Summary
Invasive lobular carcinoma (ILC) is the second most prevalent histologic subtype of invasive breast cancer. Here, we comprehensively profiled 817 breast tumors, including 127 ILC, 490 ductal (IDC), and 88 mixed IDC/ILC. Besides E-cadherin loss, the best known ILC genetic hallmark, we identified mutations targeting PTEN, TBX3 and FOXA1 as ILC enriched features. PTEN loss associated with increased AKT phosphorylation, which was highest in ILC among all breast cancer subtypes. Spatially clustered FOXA1 mutations correlated with increased FOXA1 expression and activity. Conversely, GATA3 mutations and high expression characterized Luminal A IDC, suggesting differential modulation of ER activity in ILC and IDC. Proliferation and immune-related signatures determined three ILC transcriptional subtypes associated with survival differences. Mixed IDC/ILC cases were molecularly classified as ILC-like and IDC-like revealing no true hybrid features. This multidimensional molecular atlas sheds new light on the genetic bases of ILC and provides potential clinical options.
INTRODUCTION
Invasive lobular carcinoma (ILC) is the second most frequently diagnosed histologic subtype of invasive breast cancer, constituting ~10–15% of all cases. The classical form (Foote and Stewart, 1946) is characterized by small discohesive neoplastic cells invading the stroma in a single-file pattern. The discohesive phenotype is due to dysregulation of cell–cell adhesion, primarily driven by lack of E-cadherin (CDH1) protein expression observed in ~90% of ILCs (McCart Reed et al., 2015; Morrogh et al., 2012). This feature is the ILC hallmark and immunohistochemistry (IHC) scoring for CDH1 expression is often used to discriminate between lesions with borderline ductal versus lobular histological features. ILC variants have also been described, yet all display loss of E-cadherin expression (Dabbs et al., 2013).
Classic ILCs are typically of low histologic grade and low to intermediate mitotic index. They express estrogen and progesterone receptors (ER and PR) and rarely show HER2 protein overexpression or amplification. These features are generally associated with a good prognosis, yet some studies suggest that long-term outcomes of ILC are inferior to stage-matched invasive ductal carcinoma (IDC) (Pestalozzi et al., 2008). Importantly, ILC infiltrative growth pattern complicates both physical exam and mammographic findings and its patterns of metastatic spread often differ from those of IDC (Arpino et al., 2004).
To date, genomic studies of ILC have provided limited insight into the biologic underpinnings of this disease, mostly focusing on mRNA expression and DNA copy number analysis (McCart Reed et al., 2015). The first TCGA breast cancer study (Cancer Genome Atlas, 2012) reported on 466 breast tumors assayed on six different technology platforms. ILC was represented by only 36 samples and, no lobular-specific features were noted besides mutations and decreased mRNA and protein expression of CDH1. Here, we analyzed nearly twice as many breast tumors from TCGA (n=817), including 127 ILC. This study identified multiple genomic alterations that discriminate between ILC and IDC demonstrating at the molecular level that ILC is a distinct breast cancer subtype and providing new insight into ILC tumor biology and therapeutic options.
RESULTS
Genetic determinants of Invasive Lobular Cancer (ILC)
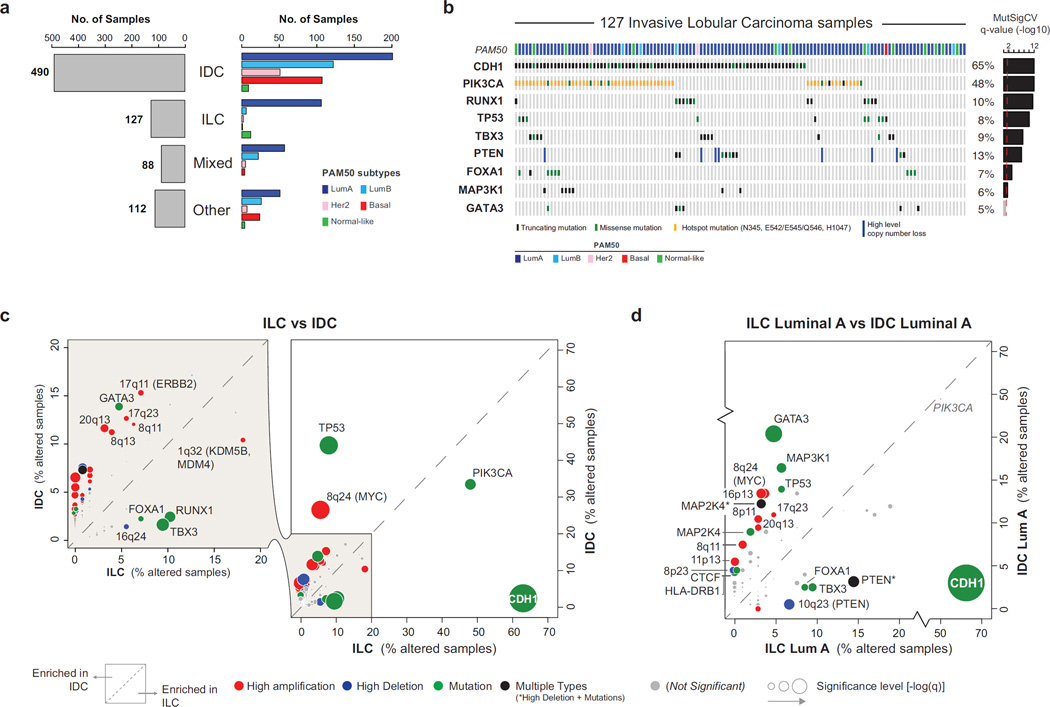
A total of 817 breast tumor samples were profiled with 5 different platforms as previously described (Cancer Genome Atlas Research, 2014) and 633 cases were also profiled by reverse-phase protein array (RPPA). A pathology committee reviewed and classified all tumors into 490 IDC, 127 ILC, 88 cases with mixed IDC and ILC features, and 112 with other histologies (Table S1). As expected, lobular tumors were predominantly classified as Luminal A (LumA) (Figure 1A) and being typically ER+ tumors characterized by low levels of proliferation markers (Table S1). ER status was clinically determined by immunohistochemistry on 120 of 127 ILC cases, with 94% (n=113) scoring positively
Figure 1. Molecular determinants of invasive lobular breast cancer.
A) Histopathological breast cancer subtypes: invasive ductal (IDC), invasive lobular (ILC), mixed ductal/lobular (Mixed), and other-type (Other) carcinoma. PAM50 intrinsic subtypes are not equally distributed across breast cancer subtypes. B) Recurrently mutated genes (MutSigCV2) in ILC. C) Comparison of the alteration frequency for 153 recurrent genomic alterations in ILC versus IDC. D) Comparison of the alteration frequency for 153 recurrent genomic alterations in ILC LumA versus IDC LumA.
Within 127 ILC, we identified 8173 total coding mutations, integrating information from both DNA and RNA sequencing (Wilkerson et al., 2014). Recurrently mutated genes in ILC were identified by MutSigCV2 (Lawrence et al., 2013) and included many genes previously implicated in breast cancer (Figure 1B, Table 1) (Cancer Genome Atlas, 2012). Similarly, recurrent copy number alterations in ILC estimated by GISTIC (Mermel et al., 2011) recapitulated known breast cancer gains and losses, in particular those observed in ER+/Luminal tumors (Figure S1A). However, the frequency of these alterations (both mutations and copy number changes) often differed significantly between IDC and ILC.
Table 1.
Recurrently mutated genes in breast cancer
| ILC (n=127) | ILC Luminal A (106) |
IDC (490) | IDC Luminal A (201) |
IDC Luminal B (122) |
IDC Her2- enriched (51) |
IDC Basal-like (107) |
ALL Breast Cancer (817) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | n | q | n | q | n | q | n | q | n | q | n | q | n | q | n | q |
| PIK3CA | 61 | 1.02E-12 | 54 | 9.18E-13 | 164 | 6.09E-13 | 93 | 6.79E-13 | 43 | 6.76E-13 | 19 | 9.14E-13 | 7 | 6.22E-02 | 282 | 2.54E-13 |
| RUNX1 | 13 | 1.02E-12 | 9 | 9.18E-13 | 13 | n.s. | 9 | 1.32E-05 | 3 | n.s. | 0 | n.s. | 1 | n.s. | 32 | 2.54E-13 |
| CDH1 | 80 | 3.40E-12 | 68 | 6.12E-12 | 10 | 7.63E-03 | 7 | 5.33E-02 | 2 | n.s. | 0 | n.s. | 0 | n.s. | 107 | 2.54E-13 |
| TP53 | 10 | 8.26E-11 | 6 | 2.22E-04 | 215 | 6.09E-13 | 28 | 6.79E-13 | 52 | 6.76E-13 | 37 | 9.14E-13 | 92 | 1.83E-12 | 280 | 2.54E-13 |
| TBX3 | 12 | 2.54E-08 | 10 | 4.01E-06 | 8 | n.s. | 5 | n.s. | 2 | n.s. | 0 | n.s. | 1 | n.s. | 26 | 1.11E-08 |
| PTEN | 9 | 8.43E-08 | 8 | 8.86E-09 | 27 | 5.61E-11 | 6 | 5.63E-03 | 11 | 9.64E-12 | 4 | 3.21E-02 | 6 | 4.65E-03 | 42 | 2.54E-13 |
| FOXA1 | 9 | 5.52E-04 | 9 | 6.53E-04 | 11 | n.s. | 5 | n.s. | 3 | n.s. | 2 | n.s. | 1 | n.s. | 30 | 4.52E-13 |
| MAP3K1 | 7 | 2.95E-02 | 6 | 7.54E-02 | 40 | 4.06E-12 | 33 | 1.25E-11 | 2 | n.s. | 1 | n.s. | 4 | n.s. | 69 | 2.54E-13 |
| GATA3 | 6 | n.s. | 5 | n.s. | 66 | 6.09E-13 | 40 | 6.79E-13 | 22 | 6.76E-13 | 3 | n.s. | 0 | n.s. | 96 | 2.54E-13 |
| AKT1 | 3 | n.s. | 3 | n.s. | 15 | 5.61E-11 | 11 | 1.25E-11 | 3 | 5.91E-02 | 1 | n.s. | 0 | n.s. | 20 | 2.54E-13 |
| NBL1 | 3 | n.s. | 2 | n.s. | 10 | 1.08E-10 | 8 | 2.04E-12 | 0 | n.s. | 1 | n.s. | 1 | n.s. | 16 | 5.24E-11 |
| KMT2C | 9 | n.s. | 8 | n.s. | 37 | 1.49E-08 | 17 | 2.94E-02 | 12 | n.s. | 3 | n.s. | 5 | n.s. | 64 | 4.89E-06 |
