Abstract
The genome of all organisms is constantly being challenged by endogenous and exogenous sources of DNA damage. Errors like base:base mismatches or small insertions and deletions, primarily introduced by DNA polymerases during DNA replication are repaired by an evolutionary conserved DNA mismatch repair (MMR) system. The MMR system, together with the DNA replication machinery, promote repair by an excision and resynthesis mechanism during or after DNA replication, increasing replication fidelity by upto-three orders of magnitude. Consequently, inactivation of MMR genes results in elevated mutation rates that can lead to increased cancer susceptibility in humans. In this review, we summarize our current understanding of MMR with a focus on the different MMR protein complexes, their function and structure. We also discuss how recent findings have provided new insights in the spatio-temporal regulation and mechanism of MMR.
Introduction
The integrity of the genetic information largely depends on the accuracy of the DNA replication process, but also on surveil-lance mechanisms that increase DNA replication fidelity. On one hand, the major replicative DNA polymerases proofread their own errors by excision of misincorporated nucleotides. Moreover, cells possess a second quality control mechanism, represented by the DNA mismatch repair (MMR) pathway. This DNA repair mechanism corrects not only errors that escaped the DNA polymerase proofreading function, but also recognizes mispaired bases and branched DNAs formed during recombination, as well as chemically modified bases in DNA, introduced for example, by chemotherapeutic agents. In fact, components of the MMR machinery are required by many aspects of DNA metabolism and affect processes such as cell cycle checkpoint control, apoptosis, somatic hypermutation of immunoglobulin genes, triplet-repeat expansion, among others (Harfe and Jinks-Robertson 2000; Li 1999, 2008; Pena-Diaz and Jiricny 2012).
Many features of MMR reactions are conserved from bacteria to humans. The core MMR machinery involves two conserved families of protein complexes, including the MutS and MutL complexes (where Mut stands for mutator) or their homologs. The basic reaction involves an excision-repair reaction in which a region of the newly synthesized strand containing the incorrect base is excised and resynthesized. Thus, MMR involves several proteins, including some components of the DNA replication machinery. Together, the core and accessory MMR proteins impart mechanistic differences specific to each organism or specific to alternative MMR sub-pathways. Although we know some of the sequential steps of the MMR reaction and have identified most of the proteins that function in MMR, some mechanistic aspects of the MMR reaction are still not well understood.
Inactivation of the MMR system leads to an increase of upto-three orders of magnitude in the overall mutation rate (reviewed in Iyer et al. 2006). Moreover, in humans, loss of MMR function results in Lynch Syndrome (previously referred to as hereditary non-polyposis colorectal cancer or HNPCC) (Boland and Goel 2010; Boland and Lynch 2013). Lynch syndrome is characterized by early-onset of cancer and the accumulation of mutations, often including frameshift mutations in repetitive sequences, a phenotype called microsatellite instability (MSI). Most Lynch syndrome patients are heterozygous carriers of germline-inactivating mutations in MMR genes, most frequently MSH2 and MLH1 (Peltomaki and Vasen 2004). Cells in these patients subsequently lose MMR function due to a mutagenic/epigenetic event that inactivates the remaining functional allele (Boland and Goel 2010; Huang et al. 1996; Kinzler and Vogelstein 1996; Markowitz et al. 1995). In this way, loss of MMR activity increases the rate of accumulation of mutations in genes encoding essential regulators of cell proliferation and apoptosis, thereby accelerating cancer progression. In addition, a proportion of sporadic tumors have acquired MMR defects, most notably the epigenetic silencing of MLH1, resulting in increased mutation rates that drive the development of cancer (Peltomaki 2014). The reader is referred to other review articles (Boland and Goel 2010; Edelmann and Edelmann 2004; Hsieh and Yamane 2008; Peltomaki and Vasen 2004) that have been published on MMR defects and cancer, as this topic will not be covered in detail in this article.
In the last few years, there have been a number of new developments in the MMR field. Important progress has been made toward understanding how the newly synthesized strand is identified in eukaryotes. Furthermore, new discoveries made using live-cell imaging to visualize MMR components have expanded our knowledge of the spatio-temporal kinetics of MMR. In addition, accessory proteins with important functions in MMR had been revisited and their involvement in MMR has been pinpointed to specific steps of MMR reactions, increasing our understanding of the mechanistic aspects of MMR and the function of specific MMR proteins. We also have an idea, albeit incomplete, of how epigenetic changes and posttranslational modifications might regulate the interaction of MMR with chromatin.
The aim of this review is to summarize some of the latest discoveries on MMR, focusing on protein complexes that function in MMR and highlighting the existence of MMR sub-pathways where previously identified MMR proteins have been found to have dedicated functions. The focus of this review is on MMR in Saccharomyces cerevisiae and humans; because of some inconsistencies in the names of individual proteins, when the equivalent S. cerevisiae and human proteins have different names, the prefixes sc and h, respectively, will be used to avoid potential confusion.
The basic MMR reaction
The basic MMR reaction is best understood in Escherichia coli, where MMR is initiated by the MutS homodimer, which recognizes mismatches and then recruits MutL (also a homodimeric complex) to the mispair site. MutL interacts with a third MMR component called MutH, resulting in the activation of the latent endonuclease activity of MutH and nicking of the newly replicated strand. Strand discrimination, an essential feature of MMR, restricts the use of the parental strand as the template for DNA resynthesis thus eliminating misincorporation errors. In E. coli, strand discrimination relies on the hemi-methylated status of the newly synthesized DNA; the DNA is methylated at d(GATC) sites by the Dam methyltransferase and because this is a post-replication modification, the newly synthesized DNA strand is transiently undermethylated. MutH is a methylation-sensitive endonuclease that exclusively nicks the unmethylated, newly synthesized DNA strand at d(GATC) sites. This nick is then used as entry point for the excision reaction, targeting it exclusively to the newly synthesized strand. During the excision reaction, the DNA is unwound by DNA helicase II starting at the nick and the error-containing strand is degraded by a ssDNA specific exonuclease (ExoI, ExoVII, and ExoX act in 3′ to 5′ excision; RecJ and ExoVII act in the 5′ to 3′ excision) and resynthesized by DNA polymerase III (Table 1) (Iyer et al. 2006; Kunkel and Erie 2005). It should be noted that DNA methylation asymmetry is not used as a strand specificity signal in eukaryotes or in bacteria outside of a subset of gammaproteobacteria that includes E. coli (Gao et al. 2009).
Table 1.
MMR proteins in E. coli, S. cerevisiae and H. sapiens
| E. coli | S. cerevisiae | H. sapiens | Comments |
|---|---|---|---|
| MutS-MutS | Msh2-Msh6 (MutSα) Msh2-Msh3 (MutSβ) |
Msh2-Msh6 (MutSα) Msh2-Msh3 (MutSβ) |
Mispair recognition complex—homodimer in E. coli and a heterodimer in eukaryotes. MutSα and MutSβ have overlapping mispair recognition specificity and are partially redundant |
| Mlh1-Pms1 (MutLα) | Mlh1-Pms2 (MutLα) | Homodimer in E. coli and heterodimer in eukaryotes. MutL (E. coli) and MutLα (eukaryotes) play a central role during MMR. In E. coli, MutL promote the nicking reaction via MutH, whereas in eukaryotes MutLα possess an intrinsic endonuclease activity | |
| MutL-MutL | Mlh1-Mlh2 (MutLβ) | Mlh1-Pms1 (MutLβ) | MutLβ is an accessory factor for MMR |
| Mlh1-Mlh3 (MutLγ) | Mlh1-Mlh3 (MutLγ) | MutLγ substitutes for MutLα in the repair of a minor fraction of mispairs, but primarily acts in the resolution of meiotic recombination intermediates | |
| Dam methylase | Absent | Absent | Promotes N6-adenine methylation at d(GATC) sites, serves as strand discrimination signal in E. coli |
| MutH | Absenta | Absenta | Endonuclease, nicks daughter strand using d(GATC) hemi-methylated sites as strand discrimination signal |
| none | Exo1 | Exo1 | 5′-3′ dsDNA exonuclease, acts in the excision reaction |
| RecJ, ExoVII | None | None | 5′-3′ ssDNA exonuclease, acts in the excision reaction |
| ExoI, ExoVII, ExoX | None | None | 3′-5′ ssDNA exonuclease, acts in the excision reaction |
| UvrD | None or unknown | None or unknown | DNA helicase II, promotes excision reaction, activated by MutS |
| β-clamp | PCNA | PCNA | DNA polymerase processivity factor. In eukaryotes stimulates Mutα endonuclease activity. The gene encoding PCNA in S. cerevisiae is POL30 |
| γ-Complex | RFC | RFC | Loading of β-clamp/PCNA |
| SSB | RPA1-3 | RPA1-3 | ssDNA binding protein, acts in the excision and DNA resynthesis reactions. The genes encoding RPA subunits in S. cerevisiae are RFA1, 2 and, 3 |
| DNA Pol III | Polδ | Pol delta | DNA polymerase that acts in the gap-filling step |
| DNA ligase | Unknown | Ligase I | Seals nicks after DNA resynthesis |
This endonuclease function appears to be present in MutLα and MutLγ
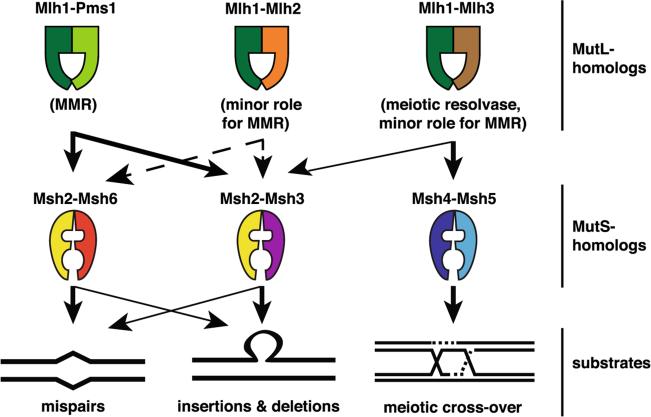
Eukaryotic MMR is mechanistically similar to bacterial MMR. However, mispair recognition utilizes two MutS homolog (MSH) complexes, the Msh2-Msh6 (sometimes called MutSα) and Msh2-Msh3 (sometimes called MutSβ) complexes that have a partial overlapping mispair recognition specificity, instead of a single MutS homodimer (Kolodner and Marsischky 1999) (Fig. 1). A third MSH complex (Msh4-Msh5) does not have an MMR-related function, but rather is important during meiotic recombination. In addition, instead of a MutL homodimer, eukaryotic MMR primarily utilizes a heterodimeric MutL homolog (MLH) complex, sometimes also referred as MutLα, which in S. cerevisiae is Mlh1-Pms1 and in humans is Mlh1-Pms2 (scPms1 is the homolog of hPms2). Two additional MLH heterodimers have been identified, MutLβ which in S. cerevisiae is Mlh1-Mlh2 and in humans is Mlh1-Pms1 (scMlh2 is the homolog of hPms1) and MutLγ which is Mlh1-Mlh3; MutLβ and MutLγ appear to have a minor role in MMR (Table 1 and Fig. 1). In contrast to MMR in E. coli, eukaryotic MMR does not utilize a MutH homolog protein, as the nicking function of MutH appears to be integral to the MutLα and MutLγ complexes (Kadyrov et al. 2006), whereas the MutLβ complex is predicted to lack endonuclease activity.
Fig. 1.
S. cerevisiae MSH and MLH complexes. The arrows indicate their functional interactions and potential functions in vivo. Arrows denote major roles (thick arrows) and minor roles (thin arrows) in the indicated processes. Dashed lanes represent interactions that are biologically relevant apparently only in specific genetic backgrounds
Biochemical studies have demonstrated two types of MMR reactions in which human MMR proteins catalyze the repair of nicked, mispair containing substrates, similar to those catalyzed by E. coli MMR proteins. In one reaction, a combination of Msh2-Msh6 or Msh2-Msh3, Replication Protein A (RPA) and Exonuclease 1 (Exo1), a 5′ to 3′ dsDNA specific exonuclease that also has flap endonuclease activity, promotes 5′ to 3′ excision of the mispaired strand from a 5′ nick, resulting in a gap that is filled in by a combination of DNA polymerase Polδ (also called Pol3), Proliferating Cell Nuclear Antigen (PCNA), Replication Factor C (RFC), and RPA (Constantin et al. 2005; Zhang et al. 2005). In a second type of reaction, a combination of Msh2-Msh6, Mlh1-Pms2, PCNA, and RFC promotes the introduction of nicks 5′ to a mispair in a substrate containing a nick 3′ to the mispair (Constantin et al. 2005; Kadyrov et al. 2006); this incised product is then subject to excision and gap filling essentially as seen with substrates containing a 5′ nick (Constantin et al. 2005; Kadyrov et al. 2006). This mode of excision is distinct from that seen in E. coli where a combination of a ssDNA-specific exonuclease and a DNA helicase promote excision instead of a dsDNA specific exonuclease in eukaryotes. There are key questions raised by these studies on eukaryotic mispair excision. (1) What targets excision to the leading strand that is synthesized continuously? (2) If nicks, which are prevalent on the lagging strand due to discontinuous DNA synthesis, can target MMR to the lagging daughter strand, then why is MutLα complex absolutely required for MMR? (3) How do preexisting nicks target MutLα to incise on the already nicked strand and what role does this incision play? (4) What accounts for excision in the absence of Exo1 since this protein is not absolutely required for MMR in vivo? (Amin et al. 2001; Tishkoff et al. 1997; Wei et al. 2003).
