Summary
Nuclear Dbf2p-related (NDR) protein kinases are important for cell differentiation and polar morphogenesis in various organisms, yet some of their functions are still elusive. Dysfunction of the Neurospora crassa NDR kinase COT1 leads to cessation of tip extension and hyperbranching. NDR kinases require the physical interaction between the kinase’s N-terminal region (NTR) and the MPS1-binding (MOB) proteins for their activity and functions. To study the interactions between COT1 and MOB2 proteins, we mutated several conserved residues and a novel phosphorylation site within the COT1 NTR. The phenotypes of these mutants suggest that the NTR is required for COT1 functions in regulating hyphal elongation and branching, asexual conidiation and germination. Interestingly, while both MOB2A and MOB2B promote proper hyphal growth, they have distinct COT1-dependent roles in regulation of macroconidiation. Immunoprecipitation experiments indicate physical association of COT1 with both MOB2A and MOB2B, simultaneously. Furthermore, the binding of the two MOB2 proteins to COT1 is mediated by different residues at the COT1 NTR, suggesting a hetero-trimer is formed. Thus, although MOB2A/B may have some overlapping functions in regulating hyphal tip extension, their function is not redundant and they are both required for proper fungal development.
Introduction
Protein kinases, the enzymes responsible for protein phosphorylation, are important mediators of fungal proliferation and development (Dickman and Yarden, 1999). In recent years, protein kinases of the nuclear Dbf2p-related (NDR) Ser/Thr protein kinase family have emerged as being important for cell differentiation and morphogenesis in various organisms (Hergovich et al., 2006b), yet our understanding of their specific functions and mode of regulation is still limited.
The Neurospora crassa COT1 is the founding member of the NDR family (Yarden et al., 1992) which is a subclass of the AGC (PKA/PKG/PKC-like) serine/threonine protein kinases (Hanks and Hunter, 1995). Defects in COT1 confer pleiotropic morphological effects, indicating that COT1 is involved in the regulation of several different cellular processes that affect hyphal morphogenesis (Yarden et al., 1992; Lauter et al., 1998; Gorovits et al., 1999; 2000; Gorovits and Yarden, 2003). In other filamentous fungi, dysfunction of COT1 and its homologues result in growth arrest, hyperbranching as well as defects in conidiation and in cell wall integrity (Chen and Dickman, 2002; Scheffer et al., 2005; Johns et al., 2006; Ziv et al., 2009).
The variety of phenotypic alterations associated with dysfunctional COT1 and its homologues in other organisms (Jorgensen et al., 2002; Nelson et al., 2003; Kanai et al., 2005; Shi et al., 2008) strongly suggest that this kinase has multiple cellular functions and that the functional kinase requires additional components. Indeed, NDR kinases require post-translational modifications (PTM) as well as interactions with other proteins for proper function. Studies in human, fly, and yeast as well as in N. crassa have demonstrated that NDR kinases require phosphorylation on both the activation segment and the hydrophobic motif for their full activation and proper function in vivo (Millward et al., 1999; Tamaskovic et al., 2003b; Emoto et al., 2004; He et al., 2005; Jansen et al., 2006; Ziv et al., 2009). We have previously determined the importance of Ser-417 (activation segment) and Thr-589 (hydrophobic motif) as phosphorylation sites for N. crassa COT1 function in regulating hyphal growth, branching and conidiation by affecting cell wall integrity and actin deposition (Ziv et al., 2009).
Studies in several fungal models (including N. crassa) and in animal cells have resulted in the identification of core components of the NDR signalling network which include the NDR kinase, its binding partner and co-activator MPS1-binding (MOB) 2 and an upstream Ste20-type kinase of the germinal centre (GC) family (Yarden et al., 1992; Nelson et al., 2003; Hergovich et al., 2005; Kanai et al., 2005; Stegert et al., 2005; Seiler et al., 2006; Maerz et al., 2009; 2012; Ziv et al., 2009 and summarized in Hergovich et al., 2006b; Hergovich, 2011; Weiss, 2012). The current regulatory model of NDR kinases suggests that activation of NDR is a multi-step process involving the binding of MOB protein to the N-terminal domain of the kinase (Shi et al., 2008; Maerz et al., 2009). NDR and their MOB proteins adopters co-localize in the cell cytoplasm and plasma membrane (Nelson et al., 2003; Hergovich et al., 2005; Gutierrez-Escribano et al., 2011; Maerz et al., 2012). NDR activation occurs in vivo through rapid recruitment to the plasma membrane by MOB proteins followed by multi-site phosphorylation (Weiss et al., 2002; Hergovich et al., 2005; 2006b; Maerz et al., 2012). An initial auto-phosphorylation event (at a highly conserved activation segment Ser, within the catalytic domain) generates an enzyme with basal catalytic activity. Full activation of the NDR kinase is induced by an additional phosphorylation event within a broadly conserved hydrophobic motif at the C-terminal extension of the protein, performed by a Ste20-type kinase of the GC family (Tamaskovic et al., 2003a; Hergovich et al., 2006b; Ziv et al., 2009; Maerz et al., 2012).
Another phosphorylation site (Thr74 of hNDR1), within the N-terminal region (NTR), has been proposed to be important for NDR regulation (Tamaskovic et al., 2003a,b). This phosphorylation site is less conserved and is not present in some of the NDR kinases. COT1 lacks this residue at the N terminus, but a neighbouring Glu residue (Glu198) was suggested as the candidate to replace this conserved phosphorylation site (Tamaskovic et al., 2003a). By substituting this Glu198 to Ala we determined that this residue is, indeed, involved in determining COT1-dependent fungal morphology yet its role in COT1 activation has not been determined (Ziv et al., 2009). In addition to the Thr74 (or its potential Glu198 substitute), in vitro analysis has shown that conserved arginine residues within the NTR are important for COT1-MOB interactions (Bichsel et al., 2004).
In N. crassa there are 3 putative MOB proteins and an additional MOB-related protein. Two of them: MOB2A (NCU03314) and MOB2B (NCU07460) are structurally very similar and both physically associate with COT1 (Maerz et al., 2009). Interestingly, while each of the single mob-2 deletion mutants grows rather normally, Δmob-2a;Δmob-2b mutants grow very poorly, forming tight hyperbranching colonies with extension-arrested tips, a phenotypic trait highly reminiscent to conditional or deletion mutants of both kinases of the COT1 complex (cot-1 and pod-6) germinating at restrictive temperature (Maerz et al., 2009).
MOB1 (NCU01605) has been shown to function within to the DBF2/LATS1/2 pathway (Dvash et al., 2010) and is apparently specific to that pathway, as no interactions were detected between COT1 and MOB1 or between DBF2 and MOB2A. The fourth protein, MOB3, (NCU07674) is required for vegetative cell–cell fusion and during sexual development and its function is unrelated to NDR signalling (Maerz et al., 2009).
The fact that both MOB2A and MOB2B physically interact with COT1 and based on the phenotypic similarities between Δmob-2a;Δmob-2b and the cot-1(ts) mutant (Maerz et al., 2009), establishes a functional link between these proteins, and may suggest that MOB2A and MOB2B have overlapping/redundant functions in the regulation of hyphal growth. Nevertheless, small differences in the phenotypic characteristics of the single mob-2 deletion mutants as well as different binding strength with COT1 may indicate differential roles for MOB2A and MOB2B in determining COT1-dependent morphology.
To further investigate COT1-MOB interactions, we have mutated several residues within the COT1 NTR and compared the resulting alleles with regard to their effect on COT1 function in vivo and on COT1-MOB2 interactions. By analysing mutations in the COT1 NTR we determined that the NTR is important for proper fungal morphology in different developmental processes and we show that on the bases of different interactions between the two MOB2 proteins and mutated COT1 NTR, MOB2A and MOB2B perform distinct functions in N. crassa and although both function in the regulation of hyphal growth, their role is not redundant.
Results
Mutations at the COT1 N terminal region affect fungal development and colony morphology
Phosphorylation of COT1, at Ser417 and Thr589 has been shown to play a critical role in determining COT1-dependent fungal morphology and development (Ziv et al., 2009). Two COT1 isoforms (67 kDa and 73 kDa) are similarly immuno-detected with anti-COT1 antibodies (Gorovits et al., 1999) as well as with anti-pSer417 antibodies (Maerz et al., 2012) indicating that both COT1 isoforms are Ser417 phosphorylated. Using mass-spec analysis we have identified Ser71 as a new phosphorylation site within the short COT1 isoform NTR (Fig. S1). This residue corresponds to Ser189 in the long isoform of the protein. This part of the NDR protein is conserved but in most fungi Ser189 is substituted with either Glutamine or Alanine.
The location of this newly identified phosphorylation site within the NTR suggests it may have significance in regulation of COT1 dimerization and interactions with the two MOB2 proteins, in light of the importance of this region for these processes (Maerz et al., 2009). In order to further dissect the significance of Ser189 and additional specific residues within the COT1 NTR, we used site directed mutagenesis to create 4 different mutated alleles of cot-1 encoding a single amino acid substitution in the COT1 NTR. The mutated residues include Ser189, as well as two conserved Arg residues (R167 and R203) present at the NTR of NDR kinases that were shown to have functional significance in other systems (Bichsel et al., 2004). Ser189 was substituted either to Ala or to Glu, mimicking non-phosphorylation and constant phosphorylation respectively. Arg167 and Arg203 were substituted with Ala. In addition, as we have previously shown that mutating the conserved Glu198 of COT1 to Ala affects fungal morphology and COT1 activity (Ziv et al., 2009), we incorporated a mutated Glu198 into the comparative analysis.