| DCTD | 0 | n.s. | 0 | n.s. | 6 | 1.02E-05 | 3 | 1.54E-02 | 1 | n.s. | 1 | n.s. | 0 | n.s. | 6 | 7.61E-04 |
| RB1 | 0 | n.s. | 0 | n.s. | 16 | 1.46E-04 | 4 | n.s. | 7 | n.s. | 1 | n.s. | 4 | n.s. | 18 | n.s. |
| SF3B1 | 4 | n.s. | 4 | n.s. | 12 | 3.20E-04 | 6 | 1.12E-03 | 4 | n.s. | 1 | n.s. | 1 | n.s. | 16 | 3.68E-04 |
| CBFB | 2 | n.s. | 2 | n.s. | 15 | 1.51E-03 | 13 | 5.68E-06 | 1 | n.s. | 1 | n.s. | 0 | n.s. | 24 | 8.14E-13 |
| ARHGAP35 | 1 | n.s. | 1 | n.s. | 13 | 1.62E-03 | 5 | n.s. | 5 | 1.24E-02 | 0 | n.s. | 3 | n.s. | 18 | 7.74E-03 |
| OR9A2 | 0 | n.s. | 0 | n.s. | 5 | 1.77E-03 | 2 | n.s. | 1 | n.s. | 1 | n.s. | 1 | n.s. | 5 | 6.47E-03 |
| NCOA3 | 6 | n.s. | 4 | n.s. | 24 | 1.77E-03 | 7 | n.s. | 7 | n.s. | 4 | n.s. | 6 | n.s. | 40 | 3.25E-07 |
| RBMX | 2 | n.s. | 2 | n.s. | 10 | 2.83E-03 | 3 | n.s. | 2 | n.s. | 1 | n.s. | 4 | 6.27E-02 | 12 | 4.01E-08 |
| MAP2K4 | 2 | n.s. | 2 | n.s. | 24 | 2.83E-03 | 18 | 5.29E-12 | 5 | n.s. | 1 | n.s. | 0 | n.s. | 30 | 1.37E-05 |
| TROVE2 | 0 | n.s. | 0 | n.s. | 6 | 4.51E-03 | 1 | n.s. | 2 | n.s. | 1 | n.s. | 2 | n.s. | 8 | 2.77E-03 |
| NADK | 0 | n.s. | 0 | n.s. | 4 | 4.51E-03 | 0 | n.s. | 4 | 5.85E-05 | 0 | n.s. | 0 | n.s. | 6 | 3.61E-03 |
| CASP8 | 1 | n.s. | 1 | n.s. | 9 | 6.00E-03 | 2 | n.s. | 3 | n.s. | 0 | n.s. | 3 | n.s. | 11 | 1.81E-03 |
| CTSS | 0 | n.s. | 0 | n.s. | 5 | 6.00E-03 | 1 | n.s. | 2 | n.s. | 0 | n.s. | 2 | n.s. | 5 | 8.91E-02 |
| ACTL6B | 2 | n.s. | 1 | n.s. | 5 | 7.63E-03 | 2 | n.s. | 1 | n.s. | 0 | n.s. | 2 | n.s. | 10 | 7.33E-05 |
| LGALS1 | 0 | n.s. | 0 | n.s. | 4 | 9.78E-03 | 2 | n.s. | 2 | n.s. | 0 | n.s. | 0 | n.s. | 5 | 6.34E-03 |
| KRAS | 2 | n.s. | 1 | n.s. | 4 | 1.54E-02 | 3 | 7.32E-03 | 0 | n.s. | 0 | n.s. | 1 | n.s. | 7 | 2.49E-04 |
| KCNN3 | 2 | n.s. | 2 | n.s. | 8 | 1.81E-02 | 1 | n.s. | 2 | n.s. | 2 | n.s. | 3 | 2.45E-02 | 16 | 4.53E-02 |
| FBXW7 | 2 | n.s. | 2 | n.s. | 6 | 2.19E-02 | 0 | n.s. | 0 | n.s. | 0 | n.s. | 6 | 8.28E-04 | 11 | n.s. |
| LRIG2 | 0 | n.s. | 0 | n.s. | 4 | 3.08E-02 | 2 | n.s. | 0 | n.s. | 1 | n.s. | 1 | n.s. | 6 | n.s. |
| PIK3R1 | 0 | n.s. | 0 | n.s. | 9 | 3.08E-02 | 2 | n.s. | 3 | n.s. | 2 | n.s. | 2 | n.s. | 13 | 1.56E-03 |
| PARP4 | 3 | n.s. | 3 | n.s. | 7 | 3.08E-02 | 3 | n.s. | 4 | n.s. | 0 | n.s. | 0 | n.s. | 12 | n.s. |
| ZNF28 | 3 | n.s. | 3 | n.s. | 7 | 3.25E-02 | 1 | n.s. | 5 | n.s. | 0 | n.s. | 1 | n.s. | 11 | 1.72E-02 |
| HLA-DRB1 | 0 | n.s. | 0 | n.s. | 13 | 3.52E-02 | 9 | 1.49E-02 | 2 | n.s. | 0 | n.s. | 2 | n.s. | 16 | n.s. |
| ERBB2 | 5 | n.s. | 4 | n.s. | 7 | 6.42E-02 | 3 | n.s. | 1 | n.s. | 2 | n.s. | 1 | n.s. | 18 | 3.36E-06 |
| ZMYM3 | 0 | n.s. | 0 | n.s. | 9 | 8.83E-02 | 3 | n.s. | 1 | n.s. | 1 | n.s. | 4 | n.s. | 11 | n.s. |
| RAB42 | 1 | n.s. | 1 | n.s. | 2 | n.s. | 0 | n.s. | 0 | n.s. | 0 | n.s. | 2 | 6.27E-02 | 4 | 1.82E-03 |
| CTCF | 0 | n.s. | 0 | n.s. | 12 | n.s. | 9 | 7.05E-08 | 1 | n.s. | 1 | n.s. | 1 | n.s. | 18 | 1.93E-03 |
| ATAD2 | 0 | n.s. | 0 | n.s. | 9 | n.s. | 2 | n.s. | 4 | 7.32E-02 | 2 | n.s. | 1 | n.s. | 12 | n.s. |
| CDKN1B | 3 | n.s. | 2 | n.s. | 5 | n.s. | 4 | 9.59E-02 | 1 | n.s. | 0 | n.s. | 0 | n.s. | 11 | 1.14E-03 |
| GRIA2 | 0 | n.s. | 0 | n.s. | 6 | n.s. | 5 | 5.33E-02 | 0 | n.s. | 0 | n.s. | 1 | n.s. | 6 | n.s. |
| NCOR1 | 8 | n.s. | 8 | n.s. | 23 | n.s. | 12 | 4.81E-03 | 7 | n.s. | 1 | n.s. | 3 | n.s. | 39 | 3.61E-03 |
| HRNR | 4 | n.s. | 4 | n.s. | 13 | n.s. | 3 | n.s. | 3 | n.s. | 3 | n.s. | 4 | n.s. | 23 | 7.65E-02 |
| GPRIN2 | 1 | n.s. | 1 | n.s. | 6 | n.s. | 3 | n.s. | 1 | n.s. | 0 | n.s. | 2 | n.s. | 11 | 1.16E-05 |
| PAX2 | 1 | n.s. | 0 | n.s. | 2 | n.s. | 2 | n.s. | 0 | n.s. | 0 | n.s. | 0 | n.s. | 4 | 4.80E-02 |
| ACTG1 | 1 | n.s. | 1 | n.s. | 4 | n.s. | 2 | n.s. | 0 | n.s. | 0 | n.s. | 2 | n.s. | 8 | 9.39E-02 |
| AQP12A | 0 | n.s. | 0 | n.s. | 3 | n.s. | 1 | n.s. | 1 | n.s. | 0 | n.s. | 1 | n.s. | 5 | 2.69E-02 |
| PIK3C3 | 2 | n.s. | 2 | n.s. | 5 | n.s. | 2 | n.s. | 0 | n.s. | 1 | n.s. | 2 | n.s. | 11 | 3.23E-02 |
| MYB | 1 | n.s. | 1 | n.s. | 7 | n.s. | 3 | n.s. | 2 | n.s. | 0 | n.s. | 2 | n.s. | 12 | 8.91E-02 |
| IRS4 | 1 | n.s. | 1 | n.s. | 6 | n.s. | 3 | n.s. | 0 | n.s. | 0 | n.s. | 3 | n.s. | 8 | 9.38E-02 |
| TBL1XR1 | 3 | n.s. | 2 | n.s. | 3 | n.s. | 1 | n.s. | 1 | n.s. | 1 | n.s. | 0 | n.s. | 12 | 4.71E-04 |
| RPGR | 4 | n.s. | 3 | n.s. | 11 | n.s. | 3 | n.s. | 3 | n.s. | 2 | n.s. | 3 | n.s. | 19 | 1.26E-03 |
| CCNI | 1 | n.s. | 1 | n.s. | 2 | n.s. | 0 | n.s. | 2 | n.s. | 0 | n.s. | 0 | n.s. | 3 | 6.93E-02 |
| ARID1A | 7 | n.s. | 5 | n.s. | 16 | n.s. | 7 | n.s. | 4 | n.s. | 3 | n.s. | 2 | n.s. | 33 | 7.91E-09 |
| CD3EAP | 1 | n.s. | 0 | n.s. | 2 | n.s. | 0 | n.s. | 0 | n.s. | 0 | n.s. | 2 | n.s. | 5 | 1.29E-02 |
| ADAMTS6 | 1 | n.s. | 1 | n.s. | 3 | n.s. | 1 | n.s. | 0 | n.s. | 0 | n.s. | 2 | n.s. | 8 | 1.81E-03 |
| OR2D2 | 0 | n.s. | 0 | n.s. | 4 | n.s. | 0 | n.s. | 3 | n.s. | 0 | n.s. | 1 | n.s. | 5 | 5.67E-02 |
| TMEM199 | 0 | n.s. | 0 | n.s. | 3 | n.s. | 0 | n.s. | 2 | n.s. | 1 | n.s. | 0 | n.s. | 4 | 3.36E-02 |
| MST1 | 0 | n.s. | 0 | n.s. | 5 | n.s. | 2 | n.s. | 2 | n.s. | 0 | n.s. | 1 | n.s. | 7 | 9.46E-02 |
| RHBG | 0 | n.s. | 0 | n.s. | 3 | n.s. | 0 | n.s. | 0 | n.s. | 1 | n.s. | 2 | n.s. | 4 | 7.91E-02 |
| ZFP36L1 | 1 | n.s. | 1 | n.s. | 5 | n.s. | 2 | n.s. | 2 | n.s. | 0 | n.s. | 1 | n.s. | 8 | 3.37E-02 |
| TCP11 | 2 | n.s. | 0 | n.s. | 3 | n.s. | 2 | n.s. | 0 | n.s. | 0 | n.s. | 1 | n.s. | 6 | 4.80E-02 |
| CASZ1 | 4 | n.s. | 4 | n.s. | 3 | n.s. | 0 | n.s. | 0 | n.s. | 1 | n.s. | 2 | n.s. | 11 | 2.03E-02 |
| GAL3ST1 | 1 | n.s. | 1 | n.s. | 2 | n.s. | 0 | n.s. | 1 | n.s. | 0 | n.s. | 1 | n.s. | 4 | 7.74E-03 |
| FRMPD2 | 1 | n.s. | 1 | n.s. | 7 | n.s. | 2 | n.s. | 4 | n.s. | 0 | n.s. | 1 | n.s. | 9 | 8.91E-02 |
| GPS2 | 1 | n.s. | 1 | n.s. | 4 | n.s. | 3 | n.s. | 0 | n.s. | 1 | n.s. | 0 | n.s. | 8 | 8.91E-02 |
| ZNF362 | 0 | n.s. | 0 | n.s. | 3 | n.s. | 3 | n.s. | 0 | n.s. | 0 | n.s. | 0 | n.s. | 3 | 8.91E-02 |
n: number of mutations q: MutSigCV2 q-value
To investigate these differences, we identified recurrent alterations across all 817 samples and separately in ILC and IDC PAM50 subtypes (Luminal A, n=201, Luminal B, n=122, HER2-enriched, n=51, and Basal-like, n=107). In total, we identified 178 events, including 68 mutated genes, 47 regions of gain and 63 regions of loss (Table 1 and Table S2). Several of these had different incidence in ILC and in IDC (Figure 1C, Table S3). ILC cases were significantly enriched for CDH1 mutations, (63% in ILC vs. 2% in IDC, q=3.94E-53), most of them truncating, and mutations affecting TBX3 (9% vs. 2%, q=0.003), RUNX1 (10% vs. 3%, q=0.008), PIK3CA (48% vs. 33%, q=0.02) and FOXA1 (7% vs. 2%, q=0.08). By contrast, alterations typically observed in ER−/Basal-like tumors were less frequent in ILC including TP53 mutations (8% in ILC vs. 44% in IDC, q=1.9E-14) and focal amplification of MYC (6% vs. 27%, q=7.42E-7) and CCNE1 (0% vs. 7%, q=0.01). These results partly reflect genetic differences between ER+/Luminal and ER−/Basal-like breast cancer, given that ILC tumors were predominantly LumA. Nonetheless, unexpected differences did emerge including a lower incidence of GATA3 mutations in ILC compared to IDC (5% in ILC vs. 13% in IDC, q=0.03) (Figure 1C).
