ABSTRACT
The cucumber anthracnose fungus Colletotrichum orbiculare forms specialized cells called appressoria for host penetration. We identified a gene, FAM1, encoding a novel peroxin protein that is essential for peroxisome biogenesis and that associates with Woronin bodies (WBs), dense-core vesicles found only in filamentous ascomycete fungi which function to maintain cellular integrity. The fam1 disrupted mutants were unable to grow on medium containing oleic acids as the sole carbon source and were nonpathogenic, being defective in both appressorium melanization and host penetration. Fluorescent proteins carrying peroxisomal targeting signals (PTSs) were not imported into the peroxisomes of fam1 mutants, suggesting that FAM1 is a novel peroxisomal biogenesis gene (peroxin). FAM1 did not show significant homology to any Saccharomyces cerevisiae peroxins but resembled conserved filamentous ascomycete-specific Pex22-like proteins which contain a predicted Pex4-binding site and are potentially involved in recycling PTS receptors from peroxisomes to the cytosol. C. orbiculare FAM1 complemented the peroxisomal matrix protein import defect of the S. cerevisiae pex22 mutant. Confocal microscopy of Fam1-GFP (green fluorescent protein) fusion proteins and immunoelectron microscopy with anti-Fam1 antibodies showed that Fam1 localized to nascent WBs budding from peroxisomes and mature WBs. Association of Fam1 with WBs was confirmed by colocalization with WB matrix protein CoHex1 (C. orbiculare Hex1) and WB membrane protein CoWsc (C. orbiculare Wsc) and by subcellular fractionation and Western blotting with antibodies to Fam1 and CoHex1. In WB-deficient cohex1 mutants, Fam1 was redirected to the peroxisome membrane. Our results show that Fam1 is a WB-associated peroxin required for pathogenesis and raise the possibility that localized receptor recycling occurs in WBs.
IMPORTANCE
Colletotrichum orbiculare is a fungus causing damaging disease on Cucurbitaceae plants. In this paper, we characterize a novel peroxisome biogenesis gene from this pathogen called FAM1. Although no genes with significant homology are present in Saccharomyces cerevisiae, FAM1 contains a predicted Pex4-binding site typical of Pex22 proteins, which function in the recycling of PTS receptors from peroxisomes to the cytosol. We show that FAM1 complements the defect in peroxisomal matrix protein import of S. cerevisiae pex22 mutants and that fam1 mutants are completely defective in peroxisome function, fatty acid metabolism, and pathogenicity. Remarkably, we found that this novel peroxin is specifically localized on the bounding membrane of Woronin bodies, which are small peroxisome-derived organelles unique to filamentous ascomycete fungi that function in septal pore plugging. Our finding suggests that these fungi have coopted the Woronin body for localized receptor recycling during matrix protein import.
INTRODUCTION
Peroxisomes are single-membrane-bound organelles in eukaryotic cells that function in diverse metabolic processes such as β-oxidation, the glyoxylate cycle, cholesterol metabolism, and methanol assimilation (1). Among filamentous fungi, peroxisomes have been implicated in sexual reproduction (2), biosynthesis of secondary metabolites (3), biotin synthesis (4), and plant pathogenicity, notably in the anthracnose fungi, Colletotrichum species, and the rice blast fungus Magnaporthe oryzae (5, 6). These pathogens elaborate highly differentiated infection structures called appressoria, which develop thick, melanized cell walls and mediate the initial penetration of host cells. The key metabolic pathways involved in appressorium-mediated penetration were extensively studied (7, 8). Lipid bodies are mobilized in appressoria, and lipolysis provides triacylglycerol and fatty acids that are subjected to β-oxidation in peroxisomes (9, 10). The production of acetyl coenzyme A (acetyl-CoA) via β-oxidation and the glyoxylate cycle is critical for pathogenesis by providing an energy source, osmolytes for turgor generation, and substrate for the synthesis of 1,8-dihydroxynaphthalene-derived melanin (6).
The life cycle of the peroxisome involves the following steps: peroxisomal membrane formation and sorting of membrane proteins, import of peroxisomal matrix proteins from the cytosol, peroxisome division, and peroxisome degradation by pexophagy, which is a type of autophagy (11, 12). Pexophagy was shown to be essential for pathogenesis of both Colletotrichum and Magnaporthe (13, 14). Proteins required for peroxisome biogenesis are collectively called peroxins, and to date, more than 32 peroxins have been identified (12, 15). The import of peroxisomal matrix proteins is mediated by Pex5 and Pex7, which are receptors for type I and type II peroxisomal targeting signals (PTSs) (peroxisomal targeting signal 1 [PTS1] and PTS2), respectively. Pex13 and Pex14 function as docking proteins for Pex5 and Pex7, respectively (16, 17), and we previously showed that Pex13 is essential for peroxisome function and pathogenesis in Colletotrichum orbiculare (10).
The recycling of PTS receptors to the cytosol involves Pex4, a ubiquitin-conjugating enzyme, together with its membrane anchor Pex22 (18, 19) and a complex containing two ATPases associated with diverse cellular activities (AAA ATPases), Pex1 and Pex6, and the Pex6 membrane anchor Pex15/Pex26 (20, 21). Kimura et al. (22) showed that the Pex6 AAA ATPase is essential for appressorium function and pathogenesis in C. orbiculare. Although the membrane-associated components Pex1, Pex4, and Pex6 are all conserved from yeast to humans, the proposed membrane anchors (Pex22 and Pex15/Pex26) are less well-conserved (15).
The Woronin body (WB) is a peroxisome-derived organelle that is unique to filamentous ascomycete fungi and that functions to maintain cellular integrity after hyphal damage by sealing septal pores, thereby preventing cytoplasmic leakage (23). In Neurospora crassa, WB formation proceeds via a series of steps: the major WB protein HEX1 is preferentially expressed at hyphal apices and is imported into the peroxisome matrix, where it self-assembles into extremely large protein complexes that form the dense core of the WB (23). The HEX1 assemblies recruit WB sorting complex protein (WSC) to the peroxisome membrane, where WSC self-assembles and assists budding of the nascent WB (24). In addition, WSC recruits the tethering protein LAH-1 that mediates fission of the WB from the mother peroxisome (25). Although many studies have shown that WBs develop from peroxisomes, so far only one report has implicated a peroxin protein in WB biogenesis, namely, Pex26, which mediates recycling of Pex5 in N. crassa (26). By binding to Hex1 oligomers, Pex26 becomes enriched in the membranes of a subset of peroxisomes destined to differentiate WBs, where it likely promotes the import of further HEX1 through a positive-feedback mechanism.
We previously used Agrobacterium tumefaciens-mediated transformation to isolate C. orbiculare mutants deficient in fatty acid metabolism and identified several new genes essential for peroxisome biogenesis (27). In this study, we report that FAM1 (fatty acid metabolism 1), a gene unique to filamentous ascomycetes, is required for peroxisome biogenesis, infection-related morphogenesis, and pathogenicity of C. orbiculare. We show that FAM1 is the functional ortholog of Saccharomyces cerevisiae PEX22, required for the import of peroxisomal matrix proteins. Despite this peroxisome-related function, we present cytological and biochemical evidence that Fam1 protein is predominantly associated with WB membranes, rather than peroxisome membranes. Our results demonstrate that a WB-localized protein functions as a peroxin and is critical for fungal pathogenesis.
RESULTS
Identification of a Pex22-like peroxin unique to filamentous ascomycetes.
Random insertional mutagenesis using Agrobacterium tumefaciens-mediated transformation (27) was performed to isolate Colletotrichum orbiculare mutants defective in fatty acid metabolism. Approximately 3,000 transformants were tested for their ability to utilize fatty acids using a medium containing oleic acids as the sole carbon source. Based on this screen, a mutant defective in fatty acid utilization was identified and named Hi2049 (see Fig. S1A in the supplemental material). Hi2049 was defective in lesion formation on intact cucumber cotyledons but retained pathogenicity on wounded tissues (Fig. S1B). These phenotypes resembled those of the C. orbiculare peroxisome-defective pex6 and pex13 mutants (10, 22).
To isolate genomic DNA segments adjacent to the T-DNA insertion site of the Hi2049 mutant, TAIL-PCR (thermal asymmetric interlaced PCR) was applied to the mutant genomic DNA. The amplified TAIL-PCR products were sequenced, and primer pairs were designed based on the obtained sequence to isolate genomic clones from a cosmid library of C. orbiculare. To identify the disrupted gene in Hi2049, the full genomic DNA sequence was obtained by primer walking, and the positions of introns and exons were confirmed by sequencing cDNA amplified by reverse transcription-PCR. The predicted open reading frame (ORF) consisted of 1,261 bp with two exons separated by one intron (49 bp), and the gene was named FAM1 (for fatty acid metabolism).
Motif searches using the Phobius program (28) identified a putative transmembrane domain between amino acids 12 and 36 of Fam1, and the amino acid sequence showed high homology to Magnaporthe oryzae A4R2Z0 (2e−177), N. crassa (GenBank accession no. Q8WZV7) (9e−110), and Aspergillus oryzae (GenBank accession no. Q2UQZ8) (5e−95) (see Fig. S2 in the supplemental material). PSI-BLAST previously revealed that these three proteins contain a predicted Pex4p-binding sequence, and based on in silico analysis only, they were previously described as filamentous ascomycete Pex22-like proteins (15). Nevertheless, amino acid sequence alignment did not show significant homology between Pex22-like proteins from filamentous ascomycetes and Saccharomyces cerevisiae Pex22 (Fig. S2), and there is only 5% amino acid identity between the C. orbiculare and S. cerevisiae proteins, with several of the conserved amino acid residues likely to be involved in interface formation with Pex4 (19). Furthermore, a stretch of positively charged amino acids is located ahead of an N-terminal transmembrane domain similar to that of Pichia pastoris Pex22 (29) (Fig. S2). Until now, there was no experimental evidence to show that these proteins perform a function similar to Pex22. In S. cerevisiae, Pex22 acts as a docking protein for the Pex4 ubiquitin-conjugating enzyme at the peroxisomal membrane and is involved in Pex5 receptor recycling (19). Therefore, to test whether C. orbiculare FAM1 possesses PEX22 function, we used a yeast complementation assay.
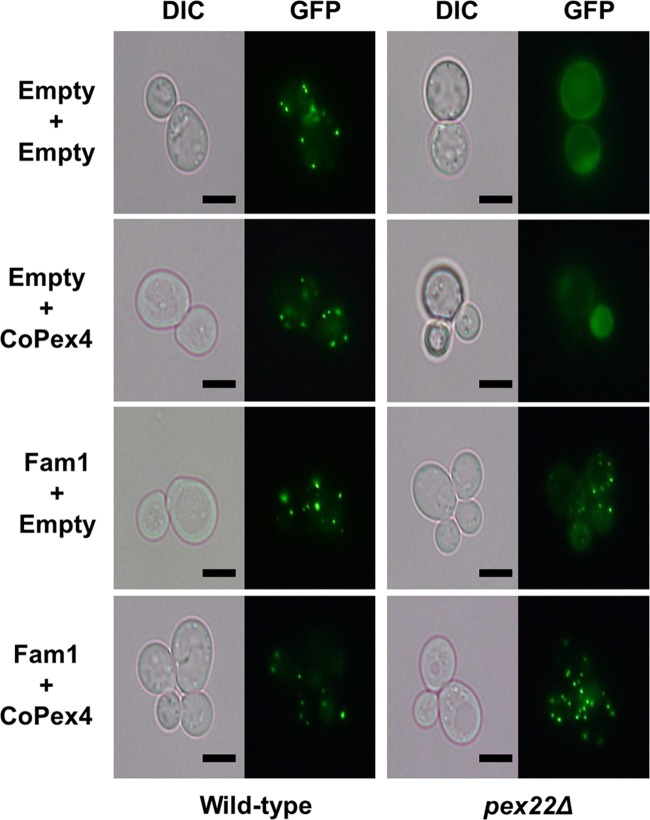
The green fluorescent protein (GFP)-peroxisome targeting signal 1 (PTS1) construct was introduced into wild-type and Δpex22 S. cerevisiae strains as a reporter to evaluate complementation of peroxisome biogenesis. In the wild-type cells, GFP-PTS1 was imported into peroxisomes, producing a punctate labeling pattern, but in Δpex22 mutant cells, no punctate localization was detected, indicating a defect in peroxisome biogenesis (Fig. 1). Then, we expressed the FAM1 and C. orbiculare PEX4 (CoPEX4) genes, either individually or together, in the Δpex22 yeast mutant. Introduction of the FAM1 gene alone into the Δpex22 mutant partially restored the transport of GFP-PTS1 to peroxisomes, producing a faint punctate localization, while simultaneous introduction of FAM1 and CoPEX4 fully restored the transport of GFP-PTS1 into peroxisomes, reproducing the wild-type GFP-PTS1 localization (Fig. 1). These results indicate that FAM1 is the functional ortholog of PEX22. On the other hand, the only partial restoration of GFP-PTS1 import by FAM1 alone suggests that Fam1 interacts better with C. orbiculare Pex4 (CoPex4) than with yeast Pex4, producing a fully functional protein complex for complete complementation of the Δpex22 mutant.
