begin to address this question, we measured mRNA levels in several of our mutants using microarrays after growth in three carbon sources: glucose, galactose, and raffinose (data not shown). Our results showed that all of the mutants have altered mRNA levels for a large number of genes, including both increased and decreased levels, and that the set of affected genes significantly overlaps between the three lineages of polygenic mutants (Reavey 2013) (Table S6). As our reporter uses the Gal4 activator, we specifically looked at the genes adjacent to the GAL genes for elevated levels in galactose-grown cells, but did not observe any increase. However, our results show that the number of adjacent gene pairs with increased mRNA levels is significantly greater than would be predicted if the effects were random (Table S7), suggesting possible cases of long-distance activation.
Our isolation and analysis of polygenic mutants that allow long-distance activation, as well as other studies that have taken a similar polygenic approach (for example, Gresham et al. 2008; Romano et al. 2010; Koschwanez et al. 2013), shows that this is a feasible genetic approach to study topics in yeast biology that might not otherwise be accessible. In our case, we were able to find a phenotype that is apparently not possible in single mutants. Furthermore, we identified genes relevant to the regulation of transcription activation distance that would not have been identified by limiting our studies to single mutants. Given the straightforward methods available to identify causative mutations in yeast, we believe that this type of approach will be useful for studying other aspects of gene regulation and yeast biology.
Supplementary Material
Acknowledgments
We thank Patrick Gibney, Burak Alver, and Peter Park for help with the microarray experiments and data analysis, and Dan Spatt for help with experiments. We also thank Selena Gell and Rajaraman Gopalakrishnan for helpful comments on the manuscript. This work was supported by National Institutes of Health (NIH) grant GM045720 to F.W. and by NIH grants GM046406 and GM071508 to D.B.
Footnotes
Communicating editor: M. Hampsey
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.181164/-/DC1.
Literature Cited
- Ansari S. A., Morse R. H., 2012. Selective role of Mediator tail module in the transcription of highly regulated genes in yeast. Transcription 3: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari S. A., Morse R. H., 2013. Mechanisms of Mediator complex action in transcriptional activation. Cell. Mol. Life Sci. 70: 2743–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari S. A., He Q., Morse R. H., 2009. Mediator complex association with constitutively transcribed genes in yeast. Proc. Natl. Acad. Sci. USA 106: 16734–16739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari S. A., Ganapathi M., Benschop J. J., Holstege F. C., Wade J. T., et al. , 2012. Distinct role of Mediator tail module in regulation of SAGA-dependent, TATA-containing genes in yeast. EMBO J. 31: 44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe H. L., Monks J., Wijgerde M., Fraser P., Proudfoot N. J., 1997. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 11: 2494–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G. et al. (Editors), 1991. Current Protocols in Molecular Biology, Greene Publishing Associates and Wiley-Interscience, New York. [Google Scholar]
- Ben-Ari G., Zenvirth D., Sherman A., David L., Klutstein M., et al. , 2006. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2: e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D. B., Gasch A. P., 2008. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell 19: 4580–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M. J., Christianson C. M., Pai D. A., Dunham M. J., 2006. Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics 173: 1813–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant G. O., Ptashne M., 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 11: 1301–1309. [DOI] [PubMed] [Google Scholar]
- Buecker C., Wysocka J., 2012. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 28: 276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., Groudine M., 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain C. W., Lohse M. B., Homann O. R., Sil A., Johnson A. D., 2012. A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics 190: 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M., Lin Y. S., Green M. R., Ptashne M., 1990. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature 345: 361–364. [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., et al. , 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- de Hoon M. J. L., Imoto S., Nolan J., Miyano S., 2004. Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., et al. , 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 8: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer A. M., Davis R. W., 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 37: 1333–1340. [DOI] [PubMed] [Google Scholar]
- Dobi K. C., Winston F., 2007. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 5575–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Gerke J. P., Kruglyak L., 2009. Genetic dissection of complex traits in yeast: insights from studies of gene expression and other phenotypes in the BYxRM cross. Cold Spring Harb. Symp. Quant. Biol. 74: 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erokhin M., Vassetzky Y., Georgiev P., Chetverina D., 2015. Eukaryotic enhancers: common features, regulation, and participation in diseases. Cell. Mol. Life Sci. 72: 2361–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg H., Ljungdahl P. O., 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21: 814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg H., Hammar M., Andreasson C., Moliner A., Ljungdahl P. O., 2001. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 158: 973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi M., Hochstrasser M., 2009. Small epitope-linker modules for PCR-based C-terminal tagging in Saccharomyces cerevisiae. Yeast 26: 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison, E., and G. Marth, 2012 Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907v2 [q-bio.GN].