To better identify ILC discriminatory features, we limited our analyses to LumA samples, representing 41% of IDC (n=201) and 83% of ILC (n=106) (Figure 1D). This analysis confirmed a high incidence of CDH1 (q=1.4E-30), TBX3 (q=0.05) and FOXA1 (q=0.065) mutations in ILC, while the frequency of RUNX1 and PIK3CA mutations was no longer significantly different. GATA3 mutations (5% ILC vs. 20% IDC, q=0.003) were the second most discriminant event after CDH1 mutations, mostly affecting IDC tumors. Interestingly, both FOXA1 and GATA3 are key regulators of ER activity (Liu et al., 2014), suggesting IDC and ILC may preferentially rely on different mechanisms to mediate the ER transcriptional program. Finally, homozygous losses of the PTEN locus (10q23) were more frequent in ILC (q=0.035) as were PTEN mutations (8% vs. 3%). Collectively, PTEN inactivating alterations were identified in 14% of LumA ILC versus 3% of LumA IDC (p=9E-4), making this the third most discriminant feature between LumA IDC and LumA ILC (Figure 1D).
E-cadherin loss in invasive lobular carcinoma
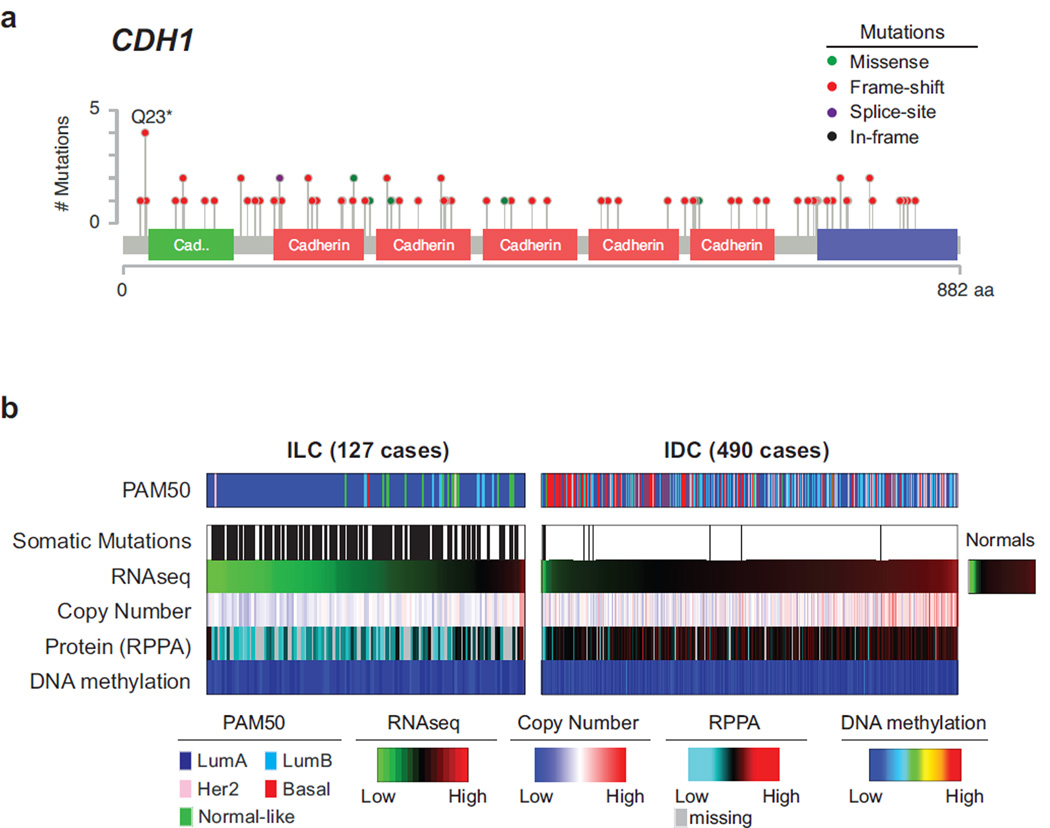
Loss of the epithelial specific cell-cell adhesion molecule E-cadherin (CDH1) is the key hallmark of ILC (Dabbs et al., 2013; Moll et al., 1993). CDH1 loss is believed to confer the highly discohesive morphology characteristic of this tumor subtype and is often associated with tumor invasion and metastasis in other tumor types, including diffuse gastric cancer (Brinck et al., 2004; Cancer Genome Atlas Research, 2014; Richards et al., 1999). Loss-of-function mutations targeting CDH1 are present in 50–60% of ILC and are believed to be an early event often observed in matching lobular carcinoma in situ (LCIS) (McCart Reed et al., 2015). CDH1 mutations typically occur in combination with chromosome 16q loss, where CDH1 is located, thus inducing complete loss of the protein.
We identified 108 mutations in the coding sequence of CDH1 in 107/817 patients (63%); 80 of these occurred in ILC cases. These mutations were rather uniformly distributed along the coding sequence and 83% of them were predicted to be truncating (Figure 2A). CDH1 mutations almost invariably co-occurred with heterozygous loss of 16q (affecting 89% of ILC cases), and were associated with down-regulation of both CDH1 transcript and protein levels (Figures 2B and S2A). By combining somatic mutations, copy number losses, and mRNA and low protein expression (the latter when available), we identified E-cadherin alterations in 120/127 (95%) cases with DNA and RNA data, and in all 79 cases with DNA, RNA, and protein data (Figure S2A–C).
Figure 2. E-cadherin loss in ILC.
A) Mutations targeting the CDH1 gene target residues across the whole sequence and are mostly predicted to be truncating (red). B) Comparison of E-cadherin status between ILC and IDC reveals frequent hemizygous copy number losses at the CDH1 locus and downregulation of both mRNA [log2(RSEM)] and protein levels. See also Figure S2A–C. Average DNA methylation level of 6 probes at the CDH1 promoter shows no change in DNA methylation in both ILC and IDC samples. See also Figure S2D–I.
Previous studies reported sporadic cases of multiple cancer types with high DNA methylation levels at the CDH1 promoter, suggesting epigenetic silencing as an alternative mechanism for down-regulation of CDH1 (Graff et al., 1997; Richards et al., 1999; Sarrio et al., 2003; Zou et al., 2009). We analyzed the DNA methylation levels in breast tumors at CpG sites spanning from upstream of the CDH1 promoter, across the promoter CpG island, and extending into the first intron (Figure S2D). Despite four of these probes matching DNA positions previously reported as methylated in ILC (Graff et al., 1997; Sarrio et al., 2003; Zou et al., 2009), we did not detect significant DNA hyper-methylation at these probes (Figure S2E), nor in any of the other CDH1 associated probes analyzed (Figures 2B and S2F–G). Moderately increased methylation was observed in a few cases near exon 2, however DNA methylation at this site correlated with lower tumor purity and increased leukocyte infiltration, and indeed it mimicked methylation levels at this CpG site in normal leukocytes (Figure S2F–G). Infinium DNA methylation results were validated by whole genome bisulfite sequencing in 5 samples (Figure S2H). Altogether these data confirm that CDH1 expression was substantially lower in ILC than in IDC, and that this expression difference did not appear associated with DNA methylation at the CDH1 promoter. Our results on 817 invasive breast tumors thus confirmed Ecadherin loss as ILC defining molecular feature, but do not support the reported occurrence of CDH1 epigenetic silencing in invasive breast cancer. The discrepancy with prior literature may be attributable in part to the reliance on highly sensitive, but non-quantitative methylation-specific PCR assays in past studies (Herman et al., 1996) and will require further investigation.
