Abstract
Achromatopsia (ACHM) is an autosomal recessive disorder characterized by color blindness, photophobia, nystagmus and severely reduced visual acuity. Using homozygosity mapping and whole-exome and candidate gene sequencing, we identified ten families carrying six homozygous and two compound-heterozygous mutations in the ATF6 gene (encoding activating transcription factor 6A), a key regulator of the unfolded protein response (UPR) and cellular endoplasmic reticulum (ER) homeostasis. Patients had evidence of foveal hypoplasia and disruption of the cone photoreceptor layer. The ACHM-associated ATF6 mutations attenuate ATF6 transcriptional activity in response to ER stress. Atf6−/− mice have normal retinal morphology and function at a young age but develop rod and cone dysfunction with increasing age. This new ACHM-related gene suggests a crucial and unexpected role for ATF6A in human foveal development and cone function and adds to the list of genes that, despite ubiquitous expression, when mutated can result in an isolated retinal photoreceptor phenotype.
Achromatopsia (synonymous with rod monochromatism; ACHM2 (MIM 216900), ACHM3 (MIM 262300), ACHM4 (MIM 613856), ACHM5/COD4 (MIM 613093) and ACHM6/RCD3A (MIM 610024)) is an autosomal recessively inherited, genetically heterogeneous disorder characterized by a lack of cone photoreceptor function. Affected individuals present from birth or early infancy with severely reduced visual acuity, nystagmus, marked photophobia, and absent or severely reduced color vision. A less common form of ACHM (incomplete ACHM) is associated with psychophysical and/or electrophysiological evidence of residual cone function resulting in patients having residual color vision and slightly better visual acuity. Although ACHM is a predominantly stationary disorder, limited progression can be seen in a subset of patients1–5. Thus far, five genes encoding components of the cone-specific phototransduction cascade have been implicated in ACHM: GNAT2 (guanine nucleotide–binding protein G(t) subunit α2; ACHM4, MIM 139340)6,7, PDE6C (cone cyclic GMP–specific 3′,5′ cyclic phosphodiesterase α; ACHM5, MIM 600827 and COD4, MIM 613093)8,9, PDE6H (cone cyclic GMP–specific phosphodiesterase γ; MIM 601190; ACHM6 or RCD3A, MIM 610024)10, CNGA3 (cyclic nucleotide–gated cation channel α3; MIM 600053; ACHM2, MIM 216900)11 and CNGB3 (cyclic nucleotide–gated cation channel β3, MIM 605080; ACHM3, MIM 262300)12,13.
We report here the identification of homozygous and compound-heterozygous disease-causing variants in ATF6 (activating transcription factor 6A; MIM 605537) in patients affected by ACHM. ATF6 is a member of the ATF/CREB (cAMP response element–binding protein) family of basic leucine-zipper (bZIP) transcription factors encoded by two paralogs, ATF6 and ATF6B. ATF6A is a key regulator of the UPR pathway necessary to maintain ER function and cellular homeostasis14–18. The results herein imply an unexpected crucial role for ATF6A in cone photoreceptors and foveal development in the human retina and link ATF6 to human disease.
RESULTS
Genetic analysis
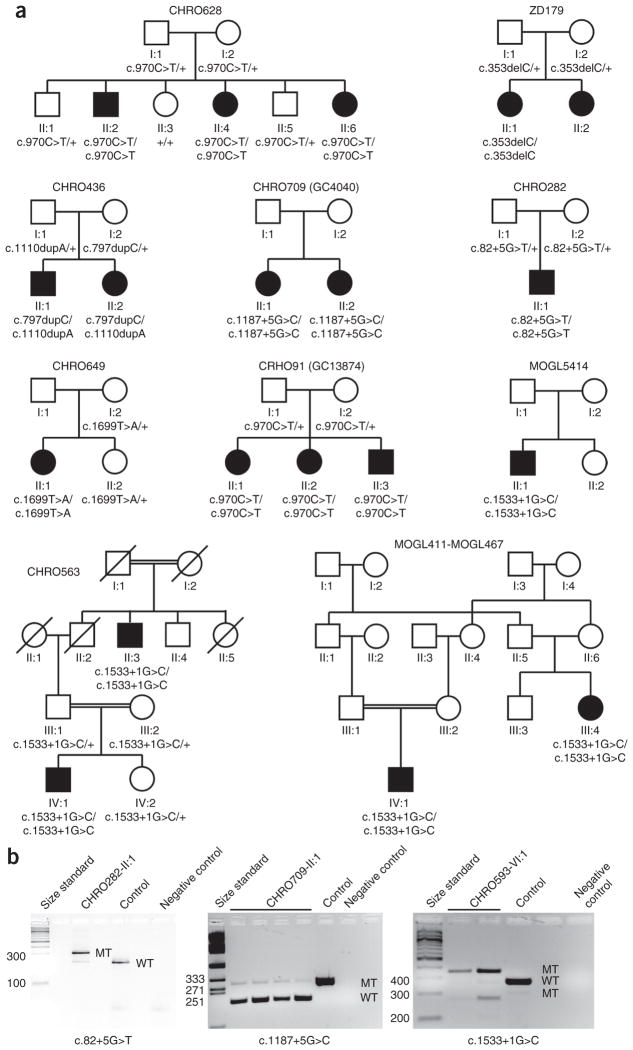
Family CHRO628 is a non-consanguineous family of Irish descent with three children affected by ACHM (Fig. 1a). Haplotype analysis applying the easyLINKAGE package indicated five potentially disease-related regions, with a maximum logarithm of odds (LOD) score of 1.57, on chromosomes 1, 7, 9, 10 and 11. On chromosomes 9 and 10, two genes known to be related to cone photoreceptor disorders, KCNV2 and PDE6C, respectively, were located within the linkage intervals, and disease-causing variants were excluded by Sanger sequencing in patient CHRO628-II:4. To narrow the list of candidate genes and intervals, we performed further genetic testing. We used homozygosity mapping with Homozygosity Mapper software because our previous study on mutations in PDE6H as a rare cause of ACHM had shown that, in such rare disorders, patients tend to harbor private homozygous mutations due to distant identity by descent10. This approach reduced the number of candidate intervals to two—one interval on chromosome 1 and another on chromosome 7—with the interval on chromosome 1p13.2–1q23.3 being the largest, spanning 50.7 Mb. The 1p13.2–1q23.3 homozygosity interval mapped within our previously identified linkage interval and consisted of 624 consecutive SNPs spanning a 2.4-Mb region flanked by SNPs rs4656862 and rs164418 (coordinates (GRCh37 Build 37.1): 159,953,102–162,354,150). To analyze this region, we performed whole-exome sequencing in patient CHRO628-II:4. The single-nucleotide variant (SNV) list from whole-exome sequencing was filtered for the homozygosity interval on chromosome 1, resulting in the identification of a homozygous missense variant, c.970C>T (p.Arg324Cys), in ATF6 that segregated in the family (Fig. 1a and Supplementary Fig. 1). The ATF6 variant altered a highly conserved arginine residue in the bZIP domain necessary for dimerization (Supplementary Fig. 2), which is not only conserved in the ATF protein family but also in the entire activator protein (AP)-1 protein family19 (data not shown).
