Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by complex interplay among immune cell types. SLE activity is experimentally assessed by several blood tests, including gene expression profiling of heterogeneous populations of cells in peripheral blood. To better understand the contribution of different cell types in SLE pathogenesis, we applied the two methods in cell-type-specific differential expression analysis, csSAM and DSection, to identify cell-type-specific gene expression differences in heterogeneous gene expression measures obtained using RNA-seq technology. We identified B-cell-, monocyte-, and neutrophil-specific gene expression differences. Immunoglobulin-coding gene expression was altered in B-cells, while a ribosomal signature was prominent in monocytes. On the contrary, genes differentially expressed in the heterogeneous mixture of cells did not show any functional enrichment. Our results identify antigen binding and structural constituents of ribosomes as functions altered by B-cell- and monocyte-specific gene expression differences, respectively. Finally, these results position both csSAM and DSection methods as viable techniques for cell-type-specific differential expression analysis, which may help uncover pathogenic, cell-type-specific processes in SLE.
Keywords: cell-type-specific, deconvolution, RNA-seq, SLE, csSAM, DSection
Introduction
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease characterized by multiorgan involvement and tissue damage.1 Although SLE is traditionally recognized as a B-cell-driven disease manifested by production of autoantibodies,2 contributions of T-cells and other immune cell types have also been described in SLE.3 A better understanding of the contributions of various immune cell types to SLE pathogenesis may aid in the development of better therapeutic regimens targeting cell-type-specific molecular mechanisms of SLE.1
Sequencing technologies are rapidly establishing their niche in clinical practice.4 The microarray revolution led to the adoption of RNA-seq as a useful tool for assessing an individual patient’s gene expression profile.5,6 However, clinical samples are often derived from peripheral blood or other heterogeneous tissues containing mixed populations of cells. Such heterogeneity makes it difficult to compare gene expression profiles between samples. Therefore, gene expression analysis requires methods that account for cell-type heterogeneity.7–9
Statistical methods developed with the advent of microarray technologies allow extraction of cell-type-specific expression profiles from a gene expression matrix obtained from heterogeneous tissues.10 The methods for estimating cell-type-specific gene expression generally use one of three measures: (1) sample-specific cell proportions7,9; (2) gene expression profiles thought to uniquely define cell types, called cell signatures3,11; or (3) inherent properties of heterogeneous gene expression mixtures,10 reviewed in Ref. 8 While the second and third approaches are highly dependent on the prerequisites, the use of the sample-specific cell proportions together with the sample-specific gene expression data is a viable approach for the cell-type-specific differential expression analysis, as both measures are biologically linked. The use of cell proportions is especially applicable to clinical practice, where complete blood cell counts are routinely collected and can readily be combined with the same patient’s heterogeneous gene expression data obtained from peripheral blood using microarray or RNA-seq.
One of the goals of cell-type-specific gene expression analysis is to be able to identify cell-type-specific gene expression differences in heterogeneous gene expression measures. Among other applications, this ability could assist in diagnosis or help monitor responses to treatment.3 To date, two methods have been developed for cell-type-specific differential gene expression analysis of microarrays using cell proportions: csSAM7 and DSection.9 While csSAM uses linear regression to estimate cell-type-specific gene expression using heterogeneous gene expression measures and cell proportions, the DSection method uses a Bayesian approach, which incorporates uncertainty in the initial proportions into estimating cell-type-specific differentially expressed genes. Well tested in microarray settings, application of such approaches to omics data, such as gene expression data obtained using RNA-seq, remains less explored.
This study was designed to apply methods for cell-type-specific differential gene expression analysis to experimentally obtained RNA-seq data and cell proportions to better understand the functional significance of cell-type-specific gene expression differences in matched cohorts of European derived healthy female subjects and patients diagnosed with SLE. We compared the results of cell-type-specific differential expression analysis using csSAM and DSection methods as well as the results of non-cell-type-specific differential expression analysis using several established methods for differential expression analysis in RNA-seq data.12–16 At each step, we validated the results of the analysis of experimentally obtained RNA-seq data by comparing them with the analyses of a simulated dataset generated using negative binomial (NB) distribution14,17,18 to capture the properties of experimental data. Finally, we provide a brief biological overview of the molecular mechanisms altered by cell-type-specific differentially expressed genes in SLE.
Methods
Study population
Experiments were performed in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (IRB) of the Oklahoma Medical Research Foundation. Written informed consent was obtained from all subjects upon enrollment under protocols approved by the IRB of the Oklahoma Medical Research Foundation. The study population included 10 SLE patients, defined based on the presence of ≥4 ACR criteria, and 10 unrelated, unaffected healthy controls. All participants were European American females. The median age at enrollment was 41.5 years for controls and 40.5 years for SLE patients. The median SLE disease activity index score for SLE patients at enrollment was 4, with a range of 0–6.
RNA-seq data
Briefly, peripheral blood was drawn by venipuncture into PAXgene tubes (BD Diagnostics), and RNA was isolated using standard protocols (Qiagen, Inc.). Globin transcripts were removed using GLOBINclear™ kit (Life Technologies), and samples were prepared for sequencing using the Encore complete kit (NuGEN). Paired-end sequencing of 100-bp reads was performed using the Illumina HiSeq 2000 by standard procedures. The quality of sequencing data was checked using the FastQC v0.11.2 tool (Supplementary Table 7); sequencing adapters were removed using Trimmomatic v0.33.19 Short reads were aligned to the GRCh38/hg38 human genome assembly using TopHat v2.0.9.20 A matrix of raw counts per gene was assembled using htseq-count21 and Homo_sapiens.GRCh38.80.gtf gene models obtained from the Ensembl web site. A matrix of quartile-normalized fragments per kilobase of exons per million reads mapped (FPKM) counts was calculated using the cuffnorm v2.2.1 tool.20 To filter out low expressed signals, as recommended by several method comparison studies,14,22–24 genes having zero FPKM expression in at least one sample were removed, keeping 18,697 expressed genes out of 65,217 total gene models.
