Abstract
Prion diseases are fatal transmissible neurodegenerative disorders that affect animals including humans. The kinetics of prion infectivity and PrPSc accumulation can differ between prion strains and within a single strain in different tissues. The net accumulation of PrPSc in animals is controlled by the relationship between the rate of PrPSc formation and clearance. Protein misfolding cyclic amplification (PMCA) is a powerful technique that faithfully recapitulates PrPSc formation and prion infectivity in a cell-free system. PMCA has been used as a surrogate for animal bioassay and can model species barriers, host range, strain co-factors and strain interference. In this study we investigated if degradation of PrPSc and/or prion infectivity occurs during PMCA. To accomplish this we performed PMCA under conditions that do not support PrPSc formation and did not observe either a reduction in PrPSc abundance or an extension of prion incubation period, compared to untreated control samples. These results indicate that prion clearance does not occur during PMCA. These data have significant implications for the interpretation of PMCA based experiments such as prion amplification rate, adaptation to new species and strain interference where production and clearance of prions can affect the outcome.
Keywords: prion clearance, prion disease, protein misfolding cyclic amplification
Introduction
Prion diseases are inevitably fatal neurodegenerative diseases affecting animals. The prion agent is likely solely comprised of a misfolded isoform of the prion protein, PrPSc, which is post-translationally derived from the host encoded normal isoform of the prion protein, PrPC.1,2,6 PrPSc can adopt a wide range of different conformations, which are thought to account for the variety of prion strains that can exist in a single PrP genotype in a given species.10-12
Formation of PrPSc is thought to occur in a 3-step process. First, PrPC binds to PrPSc likely through the N-terminal and central polybasic domains.15-16 Although other docking sites have been identified, they do not support formation of infectious prions.18 Second, PrPSc directs the conversion of PrPC to the PrPSc conformation by an unknown mechanism resulting in elongation of the PrPSc fibril. Finally, fragmentation of the PrPSc fibril results in the formation of new free ends for additional PrPC binding to take place, thus completing the cycle. The rate of increase in prion infectivity and PrPSc accumulation can differ between strains and within a strain in different tissues.20-22
Fragmentation of PrPSc may contribute to the rate of prion formation. Initial studies of Sup35 fibrils, from the yeast prion [PSI+], have low conformational stability corresponding with high conversion rates.24 Murine prion strains with short incubation periods correspond with PrPSc with low conformational stability compared to strains with longer incubation periods.25,27 These data suggest that a decrease in conformational stability of the protein aggregate results in an increase in fragmentation that leads to an increase in prion formation. In both mammalian and yeast prions, however, other studies indicate that this relationship is not always true suggesting that additional factors are involved in fragmentation.29-32
The net accumulation of PrPSc is controlled by the relationship between the rate of PrPSc formation and degradation. Elimination of neuronal PrPC expression during the course of prion infection results in clearance of PrPSc in these cells and subsequent reversal of a subset of prion-induced neuropathology.44,46 Overexpression of PrPC results in shortened incubation periods, however, due to the dual role of PrPC in both PrPSc formation and neurodegeneration, the exact mechanism responsible for shortening of the incubation period is unclear.1,3 The mechanism of PrPSc clearance is not fully understood, but may occur in lysosomes and can be accelerated by autophagy.5-8
Protein misfolding cyclic amplification (PMCA) is a powerful technique that faithfully recapitulates PrPSc formation and prion infectivity in a cell-free system. PMCA uses a repeated series of incubation periods and sonication that leads to formation of PrPSc and prion infectivity.9,11,13,32 The PMCA incubation period is thought to allow for PrPC binding to PrPSc and subsequent conversion of PrPC to PrPSc. The sonication step is thought to shear the growing PrPSc fibril providing additional free ends for PrPC to bind and convert to PrPSc.14 This process of repeated elongation and fragmentation of PrPSc fibrils leads to exponential PrPSc production.17
Relatively little is known about PMCA mediated prion clearance. Several factors of the PMCA procedure have the potential to inactivate prions. Sonication produces cavitation microbubbles that can have extreme localized pressure and temperature that can denature protein and has the potential to reduce prion infectivity.19 Additionally, endogenous proteases in brain homogenate can truncate and degrade PrPSc over time.23 The effect of the combination of heat, protease activity and detergent that is present in the PMCA conversion buffer on prion degradation is unknown.
In this study we investigated if PrPSc and/or prion infectivity degradation occurred as a result of PMCA. To accomplish this we performed PMCA under conditions that do not support PrPSc formation. PrPSc degradation and an extension of prion incubation period were not observed under the PMCA conditions with the prion strains used. These data have significant implications on the interpretation of PMCA based experiments.