Plasmid pCZ18, containing the 6myc-cot-1 construct under the control of the cot-1 promoter (Seiler et al., 2006), was subjected to point mutagenesis (Fig. 1A). The resulting 5 constructs, as well as a myc-cot-1 control, were used to replace the endogenous copy of the cot-1 gene at the cot-1 locus (facilitated in a cot-1;Δmus-52 background) and were crossed with cot-1(ts) to obtain homokaryons lacking the Δmus-52 background. The resulting strains were designated cot-1(R167A), cot-1(R203A), cot-1(E198A), cot-1(S189A) and cot-1(S189E) (Table 2). Since the control strain myc-cot-1 that expresses the myc-tagged cot-1 allele is indistinguishable from wild-type (Ziv et al., 2009), this strain was used as a reference strain in all the experiments.
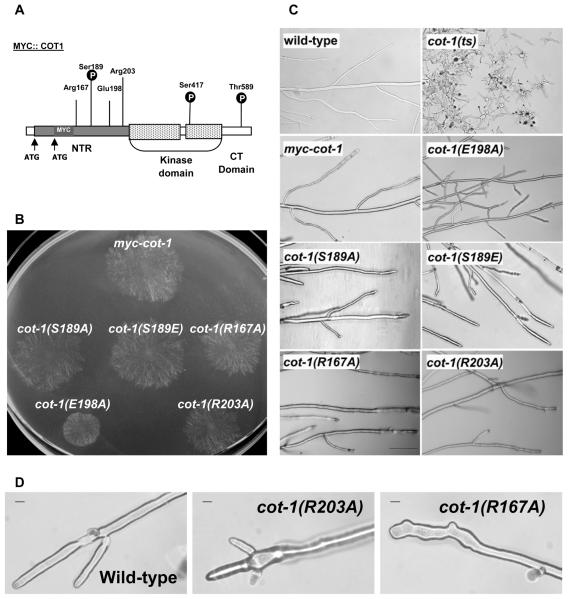
Fig. 1.
Conserved residues within the N-terminal region of COT1 affect fungal morphology.
A. Schematic summary of amino acid alterations introduced into COT1 NTR. The two highly conserved phosphorylation sites at the kinase domain (Ser417) and C Terminal domain (Thr589) are also shown. Grey box represents the NTR and a dotted box represents the kinase domain. The residues’ numbers refer to the COT1 protein (long transcript) without the Myc tag.
B. Growth of myc-cot-1 and the NTR mutants harbouring the different Myc-tagged alleles of cot-1. Cultures were grown overnight at 34°C.
C. Hyphal morphology at the edge of the colony of wild-type, cot-1(ts), myc-cot-1 and the NTR point-mutation strains cultured overnight at 34°C. Bar = 100 μm.
D. Microscopic images of myc-cot-1, cot-1(R167A) and cot-1(R203A) hyphal tips grown at 34°C. Bar represents 10 μm.
Table 2.
Neurospora crassa strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| Wild-type | 74-OR23-1 Mat A | FGSC#987 |
| Wild-type | ORS-SL6 Mat a | FGSC#4200 |
| cot-1(ts); Δ mus-52;his-3 | cot-1(C102t) Δmus-52::barR;his-3 | Ziv et al. (2009) |
| myc-cot-1 | myc-cot-1(183-4) | Ziv et al. (2009) |
| cot-1(E198A) | myc-cot-1(E198A) | Ziv et al. (2009) |
| cot-1(S189A) | myc-cot-1(S189A) | This study |
| cot-1(S189E) | myc-cot-1(S189E) | This study |
| cot-1(R167A) | myc-cot-1(R167A) | This study |
| cot-1(R203E) | myc-cot-1(R203E) | This study |
| Δ mob-2a | hph:: NCU03314.2 Δ | FGSC #11296 |
| Δ mob-2b | hph:: NCU07460.2 Δ | FGSC #13575 |
| Δ mob-2a; myc-cot-1 | hph:: NCU03314.2Δ myc-cot-1 | This study |
| Δ mob-2b; myc-cot-1 | hph:: NCU07460.2Δ myc-cot-1 | This study |
| Δ mob-2a; cot-1(E198A) | hph:: NCU03314.2Δ myc-cot-1(E198A) | This study |
| Δ mob-2b; cot-1(E198A) | hph:: NCU07460.2Δ myc-cot-1(E198A) | This study |
| Δ mob-2a; cot-1(S189A) | hph:: NCU03314.2Δ myc-cot-1(S189A) | This study |
| Δ mob-2b; cot-1(S189A) | hph:: NCU07460.2Δ myc-cot-1(S189A) | This study |
| Δ mob-2a; cot-1(S189E) | hph:: NCU03314.2Δ myc-cot-1(S189E) | This study |
| Δ mob-2b; cot-1(S189E) | hph:: NCU07460.2Δ myc-cot-1(S189E) | This study |
| Δ mob-2a; cot-1(R167A) | hph:: NCU03314.2Δ myc-cot-1(R167A) | This study |
| Δ mob-2b; cot-1(R167A) | hph:: NCU07460.2Δ myc-cot-1(R167A) | This study |
| Δ mob-2a; cot-1(R203A) | hph:: NCU03314.2Δ myc-cot-1(R203A) | This study |
| Δ mob-2b; cot-1(R203A) | hph:: NCU07460.2Δ myc-cot-1(R203A) | This study |
| cot-1 (ts); his-3 | cot-1(C102t); his-3− a | FGSC #5274 |
| myc-cot-1;his-3 | myc-cot-1(183-4) his-3− | Maerz et al. (2009) |
| cot-1(E198A); his-3 | myc-cot-1(E198A) his-3 − | This study |
| cot-1(S189A); his-3 | myc-cot-1(S189A) his-3 − | This study |
| cot-1(S189E); his-3 | myc-cot-1(S189E) his-3 − | This study |
| cot-1(R167A); his-3 | myc-cot-1(R167A) his-3 − | This study |
| cot-1(R203E); his-3 | myc-cot-1(R203E) his-3 − | This study |
|
Δ
mob-2a; HA-mob-2a;
myc-cot-1 |
hph:: NCU03314.2Δ his-3::Pccg-1-HA-mob-2a
myc-cot-1(183-4) |
This study |
|
Δ
mob-2b; HA-mob-2a;
myc-cot-1 |
hph:: NCU07460.2Δ his-3::Pccg-1-HA-mob-2a myc-cot-1(183-4) |
This study |
| myc-cot-1; HA-mob-2a | myc-cot-1(183-4) his-3::Pccg-1-HA-mob-2a | This study |
| cot-1(E198A); HA-mob-2a | myc-cot-1(E198A) his-3::Pccg-1-HA-mob-2a | This study |
| cot-1(S189A); HA-mob-2a | myc-cot-1(S189A) his-3::Pccg-1-HA-mob-2a | This study |
| cot-1(S189E); HA-mob-2a | myc-cot-1(S189E) his-3::Pccg-1-HA-mob-2a | This study |
| cot-1(R167A); HA-mob-2a | myc-cot-1(R167A) his-3::Pccg-1-HA-mob-2a | This study |
| cot-1(R203E); HA-mob-2a | myc-cot-1(R203E) his-3::Pccg-1-HA-mob-2a | This study |
The colony morphology of the strains harbouring the different point mutations at the COT1 NTR is shown in Fig. 1B. In addition, detailed morphological consequences of the strains harbouring the point mutations in the COT1 NTR are summarized in Table 1 and presented in Fig. 2 (black bars). While mutating Ser189 and Arg167 had no obvious effect on hyphal morphology and a minor effect (though significant) on growth rate, mutating Glu198 to Ala resulted in a severe reduction in growth rate. A less pronounced, although still significant, effect on growth rate was observed in cot-1(R203A) (Fig. 1B and Table 1). Microscopic analysis (Fig. 1C and Table 1) further substantiated that the hyphal width and branching pattern were also affected by the different point mutations. The most pronounced effect was observed in the cot-1 (E198A) mutant, which grew as a small dense colony. In addition, the substitution of Arg203 to Ala and of Ser189 to Ala resulted in shorter intervals between branches. Nevertheless, as the shorter length between branches of the different strains is highly correlated (R2 = 0.933) to increased hyphal width, the frequency of branching per hyphal volume unit of the different strains is probably not altered.
Table 1.
Morphological consequences of mutations within the COT1 NTR-encoding region.
| Developmental process | myc-cot-1 | cot-1(R167A) | cot-1(S189A) | cot-1(S189E) | cot-1(E198A) | cot-1(R203A) |
|---|---|---|---|---|---|---|
| Hyphal growth rate (mm h−1) | 4.99 ± 0.05 | 4.81 ± 0.03 | 4.55 ± 0.05 | 4.62 ± 0.04 | 1.75 ± 0.07 | 3.79 ± 0.04 |
| Length between branches (μm) | 169 ± 4 | 166 ± 4 | 147 ± 5 | 180 ±9 | 71 ± 1 | 143 ± 3 |
| Hyphal width (μm) | 11 ± 3.6 | 11.5 ± 4.0 | 11.5 ± 2.9 | 13.0 ± 4.6 | 5.1 ± 2.1 | 9.4 ± 3.0 |
| Total cfu (% of myc-cot-1) | 100% ± 4% | 69% ± 4% | 103% ± 8% | 136% ± 8% | 34% ± 6% | 52% ± 3% |
| Germinating conidia after 4 h of incubation (% of total) |
72% ± 0.5% | 72% ± 0.4% | 37% ± 1.1% | 45% ± 2.4% | 42% ± 4.2% | 71% ± 4.6% |
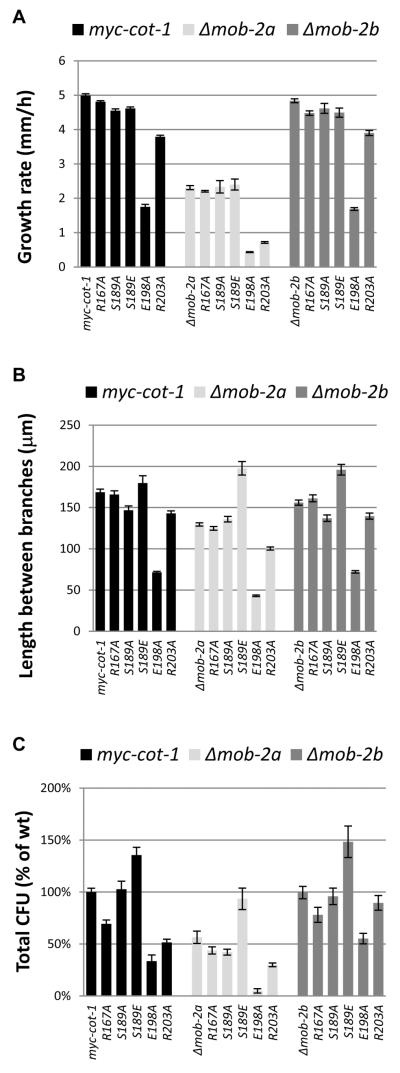
Fig. 2.