The MutS mismatch recognition complex
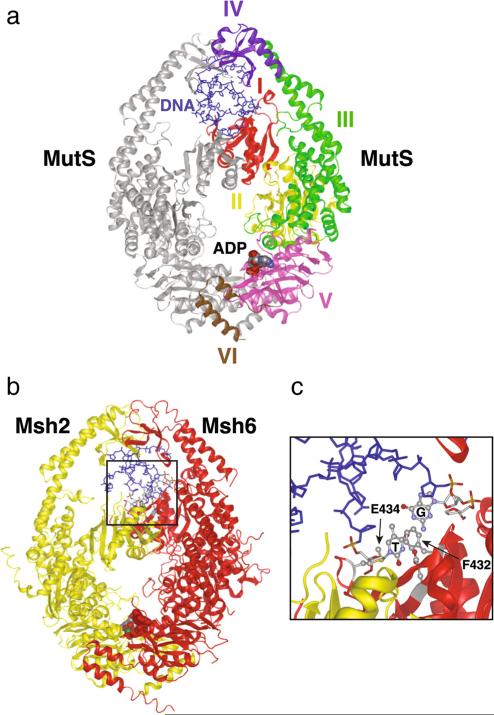
Our understanding of how a DNA mispair is recognized by the MutS family of proteins has been driven by crystallo-graphic studies of MutS from E. coli and Thermus aquaticus, in combination with different types of biochemical analysis. MutS is a homodimer, in which each monomer contains six structural domains (I to VI) with distinct functions (Fig. 2a) (Lamers et al. 2000; Mendillo et al. 2007; Obmolova et al. 2000). According to the E. coli MutS structure (Lamers et al. 2000), domain I (residues 2–115) corresponds to the mispair recognition domain; domain II (residues 116–266) or connector domain interacts with MutL (Mendillo et al. 2009); domain III (residues 267–443 and 504–567) represents the core domain that separates DNA binding and nucleotide binding sites; domain IV (residues 444–503) forms the exterior of the DNA clamp; domain V (residues 568–765) contains the ATPase domain and dimerization sites; and domain VI (residues 766–800) or helix-turn-helix domain (HTH) is required for dimerization at moderate protein concentrations (Biswas et al. 2001; Lamers et al. 2000; Mendillo et al. 2007; Obmolova et al. 2000). This section summarizes some important aspects of the structural domains of MutS and how they function during the mispair recognition reaction.
Fig. 2.
Crystal structure of E. coli MutS and human MutSα according to previous reports (Lamers et al. 2000; Warren et al. 2007). a MutS homodimer, in which the mispair-contacting subunit has been colored by domain (I–VI) with red, yellow, green, purple, pink, and brown, respectively. DNA (blue sticks) and ADP are also indicated. b Human Msh2-Msh6 complex. Msh2 is shown in yellow and Msh6 in red. c Expanded view of the region in the black box in (b) that directly interacts with the mispaired base. The conserved Msh6 residues phenylalanine (F432) and glutamic acid 434 (E434), as well as the mispaired bases guanine (G) and thymine (T) are indicated
Interaction with DNA
Biochemical and crystallography studies showed that the MutS homodimer forms a ring-like structure that encircles and sharply bends DNA when it binds a mispair (Junop et al. 2001; Lamers et al. 2000). The DNA binding complex is asymmetric; only one of the two subunits directly contacts the mispaired base via domain I, while domain I from the other subunit makes non-specific contacts with the DNA backbone. Two key amino acids in domain I that directly contact and stabilize an extra-helical base at a base:base mispair, or an extra-helical base in a +1/−1 insertion/deletion loop (IDL), are present in a Phe-X-Glu motif (corresponding in E. coli MutS to residues Phe36 and Glu38).
Phe36 contacts a base at the mispair, whereas Glu38 plays a role in stabilizing the bend in the DNA. Binding to DNA provokes conformational changes in MutS that have been revealed by deuterium exchange mass spectrometry (DXMS) studies (Mendillo et al. 2010).
ATPase domain
The domain V of MutS harbors an ATPase domain that is conserved on MutS homologs and also other members of the adenosine triphosphate (ATP)-binding cassette (ABC) transporter family (Gorbalenya and Koonin 1990). Conformational changes resulting from the binding of ATP or ADP by MutS are transduced to the DNA binding domains. Moreover, the two ATPase sites in MutS are expected to have different functions due to the intrinsic structural asymmetry of the MutS mispair-bound complex (Junop et al. 2001; Lamers et al. 2000; Obmolova et al. 2000). A key feature of MMR is that the MutS complex must bind to mismatched DNA with higher affinity or a unique conformation than when it binds matched DNA.
Based on several biochemical, structural, and genetic studies, it has been proposed two potential models for the ATP-dependent movement of MutS along the DNA, the translocation model and the molecular switch model or sliding clamp model (reviewed in Iyer et al. 2006). According to the translocation model, ATP binding reduces the affinity of MutS for the mispair and ATP hydrolysis acts as a driving force promoting movement of the MutS along DNA (Allen et al. 1997).
On the other hand, the sliding clamp model proposes that the mispair is recognized by an ADP-bound MutS, resulting in a rapid ADP-ATP exchange and a MutS conformational change that releases MutS from the mismatch allowing MutS to diffuse along the DNA backbone as a sliding clamp, in an ATP-hydrolysis-independent manner (Acharya et al. 2003; Gradia et al. 1999; Hura et al. 2013; Lee et al. 2014; Mendillo et al. 2005). DXMS studies have revealed some of these ATP binding induced conformational changes (Mendillo et al. 2010). Moreover, it has been shown that the ATP binding driven conformational change is required for the mispair-dependent recruitment of MutL (Junop et al. 2001).
MutS-MutL interacting region
Upon mismatch recognition, MutS undergoes an ATP-binding dependent conformational change that allows sliding on DNA and the interaction with MutL, resulting in the recruitment of MutL by mispair-bound MutS. This higher-order complex is dynamic and transient (Galio et al. 1999; Mendillo et al. 2005), thus the identification of the interface between MutS and MutL has been difficult to determine. However, using DXMS, it was possible to identify a region in domain II of MutS (connector domain) that specifically interacts with MutL (Mendillo et al. 2009); this finding was subsequently confirmed by crosslinking and fluorescence resonance energy transfer (FRET) studies (Winkler et al. 2011). Binding of MutL requires ATP and mispair binding by MutS, which are likely necessary for a conformational change that exposes the domain II to MutL. Consistent with this model, amino acid changes in this region of MutS (mutS-Q211S, mutS-Q212S, mutS-N214, and mutS-L215S) result in inactivation of both MutL recruitment in vitro and MMR in vivo (Mendillo et al. 2009). The MutS interface is conserved in Msh2 and analogous mutations in domain II of scMsh2 result in defects in binding of scMsh2-Msh6 to scMlh1-Pms1 in vitro and defects in MMR in vivo. Importantly, mutations in E. coli MutS and scMsh2 that block assembly of MutL and scMlh1-Pms1, respectively, do not alter mispair recognition or ATP-induced sliding on DNA (Mendillo et al. 2009).
Eukaryotic MSH complexes
In eukaryotes, mispaired bases are recognized by heterodimeric MSH complexes: the Msh2-Msh6 complex (MutSα), the Msh2-Msh3 complex (MutSβ) (Fig. 1), and in plants also by the Msh2-Msh7 complex (Culligan and Hays 2000). These complexes share a common subunit, Msh2, and possess a unique subunit that specifies a distinct but partially overlapping substrate specificity, resulting in some genetic redundancy. Similar to the structural asymmetry of the E. coli MutS homodimer, each subunit of the eukaryotic MSH heterodimer has a different role, with the Msh3, Msh6, and Msh7 subunits being the mismatched base-contacting subunit. Interestingly, replacing domain I (the mispair binding domain) of Msh6 with that of Msh3 in S. cerevisiae resulted in a functional chimeric Msh2-Msh6 complex with Msh2-Msh3-like substrate specificity, demonstrating that domain I is the major determinant of substrate specificity (Shell et al. 2007a). There are also three other eukaryotic MSH proteins that are not involved in canonical MMR and will not be discussed further in this review: Msh1, a protein that functions in mitochondria that has no homolog in mammals (Culligan et al. 2000; Reenan and Kolodner 1992a, b), and Msh4 and Msh5 that form a heterodimeric complex that functions in the resolution of meiotic recombination intermediates in conjunction with the Mlh1-Mlh3 complex (Kolas et al. 2005; Santucci-Darmanin et al. 2002; Snowden et al. 2004).
Msh2-Msh6 (MutSα)
In humans, the Msh2-Msh6 heterodimer is about 10 times more abundant than Msh2-Msh3 and is potentially the major mispair recognition complex, although in human cell lines and in mice defects in Msh6 cause weaker mutator phenotypes than defects in Msh2, suggestive of some redundancy between these two complexes. In contrast, in S. cerevisiae, Msh2-Msh6 and Msh2-Msh3 appear to play more balanced roles in MMR (Marsischky et al. 1996; Sia et al. 1997). S. cerevisiae Msh2-Msh6 appears to functionally recognize seven of the eight possible base:base mispairs (CC mispairs are poorly recognized) and +1 and +2 insertion/deletion mispairs (Srivatsan et al. 2014). Msh2-Msh6 forms an asymmetric dimer in which the Msh6 subunit resembles the structure of the mispair-contacting subunit of E. coli MutS (Fig. 2a, b) (Warren et al. 2007). Similar to MutS, Msh6 contains a conserved phenylalanine (Phe337 in S. cerevisiae and Phe432 in humans), which interacts with mismatched DNA (Fig. 2c). Accordingly, mutations that alter this phenylalanine prevent mispair recognition (Bowers et al. 1999). Interestingly, DXMS studies have shown that S. cerevisiae Msh2-Msh6 forms a ring around fully basepaired DNA in which the Msh6 mispair-contacting Phe337 contacts the DNA and have identified an Msh6 DNA backbone contacting residue Arg412, as potentially the only other residue that uniquely contacts the DNA in the mispair-bound form of Msh2-Msh6 (Mendillo et al. 2010). Overall, the mispair binding domains of MutS and Msh6 are structurally similar to each other, consistent with the high degree of sequence homology between MutS and Msh6 within this region.
Similar to MutS, domain V of Msh6 and Msh2 contains composite and asymmetric ATPase domains that bind ATP with different kinetics and affinities (Antony and Hingorani 2003; Mazur et al. 2006). Consistent with the view that both ATPase domains contribute to the ATPase activity of the complex, mutations altering the highly conserved lysine at the Walker A motif in either human Msh2 or Msh6 (msh2-K675R or msh6-K1140R) affect ATP hydrolysis and dissociation from the DNA (Iaccarino et al. 1998). As seen for the MutS homodimer, nucleotide binding by the Msh2-Msh6 complex modulates its interactions with mispaired and fully basepaired DNA in which ADP binding allows the binding of Msh2-Msh6 to a mispair, while ADP-ATP exchange converts the mispair-bound Msh2-Msh6 to the sliding clamp form that diffuses along the DNA (Gradia et al. 1999; Mazur et al. 2006). Supporting the role of this conformational change in MMR, dominant mutations have been identified in S. cerevisiae MSH6 that do not affect mispair binding but block the formation of sliding clamps and in some cases block the recruitment of Mlh1-Pms1 (Hargreaves et al. 2010; Hess et al. 2002, 2006). Furthermore, the equivalent mutations in S. cerevisiae MSH2 cause essentially the same biochemical defects (Hargreaves et al. 2010).
