Abstract
1-Deoxy-d-xylulose 5-phosphate (DXP) synthase is the first enzyme in the methylerythritol phosphate pathway to essential isoprenoids in pathogenic bacteria and apicomplexan parasites. In bacterial pathogens, DXP lies at a metabolic branchpoint, serving also as a precursor in the biosynthesis of vitamins B1 and B6 which are critical for central metabolism. Toward identifying novel bisubstrate analog inhibitors that exploit the large active site and distinct mechanism of DXP synthase, a library of aryl mixed oximes was prepared and evaluated. Trihydroxybenzaldoximes emerged as reversible, low micromolar inhibitors, competitive against d-glyceraldehyde 3-phosphate (d-GAP) and either uncompetitive or noncompetitive against pyruvate. Hydroxybenzaldoximes are the first class of d-GAP-competitive DXP synthase inhibitors offering new tools for mechanistic studies of DXP synthase and a new direction for the development of antimicrobial agents targeting isoprenoid biosynthesis.
Keywords: MEP pathway, DXP synthase, thiamin diphosphate, isoprenoid biosynthesis
Introduction
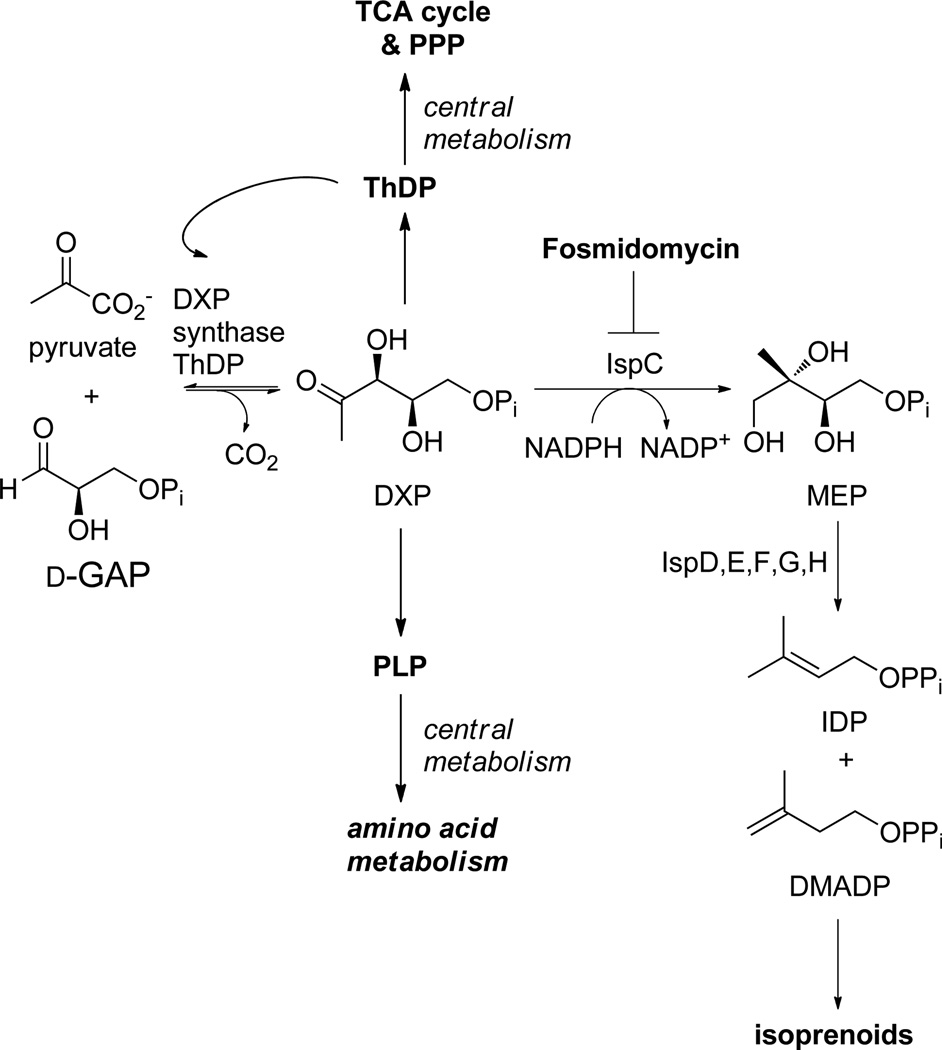
Antimicrobial-resistant infections caused over 2 million illnesses and 23,000 deaths in the United States in 2013.1 As the incidence of drug-resistant infections continues to grow, there is an increasing need for new anti-infective strategies. Combination therapy2 and targeting virulence factors,3 have shown promise for the treatment of infection. However, there is still a need for novel antimicrobial targets and new drugs to exploit them. A potential antimicrobial drug target is DXP synthase, which catalyzes the thiamin diphosphate (ThDP)-dependent condensation of pyruvate and d-GAP to form DXP. DXP lies at a metabolic branchpoint in bacterial metabolism, acting as an intermediate in the biosynthetic pathway to the essential cofactors ThDP4,5 and pyridoxal phosphate (PLP),6 and in the methylerythritol phosphate (MEP) pathway to indispensable isoprenoid precursors isopentenyl diphosphate (IDP) and dimethylallyl diphosphate (DMADP).7,8 The methylerythritol phosphate (MEP) pathway (Scheme 1) is widespread in bacterial pathogens and apicomplexan parasites and is distinct from the mevalonate pathway to isoprenoids in mammals.9,10 Selectively targeting the enzymatic steps in the MEP pathway should reduce the chance of toxicity in anti-infective therapy, as exemplified by the excellent toxicology profile of the IspC inhibitor, fosmidomycin.11 DXP synthase appears crucial to controlling flux in the MEP pathway,12 as it is subject to feedback inhibition by IDP13 and provides substrate for IspC-catalyzed synthesis of MEP, a possible feed-forward regulator of IspF.14
Scheme 1.
DXP lies at a metabolic branchpoint in bacterial metabolism, feeding into the methylerythritol phosphate (MEP) pathway to essential isoprenoids, and acting as a precursor in the biosynthetic pathways to ThDP and PLP, vitamins critical in central metabolism.
The role of DXP synthase in the biosynthesis of essential cellular cofactors, ThDP and PLP, underscores its importance in bacterial central metabolism. ThDP is required for transketolase-catalyzed production of sedoheptulose 7-phosphate and d-GAP in the pentose phosphate pathway, for production of acetyl-CoA catalyzed by the pyruvate dehydrogenase complex, and for the production of succinyl-CoA catalyzed by the α-ketoglutarate dehydrogenase complex in the citric acid cycle. Interestingly, ThDP is also required for DXP synthase catalysis itself. PLP is required for numerous enzymatic conversions in amino acid metabolism. The combined effects of inhibiting the production of essential isoprenoids and lowering cellular ThDP and PLP levels through inhibition of DXP synthase should have serious negative consequences for bacterial pathogens; however, DXP synthase remains under-developed as an antimicrobial drug target.
Selective targeting of DXP synthase is a perceived challenge as the enzyme uses a ThDP-dependent mechanism to catalyze a reaction reminiscent of mammalian ThDP-dependent pyruvate decarboxylases and carboligases (i.e., pyruvate dehydrogenase and tranksetolase), and few reports describe selective inhibitors of this enzyme.15–20 However, DXP synthase is mechanistically unique from other ThDP-dependent enzymes as it does not follow the canonical ping-pong mechanism.21,22 Instead, DXP synthase requires a ternary complex for catalysis in which d-GAP induces decarboxylation of an otherwise uniquely stable C2α-lactylthiamin diphosphate (LThDP) intermediate.21,23,24 Accordingly, the DXP synthase active-site volume is large compared to mammalian PDH or TK, and DXP synthase displays exceptionally relaxed specificity for sterically demanding aromatic acceptor substrates.15 Together, these observations suggest that DXP synthase can be selectively targeted.
Taking advantage of the distinct characteristics of DXP synthase, we have demonstrated selective inhibition of DXP synthase by sterically demanding acetylphosphonates.15,16 These inhibitors provide a basis for the development of unnatural bisubstrate inhibitors for DXP synthase, combining the indiscriminate inhibitory activity of the acetylphosphonate moiety against ThDP-dependent enzymes,25 and the lipophilic chains from known unnatural aldehyde acceptor substrates for DXP synthase26 to provide low micromolar inhibitors with reasonable selectivity.
Here we sought to extend the idea of unnatural bisubstrate inhibition of DXP synthase by covalently tethering mimics of donor and unnatural acceptor substrates through an oxime-based linker. Interestingly, our studies instead identified trihydroxybenzaldoximes exhibiting a novel mode of inhibition which is discrete from that expected of a bisubstrate analog inhibitor. Trihydroxybenzaldoximes are competitive with respect to the acceptor substrate d-GAP, but uncompetitive with respect to the donor substrate, pyruvate. Biochemical characterization of inhibitors and focused structure–activity relationship (SAR) toward identifying key structural components for inhibition are presented here. Taken together, our results suggest this new inhibitor class will offer useful mechanistic probes for this distinct ThDP-dependent enzyme, and a promising starting point for the development of novel DXP synthase inhibitors as antimicrobial agents.