FOXA1 mutations in breast cancer
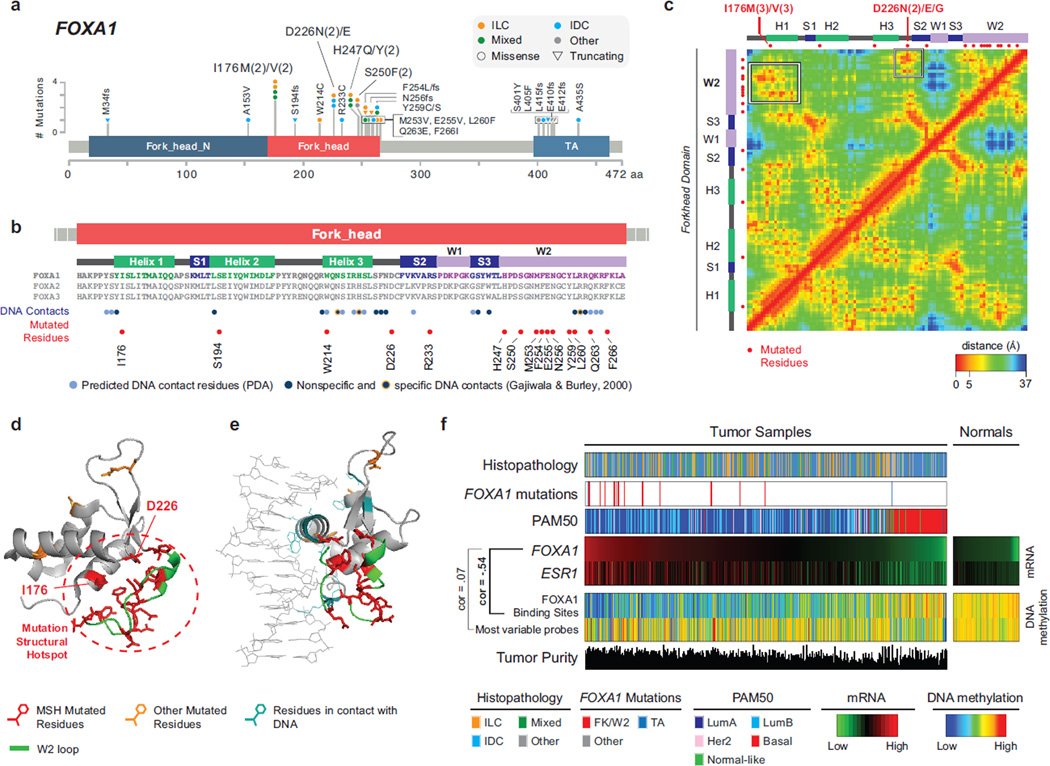
FOXA1 is a key ER transcriptional modulator (Carroll et al., 2005; Hurtado et al., 2011) coordinating ER DNA binding within a large protein complex by modifying chromatin accessibility and mediating long-range DNA interactions (Liu et al., 2014). High FOXA1 expression has been previously reported in breast and prostate cancer (Habashy et al., 2008; Sahu et al., 2011) and somatic mutations in the FOXA1 gene have been reported in these tumor types in about 3–4% of the cases (Barbieri et al., 2012; Cancer Genome Atlas, 2012; Robinson et al., 2015).
Here, we observed a total of 33 FOXA1 mutations in 30/817 (3.7%) tumors (Figure 3A) and the large sample set allowed us to identify regional hotspots in the FOXA1 mutation distribution. Mutations clustered in the fork-head DNA binding (FK) and C-terminus transactivation domains (Figure 3A). A similar mutational pattern was observed by combining multiple prostate cancer sequencing studies (Baca et al., 2013; Barbieri et al., 2012; Grasso et al., 2012; Robinson et al., 2015) (Figure S3A), and confirmed by the TCGA prostate cancer project (TCGA 2015, in press). Thus, regional FOXA1 mutation hotspots are selectively altered in a tissue-independent fashion.
Figure 3. Recurrent FOXA1 mutations cluster in the 3D space and correlate with high FOXA1 activity.
A) Recurrent FOXA1 mutations in 817 breast tumors cluster in the Fork-head DNA binding (FK) domain and in the C-terminus trans-activation (TA) domain. B) Secondary structure elements of the FK domain are not equally mutated. FOXA1 mutations cluster in the W2 loop and rarely target residues interacting with the DNA. C) Residue-residue minimum distances for all residues in the FK domain using the 3D structure of FOXA3 FK domain (PDB ID: 1VTN). Frequently mutated residues I176 and D226 are close in the 3D space (but not in the sequence) to the residues in the W2 loop. See also Figure S3C. D) 3D structure of the FK domain. Mutations in the W2 loop, in I176, and in D226 form a mutational structural hotspot (MSH). E) 3D structure of FK domain bound to the DNA molecule shows mutated residues (red) are not those in contact with the DNA (light blue). F) Across all breast cancer subtypes (histopathology and PAM50), FOXA1 mutations are associated with FOXA1 high mRNA expression. FOXA1 mRNA expression is highly correlated with ER mRNA expression [log−2(RSEM)] and anti-correlated with DNA methylation at FOXA1 binding sites consistent with FOXA1 activity. DNA methylation of randomly selected probes was used as control.
Eleven FOXA1 mutations were observed in 9/127 (7%) ILC cases. All FOXA1 mutations in ILC were in the FK domain, whereas mutations in IDC (n=11) were observed both in FK (n=6) and other structural elements (n=5), without a specific preference. The FK domain includes three α-helices (H1, H2, H3), three β-strands (S1, S2, S3) and two loops, typically referred to as “wings” (W1, W2) (Figures 3B and S3B). FOXA1 mutations in FK clustered prevalently in the W2 loop. Notable exceptions were recurrently mutated residues I176 (n=4) and D226 (n=3). These residues are far from W2 in sequence space and located in different secondary structure elements; however they are close (within 5 to 10 Angstroms) to residues in W2 in the three-dimensional (3D) space (Figures 3C–D and S3C). In total 22 out of 25 FK-mutations in our dataset fall into a restricted 3D space or “mutation structural hotspot” (MSH) (Figure 3D) indicating a selective pressure for targeting protein interactions and functions mediated by this region. Notably, 8/127 ILC cases have FOXA1 mutations within this MSH compared to 4/490 IDC cases (p=6E-4), further supporting FOXA1 selected mutations as an ILC feature.
FOXA1 DNA binding occurs mostly through helix H3, that recognizes the binding motif, and is stabilized by interactions mediated by its “wings” (Cirillo and Zaret, 2007; Gajiwala and Burley, 2000; Kohler and Cirillo, 2010). Only a few residues predicted or experimentally shown to interact with the DNA were mutated (Figure 3B–E) suggesting that these events are unlikely to affect FOXA1 DNA binding. FOXA1 is a pioneer factor that binds condensed chromatin and triggers DNA demethylation of its binding sites, making them accessible to transcription factors such as ER (Cirillo et al., 2002; Serandour et al., 2011). FOXA1 activity can therefore be estimated by the methylation status of these sites where occupied FOXA1 DNA binding sites tend to be demethylated. We analyzed DNA methylation levels of the 3976 most variable methylation probes mapping to FOXA1 binding sites (Table S3) and methylated in normal samples (Ross-Innes et al., 2012; Wang et al., 2012). DNA methylation of these sites was substantially lower when FOXA1 and ESR1 were highly expressed, while it remained high in FOXA1-negative cases and adjacent normal tissue (Figure 3F). Inverse correlation with FOXA1 expression (Pearson’s coefficient ρ=−0.54) was specifically observed for DNA methylation at FOXA1 binding sites. Indeed, no correlation was found with methylation at 2000 most variable probes with the same methylation level in normal samples as FOXA1 binding sites (ρ=0.07) (Figure 3F). These data support the hypothesis that FOXA1 mRNA expression correlates with its activity. FOXA1 mutations were positively associated with its mRNA expression (p=0.002) and maintained a similar anti-correlation with DNA methylation at FOXA1 binding sites (Figure 3F). Finally, by examining mRNA expression of FOXA1 targets, defined as genes with a FOXA1 binding motif in the promoter or matching the genomic loci covered by the 3976 methylation probes we analyzed (Table S3), no significant differences were identified and only a few genes showed moderate expression changes (Table S4). These data collectively indicate that FOXA1 mutations do not abolish protein function and, in fact, they may activate alternative mechanisms to affect ER transcriptional programs.
Differential expression analyses between FOXA1 mutant and wild type cases within distinct subsets of samples found consistent up-regulation of neuroendocrine secretory proteins SCG1 (CHGB) and SCG2, chemokine-like factor CMTM8, neuroendocrine tumor associated transcription factor NKX2-2 and Kallikrein serine proteases KLK12, KLK13, and KLK14 (Figure S3D and Table S5). While the relatively low number of FOXA1 mutations and breast cancer heterogeneity prevented the identification of strong transcriptional signals associated with FOXA1 mutations, several upregulated targets in FOXA1 mutant cases with part of them consistently found as significant suggests these lesions might drive novel binding events.
Interestingly, while ILC cases were enriched for FOXA1 mutations and in particular for those targeting the FK domain, ILC showed significantly fewer GATA3 mutations, another key ER modulator. Mutations in GATA3 were more frequent in LumA IDC (Figure 1D) and mutually exclusive with FOXA1 events. Moreover, LumA ILC tumors show lower GATA3 mRNA (p=0.007) and protein (p=2E-4) levels than LumA IDC (Figure S3E–F). Taken together, the differential expression patterns and enrichment for hotspot mutations of GATA3 in IDC and of FOXA1 in ILC, suggest a preferential requirement for distinct ER modulators in ILC and IDC.
Akt signaling is strongly activated in ILC
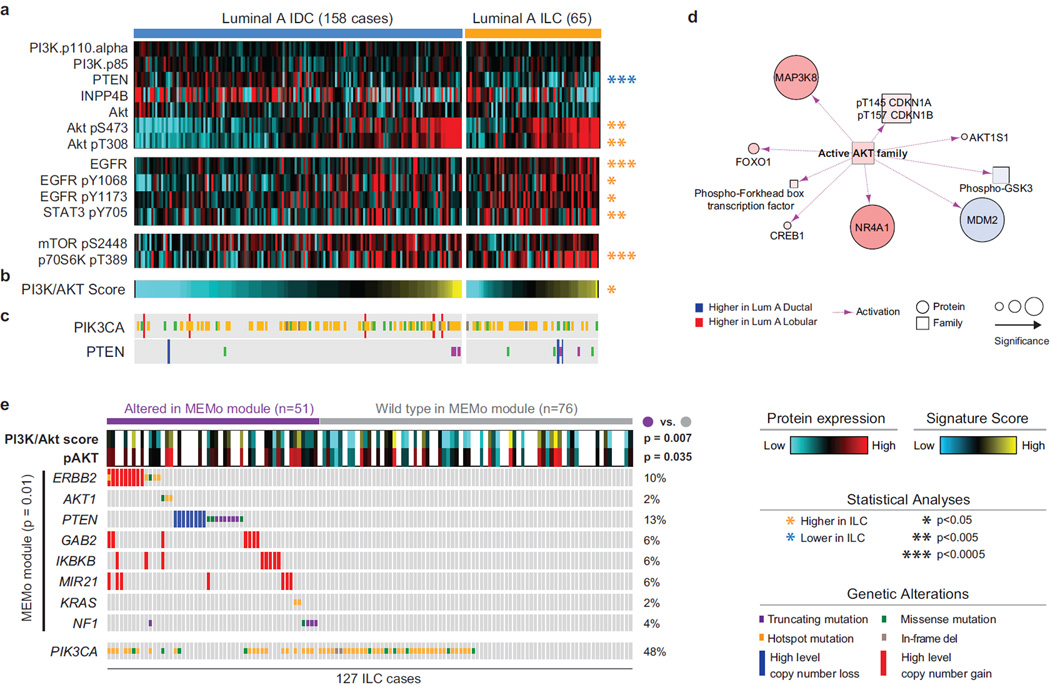
PTEN inactivation emerged as a discriminant feature between Luminal A ILC and Luminal A IDC. PTEN genetic alterations across all ILC cases included homozygous deletions (6%) and somatic mutations (7%), and were largely mutually exclusive with PIK3CA mutations (48%) (Figure S4A).