Results
PMCA formation of PrPSc
To confirm that amplification via PMCA is working within our standard parameters, either HY TME or DY TME-infected brain homogenate was diluted into uninfected hamster brain homogenate and subjected to one round of PMCA to serve as a positive control. Western blot analysis of PK treated samples demonstrated a significant (P < 0.05) increase in PK resistant PrPSc in the PMCA treated samples compared to the untreated controls and migration of the PMCA generated PrPSc maintained strain-specific patterns (Fig. 1A and B). Consistent with previous studies, HY PrPSc amplified to a significantly (P < 0.05) greater amount (Fig. 1A, lanes 5–6; Panel B) compared to DY PrPSc (Fig. 1A, lanes 9–10; Panel B). Formation of PrPSc was not observed in the mock seeded negative control reactions (Fig. 1A, lanes 11–12).
Figure 1.
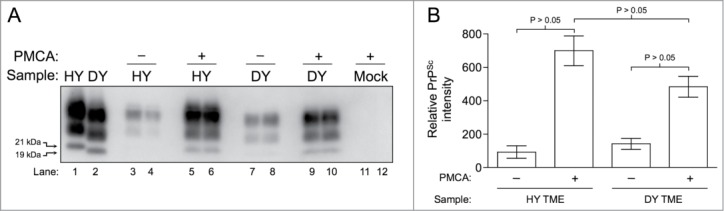
In vitro amplification of hamster adapted TME. (A) HY TME and DY TME were diluted in hamster brain homogenate and subjected to 144 cycles of 5-second sonication and 10 minute incubation. Following PK digestions, Western blot analysis show amplification of PrPSc when compared to their unsonicated controls (lanes 3–4 vs. 5-6 for HY TME and lanes 7-8 vs. 9-10 for DY TME). HY TME amplifies more efficiently than DY PrPSc (compare lanes 5-6 vs. 9-10). Mock: mock infected negative control (lanes 11–12). The migration of the 19 and 21 kDa unglycosylated PrPSc polypeptides is indicated on the left of panel A. (B) Bar graph comparing the relative PrPSc intensity before and after PMCA using HY TME or DY TME as a PrPSc seed (n = 5 per experimental group).
PMCA induced degradation of PrP is not observed
To investigate if conversion buffer and temperature induced PrP degradation, uninfected hamster brain homogenate alone and HY TME or DY TME-infected brain homogenates diluted in uninfected MoPrP0/0 brain homogenate were incubated at 37°C for 24 hours without sonication. The abundance of PrP before (Fig. 2A, lanes 1–3) and after (Fig. 2A, lanes 4–6) the 37°C incubation did not differ significantly (P > 0.05) (Fig. 2B).
Figure 2.
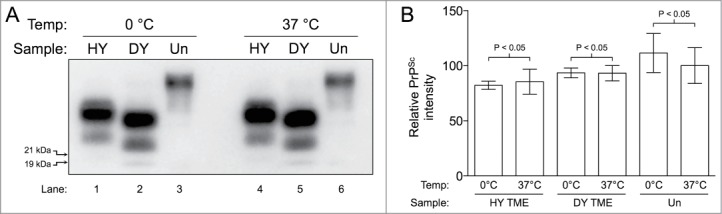
Temperature does not facilitate degradation of PrP. (A) HY TME and DY TME diluted in MoPrP0/0 brain homogenate and uninfected hamster brain homogenate incubated at 0°C without sonication (Lanes 1–3) and at 37°C without sonication (Lanes 4–6). The migration of the 19 and 21 kDa unglycosylated PrPSc polypeptides is indicated on the left of panel A. (B) Bar graph comparing the relative intensities of the Western blot analysis of each sample before and after incubation (n = 3 per experimental group).
To investigate the effect of sonication on PrP degradation, HY TME or DY TME-infected brain homogenates were diluted in either DPBS (without conversion buffer components i.e. EDTA, Triton X-100 and protease inhibitors) or uninfected MoPrP0/0 brain homogenate and subjected to one round of PMCA. The abundance of PrPSc in samples subjected to one round of PMCA or unsonicated samples in either MoPrP0/0 brain (Fig. 3A, lanes 1–4) or DPBS (Fig. 3A, lanes 5–8) did not differ significantly (P > 0.05) (Fig. 3B). To study the effect of temperature and sonication on PrPC, uninfected brain homogenate was subjected to one round of PMCA. The abundance of PrPC did not differ significantly (P < 0.05) from the unsonicated control samples. (Fig. 3C and D). As a negative control, uninfected hamster brain homogenate diluted in MoPrP0/0 brain homogenate was subjected to one round of PMCA. In these samples, Western blot analysis failed to detect PrPSc (data not shown).
Figure 3.