Results
Oxime library design and synthesis
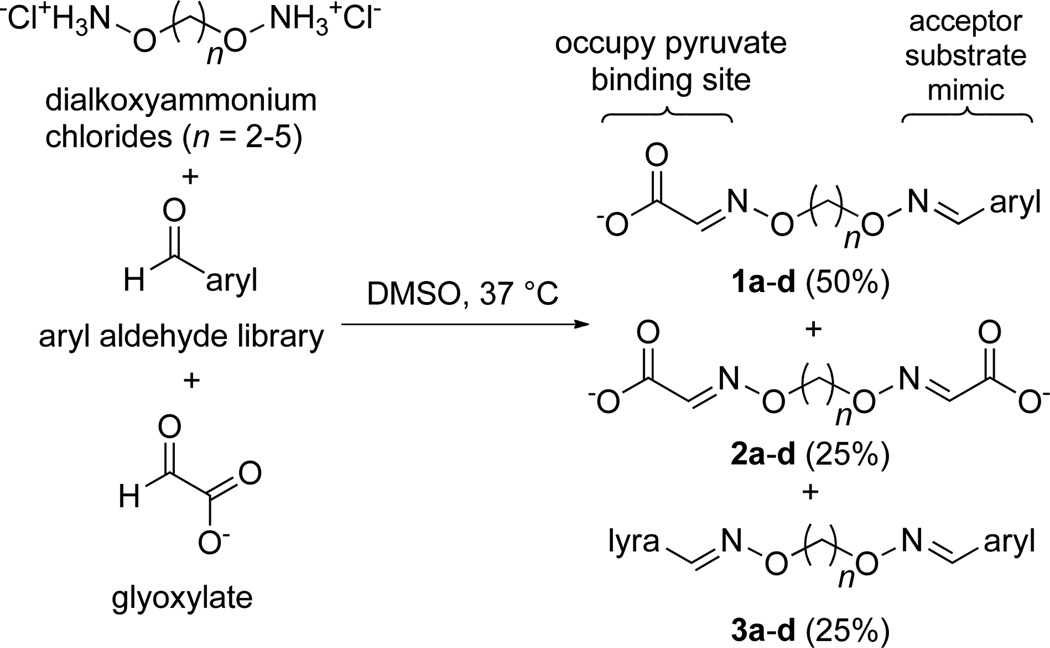
Toward identifying a bisubstrate analog inhibitor to exploit the large active site volume and relaxed specificity of DXP synthase, we prepared a library of oxime-based analogs bearing glyoxylate, designed to occupy the pyruvate-binding site, and a variety of aryl moieties to mimic unnatural aromatic acceptor substrates for DXP synthase (Scheme 2).15 The approach builds on a previous substrate-specificity study demonstrating nitroso naphthols as competent alternative substrates with comparable affinities to the natural acceptor substrate d-GAP.15 Because the structural determinants for binding of aromatic moieties to DXP synthase are largely unknown, an agnostic library approach was pursued to survey a large number of steric and electronic features around the benzene ring. Additionally, multiple oxime linker lengths were examined simultaneously to provide information about both close and distant binding partners and to permit multiple conformations.
Scheme 2.
Design of aryl oxime-based library. Mixed oximes (1) with n = 2–5 were designed as unnatural bisubstrate analogs. Oximes were generated as statistical mixtures of mixed oximes 1a–d (50%), and symmetrical oximes 2 a–d (25%) and 3 a–d (25%)
The oxime library was generated according to the procedure of Stivers and coworkers.27 Briefly, in a 96-well plate format, an equimolar mixture of glyoxylate and aryl aldehyde were combined with a pool of dialkoxyammonium hydrochloride linkers of chain lengths –(CH2)n– where n = 2–5. This method produces a statistical mixture of the desired mixed dioxime (1a–b, Scheme 2), the symmetrical diglyoxylate oxime (2a–d), and the symmetrical diaryl oxime (3a–d) in a 2:1:1 ratio for each chain length, yielding 12 compounds per well.
Identification and characterization of oxime inhibitors
Wells containing the oxime mixtures described above were tested for inhibitory activity against DXP synthase at a total oxime concentration of 100 µm, using a continuous spectrophotometric enzyme-coupled assay in which DXP synthase activity is coupled to IspC (Scheme 1), and the consumption of NADPH is monitored at 340 nm.15,21 Prior to the screening of the library at large, the diglyoxylate symmetrical dioximes 2a–d, present in all wells, were prepared individually by reacting 2 molar equivalents of glyoxylate with 1 molar equivalent of each dialkoxyammonium linker; these were tested for inhibition against DXP synthase and confirmed to be inactive up to 1 mm (data not shown). Oxime mixtures displaying > 50% inhibition at a total oxime concentration of 100 µm were evaluated further. Two hits, derived from 2,4,5-trihydroxybenaldehyde and 3,4,5-trihydroxybenzaldehyde, emerged from the screen; these showed concentration-dependent inhibition of DXP synthase (Figure S1) and are inactive against the coupling enzyme, IspC (data not shown). These mixtures exhibited IC50 values of 16.3 and 40.5 µm (total oxime concentration) for the 2,4,5- and 3,4,5-trihydroxy scaffolds, respectively.
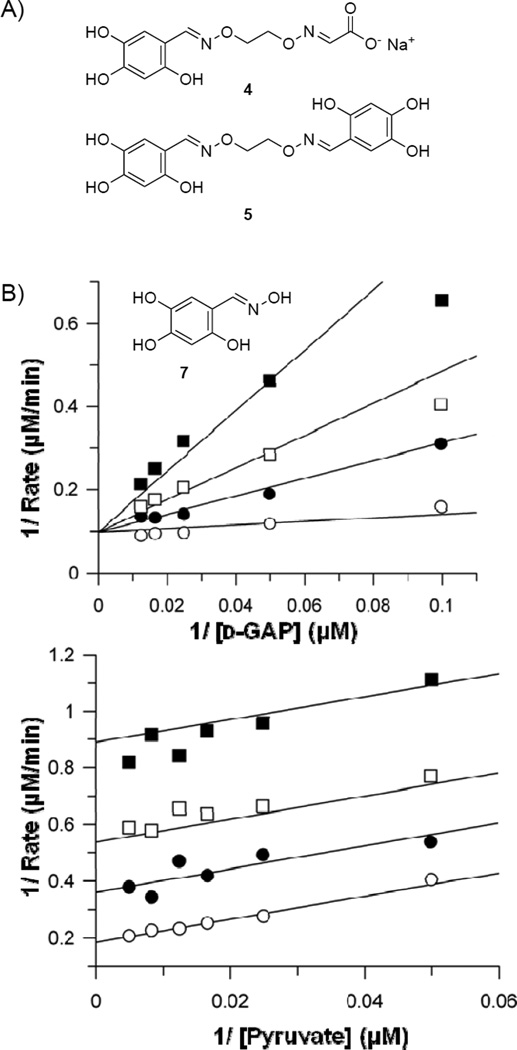
Given the more potent inhibition by the oxime mixture derived from 2,4,5-trihydroxybenzaldehyde, this scaffold was pursued further to identify active components. To determine the optimal linker length of oximes derived from 2,4,5-trihydroxybenzaldehyde scaffold, the oxime mixtures were resynthesized as described above with a single dialkoxyammonium hydrochloride linker per well, to generate the 2:1:1 statistical mixture. Evaluation of each mixture for inhibitory activity against DXP synthase revealed the most potent inhibition by oximes bearing a 2- or 3-carbon linker (n = 2 or 3, Figure S2); thus mixed oxime 4 and symmetrical oxime 5 (Figure 1A) were prepared to determine the contribution of each to the observed inhibitory activity. Mixed oxime 4 was synthesized by slow addition of sodium glyoxylate to dialkoxyamine (n = 2) and sodium acetate, followed by addition of 2,4,5-trihydroxybenzaldehyde. Trihydroxy symmetrical oxime 5 was prepared by reaction of the dialkoxyammonium linker (n = 2) with 2 equivalents of trihydroxybenzaldehyde. Inhibition analysis revealed a Ki of 18.4 ± 3.4 µm for compound 4, a competitive mode of inhibition with respect to d-GAP and noncompetitive inhibition with respect to pyruvate (Figure S3). The lack of a competitive inhibition mode against pyruvate suggests the glyoxylate moiety does not serve as a pyruvate mimic, and mixed oxime 4 does not act as a bisubstrate analog. Interestingly, compound 5 exhibits more potent competitive inhibition with respect to d-GAP (Ki of 1.0 ± 0.4 µm). In contrast to 4, oxime 5 is an uncompetitive inhibitor with respect to pyruvate (Figure S4), suggesting a distinctive inhibition mode targeting the unique LThDP complex.
Figure 1.
A) 2,4,5-Trihydroxybenzaldoxime inhibitors from library; B) Oxime 7 displays competitive inhibition against d-GAP and uncompetitive inhibition against pyruvate. Representative double reciprocal analyses are shown.