Unbiased differential protein expression analysis (Table S6) based on RPPA data revealed significant lower PTEN protein expression (p=4E-4) in LumA ILC compared to LumA IDC (Figure 4A). Consistent with PTEN function as a negative regulator of Akt activity (Cantley and Neel, 1999; Song et al., 2012), ILC tumors also showed significantly increased Akt phosphorylation at both S473 (p=0.004) and T308 (p=7E-5) (Figure 4A). Upstream of the Akt pathway, we found significant up-regulation of total EGFR (p=1E-4) and phospho-EGFR at Y1068 (p=0.005) and Y1173 (p=0.007), as well as phospho-STAT3 at Y705 (p=7E-4), supporting up-regulation of signaling axes converging on Akt activation (Wu et al., 2013). We also identified increased phospho-p27 at T157 (p=0.002), an Akt substrate, and phospho-p70S6 kinase at T389 (p=1E-4), a direct mTOR target. Notably, ILC phospho-Akt levels were comparable with those typically observed in the more aggressive HER2+ and ER−/Basal-like breast tumors (Figure S4B), which have uniformly high levels of PI3K/Akt signaling (Cancer Genome Atlas, 2012). Consistent with these results, we found significant up-regulation of a PI3K/Akt pathway-specific protein and phospho-protein expression signature (Akbani et al., 2014) in LumA ILC compared to LumA IDC (Figure 4B, Tables S1 and S6). Based on this signature, we found nearly equivalent levels of PI3K/Akt signaling in LumA ILC and Basal-like and HER2+ IDC (Figure S4C). Finally, PARADIGM analyses (Vaske et al., 2010) showed increased activation of Akt signaling in LumA ILC relative to LumA IDC (Figure 4D).
Figure 4. Akt signaling is highest in ILC tumors.
A) Differential protein and phospho-protein analysis between ILC LumA and IDC LumA reveals significant lower levels of PTEN, and higher levels of Akt, phospho-Akt, EGFR, phospho-EGFR, phopsho-STAT3, and phospho-p70S6K in ILC LumA. B) A PI3K/Akt protein expression signature is significantly up-regulated in ILC tumors. See also Figure S4B–C. C) Mutation and copy number alterations in PIK3CA and PTEN D) PARADIGM identifies increased Akt activity in LumA ILC tumors. E) MEMo identified multiple mutually exclusive alterations in ILC converging on Akt signaling and associated with increased phospho-Akt and PI3K/Akt protein signature in these tumors. Hotspot are defined as follow: PIK3CA E542, E545, Q546, and H1047; ERBB2 L755, I767, V777; AKT1 E17; KRAS G12.
PTEN protein loss and increased Akt phosphorylation were observed in association with PTEN genetic alterations, as well as in multiple ILC PTEN wild-type cases indicating that additional mechanisms contribute to the activation of the pathway. While PIK3CA mutations were frequent in LumA ILC tumors, these mutations were not associated with increased levels of phospho-Akt or pathway activity in our dataset. Using MEMo (Ciriello et al., 2012), we highlighted multiple genetic alterations converging on Akt/mTOR signaling in 45% of the samples (Figure S4D, Table S7). Among these, alterations acting upstream of Akt were identified in 40% of ILC cases and were associated with increased Akt phosphorylation and PI3K/Akt score (Figure 4E) providing an apparent molecular explanation for Akt activation in these samples. Interestingly, these events included ERBB2 amplification and mutations (Figure 4E), both of which have been identified in relapsed ILC (Ross et al., 2013).
ILC mRNA subtypes
Using mRNA-seq expression data from LumA ILC samples (n=106), we identified 3 ILC subtypes termed reactive-like, immune-related, and proliferative (Figure S5A–I, Table S8). We then used a 3-class ILC subtype classifier (60 genes, Table S13) to score all ILC samples in the TCGA (n=127) (Figure 5A) and METABRIC (Curtis et al., 2012) datasets (Table S12 to Table S8). Our analyses identified many significant genomic features that distinguished each ILC subtype at the mRNA and protein/phospho-protein level; but no distinguishing somatic mutations or DNA copy number alterations.
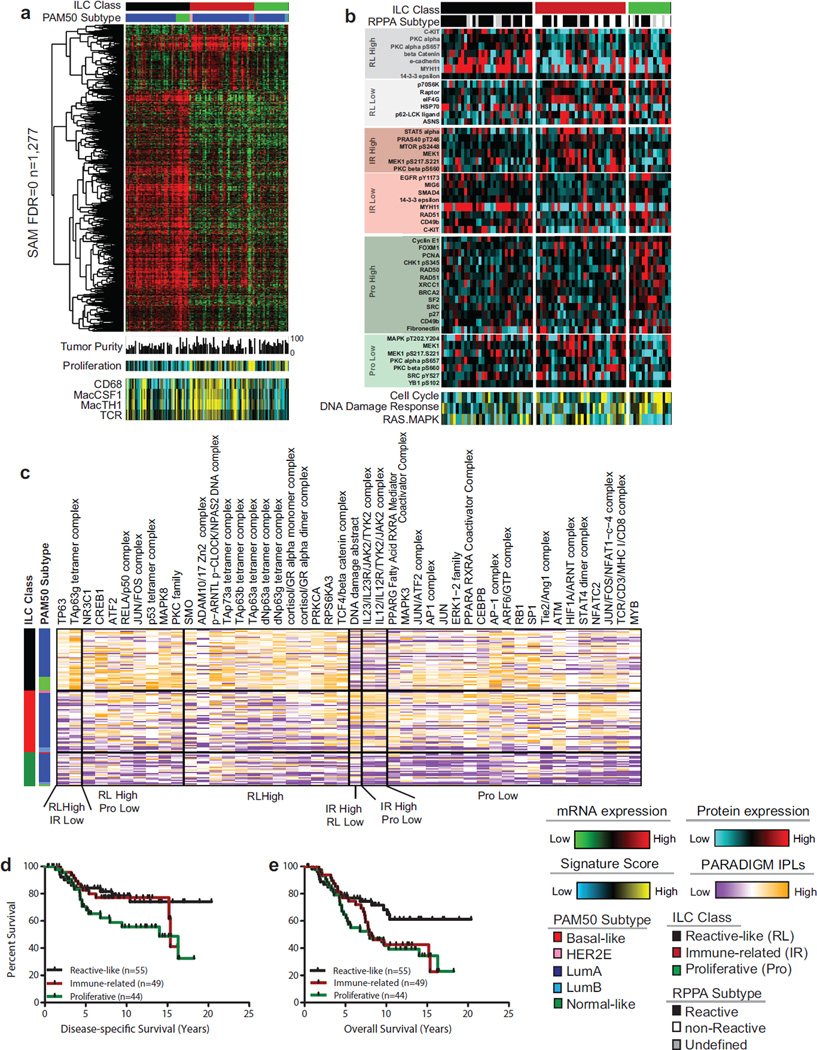
Figure 5. ILC molecular subtypes.
A) Three molecular subtype of lobular breast cancer were identified based on differential gene expression and show unique patterns highly expressed genes (n=1277, SAM FDR=0, upper panel), minor difference in tumor purity measured by ABSOLUTE, and differences in gene expression signatures measuring proliferation, CD68, Macrophage-associated CSF1, Macrophage–associated TH1, and T Cell Receptor Signaling (lower panel). Proliferation is highest in the proliferative (Pro) and immune-related (IR) subgroups; macrophage associated signaling is highest in immune-related tumors. B) Differences in protein expression profiles as determined by RPPA analysis. The reactive-like (RL) subgroup shows a significant association (p<1E-4, Fisher’s Exact test) with the RPPA-defined Reactive subgroup of breast cancer. Differences in subgroup-specific patterns of protein expression (p<0.05, t-test) for individual proteins (upper panel) as well as for protein expression signatures (lower panel) were identified. The proliferative subgroup shows higher expression of the cell cycle and DNA damage response pathways and lower levels of Ras-MAPK signaling (p<0.05). C) Subgroup-associated signaling features identified by PARADIGM. D) Reactive-like (n=55) tumors have significantly better disease specific (p=0.038, HR: 0.47, log-rank) and E) overall survival (p=0.022, HR=0.50) compared to proliferative (n=44) tumors in the METABRIC cohort. See also Figure S5.
Significant Analysis of Microarray (SAM) analysis (Tusher et al., 2001) identified 1277 genes differentially expressed between ILC subtypes (q=0) (Figure 5A, Table S8). Of these, 1005 were highly expressed in reactive-like tumors, which had lower tumor purity as determined by ABSOLUTE (Carter et al., 2012) (Figures 5A and S5P), and included genes consistent with epithelial and stromal-associated signaling including keratin, kallikrein, and claudin genes as well as the oncogenes EGFR, MET, PDGFRA and KIT (Table S14 to Table S8). The remaining 272 genes were highly expressed in immune-related tumors and include modulators of immunogenic signaling such as interleukins (IL), chemokine receptors and ligands, major histocompatibility complex, and tumor necrosis factors, as well as IDO1 and IFNG (Table S14 to Table S6). Interestingly, immune activity in this subset of tumors appears to be predominantly associated with macrophage-associated signaling as increased levels of CD68 (p<0.05), Macrophage-associated Colony Stimulating Factor (MacCSF), Macrophage-associated TH1 (MacTH1), and T-cell Receptor (TCR) gene expression signatures (Iglesia et al., 2014) were observed in both the TCGA (Figure 5A) and METABRIC (Figure S5J–K) datasets. Finally, proliferative tumors were defined by low expression of each of these 1277 genes (Figure 5A). Intriguingly, in each dataset (Figures 5 and S5K) proliferative tumors had higher levels of proliferation relative to reactive-like tumors (TCGA: p=3.3E-09; METABRIC: p=0.018) and slightly higher or equivalent levels compared to immune-related ones (TGCA: p=0.29; METABRIC: p=0.008). Regardless of ILC subtype, ILC tumor proliferation was generally lower than all IDC subtypes (Figure S5L–M).