Figure 1.
Families, ATF6 genotypes and cDNA analysis. (a) Pedigrees of all families with ACHM in this study. The results of ATF6 segregation analysis are depicted under each individual. (b) cDNA analysis of the putative splice-site mutations. RNA was extracted from PaxGene-isolated blood samples for patients as well as from a healthy control sample, reverse transcribed, PCR amplified and Sanger sequenced. In all cases, the splice-site mutations were shown to lead to aberrantly spliced products (MT), but small amounts of correctly spliced cDNA (WT) are also visible. Lanes (from left to right): size standard patient cDNA, healthy control cDNA and no-template negative control. Left, c.82+5G>T in CHRO282; sequencing of the aberrant band showed intron retention of the first 88 bp of intron 1. Middle, c.1187+5G>C in CHRO709; sequencing of the aberrant band showed exon skipping of exon 9, thereby deleting 92 bp from the ORF. Right, c.1533+1G>C in CHRO593; sequencing of the aberrant bands showed that the larger band is due to intron retention of the first 83 bp of intron 12 whereas the smaller band results from exon skipping of exon 12.
Further whole-exome sequencing and candidate gene screening of ATF6 in 301 patients with ACHM identified 15 additional patients from 9 independent families segregating disease-causing variants in ATF6, totaling 18 patients from 10 independent families (Fig. 1a, Table 1, Supplementary Fig. 1 and Supplementary Table 1). Three affected siblings from a UK family (CHRO91; also known as GC13874) were also homozygous for the original missense mutation c.970C>T (encoding a p.Arg324Cys substitution), and haplotype reconstruction from SNP chip data indicated that all carriers of this mutation share a 0.7-Mb common homozygous haplotype flanked by SNPs rs4072409 and rs16840028, suggestive of a founder mutation in the Irish and UK population or identity by descent. Five French-Canadian patients from three independent families (MOGL5414, CHRO593 and MOGL411-MOGL467) carried a homozygous variant (c.1533+1G>C) affecting the consensus splice donor sequence of exon 12 in ATF6. This finding is also suggestive of a founder mutation in the French-Canadian population, but SNP chip data were only available for the affected individuals from family MOGL411-MOGL467, who shared a 5.4-Mb homozygous region encompassing ATF6. Sanger sequencing of all coding exons and flanking intronic sequences and comparison of the sequences for all patients carrying the homozygous c.1533+1G>C allele showed that all had the same homozygous SNP genotype, at least for the ATF6 gene, as documented on the basis of homozygosity of two rare SNPs, rs371893818 and rs374093774. Two German siblings were compound heterozygous with a 1-bp duplication on each allele of ATF6 (c.797dupC; p.Asn267* and c.1110dupA; p.Val371Serfs*3). The remaining patients harbored other, private homozygous disease-causing variants, including another homozygous missense mutation, c.1699T>A (p.Tyr567Asn), in an Iranian achromat mapping to a C-terminal amino acid sequence motif that is conserved in ATF6A and ATF6B (Supplementary Figs. 1 and 2); a homozygous 1-bp deletion, c.353delC, in a Turkish achromat resulting in a frameshift after Pro118 (p.Pro118Leufs*31); and two further homozygous variants affecting canonical splice donor sites: c.82+5G>T in a patient from South Tyrol (Italy) and c.1187+5G>C in two Asian-Indian achromat sisters. In total, we identified eight different disease-causing variants (Table 1, Supplementary Fig. 1 and Supplementary Table 1). The putative disease-causing variants were not observed in our in-house databases, dbSNP or the Exome Variant Server (EVS). The c.970C>T allele was observed to be heterozygous in 3 of 120,904 genotypes called by the ExAC browser. The missense mutations were predicted to be disease causing by various prediction software packages (for example, MutationTaster, SIFT and PolyPhen).