FIG 1 .
Complementation of yeast pex22 mutant by dual expression of FAM1 and CoPEX4. Wild-type S. cerevisiae and Δpex22 mutants expressing GFP-PTS1, a fluorescent peroxisome marker, were transformed with vectors harboring FAM1 or CoPEX4 or empty vector. In the wild type, GFP localized at peroxisomes, producing a punctate pattern, while in Δpex22 mutants, GFP was uniformly distributed in the cytoplasm, indicating peroxisome deficiency. Introduction of FAM1 into the Δpex22 mutant partially restored localization of GFP-PTS1 to peroxisomes, while introduction of FAM1 together with CoPEX4 provided full restoration, producing the same punctate GFP fluorescence pattern as in the wild type. Bars, 5 µm. DIC, differential interference contrast.
FAM1 is essential for peroxisome biogenesis, appressorium maturation, and pathogenesis.
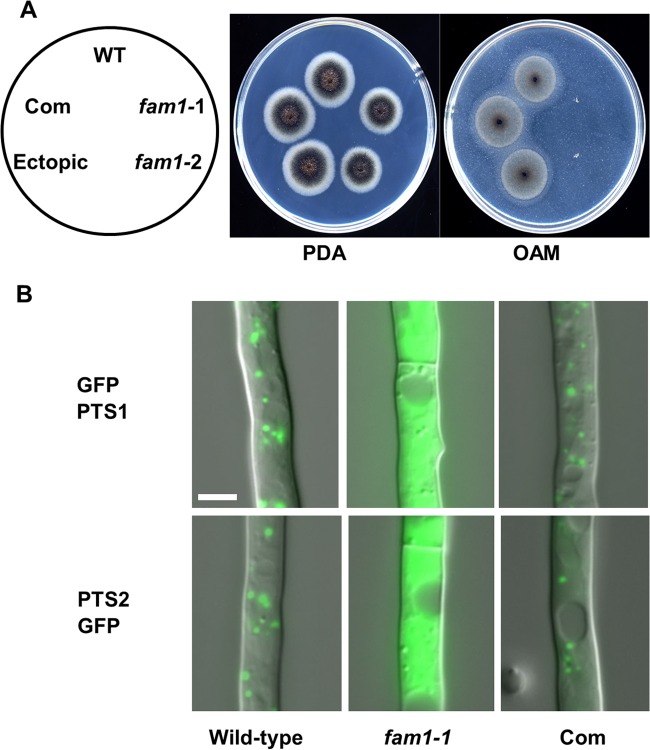
To better define the functions of FAM1 in C. orbiculare, we generated targeted fam1 mutants. Disruption of FAM1 was verified by DNA blot analysis (see Fig. S3 in the supplemental material), and the fam1 phenotype was similar to the Hi2049 phenotype. The fam1 mutants formed melanized colonies on potato dextrose agar (PDA) like the wild-type strain (Fig. 2A), but fam1 mutants could not grow on oleic acid-containing medium (OAM) (Fig. 2A), confirming that FAM1 is essential for fatty acid utilization.
FIG 2 .
C. orbiculare fam1 disruption mutants are deficient in peroxisome biogenesis. (A) The fam1 mutants are defective in fatty acid utilization. Colonies were grown for 5 days on PDA or 12 days on oleic acid-containing medium (OAM). The strains tested were the wild-type (WT) strain, fam1 mutants (fam1-1 and fam1-2), fam1-1 complemented strain (Com), and ectopic transformant. The locations of strains on the plates are indicated in the left-hand panel. (B) Mislocalization of GFP-PTS1 and PTS2-GFP proteins in the fam1 mutant. Vegetative hyphae of the wild-type, fam1-1 mutant, and the fam1-1 complemented strain (Com) expressing GFP-PTS1 or PTS2-GFP were grown for 24 h on glass slides in liquid medium and observed by differential interference contrast (DIC) and epifluorescence microscopy. Photos are merged images. Bar, 5 µm.
To investigate the involvement of FAM1 in peroxisomal function, we introduced PTS1- and PTS2-tagged GFP into the wild-type strain and fam1 mutants. In the wild-type strain expressing GFP-PTS1 or PTS2-GFP, the GFP signals were localized to peroxisomes, visible as abundant punctate organelles (Fig. 2B). In contrast, in the fam1 mutant, GFP-PTS1 and PTS2-GFP labeling was dispersed throughout the fungal cytosol, whereas peroxisome localization was restored in the fam1+FAM1 complemented strain, indicating that import of peroxisomal matrix proteins into peroxisomes was impaired by fam1 mutation. Thus, FAM1 is essential for peroxisome biogenesis in C. orbiculare.
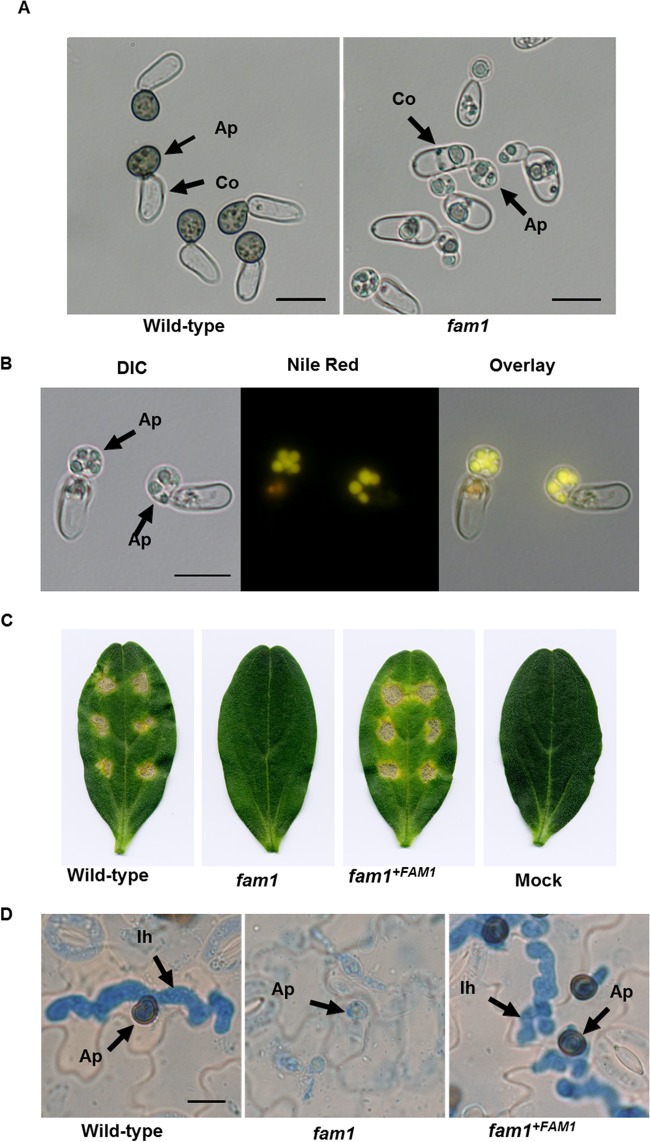
Next, we examined the effect of FAM1 gene disruption on the infection-related morphogenesis of C. orbiculare. The fam1 mutant formed nonmelanized appressoria on inductive artificial surfaces that were abnormally small and ellipsoid in shape compared with wild-type appressoria, similar to those described for the pex13 mutant (Fig. 3A) (10). Moreover, the cytoplasm of appressoria and conidia of the fam1 mutant contained large globules that were stained strongly by the lipid probe Nile red (Fig. 3B), showing that the fam1 mutant is impaired in the mobilization of storage lipid during the development of appressoria.
FIG 3 .
Appressorium development and pathogenicity of the fam1 mutant. (A) Conidia of the wild-type strain or fam1 mutant were incubated on glass slides for 24 h and observed by light microscopy. Ap, Appressoria; Co, conidia. Bars, 10 µm. (B) Distribution of lipid droplets in appressoria of the fam1 mutant. After the cells were stained with Nile red, they were viewed with differential interference contrast (DIC) microscopy, epifluorescence microscopy, and as an overlay. Bar, 10 µm. (C) The fam1 mutant showed attenuated pathogenicity on cucumber cotyledons. Cotyledons were inoculated with spores of the wild type, fam1 mutant, complemented strain, or distilled water (mock). Symptoms were observed after 6 days. (D) Histology of infection. Spores of the wild-type, fam1 mutant, and complemented strain were inoculated on cucumber cotyledons, and tissues were examined by light microscopy after 72 h. Ap, appressoria; Ih, infection hyphae. Bar, 10 µm.
To assess whether FAM1 plays a role in fungal pathogenesis, conidial suspensions of the fam1 mutant were inoculated onto detached cucumber cotyledons and observed at 6 days postinoculation (dpi). Whereas the fam1 mutant never formed necrotic lesions, the wild-type and fam1+FAM1 complemented strain formed yellow-brown lesions (Fig. 3C). To determine at what stage of infection the fam1 mutant was affected, we examined the infected cucumber leaves by light microscopy. At 3 dpi, the wild-type and fam1+FAM1 complemented strains had developed normal melanized appressoria which penetrated to form infection hyphae inside the host epidermal cells (Fig. 3D). In contrast, the fam1 mutant formed nonmelanized appressoria which never penetrated, and no infection hyphae were visible in the underlying cucumber epidermal cells (Fig. 3D).
Live-cell confocal imaging of GFP-tagged Fam1.
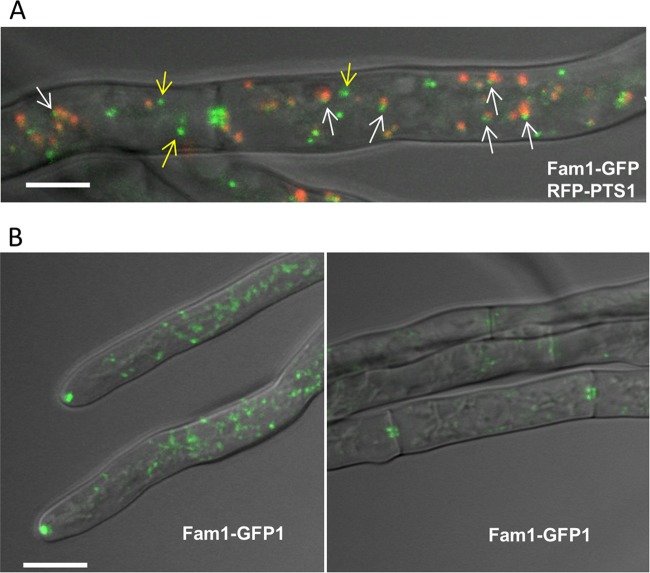
To examine the subcellular localization of Fam1 protein, we made a construct to express a Fam1-GFP fusion protein under control of the native promoter. The FAM1-GFP construct restored all the defective phenotypes of fam1, indicating that the GFP-tagged FAM1 gene was functional. Yeast Pex22 was previously shown to localize to the peroxisomal membrane (30). To evaluate the relationship between Fam1 protein and peroxisomes, we coexpressed GFP-labeled Fam1 (Fam1-GFP) together with red fluorescent protein-labeled PTS1 (RFP-PTS1) to label peroxisomes. On peroxisomes, Fam1-GFP was localized highly asymmetrically in foci at the peroxisome periphery (Fig. 4A, white arrows). Interestingly, only a fraction of Fam1-GFP localized at peroxisomes, and more often, Fam1-GFP fluorescence was observed on punctate organelles in the fungal cytoplasm that were distinct from peroxisomes (yellow arrows) (Fig. 4A). Furthermore, a subset of these labeled organelles were concentrated near growing hyphal tips and septa (Fig. 4B).