With respect to previously reported RPPA-based subtypes (Reactive or non-Reactive), reactive-like ILC largely, but not entirely, comprised tumors classified as Reactive (Figure 5B; p<1E-4), a subgroup characterized by strong microenvironment and/or cancer fibroblast signaling (Cancer Genome Atlas, 2012). Examining protein and phospho-protein expression differences between ILC subtypes identified many significant features (Table S8 to Table S6). Reactive-like tumors had higher levels of c-Kit (p=4E-4), consistent with mRNA expression, total (p=0.004) and phosphorylated PKC alpha (S657, p=0.002); beta catenin (p=0.012) and E-cadherin (p=0.011), although both beta-catenin and E-cadherin levels are significantly lower than in all IDC subtypes. Decreased levels were observed instead for p70S6 kinase (p=0.017), Raptor (p=0.027) and eIF4G (p=0.024).
Immune-related tumors had higher levels of immune modulator STAT5 alpha (p=0.019), PI3K/Akt targets phospho-PRAS40 (T246, p=0.016) and mTOR (S2448, p=0.019), and total (p=0.004) and phospho-MEK1 (S217–S221, p=0.022). Consistent with the mRNA proliferation signature, tumors in the proliferative subtype have increased expression of cell cycle proteins Cyclin E1 (p=0.036), FoxM1 (p=0.019), PCNA (p=0.019), and propsho-Chk1 (S345, p=0.038) as well as DNA repair components Rad50 (p=0.007), Rad51 (p=0.007), XRCC1 (p=0.028) and BRCA2 (p=0.038). Decreased expression was observed for total (p=0.014) and phospho-MAPK (T202-Y204, p=0.038), and phosphorylated MEK1 (S217–S221, p=0.019), PKC alpha (S657, p=0.006), PKC beta (S660, p=0.037), and Src (Y527, p=0.026). Protein pathway signatures (Akbani et al., 2014) recapitulated these findings with proliferative tumors having increased levels of the Cell Cycle (p=0.005) and DNA Damage Response (p=0.014) signatures and a lower RAS-MAPK signature (p=0.031) score (Tables S1 and S6).
Using an integrative genomics approach, PARADIGM predicted increased activation of the TP53, TP63, TP73 TCF/beta-catenin PKC, and JUN/FOS pathways in reactivelike tumors; increased activation of immune-modulators IL12, and IL23, IL12R and IL23R, JAK2 and TYK2 in immune-related ILCs (Baay et al., 2011; Duvallet et al., 2011; Strobl et al., 2011); and decreased activation of each of these pathways along with lower levels of MAPK3, RB1, and ERK1 (Figure 5C) in proliferative ILC tumors.
Lastly, we determined that reactive-like ILC patients had a significantly better disease-specific (DSS) (p=0.038, HR: 0.47) and overall survival (OS) (p=0.023, HR: 0.50) compared to proliferative ILC patients in the METABRIC dataset, which has a median follow-up of 7.2 years (compared to the TCGA median follow-up of less than 2 years), (Figure 5D). Consistent with these results, patients with more proliferative lobular tumors (i.e. greater than the median PAM50 proliferation signature score) had worse DSS (p=0.025, HR: 2.0) and a tendency towards worse OS (p=0.058, HR: 0.63) compared to patients with a lower proliferation score (Figure S5N–O). No significant differences in DSS or OS were identified between the immune-related subgroup and either the proliferative or reactive-like subgroup. These results are consistent with previous studies reporting that the reactive stromal phenotype is associated with a good prognosis in breast cancer while proliferation is one of the strongest indicators of worse outcome in luminal/ER+ breast cancers (Ciriello et al., 2013b).
Tumors with mixed ILC and IDC histology
Histologically, ~3–6% of breast tumors present both a ductal and a lobular component (Figure 6A). Pathologists currently classify these tumors as mixed ductal/lobular breast carcinoma or invasive ductal cancers with lobular features (Arps et al., 2013). There are, however, no defined criteria or uniform terminology for the classification of mixed tumors and as a consequence discordant clinical and molecular features have been reported (Bharat et al., 2009). Molecular profiling has the power to provide quantitative endpoints to compare the genetics of mixed tumors with those of pure ILC and IDC. In our dataset, 88/817 tumors (11%) were classified by as mixed ductal/lobular breast carcinomas. We characterized these mixed tumors using multiple computational approaches integrating different data-types thus to determine whether they molecularly resembled IDC (IDC-like), ILC (ILC-like), or neither.
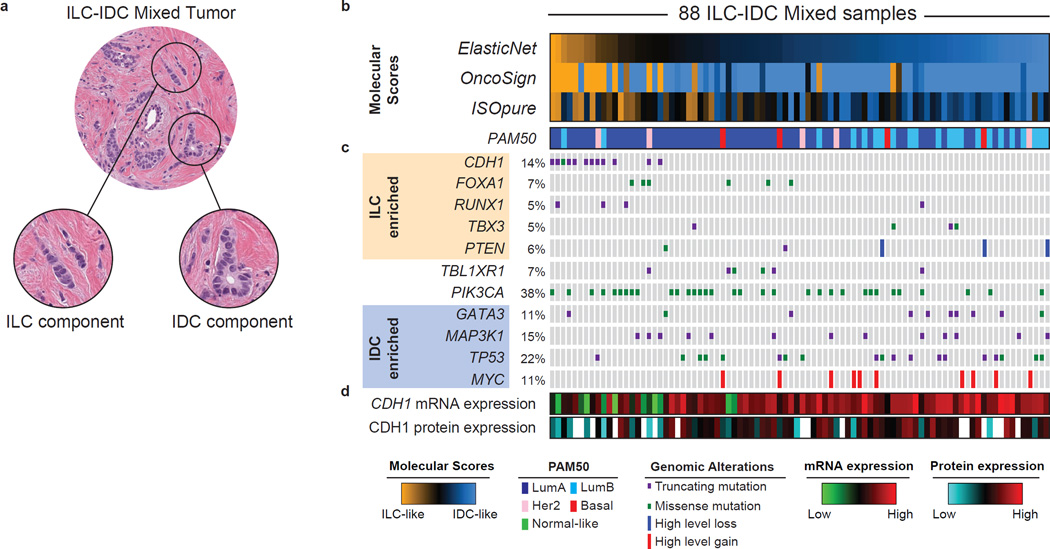
Figure 6. Molecular classification of mixed ductal/lobular carcinoma.
A) Mixed ductal/lobular tumors present at the same time both a lobular and ductal component. B) We used three algorithmic approaches (ElasticNet, OncoSign, and ISOpure) to evaluate the resemblance of mixed tumors to either ILC (ILC-like) or IDC (IDC-like) based on molecular features. ILC-IDC scores are shown for all three approached at the top. See also Figure S6B–D. C) Genetic alterations enriched in ILC tumors are frequently found in ILC-like mixed cases (in particular CDH1 mutations), whereas those enriched in IDC are more frequent in IDC-like mixed cases. D) ILC-like mixed-cases are characterized by both low E-cadherin mRNA and protein level. See also Figure S6E.
We first analyzed the transcriptional landscape of mixed tumors using the ISOpure algorithm (Quon et al., 2013), which deconvolves the transcriptional signal of each queried tumor to estimate how much of it can be explained by one or more reference populations and how much is unique. Interestingly, mRNA expression profiles of all mixed cases could almost completely be explained by either IDC or ILC reference populations, suggesting that these tumors separate into IDC-like and ILC-like cases and do not represent a molecularly distinct subtype (Figure S6A). Based on these analyses, 32/88 mixed cases received an ILC-score greater than the IDC-score, and were therefore classified as ILC-like (Figure 6B).
We next evaluated the resemblance of mixed tumors to IDC and ILC based on the previously determined selected set of copy number alterations (CNAs) and mutations (Table S2). Mixed tumors were enriched for IDC recurrent CNAs and mutations when compared to ILC, and vice versa (Figure S6B), indicating ILC and IDC genetic alterations were both present in these tumors, either simultaneously or in separate IDC-like and ILC-like subgroups. We then compared each mixed tumor to ILC and IDC based on their genomic features, by adapting the OncoSign algorithm (Ciriello et al., 2013a). This approach identified 19 ILC-like mixed samples characterized by ILC genetic features (Figures 6B and S6D). All CDH1-mutated mixed cases were classified as ILC-like, indicating CDH1 status as a dominant feature in this analysis. A few CDH1 wild-type mixed cases were also classified as ILC-like and characterized by ILC-enriched events such as mutations in RUNX1 (3/4 mutated cases), TBX3 (2/4) and FOXA1 (2/6). ILC-enriched alterations did not co-occur with IDC-enriched ones, further indicating that mixed tumors can be categorized into ILC-like or IDC-like subgroups and do not constitute a molecularly distinct subtype.
Finally, we combined 428 CNA, including focal and arm-level alterations, 409 gene expression modules (Fan et al., 2011; Gatza et al., 2014) and somatic mutations for 128 genes mutated in more than 3% of the cases into a single ElasticNet classifier (Zou and Hastie, 2005). This integrated ElasticNet predictor identified 27/88 mixed tumors as ILC-like. These were enriched for the LumA subtype, CDH1 mutations and loss of E-cadherin mRNA expression (Figures 6B and S6E).
Overall, these approaches were highly concordant (Figures 6B and S6F) with 24/88 cases (18/57 LumA cases) being called ILC-like by at least two approaches, and 64 being called IDC-like (Table S1). ILC-like and IDC-like mixed tumors when compared to pure ILC and IDC, respectively, do not show significant enrichment for specific genomic alterations, being molecularly similar to either one or the other subtype. Our analyses demonstrate that mixed histology tumors overwhelmingly tend to resemble either ILC or IDC as opposed to representing a third distinct group. Moreover, IDC and ILC discriminant molecular features, in particular CDH1 status, could be used to stratify mixed tumors into ILC-like and IDC-like tumor subgroups.
Discussion
In this study we provide the most comprehensive molecular portrait to date of ILC. E-cadherin loss was confirmed the ILC hallmark lesion and we could identify CDH1 loss at the DNA, mRNA, and protein level in almost all ILC cases. Moreover, 12/27 CDH1 mutations in non-ILC cases occurred in mixed tumors strongly resembling ILC at the molecular level. Surprisingly, we did not identify DNA hyper-methylation of the CDH1 promoter in any breast tumor, suggesting that E-cadherin loss is not epigenetically driven. In addition, ILC and IDC differed in the FOXA1 and GATA3 mutational spectra, PTEN loss, and Akt activation. The lower incidence of GATA3 mutations in ILC and lower GATA3 mRNA and protein expression suggest that in LumA ILC tumors there is a preferential occupancy of ER in FOXA1 bounded sites (Theodorou et al., 2013). Differential ER activity is also observed at the protein level where both total ER (p=0.005) and phospho-ER (p=2E-05) levels are reduced in LumA ILC vs. LumA IDC. These findings in the context of recent data suggesting an improved response to the aromatase inhibitor letrozole as compared to tamoxifen in ILC (Metzger et al., 2012; Sikora et al., 2014) warrants further investigation.