Morphological consequences of mutations within the COT1 NTR in a Δmob-2a and Δmob-2b background: Growth rate (A), branching (B) and conidiation (C) of myc-cot-1, Δmob-2a, Δmob-2b and mutants harbouring the different alleles carrying the MYC-tagged point mutations in cot-1 NTR. All experiments were conducted at 34°C, in triplicate. Black bars indicate myc-cot-1 background, white bars indicate a Δmob-2a background and grey bars indicate a Δmob-2b background. Error bars represent standard error of the mean.
Previous results indicate that COT1 is involved in the regulation of asexual conidation in fungi (Scheffer et al., 2005; Ziv et al., 2009), thus we analysed total colony-forming units (cfu) production by the different COT1 NTR mutants and determined that a marked reduction in total cfu production in both cot-1(E198A) and cot-1(R203A), as well as a smaller reduction in total cfu in cot-1(R167A), were evident (Table 1). While mutating Arg167, ArgR203 and Glu198 to Ala resulted in reduced conidiation, mutating Ser189 to Ala did not change the conidial count. However, a 40% increase in conidial production was observed when Ser189 was mutated to Glu when compared with myc-cot-1. Thus, altering all the chosen residues at the COT1 NTR affected conidiation. It is important to note that all the point-mutation strains were similar to the myc-cot-1 with regard to aerial hyphae formation, nevertheless, conidial production was altered in all of the mutants, though to a different extent.
In addition to conidiation, we also examined conidial germination in the various strains. While mutating Ser189 (either Ala or to Glu) had almost no effect on growth rate and hyphal morphology, a clear delay in conidial germination was observed in both cot-1(S189A) and cot-1(S189E) (Table 1). As this delay was not accompanied by any change in the pattern of de novo protein accumulation that is associated with conidial germination (Schmit and Brody, 1976), it is most likely that breaking of conidial dormancy was not affected by these mutations. A similar delay in conidial germination was also observed in cot-1(E198A), the most impaired strain among the COT1 NTR mutants with regard to growth rate and hyphal branching. Regardless of conidial germination rate, all strains exhibited over 98% germinating after 8 h (Fig. S2), indicating that the lower rate of germination during early stages of the process is not due to impaired conidial viability.
Taken together, the variety of phenotypic consequences observed in the different COT1 NTR mutants supports the significance of the NTR in the regulation of COT1 involvement in multiple cellular processes.
Two highly conserved Arg residues at the COT1 NTR affect hyphal tip morphology
Conserved Arg residues at the NTR of NDR kinases were shown to be important for its enzymatic activity and for its binding to MOB protein (Bichsel et al., 2004); however, their importance for NDR kinase function in vivo has not been studied. Examination of both cot-1(R167A) and cot-1(R203A) strains showed that mutations in these conserved Arg residues resulted in altered hyphal tip morphology (Fig. 1D). These aberrant hyphal tips were prevalent and observed in at least 50% of the hyphae. This was not the case when the proximal Glu198 was mutated. The specificity of this defect is further supported by the fact that cot-1(E198A), which exhibited the most severe defect in hyphal growth, produced normal hyphal tips. The two mutants that were affected in hyphal tip morphology differed in other parameters of fungal growth (e.g. substituting Arg167 to Ala had only minor effect on growth rate and branching while cot-1(R203A) grew slowly and the average length between its branches was shorter than that of the wild-type). In addition, we determined that the cot-1(R203A) mutant is temperature sensitive, as it grew faster at lower temperature (4.13 ± 0.004 mm h−1 at 30°C vs. 3.79 ± 0.04 mm h−1 at 34°C). This suggests that both highly conserved Arg residues may be important for COT1 function at the hyphal tip.
Deletion of mob-2a has a synergistic effect on the COT1 NTR mutants
Both MOB2A and MOB2B have been shown to be required for COT1 activation and the COT1 NTR was found to be critical to COT1-MOB2A/B binding (Maerz et al., 2009). To further characterize COT1-MOB2A/B interactions, the COT1-NTR mutants (as well as the myc-cot-1 control strain) were crossed with either Δmob-2a or Δmob-2b to create double mutants.
Deletion of mob-2a resulted in reduced growth rate, shorter distance between branches and reduced conidiation. When Δmob-2a was crossed with either cot-1(R167A) or with cot-1(S189A), the double mutant exhibited similar characteristics to Δmob-2a (Fig. 2, white bars). In marked contrast, deletion of mob-2a had a clear synergistic effect on strains harbouring the R203A and the E198A mutations in COT1. Both Δmob-2a;cot-1(R203A) and Δmob-2a;cot-1(E198A) showed a significant reduction in growth rate, in distance between branches and in conidiation, when compared with the parental strains. Moreover, the morphological outcome of the two double mutants was found to be temperature dependent. At 34°C (the optimal growth temperature of N. crassa) the double mutants’ morphology was drastically altered and a marked increase in aerial hyphae production was observed after 48 h of growth (Fig. S3). However, at 25°C the double mutants grew in a manner similar to their parental strains. This may suggest that the altered stability of the COT1 protein and/or the protein complex may be impaired in these mutants.
When Δmob-2a was crossed with cot-1(S189E), the resulting double mutant grew as slow as Δmob-2a. However, Δmob-2a branching and conidiation defects were suppressed by the S189E mutation (Fig. 2) resulting in longer distance between branches compared with myc-cot-1 along with normal conidial production, suggesting that phosphorylation of Ser189 bypasses, at least in part, the requirement for a functional MOB2A.
MOB2B negatively regulates COT1-dependent conidiation
Deletion of mob-2b had no effect on conidiation and showed only a minor (though statistically significant) reduction in growth rate and in the distance between branches relative to the control. When Δmob-2b was crossed with the different COT1 NTR mutants the growth rate and distance between branches in the double mutant was similar to that measured in the parental strains (Fig 2, grey bars). The Δmob-2b;cot-1(S189E) double mutant was found to be an exception with regard to its branching pattern as this mutant exhibited longer distances between branches compared with its two parents and with myc-cot-1.
When conidiation was examined in the Δmob-2b double mutants it became evident that total cfu abundance was significantly higher in the double mutants (Δmob-2b; cot-1(R167A), Δmob-2b;cot-1(S189E), Δmob-2b;cot-1(R203A) and Δmob-2b;cot-1(E198A)) compared with the less conidiating parental strains. This suggests that MOB2B has a negative regulatory role on COT1 function during macroconidiation, which is dependent on COT1 activity and, most likely, additional proteins.
In addition to the highly abundant macroconida, N. crassa produces smaller, unicellular, microconidia by budding directly of hyphae, mainly during stress conditions. Interestingly, a clear increase in microconidia production was observed in Δmob-2b;cot-1(S189A), and these comprised c. ~25% of its the total cfu produced, vs. 1–5% in all the other strains. This may suggest that in spite of apparently near-normal growth, the strain exhibits a phenotypic characteristic which is indicative of a stress response.
Based on these results we concluded that deletion of mob-2b suppresses the conidiation defect present in the COT1 NTR mutants and thus unmasks MOB2B function as a regulator of COT1-dependent conidiation in these mutants.
COT1 is expressed as three distinct protein isoforms
The cot-1 gene encodes two transcript species of about 2100 nt (short) and 2400 nt (long). The short transcript was found to be light regulated and to be sufficient for complementing the cot-1(ts) phenotype (Lauter et al., 1998). Although two COT1 isoforms are similarly immuno-detected when the protein extract is resolved on a 9% SDS-PAGE (Gorovits et al., 1999), previous attempts to correlate the two mRNA transcripts with the two COT1 protein isoforms were unsuccessful (Maerz et al., 2009).
In order to further differentiate between the two transcripts of cot-1, we generated His::MYC double epitope tag cassettes under the control of the inducible N. crassa qa-2 promoter, expressing either the long or the short transcripts (Maerz et al., 2009). These two constructs (along with the epitope tags) were able to complement cot-1(ts), indicating that both constructs encode for a functional COT1 protein. Two-step affinity purification (Fig. S4) with the two His::MYC::COT1 isoforms resulted in the co-purification of MOB2A and MOB2B (NCU03314 and NCU07460 respectively) by both COT1 long and COT1 short isoforms.
To gain additional insight concerning COT1 isoforms in vivo, we analysed a strain, in which a 6xMYC tag was inserted, in frame, at the second ATG of the endogenous cot-1 locus, which allowed the simultaneous detection of both COT1 isoforms (designated myc-cot-1). The growth characteristics of this strain were indistinguishable from wild-type (Ziv et al., 2009). Western blot analysis with anti-MYC antibodies verified the presence of two MYC-tagged COT1 isoforms, as expected. However, when the total protein extract of this strain was resolved on a 4–12% gradient SDS-PAGE gel under denaturing conditions, we detected at least three distinct MYC-tagged isoforms with estimated MWs of c. 80 kDa, 75 kDa and 70 kDa (Fig. 3, upper panel). IP with anti-MYC antibodies, followed by SDS-PAGE analysis, confirmed the presence of the same three isoforms (Fig. 3, lower panel).