Small-angle X-ray scattering (SAXS) studies for the S. cerevisiae MutSα and PCNA proteins, reveals that Msh6 contains an unstructured N-terminal region (NTR) that serves as a tether to PCNA (Shell et al. 2007b). The interaction between Msh6 and PCNA requires a conserved PCNA-interacting protein motif (PIP-box) present on the Msh6-NTR. A similar PIP-box motif is also present in Msh3-NTR, but not in Msh2; however, transferring the Msh6-NTR onto Msh2, complements deletion of the NTR on Msh6 (Shell et al. 2007b). As will be discussed later, the interaction of Msh2-Msh6 heterodimer with PCNA is necessary to recruit the mispair recognition complexes to newly replicated DNA (Hombauer et al. 2011a; Kleczkowska et al. 2001; Shell et al. 2007b). Human Msh6 (and Msh6 from other deuterostomes) but not Msh2 or Msh3 additionally contains a PWWP domain in the N-terminal region (Laguri et al. 2008), which consequences will be discussed later. Moreover, SAXS experiments done for the human MutSα and MutSβ complexes revealed that the NTR of human Msh3 and Msh6 exist rather as a globular structure, which interacts with PCNA in a well defined conformation (Iyer et al. 2008, 2010; Lang et al. 2011), and therefore differs from the unstructured NTR present on scMsh6. The potential consequences of these discrepancies between the human and the S. cerevisiae MutSα proteins remain unknown.
Msh2-Msh3 (MutSβ)
The second MSH heterodimer that participates in mispair recognition in eukaryotes is Msh2-Msh3. Genetic and biochemical studies in S. cerevisiae (Harrington and Kolodner 2007; Marsischky et al. 1996; Srivatsan et al. 2014) and in human cells (Acharya et al. 1996) have shown that the Msh2-Msh3 complex recognizes and can recruit MLH heterodimers to both small and large insertion/deletion mispairs and a limited spectrum of base:base mispairs (Acharya et al. 1996; Harrington and Kolodner 2007; Kolodner and Marsischky 1999; Marsischky et al. 1996; Srivatsan et al. 2014). However, while domain I (the mispair binding domain) of hMsh3 has a similar fold compared to domain I of hMsh6, mispair recognition occurs quite differently. Instead of using a Phe-X-Glu motif to stabilize an extra-helical base, domain I from Msh3 bends substrate DNA and separates the DNA strands at the mispair site by insertion of key side-chain residues (Dowen et al. 2010; Gupta et al. 2012). Genetic analysis done in S. cerevisiae is consistent with the need for Msh3 to induce additional stabilization to base:base mispairs relative to insertion/deletion mispairs that are intrinsically bent and strand separated (Dowen et al. 2010). While the interactions between S. cerevisiae Msh2-Msh3 and ADP and ATP have not been as well studied as for scMsh2-Msh6, the ATP binding mediated conversion of mispair-bound scMsh2-Msh3 to a sliding clamp and recruitment of scMlh1-Pms1 (Srivatsan et al. 2014) appears to be similar to that of scMsh2-Msh6.
Genetics studies in S. cerevisiae have implicated scMsh2-Msh3, in conjunction with the Rad1-Rad10 endonuclease, in the removal of non-homologous tails in recombination intermediates (Sugawara et al. 1997). This resolution event requires the amino acid residues of the scMsh3 mispair binding domain that are critical for mispair recognition but not those required for strand separation and bending of DNA (Dowen et al. 2010). In addition, studies have demonstrated that scMsh2-Msh3 binds to double-strand break-induced recombination intermediates (Sugawara et al. 1997; Surtees and Alani 2006); although whether this reflects binding to double-strand breaks or branched recombination intermediates has not yet been resolved. Furthermore, Msh2-Msh3 is known to play a role promoting trinucleotide repeat expansions, which are characteristic of several disorders including Fragile X mental retardation syndrome, Huntington's disease, myotonic dystrophy, and other types of ataxia (Lopez Castel et al. 2010; McMurray 2010; Slean et al. 2008). Initial studies in mice revealed that Msh2-Msh3 promotes genetic instability, based on the observation that wild-type as well as an MSH6 −/− mice, but not an MSH3−/− mice, accumulate expansions at trinucleotide containing regions (Owen et al. 2005; van den Broek et al. 2002). Consistent with this, it has been shown in S. cerevisiae (Surtees and Alani 2006) and with human cell extracts (Panigrahi et al. 2010; Tian et al. 2009) that Msh2-Msh3 binds branched DNAs in vitro with highest affinity for branched DNAs containing single-stranded DNA tails, as well as large insertion/deletion mispairs that might play a role in trinucleotide repeat expansions. Thus, it appears that the distinctive mode of mispair recognition has allowed Msh2-Msh3 to adopt other functions that require recognition of unusual DNA structures (Owen et al. 2005).
The MutL complex
Prokaryotic MutL is a homodimeric ATPase that belongs to the GHKL superfamily consisting of Gyrase b, Hsp90, Histidine kinases and MutL homologs (Ban and Yang 1998; Dutta and Inouye 2000). In E. coli, MutL is a critical factor that is recruited by mismatch-bound MutS and mediates the activation of both the MutH endonuclease and the UvrD DNA helicase.
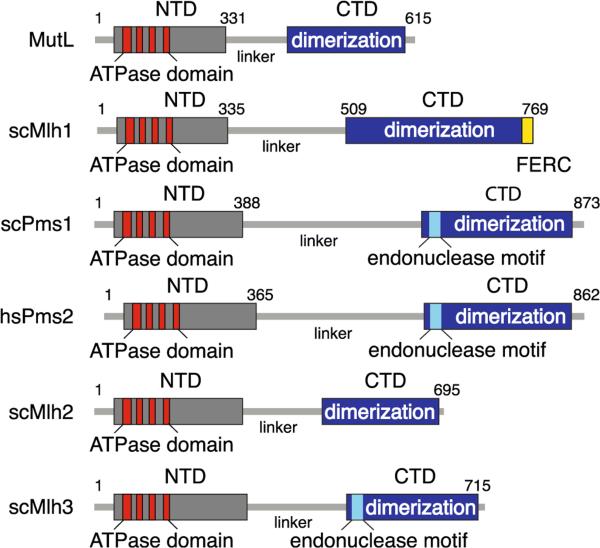
MutL as well as its eukaryotic homologs contains two distinct structural domains: a conserved N-terminal domain (NTD) and less conserved C-terminal domain (CTD), which are connected by a long proline-rich linker (Fig. 3). The NTD possesses ATPase activity (Ban and Yang 1998), whereas the CTD is required for dimerization (Guarne et al. 2004). The crystal structures of the N- and C-terminal domains of MutL have been determined (Ban and Yang 1998; Guarne et al. 2004; Kosinski et al. 2005), whereas the linker is likely to be disordered.
Fig. 3.
Structural domains of MutL and eukaryotic MutL homologs. The N-terminal domain (NTD) is indicated by a gray box that contains an ATPase domain consisting of four highly conserved motifs (shown in red). The dimerization domain and the endonuclease motif DQHA(X)2E(X)4E (light blue box) contained within the C-terminal domain (CTD) (blue box) are indicated. The conserved FERC sequence of Mlh1 is shown in yellow
The ATPase domain of E. coli MutL
The MutL NTD, which is about 300 amino acids in length, contains highly conserved residues clustered in four motifs, which are essential for ATP binding and/or hydrolysis (Fig. 3). ATP binding and hydrolysis by MutL induces major conformational changes that constitute a MutL ATPase cycle (Ban et al. 1999). In the nucleotide-unbound state, MutL exists as a dimer mediated by interactions between the two CTDs. ATP binding causes dimerization of the NTDs and compaction of the dimer (Sacho et al. 2008). ATP hydrolysis results in relaxation of the dimer structure and ADP release results in dissociation of the NTDs (Yang 2000).
MutL interactions with DNA
MutL binds DNA independently of MutS, a mismatch or a specific DNA sequence, with a binding preference toward ssDNA over dsDNA (Bende and Grafstrom 1991). The interaction has been proposed to occur in a groove created by the dimerization of both NTDs based on the effects of mutagenizing positively charged residues in this region (Ban et al. 1999). Mutations affecting this region disrupt MMR (Junop et al. 2003) and fail to activate the DNA helicase activity of UvrD (Guarne et al. 2004; Matson and Robertson 2006) but do not affect MutH activation (Ahrends et al. 2006). It has been suggested that binding of MutL to DNA might be important after MutH activation (and its release after incision) when MutL positions itself between the mismatch and the d(GATC) site facilitating the subsequent steps of the mispair excision reaction (Junop et al. 2003). On the other hand, the simultaneous binding to DNA and MutS might stabilize a repair intermediate during the search for the strand discrimination signal, triggering a conformational switch from an initiation mode to a processing mode (Ban and Yang 1998; Drotschmann et al. 2002).
Eukaryotic MLH complexes
In eukaryotes, there are three different heterodimeric MLH complexes called MutLα, MutLβ, and MutLγ, represented in S. cerevisiae by Mlh1-Pms1, Mlh1-Mlh2, and Mlh1-Mlh3 complexes, respectively. In humans, the homolog MutLα, MutLβ, and MutLγ heterodimers are formed by Mlh1-Pms2, Mlh1-Pms1, and Mlh1-Mlh3, respectively (Jiricny 2013; Kolodner and Marsischky 1999; Kunkel and Erie 2005). The nomenclature of these proteins in different species is described in Table 1 (S. cerevisiae proteins Pms1 and Mlh2 are the functional homologs of human Pms2 and Pms1, respectively). Like MutL, the eukaryotic MLH proteins have distinct NTDs and CTDs that are connected by an unstructured linker. The NTDs contain the ATPase domain, which is assembled by four conserved ATP-binding motifs characteristic of the GHKL family of ATPases (Dutta and Inouye 2000) (Fig. 3). The CTDs are not conserved in their primary sequence but are predicted to share some level of conservation with regard to their secondary structure. The CTDs contain the regions important for dimerization, and for MutLα in S. cerevisiae (Gueneau et al. 2013) or humans (Schmutte et al. 2001), the interaction domain with Exo1 (Gueneau et al. 2013; Schmutte et al. 2001). Importantly, the CTD of some of the eukaryotic MutL homologs contains a conserved endonuclease motif (DQHA(X)2E(X)4E) which is not present in E. coli MutL or MutL in other organisms that have a MutH-dependent incision system (Table 1 and Fig. 3) (Guarne 2012; Kadyrov et al. 2006).
Mlh1-Pms1 (MutLα)
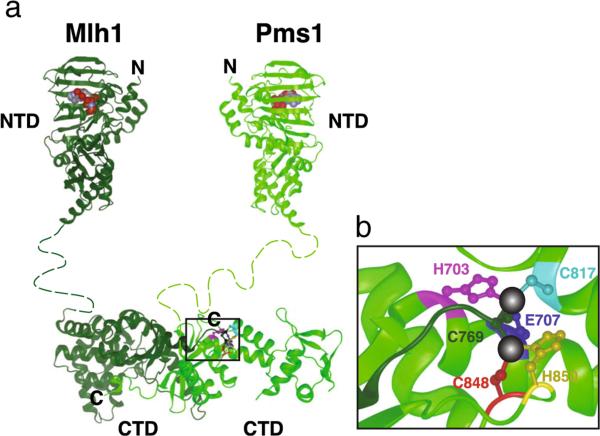
In S. cerevisiae, the Mlh1-Pms1 complex appears to be the major MutL homolog complex that functions in MMR, as mutations in MLH2 and MLH3 cause only weak MMR defects and Mlh2 and Mlh3 cannot substitute for Pms1. Importantly, MutLα possesses an endonuclease domain which resides on the CTD of the heterodimer, and which has been hypothesized to be required to make single-strand nicks in the newly synthesized DNA during MMR (Kadyrov et al. 2006). Interestingly, a similar endonuclease domain is present in Mlh3 but not in scMlh2 (or hPms1) (Fig. 3) (Kadyrov et al. 2006). The CTD of scPms1 (and hPms2) harbors in addition to the conserved DQHA(X)2E(X)4E endonuclease motif, other conserved motifs which are part of the endonuclease domain (including several residues that participate in the coordination of two metal ions present at the catalytic active site) (Fig. 4) (Gueneau et al. 2013; Smith et al. 2013). Consequently, mutations affecting these scPms1 residues (pms1-H703A, pms1-E707K, pms1-C817R, pms1-C848S, and pms1-H850R) result in loss of endonuclease function in vitro and MMR defects in vivo. In addition, according to the crystal structure of the scMlh1-Pms1 CTD, the C-terminal cysteine (Cys769) of scMlh1, present in the conserved FERC motif, is also part of this composite endonuclease domain (Fig. 4b) (reviewed in Guarne 2012). However, the role of the Cys769 remains unclear; in one study, deletion of this cysteine caused a mutator phenotype in one mutation rate assay (Gueneau et al. 2013), whereas in another study the same mutation did not affect the endonuclease activity of scMlh1-Pms1 in vitro and did not cause a mutator phenotype in three different mutation rate assays (Smith et al. 2013). In the latter study, it was necessary to delete the last three amino acids of the FERC motif of Mlh1 to affect MMR function (Smith et al. 2013).