The chromatin remodeling factor EP300, also involved in ER modulation, is able to directly acetylate FOXA1, and EP300 driven acetylation prevents FOXA1 DNA binding, but does not affect the protein when already bound (Kohler and Cirillo, 2010). Intriguingly, five acetylation sites have been identified in the wings of the fork-head domain; three of them in W2 (K264, K267 and K270) where most of our newly observed FOXA1 mutations cluster. These observations lead to the hypothesis that FOXA1 mutations could alter EP300 dependent acetylation of FOXA1 without affecting EP300 modulation of ER. While a rigorous evaluation of the role of EP300 in breast cancer and how FOXA1 mutations interfere with it goes beyond the scope of this study, FOXA1 mutations, its correlation with FOXA1 expression and lack of DNA methylation at its binding sites, and exclusivity with GATA3 mutations support these as events activating FOXA1 function and, thus, ER transcriptional program.
The PI3K/Akt pathway is among the most altered in cancer providing tumor cells with enhanced growth and survival capabilities. Integrating protein and phospho-protein data with gene expression and pathway activity signatures, we consistently identified increased Akt signaling in ILC versus IDC. Notably, E-cadherin loss has been associated with Akt activation and EGFR over-expression (Lau et al., 2011; Liu et al., 2013). Lack of E-cadherin expression, which characterizes almost all ILC tumors, may thus provide a favorable cellular context for Akt activation. Recently, PI3K and Akt inhibitors entered clinical trials for several cancer types including breast cancer. Here we showed that ILC has on average the highest levels of Akt activation, measured by phospho-Akt and PI3K/Akt signaling among all breast cancer subtypes (comparable to IDC Basal-like), making selective inhibition of this pathway in ILC a particularly attractive strategy.
Unbiased characterization of the ILC transcriptome showed a high degree of internal variability giving rise to three main subgroups: reactive-like, immune-related, and proliferative. While additional validation studies will clearly be required, we do observe increased expression of many druggable pathways/targets including increased levels of phospho-mTOR and phospho-MEK1 expression in the immunerelated subgroup as well as increased SMO and ERK pathway activity in the reactivelike subgroup. These results coupled with difference in clinical outcome suggests that these subgroups will be important for future studies focused on both the clinical and biological aspects of ILC.
Finally, we showed that mixed ILC/IDC tumors could be separated into two major groups based on their molecular resemblance to either ILC (ILC-like) or IDC (IDC-like). The ability to classify cancers with mixed phenotypes based on the underlying biology has implications for clinical practice as well as furthering our understanding of the etiology of such lesions. Indeed, ILC carcinomas often metastasize to body sites not colonized by IDCs (e.g. gastrointestinal (GI) tract and peritoneal surfaces). ILC are also typically of low histologic grade and with low to intermediate mitotic index, thus limiting their response to primary chemotherapy (Cristofanilli et al., 2005) and their ability to be detected on PET scans. As such, clinicians must be aware of non-specific symptomatology and favor diagnostic approaches such as anatomical scanning (CT scan) for ILCs. Finally, the identification of ILC enriched molecular features may ultimately lead to the design of ILC-targeted therapies. A more refined classification of mixed cancers as IDC-like or ILC-like will improve our understanding, detection and follow-up of the disease, and enable a more informed and targeted treatment selection.
This multi-platform study identified numerous molecular features discriminating between breast ILC and IDC, demonstrating different pathways underlying their pathogenesis, defining new ILC subtypes with different clinical outcomes, and pointing to previously unrecognized therapeutic possibilities. Importantly, we provided here a curated and integrated dataset for 817 breast tumors, including the largest collection to date of comprehensively profiled ILC. To facilitate the exploration of this dataset, we created a public web-service (http://cbio.mskcc.org/cancergenomics/tcga/brca_tcga) organizing all analyses and data used in this manuscript. We believe this resource will serve as a reference for many to further advance our understanding of human breast cancers.
Experimental procedures
Tumor and matched normal specimen were collected as previously described (Cancer Genome Atlas, 2012). In total 817 primary tumor samples were assayed by whole exome DNA sequencing, RNA sequencing, miRNA sequencing, SNP arrays, and DNA methylation arrays. A subset of 633 samples was assayed by reverse phase protein array (RPPA). Histological subtypes have been determined based on consensus by a pathology committee. Intrinsic breast cancer subtyping was performed on all 817 cases, using the PAM50 classifier (Parker et al., 2009). Data generation and processing were performed as previously described (Cancer Genome Atlas Research, 2014).
Enrichment analyses for selected events were performed using Fisher’s exact tests and a binary representation of copy number alterations and mutations (1 is altered, 0 is wild-type).
DNA methylation of the CDH1 promoter was assessed at probes within a window 1500 bp upstream and downstream CDH1 transcription start site using both HM27 and HM450 data. Whole genome bisulfite sequencing was performed to characterize DNA methylation levels at 157 CpGs.
Distances between of FOXA1 mutations have been determined from the tertiary structure of FOXA3 fork-head domain (PDB ID: 1VTN). Predicted DNA interactions were derived by WebPDA (http://bioinfozen.uncc.edu/webpda). Differential expression analyses on RNA-seq data were performed using the limmavoom package (Law et al., 2014).
Replication Based Normalized (RBN) RPPA data containing expression levels for 187 protein and phosphorylated proteins for 633 samples were used for protein differential expression analysis. Differential pathway activity was assessed by t-test.
ILC subtypes were determined using Consensus Cluster Plus Analysis (Wilkerson and Hayes, 2010) based on the 1000 most differentially expressed genes and a classifier was built using ClaNC (Dabney, 2006).
Detailed description of each analysis presented in this study can be found within the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Comprehensive molecular profiles of 817 invasive breast tumors.
Invasive lobular carcinoma (ILC) is a clinically and molecularly distinct disease.
ILCs show CDH1 and PTEN loss, AKT activation, and mutations in TBX3 and FOXA1.
Proliferation and immune-related gene expression signatures define 3 ILC subtypes.
Genetic features classify mixed tumors into lobular-like and ductal-like subgroups.
Acknowledgements
We wish to thank the many members of the TCGA Network, including the Tissue Source Sites, and patients, whom contributed samples to this study. This study was supported by funds from the TCGA Project (U24-CA143848), the NCI Breast SPORE program grant P50-CA58223-09A1, and the Breast Cancer Research Foundation. G.C. is supported by the Gabriella Giorgi-Cavaglieri Foundation, M.L.G. is supported by the National Cancer Institute of the US National Institutes of Health award number K99-CA166228, A.H.B. is supported by National Library of Medicine of the National Institutes of Health under Award Number K22LM011931, and A.P. is supported by Mildred-Scheel Postdoctoral Research Fellowship of the Deutsche Krebshilfe e.V. (No. 111354).
TCGA Research Network
Rehan Akbani, J. Todd Auman, Miruna Balasundaram, Saianand Balu, Thomas Barr, Andrew Beck, Christopher Benz, Stephen Benz, Mario Berrios, Rameen Beroukhim, Tom Bodenheimer, Lori Boice, Moiz S. Bootwalla, Jay Bowen, Reanne Bowlby, Denise Brooks, Andrew D. Cherniack, Lynda Chin, Juok Cho, Sudha Chudamani, Giovanni Ciriello, Tanja Davidsen, John A. Demchok, Jennifer B. Dennison, Li Ding, Ina Felau, Martin L. Ferguson, Scott Frazer, Stacey B. Gabriel, JianJiong Gao, Julie M. Gastier-Foster, Michael L. Gatza, Nils Gehlenborg, Mark Gerken,, Gad Getz, William J. Gibson, D. Neil Hayes, David I. Heiman, Katherine A. Hoadley, Andrea Holbrook, Robert A. Holt, Alan P. Hoyle, Hai Hu, Mei Huang, Carolyn M. Hutter, E. Shelley Hwang, Stuart R. Jefferys, Steven J.M. Jones, Zhenlin Ju, Jaegil Kim, Phillip H. Lai, Peter W. Laird, Michael S. Lawrence, Kristen M. Leraas, Tara M. Lichtenberg, Pei Lin, Shiyun Ling, Jia Liu, Wenbin Liu, Laxmi Lolla, Yiling Lu, Yussanne Ma, Dennis T. Maglinte, Elaine Mardis, Jeffrey Marks, Marco A. Marra, Cynthia McAllister, Michael McLellan, Shaowu Meng, Matthew Meyerson, Gordon B. Mills, Richard A. Moore, Lisle E. Mose, Andrew J. Mungall, Bradley A. Murray, Rashi Naresh, Michael S. Noble, Steffi Oesterreich, Olufunmilayo Olopade, Joel S. Parker, Charles M. Perou, Todd Pihl, Gordon Saksena, Steven E. Schumacher, Kenna R. Mills Shaw, Nilsa C. Ramirez, W. Kimryn Rathmell, Suhn K. Rhie, Jeffrey Roach, A. Gordon Robertson, Gordon Saksena, Chris Sander, Jacqueline E. Schein, Nikolaus Schultz, Hui Shen, Margi Sheth, Yan Shi, Juliann Shih, Carl Simon Shelley, Craig Shriver, Janae V. Simons, Heidi J. Sofia, Matthew G. Soloway, Carrie Sougnez, Charlie Sun, Roy Tarnuzzer, Daniel G. Tiezzi, David J. Van Den Berg, Doug Voet, Yunhu Wan, Zhining Wang, John N. Weinstein, Daniel J. Weisenberger, Matthew D. Wilkerson, Richard Wilson, Lisa Wise, Maciej Wiznerowicz, Junyuan Wu, Ye Wu, Liming Yang, Christina Yau, Travis I. Zack, Jean C. Zenklusen, Hailei Zhang, Jiashan Zhang, Erik Zmuda
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
G.C., M.L.G. and C.M.P coordinated overall study design and analyses. A.H.B., S.C.L, G.M.K.T., R.E.F., L.C.C., K.H.A., Y-Y.C., K.J., and N.B.J. coordinated pathology analyses. M.D.W., M.McL., and C.K. coordinated DNA sequencing analyses. M.L.G., K.A.H. and C.M.P coordinated RNA-seq analyses. S.K.R., H.S., and P.W.L. coordinated DNA methylation analyses. H.Z. and A.D.C. coordinated copy number analyses. R.B. and G.R. coordinated miRNA-seq analyses. G.B.M. coordinated RPPA analyses. A.P., C.Y., S.H., R.F. were involved in bioinformatics analyses. C.B. and C.S. supervised bioinformatics analyses. S.O. and T.A.K. coordinated clinical contributions. G.C. developed web-resource for breast cancer data. G.C., M.L.G and C.M.P. wrote the manuscript, which all authors reviewed.