Table 1.
Summary of patients’ genotypes and demographics
| Patient ID | ATF6 mutation, nucleotide level | ATF6 alteration, protein level or consequence | Ancestry, age, sex |
|---|---|---|---|
| CHRO282-II:1 | c.82+5G>T homozygous | Splice defect (p.Asp28Glyfs*36)a | South Tyrolean, 42, M |
| CHRO628-II:2 | c.970C>T homozygous | p.Arg324Cys | Irish, 19, M |
| CHRO628-II:4 | c.970C>T homozygous | p.Arg324Cys | Irish, 16, F |
| CHRO628-II:6 | c.970C>T homozygous | p.Arg324Cys | Irish, 9, F |
| CHRO91-II:1 | c.970C>T homozygous | p.Arg324Cys | British, 47, F |
| CHRO91-II:2 | c.970C>T homozygous | p.Arg324Cys | British, 45, F |
| CHRO91-II:3 | c.970C>T homozygous | p.Arg324Cys | British, 43, M |
| CHRO709-II:1 | c.1187+5G>C homozygous | Splice defect (p.Asn366Hisfs*12)a | Asian Indian, 27, F |
| CHRO709-II:2 | c.1187+5G>C homozygous | Splice defect (p.Asn366Hisfs*12)a | Asian Indian, 23, F |
| CHRO593-IV:1 | c.1533+1G>C homozygous | Splice defect (p.Gly512Leufs*39 and p.Gly512Valfs*11)a | French Canadian, 17, M |
| CHRO593-II:3 | c.1533+1G>C homozygous | Splice defect (p.Gly512Leufs*39 and p.Gly512Valfs*11)a | French Canadian, 94, M |
| MOGL411-MOGL467-III:4 | c.1533+1G>C homozygous | Splice defect (p.Gly512Leufs*39 and p.Gly512Valfs*11)a | French Canadian, 59, F |
| MOGL411-MOGL467-IV:1 | c.1533+1G>C homozygous | Splice defect (p.Gly512Leufs*39 and p.Gly512Valfs*11)a | French Canadian, 25, M |
| MOGL5414-II:1 | c.1533+1G>C homozygous | Splice defect (p.Gly512Leufs*39 and p.Gly512Valfs*11)a | French Canadian, 32, M |
| CHRO649-II:1 | c.1699T>A homozygous | p.Tyr567Asn | Iranian, 26, F |
| ZD179-II:1 | c.353delC homozygous | p.Pro118Leufs*31 | Turkish, 41, F |
| CHRO436-II:1 | c.797dupC/c.1110dupA compound heterozygous | p.Asn267*/p.Val371Serfs*3 | German, 22, M |
| CHRO436-II:2 | c.797dupC/c.1110dupA compound heterozygous | p.Asn267*/p.Val371Serfs*3 | German, 17, F |
Ages are shown in years. M, male; F, female.
Confirmed by cDNA analysis.
We directly assessed the effect of variants putatively affecting consensus splice-site sequences by extracting RNA from PaxGene-isolated blood samples for the patients carrying the putative disease-causing variant, synthesizing the corresponding cDNA, and evaluating the products on agarose gels and by Sanger sequencing. All three variants affecting the consensus donor splice site were shown to result in missplicing, frameshift and premature termination codons (PTCs) (Fig. 1b and Table 1). Consequently, the mutant proteins are expected to be severely truncated, or—more likely—the transcripts undergo nonsense-mediated decay. Yet, evaluation on agarose gels provided evidence that small amounts of correctly spliced ATF6 mRNA are expressed, at least in blood lymphocytes (Fig. 1b). The disease-causing variant c.82+5G>T led to exonification (intron retention) of 88 bp of intron 1, which thereby resulted in a frameshift and subsequent PTC (p.Asp28Glyfs*36) (Supplementary Table 1). The disease-causing variant c.1187+5G>C in turn caused exon skipping of exon 9 (92 bp), also resulting in a frameshift and PTC (p.Asn366Hisfs*12) (Supplementary Table 1). The disease-causing variant c.1533+1G>C showed two aberrant spliced bands: the larger band corresponded to exonification (intron retention) of 83 bp of intron 12, leading to a PTC (p.Gly512Leufs*39), and the smaller band corresponded to exon skipping of exon 12, which is 100 bp in size, also resulting in a PTC (p.Gly512Valfs*11) (Supplementary Table 1).