Fig. 4.
Model for the Mlh1-Pms1 complex in S. cerevisiae adapted from Smith et al. 2013. a Mlh1 is shown in dark green and Pms1 in brighter green. Illustration of the N-terminal domains was generated based on structure pdb.3H4L (Arana et al. 2010), whereas the C-terminal domain structure corresponds to pdb.4e4w (Gueneau et al. 2013). N- and C-terminal domains are linked by an unstructured linker (dashed line). b Expanded view of the conserved amino acids comprising the metal binding pocket (metal ions in black) of the composite endonuclease site located within the black box in (a)
It has been shown that human MutLα is an endonuclease that can nick covalently closed circular DNA in a reaction that is stimulated by PCNA and RFC (Kadyrov et al. 2006; Pluciennik et al. 2010). However, under other reaction conditions, the MutLα endonuclease specifically nicks the already nicked strand of a nicked mispair containing circular DNA in a reaction that requires a mispaired base, ATP, MutSα, PCNA, and RFC. It has been proposed that PCNA is loaded onto DNA at the preexisting nick by RFC and that the asymmetric nature of PCNA loaded onto the DNA directs the endonuclease to nick the already nicked strand by a mechanism that is not yet understood (Pluciennik et al. 2010). An intriguing question raised by these results is that if a pre-existing nick is required for MutLα to nick the already nicked strand, then does the endonuclease actually function in MMR by catalyzing nicks that act as the sites of initiation of the excision reaction during MMR or does the endonuclease play another role in MMR.
Mlh1-Mlh2 (MutLβ)
The function of the scMlh1-Mlh2 complex (hMlh1-Pms1 in humans), also called MutLβ, has remained largely unexplored, in part due to the fact that scMlh2 (or hPms1) does not contain the conserved endonuclease motifs identified on MutLα and MutLγ homologs (Fig. 3). Furthermore, deletion of MLH2 in S. cerevisiae caused only a very weak mutator phenotype (Harfe et al. 2000), deletion of PMS1 in mice did not increase cancer susceptibility (Prolla et al. 1998) and partially purified hMlh1-Pms1 showed no activity in MMR reactions in vitro (Raschle et al. 1999). However, a number of recent experiments have suggested that the scMlh1-Mlh2 heterodimer may play a direct role in MMR (Campbell et al. 2014). First, mispair-bound scMsh2-Msh6 and scMsh2-Msh3 complexes were found to recruit scMlh1-Mlh2 in vitro. Second, scMlh1-Mlh2 was found to form mispair dependent foci in vivo. And third, deletion of scMLH2 was found to cause an increased mutator phenotype when the levels of scPms1 were reduced or in the absence of Msh6. Based on these observations, it has been proposed that scMlh1-Mlh2 acts as an accessory factor that facilitates scMlh1-Pms1-dependent MMR (Campbell et al. 2014).
Mlh1-Mlh3 (MutLγ)
The Mlh1-Mlh3 heterodimer is thought to play a minor role in MMR, as evidenced by the fact that a small fraction of insertion/deletion mispairs appear to be repaired by an Mlh1-Mlh3 dependent branch of an Msh2-Msh3 dependent MMR pathway in S. cerevisiae (Flores-Rozas and Kolodner 1998) and the fact that MLH3 and PMS2 double knockout mice have greater cancer susceptibility and a stronger mutator phenotype than PMS2 single knockout mice (Chen et al. 2005). More strikingly, Mlh1-Mlh3 acts in the processing of meiotic recombination intermediates in conjunction with the Msh4-Msh5 complex (Zakharyevich et al. 2012). Mlh3 harbors an endonuclease motif in its CTD (Nishant et al. 2008) (Fig. 3) and recent studies have shown that the Mlh1-Mlh3 complex has endonuclease activity. In one study, Mlh1-Mlh3 was found to nick supercoiled circular DNA in a reaction that was stimulated by Msh2-Msh3 but was surprisingly not stimulated by ATP, PCNA, or RFC (Rogacheva et al. 2014) whereas in another study Mlh1-Mlh3 was shown to cleave branched DNAs (Ranjha et al. 2014), consistent with a role in resolution of meiotic recombination intermediates.
Visualization of MMR complexes
Fluorescence microscopy has recently been used to obtain mechanistic information related to the kinetics and spatial-temporal organization of MMR protein complexes in living cells. MMR complexes have been visualized in bacteria and in human cell lines (Elez et al. 2010; Hong et al. 2008; Kleczkowska et al. 2001; Roesner et al. 2013; Smith et al. 2001). However, in these studies, the use of overexpression systems has complicated the interpretation of the results. Recently, in B. subtilis and S. cerevisiae, MMR components have been fluorescently tagged at their endogenous loci and visualized by live-cell fluorescence microscopy, although the investigated MMR components remained functional after tagging them only for S. cerevisiae (Campbell et al. 2014; Goellner et al. 2014; Hombauer et al. 2011a; Lenhart et al. 2013a, b; Simmons et al. 2008).
Live-cell imaging of S. cerevisiae cells with functional fluorescent-tagged MMR proteins revealed the presence of nuclear Msh2-Msh6 foci in S phase that colocalized with components present at DNA replication factories, including the DNA polymerases, PCNA and RPA. Interestingly, the formation and colocalization of these Msh2-Msh6 foci was independent of the presence of a mispair or the ability of Msh2-Msh6 to recognize them (Hombauer et al. 2011a), however, was dependent on the interaction of Msh6 with PCNA, as mutations in MSH6 (msh6-F33AF34A, msh6Δ2-50, msh6Δ2-251) (Clark et al. 2000; Flores-Rozas et al. 2000; Shell et al. 2007b) or PCNA (pol30-204) (Lau et al. 2002) which disrupt Msh6-PCNA interaction in vitro, abolished the Msh6 foci. Data obtained from studies in B. subtilis and human are consistent with these findings and have also identified beta clamp-dependent coupling of MutS and PCNA-dependent coupling of human Msh2-Msh6 with replicating DNA, respectively (Kleczkowska et al. 2001; Lenhart et al. 2013a, b; Liberti et al. 2011; Simmons et al. 2008).
Mutations in S. cerevisiae MSH6 that disrupt the PIP-box motif only resulted in a 10–15% reduction in MMR in otherwise wild-type cells but completely abolished MMR in cells containing a deletion of EXO1 (Hombauer et al. 2011a). These results suggest that in S. cerevisiae, there is a pathway that can substitute for the Msh6-PCNA interaction, although this second pathway does not appear to function in Exo1-independent MMR; the mechanism of this second pathway remains to be elucidated. In contrast, disruption of the beta clamp-MutS interaction in B. subtilis appears to almost completely inactivate MMR (Lenhart et al. 2013b).
In S. cerevisiae, after mispair recognition, Msh2-Msh6 (or Msh2-Msh3) forms a ternary complex with Mlh1-Pms1 and the mispair containing DNA (Habraken et al. 1997; Mendillo et al. 2005). Interestingly, in S. cerevisiae, functional fluorescent-tagged Pms1 formed nuclear foci during S-phase that were dependent on a functional mispair recognition complex, either Msh2-Msh6 or Msh2-Msh3, and the presence of mispaired bases (Hombauer et al. 2011a). Surprisingly, Mlh1- Pms1 foci rarely colocalized with DNA replication-associated Msh2-Msh6 foci, arguing that the two types of foci represent different steps in MMR. Moreover, the half-life of the Mlh1-Pms1 foci was short (1–2 min) which is consistent with in vitro experiments arguing for a rapid repair reaction (Fang and Modrich 1993; Wang and Hays 2002). The abundance of Mlh1-Pms1 foci was increased in S. cerevisiae strains where DNA replication fidelity was compromised or by MMR defects that act downstream of Mlh1-Pms1 recruitment (i.e., Pms1-endonuclease defective mutants, pol30 mutants unable to stimulate the Pms1 endonuclease activity or an exo1Δ mutation; Table 2). These results suggest that the Pms1 foci represent active sites of MMR repair in S. cerevisiae, and might be the result of loading several Mlh1-Pms1 molecules by Msh2-Msh6 at the mispair site (Hombauer et al. 2011a).
Table 2.
Pms1-foci abundance in different MMR-related mutants in S. cerevisiae
| Biochemical defect | Relevant genotype | Mutator phenotype (frameshift-reversion assay) | % Pms1 focia | Reference |
|---|---|---|---|---|
| None (wild-type strain) | Wild-type | None | 10 | Hombauer et al. 2011a, b |
| 1. Reduced DNA replication fidelity | ||||
| (a) exonuclease defective polymerases | pol3-01; pol2-04 | None | 20–25 | Hombauer et al. 2011a |
| (b) polymerase active-site mutation | pol3-L612M | None | 20 | Hombauer et al. 2011a |
| 2. Mutations that interfere with MMR | ||||
| (a) Preventing mispair recognition | msh2Δ; msh3Δmsh6Δ | Strong | 0 | Hombauer et al. 2011a |
| (b) Msh6 dominant mutations (in an msh3Δ background) | S1036P; G1067D; G1142D | Strong | 0 | Hombauer et al. 2011a |
| (c) Excision (5′-3′ Exo1-dependent) | exo1Δ | Weak | 50 | Hombauer et al. 2011a |
| (d) Nicking reaction | ||||
| PCNA (Pol30) mutations that prevent Pms1 endonuclease activation | K13E; C22Y; K217E | Moderate | 60–95 | Goellner et al. 2014 |
| Pms1 endonuclease defective mutations | G683E, E707K, C817R, C848S | Strong | 60–85 | Smith et al. 2013 |
| Mlh1 endonuclease defective mutation | E767stop | Strong | 95 | Smith et al. 2013 |
Percentage of yeast cells containing Pms1-foci on a logarithmically growing population for the indicated genotypes
Further analysis of the alternative MutL homologs in S. cerevisiae, revealed that Mlh2 also formed nuclear foci that frequently colocalized with Mlh1-Pms1, recapitulating most of the behavior of Mlh1-Pms1 repair intermediates (Campbell et al. 2014). Importantly, Mlh2 foci were dependent on a functional Msh2-Msh6 complex and their abundance was increased by mutations affecting MMR downstream of mispair recognition or causing decreased DNA replication fidelity. Thus, even though Mlh2 lacks an endonuclease domain, these imaging studies (together with genetic and biochemical data) suggest that the Mlh1-Mlh2 complex functions during MMR in conjunction with Mlh1-Pms1. Therefore, it has been proposed that Mlh1-Mlh2 plays an accessory role during the mispair repair reaction, most likely facilitating interactions with MMR-related components (i.e., PCNA or Exo1). In contrast to Pms1, and supporting a minor role for Mlh3 in MMR, Mlh3 did not form nuclear foci in logarithmically growing S. cerevisiae cells or after DNA damaging treatments capable of inducing Mlh1-Pms1 and Mlh1-Mlh2 foci (Campbell et al. 2014).
In summary, visualization of mismatch repair components in living yeast cells revealed the existence of distinct recognition and repair MMR intermediates. Several mutations on MMR and DNA replication components have been reported to have drastic effects in the abundance of these repair intermediates. Moreover, a model for MMR in which MutSα loads catalytically several molecules of MutLα at the mispair site/proximities (described in more detail later) has been proposed (Hombauer et al. 2011a). Future studies might proof this model and potentially identify similarities in the mechanism of MMR in other higher evolved eukaryotes.
It is worth mentioning that although the visualization of fluorescently tagged proteins has proven to be a valuable tool, it is restricted by the microscopy diffraction limit (range of 100–200 nm), which is below the limit of detection required to visualize single protein complexes (which are in the range of 10 nm). This limitation might be overcome by single-molecule studies in combination of high-resolution methods, which has been already applied to investigate some mechanistic aspects of the mismatch repair reaction (Lee et al. 2014).