References
- Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, Liu W, Yang JY, Yoshihara K, Li J, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nature communications. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpino G, Bardou VJ, Clark GM, Elledge RM. Infiltrating lobular carcinoma of the breast: tumor characteristics and clinical outcome. Breast cancer research : BCR. 2004;6:R149–R156. doi: 10.1186/bcr767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arps DP, Healy P, Zhao L, Kleer CG, Pang JC. Invasive ductal carcinoma with lobular features: a comparison study to invasive ductal and invasive lobular carcinomas of the breast. Breast cancer research and treatment. 2013;138:719–726. doi: 10.1007/s10549-013-2493-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baay M, Brouwer A, Pauwels P, Peeters M, Lardon F. Tumor cells and tumor-associated macrophages: secreted proteins as potential targets for therapy. Clinical & developmental immunology. 2011;2011:565187. doi: 10.1155/2011/565187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature genetics. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat A, Gao F, Margenthaler JA. Tumor characteristics and patient outcomes are similar between invasive lobular and mixed invasive ductal/lobular breast cancers but differ from pure invasive ductal breast cancers. American journal of surgery. 2009;198:516–519. doi: 10.1016/j.amjsurg.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Brinck U, Jacobs S, Neuss M, Tory K, Rath W, Kulle B, Fuzesi L. Diffuse growth pattern affects E-cadherin expression in invasive breast cancer. Anticancer research. 2004;24:2237–2242. [PubMed] [Google Scholar]
- Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome research. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nature genetics. 2013a;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Sinha R, Hoadley KA, Jacobsen AS, Reva B, Perou CM, Sander C, Schultz N. The molecular diversity of Luminal A breast tumors. Breast cancer research and treatment. 2013b;141:409–420. doi: 10.1007/s10549-013-2699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Molecular cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS. Specific interactions of the wing domains of FOXA1 transcription factor with DNA. Journal of molecular biology. 2007;366:720–724. doi: 10.1016/j.jmb.2006.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofanilli M, Gonzalez-Angulo A, Sneige N, Kau SW, Broglio K, Theriault RL, Valero V, Buzdar AU, Kuerer H, Buchholz TA, et al. Invasive lobular carcinoma classic type: response to primary chemotherapy and survival outcomes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:41–48. doi: 10.1200/JCO.2005.03.111. [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbs DJ, Schnitt SJ, Geyer FC, Weigelt B, Baehner FL, Decker T, Eusebi V, Fox SB, Ichihara S, Lakhani SR, et al. Lobular neoplasia of the breast revisited with emphasis on the role of E-cadherin immunohistochemistry. The American journal of surgical pathology. 2013;37:e1–e11. doi: 10.1097/PAS.0b013e3182918a2b. [DOI] [PubMed] [Google Scholar]
- Dabney AR. ClaNC: point-and-click software for classifying microarrays to nearest centroids. Bioinformatics. 2006;22:122–123. doi: 10.1093/bioinformatics/bti756. [DOI] [PubMed] [Google Scholar]
- Duvallet E, Semerano L, Assier E, Falgarone G, Boissier MC. Interleukin-23: a key cytokine in inflammatory diseases. Annals of medicine. 2011;43:503–511. doi: 10.3109/07853890.2011.577093. [DOI] [PubMed] [Google Scholar]
- Fan C, Prat A, Parker JS, Liu Y, Carey LA, Troester MA, Perou CM. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics. 2011;4:3. doi: 10.1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote FW, Jr, Stewart FW. A histologic classification of carcinoma of the breast. Surgery. 1946;19:74–99. [PubMed] [Google Scholar]
- Gajiwala KS, Burley SK. Winged helix proteins. Current opinion in structural biology. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- Gatza ML, Silva GO, Parker JS, Fan C, Perou CM. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat Genet. 2014;46:1051–1059. doi: 10.1038/ng.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. The Journal of biological chemistry. 1997;272:22322–22329. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habashy HO, Powe DG, Rakha EA, Ball G, Paish C, Gee J, Nicholson RI, Ellis IO. Forkhead-box A1 (FOXA1) expression in breast cancer and its prognostic significance. European journal of cancer. 2008;44:1541–1551. doi: 10.1016/j.ejca.2008.04.020. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nature genetics. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesia MD, Vincent BG, Parker JS, Hoadley KA, Carey LA, Perou CM, Serody JS. Prognostic B-cell signatures using mRNA-seq in patients with subtype-specific breast and ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:3818–3829. doi: 10.1158/1078-0432.CCR-13-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Cirillo LA. Stable chromatin binding prevents FoxA acetylation, preserving FoxA chromatin remodeling. The Journal of biological chemistry. 2010;285:464–472. doi: 10.1074/jbc.M109.063149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau MT, Klausen C, Leung PC. E-cadherin inhibits tumor cell growth by suppressing PI3K/Akt signaling via beta-catenin-Egr1-mediated PTEN expression. Oncogene. 2011;30:2753–2766. doi: 10.1038/onc.2011.6. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Su L, Liu X. Loss of CDH1 up-regulates epidermal growth factor receptor via phosphorylation of YBX1 in non-small cell lung cancer cells. FEBS letters. 2013;587:3995–4000. doi: 10.1016/j.febslet.2013.10.036. [DOI] [PubMed] [Google Scholar]
- Liu Z, Merkurjev D, Yang F, Li W, Oh S, Friedman MJ, Song X, Zhang F, Ma Q, Ohgi KA, et al. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell. 2014;159:358–373. doi: 10.1016/j.cell.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCart Reed AE, Kutasovic JR, Lakhani SR, Simpson PT. Invasive lobular carcinoma of the breast: morphology, biomarkers and ’omics. Breast cancer research : BCR. 2015;17 doi: 10.1186/s13058-015-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome biology. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger O, Giobbie-Hurder A, Mallon E, Viale G, Winer E, Thurlimann B, Gelber RD, Colleoni M, Ejlertsen B, Bonnefoi H, et al. Abstract S1-1: Relative effectiveness of letrozole compared with tamoxifen for patients with lobular carcinoma in the BIG 1-98 trial. Thirty-Fifth CTRC-AACR San Antonio Breast Cancer Symposium (San Antonio, TX) 2012 [Google Scholar]
- Moll R, Mitze M, Frixen UH, Birchmeier W. Differential loss of Ecadherin expression in infiltrating ductal and lobular breast carcinomas. The American journal of pathology. 1993;143:1731–1742. [PMC free article] [PubMed] [Google Scholar]
- Morrogh M, Andrade VP, Giri D, Sakr RA, Paik W, Qin LX, Arroyo CD, Brogi E, Morrow M, King TA. Cadherin-catenin complex dissociation in lobular neoplasia of the breast. Breast cancer research and treatment. 2012;132:641–652. doi: 10.1007/s10549-011-1860-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestalozzi BC, Zahrieh D, Mallon E, Gusterson BA, Price KN, Gelber RD, Holmberg SB, Lindtner J, Snyder R, Thurlimann B, et al. Distinct clinical and prognostic features of infiltrating lobular carcinoma of the breast: combined results of 15 International Breast Cancer Study Group clinical trials. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:3006–3014. doi: 10.1200/JCO.2007.14.9336. [DOI] [PubMed] [Google Scholar]
- Quon G, Haider S, Deshwar AG, Cui A, Boutros PC, Morris Q. Computational purification of individual tumor gene expression profiles leads to significant improvements in prognostic prediction. Genome medicine. 2013;5:29. doi: 10.1186/gm433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards FM, McKee SA, Rajpar MH, Cole TR, Evans DG, Jankowski JA, McKeown C, Sanders DS, Maher ER. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Human molecular genetics. 1999;8:607–610. doi: 10.1093/hmg/8.4.607. [DOI] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JS, Wang K, Sheehan CE, Boguniewicz AB, Otto G, Downing SR, Sun J, He J, Curran JA, Ali S, et al. Relapsed classic E-cadherin (CDH1)-mutated invasive lobular breast cancer shows a high frequency of HER2 (ERBB2) gene mutations. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:2668–2676. doi: 10.1158/1078-0432.CCR-13-0295. [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, Sankila A, Turunen JP, Lundin M, Konsti J, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. The EMBO journal. 2011;30:3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrio D, Moreno-Bueno G, Hardisson D, Sanchez-Estevez C, Guo M, Herman JG, Gamallo C, Esteller M, Palacios J. Epigenetic and genetic alterations of APC and CDH1 genes in lobular breast cancer: relationships with abnormal E-cadherin and catenin expression and microsatellite instability. International journal of cancer Journal international du cancer. 2003;106:208–215. doi: 10.1002/ijc.11197. [DOI] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome research. 2011;21:555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora MJ, Cooper KL, Bahreini A, Luthra S, Wang G, Chandran UR, Davidson NE, Dabbs DJ, Welm AL, Oesterreich S. Invasive lobular carcinoma cell lines are characterized by unique estrogen-mediated gene expression patterns and altered tamoxifen response. Cancer research. 2014;74:1463–1474. doi: 10.1158/0008-5472.CAN-13-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nature reviews Molecular cell biology. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- Strobl B, Stoiber D, Sexl V, Mueller M. Tyrosine kinase 2 (TYK2) in cytokine signalling and host immunity. Frontiers in bioscience. 2011;16:3214–3232. doi: 10.2741/3908. [DOI] [PubMed] [Google Scholar]
- Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome research. 2013;23:12–22. doi: 10.1101/gr.139469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, Stuart JM. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome research. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson MD, Cabanski CR, Sun W, Hoadley KA, Walter V, Mose LE, Troester MA, Hammerman PS, Parker JS, Perou CM, et al. Integrated RNA and DNA sequencing improves mutation detection in low purity tumors. Nucleic acids research. 2014;42:e107. doi: 10.1093/nar/gku489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson MD, Hayes DN. Consensus Cluster Plus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Chang Q, Lu Y, Qiu P, Chen B, Thakur C, Sun J, Li L, Kowluru A, Chen F. Gefitinib resistance resulted from STAT3-mediated Akt activation in lung cancer cells. Oncotarget. 2013;4:2430–2438. doi: 10.18632/oncotarget.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Yoon HS, Perez D, Weeks RJ, Guilford P, Humar B. Epigenetic silencing in non-neoplastic epithelia identifies E-cadherin (CDH1) as a target for chemoprevention of lobular neoplasia. The Journal of pathology. 2009;218:265–272. doi: 10.1002/path.2541. [DOI] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. J Roy Stat Soc B. 2005;67:301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.