A single oxime moiety is required for potent inhibition of DXP synthase
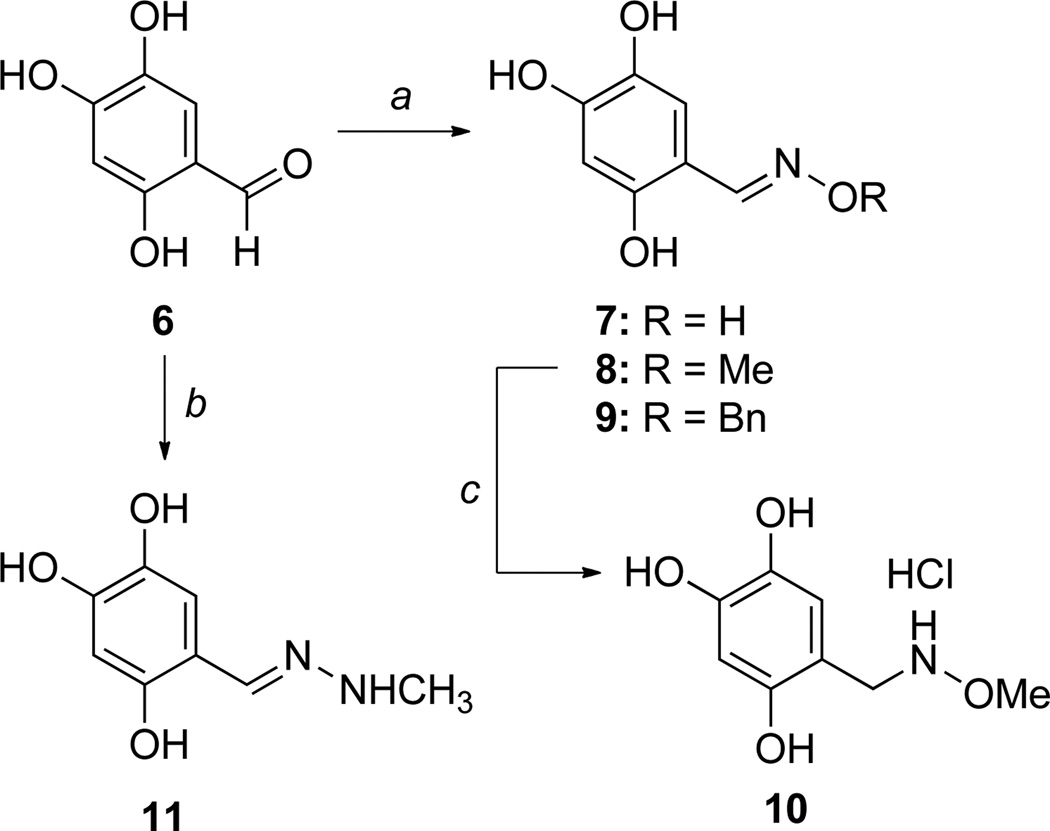
The increased potency of symmetrical oxime 5 compared to mixed oxime 4, coupled with the uncompetitive inhibition mode against pyruvate, suggested the trihydroxy scaffold may be required for inhibition but raised questions about the importance of a dioxime linking a second pharmacophore. Thus, a series of analogs bearing a single oxime moiety of varying size (7–9, Scheme 3) was synthesized in good yield by reaction of aldehyde 6 with the desired alkoxyammonium hydrochloride in the presence of sodium acetate. Inhibition analysis indicates compounds 7–9 exhibit similar low micromolar potency to symmetrical oxime 5, and maintain a competitive inhibition mode against d-GAP and uncompetitive (7, Figure 1B) or noncompetitive (8, 9) inhibition against pyruvate (Table 1, Figures S5–S7). However, aldehyde 6 is inactive up to 75 µm. Taken together, the data suggest that a single oxime moiety is sufficient for activity, and substitution at the oxime oxygen does not dramatically impact inhibitory potency, although modifications at this position appear to influence inhibition mode against pyruvate.
Scheme 3.
Synthesis of trihydroxy analogs. Reaction conditions: a) alkoxyammonium chloride, NaOAc, MeOH; b) methyl hydrazine, EtOH, reflux 2 h; c) i) NaBH3CN, EtOH 0 °C, ii) 10% HCl in EtOH, 0 °C to rt over 1.5 hours.
Table 1.
Inhibitory activity of trihydroxy analogs modified at the oxime.
| Compound | Ki |
|---|---|
| 4 | 18.4 ± 3.4 µm |
| 5 | 1.0 ± 0.2 µm |
| 6 | > 75 µm a |
| 7 | 2.6 ± 0.6 µm |
| 8 | 3.9 ± 0.6 µm |
| 9 | 9.1 ± 1.8 µm |
| 10 | > 75 µm a |
| 11 | > 75 µm a |
| Butyl Acetyl Phosphonate | 5.6 ± 0.8 µm16 |
| Ketoclomazone | 75 µm18 |
Compounds were evaluated in the presence of ascorbic acid (2.0 mm).
To further probe the oxime features required for potent inhibition, methoxyamine 10 and N-methyl hydrazone 11 were prepared (Scheme 3). Reduction of oxime 8 with sodium cyanoborohydride affords methoxyamine analog 10.28 N-Methyl hydrazone 11 was prepared by reaction of aldehyde 6 with N-methyl hydrazine.29 Both 10 and 11 lack inhibitory activity up to 75 µm. Coupled with a lack of activity observed for the aldehyde 6, these results highlight the importance of the distal oxygen atom. In particular, the inactivity of 10 suggests the distal oxygen preferentially binds in the plane of the phenyl ring, and the additional rotational freedom in 10 may be entropically costly; further, it is possible that the increased electron density in the phenyl ring of 10 compared to 8 may also factor into the loss of activity.
Hydroxyl substituents enhance inhibitor affinity
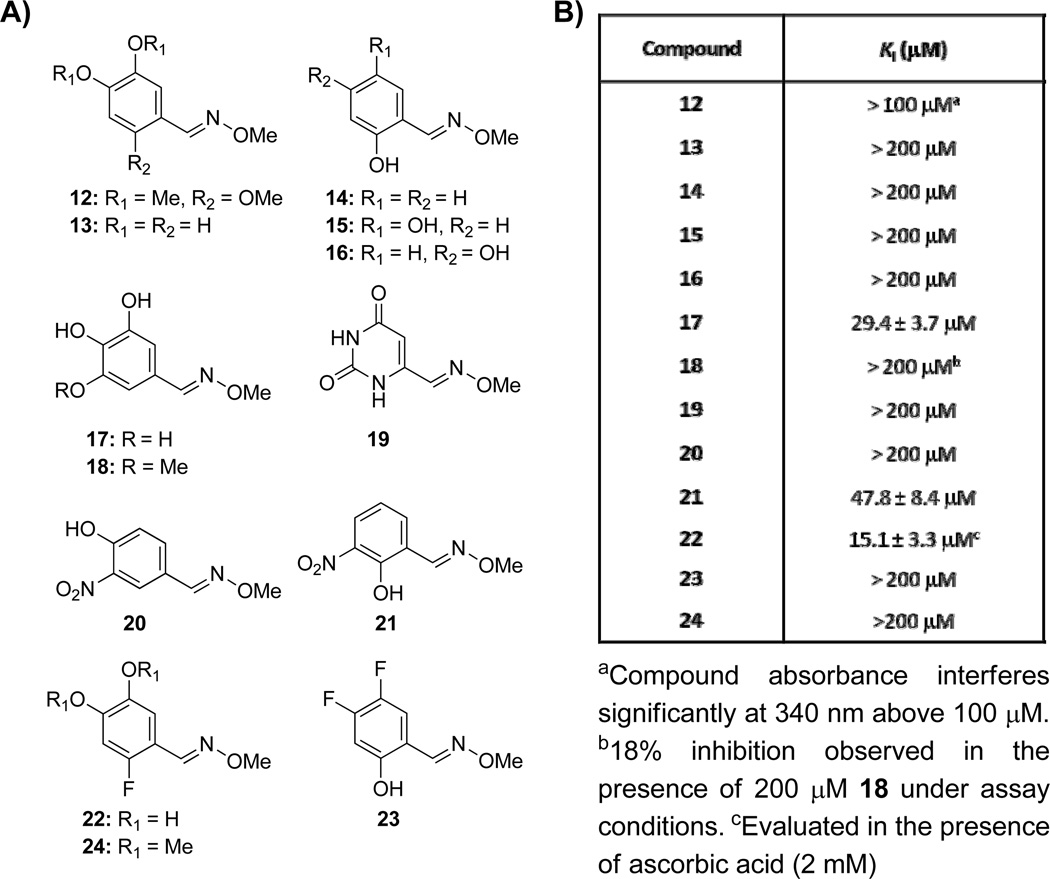
As a starting point to explore structure–activity relationships at the phenyl ring, a series of O-methyloxime analogs (12–24, Figure 2A) was prepared by reaction of the corresponding aldehyde with methoxyamine hydrochloride. Oxime analogs of 8 which are methylated (12) or that lack a hydroxyl group at the 2-, 4-, or 5-positions (13–16, Figure 2A) are inactive up to 200 µm (Figure S8). 3,4,5-Trihydroxy benzaldoxime 17 retains the competitive inhibition profile against d-GAP (Figure S9a), but exhibits lower affinity (Ki = 29.4 ± 3.7 µm, Figure 2) compared to oxime 8 (Ki = 3.9 ± 0.6 µm, Table 1). Methoxy analog 18 shows only 18% inhibition at a concentration of 200 µm, and trimethoxybenzaldoxime 12 is inactive up to 100 µm. Pyrimidine oxime 19 was prepared to mimic the delocalization of charge as might be observed for the phenolate species of 8; however 19 is inactive up to 200 µM. Interestingly, 3-nitro analogs 20 and 21 display different inhibitory activities based on the position of the adjacent hydroxyl; 4-hydroxyl-3-nitro analog 20 shows no inhibitory activity up to 200 µm (Figure S8), whereas 2-hydroxyl-3-nitro analog 21 retains the competitive inhibition profile against d-GAP (Figure S9b), but exhibits lower affinity (Ki = 47.8 ± 8.4 µm, Figure 2B) compared to oxime 8. Fluorine was also explored as an isostere for hydroxyl. 4,5-Difluoro analog 23 is minimally active up to 100 µM while 2-fluoro analog 22 maintains some activity (Ki = 15.1 ± 3.3 µm, Figure 2B). Analog 22 also displays a competitive mode of inhibition with respect to d-GAP (Figure S9c). As with methylated compound 12, the methylated analog of 22, compound 24, is also inactive up to 200 µM. These data suggest that the hydroxyl groups in positions 2, 4, and 5 all contribute to the affinity of 5 for DXP synthase; however, hydroxyl groups in positions 4 and 5 appear crucial for binding. The 2-hydroxy substituent apparently enhances affinity, but significant phenolate character must be achieved before inhibitory activity can be observed in the absence of the 4- and 5-hydroxy groups, as illustrated by comparison of 14 and 21.
Figure 2.
A) Oxime analogs modified on the phenyl ring to determine requirements for inhibition. B) Inhibitory activity of oxime analogs against DXP synthase.
Trihydroxybenzaldoximes are reversible inhibitors
Polyhydroxylated phenyl compounds are known to undergo oxidation to form highly electrophilic quinone products,30 which can interact with enzyme targets to form irreversible covalent adducts.31 It is possible that oxime inhibitors such as 8 act via a mechanism involving a reactive quinone species; thus, we have explored the reversibility of inhibition as well as the behavior of oxime inhibitor 8 in the presence of nucleophilic thiols and under non-nucleophilic reducing conditions.