- Gerke J. P., Chen C. T., Cohen B. A., 2006. Natural isolates of Saccharomyces cerevisiae display complex genetic variation in sporulation efficiency. Genetics 174: 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., et al. , 1996. Life with 6000 genes. Science 274: 546, 563–567. [DOI] [PubMed] [Google Scholar]
- Gonzalez D., Hamidi N., Del Sol R., Benschop J. J., Nancy T., et al. , 2014. Suppression of Mediator is regulated by Cdk8-dependent Grr1 turnover of the Med3 coactivator. Proc. Natl. Acad. Sci. USA 111: 2500–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D., Desai M. M., Tucker C. M., Jenq H. T., Pai D. A., et al. , 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin A. V., Rothenberg M., Downs M. A., Blumer K. J., 1998. Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signaling in Saccharomyces cerevisiae. Genetics 149: 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Hoar E., 1984. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the “TATA box”. Proc. Natl. Acad. Sci. USA 81: 7860–7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Battistella L., Morse R. H., 2008. Mediator requirement downstream of chromatin remodeling during transcriptional activation of CHA1 in yeast. J. Biol. Chem. 283: 5276–5286. [DOI] [PubMed] [Google Scholar]
- Helmlinger D., Marguerat S., Villen J., Gygi S. P., Bahler J., et al. , 2008. The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Genes Dev. 22: 3184–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Rolfs A., Bhullar B., Murthy T. V., Zhu C., et al. , 2007. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 17: 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C. J., Yang S. H., Xie Y., Zhang L., Johnston S. A., et al. , 2001. Evidence that Gal11 protein is a target of the Gal4 activation domain in the mediator. Biochemistry 40: 9421–9427. [DOI] [PubMed] [Google Scholar]
- Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., et al. , 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. K., Hemberg M., Gray J. M., Costa A. M., Bear D. M., et al. , 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S., Bohl D., Li C., Tuan D., 1997. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Mol. Cell. Biol. 17: 3955–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschwanez J. H., Foster K. R., Murray A. W., 2013. Improved use of a public good selects for the evolution of undifferentiated multicellularity. eLife 2: e00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansson E., Thorsen M., Tamas M. J., Nerman O., 2009. Evolutionary forces act on promoter length: identification of enriched cis-regulatory elements. Mol. Biol. Evol. 26: 1299–1307. [DOI] [PubMed] [Google Scholar]
- Krivega I., Dean A., 2012. Enhancer and promoter interactions-long distance calls. Curr. Opin. Genet. Dev. 22: 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L., Borggrefe T., Kornberg R. D., 2003. Association of the Mediator complex with enhancers of active genes. Proc. Natl. Acad. Sci. USA 100: 13887–13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E., Winston F., 2005. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol. Cell. Biol. 25: 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. C., Park J. M., Min S., Han S. J., Kim Y. J., 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19: 2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Louis E. J., 2012. Advances in quantitative trait analysis in yeast. PLoS Genet. 8: e1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M. B., Rosenberg O. S., Cox J. S., Stroud R. M., Finer-Moore J. S., et al. , 2014. Structure of a new DNA-binding domain which regulates pathogenesis in a wide variety of fungi. Proc. Natl. Acad. Sci. USA 111: 10404–10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., 2014. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 15: 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Stone E. A., Ayroles J. F., 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10: 565–577. [DOI] [PubMed] [Google Scholar]