FIG 4 .
Subcellular localization of Fam1 in C. orbiculare. (A) Fam1-GFP and RFP-PTS1 were coexpressed in hyphae of the wild type and observed by confocal microscopy. RFP-PTS1 localized to the peroxisome matrix, while Fam1-GFP localized asymmetrically in small foci at the peroxisome periphery (white arrows) or in punctate structures distinct from peroxisomes (yellow arrows) in the overlay projection image of bright-field, GFP, and RFP channels. Bar, 2.5 µm. (B) Fam1-GFP accumulated in punctate structures at hyphal tips (left) and near septal pores (right). Overlay projection images of bright-field and GFP channels are shown. Bar, 5 µm.
Next, to investigate whether the localization of Fam1 was affected by defects in peroxisome biogenesis, we expressed the Fam1-GFP fusion protein in copex6 and copex13 mutants (10, 22). In cells of these mutants, Fam1-GFP was detected on punctate structures that were smaller and less distinct than in wild-type cells, and there was no detectable accumulation of these structures at hyphal tips and septa (see Fig. S4 in the supplemental material). This result suggests that Fam1 localizes to an organelle derived from the peroxisome and that peroxisome function is essential for Fam1 accumulation at hyphal tips and septa.
Fam1 is localized on Woronin bodies.
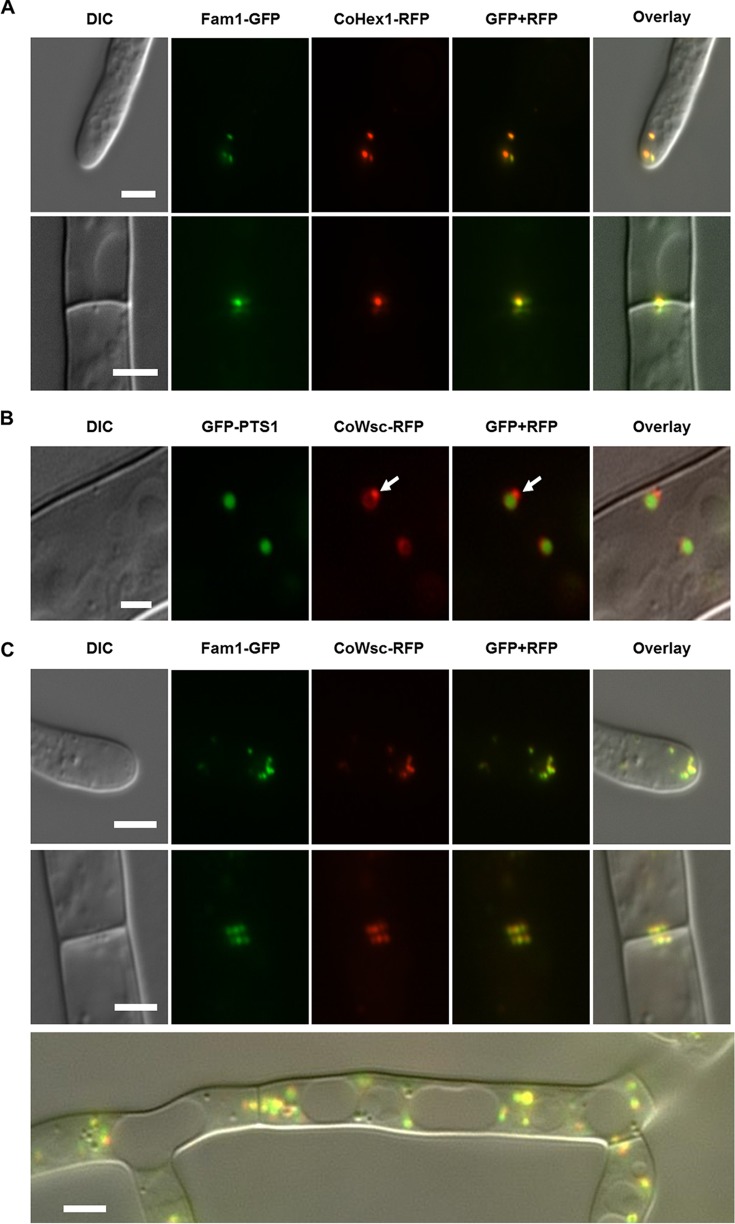
The Fam1 localization pattern described above closely resembles that reported for the Woronin body (WB), a peroxisome-derived organelle unique to filamentous ascomycete fungi (31). To verify whether Fam1 protein is present on WBs, we checked whether Fam1 colocalized with two WB marker proteins. For that purpose, we isolated C. orbiculare genes homologous to N. crassa HEX1 and WSC (Woronin body sorting complex), which encode proteins in the WB matrix and peripheral membrane, respectively. CoHEX1 comprises a 1,797-bp open reading frame, and the deduced amino acid sequence has high homology with N. crassa Hex1 (GenBank accession no. P87252), M. oryzae Hex1 (MoHex1) (GenBank accession no. Q2KGA0), and Aspergillus oryzae Hex1 (AoHex1) (I8TQ26) (see Fig. S5 in the supplemental material). CoWSC comprises an 816-bp open reading frame, and the deduced amino acid sequence has high homology with N. crassa Wsc (N4VZG1) (Fig. S5). Transformants expressing either CoHex1-RFP or CoWsc-RFP together with Fam1-GFP were then constructed. Microscopic analysis revealed that Fam1 colocalizes with CoHex1 on WBs at hyphal tips and near septa (Fig. 5A). In accordance with the Wsc localization pattern reported in N. crassa (24), CoWsc-RFP was detected on WBs and at the periphery of differentiating peroxisomes (Fig. 5B). Fam1-GFP colocalized with CoWsc-RFP on WBs situated near hyphal tips and septa and on both nascent and mature WBs in the cytoplasm of subapical cells (Fig. 5C). This suggests that in C. orbiculare new WBs are not formed exclusively in the apical cells of hyphae but also in subapical cells, in contrast to N. crassa. To examine the effect of Fam1 deletion on WB biogenesis, we examined the localization of CoHex1 in the fam1 mutant. CoHex1-RFP signals were uniformly distributed throughout the cytoplasm and were not concentrated near hyphal apices or septa (Fig. S6). Taken together, these results indicate that Fam1 is a WB-associated protein and is essential for WB formation.
FIG 5 .
Fam1 is localized in Woronin bodies. Images show vegetative hyphae viewed by differential interference contrast (DIC) microscopy, epifluorescence of GFP or RFP, merged GFP and RFP channels, and an overlay of all channels. (A) In hyphae coexpressing Fam1-GFP and CoHex1-RFP, Fam1-GFP colocalized with Woronin body (WB) matrix protein CoHex1 on WBs at hyphal tips (top panels) and septa (bottom panels). Bars, 4 µm. (B) In hyphae coexpressing GFP-PTS1 and CoWsc-RFP, the WB peripheral membrane protein CoWsc localized around the peroxisomal matrix (labeled by GFP-PTS1) and was enriched at sites where nascent WBs budded from mother peroxisomes (white arrows). Bar, 2 µm. (C) In hyphae coexpressing Fam1-GFP and CoWsc-RFP, both markers colocalized in WBs at hyphal tips (top panels), septa (middle panels), and in the cytoplasm of subapical cells (bottom panel, merge image). Bars, 2 µm.
Ultrastructural localization of Fam1 and Hex1 in Woronin bodies.
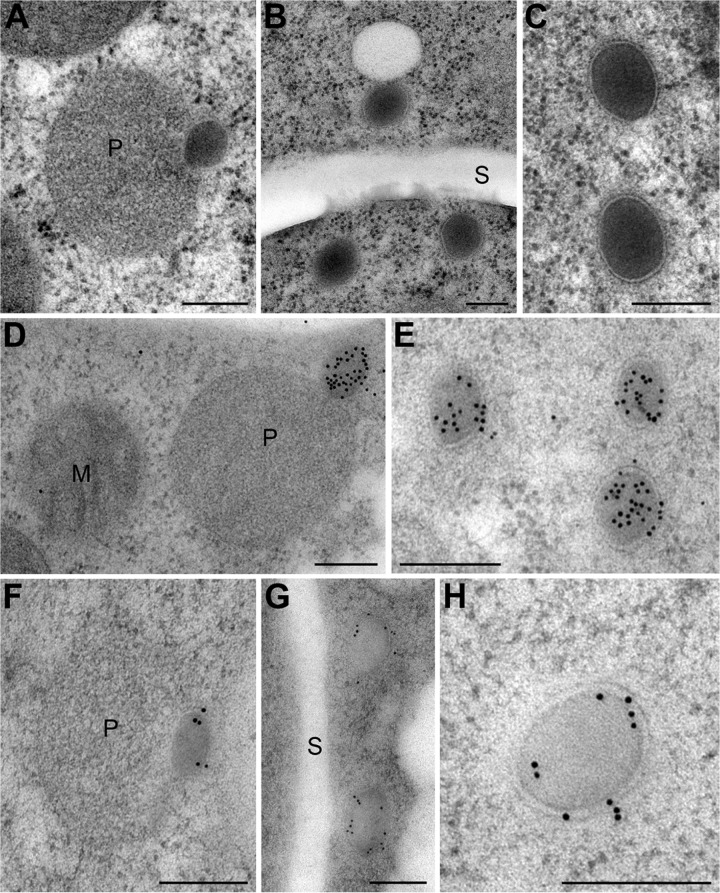
To localize Fam1 and Hex1 with greater resolution, we used immunoelectron microscopy. Thin sections of C. orbiculare vegetative hyphae were probed with either rabbit antibodies raised against recombinant Fam1 expressed in Escherichia coli or rabbit antibodies raised against N. crassa Hex1 (23). In wild-type hyphae, nascent WBs were observed budding from peroxisomes (Fig. 6A), while mature WBs were either found concentrated close to septa (Fig. 6B) or, less frequently, randomly distributed in the fungal cytoplasm (Fig. 6C). C. orbiculare WBs were on average 184 nm in diameter (n = 25; standard deviation [SD] = 37 nm), approximately fivefold smaller than those described from N. crassa (up to 1-µm diameter) (23), and with the KMnO4-uranyl acetate-lead citrate staining method (32) used here, the WB core appeared slightly more electron-opaque than the peroxisome matrix and had a homogeneous, fine-grained structure. The single membrane enclosing WBs appeared thicker than the peroxisome membrane and displayed a more clearly defined bilayer structure (Fig. 6C). Immunolabeling of wild-type hyphae showed that Hex1 was localized specifically in the dense core of nascent and mature WBs (Fig. 6D and E), while Fam1 signals were detected predominantly on the bounding membrane of nascent and mature WBs (Fig. 6F to H). However, Fam1 was barely detectable on the membranes of wild-type peroxisomes, including those to which budding WBs were attached (Fig. 6F). These immunoelectron microscopy data are consistent with those obtained from fluorescent protein tagging and confirm that Fam1 protein is predominantly located in WB membranes, rather than peroxisome membranes.
