Abstract
Candida albicans is an opportunistic human fungal pathogen that causes a variety of diseases, ranging from superficial mucosal to life-threatening systemic infections, the latter particularly in patients with defects in innate immune function. C. albicans cells phagocytosed by macrophages undergo a dramatic change in their metabolism in which amino acids are a key nutrient. We have shown that amino acid catabolism allows the cell to neutralize the phagolysosome and initiate hyphal growth. We show here that members of the 10-gene ATO family, which are induced by phagocytosis or the presence of amino acids in an Stp2-dependent manner and encode putative acetate or ammonia transporters, are important effectors of this pH change in vitro and in macrophages. When grown with amino acids as the sole carbon source, the deletion of ATO5 or the expression of a dominant-negative ATO1G53D allele results in a delay in alkalinization, a defect in hyphal formation, and a reduction in the amount of ammonia released from the cell. These strains also form fewer hyphae after phagocytosis, have a reduced ability to escape macrophages, and reside in more acidic phagolysosomal compartments than wild-type cells. Furthermore, overexpression of many of the 10 ATO genes accelerates ammonia release, and an ato5Δ ATO1G53D double mutant strain has additive alkalinization and ammonia release defects. Taken together, these results indicate that the Ato protein family is a key mediator of the metabolic changes that allow C. albicans to overcome the macrophage innate immunity barrier.
INTRODUCTION
Candida albicans is an opportunistic pathogen that colonizes the skin and gastrointestinal and genitourinary tracts of most healthy individuals but also causes a range of diseases, from nonlethal mucosal infections, such as oral thrush and vaginitis, to disseminated hematogenous candidiasis, the latter in immunocompromised individuals (1–3). As the fourth most prevalent cause of hospital-acquired infection, disseminated candidiasis is very difficult to treat, prolongs hospitalization, and has a mortality rate of ∼40% (4–6). The high mortality rates and large health care burden associated with C. albicans infection highlight the importance of understanding the physiology, virulence factors, and host-pathogen interactions of C. albicans.
The healthy immune system is able to effectively prevent systemic candidiasis; however, advances in health care have increased the population of individuals surviving despite immune dysfunctions. Conditions that predispose individuals to disseminated candidiasis include hematological malignancies, genetic immune disorders, HIV/AIDS, and iatrogenic interventions, including organ transplantation, chemotherapy, and invasive procedures (3, 7). The interaction between the innate immune system and C. albicans is a primary determinant of disease progression, as those with innate immune defects are more susceptible to serious infection (8). Macrophages, along with other professional phagocytes, are key components of the innate immune response to C. albicans (8–11). Mice in which macrophage function has been depleted are more susceptible to mucosal and disseminated candidiasis (8, 11). In counterpoint, C. albicans has evolved mechanisms to escape phagocytosis and killing by the macrophage, including differentiation into a filamentous hyphal form that facilitates escape and dissemination. This interaction is highly dynamic and is critical for C. albicans virulence and, therefore, remains the subject of intense study (12, 13).
The morphological switch alone does not fully define the response of C. albicans, as many studies have revealed large-scale transcriptional reprogramming and proteomic shifts that take place upon phagocytosis, including a shift away from glucose metabolism and toward alternative carbon assimilation (14–17). The importance of metabolic adaptation is underscored by the decreased virulence in animal models of C. albicans strains that are defective in the glyoxylate cycle and in β-oxidation (18–21). Furthermore, carbon source utilization plays a significant role in cell wall composition, stress susceptibility, and phagocyte recognition (22, 23).
Our laboratory has demonstrated that C. albicans grown in vitro in medium that mimics the nutrients predicted to be available after phagocytosis rapidly alkalinizes the extracellular environment (24). This process depends upon the utilization of amino acids as a carbon source and leads to the extrusion of ammonia from the cell, which is derived from amino acid catabolism. This secretion of ammonia counteracts the acidification of the phagolysosome, providing a neutral pH signal that induces C. albicans to undergo hyphal morphogenesis, thus facilitating escape from the macrophage (25). Furthermore, a mutant lacking the transcription factor Stp2p, which regulates amino acid permeases, fails to alkalinize both in vitro and in the phagolysosome and cannot germinate to escape the macrophage unless the phagosome is neutralized by chemical means (24, 25).
The transcript profiles of cells during alkalinization and following phagocytosis share significant similarities, including the induction of multiple genes of the ATO family, which at 10 members is greatly expanded in C. albicans relative to its size in other fungi. Though named ATO, for ammonia transport outward (YaaH in bacteria), the molecular function of these plasma membrane proteins is unknown, and there is evidence linking them to transport of acetate in Saccharomyces cerevisiae, Aspergillus nidulans, and Yarrowia lipolytica, as well as release of ammonia in S. cerevisiae (24, 26–30). Despite the potential for genetic redundancy in this large family, deletion of ATO5 alone retards alkalinization in vitro (24). This phenotype and the induction following phagocytosis suggest that the Ato proteins modulate the interaction of C. albicans with macrophages.
We report here that two alkalinization-defective strains, an ato5Δ deletion mutant and a strain expressing an ATO1G53D allele originally identified as a dominant-negative mutation that confers acetate sensitivity in Y. lipolytica (29), are impaired in several aspects of the macrophage-fungus interaction. These mutations delay alkalinization in vitro, leading to a defect in hyphal formation and a reduction in the amount of ammonia released from the cell. Furthermore, both the ato5Δ and ATO1G53D strain reside in more acidic phagolysosomes than do wild-type cells, and as a result, they form fewer hyphae after phagocytosis and have a reduced ability to escape macrophages. Both ATO genes are transcriptionally regulated by Stp2, and a double ato5Δ ATO1G53D mutant phenocopies the stp2Δ strain. Finally, overexpression of multiple ATO genes accelerates alkalinization, indicating that this gene family is an important mediator of the host-Candida interaction.
MATERIALS AND METHODS
Strains and growth media.
C. albicans strains were grown under standard conditions in YPD medium (1% yeast extract, 2% peptone, 2% dextrose). For growth on plates, 2% agar was added to the medium. To select for nourseothricin-resistant (NouR) transformants, 200 μg/ml of nourseothricin (Werner Bioagents, Jena, Germany) was added to the YPD agar plates (31). Alkalinization experiments were performed in glucose-free minimal yeast nitrogen base (YNB) medium with allantoin as the nitrogen source (0.17% yeast nitrogen base, 0.5% allantoin) supplemented with 2% Casamino acids as the sole carbon source (YAC medium). Ammonia release was measured on solid medium of the same formulation containing 2% agar. NH3 release by alkalinizing colonies was measured using an acid trap as previously described (24) on YAC plates or GM-BCP medium (27) as described above with the addition of 2% agar.
The strains used are listed in Table 1. C. albicans strains lacking ATO5 were generated using the SAT-flipper method as described previously (31). Briefly, 300 bp of homologous sequence immediately 5′ or 3′ from the ATO5 open reading frame were amplified by PCR and cloned between the KpnI/XhoI and SacI/SacII sites of pSFS1. The resulting SAT1-FLP cassette was used to transform C. albicans strain SC5314 by electroporation with selection on YPD-Nou plates. Cassette integration was confirmed in the selected candidates via PCR. To remove the nourseothricin selection marker, the mutant strain was induced to excise the deletion cassette with 1% bovine serum albumin (BSA) in YNB medium for 3 days and the NouS colonies were selected. This process was repeated to generate the independently derived homozygous deletion mutants HDC27 and HDC28 (ato5Δ::FRT/ato5Δ::FRT). Complementation of the mutant strain used plasmid pHD-9, which was generated by cloning the ATO5 open reading frame with 700 bp of 5′ untranslated region (UTR) between the MluI and XhoI sites of pAG6, a SAT1-marked version of CIp10 (25). This plasmid was linearized with StuI and used to transform SC5314 or ato5Δ mutant cells to generate the strains HDC33 (ATO5/ATO5 RPS10/rps10:CIp10-SAT1) and HDC29 and HDC30 (ato5Δ::FRT/ato5Δ::FRT RPS10/rps10::CIp10-ATO5-SAT1), respectively.
TABLE 1.