- Madison J. M., Dudley A. M., Winston F., 1998. Identification and analysis of Mot3, a zinc finger protein that binds to the retrotransposon Ty long terminal repeat (delta) in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 1879–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak J. R., Dike S., Sementchenko V., Kapranov P., Biemar F., et al. , 2006. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 38: 1151–1158. [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac R. S., Silverman S. J., McClean M. N., Gibney P. A., Macinskas J., et al. , 2011. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell 22: 4447–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers L. C., Gustafsson C. M., Hayashibara K. C., Brown P. O., Kornberg R. D., 1999. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl. Acad. Sci. USA 96: 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. M., Kim H. S., Han S. J., Hwang M. S., Lee Y. C., et al. , 2000. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol. 20: 8709–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruat J. I., Chavez S., Aguilera A., 1997. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics 147: 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reavey, C., 2013 Analysis of transcriptional activation distance as a polygenic trait in Saccharomyces cerevisae, pp. 173 in Genetics. Ph.D. thesis, Harvard University. [Google Scholar]
- Reeves W. M., Hahn S., 2005. Targets of the Gal4 transcription activator in functional transcription complexes. Mol. Cell. Biol. 25: 9092–9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano G. H., Gurvich Y., Lavi O., Ulitsky I., Shamir R., et al. , 2010. Different sets of QTLs influence fitness variation in yeast. Mol. Syst. Biol. 6: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1990. Methods in Yeast Genetics 1990: A Laboratory Course Manaual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Saeed A. I., Sharov V., White J., Li J., Liang W., et al. , 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378. [DOI] [PubMed] [Google Scholar]
- Saeed A. I., Bhagabati N. K., Braisted J. C., Liang W., Sharov V., et al. , 2006. TM4 microarray software suite. Methods Enzymol. 411: 134–193. [DOI] [PubMed] [Google Scholar]
- Salinas F., Cubillos F. A., Soto D., Garcia V., Bergstrom A., et al. , 2012. The genetic basis of natural variation in oenological traits in Saccharomyces cerevisiae. PLoS One 7: e49640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., et al. , 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. [DOI] [PubMed] [Google Scholar]
- Struhl K., 1984. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc. Natl. Acad. Sci. USA 81: 7865–7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi N., Kruglyak L., 2011. Variants in SUP45 and TRM10 underlie natural variation in translation termination efficiency in Saccharomyces cerevisiae. PLoS Genet. 7: e1002211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger J. W., Schwartz K., Sherlock G., 2010. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 6: e1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, F., 2008 EMS and UV mutagenesis in yeast. Curr. Protoc. Mol. Biol. Chapter 13: Unit 13.3B. [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L., 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55. [DOI] [PubMed] [Google Scholar]
- Womack J. E., Jang H. J., Lee M. O., 2012. Genomics of complex traits. Ann. N. Y. Acad. Sci. 1271: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Sumibcay L., Hinnebusch A. G., Swanson M. J., 2004. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell. Biol. 24: 6871–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S1 describes the code used to analyze the sequencing results. Yeast strains are listed in Table S1 and are available upon request. Table S2 lists all oligonucleotide sequences used. Table S3 lists crosses used for bulk segregant analysis. Table S4 lists sequencing coverage. Table S5 lists causative mutant candidates. Table S6 lists microarray results. Table S7 describes enrichment for transcriptional effects on adjacent gene pairs. Figure S1 shows evidence for disomy in two mutants. Figure S2 presents Western analysis of Med3 levels. The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) (Edgar et al. 2002) and are accessible through GEO series accession no. GSE72093.