FIG 6 .
Immunoelectron microscopic localization of Fam1 in Woronin bodies. TEM images showing the ultrastructure of peroxisomes and Woronin bodies in vegetative hyphae of C. orbiculare (A to C) and the immunogold localization of Hex1 (D and E) and Fam1 (F to H). (A) Peroxisome (P) with nascent WB budding from it. (B) Mature WBs in the cytoplasm near a septum (S). (C) The single membrane of mature WBs is thicker than the peroxisome membrane and has a prominent bilayer structure. (D and E) Hex1 is localized in the dense matrix of nascent and mature WBs. (F to H) Fam1 is localized in the membranes of nascent WBs (F) and mature WBs (G and H) but is not detected on the peroxisome membrane. Bars, 200 nm.
Fam1 accumulates in peroxisome membranes of mutants lacking Woronin bodies.
In N. crassa, the WB matrix protein HEX1 is essential for biogenesis of the WB (23). Therefore, to investigate the destination of Fam1 in hyphae lacking WBs, we isolated cohex1 disrupted mutants (see Fig. S3 in the supplemental material). First, we verified the role of CoHex1 in WB function in C. orbiculare. In ascomycete fungi, WBs function to seal septal pores, thereby blocking cytoplasmic leakage when adjacent hyphal compartments are damaged (23, 33, 34). Using hypotonic shock to induce hyphal tip lysis (35), we found that cytoplasm was retained in subapical hyphal compartments of wild-type and cohex1+CoHEX1 complemented mutant strains (Fig. S7A). In contrast, cytoplasmic bleeding was observed in subapical compartments of cohex1 mutant hyphae (Fig. S7A). Nevertheless, the mycelial growth rate of the cohex1 mutant on PDA and OAM did not differ from that of the wild-type strain (Fig. S7B and C). Thus, CoHEX1 is required for WB function, but not peroxisome function.
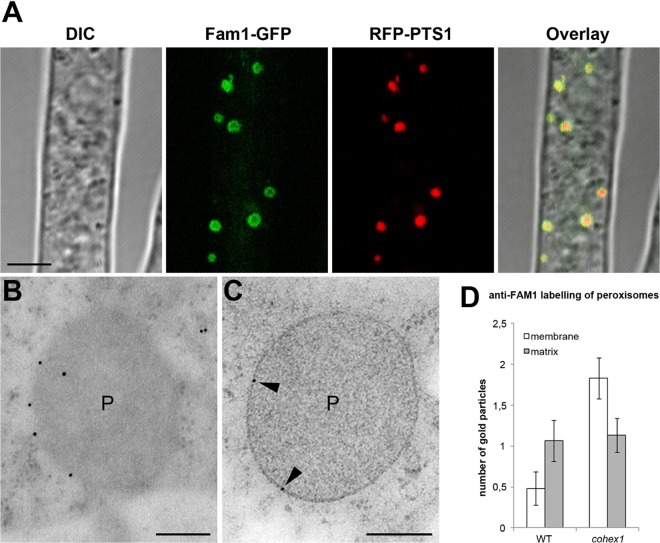
Next, we examined the subcellular localization of Fam1-GFP in the cohex1 mutant. The accumulation of Fam1-GFP on WBs near hyphal tips and septa was abolished in the absence of HEX1. Instead, most Fam1-GFP fluorescence was observed at the periphery of all peroxisomes (Fig. 7A), in striking contrast to the WB localization seen in wild-type hyphae. Consistent with this, using immunoelectron microscopy, Fam1 signals were detected on peroxisome membranes in hyphae of the cohex1 mutant (Fig. 7B and C), but almost never on those of wild-type hyphae (Fig. 6F). To verify this observation, we quantified the density of immunogold labeling on the peroxisome membrane. In wild-type hyphae, the mean number of gold particles located on the peroxisome membrane was 0.48 (n = 29; standard error [SE] = 0.2), compared to 1.83 (n = 30; SE = 0.25) in the cohex1 mutant (Fig. 7D). This nearly fourfold difference was significant in an unpaired t test (P < 0.0001). These results indicate that in the WB-deficient cohex1 mutant, Fam1 is redirected to the peroxisome membrane. Furthermore, our data suggest that translocation of Fam1 from peroxisome to WB membranes depends on the presence of CoHex1, whereas transport to the peroxisome does not.
FIG 7 .
Subcellular localization of Fam1 in Woronin body-deficient cohex1 mutants. (A) In vegetative hyphae of the cohex1 mutant coexpressing Fam1-GFP and RFP-PTS1, Fam1-GFP localized to the periphery of peroxisomes. Confocal images show differential interference contrast (DIC), GFP or RFP fluorescence, merged GFP and RFP channels, and an overlay of all channels. Bar, 5 µm. (B and C) TEM and immunogold labeling with anti-Fam1 antibodies detect Fam1 on the peroxisome membrane (black arrowheads) of the cohex1 mutant. Bars, 200 nm. (D) Quantification of immunogold labeling by anti-Fam1 antibodies. The histogram shows mean numbers of gold particles located on the membrane or matrix of wild-type (WT) peroxisomes (n = 29) and peroxisomes of the cohex1 mutant (n = 30). Each error bar depicts 1 standard error.
Subcellular fractionation confirms the association of Fam1 with Woronin bodies.
To obtain independent verification of the association of Fam1 with WBs, we used equilibrium density gradient centrifugation to fractionate organelles obtained from wild-type and cohex1 mutant hyphae expressing GFP-PTS1. In the wild-type strain, Fam1 largely cofractionated with CoHex1 in the densest region of the gradient, which contains only a minor part of the total protein, and partly overlapped with the peroxisomal matrix marker GFP-PTS1 (see Fig. S8 in the supplemental material). In contrast, in the cohex1 mutant, Fam1 largely cofractionated with the peroxisomal marker. Thus, the results from subcellular fractionation are fully consistent with the data from both fluorescent protein tagging and immunoelectron microscopy, confirming that Fam1 is a WB-associated protein.
HEX1 is not required for infection-related morphogenesis and pathogenesis in C. orbiculare.
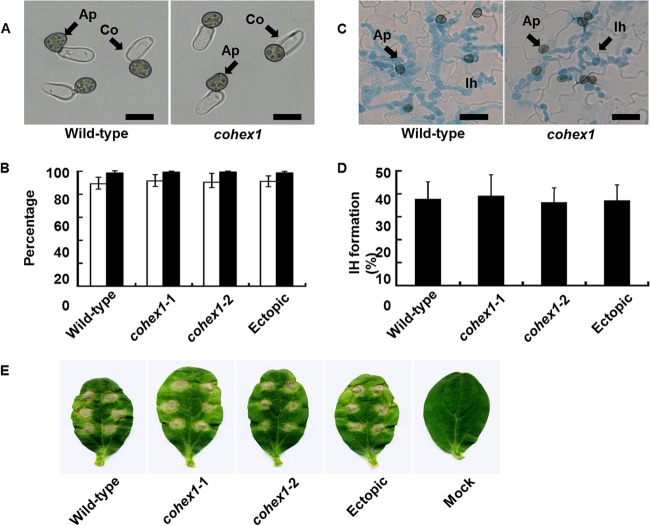
The WB was previously implicated in pathogenicity of the rice blast fungus, M. oryzae, and the causal agent of Fusarium head blight on wheat and barley, Fusarium graminearum (34, 35). To investigate the role of WBs in C. orbiculare pathogenicity, we tested the ability of cohex1 mutants to undergo infection-related morphogenesis and invade cucumber plants. The mutant showed no detectable defects in appressorium development in vitro (Fig. 8A and B). Moreover, cohex1 appressoria penetrated cucumber leaves at a frequency similar to that of the wild type (Fig. 8C and D), and the mutant produced necrotic lesions on cucumber leaves with severity similar to that of the wild type (Fig. 8E). We therefore conclude that CoHEX1 and WBs are dispensable for C. orbiculare pathogenesis.
FIG 8 .
Appressorium formation and pathogenicity of cohex1 mutants. (A) Microscopic observation of appressorium formation in vitro. Conidial suspensions of the C. orbiculare wild-type strain and cohex1 mutants were incubated on glass slides for 24 h. Ap, Appressoria; Co, conidia. Bars = 10 µm. (B) Percentage of conidia forming appressoria. A total of at least 200 conidia were scored for each genotype, based on the examination of three replicates. The wild-type strain, cohex-1-1 and cohex-1-2 CoHEX1 disruption mutants, and CoHEX1 ectopic transformant were tested. Means ± standard deviations (error bars) were calculated from three independent experiments. (C) Microscopic observation of host infection. Conidial suspensions of the wild-type strain and cohex1 mutant were inoculated on the abaxial surface of cucumber cotyledons and viewed after 72 h. Ap, appressoria on the leaf surface; Ih, infection hyphae inside epidermal cells stained with aniline blue. Bars, 10 µm. (D) Percentage of appressoria penetrating to form visible infection hyphae. A total of at least 200 appressoria were scored for each sample, based on the examination of three replicates. The wild-type strain, cohex-1-1 and cohex-1-2 CoHEX1 disruption mutants, and CoHEX1 ectopic transformant were tested. Means plus standard deviations were calculated from three independent experiments. (E) Pathogenicity assay of cohex1 mutants on intact cucumber cotyledons. Droplets of conidial suspension were inoculated onto detached cucumber cotyledons and incubated for 7 days. The wild-type strain, cohex-1-1 and cohex-1-2 CoHEX1 disruption mutants, and CoHEX1 ectopic transformant, were tested. Distilled water was used as a control (mock).
DISCUSSION
Fam1 is a Pex22-like peroxin unique to filamentous ascomycetes.
A previous analysis of the whole-genome sequences of 17 fungal species revealed that most peroxins are conserved between yeast and filamentous fungi, but among those involved in the import of peroxisome matrix proteins, the membrane anchors Pex22 and Pex15/Pex26 are much less well-conserved (15). These authors suggested that Pex22 homologs could be identified in filamentous fungi by the presence of a Pex4-binding site, which they used to define a class of Pex22-like proteins. In our study, we identified the FAM1 gene encoding a Pex22-like protein in a screen for C. orbiculare mutants impaired in fatty acid metabolism. Database searches showed that FAM1 homologs are restricted to filamentous ascomycete fungi, being absent from both basidiomycetes and ascomycete yeasts. Overall, the predicted sequence of the Fam1 protein shows very limited identity (5%) to the S. cerevisiae Pex22 sequence, with only a few conserved amino acid residues that are likely involved in interface formation with Pex4 (19), and the protein is nearly two times larger than yeast Pex22. It was therefore unclear whether Fam1 really performs a function similar to that of Pex22. Here we showed that a yeast pex22 mutant can be fully complemented by FAM1 expressed in combination with CoPEX4, demonstrating that FAM1 is a functional ortholog of PEX22. Moreover, similar to yeast pex22 mutants, the C. orbiculare fam1 mutant was deficient in peroxisome biogenesis and the import of peroxisomal matrix proteins. In S. cerevisiae, Pex22 is an integral membrane protein involved in receptor recycling that anchors Pex4 to the cytosolic face of the peroxisomal membrane (30). Fam1 contains no predicted binding site for the peroxisomal membrane protein import receptor PEX19, but it has a putative targeting signal comprising an N-terminal transmembrane domain immediately preceded by a stretch of positively charged amino acids (see Fig. S2 in the supplemental material), similar to Pichia pastoris Pex22 (29).
Fam1 is a novel Woronin body-associated peroxin.