Cell proportions data
The patient-specific cell proportions of four cell types, neutrophils, monocytes, T-cells, and B-cells, were obtained by immune phenotyping using flow cytometry. Peripheral blood was collected by venipuncture with EDTA as the anticoagulant, during the same draw used for RNA-seq analysis. Blood was layered over 1:1 Polymorphprep® (Accurate Chemical and Scientific Corporation) (1:1) and centrifuged at 650 g for 35 minutes without brakes. The plasma was drawn off, and the different layers of cells (mononuclear cells and polymorphonuclear leukocytes) were collected. Flow staining was carried out on each layer of cells using the following panel of markers: CD16-FITC (BD Pharmingen), CD19 PE (eBioscience, Inc.), CD14 PerCP (BD Biosciences), CD4 Pacific Blue (BD Biosciences), CD3 V500 (BD Biosciences), and CD8 Qdot605 (Invitrogen). The cells were read on an Attune bench top flow cytometer (Life Technologies). Analysis of data was carried out using FlowJo.
The number of T-cells, monocytes, neutrophils, and B-cell events was obtained from the FlowJo analysis. Cell-type proportions for each cell type were calculated by dividing the number of cell-type-specific events by the sum of the T-cells, monocytes, neutrophils, and B-cell events.
Detecting differential expression in the heterogeneous gene expression matrix
To detect non-cell-type-specific gene expression, we utilized several extensively benchmarked22–25 software packages that employ different strategies for detecting differentially expressed genes from the matrix of raw counts.
The DESeq2 method13 (v 1.8.1 R package) uses a NB model to model the variability of raw counts and Fisher’s exact test to estimate differences between the conditions. The default settings were used.
The edgeR14 method (v 3.10.2) also models data variability using NB distribution, applies empirical Bayes method to moderate the degree of overdispersion across genes, and determines differential expression using Fisher’s exact test. The counts were normalized by the trimmed means of M values (TMM),26 and the default settings were used to perform classical and general linear model (GLM)-based analyses.
The Limma16 approach (v 3.24.15) is based on linear modeling. Originally designed for the analysis of microarray data, it has been extended to the analysis of RNA-seq data in the form of normalized log2-transformed counts. Limma was used in conjunction with voom,27 which weighs the mean–variance relationship of the log-counts, needed for accurate modeling. The default settings as well as sample quality weights were used for the analysis.
The significance analysis of microarray (SAM) method, implemented using samr v2.0 R package, is a nonparametric method based on Wilcoxon rank statistic and a resampling procedure to account for different sequencing depths. The resp.type = “Two class unpaired” setting was used to compare heterogeneous gene expression between SLE patients and healthy controls. The assay.type = “seq” setting was used to detect differential expression using the matrix of raw counts.
The NOISeq15,18 method (v 2.14.0) is another nonparametric method designed to handle small sample size and genes with low expression level by modeling the noise distribution and contrasting fold change and absolute expression differences. The counts were normalized by TMM,26 and the default settings were used.
Overlap among gene lists identified by different methods was identified using Venny tool.28
Detecting cell-type-specific differential expression
To detect genes differentially expressed in specific cell types, we employed two methods, csSAM7 and DSection,9 implemented in the CellMix8 v 1.6.2 R package. As both methods do not apply any type of normalization methods to the raw gene expression counts, we utilized the FPKM method29,30 to normalize counts to gene length.
Multiple testing correction
Throughout the study, we used statistical measures adjusted for multiple testing. As different packages use specific metrics for reporting them, the results they provide should be interpreted accordingly. The DESeq2,13 DSection,9 Limma,16 and edgeR14 packages report Benjamini–Hochberg (BH)-adjusted P-values,31 while csSAM outputs false discovery rate (FDR)32 and SAM reports FDR in a form of q-values.33
Owing to the limited size of our cohort, we often used less significant cutoffs for measures corrected for multiple testing, at risk of selecting less reliable signals. Our rationale for this was that the downstream functional enrichment analysis would prioritize biologically relevant signal even in the presence of noise, or fail to identify functional enrichments if the noise level is too high. Thus, our reported gene lists should be considered as a proxy to identify and compare functional processes represented by them.
Simulated dataset of gene expression
Although all methods used in our study report significance measures corrected for multiple testing, we performed parallel analyses using a simulated gene expression dataset. Using the matrix of experimentally obtained FPKM measures, we simulated gene-specific expression using NB distribution.14,15,17,18 The matrix of cell proportions was simulated by randomizing the patient-specific proportions. This approach randomizes the order, ie, biological measures, of the proportions while keeping the total proportions at 100%. The vector of sample assignment to either healthy controls or SLE patients was permuted. In summary, these approaches allow preserving statistical properties of the experimental RNA-seq and cell proportion datasets while removing biological relationships.34
Functional enrichment analysis
Gene-centric functional and canonical pathway enrichment analyses were performed using the ToppGene Suite.35 Gene symbols were supplied for the analysis.
Implementation and availability
All RNA-seq data processing steps were performed in CentOS 6.6 high-performance cluster computing environment. All analyses were conducted in R/Bioconductor environment v 3.2.0.36,37 All analytical scripts are available at https://github.com/mdozmorov/deconvolution.
Results
Cell proportions of CD3+ T-cells, monocytes, neutro-phils, and B-cells are stable between healthy controls and SLE patients
To assess whether the proportions of cell types (Supplementary Table 1) were significantly different in SLE, we used the Wilcoxon test to compare proportions of cell types between healthy controls and SLE patients. Although SLE patients showed slight increases in the average proportions of CD3+ T- and B-cells, concurrent with decreases in the proportions of monocytes and neutrophils, these differences were not statistically significant between SLE patients and healthy controls (Table 1) in the limited number of subjects tested in this study.