C. albicans strains used
| Strain | Relevant genotype | Complete genotypea | Reference |
|---|---|---|---|
| SC5314 | Wild type | Prototroph | 73 |
| SVC17 | stp2Δ | stp2Δ::FRT/stp2Δ::FRT | 25 |
| HDC27 | ato5Δ | ato5Δ::FRT/ato5Δ::FRT | This study |
| HDC30 | ato5Δ+ATO5 | ato5Δ::FRT/ato5Δ::FRT RPS10/rps10::ATO5-CIp10-SAT1 | This study |
| HDC33 | Wild type SAT1 | RPS10/rps10::CIp10-SAT1 | This study |
| HDC31 | ato5Δ SAT1 | ato5Δ::FRT/ato5Δ::FRT RPS10/rps10::CIp10-SAT1 | This study |
| Can572 | ATO1G53D | ura3/ura3 RPS10/rps10::CIp10-ACT1p-ATO1G53D | 24 |
| THE1 | Wild type TetR | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 | 34 |
| HDC48 | Wild type TetR | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 RPS10/rps10::CIp10-tetR | This study |
| HDC44 | tet-ATO1 | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 RPS10/rps10::CIp10-tetR-ATO1 | This study |
| HDC38 | tet-ATO1G53D | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 RPS10/rps10::CIp10-tetR-ATO1G53D | This study |
| HDC39 | tet-ATO2 | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 RPS10/rps10::CIp10-tetR-ATO2 | This study |
| HDC40 | tet-ATO3 | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 RPS10/rps10::CIp10-tetR-ATO3 | This study |
| HDC41 | tet-ATO4 | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 RPS10/rps10::CIp10-tetR-ATO4 | This study |
| HDC42 | tet-ATO5 | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 RPS10/rps10::CIp10-tetR-ATO5 | This study |
| HDC43 | tet-ATO10 | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 RPS10/rps10::CIp10-tetR-ATO10 | This study |
| HDC45 | ato5Δ tet-CIp10 | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 ato5Δ::FRT/ato5Δ::FRT RPS10/rps10::CIp10-tetR | This study |
| HDC49 | ato5Δ tet-ATO1G53D | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 ato5Δ::FRT/ato5Δ::FRT RPS10/rps10::CIp10-tetR-ATO1G53D | This study |
FRT, FLP recombination target; HA, hemagglutinin.
Constitutive expression of the ATO1G53D allele was achieved as previously described (24). Briefly, ∼1,000 bp of the ACT1 promoter from pAU34 (32) was subcloned between the KpnI and XhoI sites in CIp10 to generate pHZ116. Then, the ATO1G53D mutation was generated by site-directed overlap PCR using complementary oligonucleotides with a single mismatch to encode a change of Gly-53 to Asp, analogous to the Y. lipolytica GPR1-1 mutant originally identified by Augstein et al. (33), and cloned into pHZ116 to generate pML341. The plasmid was digested with StuI and used to transform CAI4-F2 to uridine prototrophy. Accurate integration at the RPS10 locus was verified by PCR.
The doxycycline-repressible strains were constructed in the THE1 strain background that was generated by Nakayama (34). Briefly, the doxycycline-repressible promoter from plasmid p97-CAU1 (34) was amplified and cloned between the KpnI and XhoI restriction sites in CIp10 (35) to generate the plasmid pHD72. Subsequently, the ATO genes were PCR amplified and cloned between the XhoI and MluI restriction sites in pHD72. Plasmids were sequence verified, digested with StuI, and used to transform THE1 to uridine prototrophy. Accurate integration at the RPS10 locus was verified by PCR.
Alkalinization and ammonia release assays.
Alkalinization experiments were performed as previously described (24, 25), using YAC at pH 4.0, as described above. C. albicans cells were grown in YPD medium overnight and diluted to an optical density at 600 nm (OD600) of 0.2 in the alkalinization medium. Cells were incubated at 37°C with aeration for up to 24 h. Growth was measured via optical density at 600 nm, culture pH was measured using a standard pH electrode, and cellular morphology was scored by analyzing photomicrographs of at least 150 cells per condition. Experiments were performed at least in triplicate, and the data were analyzed using Prism 5.0 (GraphPad) software.
Ammonia release by C. albicans cells during alkalinization was assessed by using acid traps as previously described (24). In brief, cells were grown in YPD medium overnight, washed in distilled water, and resuspended at an OD600 of 1.0 in distilled water. Cells were spotted onto solid YAC medium at pH 4.0, and reservoirs containing 10% citric acid were affixed to the petri dish lid directly underneath the colonies. Cells were incubated at 37°C, and samples from the acid trap collected at 24, 48, or 72 h after initiation of the experiment. Ammonia was quantified using Nessler's reagent, as described previously (24, 36). Experiments were performed in triplicate.
Macrophage cytotoxicity assay.
C. albicans toxicity on macrophages was assessed by using the CytoTox96 nonradioactive cytotoxicity assay (Promega) as previously described (25). Briefly, RAW264.7 macrophages were seeded at 2.5 × 105 cells per well of a 96-well plate in phenol red-free RPMI medium and incubated overnight at 37°C and 5% CO2. C. albicans cells were grown to log phase in YPD medium, washed in phosphate-buffered saline (PBS), and cocultured with macrophages at a 3:1 ratio for 5 h. The release of lactate dehydrogenase (LDH) by infected macrophages relative to maximum LDH release from lysed macrophages was then calculated according to the manufacturer's protocol and corrected for spontaneous release of LDH by the macrophages or C. albicans alone. The experiment was performed in triplicate.
Hyphal formation of phagocytosed C. albicans.
To assess the interaction of single C. albicans cells with the macrophages, we seeded 2.5 × 105 RAW264.7 macrophages to glass coverslips in a 12-well plate and incubated them overnight at 37°C and 5% CO2. C. albicans cells were grown in YPD medium overnight, diluted 1:100 in fresh medium, and grown for 3 h at 30°C. Cells were then washed in distilled water and stained with 1 μM 5-carboxytetramethylrhodamine (Molecular Probes) for 15 min, washed 2 times with PBS, and resuspended in RPMI medium (HyClone). Amounts of 3 × 106 C. albicans cells were cocultured with the macrophages at 37°C for 2 h. The cocultures were then washed twice with PBS, and images of the Candida-macrophage interaction were taken using an Olympus IX81 automated inverted microscope. Images from 100 phagocytosed cells per experiment were analyzed using SlideBook 6.0 software. The percentage of hyphal morphogenesis during phagocytosis was calculated by obtaining the percentage of phagocytosed cells using the following formula: (germ tubes + hyphal cells/total amount of cells) × 100. Experiments were performed in triplicate.
Endpoint dilution assay.
C. albicans survival during interaction with the RAW264.7 macrophages was assessed as previously described (25, 37). Briefly, macrophages were seeded at 2.5 × 104 cells/well in 96-well plates and grown overnight at 37°C with 5% CO2. C. albicans cells were grown to log phase and then washed in distilled water and resuspended in fresh RPMI medium. Amounts of 1 × 104 cells/well were added to wells with or without macrophages, followed by six serial 1:5 dilutions. After 48 h, microcolonies of C. albicans in wells in which individual colonies could be distinguished were counted using an inverted microscope. The results are presented as 100 times the ratio of the number of colonies in the presence of macrophages to the number of colonies without macrophages. The experiment was performed in triplicate.
LR assay.
Assays were performed as previously reported by Vylkova and Lorenz (25). RAW264.7 macrophages were seeded onto glass coverslips in 12-well tissue culture plates at 5 × 105 cells/ml and allowed to adhere overnight 37°C in 5% CO2. Next, 1 mM LysoTracker red (LR) DM99 (Molecular Probes) was added to fresh RPMI medium and the mixture incubated for 2 h. C. albicans cells were grown overnight in YPD, diluted 1:100 in fresh YPD, and grown for 3 h at 30°C. Cells were then washed in distilled water, stained with 1 μM fluorescein isothiocyanate (FITC) for 15 min, and washed in PBS to remove excess dye. Control cells were heat killed by incubation for 60 min at 65°C. Cells were diluted to 1 × 106 cells/ml in phenol red-free RPMI medium and cocultured with macrophages for 60 min. Cultures were stained with Calcofluor white (35 μg/ml for 30 s) to label nonphagocytosed cells and fixed in 2.7% paraformaldehyde. The cocultures were then imaged at ×60 magnification. To estimate the relative phagosomal pH, the signal intensities of both FITC and tetramethyl rhodamine isocyanate (TRITC) were plotted along a line drawn transversely across the short axis of the cell for at least 50 cells per condition using Slidebook 6.0. The average LR signal intensity was calculated for a region of 10 pixels (1 μm) immediately outside the fungal cell, whose boundary was determined by the slope of the FITC signal.
In vivo virulence assay.
A murine model of disseminated C. albicans infection was performed as described in reference 19. C. albicans cells were grown to mid-log phase in YPD, washed, and resuspended in PBS at 5 × 106 cells/ml. Ten female ICR mice (weighing 21 to 25 g) per strain were inoculated via tail vein injection with 100 μl PBS containing 106 C. albicans cells. Mice were monitored at least twice daily and euthanized when moribund. All mouse experiments were performed under protocols approved by the Animal Welfare Committee of the University of Texas Health Science Center at Houston (protocol HSC-AWC-12-099).
RESULTS
The ability of C. albicans to cause invasive infections depends on its dynamic and complex interactions with cells of the innate immune system, such as macrophages (12, 38, 39). Strikingly, phagocytosed C. albicans cells form hyphae that facilitate their escape, and we have shown that this is induced by a fungus-driven neutralization of the phagolysosome (24, 25). This process is dependent on catabolism of amino acids and is regulated by Stp2p, a transcription factor that activates amino acid permeases (24, 25). We sought to identify whether additional components required for efficient alkalinization also alter interactions with macrophages.
Ato function is required for efficient environmental alkalinization response in C. albicans.