Fig. 3.
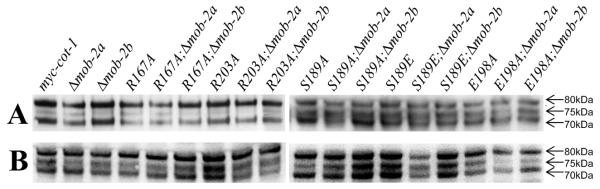
COT1 is expressed as three different isoforms: (A) Western-blot analysis of total cell extracts probed with anti-Myc antibodies. (B) Western blot analysis of IP with anti-myc detected with anti-myc antibodies. Arrows on the right indicate the three COT1 isoforms, with estimated size.
In order to verify the nature of these proteins, each of the three bands was subjected to mass spectrometric analysis, and all three were identified as COT1. The peptides identified corresponding to the 80 kDa band covered the N-terminus and the C-terminus of the expected protein encoded by the long cot-1 transcript. No peptide coverage of the long COT1 NTR was obtained in the 70 kDa and 75 kDa band preparations, yet peptides corresponding to both termini of the expected product of the short cot-1 transcript were evident in the spectra obtained. This may suggest that the two lower MW isoforms are translational products of the short cot-1 transcript, though other PTM cannot be ruled out.
Mutating the different residues of the NTR of COT1 did not affect the abundance/levels of the three COT1 isoforms (Fig. 3). Furthermore, even though deleting both mob-2a and mob-2b has been shown to affect the stability of the lower MW COT1 isoform (Maerz et al., 2009), deleting each of the mob genes had no significant effect on the three COT1 isoforms.
Auto-phosphorylation of Ser417 within the catalytic domain of COT1 is required for COT1 kinase activity and function (Ziv et al., 2009) but not for COT1-MOB2 binding (Maerz et al., 2012). All three COT1 isoforms were found to be phosphorylated at Ser417 (Fig. S5A) and this phosphorylation was not altered in any of the COT1 NTR mutants. In agreement with this, we have determined that, based on changes in COT1 mobility following phosphatase treatment, all three COT1 isoforms are indeed phosphorylated (Fig. S5B). However, as three different COT1 isoforms can still be detected after phosphatase treatment, apparently protein phosphorylation cannot provide a full explanation for mobility differences observed between the COT1 isoforms.
The three COT1 isoforms physically interact with MOB2A
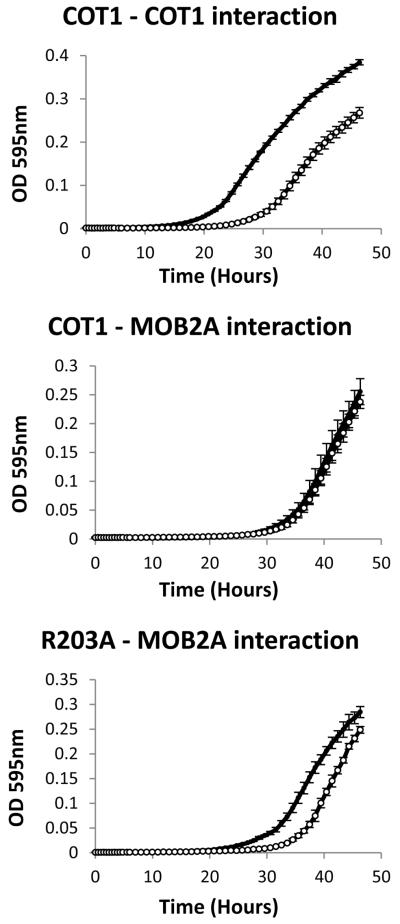
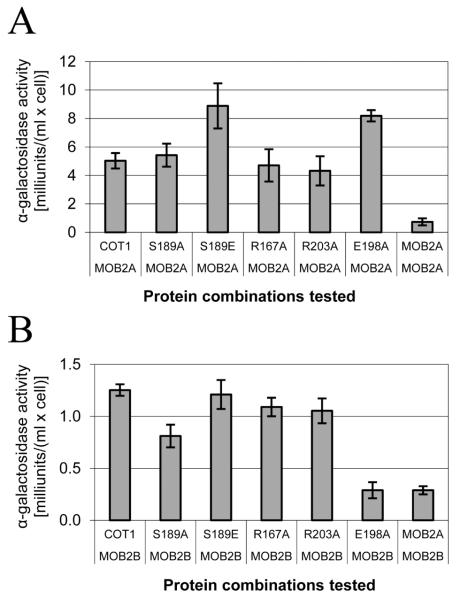
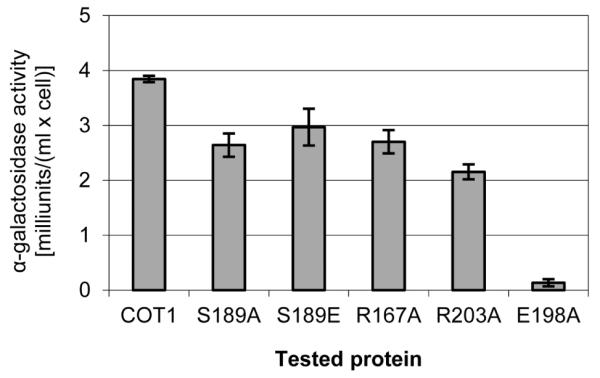
In order to determine whether mutations at the COT1 NTR affect the physical interactions between COT1 and MOB2A we produced strains carrying the COT1 NTR mutations (as well as the myc-cot-1 control strain) that express an HA-tagged version of MOB2A at the his-3 locus (see Experimental procedures). These strains were subjected to co-IP experiments in which the tagged proteins were precipitated with either anti-MYC antibodies (Fig. S6A) or with anti-HA antibodies (Fig. S6B). Western blot analysis showed that all three MYC tagged COT1 isoforms were precipitated by HA-MOB2A, indicating that all three COT1 isoforms physically interact with this protein. Within the framework of the described co-IP experiment we could not detect any differences between the different COT1 NTR mutants with regard to their binding/interactions with MOB2A. In order to increase the sensitivity of our analysis we incorporated an additional approach based on the yeast two hybrid system (Y2H). To this end, we performed site directed mutagenesis to introduce the 5 different mutations into pGBKT7, carrying the long cot-1 transcript. The five different cassettes were co-transformed into yeast along with pGADT7 which contained a mob-2a cDNA insert. All transformants were able to grow on a high selection medium (SD/-Leu/-Trp/-His/-Ade) and produced blue colonies when grown on solid SD medium supplemented with x-alpha-gal. These results clearly indicated that all cot-1 alleles expressing COT1 with the different NTR point mutations can physically interact with MOB2A, as was previously observed in the co-IP experiment. Next, we tested the different strains to determine the strength of the interaction between the different alleles of COT1 and MOB2A, by monitoring the growth of the transformed yeasts in SD-Leu-Trp compared with growth in high selection medium. All yeast strains, excluding the strain carrying the R203A mutation, grew similarly on both media, indicating a very strong interaction between the different COT1 alleles and MOB2A. The strain carrying the R203A allele of cot-1 with mob-2a showed a very minor delay in growth when grown on high selection media compared with the SD-Leu-Trp, indicating that mutating Arg203 to Ala may slightly interfere with COT1-MOB2A interactions (Fig. 4). Our previous work indicated that COT1 can dimerize to create a stable COT1-COT1 dimer (Maerz et al., 2009). The physical interactions between the two COT1 homodimer components is much weaker when compared with COT1-MOB2A interactions, as can be seen by comparing the growth curves of the relevant yeast strains (Fig. 4). While some reduction in COT1(R203A)-MOB2A interactions was evident, this interaction is still much stronger than that measured in the COT1-COT1 dimer. Results of additional experiments, in which alpha-galactosidase secretion of the different yeast strains was measured, supported the presence of a strong interaction between COT1 and MOB2A and between the proteins corresponding to different alleles of cot-1 and MOB2A (Fig. 5A). In fact, these results indicate that S189E and E198A mutations of COT1 result in an even stronger interaction with MOB2A when compared with the wild-type COT1. Even though the mentioned mutations at the NTR did not affect co-IP patterns, results obtained with the yeast two-hybrid system strongly suggest that these residues are involved in determining the nature of COT1-MOB2A interactions.
Fig. 4.
Mutating COT1 Arg203 to Ala impairs COT1-MOB2A interactions: Growth curves of yeast harbouring the indicated two-hybrid plasmids in SD-Leu/-Trp liquid medium (black) or in SD high selection liquid medium -Leu/-Trp/-His/-Ade (white). Similar growth of the yeast strains in the two different media indicates strong physical interaction between the two proteins.
Fig. 5.
Protein-protein interactions between the different alleles of COT1 NTR and the MOB2A/MOB2B proteins as determined by yeast two hybrid experiments. Different yeast transformants carrying MOB2A (A) or MOB2B (B) as a pray and the different COT1 alleles as a bait were grown overnight in SD medium lacking Leu and Trp. The supernatant of the yeast culture was assayed for α-galactosidase activity. High activity of α-galactosidase indicates strong interaction between the tested proteins. The results are an average of three different experiments that were performed in triplicate. Bar indicates standard error.