Clinical phenotype
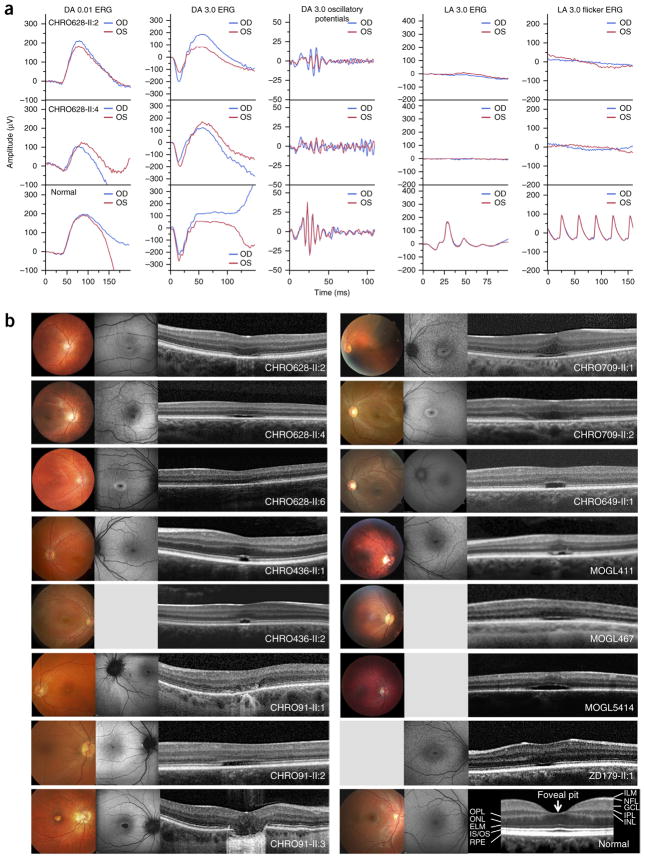
The clinical data for the 18 patients are summarized in Supplementary Table 1. All patients had a clinical diagnosis of complete (n = 16) or incomplete (n = 2) ACHM based on psychophysical and/or electrophysiological findings. Nystagmus and photophobia were present in all from birth or early infancy. All patients had very poor or absent color vision and markedly reduced visual acuity. There was no evidence of progression over time except in one patient (CHRO649-II:1). Best corrected visual acuity was severely reduced (range of 20/63 to 20/200), and refractive error ranged from −9.5 to +6.0 diopters (spherical), with variable degrees of astigmatism. Patients who underwent visual field testing (10/13) showed bilateral central scotomas. Dark adaptation was evaluated in two patients (CHRO628-II:2 and CHRO628-II:4), who showed rod-mediated function lacking the rod-cone break and cone branch, with dark-adaptation thresholds being within normal limits. Scotopic full-field electroretinography (ERG) responses were within normal limits, consistent with normal rod function, whereas photopic full-field ERG responses were either severely reduced or not detectable, in keeping with absent or markedly reduced cone system function (Fig. 2a).
Figure 2.
Functional and morphological presentation of patients carrying mutations in ATF6. (a) International Society for Clinical Electrophysiology of Vision (ISCEV) standard ERG recordings for CHRO628-II:2 (top), CHRO628-II:4 (middle) and a healthy control individual (bottom) showing intact rod function under scotopic conditions but no reproducible cone responses under photopic conditions in patients. The flash strength (in cd s m−2) and state of adaptation are given for each standard step. DA, dark adapted; LA, light adapted; blue, right eye (OD); red, left eye (OS). (b) Morphological results (color photography (left), FAF (middle) and SD-OCT (right) images) for 15 patients. Funduscopic examinations identified small, well-defined, oval-shaped RPE defects with pathological wall and foveal reflexes in each case. Defects were seen in the FAF images as parafoveal hyperfluorescent areas in the younger cases and hypofluorescent spots surrounded by a ring with increased autofluorescence in older cases. SD-OCT images showed loss of cone inner and outer segments with interruption of the ciliary layer and disruption of the RPE layer in the foveal area. All patients presented with almost missing foveal pits, which may be characteristic of the retinal disorder caused by ATF6 mutations. Pictures were not available for the patients from families CHRO593 and CHRO282. Color photography, FAF and SD-OCT images for a healthy control individual, together with labeling of the retinal layers in the SD-OCT image, are provided at the bottom right. ELM, external limiting membrane; GCL, ganglion cell layer; ILM, internal limiting membrane; INL, inner nuclear layer; IPL, inner plexiform layer; IS/OS, photoreceptor inner and outer segments; NFL, nerve fiber layer; ONL, outer nuclear layer; OPL, outer plexiform layer; RPE, retinal pigment epithelium.
Findings from anterior segment examination were normal. Fundus examination and color fundus photography showed healthy optic discs, intact retinal vasculature and normal peripheral retina. The macula showed a range of changes from relatively normal areas to well-demarcated sites of atrophy that did not appear to be related to age (Fig. 2b and Supplementary Table 1). Fundus autofluorescence (FAF) imaging showed variable changes, including parafoveal areas or rings of increased autofluorescence in younger cases and areas of reduced autofluorescence corresponding to atrophy seen clinically surrounded by a ring of increased signal in some subjects (Fig. 2b). Spectral domain–optical coherence tomography (SD-OCT) scans showed foveal hypoplasia with a missing foveal pit in all patients (Fig. 2b and Supplementary Table 1). In keeping with previous detailed reports of retinal integrity in ACHM, SD-OCT scans showed variable disruption of the inner segment ellipsoid (ISe) layer (photoreceptor layer) at the fovea1,2 (Fig. 2b and Supplementary Table 1).