Table 1.
Experimentally obtained cell proportions and the difference in cell proportions between healthy controls and SLE cases. Control/case cells contain average cell proportions per cell type ± standard deviation. Difference cells contain % change in average cell proportions between cases and controls and, in parentheses, Wilcoxon P-value of the differences.
| CONTROLS | CASES | DIFFERENCE | |
|---|---|---|---|
| CD3+ Tcells | 61.19% ± 8.34 | 64.07% ± 14.85 | 2.88% (0.48) |
| Monocytes | 13.67% ± 9.75 | 10.98% ± 7.49 | −2.70% (0.58) |
| Neutrophils | 9.51% ± 3.45 | 6.97% ± 4.42 | −2.53% (0.22) |
| Bcells | 15.63% ± 3.10 | 17.99% ± 11.70 | 2.35% (0.68) |
Detection of differentially expressed genes is limited in heterogeneous cell populations
In order to identify genes differentially expressed in whole peripheral blood from SLE patients as compared with healthy controls, we quantified gene expression as raw counts (Supplementary Table 2A) and as counts normalized to gene length (FPKM; Supplementary Table 2B). The number of raw counts ranged from 0 to 17 for the first to third quantiles, while some genes, such as hemoglobin beta (HBB), had up to one million counts. The FPKM gene expression values ranged from 0 to 24.21 FPKM for the first to third quartiles, with several outliers, such as beta-2-microglobulin (B2M), expressing up to 17,330.00 FPKM (Supplementary Fig. 1). These observations illustrate a wide dynamic range of average expression and variability of both raw counts and FPKM measures in the heterogeneous gene expression data, suggesting potential difficulties in detecting differentially expressed genes.
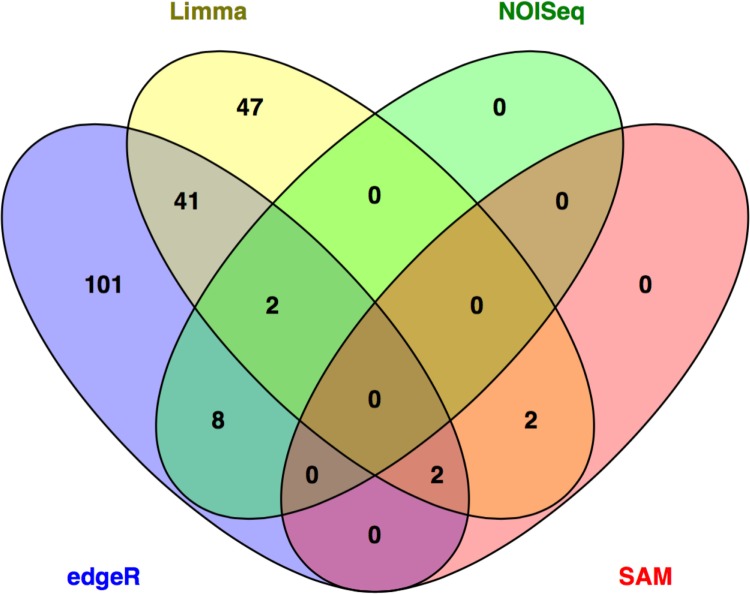
We applied several methods for detecting differentially expressed genes in RNA-seq data, keeping FDR <25% or nonadjusted P-values <0.01. Both classical and GLM-based edgeR analyses identified nearly overlapping lists of differentially expressed genes, with 154 genes detected by both methods (Supplementary Table 3A and B). The Limma–Voom approach with and without using sample weights identified 163 and 125 differentially expressed genes, respectively, with 94 genes being detected by both methods (Supplementary Table 3C and D). Only 10 genes were detected by the NOISeq method (Supplementary Table 3E), while only four and one genes were detected by SAM and DESeq2 methods (Supplementary Table 3F and G, respectively). These results highlight the fact that the selection of data analysis tools markedly affects the outcome of the analysis.24
To identify potential functions affected by differentially expressed genes, we evaluated the intersection of gene lists identified by different methods.24 A total of 55 genes were detected by at least two methods (Fig. 1 and Supplementary Table 3H). Although we observed several immune system-related genes, eg, interferon-induced protein 44 (IFI44), these genes did not show any statistically significant functional enrichments (data not shown). These results further stress difficulty in detecting differentially expressed genes in heterogeneous cell populations.
Figure 1.
Overlap among lists of differentially expressed genes detected in heterogeneous dataset using different methods. Limma and edgeR lists represent genes detected independent of method-specific settings, eg, Limma genes were detected using both default settings and sample weights (Supplementary Table 3).
csSAM method detects cell-type-specific differential gene expression in B-cells and monocytes
To define differentially expressed genes in specific cell types, we applied the csSAM method to the matrices of FPKM measures and cell-type proportions. Owing to the limited cohort size, the detection of cell-type-specific differentially expressed genes was underpowered. Therefore, instead of focusing on the most significant genes, we used minimally calculated significance thresholds and focused on comparing functional processes represented by cell-type-specific differentially expressed genes.