Coupling of MMR with DNA replication
In order to prevent mutations due to DNA replication errors that escape the proofreading activity of the major leading and lagging strand DNA polymerases, Polε and Polδ, respectively, MMR must repair the newly synthesized DNA strand prior to the next round of DNA replication. Furthermore, MMR must be able to target rare mispairs in a vast excess of basepaired DNA as replication errors are rare. Coupling of MMR to DNA replication offers a potential solution to these problems. The possibility of active coupling to replication was arguably first suggested by the observation that Msh2-Msh6 and Msh2-Msh3 interact with PCNA through a PIP-box motif located at the N-terminus of Msh6 and Msh3 potentially allowing the molecular toolbelt function of PCNA (Indiani et al. 2005) to have these mismatch recognition factors immediately available, in case they are needed (Clark et al. 2000; Flores-Rozas et al. 2000; Kleczkowska et al. 2001). The interaction between MutLα and PCNA also offers a possibility for replication coupling, although the molecular basis for this interaction, and hence its significance is not yet understood. In addition, the observation that nicks in mispaired DNA act as initiating signals in reconstituted MMR reactions in vitro suggests that DNA breaks present transiently on newly synthesized DNA strands, such as the lagging strand that is synthesized discontinuously, could passively target MMR to newly synthesized DNA during DNA replication. Consistent with this, it was initially suggested that the lagging DNA strand was more efficiently repaired by MMR than the leading DNA strand (Pavlov et al. 2003) but other subsequent studies have indicated that both leading and lagging DNA strands are efficiently repaired by MMR (Hawk et al. 2005; Hombauer et al. 2011a; Lujan et al. 2012).
As discussed above, imaging of MMR and DNA replication components revealed that the Msh2-Msh6 mispair recognition complex colocalized with DNA replication components during S-phase in human cells and in S. cerevisiae (Hombauer et al. 2011a; Kleczkowska et al. 2001). However, these observations did not formally prove that MMR actually occurred in a manner that was coupled to DNA replication. This hypothesis was further evaluated by restricting MMR protein expression to specific stages of the cell cycle in S. cerevisiae using cell cycle specific promoters to drive Msh2-Msh6 expression (Hombauer et al. 2011b). This analysis revealed a strong temporal coupling between MMR and DNA replication, and also suggested that strand discrimination in S. cerevisiae MMR relies on a transient DNA replication-associated signal.
In addition to the Msh6-PCNA interaction that targets the mismatch recognition complexes to the replication factories, it has been shown in human cells that epigenetic modifications promote the recruitment of Msh2-Msh6 to chromatin, which might facilitate a later interaction with PCNA at the replication fork (Li et al. 2013). This recruitment is mediated by histone H3K36me3 and the PWWP domain located at the N-terminus of human Msh6 (Laguri et al. 2008). Msh3 lacks a PWWP domain and hence the Msh2-Msh3 complex is unlikely to be recruited to chromatin by this mechanism. The histone H3K36me3 modification is cell cycle regulated and peaks during early S phase, and then declines until it is rarely detectable during G2/M (Bonenfant et al. 2007; Ryba et al. 2010); however, the role of H3K36me3 is best established for marking transcriptionally active chromatin and is less well understood in DNA replication and cell cycle control (Wagner and Carpenter 2012). The transition from H3K36me2 to H3K36me3 is catalyzed by the methyltransferase SetD2 (Wagner and Carpenter 2012). Human cells lacking SetD2 activity showed increased mononucleotide and dinucleotide repeat instability and an elevated mutation rate, although not as large as that caused by deletion of MSH6 (Li et al. 2013). It should be noted that cells lacking MSH6 show little if any dinucleotide repeat instability (Oki et al. 1999) because two-base insertion/deletion mispairs can still be repaired by Msh2-Msh3 (Genschel et al. 1998), which is somewhat different from the phenotype caused by loss of SetD2. In addition, cells lacking SetD2 have major alterations in transcription and chromatin structure, which could result in indirect alteration of MMR capacity. Consequently, it will be necessary to test the effect of human Msh6-PWWP domain mutations on MMR to determine if recruitment of Msh2-Msh6 to chromatin by H3K36me3 plays a direct role in MMR. The findings of Li and coworkers are not applicable to S. cerevisiae since scMsh6 and scMsh3 do not contain PWWP domains, however, deletion of the N-terminal tether of scMsh6 causes greater MMR defects than mutations that only affect binding of Msh6 to PCNA (Shell et al. 2007b), suggesting that in S. cerevisiae other features of the Msh6-NTR might regulate its recruitment/function. Additional studies are needed to address this possibility.
Mechanistic aspects of MMR
Strand discrimination during MMR in eukaryotes
In E. coli, after recognition of the mispair by MutS and recruitment of MutL, the MutH endonuclease introduces a nick into the newly synthesized DNA strand. Strand discrimination is specified by the hemi-methylated status of the newly synthesized DNA, in which the daughter strand is transiently unmethylated becoming a substrate for MutH endonuclease, which exclusively nicks the unmethylated DNA at hemimethylated d(GATC) sites (Welsh et al. 1987).
In eukaryotes, methylation does not serve as a strand discrimination signal. While, the nature of the eukaryotic strand discrimination signal has remained unknown for several decades, data have accumulated that suggest that nicks or gaps present in DNA might be used to discriminate the parental from daughter DNA strands. It has been suggested that the 5′ or 3′ ends of Okazaki fragments could initiate the MMR reaction on the lagging strand (Fang and Modrich 1993; Holmes et al. 1990; Thomas et al. 1991), a view that has been solidified by the observation that the MutLα endonuclease is targeted by pre-existing nicks to then further nick the mispair-containing strand. This finding, together with the physical coupling of MMR proteins to the replication machinery, provides a possible mechanism for how MMR can be targeted to the newly synthesized lagging strand. However, it has been also shown that MMR acts with a similar efficiency on the leading strand (Hawk et al. 2005; Hombauer et al. 2011a; Lujan et al. 2012), which is synthesized in a continuous manner and in principal lacks a high density of nicks. This observation then raises the question of what might be the strand specificity signal for leading-strand MMR.
Recently, it has been proposed that ribonucleotides, which can be misincorporated by DNA polymerases during DNA replication, might represent an additional source of nicks or gaps in the newly synthesized strand that might serve as a strand specificity signal. RNase H2 in mammals and S. cerevisiae makes single-strand breaks at the site of individual misincorporated ribonucleotides during ribonucleotide excision repair, potentially leaving behind a nick in DNA that could also be used as a strand discrimination signal (Ghodgaonkar et al. 2013; Lujan et al. 2013). However, misincorporated ribonucleotides are unlikely to provide a major strand discrimination signal during MMR for three reasons: (1) the reported weak mutator phenotype due to loss of RNase H2 was dependent upon DNA polymerase mutations that cause over a 10-fold increase in ribonucleotide misincorporation; (2) deletion of the catalytic subunit of S. cerevisiae RNase H2 results in little or no mutator general phenotype; and (3) the mutator phenotype seen in dinucleotide repeat reporters in RN-ase H2 defective mutants is dependent on nicking of the DNA by topoisomerase I and is not due to an MMR defect (Allen-Soltero et al. 2014; Ghodgaonkar et al. 2013; Kim et al. 2011; Lujan et al. 2013; Nick McElhinny et al. 2010).
PCNA is a critical replication factor that could provide a signal for DNA strand discrimination on the leading strand. PCNA is loaded onto DNA with a specific orientation relative to that of the primer termini, where it is present and acts as a processivity factor for DNA polymerases. This polarity appears to direct the strand specificity of MutLα incision (Pluciennik et al. 2010) and could in principle direct MutLα nicking of the newly synthesized leading strand if mismatch recognition in some way promoted disassembly of the replication fork allowing MutLα to then nick the DNA. However, a recent study that reconstituted the S. cerevisiae DNA replisome in vitro, has found that PCNA may not be as important for the processivity of the leading strand DNA polymerase (Polε or also called Pol2) as the interaction between Polε and the CMG complex (Cdc45, Mcm2-7, and GINS) (Georgescu et al. 2014). In contrast, PCNA is essential for promoting the processivity of the lagging strand DNA polymerase (Polδ or also called Pol3). These results suggest the possibility that either PCNA, as was initially proposed (Umar et al. 1996), or the CMG complex that is also loaded onto DNA with a distinct polarity could provide a strand discrimination signal for MMR, although this latter possibility has not been explored. Additional experimentation will be required to understand strand discrimination during MMR in eukaryotes, particularly for leading strand MMR.
Coupling mismatch recognition to downstream events
Several models have been proposed to explain how MutS recognize mispairs and how this finally leads to the excision and resynthesis of a fragment of the DNA strand that contains the mispair. These include (1) the stationary model where MutS remains bound to the mispair and interaction with MMR proteins bends and loops DNA to bring together mispair and strand discrimination signal (Guarne et al. 2004), (2) the translocation model that proposes that ATP hydrolysis is the driving force for MutS translocation along the DNA forming DNA loops that bring the mispair into the proximity of the strand discrimination signal (Allen et al. 1997), and (3) the molecular switch model also called sliding clamp model, in which ATP binding promotes a conformational change in MutS or the MSH complexes allowing them to move along the DNA followed by iterative loading of additional MutS or MSH complexes on the DNA mispair (Acharya et al. 2003). Of the three models, the translocation model and the sliding clamp model are consistent with the observation that a physical block between the mispair site and the site of an initiating nick blocks MMR (Pluciennik and Modrich 2007; Wang and Hays 2003). A general feature of these models is that they assume that one MutS or MSH complex binds one MutL or MLH complex (in a 1:1 stoichiometry) and translocates or slides away from the mispair along the DNA.
Recently, another model has been proposed in which after mispair recognition an MSH complex catalytically loads several molecules of MLH complexes at or near the site of the mispair (Hombauer et al. 2011a). This model is based on the identification in S. cerevisiae of DNA replication-associated Msh2-Msh6 foci and the observation of Mlh1-Pms1 foci that accumulate in an Msh2-Msh6 dependent fashion in response to mispaired bases but which rarely colocalize with Msh2-Msh6 foci or DNA replication factories. Since in order to be visualized as a focus, multiple molecules of these proteins must be in close proximity, and because the interaction between Msh2-Msh6 and Mlh1-Pms1 is unlikely to result in to simultaneous recruitment of multiple Mlh1-Pms1 complexes, it has been proposed that Msh2-Msh6 might be able to catalytically load Mlh1-Pms1 onto DNA in response to mispairs. This model is also supported by the observation that msh6 dominant mutations (msh6-S1036P or msh6-G1067D) in S. cerevisiae, which prevent scMlh1-Pms1 recruitment in vitro (Hess et al. 2006), were also unable to form Pms1 foci in vivo (Hombauer et al. 2011a). Remarkably, the msh6-G1142D-dominant mutation that supports scMlh1-Pms1 recruitment in vitro (Hess et al. 2006) was also defective for Pms1 foci formation, suggesting that recruitment not necessarily guarantee the catalytic loading of scMlh1-Pms1 (Hombauer et al. 2011a). Further studies will be required to demonstrate that Msh2-Msh6 actually promotes catalytic loading of scMlh1-Pms1 onto DNA.
Mismatch excision
Another feature of MMR that still remains to be further explored is how mismatch excision takes place in vivo. For a long time, it has been known that Exo1 is involved during this process; nevertheless, this exonuclease is not essential for MMR. Inactivation of EXO1 in S. cerevisiae or mice results in a very weak mutator phenotype (Amin et al. 2001; Edelmann and Edelmann 2004; Tishkoff et al. 1997; Wei et al. 2003), suggesting that other redundant mechanisms or exonucleases must exist.
In an attempt to identify redundant excision mechanisms, Amin et al. searched for mutations that confer a synergistic mutator phenotype in the absence of EXO1 (exo1Δ-dependent mutations or EDM) in S. cerevisiae (Amin et al. 2001). Interestingly, no additional exonucleases were identified but rather mutations in MMR genes (MLH1, PMS1, MSH2, and MSH3) and genes encoding DNA replication components (Pol30, Pol32, and Rnr1) were found. More recently, additional EDM mutations have been identified including mutations in the DNA polymerases Polδ (pol3-L612M) (Hombauer et al. 2011a) and Polα (pol1-L868M) (Liberti et al. 2013), Msh6-PCNA interaction defective mutations (msh6Δ2-50 or msh6-F33AF34A) (Hombauer et al. 2011a) and POL30 mutations that cause defects in activation of S. cerevisiae Mlh1-Pms1 endonuclease activity (Goellner et al. 2014). In addition, the fact that mutations affecting proteins that function in lagging strand DNA synthesis (Polα, Polδ, and Pol32) synergize with an exo1Δ mutation to a greater extent than when exo1Δ is combined with a low-fidelity leading strand polymerase mutation (pol2-M644G), suggests that Exo1 might be more important for the correction of errors generated on the lagging strand (Hombauer et al. 2011a).