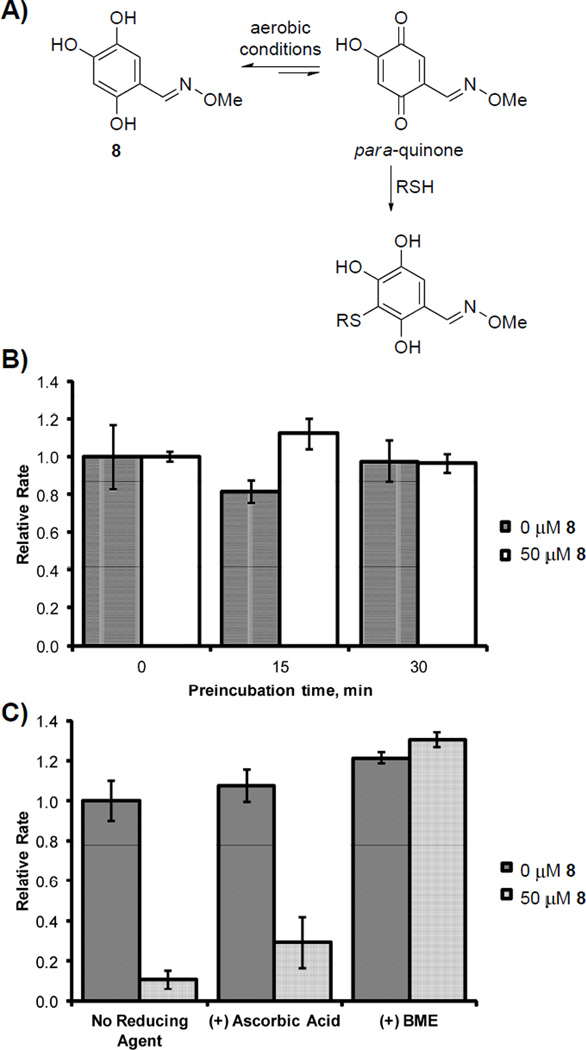
As trihydroxybenzaldoximes can, in theory, be converted to quinone species under aerobic conditions (Figure 3A), we considered the possibility of irreversible inhibition of DXP synthase by this inhibitor class. Rapid dilution experiments were performed in which DXP synthase (10 µm) was pre-incubated with 50 µm trihydroxybenzaldoxime 8 prior to dilution into assay buffer and measurement of DXP synthase activity. Pyruvate (50 µm) was included in the pre-incubation mixture, as pyruvate binding is apparently required for inhibition by 8 to occur. Mixtures were pre-incubated for a period of 15 or 30 minutes, then diluted 100-fold into assay buffer containing pyruvate, d-GAP, and ThDP to initiate enzyme-catalyzed DXP formation. Under all conditions, DXP synthase activity was comparable in the presence or absence of oxime 8 following preincubation of inhibitor with DXP synthase (Figure 3B), indicating the inhibitory activity of 8 is not time-dependent, and the effects of 8 are reversible.
Figure 3.
Oxime 8 reversibly inhibits DXP synthase in its triol form. A) A hydroquinone-quinone equilibrium is possible under aerobic assay conditions to afford a reactive para-quinone (shown) or ortho-quinone; B) Reversible inhibition by 8 shown by rapid dilution assay. C) Inhibitory activity of 8 is abolished in the presence of BME (0.5 mm) but not in the presence of ascorbic acid (2.0 mm).
β-mercaptoethanol abolishes inhibitory activity of trihydroxybenzaldoximes
Nucleophilic thiols such as β-mercaptoethanol (BME) form covalent adducts with reactive electrophiles, including quinones and other Michael acceptors. For a mechanism of inhibition by 8 involving a quinone species, addition of BME to the assay mixture is expected to reduce inhibitory activity through formation of inactive thiol adducts. Addition of BME to inhibition assay mixtures containing oxime 8 abolishes inhibitory activity (Figure 3C), suggesting an electrophilic form of oxime 8 exists under assay conditions. To determine its predominant form under assay conditions, oxime 8 was monitored by 13C NMR spectroscopy at pH 8 for 24 hours (Figure S10a). Under these conditions, there was no observable quinone species which would be apparent as new peaks emerging in the 170–180 ppm range.32 Although 13C NMR spectroscopy does not reveal the presence of quinone species at very low concentration due to the insensitivity of the method, it does suggest that the predominant form of 8 is the triol species. Addition of BME to the aqueous inhibitor solution results in the formation of new resonances in the 13C NMR spectrum, consistent with the formation of products arising from addition of BME to the phenyl ring (Figure S10). Taken together, these data suggest that while oxime 8 exists predominantly in the triol (or hydroquinone) form, there is a small population present in the quinone form (presumably via a hydroquinone-quinone equilibrium) which can react with BME to ultimately divert 8 to an inactive form. The presence of a quinone form of 8 was confirmed by mass spectrometry following incubation of 8 in triethylammonium acetate buffer at pH 8 for 10 min ((ESI) m/z: calc’d 180.03 [M-H]−; found 180.05 [M-H]−.
Trihydroxybenzaldoxime exhibits inhibitory activity in the presence of the non-nucleophilic reducing agent, ascorbic acid
The reversibility of inhibition taken together with the observation that BME reduces the concentration of triol, presumably by diverting it to an inactive thiol adduct via reaction with the quinone, suggests the triol form of oxime 8 could be the active inhibitor and the quinone form may not contribute to inhibitory activity. We tested this hypothesis by using a non-nucleophilic reducing agent, ascorbic acid, to maintain oxime 8 in its reduced triol form. Previous studies on the metabolism of dopamine have shown that thiol reducing agents quickly and efficiently add to dopaquinone to form thioether products; however, ascorbic acid reduces dopaquinone to its diol form without covalent modification.33 Addition of ascorbic acid does not abolish inhibitory activity of 8 (Figure 3C). The results are consistent with a reversible mechanism of inhibition by triol 8 and offer support to a quinone-mediated covalent inactivation of the inhibitor by BME (Figure 3A).
Discussion
The cumulative effects of depleting isoprenoid precursor pools and inhibiting biosynthesis of vitamins critical for central metabolism through inhibition of a single enzyme target, DXP synthase, should be detrimental to bacterial pathogens. Efforts to develop selective inhibitors against this intriguing enzyme target will offer opportunities to expand our armamentarium toward much needed new treatments for drug resistance infections. The original goal of this study was to identify unnatural bisubstrate analogs as inhibitors of DXP synthase to exploit the exceptionally large DXP synthase active site, relaxed substrate specificity, and unique mechanism of this target.
Toward this goal, a library of oxime-based inhibitors linking the glyoxylate carboxyl group (to occupy the pyruvate binding site) to a library of aryl moieties (to mimic unnatural aromatic acceptor substrates) was generated and evaluated for inhibitory activity against enzyme-catalyzed DXP formation. Contrary to our expectations, unnatural bisubstrate analog inhibitors were not identified; rather, trihydroxybenzaldoximes emerged as new d-GAP competitive inhibitors with the 2,4,5-trihydroxybenzaldoxime scaffold exhibiting the most potent inhibitory activity. To date, the few inhibitors of DXP synthase described15,16,18–20,34,35 exhibit micromolar inhibitory activities with Ki values ranging from ~4 µm (acetylphosphonates) to 75 µm (ketoclomazone). Here, we have shown that oxime-based inhibitors display inhibitory potencies with Ki values as low as 1 µm, placing them amongst the most potent inhibitors of DXP synthase described to date (Table 1).
A focused SAR study has revealed that a single oxime moiety is required for inhibitory activity. The oxime oxygen, which can be alkylated without significant loss of activity, contributes to inhibitor affinity as supported by the lack of inhibitory activity of aldehyde 6 and hydrazone 11 which lack a distal oxygen. On the aryl ring, all 3 phenolic hydroxyl groups contribute to binding of 2,4,5-trihydroxybenzaldoxime inhibitor 8, with the positioning of a hydroxyl group at the 2-position being favored over the 3-position for competitive inhibition against d-GAP. While a 2-hydroxy substituent appears to be optimal, incorporation of a fluorine substituent at this position (22) also imparts reasonable inhibitory activity. This observation suggests that fluorine exerts electronic effects on the 4- and 5- hydroxyl groups that are comparable to the effect of incorporating a 2-hydroxy substituent. Given the large volume of the DXP synthase active site and propensity for sterically demanding alternative acceptor substrates to adopt distinct binding modes compared to d-GAP,15 it is reasonable to suggest some oximes tested here may adopt binding modes which do not preclude binding of d-GAP.