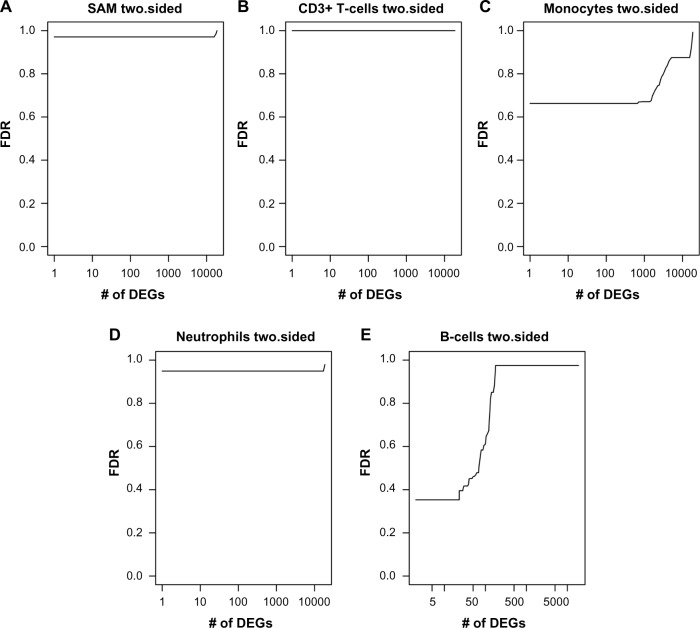
Expectedly, csSAM did not detect differentially expressed genes in heterogeneous cell populations (Fig. 2). However, 68 genes were detected as differentially expressed in B-cells at FDR ranging from 0.35 to 0.48 (Supplementary Table 4A). In addition, 800 genes were detected as differentially expressed in monocytes at FDR ranging from 0.66 to 0.67 (Supplementary Table 4B). As a negative control, we applied csSAM to the simulated dataset of random gene expression measures and did not observe any cell-type-specific differentially expressed genes (Supplementary Fig. 2). We noted that the cell-type-specific differentially expressed genes were highly expressed (average expression level 83.55 FPKM), as compared with 35.29 FPKM average expression level of all genes (P-value = 2.09E–14, Wilcoxon test). These results suggest that highly expressed B-cell-specific and, potentially, monocyte-specific differentially expressed genes can be detected from heterogeneous gene expression in SLE population.
Figure 2.
Cell-type-specific differential expression analysis results using csSAM. Each graph shows the dependence between the number of differentially expressed genes (X-axis) and FDR (Y-axis), eg, 68 genes can be detected as differentially expressed in B-cells of SLE patients at FDR <0.5. (A) SAM analysis of differentially expressed genes in heterogeneous matrix; (B) CD3+ T-cell-specific differential expression analysis; (C) monocyte-specific differential expression analysis; (D) neutrophil-specific differential expression analysis; and (E) B-cell-specific differential expression analysis.
DSection method confirms B-cell- and monocyte-specific gene expression signatures
In order to corroborate the cell-type-specific differentially expressed genes, we applied the DSection method to the FPKM data and cell-type proportions. This method identified 7 B-cell-specific differentially expressed genes, 15 neutrophil-specific genes, and 28 monocyte-specific genes (Supplementary Table 4C–E). Using the simulated dataset, between zero and three cell-type-specific differentially expressed genes were identified. These observations further strengthen the role of gene expression differences in B-cells and monocytes in SLE.
B-cell-specific differentially expressed genes in SLE influence antigen binding and the adaptive immune response
The seven B-cell-specific differentially expressed genes identified by the DSection method did not overlap with the list of genes identified by the csSAM method. Yet, both lists included genes relevant to SLE, including those encoding major histocompatibility complex class I types A, C, and F. Therefore, although the DSection and csSAM methods were underpowered to detect overlapping gene lists, they may have identified different parts of the same molecular mechanisms affected by B-cell-specific differentially expressed genes in SLE.
To test this hypothesis, we performed separate functional enrichment analyses of the two lists using the ToppGene Suite.35 Expectedly, genes in both lists were similarly enriched for several functional categories, including antigen binding (p.adjcsSAM = 2.23E–6, p.adjDSection = 5.89E–5), lymphocyte mediated immunity (p.adjcsSAM = 1.96E–4, p.adjDSection = 8.04E–6), adaptive immune response (p.adjcsSAM = 2.10E–4, p.adjDSection = 6.57E–6), and others. These results are consistent with the hypothesis that the csSAM and DSection methods identified different genetic components of the same functional mechanisms altered by B-cell-specific differentially expressed genes in SLE.
To better understand the mechanisms altered by B-cell-specific differentially expressed genes in SLE, we performed a combined functional enrichment analysis of differentially expressed genes identified by csSAM and DSection. The combined list was enriched in a set of processes similar to those enriched in the individual lists (Table 2). A set of 19 genes from the immunoglobulins gene family (p.adj = 4.72E–14) was enriched in a variety of immune-related processes, ranging from antigen binding (p.adj = 1.08E–10) and transporter associated with antigen processing (TAP) binding (p.adj = 1.23E–4) to immunoregulatory interactions between a lymphoid and a non-lymphoid cell Reactome pathway (p.adj = 3.28E–8; Table 2 and Supplementary Table 5). These results outline immunological processes altered by B-cell- specific differentially expressed genes in SLE.
Table 2.
Top functional categories enriched in B-cell-specific differential expressed genes in SLE. Category – category name; ID – function-specific ID; description – name of a function; gene counts – number of differentially expressed genes enriched in a function; q-value – significant P-value corrected for multiple testing.