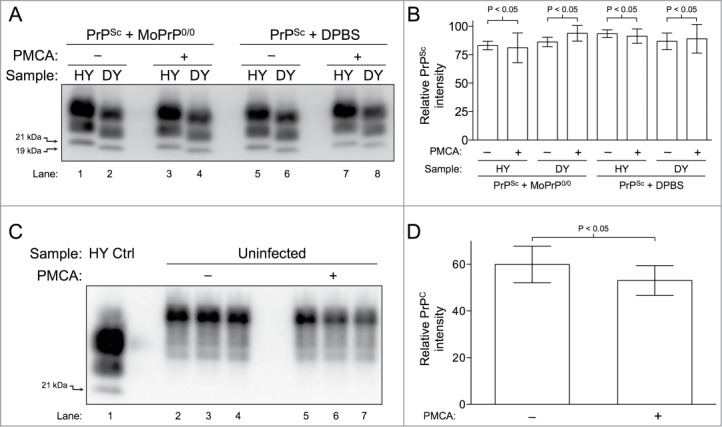
Clearance of PrP is not supported by PMCA. (A) Western blot of HY TME and DY TME showing an absence of PrPSc clearance during PMCA. HY TME and DY TME diluted in MoPrP0/0 brain homogenate without sonication (Lanes 1–2); HY TME and DY TME diluted in MoPrP0/0 brain homogenate subjected to PMCA (Lanes 3–4); HY TME and DY TME diluted in DPBS without sonication (Lanes 5–6); and HY TME and DY TME diluted in DPBS subjected to PMCA (Lanes 7–8). (B) Bar graph comparing the relative intensity of each sample before and after sonication (n = 4 per experimental group). (C) Western blot of uninfected hamster brain homogenate showing the absence of PrPC clearance during PMCA. Uninfected brain homogenate without PMCA (Lanes 2–4) and after PMCA (Lanes 5–7). (D) The relative average intensities of PrPC as quantified from the Western blot analysis of each sample before and after PMCA (n = 3 per experimental group). The migration of the 19 or 21 kDa unglycosylated PrPSc polypeptides is indicated on the left of panels A and C.
Reduction of prion infectivity is not mediated by PMCA
To investigate the effect of PMCA on prion infectivity, HY TME-infected brain homogenate was diluted in MoPrP0/0 brain homogenate and was subjected to either one round of PMCA, incubated at 37°C without sonication or left untreated as a positive control. These samples were i.c. inoculated into Syrian hamsters to determine if these treatments affected the incubation period of disease. All of the hamsters (n = 5) in each group developed clinical signs of hyperexcitability and ataxia. Animals inoculated with either the sonicated HY TME in MoPrP0/0, HY TME in MoPrP0/0 incubated at 37°C for 24 hours or the HY TME positive control had similar (P > 0.05) incubation periods of 72 ± 2, 71 ± 8 and 71 ± 3 d respectively (Fig. 4A). Western blot analysis of PK treated brain homogenates from clinically ill animals demonstrated the accumulation of PrPSc, confirming the clinical diagnosis (Fig. 4B, lanes 3–6). The electrophoretic migration of PrPSc from either the sonicated HY TME or HY TME incubated at 37°C for 24 hours were similar to that of control HY TME PrPSc (Fig. 4B). None (n = 5) of the hamsters inoculated with sonicated mock-uninfected brain homogenate developed clinical signs of prion infection up to 360 d post infection, when the experiment was terminated (Fig. 4A). This was confirmed by Western blot of the PK digested brain tissue, which failed to detect the presence of PrPSc (Fig. 4B, lanes 7 and 8).
Figure 4.
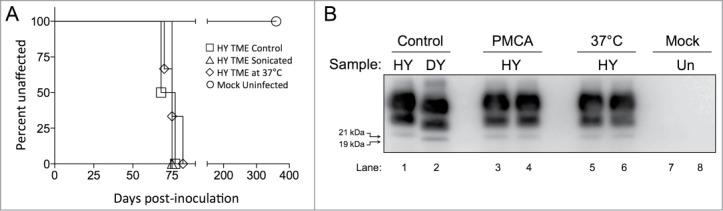
PMCA treatment of HY TME does not affect its infectivity. (A) Survival of Syrian hamsters following i.c. inoculation of HY TME control (□) HY TME in MoPrP0/0 subjected to one round of PMCA (▵) HY TME in MoPrP0/0 at 37°C without sonication (⋄) and Mock inoculated hamster (○) (n = 5 per experimental group). (B) Western blot analysis of brain homogenate from hamsters inoculated with sonicated HY TME in MoPrP0/0 (lanes 3–4); HY TME in MoPrP0/0 incubated at 37°C without sonication (lanes 5–6); and mock uninfected (lanes 7–8). The migration of the 19 and 21 kDa unglycosylated PrPSc polypeptides is indicated on the left of panel B.