Mutations in the NTR of COT1 affect its physical interaction with MOB2B
Using Y2H experiments we identified a very weak interaction between COT1 and MOB2B that was significantly different from the strong interactions between COT1 and MOB2A. The physical interaction between COT1 and MOB2B is so weak that the resulting yeast transformants were unable to grow on high selection medium and grew rather slowly on medium selection medium (SD-Leu-Trp-His), compared with the SD-Leu-Trp medium. Furthermore, secretion of alpha-galactosidase was very low (Fig 5B) in these yeast strains. Nevertheless, our results clearly demonstrate that mutating Ser189 to Ala resulted in a small, though significant, reduction in the COT1-MOB2B interaction, while the E198A mutation completely abolished the physical interaction between COT1 and MOB2B (Fig 5B). These results were confirmed by analysis of the yeast strains’ growth curves (data not shown) and by the silver stain patterns of the SDS-PAGE of proteins immunoprecipitated with anti-MYC antibodies (Fig. 6), indicating that MOB2B could not be immunopercipitated with COT1(E198A). Nevertheless, the physical interaction between the two proteins (MOB2B and COT1(E198A)) was partially resumed in a Δmob-2a-background (Fig. 6, right panel). Thus, Glu198 of the COT1 NTR is apparently required for COT1-MOB2B binding but not for COT1-MOB2A binding. These results indicate that different residues in COT1 NTR are involved in MOB2A vs. MOB2B binding and that binding of MOB2A may affect MOB2B binding (though they probably do not compete for binding sites). Interestingly, while Y2H experiments indicated that there is no physical interaction between MOB2A and MOB2B (Fig. 5B), MOB2B was immunoprecipitated with HA:MOB2A (as well as COT1) (Fig. S7), suggesting that COT1 can physically associate both MOB2 proteins simultaneously, to form a heterotrimer. These results are in agreement with our findings that different residues at the COT1 NTR are responsible for MOB2 binding.
Fig. 6.
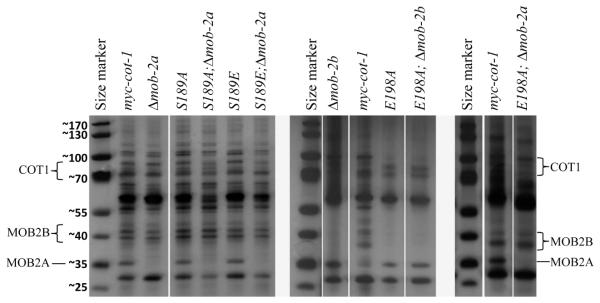
Substitution of Glu198 with Ala abolishes MYC-COT1-MOB2B physical association. Silver stain detection of MYC-immunoprecipitated proteins of myc-cot-1, Δmob-2a, Δmob-2b, cot-1(E198A) and cot-1(E198A);Δmob-2a, resolved in a polyacrylamide gel.
Glu198 at the COT1 NTR affects COT1 dimerization and MOB2B binding
Previously, we have shown that the COT1 NTR is required for COT1 dimerization and it was suggested that COT1-COT1 dimers represents part of the inactive pool of COT1 (Maerz et al., 2009; 2012). Using the yeast Y2H system we have quantified the relative extent of dimerization of each of the COT1 NTR alleles and determined that all the point mutations at the COT1 NTR resulted in reduced COT1 dimerization (Fig. 7). However, this, apparently, had no effect on COT1 protein stability as we could not correlate this reduction with reduced protein level as determined by Western blot analyses in the different mutants (Fig. 3A). While most of the mutations resulted in a minor (though significant) reduction in dimerization, mutating Glu198 to Ala completely abolished dimer formation. Nevertheless, in subsequent reciprocal co-immunoprecipitation experiments utilizing a mutant expressing both GFP::COT1 and MYC::COT1(E198A) fusion proteins, a physical interaction between the two proteins was detected, suggesting that in the presence of additional cellular components, COT1(E198A) does not completely abolish COT1 dimerization. The cot-1(E198A) was the most phenotypically-impaired strain among the COT1 NTR mutants tested here, and previous in vitro analysis indicted that this mutation results in c. 60% reduction in kinase activity of COT1 (Ziv et al., 2009). We have shown here that mutating Glu198 to Ala impaired the physical interaction between COT1 and MOB2B in vivo, but results in a very strong interaction between COT1 and MOB2A that competes with and impairs MOB2B binding. Furthermore, E198A may also have an effect on COT1 dimerization, though our data suggest that this probably does not change the COT1 inactive pool. Taken together, it seems that Glu198 is a key residue within the COT1 NTR, that participates in determining COT1-protein interactions and subsequently its kinase activity.
Fig. 7.
Mutations at the COT1 NTR affect COT1 dimerization as determined by Yeast Two Hybrid assay. Yeast transformants carrying the each of COT1 alleles, both as bait and as pray, were grown overnight in SD medium lacking Leu and Trp. The supernatant of the yeast culture was assayed for α-galactosidase activity. High α-galactosidase activity indicates strong interaction between the tested proteins. The results are an average of three independent experiments that were performed in triplicate. Bar indicates standard error.
Discussion
The regulation of COT1 kinase activity and the subsequent consequences on relevant cellular activities is dependent, among others, on PTM and interaction with COT1 binding partners and co-activators such as MOB2A/B. The MOB proteins bind to the conserved N-terminus of NDR kinases and are involved in the localization and activation of these kinases.
Phosphorylation has been shown to play an important role in the regulation of COT1 activity (Ziv et al., 2009). While phosphorylation at the kinase domain and the hydrophobic motif is highly conserved among NDR kinases (Hergovich et al., 2006a), phosphorylation at the NTR is less conserved. Some fungal NDR kinases lack the conserved human NDR Thr74 site (corresponding to Gly199 in the long COT1 isoform), while in others, like S. cerevisiae, this residue is present but mutating it to Ala conferred no morphological consequences (Jansen et al., 2006). Thus, it seems that other conserved residues at the NTR of fungal NDR kinases may be important for their activation and proper function. One example is in N. crassa, where Glu198, which is highly conserved among NDR kinases, is important for COT1 function and activity (Ziv et al., 2009). This residue was suggested to replace the mentioned human Thr74 phosphorylation site (Tamaskovic et al., 2003a) though this site is constantly negatively charged and thus cannot function as a regulatory switch.
Phosphorylation of Ser417 and Thr589 is involved in COT1-based regulation of hyphal elongation and branching as well as asexual development by altering cell wall integrity and actin organization (Ziv et al., 2009) yet does not affect MOB binding (Bichsel et al., 2004; Maerz et al., 2012). On the other hand, altering Glu198 (within the NTR) to Ala resulted in a strain defective in hyphal growth and branching, exhibited reduced conidiation, and also altered the binding of the two MOB proteins (especially MOB2B) as well as COT1 dimerization. Interestingly, Bichsel et al. (2004) reported that although both Thr74 (phosphorylation site of hNDR1 at the NTR) and Glu73 (corresponding to COT1 Glu198) are highly important for NDR activity in vitro, only Thr74 affects the MOB binding property (Bichsel et al., 2004). Our results clearly indicate that mutating Glu198 to Ala indeed reduces COT1 kinase activity in vitro (Ziv et al., 2009). However, this mutation of COT1 actually does affect MOB binding, as it abolishes MOB2B binding to COT1. Nevertheless, since deletion of mob-2b results in very mild morphological defects, the morphological defects observed in cot-1(E198A) cannot be explained by just a change in COT1-MOB2B binding. Thus it seems that Glu198 has an additional, important, role for the function of COT1 kinase.
Using MS/MS analysis we have now identified a novel phosphorylation site, Ser189, within the COT1 NTR and determined that it has a role in COT1-dependent conidiation and conidial germination. The kinase responsible for Ser189 phosphorylation has yet to be identified. Chan et al. (2005) have identified three phosphorylated Ser residues at the NTR of human LATS1 and at least one of them has been shown to be autophosphorylated (Chan et al., 2005). However, this is probably not the case of COT1 Ser189, as Ser417 is the only apparently autophosphorylated site in COT1 (Maerz et al., 2012).
Ser189 is conserved in closely related COT1 homologues (e.g. in Sordaria macrospore, Thielavia terrestris, Myceliophthora thermophila, Podospora anserine, Chaetomium thermophilum and C. globosum), yet is absent from most fungal homologues of COT1, including the functional homologues in Colletotrichum trifolii (Buhr et al., 1996) and Claviceps purpurea, where COT1 was shown to be required for proper hyphal growth (Scheffer et al., 2005). This may indicate that Ser189 phosphorylation is not required for normal, COT1-dependent, elongation/branching. In fact, to date, most of the emphasis concerning the phenotypic consequences of altered cot-1 has focused on hyphal growth and even though asexual conidiation in fungi has also been shown to be affected by alterations in cot-1 (Scheffer et al., 2005; Ziv et al., 2009), this aspect of cot-1 function has yet to be further elucidated.
The fact that mutating Ser189 had almost no effect on colony morphology and hyphal growth/branching, yet conidiation was elevated in cot-1(S189E), indicates that phosphorylation of Ser189 may be involved in determining COT1 function in macroconidial development. Furthermore, cot-1(S189E) suppressed the Δmob-2a conidiation defect and the double mutant strain conidiated in a manner similar to the wild-type. These effects were not due to a change in hyphal growth rate or aerial hyphae formation. The fact that the Δmob-2a conidiation defect is not present in the mob-2b deletion strain can be explained by the possibility that MOB2A, but not MOB2B, is required for proper conidiation. However, a more likely explanation could be that MOB2B can be involved in negative regulation of COT1-dependent conidiation (see below). Our Y2H results indicated a very strong physical interaction between COT1(S189E) and MOB2A compared with COT1(WT) and COT1(S189A). Thus, it is tempting to speculate that in vivo, perhaps at the conidiophore, phosphorylation of Ser189 promotes MOB2A binding to COT1, thus enhancing conidiation. However, such a model cannot explain the suppression of the Δmob-2a conidiation defect by cot1-(S189E). Alternatively, it could be that this mutation enables the binding of a different MOB protein when MOB2A is missing. Although MOB2B binding is at least partially dependent on Ser189 phophorylation (as S189A reduced MOB2B binding, as determined in the Y2H experiment), MOB2B probably cannot replace MOB2A as a COT1 activator, as our data suggest that MOB2B is, in fact, involved in negative regulation of conidiation under certain circumstances. Taken together, our results show that S189 plays a role in conidiation which may be mediated by MOB2A. In addition to the effect on conidiation, S189E suppresses the branching defect of both Δmob-2a and Δmob-2b and the double mutants produce longer distances between branches compared with myc-cot-1.