We previously reported that a mutant lacking ATO5 has a defect in alkalinization in medium 199, a low-glucose tissue culture medium (24). This mutant was generated using the UAU method, which uses two auxotrophic markers and selects for a chromosomal homozygosis and rearrangement (40), which may be problematic given the importance of amino acids in this phenomenon. Therefore, we constructed an ato5Δ mutant in the prototrophic SC5314 strain using the SAT-flipper methodology (31). We have subsequently defined a glucose-free minimal medium, YNB, containing 0.5% allantoin as the nitrogen source and 2% Casamino acids as the sole carbon source (YAC), which supports more robust alkalinization (24, 25), and tested both the new ato5Δ mutant and a point mutant in ATO1 (ATO1G53D) that has dominant-negative phenotypes in other systems (33).
These strains were incubated in YAC starting at pH 4.0, where all strains grew at similar rates (Fig. 1A). A rapid increase in the culture pH was observed when the wild-type strain (SC5314) was incubated in aerated culture at 37°C (Fig. 1B), with the pH rising from 4.0 to 6.9 in 8 h, while the increases in pH for the ato5Δ and the ATO1G53D mutants were significantly retarded (pH 5.19 and 5.25, respectively) in the same time frame (Fig. 1B). This lag was overcome by the 24-h time point, when all cultures had a pH near neutral. The SC5314-derived strains are more robust than those we used previously, but the magnitude of the ato5Δ defect relative to the phenotype of the control strains is similar in both backgrounds (data not shown).
FIG 1.
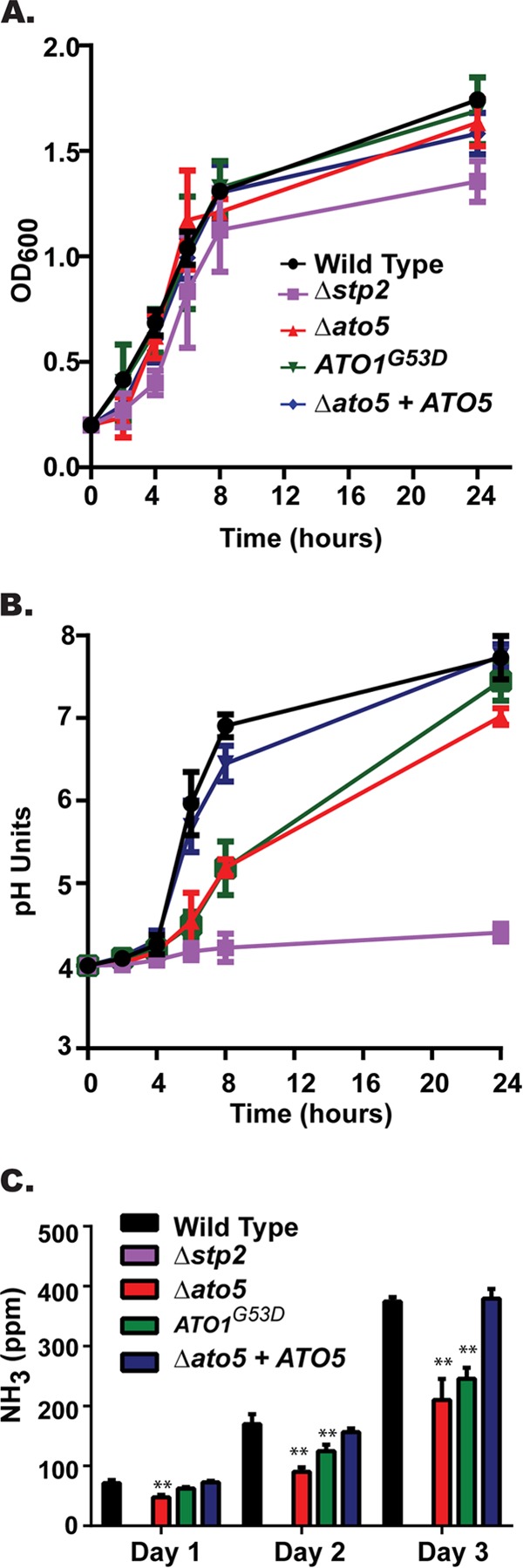
Ato proteins promote environmental alkalinization. The wild-type (SC5314), stp2Δ (SVC17), ato5Δ (HDC17), ATO1G53D (Can572), and ato5Δ+ATO5 (HDC30) strains were incubated in YAC medium initially at pH 4.0 under aerated conditions at 37°C. (A) Growth of the cells was measured by OD600 readings at the indicated time points. (B) pH of the cultures from the same experiments whose results are shown in panel A. (C) Ammonia released by C. albicans cells was collected in a citric acid trap and quantified using Nessler's reagent as described in Materials and Methods. Results are reported as mean values ± SD from triplicate assays. **, P < 0.001.
We have attributed this environmental alkalinization to the extrusion of ammonia from the cell (24), and we hypothesized that the ATO proteins, which have been proposed to facilitate ammonia export (26, 27), may be important effectors of this release. To test this hypothesis, we performed an ammonia release assay in which colonies were allowed to develop on solid defined alkalinization medium (YAC, pH 4) directly apposed across an air interface from an acid trap containing 10% citric acid. Ammonia excreted from the colony is converted to ammonium in the acid trap, where it can be quantified using Nessler's reagent (24, 41). Detectible ammonia from wild-type (SC5314) and reconstituted ato5Δ+ATO5 cells increased significantly over the 72-h period (Fig. 1C). In contrast, the levels of ammonia released from both the ato5Δ and ATO1G53D cells were significantly reduced (Fig. 1C). Ammonia excretion correlated with the degree of alkalinization, with the stp2Δ mutant completely deficient, the ato mutants intermediate, and the wild-type and complemented strains releasing abundant ammonia.
Our initial formulation of the minimal Casamino acid medium included ammonium sulfate as the nitrogen source, as is typical in defined yeast media, which we realized may affect our ammonia release results. Indeed, we found that at neutral pH, the presence of ammonium sulfate significantly increased the amount of ammonia present in the trap even in the absence of cells, while no ammonia was released from acidic medium (see Fig. S1 in the supplemental material). This may lead to a feedback loop in which ammonia generated by cellular metabolism raises the pH, which in turn liberates ammonia from the medium, thus overstating the contribution of the cells. To avoid this, we tested additional nitrogen sources and found that no ammonia was released from cell-free medium containing allantoin, urea, or amino acids as the nitrogen source, regardless of pH (see Fig. S1; also data not shown). Allantoin supported optimal growth, and all the assays reported here used allantoin as the nitrogen source. The amount of ammonia released was slightly lower on allantoin than on ammonium sulfate, but the temporal pattern and genetic phenotypes were similar to those we have previously reported and no differences in the rates of pH changes were observed (see Fig. S1; also data not shown).
Mutation of Ato proteins affects autoinduction of hyphal formation.
The ability of C. albicans to undergo a reversible morphological switch from yeast to hyphal form has been shown to be critical to virulence of the organism (42, 43). Neutral pH is a key factor that induces this switch, and we have shown that efficient environmental alkalinization is sufficient to promote this morphological switch (24). Therefore, we hypothesized that the ATO mutant strains would have a reduced ability to form hyphae due to the defect in alkalinization. The ato5Δ and ATO1G53D strains were grown in unbuffered alkalinization medium (YAC, pH 4.0) over a period of 6 h and were significantly impaired in hyphal formation compared to that of the wild-type (SC5314) or complemented strains (Fig. 2A). However, when these strains were cultured in the same medium adjusted to pH 7 for 2 h, no defects in hyphal formation were detected (see Fig. S2 in the supplemental material), indicating that the mutants were able to respond to neutral pH cues but could not autoinduce hyphal growth by changing the pH.
FIG 2.
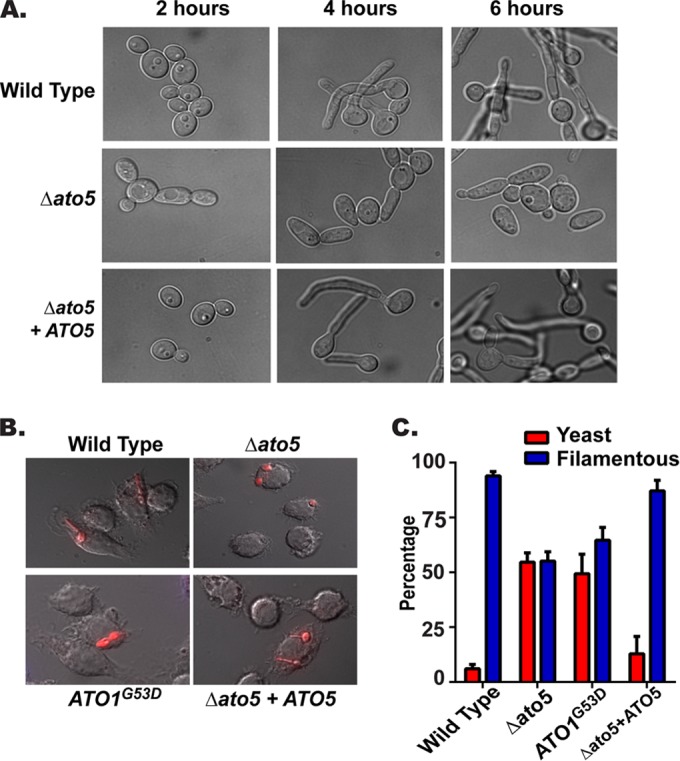
Mutation of Ato proteins reduces autoinduction of hyphal formation. The wild-type (SC5314), stp2Δ (SVC17), ato5Δ (HDC17), ATO1G53D (Can572), and ato5Δ+ATO5 (ATO5 complemented) strains were assayed for hyphal formation. (A) Strains were grown in YAC medium for 6 h and photographed at ×100 magnification. (B) Cells were labeled with 5-carboxytetramethylrhodamine, cocultured for 2 h with RAW267.4 macrophages, fixed, and photographed at ×60 magnification. (C) Filamentous cells were quantitated in the captured images by counting at least 150 cells per condition. Results are reported as mean values ± SD from triplicate experiments.