After demonstrating that FAM1 is a functional ortholog of the PEX22 peroxin, we had expected Fam1 to be located on peroxisome membranes, as in other organisms (30). Surprisingly, colocalization experiments with FP-tagged markers of the peroxisome matrix, WB matrix, and WB membranes showed that Fam1 is predominantly associated with WBs, rather than with peroxisome membranes. Moreover, a subset of the punctate organelles labeled by Fam1-GFP were concentrated near hyphal apices and septa, which is the typical distribution of WBs in filamentous ascomycetes (25, 31). The localization of Fam1 on WBs was further verified by immunoelectron microscopy, which showed that Fam1 is located predominantly on the bounding membrane of WBs. The WB association of Fam1 was further supported by cellular fractionation experiments. Thus, all our cytological and biochemical data point to Fam1 being a WB-associated protein.
Fam1 was present on the membranes around nascent (budding) WBs attached to parent peroxisomes, as well as mature (released) WBs, but it was not detected on peroxisome membranes, including those producing nascent WBs. Fam1 therefore represents a novel component of the WB membrane, although we cannot exclude the possibility that some Fam1 protein is also present in peroxisome membranes at levels below the detection limits of confocal and immunoelectron microscopy. To date, only three other proteins were shown to be associated with WB membranes, namely, the WB sorting complex protein WSC (24), the WB tethering protein Leashin (25), and TmpL, a transmembrane protein implicated in redox homeostasis (36). Thus, Fam1 is the only WB-associated protein involved in peroxisome biogenesis.
One plausible explanation for the localization of Fam1 is that after insertion into the peroxisome membrane, the protein becomes enriched in WB membranes via interaction with WB matrix or membrane proteins such as Hex1 or WSC, respectively. In N. crassa, the membrane-anchored peroxin Pex26, which like Pex22 functions in receptor recycling, is concentrated in the membranes of a subset of peroxisomes producing WBs (26). Low levels of Pex26 were also detected in WB membranes, but unlike Fam1, Pex26 did not become enriched in WB membranes (26). Pex26 was shown to physically interact with Hex1, and deletion of Hex1 resulted in the uniform targeting of Pex26 to all peroxisomes, suggesting that Hex1 directly influences Pex26 localization. In this way, Pex26 was proposed to amplify the import of additional Hex1 via a positive-feedback loop (26). We found that similar to Pex26, in the absence of CoHex1 and WBs, Fam1 was uniformly relocated to peroxisome membranes in C. orbiculare, suggesting that the subcellular localization of Fam1 depends on CoHex1.
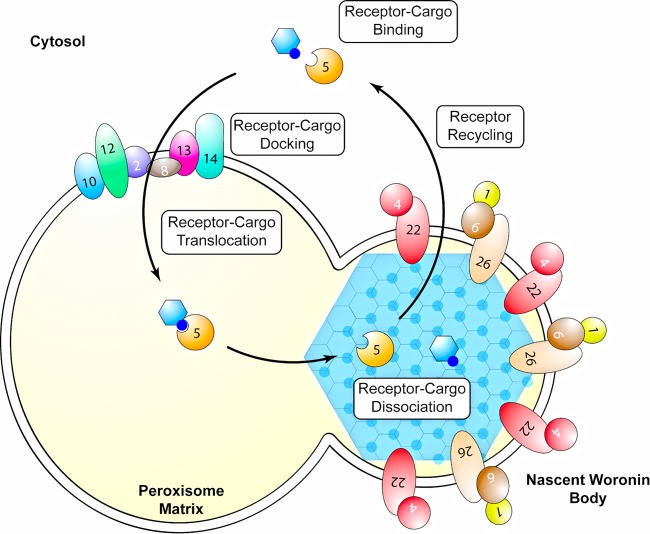
Although our results indicate that Fam1 functions like Pex22 in the import of peroxisome matrix proteins, the protein was exclusively localized in WB membranes. It is surprising that a protein required for crucial functions in peroxisomes is efficiently sorted into WBs, and this finding raises the possibility that receptor recycling is at least partly compartmentalized in WBs, perhaps before their separation from parent peroxisomes. Very high levels of Hex1 oligomer enter peroxisomes via the PTS1 import pathway and then become sequestered into WBs (26). If dissociation of the receptor-cargo complex occurs after the concentration of Hex1 into nascent WBs, it is conceivable that the release of the PTS1 receptor Pex5p back into the cytosol occurs preferentially on the surfaces of WBs. An implication of this model would be that receptor-cargo docking and translocation are spatially separated from cargo dissociation and receptor recycling in C. orbiculare, and perhaps other filamentous ascomycetes, as shown schematically in Fig. 9. Two proteins mediating earlier steps of the PTS1 import pathway, Pex13 and Pex14, are known to be absent from mature WBs (37). However, there is no information on whether components of the receptor recycling machinery, such as Pex1, Pex2, Pex4, Pex6, Pex10, and Pex12, are present in WBs. Since the cytosolic ubiquitin-conjugating enzyme Pex4 is anchored to peroxisome membranes via Pex22 (19), it would be particularly interesting to determine whether Pex4 is enriched on the surfaces of C. orbiculare WBs together with Fam1. An alternative possibility is that sequestration of Fam1 in WB membranes provides a mechanism to downregulate levels of the protein in peroxisome membranes, with every cycle of WB budding depleting more Fam1 protein. A third possibility is that Fam1 has evolved additional roles specific to the biogenesis or function of WBs that are unrelated to receptor recycling. The fact that WBs and Pex22-like proteins such as Fam1 show the same phylogenetic distribution among filamentous ascomycetes is consistent with this hypothesis.
FIG 9 .
Proposed model for the import of Woronin body matrix proteins and receptor recycling in C. orbiculare. Woronin body matrix protein Hex1 binds the Pex5 receptor in the cytosol via its C-terminal peroxisome targeting signal (PTS1). The Pex5-cargo complex first interacts with docking proteins Pex13 and Pex14 on the peroxisome surface and then translocates into the peroxisome matrix, mediated by Pex2, Pex8, Pex10, and Pex12. Dissociation of the Pex5 receptor from Hex1 occurs after sequestration of the complex into the nascent Woronin body (WB), driven by the self-assembly of Hex1. Recycling of Pex5 back to the cytosol occurs preferentially on the WB membrane, mediated by FAM1 (i.e., Pex22), Pex4, Pex26, Pex1, and Pex6.
In copex6 and copex13 mutants, which are defective in the import of peroxisome matrix proteins (10, 22), the localization of Fam1 on WBs at hyphal tips and septa was abolished, indicating that peroxisome function is required for WB formation in C. orbiculare, as in other fungi (37). Interestingly, in copex6 and copex13 mutants, Fam1-GFP localized to punctate structures that were smaller and less sharply defined than peroxisomes or WBs and not associated with hyphal tips or septa. These structures may represent empty peroxisomal membrane remnants, sometimes termed “peroxisome ghosts,” which are characteristic of mutants lacking components of the matrix protein import machinery in yeast and other organisms (38).
Fam1, but not Hex1, is essential for C. orbiculare pathogenicity.
It was previously reported that homologs of S. cerevisiae PEX6 and PEX13 are essential for C. orbiculare pathogenicity (10, 22). The fam1 mutant was defective in the import of peroxisomal matrix proteins, which would impact multiple appressorial functions depending on peroxisome activity, notably the biosynthesis of 1,8-dihydroxynaphthalene-derived melanin, the mobilization of storage lipids, and turgor generation (6). Similar to copex6 and copex13 mutants, the fam1 mutant was impaired in all these activities, which are known to be essential for appressorium-mediated mechanical penetration of plant surfaces (10). This probably accounts for the complete loss of pathogenicity in the fam1 mutant.
The self-assembling WB matrix protein HEX1 is essential for WB biogenesis in N. crassa and other fungi (39). As expected, deletion of HEX1 in C. orbiculare caused a defect in WB-mediated septal plugging upon hyphal rupture, and WBs were not visible in the cytoplasm of cohex1 mutants by transmission electron microscopy (TEM). Similar to other fungi (40), CoHEX1 was not essential for fatty acid metabolism because cohex1 mutants grew normally on oleic acid-containing medium. Moreover, the peroxisomes of cohex1 mutants retained the ability to import peroxisomal matrix proteins, showing that CoHEX1 is dispensable for peroxisome biogenesis. Interestingly, cohex1 mutants were not impaired in either appressorial function or pathogenicity. This contrasts with the situation in the rice blast fungus, M. oryzae, where deletion of HEX1 showed that WBs are required not only for appressorial morphogenesis and function but also for invasive growth in planta by enhancing resistance to nutritional stress (40). The WB-associated transmembrane protein TmpL, required for generating intracellular reactive oxygen species (ROS) and resistance to exogenous ROS, was also found to be essential for host infection by the plant pathogen Alternaria brassicicola and the human pathogen Aspergillus fumigatus (36). Overall, our data suggest that, unlike these other ascomycete fungi, CoHEX1 and WBs are dispensable for C. orbiculare pathogenicity.
Perspectives.
In this study, we present evidence that filamentous ascomycete fungi have evolved a novel peroxin that functions like Pex22 in receptor recycling during matrix protein import. In C. orbiculare, the Pex22-like protein Fam1 is essential for pathogenicity on cucumber due to the key role played by peroxisomes in multiple aspects of appressorium-mediated host penetration. Notably, we show that Fam1 is specifically enriched in WB membranes, making it the only WB-associated protein so far identified that is required for peroxisome biogenesis. In the future, it will be important to determine whether Pex22-like proteins from other filamentous ascomycetes are similarly targeted to the WB membrane and whether other components of the receptor recycling machinery become sequestered into WBs. Further work is now needed to determine why Fam1 is targeted to WBs, given that this organelle is not required for peroxisome biogenesis per se.
MATERIALS AND METHODS
Fungal and bacterial strains.
Strain 104-T (MAFF240422) of Colletotrichum orbiculare (syn. C. lagenarium) was used as the wild-type strain. Culture conditions and methods for Agrobacterium-mediated transformation were described previously (22, 27).
Cloning and sequence analysis of FAM1, CoHEX1, and CoPEX4.
Fungal genomic DNA flanking the T-DNA insert was rescued using the thermal asymmetrical interlaced PCR (TAIL-PCR) protocol and sequenced as described previously (27). Genomic clones of FAM1, CoHEX1, and CoPEX4 genes were isolated from the cosmid library of C. orbiculare by PCR using primer pairs CoHEX1-S2 and CoHEX1-AS2, CoHEX1-S2 and CoHEX1-AS2, and PEX4-S2 and PEX4-AS2, respectively (see Table S1 in the supplemental material). The obtained cosmid clones containing each gene were sequenced to obtain the entire open reading frame (ORF).
Construction of GFP or RFP fusion plasmids.