Although altering Ser189 to Ala or Glu had only a minor effect on hyphal growth, a significant reduction (60%) in conidial germination rate was observed in both mutants. Thus, timely phosphorylation of Ser189 is important for conidial germination while it is dispensable for hyphal elongation. This reflects differences in the molecular mechanisms governing polarity during conidial germination vs. hyphal extension and indicates their function as distinct processes (Ziv et al., 2008) and is supported by the differential involvement of regulatory components such as Cdc42 and Rho GTPases in controlling polarity during germination vs. hyphal elongation (Harris, 2006).
Two highly conserved Arg residues (Arg167 and Arg203) were identified at the COT1 NTR and determined to affect COT1 function in vivo. Although previous work with human NDR kinases indicated the importance of these residues to NDR kinase activity in vitro and to MOB binding (Bichsel et al., 2004; Kohler et al., 2010), this is to our knowledge, the first time by which the morphological outcome of mutating these Arg residues to Ala is characterized. Interestingly, mutating these two Arg residues resulted in different effects. While the R167A substitution had only mild observed consequences, R203A exhibited a clear hyphal growth defect. Nevertheless, both Arg point-mutated cot-1 strains exhibited abnormal hyphal tip morphology, typified by hyphal swelling and dichotomous branching. These defects were absent in cot-1(E198A) that showed a more severe hyphal growth defect, indicating that the abnormal hyphal tips were not a consequence of other growth defects and suggests that the Arg residues are important for proper COT1 function at the hyphal tip. COT1 recruitment to the membrane has been shown to be dependent on MOB2A/B binding to the COT1 NTR (Maerz et al., 2012). Furthermore, C. albicans Cbk1 requires Mob2 binding for localization to regions of polarized growth (Gutierrez-Escribano et al., 2011). Thus, the morphological outcome of mutating the two Arg residues may result from improper COT1 binding to MOB2A/B. However, our data indicate that both Arg residues have little or no effect on the physical COT1-MOB2 associations. While crossing R167A with Δmob-2a resulted in a strain exhibiting a similar morphology to that of the Δmob-2a parent, a Δmob-2a;cot-1(R203A) strain produced a colony with tighter, restricted, growth and increased formation of aerial hyphae with delayed carotenoid biosynthesis. This synergistic effect between R203A and Δmob-2a further indicates that R203 has a role in COT1 regulation which is not exclusively mediated by MOB2A binding and perhaps affects interaction with other proteins that are required for COT1-dependent polar growth. The importance of Arg203 to COT1 function is further indicated by the fact that mutation of this residue, but not the others studied here, is temperature sensitive.
Differences in MOB-NDR binding are probably a result of differences attributed to MOB protein structure and not to the NDR kinase itself, as the highly conserved NDR residues affect the binding of each of the MOB proteins to different extents. The results presented here indicate that COT1 Ser189 affects both binding to MOB2A and to MOB2B. Arg203 (R78 of hNDR1) may be involved, to some degree, in MOB2A (but not MOB2B) binding while Arg167 (R41 of hNDR1) had no effect on the binding of neither MOB2A nor MOB2B. On the other hand, Glu198 is clearly involved in MOB2 binding as mutating this residue to Ala resulted in tighter binding to MOB2A and abolished the binding to MOB2B. Moreover, although COT1 binds both MOB2A and MOB2B, the binding of the two MOB proteins is different in such that MOB2A binds to COT1 much stronger than MOB2B. These results were evident in both the co-IP as well as in the Y2H experiments, suggesting that they represent a true difference and not a result of differences in the proteins’ sensitivity to salt and/or detergents. Differential binding of MOB proteins to NDR kinases was also reported in human cells. While NDR1 and NDR2 bind both hMob2 and hMob1, the kinases’ binding mode to the two MOB proteins differ significantly and the kinases appear to bind hMOB2 more tightly than hMOB1 in cell extracts (Bichsel et al., 2004; Devroe et al., 2004; Kohler et al., 2010).
The co-purification of both MOB2 proteins with COT1 and the fact that a mob-2a;mob-2b mutant highly resembles cot-1(ts), previously led us to suspect overlapping functions of the two MOBs (Maerz et al., 2009). However, the results presented here clearly indicate a different role for MOB2A and MOB2B with regard to hyphal morphology and development. Thus, even though MOB2A/B probably have some overlapping functions during hyphal growth, MOB2A is required for normal hyphal tip extension and branching while MOB2B is less important for that process. The observed consequences of COT1 and the MOB2A/B dysfunction, with regard to hyphal tip growth are also reflected in spatial transcript analysis data, showing that cot-1 and mob-2a are expressed at higher levels in the peripheral region of the colony while mob-2b mRNA is not detected in the youngest section, but was constant throughout the remaining colony (Kasuga et al., 2005), and support functions of COT1 and MOB2A during tip growth, while MOB2B may primarily function in subapical regions, for example during conidiation.
MOB2A and MOB2B affect distinct COT1-dependent roles in regulation of macroconidiation, as deletion of mob-2a results in a conidiation defect not present in the mob-2b deletion strain. MOB2B is involved in regulation of conidiation, probably in a COT1-dependent manner. This conclusion is drawn from our observation that mutating Arg167, Arg203 and E198 to Ala resulted in reduced macroconidiation, which was suppressed (at least, partially) by deletion of mob-2b. Nonetheless, the fact that deletion of mob-2b in wild-type background did not elevate conidiation points to the involvement of additional factors in regulating conidiation. Overexpression of mob-2b may provide additional insight as to the role of MOB2B in regulating cfu production. Kohler et al. (2010) were first to describe an endogenous inhibitory function of a human MOB protein. Interestingly, hMOB2, in contrast to hMOB1A/B, is bound to unphosphorylated NDR and directly competes with hMOB1 for binding to NDR1/2 thereby negatively regulates centrosome duplication and apoptosis. On the other hand, hMOB2 cooperates positively with NDR2 in the formation of neuritis. Therefore, hMOB1 and hMOB2 appear to have opposing functions, where hMOB2 functions as an inhibitor in one process and as a co-activator in another process. (Kohler et al., 2010; Hergovich, 2011). Our results indicate that an inhibitory effect of the MOB protein on NDR function may also exist in N. crassa. The fact that the impaired binding of COT1(E198A) to MOB2B is resumed in a Δmob-2a background indicates that competition between the two MOB2 proteins on COT1 binding is also a hallmark of NDR kinases. These results, in combination with our genetic data, suggest a negative regulatory role for MOB2B and may indicate that competition between MOB2A and MOB2B on a binding site at the COT1 NTR, or via other proteins, may be a key regulator of COT1 function.
We have demonstrated that both MOB2A and MOB2B physically interact with the three COT1 isoforms. Our mass spectrometry data suggest that the 70 kDa and 75 kDa COT1 isoforms are the product of the short cot-1 transcript. However, as PTM can affect protein size and mobility on PAGE, we cannot rule out that these isoforms are the result of PTM of the long transcript translation product. We have determined that COT1 is phosphorylated, yet this PTM cannot provide a sole explanation for the size differences observed. Furthermore, our mass-spectrometry analysis (as well as bioinformatic predictions) determined that the low MW isoform is also N-acetylated (data not shown). While the long isoform is not N-terminally acetylated, we cannot determine if this PTM is isoform-specific or not. As many as 50–80% of eukaryotic proteins undergo amino-terminal N-acetylated and its functional implications includes regulation of protein–protein interactions and protein sorting and localization (Arnesen, 2011; Forte et al., 2011). This may indicate that the two isoforms of COT1 are localized differently and may differently interact with other proteins.
A search for MOB2 homologues in filamentous fungi indicates the presence of two MOB2 proteins in Trichoderma and Chaetomium species as well as in C. purpurea, S. macrospora, P. anserine and other closely related species, exhibiting a wide range of morphological characteristics and growth rates. However, a wide range among the ascomyctes analysed appears to have only a single MOB2 protein. A phylogenetic analysis of COT1 homologues in the different fungi (Fig. S8) indicates a correlation between the degree of COT1 conservation and the number of MOB2 proteins present, suggesting a link between COT1 structure and its mode of regulation. The fact that N. crassa expresses two MOB2 proteins that physically interact with COT1 while other filamentous fungi (like A. nidulans) expresses a single MOB protein that forms a complex with the COT1 homologue (Shi et al., 2008) may suggest more complex regulation of COT1 in N. crassa. It will be interesting to determine if in A. nidulans (and other fungi) the functions attributed to the different MOB proteins present in N. crassa are performed by a single protein or alternative NDR kinase complex components.