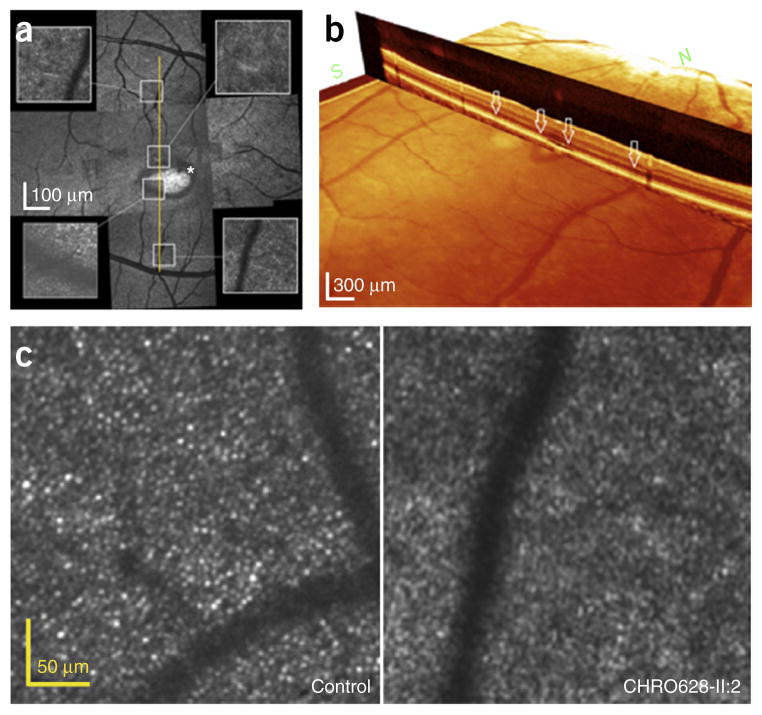
Although SD-OCT allows retinal lamination to be identified, adaptive optics scanning light ophthalmoscopy (AOSLO) allows direct visualization of cone photoreceptors. Therefore, AOSLO was performed to assess cone photoreceptor mosaic integrity in siblings of the affected individual from family CHRO628 (Fig. 3). Instead of a complete and continuous cone photoreceptor mosaic, we observed dark areas devoid of wave-guiding cones and highly reflective regions. Comparison of the AOSLO image with the SD-OCT scan showed that the dark areas observed in AOSLO correlated with absence of an ISe layer in SD-OCT, and hyper-reflective regions correlated with dome-shaped hyporeflective deposits in SD-OCT (Fig. 3a). AOSLO montage images showed the obvious ring sharp and dark area with missing or invisible cone cells, which correlated with the absence of an inner segment–outer segment junction in SD-OCT (Fig. 3b). The average cone density (18,142.25/mm2) of a 0.5-mm eccentricity area was considerably lower than for the normal group (32,554 ± 2,884/mm2). The average cone density of a 1.0-mm eccentricity area (14,997.5/mm2) was also lower than normal (19,802 ± 1,460/mm2). In contrast, the average cone density of a 1.5-mm eccentricity area (14,302.75/mm2) was higher than in the normal group (11,249 ± 1,967/mm2). According to these results, only cones in the macular area and part of the paramacular area were affected.
Figure 3.
Montages of AOSLO imaging of individual cones from the right eye of patient CHRO628-II:2. (a) Instead of a complete and continuous photoreceptor mosaic, dark areas devoid of normal wave-guiding cones and highly reflective regions are observed. There are some dark areas where cone cells are absent or damaged. AOSLO images from the left eye are superimposed on SD-OCT images (yellow line). Within this scanning area, grossly continuous and normal cone mosaic patterns are observed in some areas of the AOSLO images, but dark areas can be seen as well. Comparison of the AOSLO image with the SD-OCT scan (yellow line) shows that the dark areas that can be observed on the AOSLO image correlate with the absence of an ellipsoid zone and hyper-reflective regions, consistent with the ‘optical gap’ seen in the SD-OCT images. An asterisk marks dark areas where cone cells are absent or damaged. (b) SD-OCT imaging showed the loss of outer segments in foveal cones in the optical gap. White arrows correspond to the regions sampled for AOSLO images. (c) Only macular cones are affected in the right eye of patient CHRO628-II:2. In comparison to age-matched normal data, the average cone density is considerably lower.