For GFP labeling of peroxisomes, pEGFPPTS1 and pPTS2EGFP plasmids were used as previously described (10). For RFP labeling of peroxisomes, the full-length monomeric red fluorescent protein gene (mRFP1) was amplified by PCR with the primers mRFP1S1 and PTS1RFP-AS. The amplified product was digested with SpeI and BamHI and introduced into pBI-SCD1pGFP-TUB1S as previously described (42), and the resulting plasmid was named pBI-RFPPTS1S. For the FAM1-GFP fusion gene, we added the enhanced green fluorescent protein (EGFP) to the N terminus of FAM1. The SalI fragment containing the bialaphos resistance gene from pCB1635 was introduced into the XhoI site of pGreenII0000 (43) and designated pGr2B1. The EGFP ORF encoding glycine residues in the N-terminal region and the terminator of the glucoamylase gene of Aspergillus awamori (44) were amplified by PCR with primer EGFPSGly. The amplified product was digested with HindIII and SalI, introduced into pGr2B1, and the resulting plasmid was designated pGr2B1GlyGFP. The BamHI-KpnI fragment containing EGFP and the bialaphos resistance gene from pGr2B1GlyGFP was introduced into pBIG4MRH and designated pBI-BglyGFP. The 1,030-bp 5′ region upstream of FAM1 together with the full-length ORF of FAM1 without the stop codon was amplified by PCR with primers FAM1pro-S1 and FAM1deS-AS. The amplified product was digested with BamHI and introduced into pBI-BglyGFP. The resulting clone was named pBI-FAM1-GFPB. For RFP labeling of WBs, the CoHEX1-mRFP1 fusion gene was constructed. mRFP1 containing a PTS1 (SRL) sequence was amplified by PCR with primers glyRFP-S and RFPsrl-AS. The amplified product was digested with BamHI and EcoRI, and the α-TUB1 gene in pBI-SCD1pGFP-TUB1S was replaced with the EcoRI-BamHI fragment, and the resulting plasmid was named pBI-SCD1pGFPRFPsrlS. A 3.5-kb fragment containing the 5′ flanking region and CoHEX1 ORF without the stop codon and the PTS1 sequence was amplified by PCR with T3 and HEX1dsrl-AS and digested with XbaI and EcoRI. The short SCD1 promoter and GFP ORF in pBI-SCD1pGFPRFPsrlS were replaced with the XbaI and EcoRI fragment. The resulting clone was named pBI-HEX1-RFPsrlS. For constructing the CoWsc-RFP fusion gene, the mRFP1 ORF containing five additional glycine residues in the N-terminal region were amplified by PCR with primers glyRFP-S and mRFP1AS1. The amplified product was digested with EcoRI and BamHI and introduced into pBI-SCD1pGFPS (42) and named pBI-SCD1pGFP-RFPS. The 981-bp 5′ upstream region of the CoWSC gene with the full-length ORF of CoWSC without the stop codon was amplified by PCR with CoWSC-S1-Sp and CoWSC-AS1-EI. The amplified product was digested with SpeI and EcoRI. The short SCD1 promoter and GFP ORF in pBI-SCD1pGFP-RFPS were replaced with the SpeI and EcoRI fragment. The resulting clone was named pBI-CoWSC-RFPS.
Plasmid constructs for gene disruption and complementation in C. orbiculare.
To construct the gene replacement vector pFAM1AH3-2, a cosmid clone containing pFAM1cos was subjected to transposon mutagenesis using the EZ::TN transposase (Epicenter) and transposon (AH3 fragment) in which the kanamycin resistance gene was replaced with the ampicillin resistance and hygromycin resistance genes. After PCR and sequence analysis, a cosmid clone (pFAM1cos1AH3-2) was selected in which the transposon was inserted 541 bp downstream of the predicted start codon of FAM1. Next, a 4.7-kb PCR product, including FAM1 together with its 5′ and 3′ flanking regions and the transposon was amplified from pFAM1cos1AH3-2 with primer pair FAM1S4/AS4 and introduced into the BamHI site of the Agrobacterium tumefaciens binary vector pBIG4MRB, and named pFAM1H3-2. The gene complementation vector pFAM1C was amplified from pFAM1os1 with primer pair FAM1proS1 containing a terminal BamHI site and Hi2049-AS4 containing a terminal BamHI site. The PCR product was cloned into the BamHI site of binary vector pBIG4MRSrev containing the sulfonylurea resistance gene. To construct the CoHEX1 gene replacement vector pBI-HEX1AH3-2-7S, the pCBHEX1E plasmid was constructed by introducing a 9.0-kb EcoRI genomic fragment from a cosmid clone containing CoHEX1 into the EcoRI site of pCB1004. The pCBHEX1E plasmid was digested with BamHI, and self-ligation produced a 0.8-kb fragment containing CoHEX1, named pCBHEX1EB. The EcoRI-and-BamHI fragment from pCBHEX1EB was introduced into the pBIG4MRSrev EcoRI and BamHI site and named pBI-HEX1S. The pBI-HEX1S plasmid was used as the gene complementation vector for the cohex1 mutant. The pBI-HEX1S plasmid was subjected to mutagenesis with the EZ::TN transposon system as described above, and after PCR and sequence analysis, a clone in which the transposon was inserted 104 bp downstream of the predicted start codon of HEX1 was selected and named pBI-HEX1AH3-2-7S.
Yeast complementation assay.
For complementation of the yeast Δpex22 mutant, the full-length cDNA sequence of FAM1 was amplified from C. orbiculare mycelial cDNA with primers FAM1yesS1 and CoFAM1BamAS1. The amplified product was cloned into pTGPD containing the GPD promoter and TRP1 as a selection marker. For complementation of the yeast Δpex4 mutant, the full-length cDNA sequence of CoPEX4 was amplified from C. orbiculare mycelial cDNA with primers CoPEX4f-S2 and CoPEX4f-AS2. The amplified product was cloned into pRS-GPD and named pRS-PEX4. The plasmid pRS-PEX4 contains CoPEX4 under control of the GPD promoter and LEU2 as a selection marker. S. cerevisiae wild-type strain JD53 and Δpex4 and Δpex22 mutants (45) were transformed with pEW88 which contains GFP-PTS1 and URA3 (46) and with combinations of two plasmids as follows: pTGPD and pRS-GPD, pTGPD-FAM1 and pRS-GPD, pTGPD and pRS-PEX4, and pTGPD-FAM1 and pRS-PEX4. Transformants were selected on SD medium lacking Leu, Trp, and uracil (42). GFP-PTS1 fluorescence was observed in yeast grown on 0.3% glucose overnight.
Preparation of Fam1 antibodies.
Polyclonal antisera against Fam1 were obtained by immunizing rabbits with recombinant Fam1 purified from Escherichia coli DH5α carrying a FAM1 expression vector as previously described (27). The single 44-kDa band corresponding to Fam1 was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the eluted Fam1 protein was used for immunizing rabbits (SCRUM, Inc., Japan). The immunoglobulin G (IgG) fraction was purified with an Hi-Trap protein A affinity column according to the manufacturer’s instructions (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom).
Cellular fractionation and Western blotting.
Cellular fractionation and Western blot analysis were performed by following the procedures of Liu et al. (24). Mycelium grown in potato sucrose medium for 5 days was ground to a powder in liquid nitrogen using a mortar and pestle and used for cellular fractionation. Protein distribution in the fractionated samples was determined by enhanced chemiluminescence (ECL) and Western blotting (Millipore, Billerica, MA, USA). Primary antibodies were obtained from as follows: anti-HEX (23), anti-GFP (Clontech Laboratories, Inc., CA, USA), and anti-Fam1 (this study). Secondary antibodies were alkaline phosphatase-conjugated anti-rabbit IgG (H+L) (Jackson Immuno Research Laboratories, PA, USA).
Transmission electron microscopy and immunocytochemistry.
For high-pressure freezing, the mycelium of C. orbiculare was grown on potato dextrose agar (PDA) for 3 days. Using a biopsy punch, 2-mm-diameter discs of mycelium were cut from the colony margins, and most of the agar was cut away to produce ~0.3-mm-thick samples. Aluminum specimen carriers (3-mm diameter, 0.5-mm thickness; Leica Microsystems GmbH, Germany) were prefilled with potato dextrose broth, and mycelial discs were placed in the cavity, mycelium side downwards. A second specimen carrier was dipped in 1-hexadecene and used as a lid (flat side toward the sample), and the assembly was immediately frozen using a Leica EM HPM 100 high-pressure freezer (Leica Microsystems GmbH). Samples were freeze-substituted in acetone containing 0.5% (wt/vol) uranyl acetate as described previously (47), rinsed in ethanol, and infiltrated with medium-grade LR White resin for 8 days at −20°C. Polymerization with UV light was for 24 h at −20°C and 24 h at 0°C. After blocking, ultrathin sections were incubated at 4°C overnight in the purified IgG fraction of rabbit polyclonal anti-FAM1 antiserum (diluted 1:50) or anti-HEX1 antiserum (diluted 1:500) raised against N. crassa Hex1p (23). Primary antibodies were detected with goat anti-rabbit antibody conjugated to 10-nm colloidal gold particles (British Biocell International, Cardiff, United Kingdom) for 1 h. For a negative control, the primary antibody was omitted. Sections were stained with 0.1% (wt/vol) potassium permanganate in 0.1 N sulfuric acid for 1 min (32) followed by 2% (wt/vol) uranyl acetate for 10 min and lead citrate for 15 min and examined with a Hitachi H-7650 transmission electron microscope (TEM) at 100 kV.
Fluorescence microscopy.
For appressorium formation in vitro, conidia were harvested from 7-day-old cultures on PDA and suspended in distilled water. The conidial suspension, adjusted to 1 × 105 conidia per ml, was placed in the wells of 8-well multitest glass slides (ICN Biomedicals, Aurora, OH, USA) and incubated at 24°C for 24 h. Germlings were observed with a Nikon Eclipse E600 microscope with differential interference contrast (DIC) optics. Lipid droplets were visualized by staining with Nile red (48). Nile red fluorescence was viewed with the Nikon B-2A filter set (450- to 490-nm-wavelength excitation filter, 505-nm-wavelength dichroic mirror, and 520-nm-wavelength barrier filter). For observation of GFP fluorescence, cells were viewed with the Nikon GFP(R)-BP filter set (460- to 500-nm or 595- to 620-nm excitation filter, 400-nm dichroic mirror, and 400-nm barrier filter). Confocal images were obtained with Leica TCS SP2 or Zeiss LSM 700 confocal scanning microscopes. Excitation for imaging GFP fluorescence used the 488-nm laser line, and emission was detected at 492 to 550 nm. For imaging mRFP, excitation was at 563 nm (Leica) or 555 nm (Zeiss), and emission was detected at 566 to 620 nm (Leica) or 557 to 600 nm (Zeiss).
Hyphal tip lysis experiments.
Hypotonic shock was used to induce hyphal tip lysis. C. orbiculare mycelia were grown on Marthur's agar medium (0.12% MgSO4 × 7H2O, 0.27% KH2PO4, 0.28% glucose, and 0.22% Peptone) containing 2% sorbose at 24°C. After 2 or 3 days, distilled water was added to the fungal colony to induce hyphal tip lysis, and the hyphae were then observed using light microscopy.
Pathogenicity tests.
Conidia of C. orbiculare were collected from 7-day-old cultures grown on PDA at 24°C under constant darkness. The pathogenicity of each strain was tested by droplet inoculation on cucumber cotyledons (Cucumis sativus L. ‘Suyo’) as previously described (49).
Accession numbers.
The isolated genes have the following GenBank accession numbers, based on the recently published C. orbiculare genome annotation (42). FAM1, ENH87037.1 (N4VSY8); CoPEX4, ENH88331.1 (N4W4I0); CoHEX1, ENH76003.1 (N4UJP9); CoWSC, ENH86471.1 (N4VZG1).