The identification of exo1Δ-dependent pol30 mutations resulting in mutant PCNAs that were partially defective in activating the scMlh1-Pms1 endonuclease suggests that mispair excision becomes more Exo1-dependent when scMlh1-Pms1 endonuclease activity is reduced (Goellner et al. 2014). In addition, it was observed that an Msh6-PCNA interaction defective mutation (msh6Δ2-50) resulted in a synergistic increase in mutation rates in combination with an exo1Δ mutation or with pol30 mutations that interfere with the activation of scMlh1-Pms1 endonuclease activity but not in combination with other pol30 mutations that affect the interaction between Pol30 and Msh6. These results have suggested a model in which Msh2-Msh6 plays a role downstream of scMlh1-Pms1 recruitment by retaining/recruiting PCNA in proximity to the mispair site, which then supports activation of the scMlh1-Pms1 endonuclease (Fig. 5). Then, in the absence of Exo1, scMlh1-Pms1 performs multiple rounds of DNA nicking (stimulated by PCNA) that either directly results in nick-directed degradation of the mutant DNA strand or possibly processing by DNA Polδ-dependent strand displacement (Kadyrov et al. 2009) or by the 3′–5′ exonuclease activity of DNA polymerases (Tran et al. 1999). Further studies will be required to validate the existence of this type of Exo1-independent mispair excision.
Fig. 5.
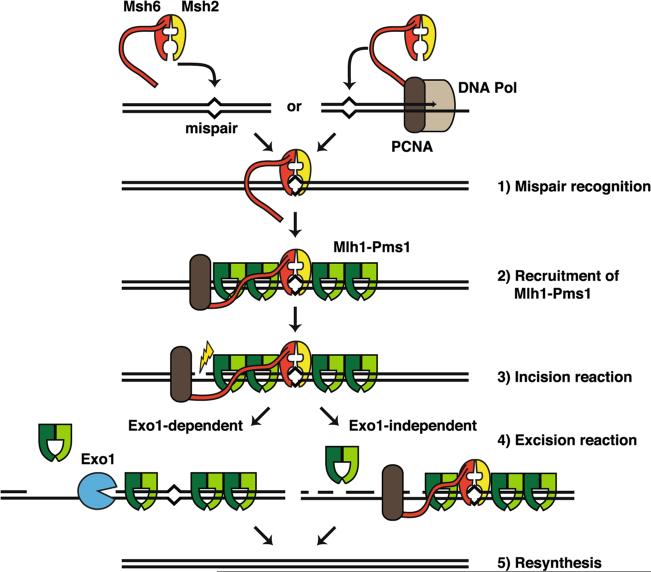
Model of alternative excision pathways that act during MMR adapted from Goellner et al. 2014. The diagram describes the sequential steps of the MMR reaction. The Msh2-Msh6 heterodimer recognizes mispairs (1). A fraction of the recognition complexes is coupled to replication machinery (PCNA-DNA Pol) through PCNA. Alternatively, the Msh2-Msh6 complex scans the DNA for mispairs in a PCNA-interaction independent manner. The mispair-bound Msh2-Msh6 complex recruits Mlh1-Pms1 (2). The loading of Mlh1-Pms1 onto DNA is catalytic whereby one mispair recognition complex recruits several Mlh1-Pms1 complexes to or near the mispair site. The Mlh1-Pms1 endonuclease is then activated by PCNA to nick the DNA comprising the incision step (3). At this step, Msh6 plays a role by retaining or recruiting PCNA in proximity to the mispair site. The excision reaction (4) can be mediated by Exo1 (Exo1-dependent) or by unknown factors (Exo1-independent) potentially including multiple rounds of incision by Mlh1-Pms1, strand displacement by the DNA replication machinery or other exonucleases followed by resynthesis of the DNA (5)
Non-canonical mismatch repair functions
In addition to the antimutator role of MMR, which includes the correction of DNA replication errors and the suppression of homeologous recombination events, there is substantial evidence that indicate that MMR can also play a mutagenic role promoting genome instability. As was previously mentioned, one of these functions is related to trinucleotide repeats, in which Msh2-Msh3 complexes promote expansions at these regions, by a mechanism which is still not completely understood.
In addition, MMR plays mutagenic functions also referred as non-canonical mismatch repair (ncMMR) during antibody diversification, which takes place during somatic hypermutation (SHM) and class switch recombination (CSR) of immunoglobulin genes (Bak et al. 2014; Slean et al. 2008). The mutagenic role of ncMMR is driven by the correction of mispairs uncoupled from DNA replication; repair events that occurs in this manner are expected to be mutagenic due to the absence of a DNA strand discrimination signal (Hombauer et al. 2011b), which results in an unbiased and therefore mutagenic repair.
SHM and CSR require the action of the activation-induced deamination (AID) enzyme, as well as the base excision machinery (BER). During SHM, AID converts cytosine into uracil at regions that are actively transcribed, resulting in U:G mismatches at the immunoglobulin genes, that are then recognized by BER and/or MMR. Strand excision is followed by MMR-dependent PCNA ubiquitylation and recruitment of error-prone polymerases (Polη), which introduces mutations at the repaired track. During CSR, incisions generated by BER and MMR lead to double-strand breaks at the Ig constant region genes, which are then repaired by recombination (non-homologous end-joining and alternative end-joining) with the upstream variable Ig region.
It has been also proposed that ncMMR might not be restricted to the process of immunoglobulin diversification occurring in B cells, but rather might occur in other cell types and potentially drive in some cases carcinogenesis of somatic and non-diving cells (MacPhee 1995). This hypothesis is supported by two recent articles that came to a similar conclusion, though having used different approaches. In a first report (Rodriguez et al. 2012), the authors used single-strand oligo-nucleotides to introduce mutations and revert a non-functional allele of the TRP5 gene in S. cerevisiae. Analysis of the recovery of Trp+ revertants after transforming oligos targeting the transcribed or non-transcribed strand, in MMR-proficient or MMR-deficient strains, revealed that when mispair correction is uncoupled from DNA replication, it uses as template for repair either one of the two DNA strands, most likely due to the absence of a DNA strand discrimination signal and is therefore mutagenic. In agreement with mutagenic role for MMR, a second report showed that treatment of human cells with the alkylating agent MNNG results in Mlh1-dependent PCNA ubiquitylation and recruitment of Polη, independent of DNA replication (Pena-Diaz et al. 2012). Moreover, this study also provided evidence that MMR can promote MNNG-induced mutagenesis as detected using the HPRT gene inactivation assay. In summary, these results indicate that MMR participates in recognition of MNNG-induced DNA damage (during or outside S-phase) promoting mutagenesis due to the lack of a DNA strand directionality signal and the involvement of low-fidelity polymerases, resulting in mutagenic repair (Pena-Diaz et al. 2012).
Identification of new genes/mechanisms preventing mutator phenotypes
It is well known that germline inactivating mutations affecting the major MMR genes cause inherited cancer susceptibility, and that sporadic cancers showing increased mutation rates, often seen as increased microsatellite instability, have acquired MMR defects, which in most cases are consequence of epigenetic silencing of hMLH1 (Kinzler and Vogelstein 1996; Kolodner et al. 1999; Peltomaki and Vasen 2004). However, not all microsatellite unstable tumors and tumors with increased mutation rates are associated with mutation or altered expression of the genes encoding the essential MMR components, suggesting that additional factors may contribute to mutator phenotypes linked to the development of cancer (Peltomaki 2003).
Several recent studies have demonstrated a potential role for interactions of chromatin modifications with MMR. These include the observation that the activity of H3K36 trimethyltransferase SetD2 plays a role in the recruitment of MutSα to chromatin in humans (Li et al. 2013). The observation that depletion of SetD2 causes a weak mutator phenotype and that SETD2 may be rarely mutated in some cancers raises the possibility that defects which affect the crosstalk between MMR and chromatin can result in a mutator phenotype and cancer susceptibility, although this remains to be definitively proven. In addition, in S. cerevisiae, a second histone modification, the S-phase specific H3K56 acetylation, has been suggested to play a role in genome maintenance, together with MMR and DNA Polε and Polδ proofreading activity (Kadyrova et al. 2013). Intriguingly, deletion of MSH2 synergizes with defects in both acetylation and deacetylation, suggesting that alteration of H3K56 acetylation is distinct from a MMR defect, but causes genome instability due to the interaction of MMR defects with damage caused by deregulation of histone modification.
A second mechanism that could lead to increased mutation rates and the development of cancer is an imbalance in the expression of MMR proteins. In S. cerevisiae, it is known that overexpression of wild-type scMlh1, scMlh2, scMlh3, or scPms1 results in a mutator phenotype (Campbell et al. 2014; Shcherbakova et al. 2001; Shcherbakova and Kunkel 1999). This suggests that the overexpression of some MMR proteins can titrate out other MMR proteins resulting in an MMR defect; an example is the titration of scMlh1 by increased expression of either scMlh2 or scMlh3 that then interferes with the formation of sufficient scMlh1-Pms1 complex to support MMR (Campbell et al. 2014). Similarly, overexpression of Msh3 in mammalian cell lines (Drummond et al. 1997; Marra et al. 1998) and overexpression of hPms2 in mouse cells can result in elevated mutation frequencies (Gibson et al. 2006). In line with these findings, immunohisto-chemical staining of prostate cancer samples identified elevated levels of hPms2, which correlated with MSI (Norris et al. 2007) and increased recurrence after prostatectomy (Norris et al. 2009). More recently, a comparative study of genome databases available for head and neck squamous cell carcinomas (HNSCC) and human HNSCC cell lines, identified hPMS1 as one of eight genes that were consistently found to have increased copy number (Li et al. 2014). Yet another study that analyzed chromosomal genomic hybridization databases for human cancer identified multiple chromosomal hot spots for genome amplification that contained different oncogenes as well as MMR genes including hMSH2, hPMS1, and hPMS2 (Myllykangas et al. 2006). These latter studies provide some support for the idea that increased copy number and increased expression of MMR genes might be linked to increased mutation rates and development of cancer although additional analysis will be required to provide definitive proof of this.
Finally, it has been known for many years that mutations affecting the active site of DNA polymerases or their exonuclease domain (reviewed in McCulloch and Kunkel 2008; Prindle and Loeb 2012; Reha-Krantz 2010) can result in increased accumulation of mutations. The high levels of misincorporation of errors likely results in saturation of MMR, such that some of the resulting mispairs escape repair (Damagnez et al. 1989; Schaaper and Radman 1989). Consistent with this, a number of studies have recently identified both inherited and acquired mutations in human leading and lagging strand polymerases (POLE and POLD1, respectively) affecting conserved residues, which are part of the proofreading domain of these DNA polymerases in colorectal and endometrial cancers (Church et al. 2013; Kane and Shcherbakova 2014; Palles et al. 2013). These defects cause a hyper-mutator phenotype characterized by the accumulation of base substitution mutations, but do not appear to result in increased microsatellite instability.
Acknowledgments
The authors thank Dr. Christopher Putnam for critical reading of this review.
Funding This work was supported by the Harald zur Hausen Fellowship from the Deutsches Krebsforschungszentrum (DKFZ) and the Marie Curie Career Integration Grant “iMMR” (both granted to H.H.). R.K. was supported by the NIH grant GM50006 and the Ludwig Institute for Cancer Research.
Footnotes
Compliance with ethical standards
Conflict of interest statement The authors declare that they have no conflict of interest.