We have shown that C. albicans blocks the normal acidification of the phagolysosome and that, as in vitro, the resulting neutral pH induces hyphal growth; in contrast, an stp2Δ mutant does not neutralize this compartment and as a result does not germinate postphagocytosis (25). We hypothesized that the ato mutants would also have a defect in the autoinduction of hyphal formation inside the macrophage. In order to test this, strains were cocultured with RAW264.7 macrophages for 2 h, fixed, and assessed for hyphal formation microscopically (Fig. 2B). As expected, the ATO mutant strains showed a significant reduction in hyphal formation (∼50%), while the wild-type (SC5314) and reconstituted strain were nearly all hyphal (94% and 87%, respectively) (Fig. 3C). Taken together, these results confirm that the ability to efficiently alkalinize the phagolysosome is an important signal for hyphal formation and that the Ato proteins are important effectors of this signaling.
FIG 3.
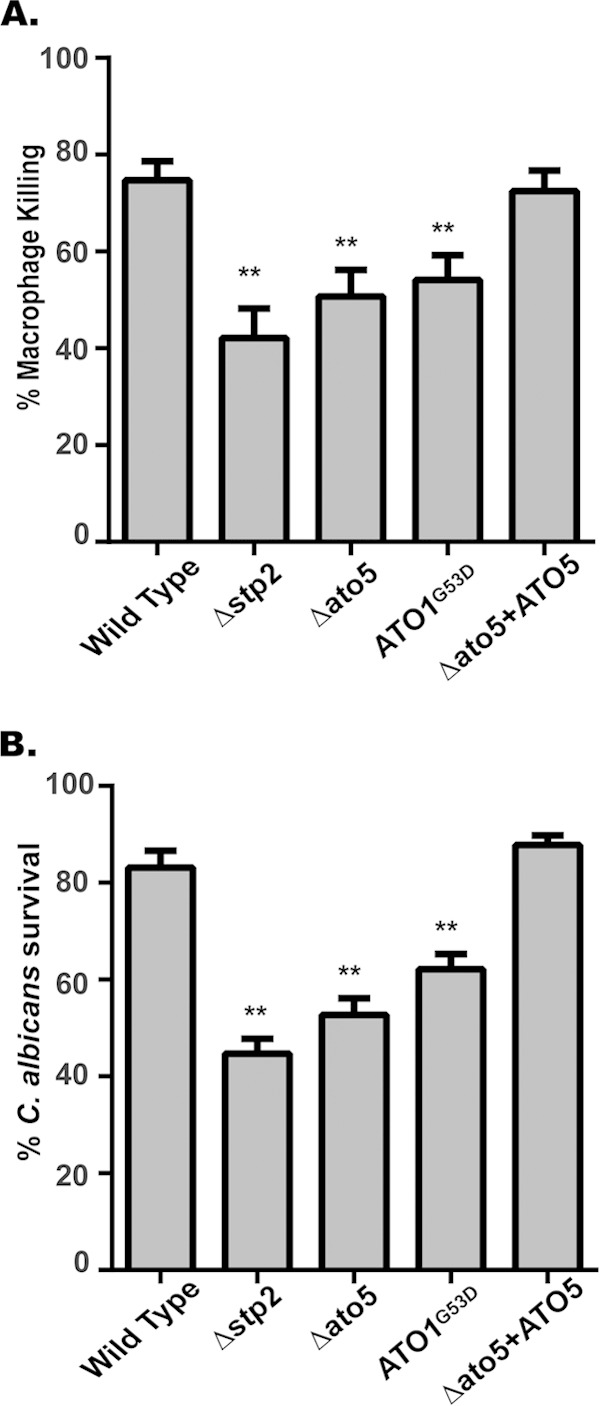
Ato proteins are required for efficient lysis of macrophages and proliferation after phagocytosis. (A) The wild-type (SC5314), stp2Δ (SVC17), ato5Δ (HDC17), ATO1G53D (Can572), and ato5Δ+ATO5 (ATO5 complemented) strains were cocultured with RAW264.7 macrophages. Macrophage death was assessed based upon lactate dehydrogenase (LDH) release. (B) Strains were cocultured in the presence and absence of RAW264.7 macrophages for 24 h. Fungal survival was calculated as the ratio of microcolonies in the presence versus the absence of macrophages. Results are reported as mean values ± SD from triplicate experiments. **, P < 0.001.
Ato proteins are required for efficient macrophage lysis and escape.
We have shown that the ability to alkalinize the phagosome significantly contributes to the ability of C. albicans to escape the macrophage (25). As a result of the alkalinization and hyphal formation defects, we predicted that the ATO mutant strains would be impaired in the ability to pierce the macrophage and escape. In order to assess this, we utilized a lactate dehydrogenase (LDH) release assay to assess macrophage membrane damage after 5 h of coculture with RAW267.4 macrophages (25). Coculture of macrophages with wild-type (SC5314) and ATO5-complemented strains resulted in ∼75% maximal LDH release (relative to the results for chemically lysed macrophages). In contrast, the ato5Δ and ATO1G53D mutants were less able to damage macrophages, with only 50% and 52% of the maximal LDH release, while the stp2Δ strain released only 42% of the maximum (Fig. 3A). These data suggest that functional Ato proteins are necessary for efficient escape from the phagosome.
A reduced ability to escape the macrophage might predict that the ATO mutant strains are more susceptible to macrophage killing. To address this, we utilized an established endpoint dilution assay to assess C. albicans survival after phagocytosis (37). In good agreement with the LDH release assay, significantly reduced survival was seen in the ato5Δ (52%) and ATO1G53D (62%) strains, as well as the stp2Δ control strain (45%); in contrast, more than 80% of the wild-type cells survived this interaction (Fig. 3B). Thus, we conclude that a defect in macrophage escape also leads to an increased ability of the macrophage to clear the pathogen.
Ato5p is necessary for efficient alkalinization of the phagosome.
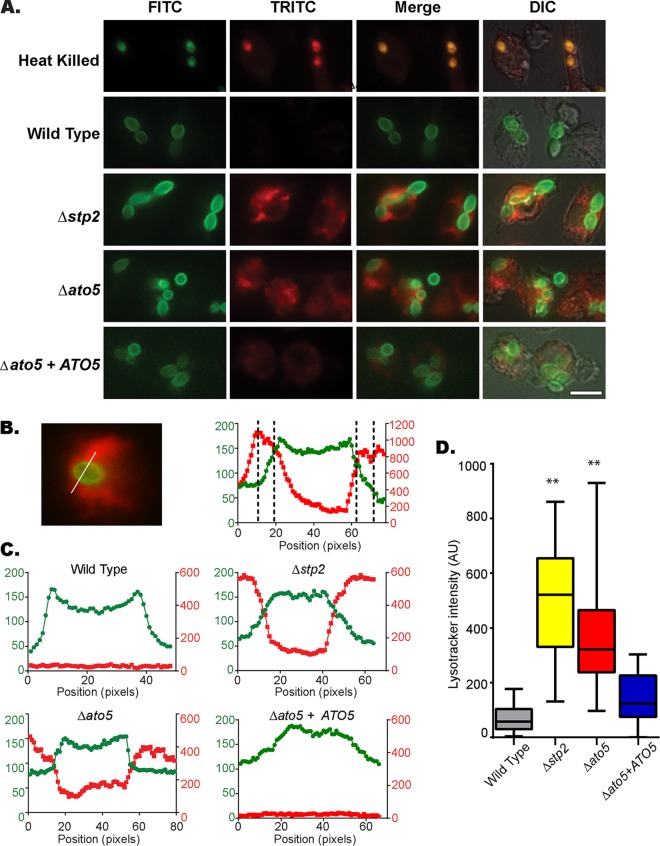
Taken together, the phenotypes of the ato mutant strains strongly suggest that they may have a defect in neutralization of the phagolysosome. To test our hypothesis, we preloaded RAW267.4 macrophages with the acidophilic dye LysoTracker red (LR), which accumulates and fluoresces in acidic organelles, and then cocultured these cells with FITC-labeled C. albicans cells. As expected, heat-killed cells strongly colocalized with LR after 60 min of coculture (Fig. 4A). Conversely, wild-type (SC5314) cells were surrounded by a low-level diffuse LR signal, suggesting a more neutral pH (Fig. 4A). stp2Δ and ato5Δ cells were both frequently contained in acidic compartments (Fig. 4A). To estimate the difference in the pH of phagosomes containing wild-type versus ato5Δ or stp2Δ cells, we utilized the Slidebook 6 image software to quantitate the signal intensity in the FITC and LR channels along a line drawn through the middle of the C. albicans cell on the short axis (Fig. 4B). Plotting the FITC fluorescence intensity clearly delineated the fungal cell (Fig. 4B and C, green). The phagolysosomal membrane is tightly apposed to the fungal cell wall, so LR was confined to the lumen in a narrow band immediately outside the cell (notably, in the heat-killed controls, LR accumulated both in the phagosome lumen and in the permeabilized fungal cell), as seen by the sharp rise in LR intensity as the FITC signal decreased (Fig. 4B and C). This lumenal fluorescence was absent from wild-type and complemented cells but was readily apparent in stp2Δ and ato5Δ mutants (Fig. 4C). We quantitated the average LR intensity over 10 pixels (1 μm) on each side of the fungal cell (indicated by dashed lines in Fig. 4B). Both the stp2Δ and the ato5Δ mutant resided in more acidic compartments, as indicated by the LysoTracker red signal being stronger than those of the wild-type and complemented strains (Fig. 4D). From these results, we conclude that ato mutants occupy an acidic phagolysosome, indicating that the in vitro alkalinization defect we have described extends to the phagocyte as well.