The mode of inhibition by trihydroxybenzaldoximes is unique amongst inhibitor classes targeting DXP synthase, with important implications for the design of novel antimicrobial agents. DXP synthase inhibitors identified thus far include acetylphosphonates and acetylphosphinates which are competitive against pyruvate,16,36 ketoclomazone which is uncompetitive with respect to pyruvate and noncompetitive against d-GAP,18 and inhibitors which compete for the ThDP cofactor binding site.13,37 In contrast, trihydroxybenzaldoximes are the first inhibitor class to act competitively against d-GAP and, interestingly, show an uncompetitive or noncompetitive inhibition mode with respect to pyruvate. Thus, the distinctive DXP synthase-LThDP complex is the inhibitor target in some cases. This form of inhibition is unprecedented for this enzyme target, and is rare in drug discovery.38 Uncompetitive inhibition represents an appealing strategy for intervention given that the cellular response to enzyme inhibition is to increase substrate concentration.39 Theoretical models of metabolic pathways have shown that competitive inhibitors are eventually outcompeted at the active site by accumulated substrate, resulting in little change in pathway flux. In contrast, uncompetitive inhibitors are able to maintain inhibition in the face of high substrate concentration, ultimately crippling metabolic flux.38 In this circumstance, an increase in pyruvate concentration would lead to increased enzyme-LThDP complex levels and, therefore, enhanced inhibitory potency.40
In addition to important implications for uncompetitive DXP synthase inhibitors in antimicrobial drug design, inhibitors that bind to the enzyme-LThDP complex represent new probes for studies to elucidate the unique LThDP decarboxylation mechanism on DXP synthase. Our recent mechanistic studies have shown that the LThDP intermediate, formed via reaction of the cofactor with pyruvate, is remarkably stable on DXP synthase. LThDP decarboxylation is triggered by the acceptor substrate d-GAP, by an unknown mechanism.17 As noted, the uncompetitive mode of inhibition by 5 and 7 suggest these inhibitors preferentially bind to the DXP synthase-LThDP complex. It is unknown whether trihydroxybenzaldoximes stabilize the enzyme-LThDP complex to block LThDP decarboxylation, or if they act as triggers of LThDP decarboxylation and prevention of carboligation by blocking the d-GAP binding site. Circular dichroism and NMR studies are required to elucidate the finer details of this inhibition mechanism. Nevertheless, inhibition via either mechanism will offer a new tool to study the active site in its pre-decarboxylation or post-decarboxylation conformations, toward the long-term goal to develop selective inhibitors against DXP synthase.
Finally, although we have shown that oxidation of our inhibitors does not likely contribute to DXP synthase inhibition in vitro (Figure 3), it is possible that production of quinone forms, through oxidation of the polyhydroxy phenyl moiety, could be a potential source of toxicity in vivo. Thus, future medicinal chemistry efforts should aim to optimize these inhibitor scaffolds to reduce this liability.
Overall, our results are of interest because they describe a new class of inhibitors of a highly promising, yet under-developed antimicrobial target. Trihydroxybenzaldoximes display a novel mode of inhibition against DXP synthase and are amongst the most potent inhibitors of the enzyme reported to date, suggesting an excellent starting point for SAR studies to enhance potency, impart selectivity of inhibition and circumvent toxicity of this new inhibitor class.
Experimental Section
General
Unless otherwise noted, all reagents were obtained from commercial suppliers and used without further purification. Dichloromethane was distilled after drying on CaH2 then stored over 3Å molecular sieves. Yields of all reactions refer to the purified products. Dynamic Adsorbents 32 – 63 µm silica gel was used for flash column chromatography and 250 µm w/h F254 plates were used for thin layer chromatography (TLC). 1H and 13C NMR spectra were acquired on a Bruker Avance III 500 spectrometer operating at 500 MHz for 1H and 126 MHz for 13C. Chemical shift values are reported as δ (ppm) relative to CHCl3 at δ 7.27 ppm, MeOH at δ 3.31 ppm, and DMSO at δ 2.50 ppm for 1H NMR and CHCl3 at δ 77.0 ppm, MeOH at δ 49.15 ppm, and DMSO at δ 39.51 ppm for 13C NMR. Mass spectrometry analysis was carried out at University of Illinois at Urbana-Champaign, School of Chemical Sciences, Mass Spectrometry Laboratory (ESI ionization) and the Mass Spectrometry Laboratory, The Johns Hopkins University, Baltimore, MD (EI ionization). The purity of synthesized compounds was ≥95% as analyzed by HPLC (Beckman Gold Nouveau System Gold) on a C18 column (Grace Altima, 3 µm C18 analytical Rocket® column, 53 mm × 7 mm) using the following method: 5% to 100% B over 10 minutes at a flow rate of 3 mLmin−1 (solvent A: Et3NHOAc (50 mM, pH 8), solvent B: acetonitrile).
Both E and Z stereoisomers are theoretically possible for all oximes synthesized; however, we observed a strong preference for the formation of a single product in agreement with previous reports.27,41 Only compounds 12 and 13 yielded a mixture of isomers and in both cases, the oxime proton of the major product possessed a downfield chemical shift compared to the minor product suggesting the thermodynamically favorable E stereoisomer is the major product.42
All enzyme reaction mixtures contained 10% DMSO, added to solubilize lipophilic inhibitors. These conditions only have a minimal effect on the uninhibited reaction.15 Recombinant DXP synthase26 and IspC21 was expressed, purified, and characterized as previously described.
Chemistry
Synthesis Oxime-Based Aryl Carboxylate library.41
To each 0.3-mL well of a 96-well microtiter plate was added a DMSO stock solution of AcOH (17 µL of a 150 mm stock), glyoxylate (20.4 µL of a 150 mm stock), and a single aryl aldehyde (20.4 µL of a 150 mm stock). The plate was carefully agitated until the solutions were homogeneous. To each of the glyoxylate-aryl aldehyde mixtures was added a DMSO solution of the O,O′-diaminoalkanediol-containing mixture that contained four linker lengths in equal proportion (19.1 µL of a 160 mm stock of each). The plate was sealed, further agitated, and incubated for 12 hours at 37 °C.
Sodium (1E,7E)-1-(2,4,5-trihydroxyphenyl)-3,6-dioxa-2,7-diazanona-1,7-dien-9-oate (Mixed glyoxyl 2,4,5-trihydroxybenzalde dioxime) (4)
To a stirred solution of O,O’-diaminoethanediol dihydrochloride (0.100 g, 0.606 mmol) in methanol (2.0 mL) was added sodium glyoxylate monohydrate (0.070 g, 0.606 mmol) followed by sodium bicarbonate (0.051 g, 0.606 mmol). The mixture was stirred at ambient temperature for 40 minutes during which a precipitate formed. Aldehyde 6 (0.094 g, 0.606 mmol) was then added, and the mixture was stirred at ambient temperature for an additional 90 minutes. The solution was filtered to remove the byproduct, symmetrical dioxime 5. The filtrate was condensed, and the resulting solid was suspended in water, filtered, and further washed with water. The solid was collected and dissolved in 25% saturated NaHCO3 (1 mL) and loaded onto a C18 solid phase extraction column. The column was washed with water (1 mL), and the product was eluted with acetonitrile. Solvent was removed under reduced pressure to give a purple solid (0.025 g, 14% yield). RT = 2.29 min λmax = 324 nm. 1H NMR (500 MHz, DMSO-d6) δ 8.21 (s, 1H), 7.35 (s, 1H), 6.90 (s, 1H), 6.31 (s, 1H), 4.20 (s, 4H) 13C NMR (126 MHz, DMSO-d6) δ 164.3, 150.3, 149.5, 149.1, 147.0, 138.7, 112.7, 107.7, 103.6, 71.6, 71.5 HRMS (ESI) m/z: calc’d 307.0542 (M+H+); found 307.0540 (M+H+)
(1E,1'E)-O,O'-(Ethane-1,2-diyl)bis(1-(2,4,5-trihydroxyphenyl)-2,4,5-trihydroxybenzaldehyde oxime) (Symmetrical bis(2,4,5-trihydroxybenzaldoxime)) (5)
To a stirred solution of O,O’-diaminoethanediol dihydrochloride (0.054 g, 0.32 mmol), aldehyde 6 (0.100 g, 0.65 mmol), and sodium acetate (0.267 g, 1.04 mmol) in methanol (1.0 mL) was added water (~15 drops) until a homogenous solution was obtained. The solution was stirred at ambient temperature for 90 minutes during which a tan precipitate formed. Solvent was removed under reduced pressure and the resulting solid was suspended in ethyl acetate (20 mL) and washed with water (20 mL). This biphasic suspension was filtered and washed with water to give a tan solid after drying in vacuo (0.077 g, 66% yield). RT = 3.14 min λmax = 324 nm. 1H NMR (500 MHz, DMSO-d6) δ 9.38 (br. s., 2H), 9.21 (s, 2H), 8.50 (br. s., 2H), 8.23 (s, 2H), 6.89 (s, 2H), 6.31 (s, 2H), 4.26 (s, 4H) 13C NMR (126 MHz, DMSO-d6) δ 150.2, 148.9, 147.0, 138.6, 112.7, 107.8, 103.5, 71.7 HRMS (ESI) m/z: calc’d 365.0985 (M+H+); found 365.0981 (M+H+)
(E)-5-((2-Methylhydrazono)methyl)benzene-1,2,4-triol (10)
A solution of aldehyde 6 (0.100 g, 0.649 mmol) and methyl hydrazine (0.035 mL, 0.649 mmol) in ethanol (1.0 mL) was heated to reflux for 90 minutes. The solution was allowed to cool to ambient temperature, and the solvent was removed under reduced pressure. The resulting solid was purified via silica flash chromatography (ethyl acetate/hexanes/methanol 60:39:1) to yield an orange powder (0.088 g, 74% yield). RT = 1.56 min λmax = 270 nm. 1H NMR (500 MHz, MeOD-d4) δ 7.63 (s, 1H), 6.61 (s, 1H), 6.32 (s, 1H), 2.82 (s, 3H) 13C NMR (126 MHz, MeOD-d4) δ 152.8, 148.1, 143.0, 139.1, 116.5, 112.3, 104.2, 35.4 HRMS (ESI) m/z: calc’d 186.0766 (M+H+); found 186.0775 (M+H+)
5-((Methoxyamino)methyl)benzene-1,2,4-triol hydrochloride (11)
Oxime 8 (0.044 g, 0.24 mmol) was dissolved in ethanol (1.0 mL) and cooled to 0 °C. Sodium cyanoborohydride (0.068 g, 1.08 mmol) was added followed by 10% HCl in ethanol (0.38 mL). The solution was allowed to warm to ambient temperature and stirred for 90 minutes. Solids were removed by filtration through celite. The filtrate was condensed under reduced pressure and the resulting residue was dissolved in a minimal volume of 10% HCl in ethanol and precipitated from diethyl ether to give a tan solid after filtration (0.011 g, 21% yield). RT = 2.15 min λmax = 276 nm. 1H NMR (500 MHz, MeOD-d4) δ 6.76 (s, 1H), 6.45 (s, 1H), 4.33 (s, 2H), 3.94 (s, 3H) 13C NMR (126 MHz, MeOD-d4) δ 151.5, 149.6, 139.6, 119.4, 105.5, 104.2, 62.1, 49.9 HRMS (ESI) m/z: calc’d 183.0770 (M+H+); found 183.0772 (M+H+)
General Procedure for Synthesis of Oximes
With the exception of compound 19, all oximes were synthesized in the following manner. The appropriate aldehyde (0.050 g, 1 eq.), methoxyamine·HCl (1.1 eq.), and sodium acetate (1.6 eq.) were suspended in methanol (0.7 mL). Deionized water (10–20 drops) was added to the mixture dropwise until a homogeneous solution was obtained. The mixture was stirred at ambient temperature for 4 hours, and monitored by TLC. The solvent was removed under reduced pressure. The resulting crude mixture was purified via silica flash chromatography.