| CATEGORY | ID | DESCRIPTION | GENE COUNTS | GENE NAMES | Q-VALUE |
|---|---|---|---|---|---|
| GO: Molecular function | GO:0003823 | Antigen binding | 10 | HLA-A, HLA-C, IGHA1, IGHG1, IGKC, HLA-F, IGLC1, IGLC2, IGKV4-1, IGKV3-20 | 1.08E-10 |
| GO:0046977 | TAP binding | 3 | HLA-A, HLA-C, HLA-F | 1.23E-04 | |
| GO:0005506 | Iron ion binding | 6 | FECH, LTF, LCN2, SNCA, HBA2, HBM | 1.10E-03 | |
| GO:0031720 | Haptoglobin binding | 2 | HBA1, HBA2 | 1.83E-03 | |
| GO:0070051 | Fibrinogen binding | 2 | ITGA2B, ITGB3 | 2.92E-03 | |
| GO: Biological process | GO:0002449 | Lymphocyte mediated immunity | 11 | HLA-A, HLA-C, IGHG1, B2M, IGKC, HLA-F, HPX, IGLC1, IGLC2, IGKV4-1, IGKV3-20 | 1.27E-07 |
| GO:0002460 | Adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 11 | HLA-A, HLA-C, IGHG1, B2M, IGKC, HLA-F, HPX, IGLC1, IGLC2, IGKV4-1, IGKV3-20 | 1.27E-07 | |
| GO:0006959 | Humoral immune response | 10 | DEFA4, IGHA1, IGHG1, IGKC, HPX, IGLC1, IGLC2, LTF, IGKV4-1, IGKV3-20 | 1.27E-07 | |
| GO:0002250 | Adaptive immune response | 11 | HLA-A, HLA-C, IGHG1, B2M, IGKC, HLA-F, HPX, IGLC1, IGLC2, IGKV4-1, IGKV3-20 | 3.65E-07 | |
| GO:0002443 | Leukocyte mediated immunity | 11 | HLA-A, HLA-C, IGHG1, B2M, IGKC, HLA-F, HPX, IGLC1, IGLC2, IGKV4-1, IGKV3-20 | 3.65E-07 | |
| Human phenotype | HP:0010973 | Abnormality of erythroid lineage cell | 12 | GLRX5, BPGM, HLA-A, FECH, GPX1, CYB5R3, ALAS2, ITGA2B, HBA1, HBA2, ITGB3, SLC4A1 | 2.60E-06 |
| HP:0001877 | Abnormality of erythrocytes | 12 | GLRX5, BPGM, HLA-A, FECH, GPX1, CYB5R3, ALAS2, ITGA2B, HBA1, HBA2, ITGB3, SLC4A1 | 2.60E-06 | |
| HP:0001903 | Anemia | 11 | GLRX5, BPGM, HLA-A, FECH, GPX1, ALAS2, ITGA2B, HBA1, HBA2, ITGB3, SLC4A1 | 8.96E-06 | |
| HP:0001930 | Nonspherocytic hemolytic anemia | 3 | BPGM, HBA1, HBA2 | 3.37E-04 | |
| HP:0001878 | Hemolytic anemia | 6 | BPGM, FECH, GPX1, HBA1, HBA2, SLC4A1 | 3.37E-04 | |
| Pathway | 771600 | Scavenging of Heme from Plasma | 9 | IGHA1, IGKC, HPX, IGLC1, IGLC2, HBA1, HBA2, IGKV4-1, IGKV3-20 | 3.96E-12 |
| 771599 | Binding and Uptake of Ligands by Scavenger Receptors | 10 | SPARC, IGHA1, IGKC, HPX, IGLC1, IGLC2, HBA1, HBA2, IGKV4-1, IGKV3–20 | 1.53E-11 | |
| 106413 | Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell | 9 | HLA-A, HLA-C, B2M, IGKC, HLA-F, IGLC1, IGLC2, IGKV4-1, IGKV3-20 | 3.28E-08 | |
| M19553 | Hemoglobin’s Chaperone | 5 | FECH, ALAS2, HBA1, HBA2, AHSP | 1.43E-07 | |
| 106409 | Classical antibody-mediated complement activation | 6 | IGHG1, IGKC, IGLC1, IGLC2, IGKV4-1, IGKV3-20 | 1.43E-07 | |
| Gene family | IG | Immunoglobulins | 19 | IGLV3-10, IGKV1-17, IGKV1-6, IGLV1-44, IGHV3-15, IGHV3-7, IGHA1, IGHV1-69-2, IGHG1, IGKC, IGLC1, IGKV2D-29, IGLC2, IGHV5-51, IGLV6-57, IGKV4-1, IGKV3-20, IGLV3-21, IGLV3-19 | 4.72E-14 |
| HLA | Histocompatibility complex genes | 3 | HLA-A, HLA-C, HLA-F | 3.72E-03 | |
| MARCH | Membrane-associated ring fingers | 2 | MARCH8, MARCH2 | 3.72E-03 | |
| CD | CD molecules | 5 | GYPC, CD24, ADGRE2, ITGB3, SLC4A1 | 2.30E-02 |
Monocyte-specific differentially expressed genes comprise a ribosomal signature in SLE
The csSAM and DSection methods identified vastly different numbers of differentially expressed monocyte-specific genes (800 and 28, respectively). Twenty-four genes were detected by both methods. For reasons previously described,22 we focused our subsequent functional enrichment analysis on the 24 monocyte-specific genes identified by both methods. A total of 16 of the 24 monocyte- specific differentially expressed genes encoded ribosomal proteins. They were nearly exclusively enriched in ribosome-related functions, such as structural constituent of ribosome (p.adj = 2.71E–26), translational elongation (p.adj = 1.26E–29), and the like processes (Supplementary Table 6). These results suggest that the well-known altered ribosomal signature in SLE38 may be monocyte specific.
Neutrophil-specific differentially expressed genes in SLE show no functional enrichments
While the DSection method identified 15 genes as differentially expressed in neutrophils (Supplementary Table 4D), the csSAM method did not identify neutrophil-specific gene expression differences (Fig. 1). To test whether the DSection method was able to detect additional molecular mechanisms driven by neutrophil-specific differentially expressed genes in SLE, we performed functional enrichment analysis of these genes. Surprisingly, we did not observe any functional enrichment of the neutrophil-specific differentially expressed genes (data not shown). Combined with the lack of neutrophil-specific differences identified by the csSAM method, these results suggest a relatively minor contribution of neutrophil-specific differentially expressed genes to the pathogenesis of SLE in the cohort tested.
Discussion
We performed cell-type-specific differential gene expression analysis, implemented by the csSAM and DSection methods, to characterize the functional significance of cell-type-specific gene expression differences in SLE. Both csSAM and DSection methods were able to identify B-cell- and monocyte-specific genes differentially expressed in SLE. The B-cell-specific functional signature included immunoglobulins and major histocompatibility complex genes, enriched in antigen binding molecular function. In contrast, the monocyte-specific functional signature comprised ribosomal genes enriched in ribosomal-related functions, such as translational elongation. In summary, our results suggest that cell-type-specific differential gene expression analysis may provide additional insights into the cell-type specificity of gene expression changes in SLE.