FIG 4.
Ato5p is necessary for neutralization of the macrophage phagosome. (A) FITC-stained C. albicans cells were cocultured with RAW264.7 macrophages preloaded with LysoTracker red (LR) for 1 h. The cocultures were then fixed and imaged at ×60 magnification. DIC, differential interference contrast. (B) Image analysis of the cocultures shown in panel A was performed using Slidebook 6.0 software. Left, fluorescence intensity for both the FITC and LR channels was plotted along a line drawn through the middle of the C. albicans cell on the short axis. Right, dashed lines indicate the regions adjacent to the fungal cell used to quantitate the LR signal (10 pixels = 1 μm). (C) Representative plots from each strain. The FITC signal is plotted on the left axis in green, and the TRITC signal is plotted on the right axis in red. (D) Box (25% to 75%)-and-whisker (minimum to maximum) plot of the average TRITC intensity. At least 50 cells were counted per strain. All assays were performed in triplicate. **, P value < 0.001. AU, arbitrary units.
Loss of only ATO5 does not compromise virulence.
The results of the macrophage coculture experiments indicate that ATO mutants are less able to tolerate phagocytosis and suggest that this may be an important determinant for virulence of C. albicans. We had previously shown that the deletion of STP2 resulted in a modest but significant attenuation of virulence in this model (25), so we tested whether this was true of an ATO5 deletion strain by using the standard mouse tail-vein model of disseminated hematogenous candidiasis (see Fig. S3 in the supplemental material) and found no statistically significant attenuation in virulence. This result may be explained by the modest alkalinization phenotype and the potential for functional redundancy with other Ato proteins, whose expression may compensate for the loss of ATO5 in vivo.
Expression of ATO genes is dependent upon the transcription factor STP2.
Stp2p is a transcription factor that regulates amino acid permeases (25, 44). Given the similar phenotypes of the stp2Δ and ato5Δ mutants, we asked whether Stp2p had any role in the regulation of the ATO genes by using quantitative real-time PCR to assess the transcript abundance of ATO1 and ATO5 in wild-type and stp2Δ strains under alkalinizing conditions. ATO1, which is highly induced following phagocytosis and in YAC (14, 24), was upregulated 279-fold in wild-type cells compared to its induction under nonalkalinizing conditions. This induction was almost completely abolished in cells lacking STP2 (Fig. 5A). Similarly, ATO5 was upregulated 7.5-fold in alkalinizing wild-type cells (Fig. 5B) but only 2-fold in the stp2Δ mutant. Taken together, these results indicate that the ATO genes are regulated by STP2.
FIG 5.
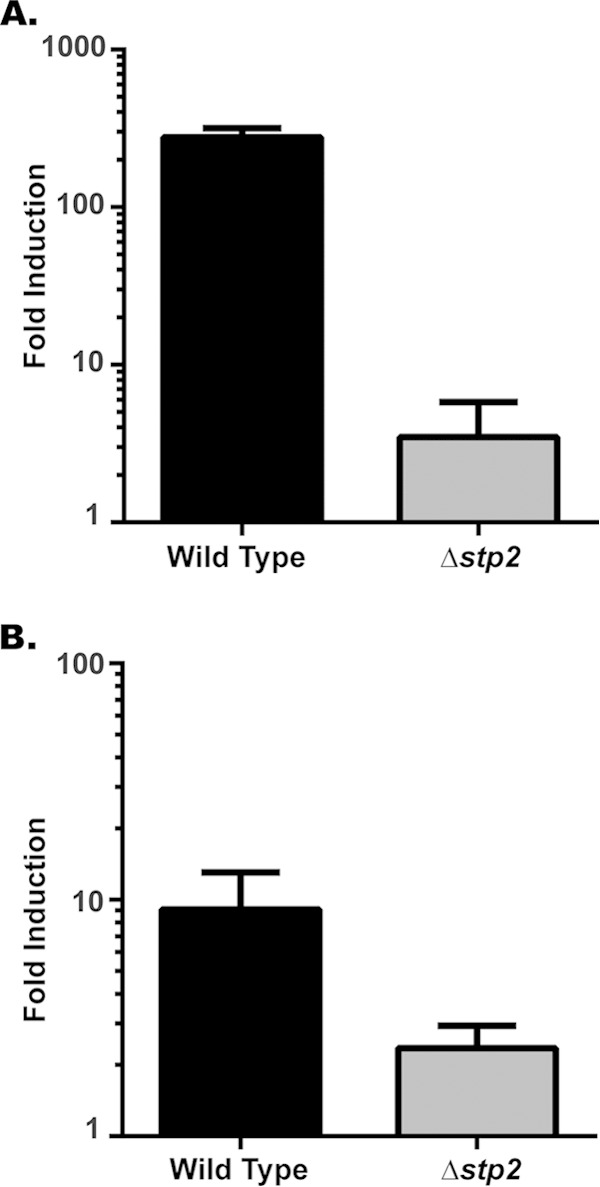
ATO gene expression is dependent upon STP2. Quantitative real-time PCR analysis of cells grown in alkalinization medium. Cells of both the wild-type (SC5314) and stp2Δ (SVC17) strains were grown in YAC (alkalinizing conditions) or the same medium with glucose added (nonalkalinizing conditions), and the transcript levels of ATO1 (A) and ATO5 (B) expression were determined. Transcript abundance was normalized to that of ACT1, and the expression in glucose was set to 1. Data are expressed as mean values ± SD from triplicate experiments.
Many ATO genes affect C. albicans alkalinization.
The Stp2-dependence of ATO gene expression raised the question of whether heterologous ATO expression might suppress stp2Δ mutant phenotypes. To test this, we generated ATO alleles under the control of the constitutive ACT1 promoter. Surprisingly, stp2Δ strains expressing these alleles failed to grow in YAC medium, although growth was unaffected in medium containing glucose (see Fig. S4 in the supplemental material). This suggests that the dysregulation conferred by deletion of STP2 cannot be suppressed (and might be exacerbated) by overexpression of target genes, perhaps indicative of a careful stoichiometry between Atos and other cellular proteins. To address this, we constructed strains with ATO genes under the control of a doxycycline (Dox)-repressible promoter. Overexpression of many ATO genes accelerated alkalinization of the environment compared to the rate of alkalinization for the wild-type control (Fig. 6A), while these strains also released more ammonia (Fig. 6B and C). Conversely, overexpression of ATO3 or the dominant-negative ATO1G53D inhibited alkalinization and ammonia release. These data indicate that the ATO gene family is broadly involved in the ability of C. albicans to alkalinize the extracellular space but that there might be specific interactions between Ato and other proteins that regulate this phenomenon.
FIG 6.