Experimental procedures
Strain, media and growth conditions
General procedures and media used in the handling of N. crassa have been previously described (Davis, 2000) or are available through the Fungal Genetics Stock Center (http://www.fgsc.net/Neurospora/NeurosporaProtocolGuide.htm). N. crassa strains used in this study are listed in Table 2. Strains were grown in either liquid or solid (supplemented with 1.5% agar) Vogel’s minimal medium with 1.5% (w/v) sucrose. When required, the medium was supplemented with 10 μg ml−1 hygromycin B (Calbiochem, Riverside, CA).
cot-1 alleles with point mutations at the N terminal region were constructed using the QuikChange site-directed mutagenesis kit (Stratagen, La Jolla, CA) according to the manufacturer’s protocol. The mutation-harbouring oligonucleotides used are listed in Table 3. In all cases their complement (not specified) was also used in the mutagenesis process. Mutagenesis was performed on pCZ18 [a cot-1 genomic fragment which includes the 6Myc tag at the NspV site (Seiler et al., 2006)] to create the plasmids that express the different alleles of cot-1. The resulting plasmids were linearized with XhoI and co-transformed into the cot-1;Δmus-52;his-3 strain along with the HindIII/BamHI genomic fragment that can complement his-3 auxotrophy. Transformants were screened for their ability to grow in medium lacking L-His, at 34°C. For each construct, two strains that were found to include the myc-tagged cot-1 point-mutation allele (verified by PCR) were further crossed with N. crassa myc-cot-1 (FGCS#4200), to obtain the COT1 phosphorylation mutations in a myc-cot-1 background. Progeny from these crosses were screened for their ability to grow without L-His and for their sensitivity to Basta (to eliminate the Δmus-52::barR+ background). The presence of the myc-tagged allele (but not the cot-1(ts) allele) in the resulting progeny of the crosses was verified by PCR. The presence of the expected point mutations within the different constructs were verified by direct sequencing of PCR products amplified using transformant DNA as template. Western-blot analysis using monoclonal c-MYC antibodies (Santa Cruz, CA) verified the presence of MYC-tagged COT1 with the expected molecular mass.
Table 3.
Sequence of oligonucleotides used for introducing point mutations.
| Name | Sequence |
|---|---|
| Ser189Ala | GCCGTCGCGAAgCGATCTGGTCAACCG |
| Ser189Glu | CCCGCCGTCGCGAAgaGATCTGGTCAACCG |
| Arg167Ala | GCGCGCCAGGGAGgcCAACCAGAGGTG |
| Arg203Ala | GGAGGGCCAGTATTTGgcCTTCCTGAGAACCAAG |
Producing COT1 NTR mutants that express HA-MOB2A
To produce strains carrying the COT1 NTR mutations that express an HA tag version of MOB2A at the his-3 locus of N. crassa, we crossed myc-cot-1 as well as the other mutants carrying the NTR point mutations with the his3-strain. The histidine auxotrophs carrying the myc-tagged NTR mutation of COT1 were transformed with an HA-MOB2A expressing cassettes (Maerz et al., 2009). The resulting transformants were screened for growth without L-His in the growth medium. The presence of the HA-mob-2a cassette and the myc-tagged cot-1 in all the strains was verified by PCR. Furthermore, the NTR of all the strains was sequenced to verify the presence of all the desired point mutations. The expression of the tagged MOB2A and COT1 in the transformants was verified by Western blot analysis, using anti-HA and anti-MYC and antibodies respectively.
Determining growth rate, length between branches, conidiation rate and conidial germination
For growth-rate measurements, race tubes containing Vogel’s sucrose minimal medium were inoculated with 10 μl of a conidial suspension (2 × 106 conidia ml−1). The race tubes were incubated for several days at 34°C and growth was measured twice daily.
To determine the length between branches, conidia of the different N. crassa strains were inoculated on glass slides covered with Vogel’s sucrose minimal medium and incubated overnight at 25°C in a high humidity chamber and then shifted to 34°C for 6 h. The edges of the growing colonies were observed microscopically and the entire colony edge was documented in c. 20 photographs. Each of the photographs was analysed using ImageJ 1.37V (Rasband, W.S., U.S. National Institutes of Health, Bethesda, MD, http://rsb.info.nih.gov/ij/, 1997–2006). The length between each two successive branches of leading hyphae was determined and at least 400 measurements were performed for each colony. The entire data set of each strain was used to calculate the average length between branches. The presented data (and calculated standard error) is based on the averages of at least three independent experiments performed with each strain.
To determine conidiation rate, strains were grown in a 250 ml Erlenmeyer flasks containing 50 ml solid Vogel’s sucrose minimal medium. The flasks were incubated for several days at 34°C until conidia were produced and matured. Conidia were collected by adding 50 ml of distilled water and vigorously shaking the flasks. The conidial suspensions were filtered through cheesecloth and subsequently centrifuged for 5 min at 3000 g and the supernatant completely removed. The conidia were then re-suspended in 50 ml of distilled water and the number of total cfu was determined by use of a hemocytometer, while distinguishing between macro- and arthro-conidia. To determine the number of microconidia, the suspensions were filtered through a 5 μM filter and were counted separately. The results presented are the average of at least three independent experiments. In each experiment duplicate counts were performed.
To determine conidial germination rate of the different strains, pre-warmed Vogel sucrose minimal medium was inoculated with a conidial suspension (final concentration of 107 conidia ml−1) and incubated (34°C) at 150 r.p.m. for several hours. Cultures were sampled every 1–2 h for 8 h and examined by light microscopy.
Protein extraction and immunoblotting
Mycelial samples were frozen in liquid nitrogen, pulverized, and suspended in lysis buffer [1 M sorbitol, 10 mM HEPES (pH 7.5), 5 mM EDTA, 5 mM EGTA, 5 mM NaF, 0.1 M KCl, 0.2% Triton X-100, complete protease inhibitor mixture (Roche Applied Science, Mannheim, Germany)]. The samples were homogenized by 10 strokes of pestle A in a Dounce homogenizer. The homogenates were centrifuged for 40 min at 10 000 g, and the supernatants were recovered and stored at −70°C until analysis. Proteins were separated by NuPAGE® Bis-Tris 4–12% SDS-PAGE (Invitrogen, Carlsbad, CA). Western blotting was performed by standard procedures (Sambrook et al., 1989). Antibodies used throughout this study included anti-cMYC (Santa Cruz, CA) and anti-HA (Sigma-Aldrich) or anti-Phosph-Ser CSK [the S. cereviciae homologue of COT1 (Jansen et al., 2006)]. The secondary antibodies used were either goat anti-rabbit IgG-HRP (Santa Cruz, CA) for anti-Phosph-Ser CSK or goat anti-mouse IgG-HRP (Santa Cruz, CA) for anti-cMYC and anti-HA.
Immunoprecipitation of the tagged proteins by either anti-cMYC or anti-HA antibodies was performed according to (Maerz et al., 2009). For protein detection in polyacrylamide gels the Pierce Silver Stain Kit (Rockford, IL) was used, according to the manufacturer’s protocol.
De-Phosphorylation of N. crassa proteins was performed by incubating the total protein extracts with lambda protein phosphatase (Lambda PP – New England Biolabs) for 1 h at 30°C, followed by a Western blot.
Mass spectrometric analysis
Products of protein immunoprecipitation (IP) using anti-myc antibodies were resolved by SDS-PAGE, and specific bands were excised from gels and analysed by mass spectrometry. The tryptic peptides resulting from in-gel digestion were separated by a homemade analytical capillary column (50 μm × 10 cm) packed with 5 μm spherical C18 reverse phase material (YMC, Kyoyo, Japan). An Agilent 1100 binary pump was used to generate HPLC gradient as follows: 0–5% B in 5 min, 5–40% B in 25 min, 40–100% B in 15 min (A = 0.1 M acetic acid in water, B = 0.1 M acetic acid/70% methanol). The eluted peptides were sprayed directly into a QSTAR XL mass spectrometer (MDS SCIEX, Toronto, Canada) equipped with a nano-ESI ion source. The mass spectrometer was operated in data dependent mode with one MS scan followed by three MS/MS scans for each cycle. Database searches were performed on an in-house Mascot server (Matrix Science, London, UK). Methionine oxidation, phosporylation of serine, threonine and tyrosine were set as variable modifications. All identified phosphorylated peptides were manually checked to exclude false-positives.
Yeast two hybrid assay
The Matchmaker Two-Hybrid system 3 (Clontech, USA) was used according to the manufacturer’s instructions. The construction of pGADT7 vector (containing the GAL4 activation domain) or the pGBKT7 (containing the DNA-binding domain) expressing either COT1 (cDNA of the long transcript), MOB2A or MOB2B was described in Maerz et al. (2009). The pGADT7-COT1 and pGBKT7-COT1 cassettes were mutagenized to introduce the NTR point mutations using the primers described in Table 3 or in Ziv et al. (2009), using the QuikChange site-directed mutagenesis kit (Stratagen, La Jolla, CA) according to the manufacturer’s protocol. The fusion proteins were (co)expressed in S. cerevisiae AH109 and potential interactions determined by activation of lacZ or his3 and ade2 reporter constructs that discriminate positive interactions on the basis of colour in the presence of X-α-galactopyranoside or by selection for viability and monitoring growth on SD medium lacking adenine and histidine. To quantify the strength of the physical interaction between the tested proteins the α-Gal quantitative assay was performed, according to the manufacturer’s instructions. This is a sensitive colorimetric method for the detection and quantification of yeast α-galactosidase activity resulting from expression of the MEL1reporter gene in the GAL4-based Matchmaker two-hybrid systems. The quantitative nature of the α-Gal Assay makes it possible to compare the degree of MEL1 expression in different Matchmaker two-hybrid host cell populations containing different pairs of interacting proteins, or to measure differences in the relative strength of binding between mutant forms of interacting proteins. At least 3 independent transformants were analysed for each interaction that was tested.
Microscopy
Light microscopy was performed with a Zeiss Axioscope microscope equipped with a Nikon DXM1200F digital camera.
Statistical analysis
Statistical analysis was performed by Student’s t-test, using JMP statistical package (version 7). A two-tailed P-value less than 0.05 was considered significant.
Bioinformatics
Neurospora crassa COT1 protein sequence was used to identify similar sequences from NCBI databases. Phylogenetic and molecular evolutionary analyses of the fungal COT1 proteins were conducted using MEGA version 5 (Tamura et al., 2011). Three NCBI BlastP searches were performed for each fungal species against N. crassa MOB2A, MOB2B and MOB1. Only fungi with hits for the MOB proteins are shown in the COT1 Phylogenetic tree.