The main limitation of the current study is its small sample size. Although some software methods, such as edgeR14 and NOISeq,15,18 have been specifically designed to deal with minimal or no replicates, the majority of the studies agree on the need for increasing the number of samples to improve detection power of differentially expressed genes.23,25
Despite insufficient sample size, analysis of the heterogeneous gene expression matrix was able to identify several genes previously associated with SLE activity and autoantibody production, such as genes encoding MER receptor tyrosine kinase and CD163, IRF2, IL1R2, and IFI44. For example, ribosomal RNA genes are differentially methylated and differentially expressed in monozygotic twins who are discordant for SLE.39 In addition, levels of CD163 and soluble MER receptor tyrosine kinase correlate with monocyte/macrophage activation, autoantibody specificities, and disease activity.40 IRF2, part of the interferon regulatory pathway, contains two SLE-associated single nucleotide polymorphisms that increase IRF2 transcription upon interferon stimulation.41 Whole-genome methylation analysis has identified associations between ILIR2 promoter hypomethylation and SLE risk as well as disease activity.42 Finally, the interferon-inducible gene IFI44 is known to be associated with the type I interferon signature in lupus, which in turn correlates with levels of anti-RNA binding protein autoantibodies.43 Therefore, the functional enrichments observed here support previous studies, implicating ribosomal genes, immunoglobulins, and major histocompatibility complex genes in SLE pathogenesis.
Although we expected to detect cell-type-specific differentially expressed genes across the whole range of average gene expression levels, our results appeared biased toward detecting cell-type-specific differential expression of highly expressed genes. Thus, many important genes with lower expression, such as those encoding transcription factors44 or noncoding transcripts,45 may have been missed. For example, a recent study demonstrated dysregulation of interferon signature genes in the neutrophils of lupus patients, but no neutrophil-specific functional enrichment was observed in the present study.46 Furthermore, we noted relatively poor overlap between the results produced by both csSAM and DSection methods. However, the complementary functional enrichment analysis results suggest that both methods were able to capture parts of the same molecular mechanisms altered in SLE by cell-type-specific SLE gene signatures. The bias toward highly expressed genes and differences in the detection of cell-type-specific differentially expressed genes can be attributed to the need for a larger sample size and/or increased variability in cell proportions, which would be expected to have a positive effect on linear regression. Our future work will address the relationship between sample size and the ability of linear regression to detect cell-type-specific differentially expressed genes across the whole spectrum of gene expression levels.
In our study, we considered a commonly used FPKM measure of gene expression obtained from RNA-seq data. Multiple methods are used for quantifying gene expression, such as raw counts of reads per gene through RNA-Seq by Expectation Maximization quantification47 and transcripts per million,48 reviewed in Ref. 49; however, for the sake of clarity, we did not test the performance of all methods for quantifying gene expression. Our future studies will address the effect of measuring gene expression levels in RNA-seq data for the purpose of cell-type-specific differential expression analysis.
The csSAM and DSection methods were originally designed for microarray data, while we have been applying them to RNA-seq data. It should be noted that although data preprocessing steps for microarray and RNA-seq data differ, a strong linear relationship between RNA concentration and short read counts has been reported.50 On the contrary, because of technological limitations, microarrays show less linear relationships between RNA concentration and signal intensity.51 Therefore, methods for cell-type-specific differential expression analysis can be expected to perform well in RNA-seq settings, as illustrated by our study.
The use of cell-type-specific differential expression analysis goes well beyond gene expression data obtained with microarray or RNA-seq technologies. Cell-type-specific differential expression analysis will ultimately benefit many epigenomics-oriented sequencing technologies, such as DNA methylation, ChIP-seq, and histone modification profiling, thereby providing a deeper understanding of the molecular mechanisms operating at the level of specific cell types. Although such work has begun,52–55 many issues in detecting cell-type-specific epigenomic differences remain unexplored. Our current work extends efforts to better understand the contribution of cell-type-specific differential expression analysis to understand the SLE pathogenesis on a cell-specific level.3
Supplementary Materials
Supplementary Figure 1. Frequency histogram of gene expression measured as FPKM measures. X-axis – average FPKM measures and Y-axis – frequency of genes at any given average FPKM. FPKM range from zero to the third quartile is displayed for clarity.
Supplementary Figure 2. Cell-type-specific differential expression analysis results using simulated dataset. Each graph shows the dependence between the number of differentially expressed genes (X-axis) and FDR (Y-axis). For example, no genes can be called differentially expressed in any cell-type-specific analysis at FDR <1.
Supplementary Table 1. A matrix of patient-specific proportions for four cell types. Each cell represents the proportion of a given cell type in a given sample. Group – disease status; control/case – healthy/SLE patients; subject – internal sample identifier.
Supplementary Table 2. (A) A matrix of raw counts of genes. Group – disease status; control/case – healthy/SLE patients; subject – internal sample identifier. (B) A matrix of FPKM measures of genes expressed above zero across all samples. Group – disease status; control/case – healthy/SLE patients; subject – internal sample identifier.
Supplementary Table 3. Genes differentially expressed in heterogeneous group. (A) edgeR classic; (B) edgeR GLM; (C) Limma–Voom; (D) Limma–Voom weighted; (E) NOISeq; (F) SAM; and (G) DESeq2 methods. logFC – log2 fold change between SLE patients and healthy controls; P.value/adj.p.val – noncorrected/corrected for multiple testing significant P-value (H) Genes differentially expressed in heterogeneous group, identified by at least two methods (Fig. 1).