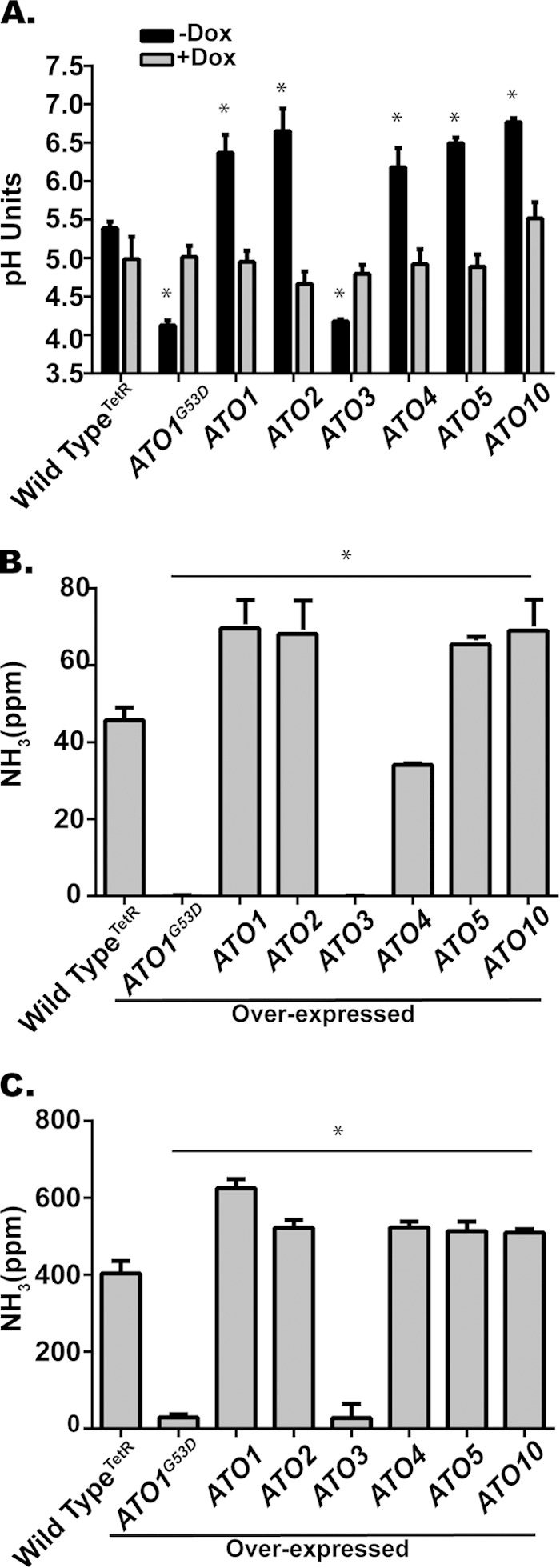
ATO overexpression alters alkalinization. (A) The wild type (THE1) and strains with ATO genes under the control of a doxycycline (Dox)-repressible promoter were incubated in YAC initially at pH 4.0 under aerated conditions at 37°C with and without Dox. (A) The pH of the cultures after 8 h is shown. The results for all overexpression strains (−Dox) are statistically different from the results for the wild-type control (P < 0.001). (B to C) Ammonia released by C. albicans cells during alkalinization on solid medium in the absence of Dox after 24 h (B) or 72 h (C). Data are expressed as mean values ± SD from triplicate experiments.
Double ATO mutation results in additive alkalinization defects.
Mutations in ATO5 (ato5Δ) and ATO1 (ATO1G53D) result in strikingly similar phenotypes, and we sought to determine whether a double mutant strain would confer a synthetic phenotype. Indeed, the in vitro alkalinization of the double mutant was drastically inhibited compared to that of either single mutant (Fig. 7), despite near-normal growth under these conditions (data not shown). This phenotype is very similar to that of an stp2Δ strain (Fig. 7); consistent with this, the double mutant also fails to release ammonia (data not shown). These results provide strong evidence that multiple Ato proteins can facilitate ammonia release, although further studies will be necessary to determine the molecular mechanism through which this process occurs.
FIG 7.
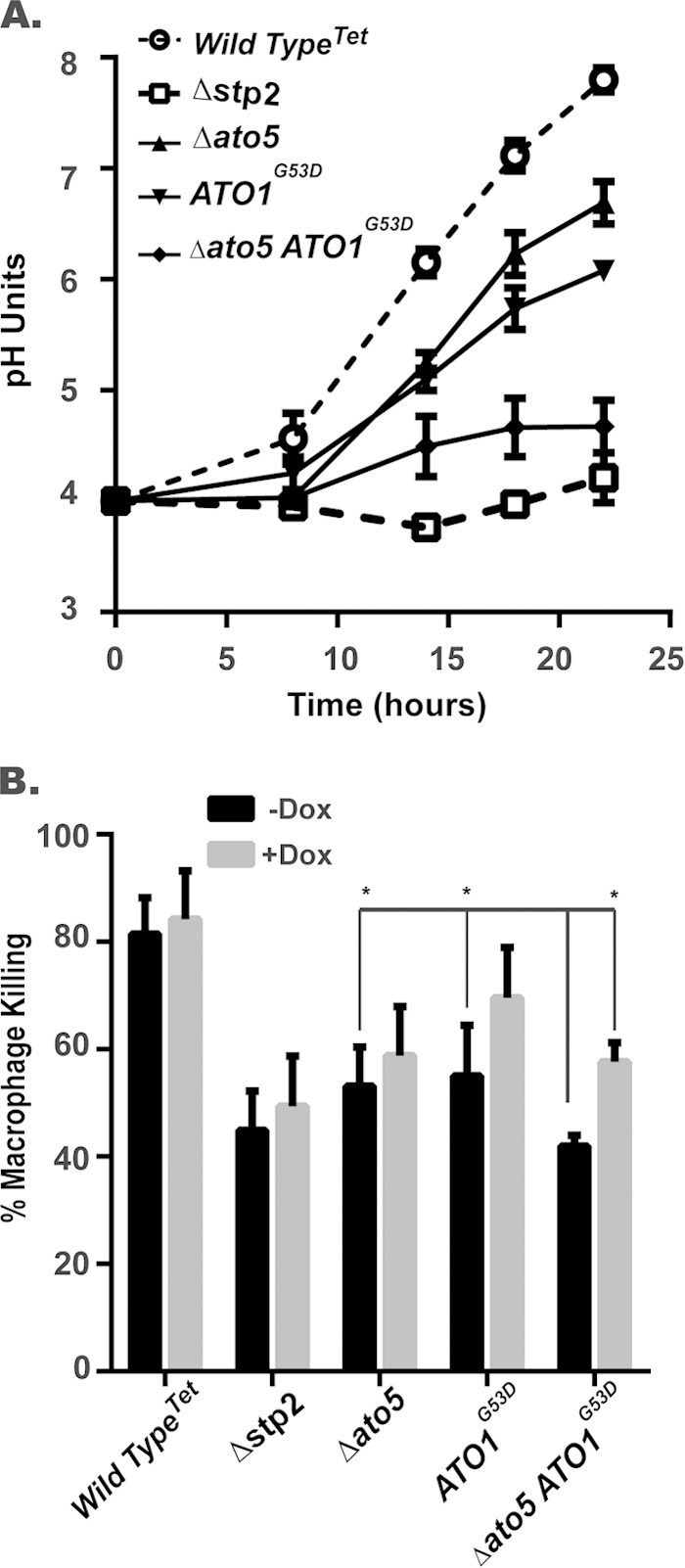
Double ato mutation results in additive alkalinization defects. (A) Wild-type (SC5314), stp2Δ (SVC17), ato5ΔTet (HDC45), and ato5Δ ATO1G53D (HDC49) strains were incubated in YAC with an initial pH of 4.0 under aerated conditions at 37°C without Dox. The pH of the cultures was determined at the indicated time points. (B) Indicated strains were cocultured with RAW264.7 macrophages in the presence and absence of 50 μg/ml of Dox in RPMI tissue culture medium. Macrophage death was assessed based upon lactate dehydrogenase (LDH) release. An asterisk indicates a significant difference (P < 0.01) relative to the result for the double ATO1G53D ato5Δ strain grown in the absence of Dox.
DISCUSSION
We show here that members of the ATO gene family are important mediators of the ability of C. albicans to neutralize its surrounding pH, both in vitro and in the macrophage phagolysosome. This conclusion is supported by evidence that mutants with single mutations of ATO1 (ATO1G53D) or ATO5 (ato5Δ) raise the extracellular pH more slowly during in vitro growth under amino acid-rich conditions, release less ammonia, and are slower to germinate. In contact with macrophages, ato mutants occupy an acidic phagolysosome, which reduces hyphal growth and fungal survival while increasing macrophage integrity. While it is reasonable to infer that the defect in germination is directly responsible for the reduction in macrophage damage, recent publications attribute some of this damage to fungus-dependent induction of pyroptosis (45, 46). It is possible that the aberrant maturation of the C. albicans-containing phagosome resulting from the failure to acidify might reduce pyroptosis and, thus, also contribute to improved macrophage survival. Indeed, we have evidence that this is the case (S. Vylkova, H. Danhof, and M. Lorenz, unpublished observations). The phenotypes of a double ATO1G53D ato5Δ mutant are additive and are similar to those of the previously reported stp2Δ strain (24, 25). ATO1 and ATO5 are both transcriptionally regulated by Stp2p, though this could be direct or indirect. Furthermore, overexpression of multiple ATO genes enhances in vitro alkalinization, suggesting that there is a broad role for this family in the pH alteration phenomenon.
Acquisition and utilization of available nutrients from the host is fundamental to the survival and pathogenicity of microorganisms, and the host uses nutrient deprivation and sequestration as a defense mechanism to limit microbial growth. Pathogens have to identify ready sources of nutrients in a host, and it is clear that some are scarce, such as nucleotides and iron, since mutants impaired in the synthesis or uptake of these compounds are avirulent in many pathogens (47–49). In contrast, amino acids appear abundant, since many (but not all) auxotrophic mutants retain full virulence (4, 50, 51). We and others have shown that pathways needed to assimilate amino acids, fatty acids, and other alternative carbon sources are required for virulence (19, 52, 53). We have shown that C. albicans efficiently catabolizes amino acids to satisfy carbon requirements and that this results in the neutralization of acidic environments, which has important effects on host-pathogen interactions.