Hearing loss was observed in four patients. Patient CHRO649-II:1, homozygous for the mutation encoding p.Tyr567Asn, had bilateral sensorineural hearing loss diagnosed at 1 year of age, hypothyroidism since childhood and celiac disease. However, her elder sister (CHRO649-II:2), who also had bilateral early-onset sensorineural hearing loss, had a normal eye examination and is heterozygous for the mutation in ATF6 encoding p.Tyr567Asn. All three siblings of family CHRO91 (GC13874), homozygous for the mutation encoding p.Arg324Cys, presented with sensorineural hearing loss with age of onset between 25 and 35 years. All other patients with ACHM were otherwise healthy, with the youngest patient being 8 years old and the oldest 94 years old. The possibility cannot be excluded that the finding of ACHM and sensorineural hearing loss in these individuals may be related to the ATF6 mutations, yet the situation in family CHRO649 suggests that this co-occurrence is a coincidence.
Functional studies on patient fibroblasts
Human ATF6 disease-causing variant causes loss of ATF6 transcriptional activity
To determine how human ATF6 disease-causing variants affected ATF6A function, we collected fibroblasts from individuals in family CHRO628 homo- or heterozygously carrying the ATF6 variant encoding p.Arg324Cys. We exposed fibroblasts from the heterozygous mother (CHRO628-I:2) and the three homozygous, affected children (CHRO628-II:2, CHRO628-II:4 and CHRO628-II:6) in medium to tunicamycin, an inhibitor of N-linked glycosylation that causes protein misfolding in the ER and potently activates ATF6A. As HSPA5 (encoding GRP98; also known as BiP) is one of the most highly induced transcriptional targets of ATF6A15,16, we tested the effect of the variant on transcriptional activation. All of the patient-derived fibroblasts homozygous for the ATF6 mutation encoding p.Arg324Cys showed significantly reduced HSPA5 mRNA and GRP78 (BiP) protein induction after tunicamycin exposure in comparison to fibroblasts derived from the heterozygous carrier mother (Fig. 4a,b). Analysis of additional ATF6A downstream transcriptional targets, including HERPUD1, SEL1L and EDEM1, also showed significantly reduced mRNA induction for these genes after tunicamycin exposure in all patient-derived fibroblasts homozygous for the ATF6 mutation encoding p.Arg324Cys (Fig. 4b). These findings suggested that the fibroblasts with homozygous ATF6 mutation were defective in ATF6 transcriptional activity. To rule out the possibility that transcriptional differences arose from genetic variation between the different fibroblast cell lines, we used a system in which wild-type ATF6A or Arg324Cys ATF6A was fused to the dihydrofolate reductase (DHFR) protein to create fusion ATF6A transcription factors whose activities could be specifically regulated by the addition of trimethoprim. This strategy has previously been shown to enable potent activation of ATF6A transcription independently of ER stress and activation of other UPR signaling pathways20. We transfected HEK293 cells with construct for wild-type or Arg324Cys ATF6 fusion proteins and confirmed expression of the human DHFR-ATF6A fusion proteins via immunoblotting (Fig. 4c). After addition of trimethoprim, HSPA5 mRNA and GRP78 (BiP) protein levels were significantly induced in cells expressing DHFR fused to wild-type ATF6A (Fig. 4c). By contrast, HSPA5 mRNA and GRP78 (BiP) protein levels were not induced in cells expressing DHFR fused to the Arg324Cys ATF6A mutant (Fig. 4c). Taken together, our studies demonstrate that the ATF6 mutation encoding p.Arg324Cys compromises ATF6A transcriptional activity.
Figure 4.
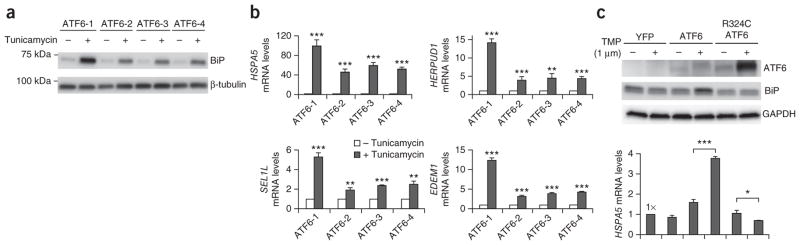
Functional analysis in human fibroblasts and HEK293 cells shows defective transcriptional activity for the Arg324Cys ATF6A mutant. (a,b) Fibroblasts from a heterozygous carrier of the ATF6 mutation encoding p.Arg324Cys (mother; ATF6-1) and children homozygous for the mutation (ATF6-2, ATF6-3 and ATF6-4) were challenged with tunicamycin (5 μg/ml) for 24 h. (a) Cells were collected and lysed. BiP protein levels were detected by immunoblotting. β-tubulin protein levels served as a loading control. Representative blots of the three independent experiments are shown. (b) HSPA5 (encoding GRP78 (BiP)), HERPUD1, SEL1L and EDEM1 mRNA levels were measured by quantitative PCR (qPCR) and are shown relative to the mRNA levels in untreated cells. Data are presented as means ± s.d. from three independent experiments. (c) HEK293 cells were transfected with constructs for DHFR fused to YFP, wild-type ATF6A or Arg324Cys ATF6A, and trimethoprim (TMP) was added as indicated. After 24 h, cell lysates were probed for ATF6A and BiP protein by immunoblotting. GAPDH protein levels were used as a loading control. Representative blots of the three independent experiments are shown. HSPA5 (encoding GRP78) mRNA levels were also measured by qPCR and are shown relative to levels in cells transfected with YFP construct. Data are presented as means ± s.d. from three independent experiments. For b and c, a P value of <0.05 was considered significant (*P < 0.05, **P < 0.01, ***P < 0.001, Student’s two-tailed t test).