Discussion
PMCA can efficiently amplify large amounts of PrPSc and prion infectivity in an in vitro system.11,26 Several aspects of prion biology have been accurately recapitulated using PMCA such as interspecies transmission, adaptation, replication co-factors, strain formation and environmental interactions.18,26,28,30,32-42 While PMCA effectively recapitulates in vivo prion formation in many respects, little is known about the role of prion clearance during PMCA. Under conditions that do not support prion formation, we failed to find evidence of a reduction of PrPSc abundance or a change in incubation period due to the PMCA process. These observations are consistent with reports indicating that PrPSc can survive high levels of heat and proteolytic activity.43,45,47 While not directly measured, it is likely that protease sensitive forms of PrPSc survive the PMCA process since the abundance of PrPC, which is also sensitive to protease digestion, is unaltered. Overall, this data indicates that the net increase in PrPSc and infectivity in PMCA is due solely to PrPSc formation, and is not the result of a dynamic balance between formation and clearance in contrast to what occurs in vivo. This observation affects the interpretation of PMCA based experiments and might contribute to the ability of PMCA to amplify PrPSc amounts that are below the limit of detection in bioassay.
Accurate measurements of prion formation in vivo have not been made. In animals both prion formation and clearance contribute to the net accumulation of PrPSc and infectivity.48-50 Studies utilizing PMCA have been used to calculate the efficiency of PrPSc formation, however, the possibility that prion clearance contributed to the net accumulation of PrPSc and infectivity could not be excluded.13,32,38,40 The results of this study indicate that PMCA provides an accurate measurement of prion formation. Strain specific differences in PMCA PrPSc formation efficiency have been observed and must be due to strain-specific differences in prion formation and not clearance.32
Adaptation of prions to a new host species using PMCA can effectively overcome the species barrier. This process can also result in the identification of novel prion strains when transmitted to the new host species.33,51-54 One possibility for this observation is that PMCA allows for the formation of PrPSc conformations (i.e., sub strains) that are not favored in vivo. Alternatively, identical populations of PrPSc conformations are produced both in vivo and in PMCA, but a subpopulation of PrPSc conformations are cleared in vivo that are not cleared in PMCA. This could result in the evolution of PrPSc conformations that gain the ability to survive in vivo clearance mechanisms or can alter the balance of prion strains in a mixture, which has been shown to influence strain emergence consistent with the prion quasispecies hypothesis.2,4,55-61
Prion strains in a mixture do not act independently but rather interfere with each other.6,62 The mechanism(s) for strain interference are only beginning to be understood.10,12,30,63 One unexplored possibility is that infection with one prion strain can facilitate the clearance of a second strain and contribute to strain interference. This has not been possible to investigate in vivo. Strain interference can occur using PMCA and the same experimental parameters that influence strain interference in vivo apply to PMCA strain interference.15,16,30 Based on the observation of a lack of clearance of PrPSc in PMCA in combination with the ability of PMCA to accurately model strain interference in vivo, we hypothesize that prion clearance is not involved in strain interference. Overall, the inability of PMCA to support prion clearance is an important variable to fully interpret PMCA based studies and can be exploited to explore the relationship between prion formation and clearance.
Materials and Methods
Prion strains and negative controls
Brains from terminally-ill hamsters inoculated with either biologically cloned HY or DY TME agents were homogenized to 10% w/v in Dulbecco's phosphate buffered saline (DPBS) (Mediatech, Herndon, VA). Uninfected hamster brain or PrPC knock-out mice brain MoPrP0/0 was homogenized to 10% w/v in PMCA conversion buffer (phosphate buffer saline containing 1% v/v Triton-X100, 5 mM EDTA with protease inhibitors).64 All homogenates were stored at −80ºC prior to use.
Protein misfolding cyclic amplification
PMCA was performed as described previously.65 Briefly, we used a Misonix 3000 sonicator (Farmingdale, NY) coupled to a 96-well plate titanium cup-horn. The sonicator output was set to level 6. With the sonicator cup horn filled with distilled water at 37°C the average power output was 160 W. One round of PMCA consisted of 144 cycles of 5 seconds sonication and 10 minutes incubation per cycle. The PMCA reactions contain 20 μl of 10% w/v brain homogenates from either HY TME or DY TME-infected hamsters diluted into 80 μl of 10% w/v uninfected hamster or mouse PrP0/0 brain homogenate in PMCA conversion buffer or DPBS and were subjected to one round of PMCA. The negative control PMCA reaction consisted of uninfected hamster brain homogenate. All PMCA reactions were replicated a minimum of 3 times.
SDS-PAGE and Western blot analysis
The PMCA reactions were digested with 0.4 U/ml of proteinase K (PK) at 37°C for 30 minutes with constant agitation (Roche Diagnostics Corporation, Indianapolis, IN). The PK digestion was terminated by incubating the samples at 100°C for 10 minutes in gel loading buffer (4% SDS, 2% β-mercapto ethanol, 40% glycerol, 0.004% Bromophenol blue, and 0.5 M Tris buffer pH 6.8). SDS-PAGE and Western blot analysis were performed as described previously using the anti-PrP antibody 3F4 (final concentration of 0.1 μg/ml; Chemicon; Billerica, MA) to recognize hamster prion protein.30 The Western blot was developed with Pierce supersignal west femto maximum sensitivity substrate according to manufacturer instructions (Pierce, Rockford, IL) and imaged on a Kodak 4000R Imaging Station (Kodak, Rochester, NY). The abundance of PK resistant PrPSc was determined using the Kodak molecular imaging software v.5.0.1.27 (New Haven, CT) as described previously.30
Animal bioassay
All procedures involving animals were approved by the Creighton University Institutional Animal Care and Use Committee and were in compliance with the Guide for the Care and Use of Laboratory Animals. Intracerebral inoculations were performed on 3–4 week old male Syrian hamsters (Harlan-Sprague-Dawley, Indianapolis, IN) as previously described.66 Groups of 5 hamsters were intracerebrally inoculated with 25 μl of the PMCA treated and unsonicated samples diluted to 1% w/v in DPBS. Hamsters were observed 3 times per week for the onset of clinical signs of prion disease and the incubation period was calculated as the number of days between inoculation and onset of clinical signs.67
Statistical analysis
Student's t test analysis was performed using Prism 6 for Mac (GraphPad Software Inc., La Jolla, CA).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Anthony Kincaid for critical reading of the manuscript.