SUPPLEMENTAL MATERIAL
Isolation of C. orbiculare mutant deficient in fatty acid metabolism. (A) Colonies of the wild-type (WT) and Hi2049 mutant strain grown on PDA and OAM for 5 and 12 days, respectively. (B) Pathogenicity of the C. orbiculare wild-type strain and the Hi2049 mutant strain on detached cucumber cotyledons. The leaves were inoculated with droplets of conidial suspension and incubated for 7 days. The right half of the leaves was intact, while the left half was wounded by toothpick scratching. Download
Protein sequence alignment of C. orbiculare Fam1 with S. cerevisiae Pex22 and Pex22-like proteins from filamentous ascomycetes. Amino acid sequence of C. orbiculare Fam1 (N4VSY8) was aligned with Pex22-like proteins of M. oryzae (UniProtKB accession no. A4R2Z0), N. crassa (UniProtKB accession no. Q8WZV7), A. oryzae (UniProtKB accession no. Q2UQZ8), and S. cerevisiae Pex22 (PEX22 YAL055W) using the Clustal W program. Identical and similar amino acids are indicated. Residues involved in interface formation between Pex22 and Pex4 in S. cerevisiae (19) are underlined with a red line. Residues comprising a putative N-terminal transmembrane domain are indicated by a blue line above the sequences. Positively charged residues (PC) immediately upstream of the transmembrane domain (29) are highlighted in green. Download
Targeted gene disruption of C. orbiculare FAM1 and CoHEX1. (A) Schematic representation of the FAM1 gene disruption by homologous recombination. Binary plasmid pBIFAM1AH3-2 contains the AMP::HPH (ampicillin-hygromycin resistance) cassette inserted into the FAM1 gene between the left-border (LB) and right-border (RB) sequences. Through double crossing over, a 3.9-kb XhoI fragment of the wild-type strain is predicted to be replaced by a 6.6-kb XhoI fragment containing the AMP::HPH cassette. (B) DNA blot analysis of FAM1 disrupted transformants. Genomic DNA was digested with XhoI and probed with an internal fragment of FAM1 indicated in the pBIFAM1AH3-2 vector map. The wild-type (WT) strain, fam1-1, fam1-2, and fam1-3 FAM1 disruption mutants, FAM1 ectopic transformant, and FAM1 complemented transformant of the FAM1 mutant (FAM1-C) were studied. (C) Schematic representation of the CoHEX1 gene disruption by homologous recombination. The binary plasmid pBII-HEX1AH3-2-7 contains the AMP::HPH cassette inserted into the CoHEX1 gene between the left-border (LB) and right-border (RB) sequences. Through double crossing over, an ~9.0-kb BamHI fragment of the wild type is predicted to be replaced by a 12-kb BamHI fragment containing the AMP::HPH cassette. (D) DNA blot analysis of CoHEX1 disrupted transformants. Genomic DNA was digested with XhoI and probed with an internal fragment of CoHEX1 indicated in the pBII-HEX1AH3-2-7 vector map. The wild-type (WT) strain, cohex1-1, cohex1-2, and cohex1-3 CoHEX1 disruption mutants, and CoHEX1 ectopic transformant were examined. Download
Subcellular localization of Fam1 in peroxisome-deficient mutants. Fam1-GFP was expressed in vegetative hyphae of peroxisome-deficient copex13 and copex6 mutants growing on PDA. Confocal microscopy showed Fam1-GFP neither localized at hyphal tips nor septa in these peroxisome mutants. Numerous small punctate structures labeled by Fam1-GFP were randomly distributed through the cytoplasm. Images represent overlay projections of bright-field and GFP fluorescence channels. Bars, 5 µm. Download
Alignment of C. orbiculare CoHex1, CoWsc, and CoPeX4 with homologs of other ascomycete fungi. (A) The amino acid sequence of C. orbiculare CoHex1 (N4UJP9) was aligned with the amino acid sequences of N. crassa Hex1 (P87252), A. oryzae AoHex (I8TQ26), and M. oryzae MgHex (Q2KGA0) using the Clustal W program. (B) Amino acid sequence of C. orbiculare CoWsc (N4VZG1) was aligned with the amino acid sequence of N. crassa Wsc (U9W802) using the Clustal W program. (C) Alignment of C. orbiculare CoPex4 with S. cerevisiae Pex4. Amino acid sequence of C. orbiculare CoPex4 (N4W4I0) was aligned with Colletotrichum graminicola Pex4 (E3Q679), Colletotrichum higginsianum Pex4 (H1UZJ1), and S. cerevisiae Pex4 (C7GYG2) using the Clustal W program. Shading of residues represents percentage amino acid conservation: black, 100%; dark grey, 75%; light grey, 50%. Download
Subcellular localization of CoHex1 in the fam1 mutant. CoHex1-RFP was expressed in vegetative hyphae of the peroxisome-deficient fam1 mutant. CoHex1-RFP was not localized at hyphal tips or at septa, but instead it was uniformly distributed throughout the cytoplasm. Images represent overlay projections of bright-field and GFP fluorescence channels. Bars, 5 µm. Download
The cohex1 mutant is defective in WB function. (A) Bright-field microscopy images showing the blockage of cytosolic bleeding from subapical cells following hyphal tip rupture. When distilled water was placed onto hyphae growing on Mathur's agar medium containing 2% sorbose, hyphal tips swelled and burst. The cytoplasm leaked from subapical cells of the cohex1 mutant but was retained in subapical cells of the wild-type and cohex1+CoHex1 complemented strain. Arrows indicate the first septum behind the hyphal tip. Bars, 3 µm. (B) Mycelial radial growth of the C. orbiculare wild-type strain, fam1 mutant, and cohex1 mutants after 12 days on OAM. (C) Radial growth rate of the wild type and cohex1 mutant on PDA or OAM. The increase in colony diameters was measured between 3 and 7 days on PDA and between 4 and 14 days on OAM. The mean and standard deviation were calculated from six colonies of each strain. Download
Fam1 cofractionates with WB marker proteins after separation of organelles. Fungal organelles were separated by centrifugation on a 17 to 60% Nycodenz density gradient, and fractions were tested for the presence of Fam1 and CoHex1 by Western blot analysis. The graph shows the shape of the gradient and total protein distribution across the gradient. PTS1-GFP provided a marker of the peroxisome matrix, detected with anti-GFP antibodies. Download
Primers used in this study
ACKNOWLEDGMENTS
We are grateful to Elmon Schmelzer for expert assistance with confocal microscopy. We thank Gregory Jedd for the supply of Hex1 antibody, Nils Johnson for S. cerevisiae pex4 and pex22 mutants, Bonnie Bartel for yeast transformation plasmids, Jan Kiel for the information on Pex22-like peroxin, and Yoshitaka Takano for the copex6 mutant.
This work was supported by Grants-in-Aid for Scientific Research (19380029, 24248009, and 15H05780) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Max Planck Society, Germany. G. P. Robin and R. O'Connell are supported by funding from the Agence Nationale de la Recherche (grant number ANR-12-CHEX-0008-01).
Footnotes
Citation Kubo Y, Fujihara N, Harata K, Neumann U, Robin GP, O’Connell R. 2015. Colletotrichum orbiculare FAM1 encodes a novel Woronin body-associated Pex22 peroxin required for appressorium-mediated plant infection. mBio 6(5):e01305-15. doi:10.1128/mBio.01305-15.
REFERENCES
- 1.Smith JJ, Aitchison JD. 2013. Peroxisomes take shape. Nat Rev Mol Cell Biol 14:803–817. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet C, Espagne E, Zickler D, Boisnard S, Bourdais A, Berteaux-Lecellier V. 2006. The peroxisomal import proteins PEX2, PEX5 and PEX7 are differently involved in Podospora anserina sexual cycle. Mol Microbiol 62:157–169. doi: 10.1111/j.1365-2958.2006.05353.x. [DOI] [PubMed] [Google Scholar]
- 3.Imazaki A, Tanaka A, Harimoto Y, Yamamoto M, Akimitsu K, Park P, Tsuge T. 2010. Contribution of peroxisomes to secondary metabolism and pathogenicity in the fungal plant pathogen Alternaria alternata. Eukaryot Cell 9:682–694. doi: 10.1128/EC.00214-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magliano P, Flipphi M, Arpat BA, Delessert S, Poirier Y. 2011. Contributions of the peroxisome and the β-oxidation cycle to biotin synthesis in fungi. J Biol Chem 286:42133–42140. doi: 10.1074/jbc.M111.279687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubo Y. 2012. Appressorium function in Colletotrichum orbiculare and prospect for genome based analysis. Top Curr Genet 22:115–131. doi: 10.1007/978-3-642-22916-9_7. [DOI] [Google Scholar]
- 6.Kubo Y. 2013. Function of peroxisomes in plant-pathogen interactions. Subcell Biochem 69:329–345. doi: 10.1007/978-94-007-6889-5_18. [DOI] [PubMed] [Google Scholar]
- 7.Wilson RA, Talbot NJ. 2009. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol 7:185–195. doi: 10.1038/nrmicro2032. [DOI] [PubMed] [Google Scholar]
- 8.Kubo Y, Takano Y. 2013. Dynamics of infection-related morphogenesis and pathogenesis in Colletotrichum orbiculare. J Gen Plant Pathol 79:233–242. doi: 10.1007/s10327-013-0451-9. [DOI] [Google Scholar]
- 9.Wang ZY, Soanes DM, Kershaw MJ, Talbot NJ. 2007. Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid β-oxidation during appressorium-mediated plant infection. Mol Plant Microbe Interact 20:475–491. doi: 10.1094/MPMI-20-5-0475. [DOI] [PubMed] [Google Scholar]
- 10.Fujihara N, Sakaguchi A, Tanaka S, Fujii S, Tsuji G, Shiraishi T, O’Connell R, Kubo Y. 2010. Peroxisome biogenesis factor PEX13 is required for appressorium-mediated plant infection by the anthracnose fungus Colletotrichum orbiculare. Mol Plant Microbe Interact 23:436–445. doi: 10.1094/MPMI-23-4-0436. [DOI] [PubMed] [Google Scholar]
- 11.Farré JC, Subramani S. 2004. Peroxisome turnover by micropexography: an autophagy-related process. Trends Cell Biol 14:515–523. doi: 10.1016/j.tcb.2004.07.014 [DOI] [PubMed] [Google Scholar]
- 12.Platta HW, Erdmann R. 2007. Peroxisomal dynamics. Trends Cell Biol 17:474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Asakura M, Ninomiya S, Sugimoto M, Oku M, Yamashita S, Okuno T, Sakai Y, Takano Y. 2009. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell 21:1291–1304. doi: 10.1105/tpc.108.060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kershaw MJ, Talbot NJ. 2009. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci U S A 106:15967–15972. doi: 10.1073/pnas.0901477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiel JA, Veenhuis M, van der Klei IJ. 2006. PEX genes in fungal genomes: common, rare or redundant. Traffic 7:1291–1303. doi: 10.1111/j.1600-0854.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- 16.Erdmann R, Blobel G. 1996. Identification of Pex13p, a peroxisomal membrane receptor for the PTS1 recognition factor. J Cell Biol 135:111–121. doi: 10.1083/jcb.135.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould SJ, Kalish JE, Morrell JC, Bjorkman J, Urquhart AJ, Crane DI. 1996. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J Cell Biol 135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. 2005. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17:3422-3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams C, van den Berg M, Panjikar S, Stanley WA, Distel B, Wilmanns M. 2012. Insights into ubiquitin-conjugating enzyme/co-activator interactions from the structure of the Pex4p:Pex22p complex. EMBO J 31:391–402. doi: 10.1038/emboj.2011.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Titorenko VI, Rachubinski RA. 2000. Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J Cell Biol 150:881–886. doi: 10.1083/jcb.150.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birschmann I, Stroobants AK, van den Berg M, Schäfer A, Rosenkranz K, Kunau WH, Tabak HF. 2003. Pex15p of Saccharomyces cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6 to peroxisomal membranes. Mol Biol Cell 14:2226–2236. doi: 10.1091/mbc.E02-11-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura A, Takano Y, Furusawa I, Okuno T. 2001. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13:1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jedd G, Chua NH. 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat Cell Biol 2:226–231. doi: 10.1038/35008652. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Ng SK, Lu Y, Low W, Lai J, Jedd G. 2008. Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J Cell Biol 180:325–339. doi: 10.1083/jcb.200705049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng SK, Liu F, Lai J, Low W, Jedd G. 2009. A tether for Woronin body inheritance is associated with evolutionary variation in organelle positioning. PLoS Genet 5:e1000521. doi: 10.1371/journal.pgen.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Lu Y, Pieuchot L, Dhavale T, Jedd G. 2011. Import oligomers induce positive feedback to promote peroxisome differentiation and control organelle abundance. Dev Cell 21:457–468. doi: 10.1016/j.devcel.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Tsuji G, Fujii S, Fujihara N, Hirose C, Tsuge S, Shiraishi T, Kubo Y. 2003. Agrobacterium tumefaciens-mediated transformation for random insertional mutagenesis in Colletotrichum lagenarium. J Gen Plant Pathol 69:230–239. doi: 10.1007/s10327-003-0040-4. [DOI] [Google Scholar]