Supplementary Table 4. (A) Genes differentially expressed in B-cells, csSAM method. Gene.Name – Ensembl ID; FDR – false discovery rate; AVEXP/SD – average gene expression/standard deviation; hgnc_symbol/description – gene symbol/description. (B) Genes differentially expressed in monocytes, csSAM method. Gene.Name – Ensembl ID; FDR – false discovery rate; AVEXP/SD – average gene expression/standard deviation; hgnc_symbol/description – gene symbol/description. (C) Genes differentially expressed in B-cells, DSection method. Gene.Name – Ensembl ID; q-value – significant P-value corrected for multiple testing; AVEXP/SD – average gene expression/standard deviation; hgnc_symbol/description – gene symbol/description. (D) Genes differentially expressed in neutrophils, DSection method. Gene.Name – Ensembl ID; q-value – significant P-value corrected for multiple testing; AVEXP/SD – average gene expression/standard deviation; hgnc_symbol/description – gene symbol/description. (E) Genes differentially expressed in monocytes, DSection method. Gene.Name – Ensembl ID; q-value – significant P-value corrected for multiple testing; AVEXP/SD – average gene expression/standard deviation; hgnc_symbol/description – gene symbol/description.
Supplementary Table 5. Functional enrichment analysis of the B-cell-specific differentially expressed genes in SLE. Category – category name; ID – function-specific ID; description – name of a function; P-value/q-value – noncorrected/corrected for multiple testing significant P-value; hit count in query list – number of differentially expressed genes enriched in a function; hit count in genome – total number of genes in a function; hit in query list – names of differentially expressed genes enriched in a function.
Supplementary Table 6. Functional enrichment analysis of the monocyte-specific differentially expressed genes in SLE. Category – category name; ID – function-specific ID; description – name of a function; P-value/q-value – noncorrected/corrected for multiple testing significance P-value; hit count in query list – number of differentially expressed genes enriched in a function; hit count in genome – total number of genes in a function; hit in query list – names of differentially expressed genes enriched in a function.
Supplementary Table 7. Quality metrics of RNA-seq data, obtained with FastQC tool.
Acknowledgments
The authors thank Rebecka Bourn for editorial assistance.
Footnotes
ACADEMIC EDITOR: J.T. Efird, Associate Editor
PEER REVIEW: Seven peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,838 words, excluding any confidential comments to the academic editor.
FUNDING: Funding for this work was provided by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (P30AR053483), the National Institute of General Medical Sciences (P30GM103510, P30GM110766, U54GM104938), the National Institutes of Health (P20RR020143), and the National Center for Advancing Translational Sciences (UL1TR000058). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of any agency involved in the funding of the work. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: MGD, JMG, JAJ. Contributed to the experimental parts: ND, KB, SRM, VR, EG, JMG, JAJ. Wrote the manuscript: MGD. Made critical revisions and approved the final version: JAJ, JMG. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Frieri M. Mechanisms of disease for the clinician: systemic lupus erythematosus. Ann Allergy Asthma Immunol. 2013;110(4):228–32. doi: 10.1016/j.anai.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 2.Jenks SA, Sanz I. Altered B cell receptor signaling in human systemic lupus erythematosus. Autoimmun Rev. 2009;8(3):209–13. doi: 10.1016/j.autrev.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One. 2009;4(7):e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunham LR, Hayden MR. Medicine. Whole-genome sequencing: the new standard of care? Science. 2012;336(6085):1112–3. doi: 10.1126/science.1220967. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Yu Y, Hertwig F, et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 2015;16(1):133. doi: 10.1186/s13059-015-0694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frangou EA, Bertsias GK, Boumpas DT. Gene expression and regulation in systemic lupus erythematosus. Eur J Clin Invest. 2013;43(10):1084–96. doi: 10.1111/eci.12130. [DOI] [PubMed] [Google Scholar]
- 7.Shen-Orr SS, Tibshirani R, Khatri P, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7(4):287–9. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaujoux R, Seoighe C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics. 2013;29(17):2211–2. doi: 10.1093/bioinformatics/btt351. [DOI] [PubMed] [Google Scholar]
- 9.Erkkila T, Lehmusvaara S, Ruusuvuori P, Visakorpi T, Shmulevich I, Lahdesmaki H. Probabilistic analysis of gene expression measurements from heterogeneous tissues. Bioinformatics. 2010;26(20):2571–7. doi: 10.1093/bioinformatics/btq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venet D, Pecasse F, Maenhaut C, Bersini H. Separation of samples into their constituents using gene expression data. Bioinformatics. 2001;17(suppl 1):S279–87. doi: 10.1093/bioinformatics/17.suppl_1.s279. [DOI] [PubMed] [Google Scholar]
- 11.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21(12):2213–23. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 17.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarazona S, Furió-Tarí P, Turrà D, et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv711. pii:gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics. 2014;1(30(15)):2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. 2012;7(3):562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;15(31(2)):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang ZH, Jhaveri DJ, Marshall VM, et al. A comparative study of techniques for differential expression analysis on RNA-Seq data. PLoS One. 2014;9(8):e103207. doi: 10.1371/journal.pone.0103207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soneson C, Delorenzi M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics. 2013;14:91. doi: 10.1186/1471-2105-14-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seyednasrollah F, Laiho A, Elo LL. Comparison of software packages for detecting differential expression in RNA-seq studies. Brief Bioinform. 2015;16(1):59–70. doi: 10.1093/bib/bbt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapaport F, Khanin R, Liang Y, et al. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2013;14(9):R95. doi: 10.1186/gb-2013-14-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveros JC. VENNY: An Interactive Tool for Comparing Lists with Venn Diagrams. 2007–2015. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- 29.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 32.Efron B, Tibshirani R. Empirical Bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23(1):70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- 33.Storey JD. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol. 2002;64(3):479–98. [Google Scholar]