(E)-2,4,5-Trihydroxybenzaldehyde oxime (7)
Purified via silica flash chromatography (ethyl acetate/hexanes 1:1). Yield 66%. RT = 2.27 min λmax = 255 nm. 1H NMR (500 MHz, MeOD-d4): δ 8.04 (s, 1H); 6.65 (s, 1H); 6.33 (s, 1H). 13C NMR (126 MHz, MeOD-d4) δ 153.0, 152.1, 149.4, 139.5, 116.7, 109.9, 104.3 HRMS (ESI) m/z: calc’d 170.0453 (M+H+); found 170.0453 (M+H+)
(E)-2,4,5-Trihydroxybenzaldehyde O-methyl oxime (8)
Product was purified by silica flash chromatography (ethyl acetate/hexanes 1:1). Yield 81%. RT = 2.81 min λmax = 260 nm. 1H NMR (500 MHz, MeOD-d4): δ 8.09 (s, 1H); 6.69 (s, 1H); 6.34 (s, 1H); 3.88 (s, 3H) 13C NMR (126 MHz, MeOD-d4) δ 153.2, 151.7, 150.2, 139.8, 116.6, 109.2, 104.3, 62.5 HRMS (ESI) m/z: calc’d 184.0610 (M+H+); found 184.0611 (M+H+)
(E)-2,4,5-Trihydroxybenzaldehyde O-benzyl oxime (9)
Purified via silica flash chromatography (ethyl acetate/hexanes 4:6). Yield 85%. RT = 4.49 min λmax = 263 nm. 1H NMR (500 MHz, MeOD-d4): δ 8.14 (s, 1H); 7.40-7.30 (m, 5H); 6.68 (s, 1H); 6.32 (s, 1H); 5.10 (s, 2H) 13C NMR (126 MHz, MeOD-d4) δ 153.1, 152.2, 150.2, 139.8, 138.9, 129.6, 129.6, 129.2, 116.8, 109.2, 104.3, 77.4 HRMS (ESI) m/z: calc’d 260.0923 (M+H+); found 260.0924 (M+H+)
(E,Z)-2,4,5-Trimethoxybenzaldehyde O-methyl oxime (12)
In addition to water, THF (15 drops) was added to the suspension prior to addition of water. Purified via silica flash chromatography (ethyl acetate/hexanes 1:1). Yield 83% (81% (E isomer)). (E isomer) RT = 5.69 min λmax = 273 nm; (Z isomer) RT = 5.34 min λmax = 270. 1H NMR (500 MHz, CDCl3) δ (E isomer) 8.40 (s, 1H), 7.32 (s, 1H), 6.49 (s, 1H), 3.96 (s, 3H), 3.92 (s, 3H), 3.90 (s, 3H), 3.83 (s, 3H); (Z isomer) 7.98 (s, 1H), 7.68 (s, 1H), 6.50 (s, 1H), 4.00 (s, 3H), 3.93 (s, 3H), 3.85 (s, 3H), 3.85 (s, 3H); 13C NMR (126 MHz, CDCl3) δ (E isomer) 152.8, 151.5, 144.5, 143.5, 112.4, 108.2, 97.1, 61.8, 56.7, 56.3, 56.0; (Z isomer) 153.7, 151.3, 142.1, 140.4, 115.2, 111.4, 96.4, 62.4, 56.6, 56.5, 55.9. HRMS (ESI) m/z: calc’d 226.1079 (M+H+); found 226.1079 (M+H+)
(E,Z)-3,4-Dihydroxybenzaldehyde O-methyl oxime (13)
Purified via silica flash chromatography (ethyl acetate/hexanes 1:1). Yield 80% (86% (E isomer)). RT = 3.46 min λmax = 277 nm, (Z isomer) RT = 5.47 min λmax = 274 nm. 1H NMR (500 MHz, MeOD-d4) (E isomer) δ 7.91 (s, 1H), 7.09 (d, J = 1.89 Hz, 1H), 6.86 (dd, J = 1.96, 8.10 Hz, 1H), 6.76 (d, J = 8.02 Hz, 1H), 3.86 (s, 3H); (Z isomer) 7.56 (d, J = 2.04 Hz, 1H), 7.18 (dd, J = 2.04, 8.33 Hz, 1H), 7.11 (s, 1H), 6.78 (d, J = 8.33 Hz, 1H), 3.94 (s, 3H); 13C NMR (126 MHz, CD3OD) δ (E isomer) 150.2, 148.9, 146.8, 125.5, 121.6, 116.4, 114.0, 62.0; (Z isomer) 149.0, 147.4, 146.0, 125.7, 124.4, 119.4, 116.0, 62.7. HRMS (ESI) m/z: calc’d 168.0661 (M+H+); found 168.0661 (M+H+)
(E)-2-Hydroxybenzaldehyde O-methyl oxime (14)
Purified via silica flash chromatography (ethyl acetate/hexanes 1:9). Yield 53%. RT = 6.03 min λmax = 265 nm. 1H NMR (500 MHz, CDCl3) δ 9.88 (s, 1H), 8.16 (s, 1H), 7.29 (dt, J = 1.60, 8.00 Hz, 1H), 7.16 (dd, J = 1.57, 7.70 Hz, 1H), 7.01 (dd, J = 1.00, 8.00 Hz, 1H), 6.92 (dt, J = 1.02, 7.70 Hz, 1H), 4.00 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 157.4, 151.4, 131.1, 130.7, 119.5, 116.7, 116.2, 62.4. HRMS (ESI) m/z: calc’d 152.0712 (M+H+); found 152.0716 (M+H+)
(E)-2,5-Dihydroxybenzaldehyde O-methyl oxime (15)
Purified via silica flash chromatography (ethyl acetate/hexanes 1:1). Yield 80%. RT = 4.20 min λmax = 268 nm. 1H NMR (500 MHz, DMSO-d6) δ 9.26 (br. s, 1H), 8.91 (br. s, 1H), 8.28 (s, 1H), 6.95 (d, J = 2.67 Hz, 1H), 6.71 (d, J = 8.80 Hz, 1H), 6.67 (dd, J = 2.70, 8.80 Hz, 1H), 3.86 (s, 3H); 13C NMR (126 MHz, MeOD-d4) δ 151.7, 151.5, 151.3, 119.9, 118.6, 118.2, 116.3, 62.9. HRMS (EI) m/z: calc’d 167.05824 (M +); found 167.05839 (M +)
(E)-2,4-Dihydroxybenzaldehyde O-methyl oxime (16)
Purified via silica flash chromatography (ethyl acetate/hexanes 4:6). Yield 87%. RT = 4.52 min λmax = 280 nm. 1H NMR (500 MHz, MeOD-d4) δ 8.16 (s, 1H), 7.10 (d, J = 8.33 Hz, 1H), 6.34 (dd, J = 2.36, 8.33 Hz, 1H), 6.31 (d, J = 2.36 Hz, 1H), 3.90 (s, 3H). 13C NMR (126 MHz, MeOD-d4) δ 162.0, 160.4, 152.1, 132.8, 110.4, 108.9, 103.8, 62.6. HRMS (ESI) m/z: calc’d 168.0661 (M+H+); found 168.0660 (M+H+)
(E)-3,4,5-Trihydroxybenzaldehyde O-methyl oxime (17)
Purified via silica flash chromatography (ethyl acetate/hexanes 1:1). Yield 51%. RT = 2.83 min λmax = 295 nm. 1H NMR (500 MHz, MeOD-d4) δ 7.74 (s, 1H), 6.52 (s, 2H), 3.77 (s, 3H); 13C NMR (126 MHz, MeOD-d4) δ 150.6, 147.3, 136.9, 124.7, 107.7, 62.2. HRMS (ESI) m/z: calc’d 184.0610 (M+H+); found 184.0611 (M+H+)
(E)-3,4-Dihydroxy-5-methoxybenzaldehyde O-methyl oxime (18)
Purified via silica flash chromatography (ethyl acetate/hexanes 1:1). Yield 79% (95% (E isomer)). RT = 3.49 min λmax = 297 nm. 1H NMR (500 MHz, MeOD-d4) δ 7.92 (s, 1H), 6.78 (d, J = 1.89 Hz, 1H), 6.72– 6.74 (m, 1H), 3.89 (s, 3H), 3.85 (s, 3H); 13C NMR (126 MHz, MeOD-d4) δ 150.4, 149.9, 146.9, 137.7, 124.5, 109.6, 103.3, 62.1, 56.7. HRMS (ESI) m/z: calc’d 198.0766 (M+H+); found 198.0786 (M+H+)
(E)-Uracil-6-carbaldehyde O-methyl oxime (19)
A vial was charged with uracil-6-carbaldehyde (0.050 g, 0.316 mmol), methoxyamine·HCl (0.029 g, 0.348 mmol), and sodium acetate (0.042 mg, 0.506 mmol). DMF (0.7 mL) was then added to the flask. While stirring, deionized water (~18 drops) was added until all solids dissolved. The mixture was left to stir at ambient temperature overnight. The solvent was evaporated from the resulting heterogeneous mixture under reduced pressure. The remaining solids were washed with deionized water and collected via vacuum filtration. Yield 24%. RT = 1.89 min λmax = 294 nm. 1H NMR (500 MHz, DMSO-d6) δ 7.91 (s, 1H), 5.77 (s, 1H), 3.96 (s, 3H); 13C NMR (126 MHz, DMSO-d6) δ 163.8, 151.0, 144.4, 142.1, 101.4, 62.8. HRMS (ESI) m/z: calc’d 170.0566 (M+H+); found 170.0565 (M+H+)
(E)-4-Hydroxy-3-nitrobenzaldehyde O-methyl oxime (20)
Compound 20 was prepared according to the general procedure outlined above with the following minor modification. Dichloromethane (1.5 mL) was added to the methanol suspension prior to the addition of water. The biphasic mixture was stirred at room temperature for 2 hours. The crude solids were purified via silica flash chromatography (ethyl acetate/hexanes 1:3). Yield 73%. RT = 3.59 min λmax = 302 nm. 1H NMR (500 MHz, CDCl3) δ 10.72 (s, 1H), 8.24 (d, J = 2.04 Hz, 1H), 8.02 (s, 1H), 7.92 (dd, J = 2.04, 8.80 Hz, 1H), 7.19 (d, J = 8.80 Hz, 1H), 3.99 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 155.9, 145.6, 135.0, 133.5, 125.2, 123.8, 120.6, 62.3. HRMS (EI) m/z: calc’d 196.04841 (M+); found 196.04839 (M+)
(E)-2-Hydroxy-3-nitrobenzaldehyde O-methyl oxime (21)