Ethical approval This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- Ahrends R, Kosinski J, Kirsch D, Manelyte L, Giron-Monzon L, Hummerich L, Schulz O, Spengler B, Friedhoff P. Identifying an interaction site between MutH and the C-terminal domain of MutL by crosslinking, affinity purification, chemical coding and mass spectrometry. Nucleic Acids Res. 2006;34:3169–3180. doi: 10.1093/nar/gkl407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen-Soltero S, Martinez SL, Putnam CD, Kolodner RD. A Saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol Cell Biol. 2014;34:1521–1534. doi: 10.1128/MCB.00960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21:5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony E, Hingorani MM. Mismatch recognition-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry. 2003;42:7682–7693. doi: 10.1021/bi034602h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana ME, Holmes SF, Fortune JM, Moon AF, Pedersen LC, Kunkel TA. Functional residues on the surface of the N-terminal domain of yeast Pms1. DNA Repair (Amst) 2010;9:448–457. doi: 10.1016/j.dnarep.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak ST, Sakellariou D, Pena-Diaz J. The dual nature of mismatch repair as antimutator and mutator: for better or for worse. Front Genet. 2014;5:287. doi: 10.3389/fgene.2014.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- Bende SM, Grafstrom RH. The DNA binding properties of the MutL protein isolated from Escherichia coli. Nucleic Acids Res. 1991;19:1549–1555. doi: 10.1093/nar/19.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Obmolova G, Takahashi M, Herr A, Newman MA, Yang W, Hsieh P. Disruption of the helix-u-turn-helix motif of MutS protein: loss of subunit dimerization, mismatch binding and ATP hydrolysis. J Mol Biol. 2001;305:805–816. doi: 10.1006/jmbi.2000.4367. [DOI] [PubMed] [Google Scholar]
- Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(2073–2087):e2073. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland CR, Lynch HT. The history of Lynch syndrome. Familial Cancer. 2013;12:145–157. doi: 10.1007/s10689-013-9637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonenfant D, Towbin H, Coulot M, Schindler P, Mueller DR, van Oostrum J. Analysis of dynamic changes in posttranslational modifications of human histones during cell cycle by mass spectrometry. Mol Cell Proteomics: MCP. 2007;6:1917–1932. doi: 10.1074/mcp.M700070-MCP200. [DOI] [PubMed] [Google Scholar]
- Bowers J, Sokolsky T, Quach T, Alani E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J Biol Chem. 1999;274:16115–16125. doi: 10.1074/jbc.274.23.16115. [DOI] [PubMed] [Google Scholar]
- Campbell CS, Hombauer H, Srivatsan A, Bowen N, Gries K, Desai A, Putnam CD, Kolodner RD. Mlh2 is an accessory factor for DNA mismatch repair in Saccharomyces cerevisiae. PLoS Genet. 2014;10:e1004327. doi: 10.1371/journal.pgen.1004327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PC, Dudley S, Hagen W, Dizon D, Paxton L, Reichow D, Yoon SR, Yang K, Arnheim N, Liskay RM, Lipkin SM. Contributions by MutL homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005;65:8662–8670. doi: 10.1158/0008-5472.CAN-05-0742. [DOI] [PubMed] [Google Scholar]
- Church DN, Briggs SE, Palles C, Domingo E, Kearsey SJ, Grimes JM, Gorman M, Martin L, Howarth KM, Hodgson SV, Collaborators N, Kaur K, Taylor J, Tomlinson IP. DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Hum Mol Genet. 2013;22:2820–2828. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J Biol Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Hays JB. Arabidopsis MutS homologs-AtMSH2, AtMSH3, AtMSH6, and a novel AtMSH7-form three distinct protein heterodimers with different specificities for mismatched DNA. Plant cell. 2000;12:991–1002. doi: 10.1105/tpc.12.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Meyer-Gauen G, Lyons-Weiler J, Hays JB. Evolutionary origin, diversification and specialization of eukaryotic MutS homolog mismatch repair proteins. Nucleic Acids Res. 2000;28:463–471. doi: 10.1093/nar/28.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damagnez V, Doutriaux MP, Radman M. Saturation of mismatch repair in the mutD5 mutator strain of Escherichia coli. J Bacteriol. 1989;171:4494–4497. doi: 10.1128/jb.171.8.4494-4497.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Putnam CD, Kolodner RD. Functional studies and homology modeling of Msh2-Msh3 predict that mispair recognition involves DNA bending and strand separation. Mol Cell Biol. 2010;30:3321–3328. doi: 10.1128/MCB.01558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drotschmann K, Hall MC, Shcherbakova PV, Wang H, Erie DA, Brownewell FR, Kool ET, Kunkel TA. DNA binding properties of the yeast Msh2-Msh6 and Mlh1-Pms1 heterodimers. Biol Chem. 2002;383:969–975. doi: 10.1515/BC.2002.103. [DOI] [PubMed] [Google Scholar]
- Drummond JT, Genschel J, Wolf E, Modrich P. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proc Natl Acad Sci U S A. 1997;94:10144–10149. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Edelmann W. Loss of DNA mismatch repair function and cancer predisposition in the mouse: animal models for human hereditary nonpolyposis colorectal cancer. Am J Med Genet Part C, Seminars Med Genet. 2004;129C:91–99. doi: 10.1002/ajmg.c.30021. [DOI] [PubMed] [Google Scholar]
- Elez M, Murray AW, Bi LJ, Zhang XE, Matic I, Radman M. Seeing mutations in living cells. Curr Biol. 2010;20:1432–1437. doi: 10.1016/j.cub.2010.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang WH, Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frame-shift mutations. Proc Natl Acad Sci U S A. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- Galio L, Bouquet C, Brooks P. ATP hydrolysis-dependent formation of a dynamic ternary nucleoprotein complex with MutS and MutL. Nucleic Acids Res. 1999;27:2325–2331. doi: 10.1093/nar/27.11.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Mohan R, Gupta RS. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int J Syst Evol Microbiol. 2009;59:234–247. doi: 10.1099/ijs.0.002741-0. [DOI] [PubMed] [Google Scholar]
- Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- Georgescu RE, Langston L, Yao NY, Yurieva O, Zhang D, Finkelstein J, Agarwal T, O'Donnell ME. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodgaonkar MM, Lazzaro F, Olivera-Pimentel M, Artola-Boran M, Cejka P, Reijns MA, Jackson AP, Plevani P, Muzi-Falconi M, Jiricny J. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SL, Narayanan L, Hegan DC, Buermeyer AB, Liskay RM, Glazer PM. Overexpression of the DNA mismatch repair factor, PMS2, confers hypermutability and DNA damage tolerance. Cancer Lett. 2006;244:195–202. doi: 10.1016/j.canlet.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Goellner EM, Smith CE, Campbell CS, Hombauer H, Desai A, Putnam CD, Kolodner RD. PCNA and Msh2-Msh6 activate an Mlh1-Pms1 endonuclease pathway required for Exo1-independent mismatch repair. Mol Cell. 2014;55:291–304. doi: 10.1016/j.molcel.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Superfamily of UvrA-related NTP-binding proteins. Implications for rational classification of recombination/repair systems. J Mol Biol. 1990;213:583–591. doi: 10.1016/S0022-2836(05)80243-8. [DOI] [PubMed] [Google Scholar]
- Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- Guarne A. The functions of MutL in mismatch repair: the power of multitasking. Prog Mol Biol Transl Sci. 2012;110:41–70. doi: 10.1016/B978-0-12-387665-2.00003-1. [DOI] [PubMed] [Google Scholar]
- Guarne A, Ramon-Maiques S, Wolff EM, Ghirlando R, Hu X, Miller JH, Yang W. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO J. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueneau E, Dherin C, Legrand P, Tellier-Lebegue C, Gilquin B, Bonnesoeur P, Londino F, Quemener C, Le Du MH, Marquez JA, Moutiez M, Gondry M, Boiteux S, Charbonnier JB. Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nat Struct Mol Biol. 2013;20:461–468. doi: 10.1038/nsmb.2511. [DOI] [PubMed] [Google Scholar]
- Gupta S, Gellert M, Yang W. Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops. Nat Struct Mol Biol. 2012;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken Y, Sung P, Prakash L, Prakash S. Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Curr Biol. 1997;7:790–793. doi: 10.1016/s0960-9822(06)00337-x. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Minesinger BK, Jinks-Robertson S. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr Biol. 2000;10:145–148. doi: 10.1016/s0960-9822(00)00314-6. [DOI] [PubMed] [Google Scholar]
- Hargreaves VV, Shell SS, Mazur DJ, Hess MT, Kolodner RD. Interaction between the Msh2 and Msh6 nucleotide-binding sites in the Saccharomyces cerevisiae Msh2-Msh6 complex. J Biol Chem. 2010;285:9301–9310. doi: 10.1074/jbc.M109.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol Cell Biol. 2007;27:6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk JD, Stefanovic L, Boyer JC, Petes TD, Farber RA. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc Natl Acad Sci U S A. 2005;102:8639–8643. doi: 10.1073/pnas.0503415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess MT, Gupta RD, Kolodner RD. Dominant Saccharomyces cerevisiae msh6 mutations cause increased mispair binding and decreased dissociation from mispairs by Msh2-Msh6 in the presence of ATP. J Biol Chem. 2002;277:25545–25553. doi: 10.1074/jbc.M202282200. [DOI] [PubMed] [Google Scholar]
- Hess MT, Mendillo ML, Mazur DJ, Kolodner RD. Biochemical basis for dominant mutations in the Saccharomyces cerevisiae MSH6 gene. Proc Natl Acad Sci U S A. 2006;103:558–563. doi: 10.1073/pnas.0510078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes J, Jr, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci U S A. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011a;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science. 2011b;334:1713–1716. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jiang J, Hashiguchi K, Hoshi M, Lan L, Yasui A. Recruitment of mismatch repair proteins to the site of DNA damage in human cells. J Cell Sci. 2008;121:3146–3154. doi: 10.1242/jcs.026393. [DOI] [PubMed] [Google Scholar]
- Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Papadopoulos N, McKinley AJ, Farrington SM, Curtis LJ, Wyllie AH, Zheng S, Willson JK, Markowitz SD, Morin P, Kinzler KW, Vogelstein B, Dunlop MG. APC mutations in colorectal tumors with mismatch repair deficiency. Proc Natl Acad Sci U S A. 1996;93:9049–9054. doi: 10.1073/pnas.93.17.9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Tsai CL, Claridge SA, Mendillo ML, Smith JM, Williams GJ, Mastroianni AJ, Alivisatos AP, Putnam CD, Kolodner RD, Tainer JA. DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proc Natl Acad Sci U S A. 2013;110:17308–17313. doi: 10.1073/pnas.1308595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Iyer RR, Pohlhaus TJ, Chen S, Hura GL, Dzantiev L, Beese LS, Modrich P. The MutSalpha-proliferating cell nuclear antigen interaction in human DNA mismatch repair. J Biol Chem. 2008;283:13310–13319. doi: 10.1074/jbc.M800606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Genschel J, Tsai MS, Beese LS, Modrich P. MutLalpha and proliferating cell nuclear antigen share binding sites on MutSbeta. J Biol Chem. 2010;285:11730–11739. doi: 10.1074/jbc.M110.104125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. Postreplicative mismatch repair. Cold Spring Harbor Perspectives Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- Junop MS, Yang W, Funchain P, Clendenin W, Miller JH. In vitro and in vivo studies of MutS, MutL and MutH mutants: correlation of mismatch repair and DNA recombination. DNA Repair (Amst) 2003;2:387–405. doi: 10.1016/s1568-7864(02)00245-8. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci U S A. 2009;106:8495–8500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova LY, Mertz TM, Zhang Y, Northam MR, Sheng Z, Lobachev KS, Shcherbakova PV, Kadyrov FA. A reversible histone H3 acetylation cooperates with mismatch repair and replicative polymerases in maintaining genome stability. PLoS Genet. 2013;9:e1003899. doi: 10.1371/journal.pgen.1003899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase epsilon mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014;74:1895–1901. doi: 10.1158/0008-5472.CAN-13-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Svetlanov A, Lenzi ML, Macaluso FP, Lipkin SM, Liskay RM, Greally J, Edelmann W, Cohen PE. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J Cell Biol. 2005;171:447–458. doi: 10.1083/jcb.200506170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Tytell JD, Schmeits JL, Kane MF, Gupta RD, Weger J, Wahlberg S, Fox EA, Peel D, Ziogas A, Garber JE, Syngal S, Anton-Culver H, Li FP. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
- Kosinski J, Steindorf I, Bujnicki JM, Giron-Monzon L, Friedhoff P. Analysis of the quaternary structure of the MutL C-terminal domain. J Mol Biol. 2005;351:895–909. doi: 10.1016/j.jmb.2005.06.044. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Laguri C, Duband-Goulet I, Friedrich N, Axt M, Belin P, Callebaut I, Gilquin B, Zinn-Justin S, Couprie J. Human mismatch repair protein MSH6 contains a PWWP domain that targets double stranded DNA. Biochemistry. 2008;47:6199–6207. doi: 10.1021/bi7024639. [DOI] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- Lang WH, Coats JE, Majka J, Hura GL, Lin Y, Rasnik I, McMurray CT. Conformational trapping of mismatch recognition complex MSH2/MSH3 on repair-resistant DNA loops. Proc Natl Acad Sci U S A. 2011;108:E837–844. doi: 10.1073/pnas.1105461108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PJ, Flores-Rozas H, Kolodner RD. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol Cell Biol. 2002;22:6669–6680. doi: 10.1128/MCB.22.19.6669-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JB, Cho WK, Park J, Jeon Y, Kim D, Lee SH, Fishel R. Single-molecule views of MutS on mismatched DNA. DNA Repair (Amst) 2014;20:82–93. doi: 10.1016/j.dnarep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart JS, Pillon MC, Guarne A, Simmons LA. Trapping and visualizing intermediate steps in the mismatch repair pathway in vivo. Mol Microbiol. 2013a;90:680–698. doi: 10.1111/mmi.12389. [DOI] [PubMed] [Google Scholar]