Funding
This work was supported by the National Institutes of Health National P20 RR0115635–6, C06 RR17417–01 and G200RR02400.
References
- 1. Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, Marino S, Weissmann C, Aguzzi A. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature 1996; 379:339-43; PMID:8552188; http://dx.doi.org/ 10.1038/379339a0 [DOI] [PubMed] [Google Scholar]
- 2. Miller MB, Wang DW, Wang F, Noble GP, Ma J, Woods VL, Li S, Supattapone S. Cofactor molecules induce structural transformation during infectious prion formation. Structure 2013; 21:2061-8; PMID:24120764; http://dx.doi.org/ 10.1016/j.str.2013.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Westaway D, Mirenda C, Foster D, Zebarjadian Y, Scott M, Torchia M, Yang S, Serban H, DeArmond S, Ebeling C, et al. . Paradoxical shortening of scrapie incubation times by expression of prion protein transgenes derived from long incubation period mice. Neuron 1991; 7:59-68; PMID:1676894; http://dx.doi.org/ 10.1016/0896-6273(91)90074-A [DOI] [PubMed] [Google Scholar]
- 4. Smirnovas V, Baron GS, Offerdahl DK, Raymond GJ, Caughey B, Surewicz WK. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat Struct Mol Biol 2011; 18:504-6; PMID:21441913; http://dx.doi.org/ 10.1038/nsmb.2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marzo L, Marijanovic Z, Browman D, Chamoun Z, Caputo A, Zurzolo C. 4-hydroxytamoxifen leads to PrPSc clearance by conveying both PrPC and PrPSc to lysosomes independently of autophagy. J Cell Sci 2013; 126:1345-54; PMID:23418355; http://dx.doi.org/ 10.1242/jcs.114801 [DOI] [PubMed] [Google Scholar]
- 6. Bolton D, Bendheim P. A modified host protein model of scrapie. Ciba Found Symp 1988; 135:164-81; PMID:3136999 [DOI] [PubMed] [Google Scholar]
- 7. Joshi-Barr S, Bett C, Chiang W-C, Trejo M, Goebel HH, Sikorska B, Liberski P, Raeber A, Lin JH, Masliah E, et al. . De novo prion aggregates trigger autophagy in skeletal muscle. J Virol 2013; PMID:24307586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heiseke A, Aguib Y, Riemer C, Baier M, Schätzl HM. Lithium induces clearance of protease resistant prion protein in prion-infected cells by induction of autophagy. J Neurochem 2009; 109:25-34; PMID:19183256; http://dx.doi.org/ 10.1111/j.1471-4159.2009.05906.x [DOI] [PubMed] [Google Scholar]
- 9. Soto C, Saborio GP, Anderes L. Cyclic amplification of protein misfolding: application to prion-related disorders and beyond. Trends Neurosci 2002; 25:390-4; PMID:12127750; http://dx.doi.org/ 10.1016/S0166-2236(02)02195-1 [DOI] [PubMed] [Google Scholar]
- 10. Bessen R, Marsh R. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol 1992; 66:2096-101; PMID:1347795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castilla J, Saa P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell 2005; 121:195-206; PMID:15851027; http://dx.doi.org/ 10.1016/j.cell.2005.02.011 [DOI] [PubMed] [Google Scholar]
- 12. Caughey B, Raymond GJ, Bessen RA. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J Biol Chem 1998; 273:32230-5; PMID:9822701; http://dx.doi.org/ 10.1074/jbc.273.48.32230 [DOI] [PubMed] [Google Scholar]
- 13. Shikiya RA, Bartz JC In vitro generation of high titer prions. J Virol 2011; 85:13439-13442; http://dx.doi.org/ 10.1128/JVI.06134-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gonzalez-Montalban N, Makarava N, Savtchenko R, Baskakov IV. Relationship between conformational stability and amplification efficiency of prions. Biochemistry 2011; 50:6815-23; PMID:21749158; http://dx.doi.org/ 10.1021/bi200950v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moroncini G, Kanu N, Solforosi L, Abalos G, Telling GC, Head M, Ironside J, Brockes JP, Burton DR, Williamson RA. Motif-grafted antibodies containing the replicative interface of cellular PrP are specific for PrPSc. Proc Natl Acad Sci U S A 2004; 101:10404-9; PMID:15240877; http://dx.doi.org/ 10.1073/pnas.0403522101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Solforosi L, Bellon A, Schaller M, Cruite JT, Abalos GC, Williamson RA. Toward molecular dissection of PrPC-PrPSc interactions. J Biol Chem 2007; 282:7465-71; PMID:17218310; http://dx.doi.org/ 10.1074/jbc.M610051200 [DOI] [PubMed] [Google Scholar]