Compound 21 was prepared according to the general procedure outlined above with the following minor modification.THF (1.1 mL) was added to the methanol suspension prior to the addition of water. The crude solids were purified via silica flash chromatography (ethyl acetate/hexanes 1:2); Yield 78%. RT = 3.76 min λmax = 306 nm. 1H NMR (500 MHz, CDCl3) δ 11.07 (s, 1H), 8.43 (s, 1H), 8.10 (dd, J = 1.65, 8.41 Hz, 1H), 7.96 (dd, J = 1.65, 7.62 Hz, 1H), 7.02 (t, J = 8.02 Hz, 1H), 4.02 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 152.6, 144.3, 135.1, 134.3, 126.4, 122.3, 119.6, 62.5. HRMS (ESI) m/z: calc’d 197.0562 (M+H+); found 197.0562 (M+H+)
(E)-2-Fluoro-4,5-dihydroxybenzaldehyde O-methyl oxime (22)
Purified via silica flash chromatography (ethyl acetate/hexanes 1:1). Yield 51%. RT = 2.69 min λmax = 331 nm. 1H NMR (500 MHz, MeOD-d4) δ 8.13 (s, 1H), 7.16 (d, J = 7.07 Hz, 1H), 6.54 (d, J = 11.48 Hz, 1H), 3.91 (s, 3H); 13C NMR (126 MHz, MeOD-d4) δ 156.7 (d, JCF = 242.5 Hz), 150.3 (d, JCF = 11.8 Hz), 143.5, 143.4 (d, JCF = 3.6 Hz), 112.0 (d, JCF = 3.6 Hz), 111.5 (d, JCF = 11.8 Hz), 103.7 (d, JCF = 26 Hz), 62.3. HRMS (ESI) m/z: calc’d 186.0566 (M+H+); found 186.0570 (M+H+)
(E)-4,5-Difluoro-2-hydroxybenzaldehyde O-methyl oxime (23)
Purified via silica flash chromatography (ethyl acetate/hexanes 1:1). Yield 52%. RT = 6.53 min λmax = 262 nm. 1H NMR (500 MHz, CDCl3) δ 9.96 (d, J = 1.41 Hz, 1H), 8.06 (s, 1H), 6.96 (dd, J = 8.65, 10.22 Hz, 1H), 6.79 (dd, J = 6.76, 11.63 Hz, 1H), 4.00 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 154.4 (d, JCF = 10.9 Hz), 151.5 (dd, JCF = 13.9, 253.3 Hz), 149.7, 144.0 (dd, JCF = 13.9, 240 Hz), 117.7 (dd, JCF = 2.3, 19.5 Hz), 112.0 (dd, JCF = 3.6, 5.4 Hz), 105.9 (d, JCF = 20 Hz), 62.7. HRMS (ESI) m/z: calc’d 188.0523 (M+H+); found 188.0526 (M+H+)
(E)-2-Fluoro-4,5-dimethoxybenzaldehyde O-methyl oxime (24)
Purified via silica flash chromatography (ethyl acetate/hexanes 1:1). Yield 36%. RT = 5.96 min λmax = 269 nm. 1H NMR (500 MHz, CDCl3) δ 8.26 (s, 1H), 7.27 (d, J = 6.80 Hz, 1H), 6.61 (d, J = 11.48 Hz, 1H), 3.98 (s, 3H), 3.91 (s, 3H), 3.89 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 155.8 (d, JCF = 245 Hz), 151.5 (d, JCF = 10 Hz), 145.6 (d, JCF = 2.7 Hz), 142.0 (d, JCF = 4.5 Hz), 110.9 (d, JCF = 11.8 Hz), 106.9 (d, JCF = 3.6 Hz), 99.6 (d, JC = 26 Hz), 62.0, 56.3, 56.1. HRMS (ESI) m/z: calc’d 214.0879 (M+H+); found 214.0874 (M+H+)
Biochemical Evaluation
DXP synthase inhibition assays
Library Screen
The initial screen of unnatural bisubstrate analogs produced through the oxime coupling was performed in a 96-well plate format. Each well contained HEPES (100 mm, pH 8.0), MgCl2 (2 mm), NaCl (5 mm), ThDP (1 mM), BSA (1 mg/mL), IspC (1.0 µm), NADPH (160 µm), DMSO (10 %, v/v), inhibitor (100 µm total oxime), and DXP synthase (50 nm). Mixtures were preincubated at 37 °C for 5 min. Total oxime concentration refers to the concentration of all dioxime products irrespective of oxime derivatization or length of the dialkyoxyamine portion. Pyruvate and d-GAP were added together to give final concentrations of 150 µm and 75 µm, respectively, to initiate the reaction. Rates were determined by following the depletion of NADPH at 340 nm.
Kinetic Analysis of Inhibitors
In order to evaluate the inhibitory activity against DXP synthase, a continuous spectrophotometric coupled assay was used to measure formation of DXP by monitoring IspC catalyzed consumption of NADPH (340 nm) in the conversion of DXP to MEP.16,21 DXP synthase reaction mixtures containing HEPES (100 mm, pH 8.0), MgCl2 (2 mm), NaCl (5 mm), ThDP (1 mm), BSA (1 mg/mL), pyruvate (12.5–250 µm), d-GAP (5–200 µm), IspC (1.7 µm), and NADPH (160 µm) and DMSO (10 %, v/v), and inhibitor (0–300 µm) were preincubated at 37 °C for 5 min. Enzymatic reactions were initiated by addition of DXP synthase (50 – 100 nm). Initial rates were measured upon addition of DXP synthase. The majority of inhibitors evaluated are stable under the conditions of the assay, as determined by HPLC analysis (data not shown). Compounds 6, 10, 11 and 22 are unstable under these conditions, presumably a consequence of catechol oxidation. Addition of the non-nucleophilic reducing agent ascorbic acid was found to stabilize these compounds for inhibition assays. In addition, inhibitors were monitored spectrophotometrically in the absence of DXP synthase, to confirm there are minimal DXP synthase-independent changes in absorbance at 340 nm under the assay conditions. At the highest inhibitor concentration, small DXP synthase-independent background rates were observed, indicated by a decrease in absorbance at 340 nm over time (< 1 µm/min), and in these cases the DXP synthase reaction rate was corrected arithmetically for the non-enzymatic interference. All compounds were evaluated for inhibitory activity against the coupling enzyme (IspC) using the method described below; inhibition of IspC was not observed for any inhibitors at the highest concentration analyzed for inhibition of DXP synthase under assay conditions. All experiments were performed in triplicate. Ki values were determined by non-linear regression analysis using GraphPad Prism 6.0. The mode of inhibition in each case was determined by model discrimination analysis in GraphPad Prism, using Akaike's information criterion (AIC) to differentiate between competitive, uncompetitive, and noncompetitive inhibition.
Testing inhibitory activity against coupling enzyme, IspC
To ensure that inhibitors evaluated via the spectrophotometric enzyme-coupled assay do not inhibit the coupling enzyme IspC, compounds were tested against IspC directly in the absence of DXP synthase. Measurements were made spectrophotometrically by monitoring the consumption of NADPH at 340 nm. DXP was synthesized enzymatically as follows: A DXP synthase reaction mixture including HEPES (100 mm, pH 8.0), MgCl2 (2 mm), NaCl (5 mm), ThDP (1 mm), BSA (1 mg/mL), pyruvate (4 mm), and d-GAP (4 mm) was preincubated at 37 °C for 5 min. The reaction was initiated by addition of DXP synthase (1 µm). The reaction mixture was incubated at 37 °C for 90 min. After 90 min, the reaction mixture was centrifuged for 5 min at 13,000 rpm and then incubated on ice. Separately, IspC reaction mixtures were prepared by adding HEPES (100 mm, pH 8.0), MgCl2 (2 mm), NADPH (160 µm), DXP from the aforementioned DXP synthase reaction (140 µm), DMSO (10 %, v/v), and inhibitor (20–200 µm). The mixture was preincubated at 37 °C for 5 min, and the reaction was initiated by addition of IspC (50 nm). Initial rates were measured upon addition of IspC. A reduction in the rate of IspC-catalyzed NADPH depletion was not observed for any compound tested in the absence of DXP synthase (data not shown).