Young Atf6−/− mouse retinas appear normal
We investigated whether retinal protein expression was altered in 3-month-old Atf6−/− homozygous-knockout, Atf6+/− heterozygous and Atf6+/+ wild-type mouse littermates. In retinas collected from these mice under ambient conditions, there was no obvious difference in the steady-state expression levels of the cone-specific M-opsin, S-opsin and glycogen phosphorylase proteins (Supplementary Fig. 3b). Similarly, there were no differences in expression of rod-specific proteins, including rhodopsin (Rho) and rod α-transducin (Gαt1; encoded by Gnat1) (Supplementary Fig. 3c). Lastly, we observed no significant upregulation of ER stress–induced proteins, including PDI, calreticulin and BiP, among Atf6−/−, Atf6+/− and Atf6+/+ littermates (Supplementary Fig. 3d). Taken together, our findings indicate that cones are present and functional in 3-month-old Atf6−/− mice, and that there are no significant differences in protein levels among Atf6−/−, Atf6+/− and Atf6+/+ littermates.
Retinal alterations in aged Atf6−/− mice
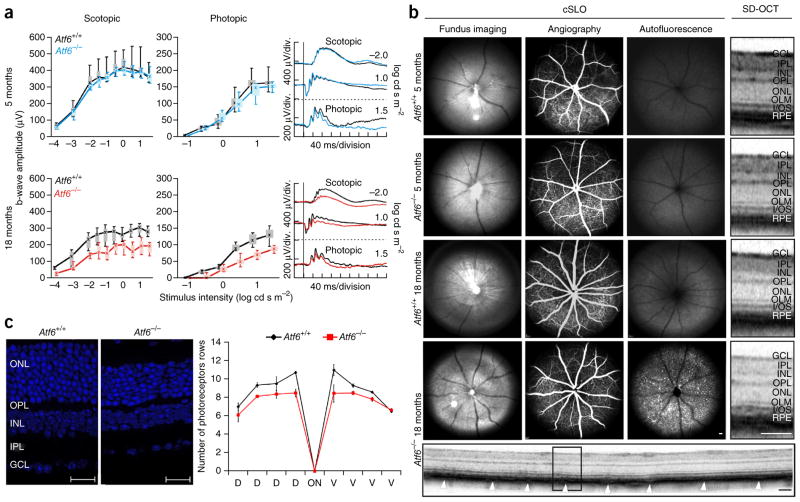
We performed functional studies based on ERG of Atf6−/− mice and respective Atf6+/+ controls at the ages of 3, 5 and 18 months. Full-field ERG measurements allowed the assessment of both dark-adapted (scotopic), rod-dominated responses and light-adapted (photopic), cone-driven responses21. ERG recordings of younger Atf6−/− mice showed normal rod and cone activity, when compared to Atf6+/+ control littermates (Fig. 5a, top). However, functional analysis of 18-month-old Atf6−/− mice and corresponding Atf6+/+ controls showed that both rod and cone single-flash ERG responses were markedly reduced in Atf6−/− mice (Fig. 5a, bottom). In contrast to human ACHM, the functional deficits in mice show that the rod and cone systems are affected only at an older age.
Figure 5.
Functional and morphological presentation of Atf6−/− and Atf6+/+ mice. (a) Quantitative evaluation of scotopic and photopic b-wave amplitude data, shown as box-and-whiskers plots (boxes, 25–75% quantile range; whiskers, 5% and 95% quantiles; asterisks, medians), for 5-month-old (blue) and 18-month-old (red) Atf6−/− mice and for age-matched Atf6+/+ mice (black). Selected ERG traces for each group are shown to the right. Lack of Atf6a impairs retinal function in aged but not younger mice (5 months: Atf6−/−, n = 4; Atf6+/+, n = 3; 18 months: Atf6−/−, n = 3; Atf6+/+, n = 4). (b) Retinal structure of Atf6−/− and Atf6+/+ mice. Fundus autofluorescence imaging showed hyperfluorescent spots in older Atf6−/− mice in comparison to age-matched controls, indicating photoreceptor degeneration. SD-OCT analysis showed a disruption of layers corresponding to the inner segment–outer segment junction (SD-OCT magnification) and the RPE (arrowheads). Scale bars: cSLO, 200 μm; SD-OCT magnification, 100 μm; SD-OCT scan, 100 μm. (c) Retinal nuclear staining of 18-month-old ATF6+/+ (left) and ATF6−/− (middle) mice (scale bars, 20 μm). The spider diagram displays the number of photoreceptor nuclei rows in the outer nuclear layer (right). Preliminary statistics suggest a reduced number of rows in the central retina, indicating a mild degenerative process. Data are presented as means ± s.e.m. (Atf6−/−, n = 3; Atf6+/+, n = 2). GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; OLM, outer limiting membrane; I/OS, inner/outer segment; RPE, retinal pigment epithelium; D, dorsal; V, ventral.