- 17. Gonzalez-Montalban N, Makarava N, Ostapchenko VG, Savtchenk R, Alexeeva I, Rohwer RG, Baskakov IV. Highly efficient protein misfolding cyclic amplification. PLoS Pathog 2011; 7:e1001277; PMID:21347353; http://dx.doi.org/ 10.1371/journal.ppat.1001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller MB, Geoghegan JC, Supattapone S. Dissociation of infectivity from seeding ability in prions with alternate docking mechanism. PLoS Pathog 2011; 7:e1002128; PMID:21779169; http://dx.doi.org/ 10.1371/journal.ppat.1002128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gong C, Hart DP. Ultrasound induced cavitation and sonochemical yields. J Acoust Soc Am 1998; 104:2675-82; http://dx.doi.org/ 10.1121/1.423851 [DOI] [Google Scholar]
- 20. Mulcahy ER, Bessen RA. Strain-specific kinetics of prion protein formation in vitro and in vivo. J Biol Chem 2004; 279:1643-9; PMID:14573620; http://dx.doi.org/ 10.1074/jbc.M307844200 [DOI] [PubMed] [Google Scholar]
- 21. Kimberlin R, Walker C. Pathogenesis of mouse scrapie: dynamics of agent replication in spleen, spinal cord and brain after infection by different routes. J Comp Pathol 1979; 89:551-62; PMID:120379; http://dx.doi.org/ 10.1016/0021-9975(79)90046-X [DOI] [PubMed] [Google Scholar]
- 22. Lasmezas C, Deslys J, Demaimay R, Adjou K, Hauw J, Dormont D. Strain specific and common pathogenic events in murine models of scrapie and bovine spongiform encephalopathy. J Gen Virol 1996; 77:1601-9; PMID:8758005; http://dx.doi.org/ 10.1099/0022-1317-77-7-1601 [DOI] [PubMed] [Google Scholar]
- 23. Saunders SE, Bartz JC, Telling GC, Bartelt-Hunt SL. Environmentally-relevant forms of the prion protein. Environ Sci Technol 2008; 42:6573-9; PMID:18800532; http://dx.doi.org/ 10.1021/es800590k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanaka M, Collins S, Toyama B, Weissman J. The physical basis of how prion conformations determine strain phenotypes. Nature 2006; 442:585-9; PMID:16810177; http://dx.doi.org/ 10.1038/nature04922 [DOI] [PubMed] [Google Scholar]
- 25. Legname G, Nguyen H, Peretz D, Cohen F, DeArmond S, Prusiner S. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci U S A 2006; 103:19105-10; PMID:17142317; http://dx.doi.org/ 10.1073/pnas.0608970103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deleault N, Harris B, Rees J, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci USA 2007; 104:9741-6; PMID:17535913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Colby DW, Giles K, Legname G, Wille H, Baskakov IV, Dearmond SJ, Prusiner SB. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci USA 2009; 106:20417-22; PMID:19915150; http://dx.doi.org/ 10.1073/pnas.0910350106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saunders SE, Shikiya RA, Langenfeld K, Bartelt-Hunt SL, Bartz JC. Replication efficiency of soil-bound prions varies with soil type. J Virol 2011; 85:5476-82; PMID:21430062; http://dx.doi.org/ 10.1128/JVI.00282-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stein KC, True HL. Extensive diversity of prion strains is defined by differential chaperone interactions and distinct amyloidogenic regions. PLoS Genet 2014; 10:e1004337; PMID:24811344; http://dx.doi.org/ 10.1371/journal.pgen.1004337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shikiya RA, Ayers JI, Schutt CR, Kincaid AE, Bartz JC. Coinfecting prion strains compete for a limiting cellular resource. J Virol 2010; 84:5706-14; PMID:20237082; http://dx.doi.org/ 10.1128/JVI.00243-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peretz D, Scott MR, Groth D, Williamson RA, Burton DR, Cohen FE, Prusiner SB. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci 2001; 10:854-63; PMID:11274476; http://dx.doi.org/ 10.1110/ps.39201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ayers JI, Schutt CR, Shikiya RA, Aguzzi A, Kincaid AE, Bartz JC. The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease. PLoS Pathog 2011; 7:e1001317; PMID:21437239; http://dx.doi.org/ 10.1371/journal.ppat.1001317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Green KM, Castilla J, Seward TS, Napier DL, Jewell JE, Soto C, Telling GC. Accelerated high fidelity prion amplification within and across prion species barriers. PLoS Pathog 2008; 4:e1000139; PMID:18769716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barria MA, Telling GC, Gambetti P, Mastrianni JA, Soto C. Generation of a new form of human PrPSc in vitro by interspecies transmission from cervid