- 28.Käll L, Krogh A, Sonnhammer EL. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Van Ael E, Fransen M. 2006. Targeting signals in peroxisomal membrane proteins. Biochim Biophys Acta 1763:1629–1638. doi: 10.1016/j.bbamcr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 30.Koller A, Snyder WB, Faber KN, Wenzel TJ, Rangell L, Keller GA, Subramani S. 1999. Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J Cell Biol 146:99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Momany M, Richardson EA, Van Sickle C, Jedd G. 2002. Mapping Woronin body function in Aspergillus nidulans. Mycologia 94:260–266. doi: 10.2307/3761802. [DOI] [PubMed] [Google Scholar]
- 32.Sawaguchi A, Ide S, Goto Y, Kawano J, Oinuma T, Suganuma T. 2001. A simple contrast enhancement by potassium permanganate oxidation for Lowicryl K4M ultrathin sections prepared by high pressure freezing/freeze substitution. J. Microsc 201:77–83. doi: 10.1046/j.1365-2818.2001.00787.x. [DOI] [PubMed] [Google Scholar]
- 33.Trinci AP, Collinge AJ. 1974. Occlusion of the septal pores of damaged hyphae of Neurospora crassa by hexagonal crystals. Protoplasma 80:57–67. doi: 10.1007/BF01666351. [DOI] [PubMed] [Google Scholar]
- 34.Markham P, Collinge AJ. 1987. Woronin bodies in filamentous fungi. FEMS Microbiol Lett 46:1–11. doi: 10.1111/j.1574-6968.1987.tb02448.x. [DOI] [Google Scholar]
- 35.Maruyama J, Juvvadi PR, Ishi K, Kitamoto K. 2005. Three-dimensional image analysis of plugging at the septal pore by Woronin body during hypotonic shock inducing hyphal tip bursting in the filamentous fungus Aspergillus oryzae. Biochem Biophys Res Commun 331:1081–1088. doi: 10.1016/j.bbrc.2005.03.233. [DOI] [PubMed] [Google Scholar]
- 36.Kim KH, Willger SD, Park SW, Puttikamonkul S, Grahl N, Cho Y, Mukhopadhyay B, Cramer RA Jr, Lawrence CB. 2009. TmpL, a transmembrane protein required for intracellular redox homeostasis and virulence in a plant and an animal fungal pathogen. PLoS Pathog 5:e1000653. doi: 10.1371/journal.ppat.1000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Managadze D, Würtz C, Sichting M, Niehaus G, Veenhuis M, Rottensteiner H. 2007. The peroxin PEX14 of Neurospora crassa is essential for the biogenesis of both glyoxysomes and Woronin bodies. Traffic 8:687–701. doi: 10.1111/j.1600-0854.2007.00560.x. [DOI] [PubMed] [Google Scholar]
- 38.Hettema EH, Girzalsky W, van den Berg M, Erdmann R, Distel B. 2000. Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J 19:223–233. doi: 10.1093/emboj/19.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenney K, Hunt I, Sweigard J, Pounder JI, McClain C, Bowman EJ, Bowman BJ. 2000. Hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet Biol 31:205-217. doi: 10.1006/fgbi.2000.1230. [DOI] [PubMed] [Google Scholar]
- 40.Soundararajan S, Jedd G, Li X, Ramos-Pamploña M, Chua NH, Naqvi NI. 2004. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell 16:1564–1574. doi: 10.1105/tpc.020677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gan P, Ikeda K, Irieda H, Narusaka M, O’Connell RJ, Narusaka Y, Takano Y, Kubo Y, Shirasu K. 2013. Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol 197:1236–1249. doi: 10.1111/nph.12085. [DOI] [PubMed] [Google Scholar]
- 42.Sakaguchi A, Miyaji T, Tsuji G, Kubo Y. 2008. Kelch repeat protein Clakel2p and calcium signaling control appressorium development in Colletotrichum lagenarium. Eukaryot Cell 7:102–111. doi: 10.1128/EC.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42:819–832. doi: 10.1023/A:1006496308160. [DOI] [PubMed] [Google Scholar]
- 44.Gouka RJ, Punt PJ, van den Hondel CA. 1997. Glucoamylase gene fusions alleviate limitations for protein production in Aspergillus awamori at the transcriptional and (post)translational levels. Appl Environ Microbiol 63:488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckert JH, Johnsson N. 2003. Pex10p links the ubiquitin conjugating enzyme Pex4p to the protein import machinery of the peroxisome. J Cell Sci 116:3623–3634. doi: 10.1242/jcs.00678. [DOI] [PubMed] [Google Scholar]
- 46.Hettema EH, Ruigrok CC, Koerkamp MG, van den Berg M, Tabak HF, Distel B, Braakman I. 1998. The cytosolic DnaJ-like protein Djp1p is involved specifically in peroxisomal protein import. J Cell Biol 142:421–434. doi: 10.1083/jcb.142.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Micali CO, Neumann U, Grunewald D, Panstruga R, O’Connell R. 2011. Biogenesis of a specialized plant–fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell Microbiol 13:210–226. doi: 10.1111/j.1462-5822.2010.01530.x. [DOI] [PubMed] [Google Scholar]
- 48.Thines E, Weber RW, Talbot NJ. 2000. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12:1703–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harata K, Kubo Y. 2014. Ras GTPase activating protein CoIra1 is involved in infection-related morphogenesis by regulating cAMP and MAPK signaling pathways through CoRas2 in Colletotrichum orbiculare. PLoS One 9:e109045. doi: 10.1371/journal.pone.0109045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolation of C. orbiculare mutant deficient in fatty acid metabolism. (A) Colonies of the wild-type (WT) and Hi2049 mutant strain grown on PDA and OAM for 5 and 12 days, respectively. (B) Pathogenicity of the C. orbiculare wild-type strain and the Hi2049 mutant strain on detached cucumber cotyledons. The leaves were inoculated with droplets of conidial suspension and incubated for 7 days. The right half of the leaves was intact, while the left half was wounded by toothpick scratching. Download
Protein sequence alignment of C. orbiculare Fam1 with S. cerevisiae Pex22 and Pex22-like proteins from filamentous ascomycetes. Amino acid sequence of C. orbiculare Fam1 (N4VSY8) was aligned with Pex22-like proteins of M. oryzae (UniProtKB accession no. A4R2Z0), N. crassa (UniProtKB accession no. Q8WZV7), A. oryzae (UniProtKB accession no. Q2UQZ8), and S. cerevisiae Pex22 (PEX22 YAL055W) using the Clustal W program. Identical and similar amino acids are indicated. Residues involved in interface formation between Pex22 and Pex4 in S. cerevisiae (19) are underlined with a red line. Residues comprising a putative N-terminal transmembrane domain are indicated by a blue line above the sequences. Positively charged residues (PC) immediately upstream of the transmembrane domain (29) are highlighted in green. Download
Targeted gene disruption of C. orbiculare FAM1 and CoHEX1. (A) Schematic representation of the FAM1 gene disruption by homologous recombination. Binary plasmid pBIFAM1AH3-2 contains the AMP::HPH (ampicillin-hygromycin resistance) cassette inserted into the FAM1 gene between the left-border (LB) and right-border (RB) sequences. Through double crossing over, a 3.9-kb XhoI fragment of the wild-type strain is predicted to be replaced by a 6.6-kb XhoI fragment containing the AMP::HPH cassette. (B) DNA blot analysis of FAM1 disrupted transformants. Genomic DNA was digested with XhoI and probed with an internal fragment of FAM1 indicated in the pBIFAM1AH3-2 vector map. The wild-type (WT) strain, fam1-1, fam1-2, and fam1-3 FAM1 disruption mutants, FAM1 ectopic transformant, and FAM1 complemented transformant of the FAM1 mutant (FAM1-C) were studied. (C) Schematic representation of the CoHEX1 gene disruption by homologous recombination. The binary plasmid pBII-HEX1AH3-2-7 contains the AMP::HPH cassette inserted into the CoHEX1 gene between the left-border (LB) and right-border (RB) sequences. Through double crossing over, an ~9.0-kb BamHI fragment of the wild type is predicted to be replaced by a 12-kb BamHI fragment containing the AMP::HPH cassette. (D) DNA blot analysis of CoHEX1 disrupted transformants. Genomic DNA was digested with XhoI and probed with an internal fragment of CoHEX1 indicated in the pBII-HEX1AH3-2-7 vector map. The wild-type (WT) strain, cohex1-1, cohex1-2, and cohex1-3 CoHEX1 disruption mutants, and CoHEX1 ectopic transformant were examined. Download
Subcellular localization of Fam1 in peroxisome-deficient mutants. Fam1-GFP was expressed in vegetative hyphae of peroxisome-deficient copex13 and copex6 mutants growing on PDA. Confocal microscopy showed Fam1-GFP neither localized at hyphal tips nor septa in these peroxisome mutants. Numerous small punctate structures labeled by Fam1-GFP were randomly distributed through the cytoplasm. Images represent overlay projections of bright-field and GFP fluorescence channels. Bars, 5 µm. Download
Alignment of C. orbiculare CoHex1, CoWsc, and CoPeX4 with homologs of other ascomycete fungi. (A) The amino acid sequence of C. orbiculare CoHex1 (N4UJP9) was aligned with the amino acid sequences of N. crassa Hex1 (P87252), A. oryzae AoHex (I8TQ26), and M. oryzae MgHex (Q2KGA0) using the Clustal W program. (B) Amino acid sequence of C. orbiculare CoWsc (N4VZG1) was aligned with the amino acid sequence of N. crassa Wsc (U9W802) using the Clustal W program. (C) Alignment of C. orbiculare CoPex4 with S. cerevisiae Pex4. Amino acid sequence of C. orbiculare CoPex4 (N4W4I0) was aligned with Colletotrichum graminicola Pex4 (E3Q679), Colletotrichum higginsianum Pex4 (H1UZJ1), and S. cerevisiae Pex4 (C7GYG2) using the Clustal W program. Shading of residues represents percentage amino acid conservation: black, 100%; dark grey, 75%; light grey, 50%. Download
Subcellular localization of CoHex1 in the fam1 mutant. CoHex1-RFP was expressed in vegetative hyphae of the peroxisome-deficient fam1 mutant. CoHex1-RFP was not localized at hyphal tips or at septa, but instead it was uniformly distributed throughout the cytoplasm. Images represent overlay projections of bright-field and GFP fluorescence channels. Bars, 5 µm. Download
The cohex1 mutant is defective in WB function. (A) Bright-field microscopy images showing the blockage of cytosolic bleeding from subapical cells following hyphal tip rupture. When distilled water was placed onto hyphae growing on Mathur's agar medium containing 2% sorbose, hyphal tips swelled and burst. The cytoplasm leaked from subapical cells of the cohex1 mutant but was retained in subapical cells of the wild-type and cohex1+CoHex1 complemented strain. Arrows indicate the first septum behind the hyphal tip. Bars, 3 µm. (B) Mycelial radial growth of the C. orbiculare wild-type strain, fam1 mutant, and cohex1 mutants after 12 days on OAM. (C) Radial growth rate of the wild type and cohex1 mutant on PDA or OAM. The increase in colony diameters was measured between 3 and 7 days on PDA and between 4 and 14 days on OAM. The mean and standard deviation were calculated from six colonies of each strain. Download
Fam1 cofractionates with WB marker proteins after separation of organelles. Fungal organelles were separated by centrifugation on a 17 to 60% Nycodenz density gradient, and fractions were tested for the presence of Fam1 and CoHex1 by Western blot analysis. The graph shows the shape of the gradient and total protein distribution across the gradient. PTS1-GFP provided a marker of the peroxisome matrix, detected with anti-GFP antibodies. Download
Primers used in this study