Transcript profiling established that many ATO genes are significantly upregulated during environmental alkalinization and that an overlapping but not identical set is also upregulated during phagocytosis (14, 24). We show here that the induction of at least some ATO genes is largely or entirely Stp2p dependent, emphasizing the central role of this transcription factor in regulating the metabolic changes that support the fitness of C. albicans in contact with phagocytic cells. The dramatic expansion of this gene family strongly suggests that differential functions and/or regulation exists between them, given the potential for redundancy. Indeed, we found this to be the case, as overexpression analysis showed that many but not all ATO genes promote environmental alkalinization under the conditions tested.
We attempted to suppress the phenotypes of the stp2Δ mutant by constitutively expressing individual ATO genes. We were surprised to find that these overexpression strains failed to grow on medium in which amino acids were the sole carbon source, though they were viable when glucose was present, which suppressed alkalinization. There is evidence for homomultimeric and heteromultimeric interactions between Ato homologs in yeast (54, 55), and our data would support the idea that these proteins form one or more functional complexes. In the stp2Δ mutant, then, either the correct stoichiometry is not maintained or a key non-ATO target of Stp2p is missing. Elucidating the details of the Ato molecular machine will require further study.
Disruption of Ato function compromises interactions with macrophages but does not attenuate virulence in our mouse model. It stands to reason that pathogens would expand gene families that mediate specific interactions with the host, but this has long been very difficult to demonstrate experimentally because of technical limitations in knocking out multigene families. C. albicans has 10 secreted aspartyl proteases (SAPs) with different expression profiles and pH optima, but conclusive evidence of a role in pathogenesis has been elusive (56–58). In C. glabrata, a compelling virulence phenotype is not observed until one deletes many members of a cell wall-associated protease family (yapsins) (59). Als3, one of a family of eight adhesins, is required in vitro for adhesion, cadherin binding, and iron uptake but is not required for animal virulence (60–62). Thus, we expect that functional redundancy explains the lack of in vivo effects and that a more ambitious deletion effort may be necessary; new tools, such as the recent adaptation of the Cas9/CRISPR technology (63), may facilitate this in the future.
The CUG clade of fungi includes the most clinically relevant fungal species, C. albicans, C. tropicalis, and C. parapsilosis, which are predicted to have expanded ATO families, and there is a positive correlation between the number of ATO homologs and the robustness of alkalinization (24, 64). The correlation does not hold with C. glabrata, which is not a CUG species and has only three ATO genes but has been shown to alkalinize its environment in vitro in the presence of amino acids, as well as in the phagolysosome (65). The only evidence for a connection between Ato function and ammonia export or pH modulation outside our work in C. albicans is in S. cerevisiae, in which the rudimentary ability of this species to alter extracellular pH is further degraded by deletion of one of the three ATO homologs (26, 27). Similar to our findings presented here, the S. cerevisiae ATO3 is regulated by the SPS plasma membrane amino acid sensor, the downstream target of which is Stp2p (66).
In contrast, there is significantly more support for a role for the Ato protein family in acetate transport. The dominant-negative ATO1G53D mutation we use here was originally identified in Y. lipolytica, in which it confers sensitivity to acetic acid, as do similar mutations in S. cerevisiae (29, 30). Ato proteins are required for active import of acetate in yeast and Aspergillus nidulans (28, 67), and the E. coli SatP/YaaH is a succinate/acetate transporter (68). Reconciling the biochemical and physiological functions of these proteins will clearly require further study.
All pathogens must identify sources of nutrients before they can elaborate specific virulence attributes within the host; in C. albicans, it has become clear that nonfermentable compounds are a key carbon source in vitro and that at least some of the pathways required to metabolize fatty acids, amino acids, lactate, acetate, and others are required for full virulence (14, 19, 20, 69, 70). Moreover, the available carbon source regulates morphogenesis, cell wall structure and stress responses (22, 71, 72). Catabolism of amino acids, which appear to be readily available in many host niches, also strongly affects the interaction of C. albicans with the host, by allowing the pathogen to modulate the surrounding pH. We demonstrate here that this phenomenon requires the Ato protein family, whose members warrant further study as key mediators of the host-pathogen interaction.
Supplementary Material
ACKNOWLEDGMENTS
We thank other members of the Lorenz laboratory, particularly S. Vylkova, for advice and helpful discussions.
This work was supported by award R01AI075091 (to M.C.L.) and award T32AI055449-09 (to H.A.D.) from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00984-15.
REFERENCES
- 1.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfaller MA, Diekema DJ. 2012. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J Clin Microbiol 50:2846–2856. doi: 10.1128/JCM.00937-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 4.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream Infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 5.Beck-Sagué CM, Jarvis WR. 1993. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980-1990. National Nosocomial Infections Surveillance System J Infect Dis 167:1247–1251. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein MP, Towns ML, Quartey SM, Mirrett S, Reimer LG, Parmigiani G, Reller LB. 1997. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin Infect Dis 24:584–602. doi: 10.1093/clind/24.4.584. [DOI] [PubMed] [Google Scholar]
- 7.Hajjeh RA, Sofair AN, Harrison LH, Lyon GM, Arthington-Skaggs BA, Mirza SA, Phelan M, Morgan J, Lee-Yang W, Ciblak MA, Benjamin LE, Thomson Sanza L, Huie S, Yeo SF, Brandt ME, Warnock DW. 2004. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J Clin Microbiol 42:1519–1527. doi: 10.1128/JCM.42.4.1519-1527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen J, Warner T, Balish E. 1994. The role of phagocytic cells in resistance to disseminated candidiasis in granulocytopenic mice. J Infect Dis 170:900–905. doi: 10.1093/infdis/170.4.900. [DOI] [PubMed] [Google Scholar]
- 9.Romani L. 2000. Innate and adaptive immunity in Candida albicans infections and saprophytism. J Leukocyte Biol 68:175–179. [PubMed] [Google Scholar]
- 10.Shoham S, Levitz SM. 2005. The immune response to fungal infections. Br J Haematol 129:569–582. doi: 10.1111/j.1365-2141.2005.05397.x. [DOI] [PubMed] [Google Scholar]
- 11.Qian Q, Jutila MA, Van Rooijen N, Cutler JE. 1994. Elimination of mouse splenic macrophages correlates with increased susceptibility to experimental disseminated candidiasis. J Immunol 152:5000–5008. [PubMed] [Google Scholar]
- 12.Jiménez-López C, Lorenz MC. 2013. Fungal immune evasion in a model host-pathogen interaction: Candida albicans versus macrophages. PLoS Pathog 9:e1003741. doi: 10.1371/journal.ppat.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seider K, Heyken A, Lüttich A, Miramón P, Hube B. 2010. Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr Opin Microbiol 13:392–400. doi: 10.1016/j.mib.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin-Bejerano I, Abeijon C, Magnelli P, Grisafi P, Fink GR. 2007. Phagocytosis by human neutrophils is stimulated by a unique fungal cell wall component. Cell Host Microbe 2:55–67. doi: 10.1016/j.chom.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández-Arenas E, Cabezón V, Bermejo C, Arroyo J, Nombela C, Diez-Orejas R, Gil C. 2007. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol Cell Proteomics 6:460–478. doi: 10.1074/mcp.M600210-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, Hube B. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol 56:397–415. doi: 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- 18.Lorenz MC, Fink GR. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- 19.Ramírez MA, Lorenz MC. 2007. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot Cell 6:280–290. doi: 10.1128/EC.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barelle CJ, Priest CL, MacCallum DM, Gow NAR, Odds FC, Brown AJP. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol 8:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piekarska K, Mol E, van den Berg M, Hardy G, van den Burg J, van Roermund C, MacCallum D, Odds F, Distel B. 2006. Peroxisomal fatty acid β-oxidation is not essential for virulence of Candida albicans. Eukaryot Cell 5:1847–1856. doi: 10.1128/EC.00093-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ene IV, Adya AK, Wehmeier S, Brand AC, MacCallum DM, Gow NAR, Brown AJP. 2012. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol 14:1319–1335. doi: 10.1111/j.1462-5822.2012.01813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ene IV, Cheng S-C, Netea MG, Brown AJP. 2013. Growth of Candida albicans cells on the physiologically relevant carbon source lactate affects their recognition and phagocytosis by immune cells. Infect Immun 81:238–248. doi: 10.1128/IAI.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. 2011. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio 2:e00055-11. doi: 10.1128/mBio.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vylkova S, Lorenz MC. 2014. Modulation of phagosomal pH by Candida albicans promotes hyphal morphogenesis and requires Stp2p, a regulator of amino acid transport. PLoS Pathog 10:e1003995. doi: 10.1371/journal.ppat.1003995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palkova Z, Janderova B, Gabriel J, Zikanova B, Pospisek M, Forstova J. 1997. Ammonia mediates communication between yeast colonies. Nature 390:532–536. doi: 10.1038/37398. [DOI] [PubMed] [Google Scholar]