Rapid dilution experiments
DXP synthase (260 µm, purified without BME) was added to a solution of HEPES (100 mm, pH 8), MgCl2 (2 mm), NaCl (5 mm), ThDP (1 mm), pyruvate (50 µm), and 8 (50 µm) or DMSO for the no inhibitor control. All solutions contained 10% DMSO (v/v). Solutions were then incubated at 37 °C for 0, 15, and 30 minutes. Following incubation, the DXP synthase-containing solutions were diluted 100-fold into a solution containing HEPES (100 mm, pH 8), MgCl2 (2 mm), NaCl (5 mm), ThDP (1 mm), DMSO (10% v/v), pyruvate (50 µm), d-GAP (30 µm), NADPH (160 µm), and IspC (1.7 µm) giving a final DXPS concentration of 100 nm and inhibitor concentration of 0.5 µm. Upon dilution, initial rates of DXP synthase-catalyzed DXP formation were determined spectrophotometrically by following the depletion of NADPH at 340 nm as previously described.
Determination of inhibitory activity in the presence of BME or ascorbic acid
To a solution of HEPES (100 mm, pH 8), BSA (1 mg/mL), MgCl2 (2 mm), NaCl (5 mm), ThDP (1 mm), pyruvate (125 µm), and d-GAP (50 µm) was added BME (0.5 mm) or freshly prepared ascorbic acid (2.0 mm) followed by addition of 8 (50 µm) or DMSO. NADPH (160 µm) was then added, followed by IspC (1.7 µm). After a 5 minute pre-incubation at 37 °C, the reaction was initiated by the addition of DXP synthase (100 nm) and initial rates were determined spectrophotometrically by monitoring the depletion of NADPH at 340 nm.
Supplementary Material
Acknowledgements
We gratefully acknowledge Dr. James Stivers for providing the aldehyde library and Dr. Theresa Shapiro for the use of instrumentation for the library screen. This work was supported by funding from The Johns Hopkins Malaria Research Institute Pilot Grant (F.M.M. and C.F.M.), and the National Institutes of Health (GM084998 for C.F.M., F.M.M., D.B..; T32GM08018901 for F.M.M., D. B.; T32GM008763 for A. A-k., and AI094967 for F.M.M.)
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- 1.Antibiotic resistance threats in the United States, 2013. Washington, DC: U.S. Department of Health and Human Services. Center for Disease Control and Prevention. U.S. Government Printing Office; 2014. [Google Scholar]
- 2.Tamma PD, Cosgrove SE, Maragakis LL. Clinical Microbiology Reviews. 2012;25:450–470. doi: 10.1128/CMR.05041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen RC, Popat R, Diggle SP, Brown SP. Nat Rev Micro. 2014;12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 4.Julliard JH, Douce R. Proceedings of the National Academy of Sciences. 1991;88:2042–2045. doi: 10.1073/pnas.88.6.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sprenger GA, Schörken U, Wiegert T, Grolle S, de Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H. Proceedings of the National Academy of Sciences. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill RE, Sayer BG, Spenser ID. J Am. Chem. Soc. 1989;111:1916–1917. [Google Scholar]
- 7.Takahashi S, Kuzuyama T, Watanabe H, Seto H. Proceedings of the National Academy of Sciences. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohmer M, Seemann M, Horbach S, Bringer-Meyer S, Sahm H. J Am. Chem. Soc. 1996;118:2564–2566. [Google Scholar]
- 9.Cassera MB, Gozzo FC, D'Alexandri FL, Merino EF, del Portillo HA, Peres VJ, Almeida IC, Eberlin MN, Wunderlich G, Wiesner J, Jomaa H, Kimura EA, Katzin AM. Journal of Biological Chemistry. 2004;279:51749–51759. doi: 10.1074/jbc.M408360200. [DOI] [PubMed] [Google Scholar]
- 10.Heuston S, Begley M, Gahan CGM, Hill C. Microbiology. 2012;158:1389–1401. doi: 10.1099/mic.0.051599-0. [DOI] [PubMed] [Google Scholar]
- 11.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Türbachova I, Eberl M, Zeidler J, Lichtenthaler HK, Soldati D, Beck E. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 12.Wright LP, Rohwer JM, Ghirardo A, Hammerbacher A, Ortiz-Alcaide M, Raguschke B, Schnitzler J, Gershenzon J, Phillips MA. Plant Physiology. 2014;165:1488–1504. doi: 10.1104/pp.114.245191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee A, Wu Y, Banerjee R, Li Y, Yan H, Sharkey TD. Journal of Biological Chemistry. 2013;288:16926–16936. doi: 10.1074/jbc.M113.464636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bitok JK, Meyers CF. ACS Chem. Biol. 2012;7:1702–1710. doi: 10.1021/cb300243w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris F, Vierling R, Boucher L, Bosch J, Freel Meyers CL. ChemBioChem. 2013;14:1309–1315. doi: 10.1002/cbic.201300187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith JM, Vierling RJ, Meyers CF. Med. Chem. Commun. 2012;3:65–67. doi: 10.1039/C1MD00233C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi D, Kato N, Kuzuyama T, Sato Y, Ohkanda J. Chem. Commun. 2013;49:5535–5537. doi: 10.1039/c3cc40758f. [DOI] [PubMed] [Google Scholar]
- 18.Matsue Y, Mizuno H, Tomita T, Asami T, Nishiyama M, Kuzuyama T. J Antibiot. 2010;63:583–588. doi: 10.1038/ja.2010.100. [DOI] [PubMed] [Google Scholar]
- 19.Witschel M, Röhl F, Niggeweg R, Newton T. Pest Manag. Sci. 2013;69:559–563. doi: 10.1002/ps.3479. [DOI] [PubMed] [Google Scholar]
- 20.Masini T, Pilger J, Kroezen BS, Illarionov B, Lottmann P, Fischer M, Griesinger C, Hirsch AKH. Chem. Sci. 2014;5:3543–3551. [Google Scholar]
- 21.Brammer LA, Smith JM, Wade H, Meyers CF. Journal of Biological Chemistry. 2011;286:36522–36531. doi: 10.1074/jbc.M111.259747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eubanks LM, Poulter CD. Biochemistry (N. Y.) 2003;42:1140–1149. doi: 10.1021/bi0205303. [DOI] [PubMed] [Google Scholar]
- 23.Patel H, Nemeria NS, Brammer LA, Freel Meyers CL, Jordan F. J Am. Chem. Soc. 2012;134:18374–18379. doi: 10.1021/ja307315u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brammer Basta LA, Patel H, Kakalis L, Jordan F, Freel Meyers CL. FEBS Journal. 2014;281:2820–2837. doi: 10.1111/febs.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Brien TA, Kluger R, Pike DC, Gennis RB. Biochimica et Biophysica Acta (BBA) - Enzymology. 1980;613:10–17. doi: 10.1016/0005-2744(80)90186-2. [DOI] [PubMed] [Google Scholar]
- 26.Brammer LA, Meyers CF. Org. Lett. 2009;11:4748–4751. doi: 10.1021/ol901961q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang YL, Krosky DJ, Seiple L, Stivers JT. J Am. Chem. Soc. 2005;127:17412–17420. doi: 10.1021/ja055846n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirokane K, Kurosaki Y, Sato T, Chida N. Angewandte Chemie International Edition. 2010;49:6369–6372. doi: 10.1002/anie.201001127. [DOI] [PubMed] [Google Scholar]
- 29.Turbiak AJ, Showalter HDH. Synthesis. 2009;2009:4022–4026. [Google Scholar]
- 30.Schweigert N, Zehnder AJB, Eggen RIL. Environ. Microbiol. 2001;3:81–91. doi: 10.1046/j.1462-2920.2001.00176.x. [DOI] [PubMed] [Google Scholar]
- 31.Saner A, Thoenen H. Molecular Pharmacology. 1971;7:147–154. [PubMed] [Google Scholar]
- 32.Mure M, Klinman JP. J Am. Chem. Soc. 1993;115:7117–7127. [Google Scholar]
- 33.Tse DCS, McCreery RL, Adams RN. J Med. Chem. 1976;19:37–40. doi: 10.1021/jm00223a008. [DOI] [PubMed] [Google Scholar]
- 34.Masini T, Hirsch AKH. J Med. Chem. 2014;57:9740–9763. doi: 10.1021/jm5010978. [DOI] [PubMed] [Google Scholar]
- 35.Smith JM, Warrington NV, Vierling RJ, Kuhn ML, Anderson WF, Koppisch AT, Freel Meyers L C. J Antibiot. 2014;67:77–83. doi: 10.1038/ja.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nemeria NS, Korotchkina LG, Chakraborty S, Patel MS, Jordan F. Bioorg. Chem. 2006;34:362–379. doi: 10.1016/j.bioorg.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao J, Eoh H, He R, Wang Y, Wan B, Franzblau SG, Crick DC, Kozikowski AP. Bioorg. Med. Chem. Lett. 2008;18:5320–5323. doi: 10.1016/j.bmcl.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cornish-Bowden A. FEBS Lett. 1986;203:3–6. doi: 10.1016/0014-5793(86)81424-7. [DOI] [PubMed] [Google Scholar]
- 39.Westley AM, Westley J. Journal of Biological Chemistry. 1996;271:5347–5352. doi: 10.1074/jbc.271.10.5347. [DOI] [PubMed] [Google Scholar]
- 40.Copeland R. Evaluation of enzyme inhibitors in drug discovery: A guide for medicinal chemists and pharmacologists. New York, NY: Wiley-Interscience; 2005. pp. 178–213. [PubMed] [PubMed] [Google Scholar]
- 41.Winans KA, Bertozzi CR. Chem. Biol. 2002;9:113–129. doi: 10.1016/s1074-5521(02)00093-5. [DOI] [PubMed] [Google Scholar]
- 42.Karabatsos GJ, Hsi N. Tetrahedron. 1967;23:1079–1095. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.