Supplementary Material
Acknowledgements
We thank E. L. Weiss, Northwestern University, Illinois, USA, for providing the anti-Phosph-Ser CBK1 antibodies. This research project was supported by DFG research grant SE1054/3-2, by The Israel Science Foundation and by the German-Israel Fund.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Arnesen T. Towards a functional understanding of protein N-terminal acetylation. PLoS Biol. 2011;9:e1001074. doi: 10.1371/journal.pbio.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichsel SJ, Tamaskovic R, Stegert MR, Hemmings BA. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J Biol Chem. 2004;279:35228–35235. doi: 10.1074/jbc.M404542200. [DOI] [PubMed] [Google Scholar]
- Buhr TL, Oved S, Truesdell GM, Huang C, Yarden O, Dickman MB. A kinase-encoding gene from Colletotrichum trifolii complements a colonial growth mutant of Neurospora crassa. Mol Gen Genet. 1996;251:565–572. doi: 10.1007/BF02173646. [DOI] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillie HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- Chen C, Dickman MB. Colletotrichum trifolii TB3 kinase, a COT1 homolog, is light inducible and becomes localized in the nucleus during hyphal elongation. Eukaryot Cell. 2002;1:626–633. doi: 10.1128/EC.1.4.626-633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RH. Neurospora: Contributions of a Model Organism. Oxford University Press; Oxford: 2000. [Google Scholar]
- Devroe E, Erdjument-Bromage H, Tempst P, Silver PA. Human MOB proteins regulate the NDR1 and NDR2 serine-threonine kinases. J Biol Chem. 2004;279:24444–24451. doi: 10.1074/jbc.M401999200. [DOI] [PubMed] [Google Scholar]
- Dickman MB, Yarden O. Serine/threonine protein kinases and phosphatases in filamentious fungi. Fungal Genet Biol. 1999;26:99–117. doi: 10.1006/fgbi.1999.1118. [DOI] [PubMed] [Google Scholar]
- Dvash E, Kra-Oz G, Ziv C, Carmeli S, Yarden O. The NDR kinase DBF-2 is involved in regulation of mitosis, conidial development and glycogen metabolism in Neurospora crassa. Eukaryot Cell. 2010;9:502–513. doi: 10.1128/EC.00230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Forte GMA, Pool MR, Stirling CJ. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 2011;9:e1001073. doi: 10.1371/journal.pbio.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovits R, Yarden O. Environmental suppression of Neurospora crassa cot-1 hyperbranching: a link between COT1 kinase and stress-sensing. Eukaryot Cell. 2003;2:699–707. doi: 10.1128/EC.2.4.699-707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovits R, Propheta O, Kolot M, Dombradi V, Yarden O. A mutation within the catalytic domain of COT1 kinase confers changes in the presence of two COT1 isoforms and in Ser/Thr protein kinase and phosphatase activities in Neurospora crassa. Fungal Genet Biol. 1999;27:264–274. doi: 10.1006/fgbi.1999.1152. [DOI] [PubMed] [Google Scholar]
- Gorovits R, Sjollema KA, Sietsma JH, Yarden O. Cellular distribution of COT1 kinase in Neurospora crassa. Fungal Genet Biol. 2000;30:63–70. doi: 10.1006/fgbi.2000.1198. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Escribano P, Gonzalez-Novo A, Suarez MB, Li CR, Wang Y, de Aldana CRV, Correa-Bordes J. CDK-dependent phosphorylation of Mob2 is essential for hyphal development in Candida albicans. Mol Biol Cell. 2011;22:2458–2469. doi: 10.1091/mbc.E11-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Harris SD. Cell polarity in filamentous fungi: shaping the mold. Int Rev Cytol. 2006;251:41–77. doi: 10.1016/S0074-7696(06)51002-2. [DOI] [PubMed] [Google Scholar]
- He Y, Fang XL, Emoto K, Jan YN, Adler PN. The tricornered Ser/Thr protein kinase is regulated by phosphorylation and interacts with furry during Drosophila wing hair development. Mol Biol Cell. 2005;16:689–700. doi: 10.1091/mbc.E04-09-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A. MOB control: reviewing a conserved family of kinase regulators. Cell Signal. 2011;23:1433–1440. doi: 10.1016/j.cellsig.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Bichsel SJ, Hemmings BA. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol Cell Biol. 2005;25:8259–8272. doi: 10.1128/MCB.25.18.8259-8272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Schmitz D, Hemmings BA. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem Biophys Res Commun. 2006a;345:50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006b;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Jansen JM, Barry MF, Yoo CK, Weiss EL. Phosphoregulation of Cbk1 is critical for RAM network control of transcription and morphogenesis. J Cell Biol. 2006;175:755–766. doi: 10.1083/jcb.200604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns SA, Leeder AC, Safaie M, Turner G. Depletion of Aspergillus nidulans cotA causes a severe polarity defect which is not suppressed by the nuclear migration mutation nudA2. Mol Genet Genomics. 2006;275:593–604. doi: 10.1007/s00438-006-0113-0. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Nelson B, Robinson MD, Chen YQ, Andrews B, Tyers M, Boone C. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics. 2002;162:1091–1099. doi: 10.1093/genetics/162.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, Kume K, Miyahara K, Sakai K, Nakamura K, Leonhard K, et al. Fission yeast MO25 protein is localized at SPB and septum and is essential for cell morphogenesis. EMBO J. 2005;24:3012–3025. doi: 10.1038/sj.emboj.7600782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga T, Townsend JP, Tian C, Gilbert LB, Mannhaupt G, Taylor JW, Glass NL. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res. 2005;33:6469–6485. doi: 10.1093/nar/gki953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler RS, Schmitz D, Cornils H, Hemmings BA, Hergovich A. Differential NDR/LATS interactions with the human MOB family reveal a negative role for human MOB2 in the regulation of human NDR kinases. Mol Cell Biol. 2010;30:4507–4520. doi: 10.1128/MCB.00150-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter FR, Marchfelder U, Russo VE, Yamashiro CT, Yatzkan E, Yarden O. Photoregulation of cot-1, a kinase-encoding gene involved in hyphal growth in Neurospora crassa. Fungal Genet Biol. 1998;23:300–310. doi: 10.1006/fgbi.1998.1038. [DOI] [PubMed] [Google Scholar]
- Maerz S, Dettmann A, Ziv C, Liu Y, Valerius O, Yarden O, Seiler S. Two NDR kinase-MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa. Mol Microbiol. 2009;74:707–723. doi: 10.1111/j.1365-2958.2009.06896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerz S, Dettmann A, Seiler S. Hydrophobic motif phosphorylation coordinates activity and polar localization of the Neurospora crassa nuclear Dbf2-related kinase COT1. Mol Cell Biol. 2012;32:2083–2098. doi: 10.1128/MCB.06263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward TA, Hess D, Hemmings BA. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J Biol Chem. 1999;274:33847–33850. doi: 10.1074/jbc.274.48.33847. [DOI] [PubMed] [Google Scholar]
- Nelson B, Kurischko C, Horecka J, Mody M, Nair P, Pratt L, et al. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell. 2003;14:3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. CSH Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Scheffer J, Ziv C, Yarden O, Tudzynski P. The COT1 homologue CPCOT1 regulates polar growth and branching and is essential for pathogenicity in Claviceps purpurea. Fungal Genet Biol. 2005;42:107–118. doi: 10.1016/j.fgb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Schmit JC, Brody S. Biochemical genetics of Neurospora crassa conidial germination. Bacteriol Rev. 1976;40:1–41. doi: 10.1128/br.40.1.1-41.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S, Vogt N, Ziv C, Gorovits R, Yarden O. The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol Biol Cell. 2006;17:4080–4092. doi: 10.1091/mbc.E06-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Chen W, Liu Q, Chen S, Hu H, Turner G, Lu L. Depletion of the MobB and CotA complex in Aspergillus nidulans causes defects in polarity maintenance that can be suppressed by the environment stress. Fungal Genet Biol. 2008;45:1570–1581. doi: 10.1016/j.fgb.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Stegert MR, Hergovich A, Tamaskovic R, Bichsel SJ, Hemmings BA. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol Cell Biol. 2005;25:11019–11029. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaskovic R, Bichsel SJ, Hemmings BA. NDR family of AGC kinases – essential regulators of the cell cycle and morphogenesis. FEBS Lett. 2003a;546:73–80. doi: 10.1016/s0014-5793(03)00474-5. [DOI] [PubMed] [Google Scholar]
- Tamaskovic R, Bichsel SJ, Rogniaux H, Stegert MR, Hemmings BA. Mechanism of Ca2+-mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J Biol Chem. 2003b;278:6710–6718. doi: 10.1074/jbc.M210590200. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EL. Mitotic exit and separation of mother and daughter cells. Genetics. 2012;192:1165–1202. doi: 10.1534/genetics.112.145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, Luca FC. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J Cell Biol. 2002;158:885–900. doi: 10.1083/jcb.200203094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden O, Plamann M, Ebbole DJ, Yanofsky C. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 1992;11:2159–2166. doi: 10.1002/j.1460-2075.1992.tb05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv C, Gorovits R, Yarden O. Carbon source affects PKA-dependent polarity of Neurospora crassa in a CRE-1-dependent and independent manner. Fungal Genet Biol. 2008;45:103–116. doi: 10.1016/j.fgb.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Ziv C, Kra-Oz G, Gorovits R, Marz S, Seiler S, Yarden O. Cell elongation and branching are regulated by differential phosphorylation states of the nuclear Dbf2-related kinase COT1 in Neurospora crassa. Mol Microbiol. 2009;74:974–989. doi: 10.1111/j.1365-2958.2009.06911.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.