- 27.Palkova Z, Devaux F, Ricicova M, Minarikova L, Le Crom S, Jacq C. 2002. Ammonia pulses and metabolic oscillations guide yeast colony development. Mol Biol Cell 13:3901–3914. doi: 10.1091/mbc.E01-12-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paiva S, Devaux F, Barbosa S, Jacq C, Casal M. 2004. Ady2p is essential for the acetate permease activity in the yeast Saccharomyces cerevisiae. Yeast 21:201–210. doi: 10.1002/yea.1056. [DOI] [PubMed] [Google Scholar]
- 29.Tzschoppe K, Augstein A, Bauer R, Kohlwein SD, Barth G. 1999. trans-dominant mutations in the GPR1 gene cause high sensitivity to acetic acid and ethanol in the yeast Yarrowia lipolytica. Yeast 15:1645–1656. doi:. [DOI] [PubMed] [Google Scholar]
- 30.Gentsch M, Kuschel M, Schlegel S, Barth G. 2007. Mutations at different sites in members of the Gpr1/Fun34/YaaH protein family cause hypersensitivity to acetic acid in Saccharomyces cerevisiae as well as in Yarrowia lipolytica. FEMS Yeast Res 7:380–390. doi: 10.1111/j.1567-1364.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 31.Reuss O, Vik A, Kolter R, Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 32.Uhl MA, Johnson AD. 2001. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology 147:1189–1195. doi: 10.1099/00221287-147-5-1189. [DOI] [PubMed] [Google Scholar]
- 33.Augstein A, Barth K, Gentsch M, Kohlwein SD, Barth G. 2003. Characterization, localization and functional analysis of Gpr1p, a protein affecting sensitivity to acetic acid in the yeast Yarrowia lipolytica. Microbiology 149:589–600. doi: 10.1099/mic.0.25917-0. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama H, Mio T, Nagahashi S, Kokado M, Arisawa M, Aoki Y. 2000. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect Immun 68:6712–6719. doi: 10.1128/IAI.68.12.6712-6719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325–327. doi:. [DOI] [PubMed] [Google Scholar]
- 36.Butler G, Rasmussen MD, Lin MF, Santos MAS, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJP, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PWJ, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KAT, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MPH, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NAR, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocha CRC, Schröppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell 12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netea MG, Brown GD, Kullberg BJ, Gow NAR. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol 6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert AS, Wheeler RT, May RC. 2014. Fungal pathogens: survival and replication within macrophages. Cold Spring Harb Perspect Med 5:a019661. doi: 10.1101/cshperspect.a019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enloe B, Diamond A, Mitchell AP. 2000. A single-transformation gene function test in diploid Candida albicans. J Bacteriol 182:5730–5736. doi: 10.1128/JB.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gori K, Mortensen HD, Arneborg N, Jespersen L. 2007. Ammonia production and its possible role as a mediator of communication for Debaryomyces hansenii and other cheese-relevant yeast species. J Dairy Sci 90:5032–5041. doi: 10.3168/jds.2006-750. [DOI] [PubMed] [Google Scholar]
- 42.Lo H-J, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. doi: 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
- 43.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez P, Ljungdahl PO. 2005. Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol Cell Biol 25:9435–9446. doi: 10.1128/MCB.25.21.9435-9446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, Cowen LE. 2015. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun 6:6741. doi: 10.1038/ncomms7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wellington M, Koselny K, Sutterwala FS, Krysan DJ. 2014. Candida albicans triggers NLRP3-mediated pyroptosis in macrophages. Eukaryot Cell 13:329–340. doi: 10.1128/EC.00336-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleck CB, Schöbel F, Brock M. 2011. Nutrient acquisition by pathogenic fungi: nutrient availability, pathway regulation, and differences in substrate utilization. Int J Med Microbiol 301:400–407. doi: 10.1016/j.ijmm.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Ene IV, Brunke S, Brown AJP, Hube B. 2014. Metabolism in Fungal pathogenesis. Cold Spring Harb Perspect Med 4:a019695. doi: 10.1101/cshperspect.a019695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vylkova SL, Lorenz MC. 2012. Encounters with mammalian cells: survival strategies of Candida species, p 261–282. In Calderone RA. (ed), Candida and candidiasis, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 50.Kirsch DR, Whitney RR. 1991. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect Immun 59:3297–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brand A, MacCallum DM, Brown AJP, Gow NAR, Odds FC. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell 3:900–909. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cole MF, Bowen WH, Zhao X-J, Cihlar RL. 1995. Avirulence of Candida albicans auxotrophic mutants in a rat model of oropharyngeal candidiasis. FEMS Microbiology Lett 126:177–180. doi: 10.1111/j.1574-6968.1995.tb07413.x. [DOI] [PubMed] [Google Scholar]
- 54.Řičicová M, Kučerová H, Váchová L, Palková Z. 2007. Association of putative ammonium exporters Ato with detergent-resistant compartments of plasma membrane during yeast colony development: pH affects Ato1p localisation in patches. Biochim Biophys Acta 1768:1170–1178. doi: 10.1016/j.bbamem.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 55.Strachotová D, Holoubek A, Kučerová H, Benda A, Humpolíčková J, Váchová L, Palková Z. 2012. Ato protein interactions in yeast plasma membrane revealed by fluorescence lifetime imaging (FLIM). Biochim Biophys Acta 1818:2126–2134. doi: 10.1016/j.bbamem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lermann U, Morschhäuser J. 2008. Secreted aspartic proteases are not required for invasion of reconstituted human epithelia by Candida albicans. Microbiology 154:3281–3295. doi: 10.1099/mic.0.2008/022525-0. [DOI] [PubMed] [Google Scholar]
- 58.Correia A, Lermann U, Teixeira L, Cerca F, Botelho S, Gil da Costa RM, Sampaio P, Gärtner F, Morschhäuser J, Vilanova M, Pais C. 2010. Limited role of secreted aspartyl proteinases Sap1 to Sap6 in Candida albicans virulence and host immune response in murine hematogenously disseminated candidiasis. Infect Immun 78:4839–4849. doi: 10.1128/IAI.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaur R, Ma B, Cormack BP. 2007. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci U S A 104:7628–7633. doi: 10.1073/pnas.0611195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cleary IA, Reinhard SM, Miller CL, Murdoch C, Thornhill MH, Lazzell AL, Monteagudo C, Thomas DP, Saville SP. 2011. Candida albicans adhesin Als3p is dispensable for virulence in the mouse model of disseminated candidiasis. Microbiology 157:1806–1815. doi: 10.1099/mic.0.046326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE Jr, Filler SG, Hube B. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog 4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE Jr, Filler SG. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol 5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vyas VK, Barrasa MI, Fink GR. 2015. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv 1:e1500248. doi: 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Priest SJ, Lorenz MC. 2015. Characterization of virulence-related phenotypes in Candida species of the CUG clade. Eukaryot Cell 14:931–940. doi: 10.1128/EC.00062-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kasper L, Seider K, Gerwien F, Allert S, Brunke S, Schwarzmüller T, Ames L, Zubiria-Barrera C, Mansour MK, Becken U, Barz D, Vyas JM, Reiling N, Haas A, Haynes K, Kuchler K, Hube B. 2014. Identification of Candida glabrata genes involved in pH modulation and modification of the phagosomal environment in macrophages. PLoS One 9:e96015. doi: 10.1371/journal.pone.0096015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guaragnella N, Butow RA. 2003. ATO3 encoding a putative outward ammonium transporter is an RTG-independent retrograde responsive gene regulated by GCN4 and the Ssy1-Ptr3-Ssy5 amino acid sensor system. J Biol Chem 278:45882–45887. doi: 10.1074/jbc.M309301200. [DOI] [PubMed] [Google Scholar]
- 67.Robellet X, Flipphi M, Pégot S, MacCabe AP, Vélot C. 2008. AcpA, a member of the GPR1/FUN34/YaaH membrane protein family, is essential for acetate permease activity in the hyphal fungus Aspergillus nidulans. Biochem J 412:485. doi: 10.1042/BJ20080124. [DOI] [PubMed] [Google Scholar]
- 68.Sá-Pessoa J, Paiva S, Ribas D, Silva IJ, Viegas SC, Arraiano CM, Casal M. 2013. SATP (YaaH), a succinate-acetate transporter protein in Escherichia coli. Biochem J 454:585–595. doi: 10.1042/BJ20130412. [DOI] [PubMed] [Google Scholar]
- 69.Ramírez MA, Lorenz MC. 2009. The transcription factor homolog CTF1 regulates β-oxidation in Candida albicans. Eukaryot Cell 8:1604–1614. doi: 10.1128/EC.00206-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Piekarska K, Hardy G, Mol E, van den Burg J, Strijbis K, van Roermund C, van den Berg M, Distel B. 2008. The activity of the glyoxylate cycle in peroxisomes of Candida albicans depends on a functional β-oxidation pathway: evidence for reduced metabolite transport across the peroxisomal membrane. Microbiology 154:3061–3072. doi: 10.1099/mic.0.2008/020289-0. [DOI] [PubMed] [Google Scholar]
- 71.Ene IV, Heilmann CJ, Sorgo AG, Walker LA, de Koster CG, Munro CA, Klis FM, Brown AJP. 2012. Carbon source-induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candida albicans. Proteomics 12:3164–3179. doi: 10.1002/pmic.201200228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simonetti N, Strippoli V, Cassone A. 1974. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature 250:344. doi: 10.1038/250344a0. [DOI] [PubMed] [Google Scholar]
- 73.Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.