prions. J Biol Chem 2011; 286:7490-5; PMID:21209079; http://dx.doi.org/ 10.1074/jbc.M110.198465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haldiman T, Kim C, Cohen Y, Chen W, Blevins J, Qing L, Cohen ML, Langeveld J, Telling GC, Kong Q, et al. . Coexistence of distinct prion types enables conformational evolution of human PrPSc by competitive selection. J Biol Chem 2013; 288(41):29846-61; PMID:23974118; http://dx.doi.org/ 10.1074/jbc.M113.500108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gonzalez-Montalban N, Lee YJ, Makarava N, Savtchenko R, Baskakov IV. Changes in prion replication environment cause prion strain mutation. FASEB J 2013; 27(9):3702-10; PMID:23729586; http://dx.doi.org/ 10.1096/fj.13-230466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Makarava N, Savtchenko R, Baskakov IV. Selective amplification of classical and atypical prions using modified protein misfolding cyclic amplification. J Biol Chem 2013; 288:33-41; PMID:23168413; http://dx.doi.org/ 10.1074/jbc.M112.419531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Castilla J, Morales R, Saá P, Barria M, Gambetti P, Soto C. Cell-free propagation of prion strains. EMBO J 2008; 27:2557-66; PMID:18800058; http://dx.doi.org/ 10.1038/emboj.2008.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gonzalez-Romero D, Barria MA, Leon P, Morales R, Soto C. Detection of infectious prions in urine. FEBS Lett 2008; 582:3161-6; PMID:18706416; http://dx.doi.org/ 10.1016/j.febslet.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klingeborn M, Race B, Meade-White KD, Chesebro B. Lower specific infectivity of protease-resistant prion protein generated in cell-free reactions. Proc Natl Acad Sci USA 2011; 108(48):E1244-53; PMID:22065744; http://dx.doi.org/ 10.1073/pnas.1111255108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, Rees JR, Supattapone S. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc Natl Acad Sci U S A 2012; 109:E1938-46; PMID:22711839; http://dx.doi.org/ 10.1073/pnas.1206999109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deleault NR, Kascsak R, Geoghegan JC, Supattapone S. Species-dependent differences in cofactor utilization for formation of the protease-resistant prion protein in vitro. Biochemistry 2010; 49:3928-34; PMID:20377181; http://dx.doi.org/ 10.1021/bi100370b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saunders SE, Bartelt-Hunt SL, Bartz JC. Resistance of soil-bound prions to rumen digestion. PLoS ONE 2012; 7:e44051; PMID:22937149; http://dx.doi.org/ 10.1371/journal.pone.0044051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Safar JG, Dearmond SJ, Kociuba K, Deering C, Didorenko S, Bouzamondo-Bernstein E, Prusiner SB, Tremblay P. Prion clearance in bigenic mice. J Gen Virol 2005; 86:2913-23; PMID:16186247; http://dx.doi.org/ 10.1099/vir.0.80947-0 [DOI] [PubMed] [Google Scholar]
- 45. Nicholson EM, Richt JA, Rasmussen MA, Hamir AN, Lebepe-Mazur S, Horst RL. Exposure of sheep scrapie brain homogenate to rumen-simulating conditions does not result in a reduction of PrP(Sc) levels. Lett Appl Microbiol 2007; 44:631-6; PMID:17576225; http://dx.doi.org/ 10.1111/j.1472-765X.2007.02124.x [DOI] [PubMed] [Google Scholar]
- 46. Mallucci G, Dickinson A, Linehan J, Klöhn P-C, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 2003; 302:871-4; PMID:14593181; http://dx.doi.org/ 10.1126/science.1090187 [DOI] [PubMed] [Google Scholar]
- 47. Brown P, Rau E, Johnson B, Bacote A, Gibbs C, Gajdusek D. New studies on the heat resistance of hamster-adapted scrapie agent: threshold survival after ashing at 600 degrees C suggests an inorganic template of replication. Proc Natl Acad Sci U S A 2000; 97:3418-21; PMID:10716712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bartz JC, Kincaid AE, Bessen RA. Retrograde transport of transmissible mink encephalopathy within descending motor tracts. J Virol 2002; 76:5759-68; PMID:11992004; http://dx.doi.org/ 10.1128/JVI.76.11.5759-5768.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Beekes M, Baldauf E, Diringer H. Sequential appearance and accumulation of pathognomonic markers in the central nervous system of hamsters orally infected with scrapie. J Gen Virol 1996; 77:1925-34; PMID:8760444; http://dx.doi.org/ 10.1099/0022-1317-77-8-1925 [DOI] [PubMed] [Google Scholar]