- Lenhart JS, Sharma A, Hingorani MM, Simmons LA. DnaN clamp zones provide a platform for spatiotemporal coupling of mismatch detection to DNA replication. Mol Microbiol. 2013b;87:553–568. doi: 10.1111/mmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM. The role of mismatch repair in DNA damage-induced apoptosis. Oncol Res. 1999;11:393–400. [PubMed] [Google Scholar]
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wawrose JS, Gooding WE, Garraway LA, Lui VW, Peyser ND, Grandis JR. Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: a rational approach to preclinical model selection. Mol Cancer Res: MCR. 2014;12:571–582. doi: 10.1158/1541-7786.MCR-13-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti SE, Andersen SD, Wang J, May A, Miron S, Perderiset M, Keijzers G, Nielsen FC, Charbonnier JB, Bohr VA, Rasmussen LJ. Bi-directional routing of DNA mismatch repair protein human exonuclease 1 to replication foci and DNA double strand breaks. DNA Repair (Amst) 2011;10:73–86. doi: 10.1016/j.dnarep.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti SE, Larrea AA, Kunkel TA. Exonuclease 1 preferentially repairs mismatches generated by DNA polymerase alpha. DNA Repair (Amst) 2013;12:92–96. doi: 10.1016/j.dnarep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Pursell ZF, Abdulovic-Cui AA, Clark AB, Nick McElhinny SA, Kunkel TA. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012;8:e1003016. doi: 10.1371/journal.pgen.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPhee DG. Mismatch repair, somatic mutations, and the origins of cancer. Cancer Res. 1995;55:5489–5492. [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc Natl Acad Sci U S A. 1998;95:8568–8573. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- Matson SW, Robertson AB. The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res. 2006;34:4089–4097. doi: 10.1093/nar/gkl450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur DJ, Mendillo ML, Kolodner RD. Inhibition of Msh6 ATPase activity by mispaired DNA induces a Msh2(ATP)-Msh6(ATP) state capable of hydrolysis-independent movement along DNA. Mol Cell. 2006;22:39–49. doi: 10.1016/j.molcel.2006.02.010. [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- Mendillo ML, Putnam CD, Kolodner RD. Escherichia coli MutS tetramerization domain structure reveals that stable dimers but not tetramers are essential for DNA mismatch repair in vivo. J Biol Chem. 2007;282:16345–16354. doi: 10.1074/jbc.M700858200. [DOI] [PubMed] [Google Scholar]
- Mendillo ML, Hargreaves VV, Jamison JW, Mo AO, Li S, Putnam CD, Woods VL, Jr, Kolodner RD. A conserved MutS homolog connector domain interface interacts with MutL homologs. Proc Natl Acad Sci U S A. 2009;106:22223–22228. doi: 10.1073/pnas.0912250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo ML, Putnam CD, Mo AO, Jamison JW, Li S, Woods VL, Jr, Kolodner RD. Probing DNA- and ATP-mediated conformational changes in the MutS family of mispair recognition proteins using deuterium exchange mass spectrometry. J Biol Chem. 2010;285:13170–13182. doi: 10.1074/jbc.M110.108894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllykangas S, Himberg J, Bohling T, Nagy B, Hollmen J, Knuutila S. DNA copy number amplification profiling of human neoplasms. Oncogene. 2006;25:7324–7332. doi: 10.1038/sj.onc.1209717. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant KT, Plys AJ, Alani E. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics. 2008;179:747–755. doi: 10.1534/genetics.108.086645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AM, Woodruff RD, D'Agostino RB, Jr, Clodfelter JE, Scarpinato KD. Elevated levels of the mismatch repair protein PMS2 are associated with prostate cancer. Prostate. 2007;67:214–225. doi: 10.1002/pros.20522. [DOI] [PubMed] [Google Scholar]
- Norris AM, Gentry M, Peehl DM, D'Agostino R, Jr, Scarpinato KD. The elevated expression of a mismatch repair protein is a predictor for biochemical recurrence after radical prostatectomy. Cancer Epidemiol, Biomarkers Prevent: Publ Am Assoc Cancer Res, Cosponsored Am Soc Preventive Oncol. 2009;18:57–64. doi: 10.1158/1055-9965.EPI-08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- Oki E, Oda S, Maehara Y, Sugimachi K. Mutated gene-specific phenotypes of dinucleotide repeat instability in human colorectal carcinoma cell lines deficient in DNA mismatch repair. Oncogene. 1999;18:2143–2147. doi: 10.1038/sj.onc.1202583. [DOI] [PubMed] [Google Scholar]
- Owen BA, Yang Z, Lai M, Gajec M, Badger JD, 2nd, Hayes JJ, Edelmann W, Kucherlapati R, Wilson TM, McMurray CT. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Humphray S, Consortium C, Consortium WGS, Lucassen A, Holmes CC, Bentley D, Donnelly P, Taylor J, Petridis C, Roylance R, Sawyer EJ, Kerr DJ, Clark S, Grimes J, Kearsey SE, Thomas HJ, McVean G, Houlston RS, Tomlinson I. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi GB, Slean MM, Simard JP, Gileadi O, Pearson CE. Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutSbeta, but clustered slip-outs are poorly repaired. Proc Natl Acad Sci U S A. 2010;107:12593–12598. doi: 10.1073/pnas.0909087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. Epigenetic mechanisms in the pathogenesis of Lynch syndrome. Clin Genet. 2014;85:403–412. doi: 10.1111/cge.12349. [DOI] [PubMed] [Google Scholar]
- Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition—update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Diaz J, Jiricny J. Mammalian mismatch repair: error-free or error-prone? Trends Biochem Sci. 2012;37:206–214. doi: 10.1016/j.tibs.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Pena-Diaz J, Bregenhorn S, Ghodgaonkar M, Follonier C, Artola-Boran M, Castor D, Lopes M, Sartori AA, Jiricny J. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol Cell. 2012;47:669–680. doi: 10.1016/j.molcel.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Pluciennik A, Modrich P. Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. Proc Natl Acad Sci U S A. 2007;104:12709–12713. doi: 10.1073/pnas.0705129104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair. Proc Natl Acad Sci U S A. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prindle MJ, Loeb LA. DNA polymerase delta in DNA replication and genome maintenance. Environ Mol Mutagen. 2012;53:666–682. doi: 10.1002/em.21745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla TA, Baker SM, Harris AC, Tsao JL, Yao X, Bronner CE, Zheng B, Gordon M, Reneker J, Arnheim N, Shibata D, Bradley A, Liskay RM. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet. 1998;18:276–279. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- Ranjha L, Anand R, Cejka P. The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J Biol Chem. 2014;289:5674–5686. doi: 10.1074/jbc.M113.533810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Marra G, Nystrom-Lahti M, Schar P, Jiricny J. Identification of hMutLbeta, a heterodimer of hMLH1 and hPMS1. J Biol Chem. 1999;274:32368–32375. doi: 10.1074/jbc.274.45.32368. [DOI] [PubMed] [Google Scholar]
- Reenan RA, Kolodner RD. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992a;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan RA, Kolodner RD. Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics. 1992b;132:963–973. doi: 10.1093/genetics/132.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz LJ. DNA polymerase proofreading: multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804:1049–1063. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Rodriguez GP, Romanova NV, Bao G, Rouf NC, Kow YW, Crouse GF. Mismatch repair-dependent mutagenesis in nondividing cells. Proc Natl Acad Sci U S A. 2012;109:6153–6158. doi: 10.1073/pnas.1115361109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesner LM, Mielke C, Fahnrich S, Merkhoffer Y, Dittmar KE, Drexler HG, Dirks WG. Stable expression of MutLgamma in human cells reveals no specific response to mismatched DNA, but distinct recruitment to damage sites. J Cell Biochem. 2013;114:2405–2414. doi: 10.1002/jcb.24591. [DOI] [PubMed] [Google Scholar]
- Rogacheva MV, Manhart CM, Chen C, Guarne A, Surtees J, Alani E. Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J Biol Chem. 2014;289:5664–5673. doi: 10.1074/jbc.M113.534644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, Erie DA. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol Cell. 2008;29:112–121. doi: 10.1016/j.molcel.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci-Darmanin S, Neyton S, Lespinasse F, Saunieres A, Gaudray P, Paquis-Flucklinger V. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum Mol Genet. 2002;11:1697–1706. doi: 10.1093/hmg/11.15.1697. [DOI] [PubMed] [Google Scholar]
- Schaaper RM, Radman M. The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. EMBO J. 1989;8:3511–3516. doi: 10.1002/j.1460-2075.1989.tb08516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem. 2001;276:33011–33018. doi: 10.1074/jbc.M102670200. [DOI] [PubMed] [Google Scholar]
- Shcherbakova PV, Kunkel TA. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Hall MC, Lewis MS, Bennett SE, Martin KJ, Bushel PR, Afshari CA, Kunkel TA. Inactivation of DNA mismatch repair by increased expression of yeast MLH1. Mol Cell Biol. 2001;21:940–951. doi: 10.1128/MCB.21.3.940-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell SS, Putnam CD, Kolodner RD. Chimeric Saccharomyces cerevisiae Msh6 protein with an Msh3 mispair-binding domain combines properties of both proteins. Proc Natl Acad Sci U S A. 2007a;104:10956–10961. doi: 10.1073/pnas.0704148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol Cell. 2007b;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Davies BW, Grossman AD, Walker GC. Beta clamp directs localization of mismatch repair in Bacillus subtilis. Mol Cell. 2008;29:291–301. doi: 10.1016/j.molcel.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slean MM, Panigrahi GB, Ranum LP, Pearson CE. Mutagenic roles of DNA “repair” proteins in antibody diversity and disease-associated trinucleotide repeat instability. DNA Repair (Amst) 2008;7:1135–1154. doi: 10.1016/j.dnarep.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Smith BT, Grossman AD, Walker GC. Visualization of mismatch repair in bacterial cells. Mol Cell. 2001;8:1197–1206. doi: 10.1016/s1097-2765(01)00402-6. [DOI] [PubMed] [Google Scholar]
- Smith CE, Mendillo ML, Bowen N, Hombauer H, Campbell CS, Desai A, Putnam CD, Kolodner RD. Dominant mutations in S. cerevisiae PMS1 identify the Mlh1-Pms1 endonuclease active site and an exonuclease 1-independent mismatch repair pathway. PLoS Genet. 2013;9:e1003869. doi: 10.1371/journal.pgen.1003869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Bowen N, Kolodner RD. Mispair-specific recruitment of the Mlh1-Pms1 complex identifies repair substrates of the Saccharomyces cerevisiae Msh2-Msh3 complex. J Biol Chem. 2014;289:9352–9364. doi: 10.1074/jbc.M114.552190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Paques F, Colaiacovo M, Haber JE. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci U S A. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surtees JA, Alani E. Mismatch repair factor MSH2-MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J Mol Biol. 2006;360:523–536. doi: 10.1016/j.jmb.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Thomas DC, Roberts JD, Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- Tian L, Hou C, Tian K, Holcomb NC, Gu L, Li GM. Mismatch recognition protein MutSbeta does not hijack (CAG)n hairpin repair in vitro. J Biol Chem. 2009;284:20452–20456. doi: 10.1074/jbc.C109.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci U S A. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Gordenin DA, Resnick MA. The 3′→5′ exonucleases of DNA polymerases delta and epsilon and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, Groenen PJ, Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hays JB. Mismatch repair in human nuclear extracts. Quantitative analyses of excision of nicked circular mismatched DNA substrates, constructed by a new technique employing synthetic oligonucleotides. J Biol Chem. 2002;277:26136–26142. doi: 10.1074/jbc.M200357200. [DOI] [PubMed] [Google Scholar]
- Wang H, Hays JB. Mismatch repair in human nuclear extracts: effects of internal DNA-hairpin structures between mismatches and excision-initiation nicks on mismatch correction and mismatch-provoked excision. J Biol Chem. 2003;278:28686–28693. doi: 10.1074/jbc.M302844200. [DOI] [PubMed] [Google Scholar]
- Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H, Jr, Kneitz B, Yang G, Kunkel TA, Kolodner RD, Cohen PE, Edelmann W. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh KM, Lu AL, Clark S, Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987;262:15624–15629. [PubMed] [Google Scholar]
- Winkler I, Marx AD, Lariviere D, Heinze RJ, Cristovao M, Reumer A, Curth U, Sixma TK, Friedhoff P. Chemical trapping of the dynamic MutS-MutL complex formed in DNA mismatch repair in Escherichia coli. J Biol Chem. 2011;286:17326–17337. doi: 10.1074/jbc.M110.187641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Structure and function of mismatch repair proteins. Mutat Res. 2000;460:245–256. doi: 10.1016/s0921-8777(00)00030-6. [DOI] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]