- 34.Dozmorov MG, Guthridge JM, Hurst RE, Dozmorov IM. A comprehensive and universal method for assessing the performance of differential gene expression analyses. PLoS One. 2010;5(9):e12657. doi: 10.1371/journal.pone.0012657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 38.Lamon EW, Bennett JC. Antibodies to ribosomal ribonucleic acid (rRNA) in patients with systemic lupus erythematosus (SLE) Immunology. 1970;19(3):439–42. [PMC free article] [PubMed] [Google Scholar]
- 39.Javierre BM, Fernandez AF, Richter J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20(2):170–9. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zizzo G, Guerrieri J, Dittman LM, Merrill JT, Cohen PL. Circulating levels of soluble MER in lupus reflect M2c activation of monocytes/macrophages, auto antibody specificities and disease activity. Arthritis Res Ther. 2013;15(6):R212. doi: 10.1186/ar4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawasaki A, Furukawa H, Nishida N, et al. Association of functional polymorphisms in interferon regulatory factor 2 (IRF2) with susceptibility to systemic lupus erythematosus: a case-control association study. PLoS One. 2014;9(10):e109764. doi: 10.1371/journal.pone.0109764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin SY, Hsieh SC, Lin YC, et al. A whole genome methylation analysis of systemic lupus erythematosus: hypomethylation of the IL10 and IL1R2 promoters is associated with disease activity. Genes Immun. 2012;13(3):214–20. doi: 10.1038/gene.2011.74. [DOI] [PubMed] [Google Scholar]
- 43.Hua J, Kirou K, Lee C, Crow MK. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 2006;54(6):1906–16. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 44.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 45.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 46.Coit P, Yalavarthi S, Ognenovski M, et al. Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J Autoimmun. 2015;58:59–66. doi: 10.1016/j.jaut.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131(4):281–5. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 49.Dillies MA, Rau A, Aubert J, et al. French StatOmique Consortium. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14(6):671–83. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 50.Fonseca NA, Marioni J, Brazma A. RNA-Seq gene profiling – a systematic empirical comparison. PLoS One. 2014;9(9):e107026. doi: 10.1371/journal.pone.0107026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 52.Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8(3):290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15(2):R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Montaño CM, Irizarry RA, Kaufmann WE, et al. Measuring cell-type specific differential methylation in human brain tissue. Genome Biol. 2013;14(8):R94. doi: 10.1186/gb-2013-14-8-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Frequency histogram of gene expression measured as FPKM measures. X-axis – average FPKM measures and Y-axis – frequency of genes at any given average FPKM. FPKM range from zero to the third quartile is displayed for clarity.
Supplementary Figure 2. Cell-type-specific differential expression analysis results using simulated dataset. Each graph shows the dependence between the number of differentially expressed genes (X-axis) and FDR (Y-axis). For example, no genes can be called differentially expressed in any cell-type-specific analysis at FDR <1.
Supplementary Table 1. A matrix of patient-specific proportions for four cell types. Each cell represents the proportion of a given cell type in a given sample. Group – disease status; control/case – healthy/SLE patients; subject – internal sample identifier.
Supplementary Table 2. (A) A matrix of raw counts of genes. Group – disease status; control/case – healthy/SLE patients; subject – internal sample identifier. (B) A matrix of FPKM measures of genes expressed above zero across all samples. Group – disease status; control/case – healthy/SLE patients; subject – internal sample identifier.
Supplementary Table 3. Genes differentially expressed in heterogeneous group. (A) edgeR classic; (B) edgeR GLM; (C) Limma–Voom; (D) Limma–Voom weighted; (E) NOISeq; (F) SAM; and (G) DESeq2 methods. logFC – log2 fold change between SLE patients and healthy controls; P.value/adj.p.val – noncorrected/corrected for multiple testing significant P-value (H) Genes differentially expressed in heterogeneous group, identified by at least two methods (Fig. 1).
Supplementary Table 4. (A) Genes differentially expressed in B-cells, csSAM method. Gene.Name – Ensembl ID; FDR – false discovery rate; AVEXP/SD – average gene expression/standard deviation; hgnc_symbol/description – gene symbol/description. (B) Genes differentially expressed in monocytes, csSAM method. Gene.Name – Ensembl ID; FDR – false discovery rate; AVEXP/SD – average gene expression/standard deviation; hgnc_symbol/description – gene symbol/description. (C) Genes differentially expressed in B-cells, DSection method. Gene.Name – Ensembl ID; q-value – significant P-value corrected for multiple testing; AVEXP/SD – average gene expression/standard deviation; hgnc_symbol/description – gene symbol/description. (D) Genes differentially expressed in neutrophils, DSection method. Gene.Name – Ensembl ID; q-value – significant P-value corrected for multiple testing; AVEXP/SD – average gene expression/standard deviation; hgnc_symbol/description – gene symbol/description. (E) Genes differentially expressed in monocytes, DSection method. Gene.Name – Ensembl ID; q-value – significant P-value corrected for multiple testing; AVEXP/SD – average gene expression/standard deviation; hgnc_symbol/description – gene symbol/description.
Supplementary Table 5. Functional enrichment analysis of the B-cell-specific differentially expressed genes in SLE. Category – category name; ID – function-specific ID; description – name of a function; P-value/q-value – noncorrected/corrected for multiple testing significant P-value; hit count in query list – number of differentially expressed genes enriched in a function; hit count in genome – total number of genes in a function; hit in query list – names of differentially expressed genes enriched in a function.
Supplementary Table 6. Functional enrichment analysis of the monocyte-specific differentially expressed genes in SLE. Category – category name; ID – function-specific ID; description – name of a function; P-value/q-value – noncorrected/corrected for multiple testing significance P-value; hit count in query list – number of differentially expressed genes enriched in a function; hit count in genome – total number of genes in a function; hit in query list – names of differentially expressed genes enriched in a function.
Supplementary Table 7. Quality metrics of RNA-seq data, obtained with FastQC tool.