- 50. Kimberlin R, Walker C. Pathogenesis of mouse scrapie: evidence for neural spread of infection to the CNS. J Gen Virol 1980; 51:183-7; PMID:6780656; http://dx.doi.org/ 10.1099/0022-1317-51-1-183 [DOI] [PubMed] [Google Scholar]
- 51. Castilla J, Gonzalez-Romero D, Saá P, Morales R, De Castro J, Soto C. Crossing the species barrier by PrP(Sc) replication in vitro generates unique infectious prions. Cell 2008; 134:757-68; PMID:18775309; http://dx.doi.org/ 10.1016/j.cell.2008.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kurt TD, Telling GC, Zabel MD, Hoover EA. Trans-species amplification of PrP(CWD) and correlation with rigid loop 170N. Virology 2009; 387:235-43; PMID:19269662; http://dx.doi.org/ 10.1016/j.virol.2009.02.025 [DOI] [PubMed] [Google Scholar]
- 53. Chianini F, Fernandez-Borges N, Vidal E, Gibbard L, Pintado B, De Castro J, Priola SA, Hamilton S, Eaton SL, Finlayson J, et al. Rabbits are not resistant to prion infection. Proc Natl Acad Sci USA 2012; 109:5080-5085; http://dx.doi.org/ 10.1073/pnas.1120076109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barria MA, Mukherjee A, Gonzalez-Romero D, Morales R, Soto C. De novo generation of infectious prions in vitro produces a new disease phenotype. PLoS Pathog 2009; 5:e1000421; PMID:19436715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bartz JC, Bessen R, McKenzie D, Marsh R, Aiken JM. Adaptation and selection of prion protein strain conformations following interspecies transmission of transmissible Mink encephalopathy. J Virol 2000; 74:5542-7; PMID:10823860; http://dx.doi.org/ 10.1128/JVI.74.12.5542-5547.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kimberlin R, Walker C. Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J Gen Virol 1978; 39:487-96; PMID:96212; http://dx.doi.org/ 10.1099/0022-1317-39-3-487 [DOI] [PubMed] [Google Scholar]
- 57. Saijo E, Kang H-E, Bian J, Bowling KG, Browning S, Kim S, Hunter N, Telling GC. Epigenetic dominance of prion conformers. PLoS Pathog 2013; 9:e1003692-2; PMID:24204258; http://dx.doi.org/ 10.1371/journal.ppat.1003692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bian J, Kang H-E, Telling GC. Quinacrine promotes replication and conformational mutation of chronic wasting disease prions. PNAS 2014; 111:6028-33; PMID:24711410; http://dx.doi.org/ 10.1073/pnas.1322377111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science 2010; 327:869-72; PMID:20044542; http://dx.doi.org/ 10.1126/science.1183218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li J, Mahal SP, Demczyk CA, Weissmann C. Mutability of prions. EMBO Rep 2011; 12:1243-1250; http://dx.doi.org/ 10.1038/embor.2011.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Daus ML, Wagenführ K, Thomzig A, Boerner S, Hermann P, Hermelink A, Beekes M, Lasch P. Infrared microspectroscopy detects protein misfolding cyclic amplification (PMCA)-induced conformational alterations in hamster scrapie progeny seeds. J Biol Chem 2013; 288(49):35068-80; PMID:24163371; http://dx.doi.org/ 10.1074/jbc.M113.497131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dickinson A, Fraser H, Meikle V, Outram G. Competition between different scrapie agents in mice. Nat New Biol 1972; 237:244-5; PMID:4624846 [DOI] [PubMed] [Google Scholar]
- 63. Bartz JC, Kramer ML, Sheehan MH, Hutter JAL, Ayers JI, Bessen RA, Kincaid AE. Prion interference is due to a reduction in strain-specific PrPSc levels. J Virol 2007; 81:689-97; PMID:17079313; http://dx.doi.org/ 10.1128/JVI.01751-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp H, DeArmond S, Prusiner S, Aguet M, Weissmann C. Normal development and behaviour of mice lacking the neuronal cell- surface PrP protein. Nature 1992; 356:577-82; PMID:1373228 [DOI] [PubMed] [Google Scholar]
- 65. Saunders SE, Bartz JC, Shikiya RA. Protein misfolding cyclic amplification of prions. J Vis Exp 2012; PMID:23168797; http://dx.doi.org/ 10.3791/4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ayers JI, Kincaid AE, Bartz JC. Prion strain targeting independent of strain-specific neuronal tropism. J Virol 2009; 83:81-7; PMID:18971281; http://dx.doi.org/ 10.1128/JVI.01745-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bessen R, Marsh R. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J Gen Virol 1992; 73:329-34; PMID:1531675; http://dx.doi.org/ 10.1099/0022-1317-73-2-329 [DOI] [PubMed] [Google Scholar]