We assessed retinal morphology in Atf6−/− and corresponding control mice at 3, 5 and 18 months of age by in vivo confocal scanning-laser ophthalmoscopy (cSLO) and SD-OCT imaging. Fundus appearance, vasculature and retinal layering in mutant mice did not show any differences in comparison to controls at 3 and 5 months of age (Fig. 5b). However, for mice at 18 months of age, we could observe retinal degeneration (Fig. 5b). Fundus imaging showed spot-like disturbances, which could be correlated to hyperfluorescent spots detected in autofluorescence cSLO imaging mode (Fig. 5b). Degeneration of photoreceptors results in the accumulation of autofluorescent material, which can be detected in cSLO imaging22,23; thus, spot-like enhanced autofluorescence clearly indicates ongoing retinal degeneration. As observed in patients, retinal vasculature also appeared unaffected in the mouse model (Fig. 5b, angiography). Further in vivo analysis of retinal layering by SD-OCT imaging showed a disruption of the layers corresponding to the inner and outer segments (Fig. 5b), which could also be observed in the human clinical phenotype; however, in mouse retinas, the retinal pigment epithelium (RPE) layer was also affected (Fig. 5b, SD-OCT magnification, arrowheads in the SD-OCT full scan).
All these findings suggest that there is a profound difference in the function and impact of Atf6a in mouse and human retinal development and maintenance. Foveas are only found in primates, birds and in one lizard species. Perhaps this explains why ATF6A does not have a role in mouse retinal development, as mice do not develop foveas.
To determine whether cones were lost in Atf6−/− mice at 12 months of age, we analyzed retina flat-mounts from heterozygous Atf6+/− and homozygous-knockout Atf6−/− mice and labeled cones with peanut agglutinin (PNA) (Supplementary Fig. 3a). We found no gross difference in the number of PNA-labeled cells between Atf6−/− and Atf6+/− littermates at 12 months of age.
Nuclear staining on retinal sections of the same 18-month-old knockout mice and wild-type controls previously assessed electrophysiologically and morphologically in vivo showed a median of 10 rows of photoreceptors in wild-type mice and 7–8 rows of nuclei in the outer nuclear layer in Atf6−/− homozygous-knockout mice (Fig. 5c). Such a reduction is suggestive of a potential mild degenerative process.
DISCUSSION
Mutations in ATF6 are a rare cause of ACHM. Extrapolating the numbers from of our patient cohort, we conclude that about 1% of European-ancestry individuals with ACHM carry mutations in this gene. Patients presented with findings consistent with either autosomal recessive complete or incomplete ACHM. Although a clear genotype-phenotype correlation could not be observed, interestingly, all our ATF6-associated patients with ACHM had marked foveal hypoplasia, including persistent inner retinal layers at the fovea and a poorly formed or absent foveal pit. Foveal hypoplasia can also be present in ACHM caused by mutations in phototransduction-associated genes (up to 70–80% of cases, depending on the definition of foveal hypoplasia)1–5, but the absence of foveal development is rare. As we found this phenotype in all 18 patients from 10 independent families, we hypothesize that severe foveal hypoplasia with a poorly formed or absent foveal pit may be a hallmark of ATF6-related disease that distinguishes it from other forms of ACHM, where the pathogenesis relates to abnormalities of the cone phototransduction pathway. It is evident that ATF6 does not primarily affect cone phototransduction but has a key role in foveal development, alongside a generalized effect on cone function. This observation is further corroborated by the adaptive optics images of individual cones showing that only central foveal cones are missing, whereas peripheral cones may be present. This finding is consistent with a foveal developmental defect in individuals with ATF6-associated ACHM. The counts of cones outside of the central 1.5-mm area in both children examined are supranormal, but the cones are non-functional, as ERGs are extinguished. The presence of peripheral cones suggests that ATF6-associated ACHM may be amenable to gene therapy, as has also been shown in animal models for ACHM caused by mutations in GNAT2, CNGA3 and CNGB3 (refs. 24–27), and clinical phase I and II safety studies are in preparation for the latter two.
ATF6 encodes a ubiquitously expressed 90-kDa ER stress–regulated transmembrane transcription factor, required for ER stress response and transcriptional induction from ER stress–response elements (ERSEs) in the promoter regions of genes encoding ER protein chaperones, ER-associated protein degradation and ER protein trafficking14,15,24,28. Upon induction of ER stress, the cytosolic ~400-residue N-terminal portion of ATF6A (N-ATF6Ap50) is released as a result of regulated intramembraneous proteolysis16,29. N-ATF6Ap50 possesses a transcriptional activation domain, a bZIP domain, a DNA-binding domain and nuclear localization signals (Fig. 6). N-ATF6Ap50 then translocates to the nucleus, where it interacts with several other proteins to form an ERSE-binding complex that is responsible for the induction of ER stress genes (ERSGs), including the ER chaperone HSPA5 (encoding glucose-regulated protein, 78 kDa (GRP78); also known as BiP)16,29,30. N-ATF6Ap50 is rapidly degraded in a proteasome-mediated process31. Thus, similar to several other potent transcription factors that exert rapid, transient effects32, the degradation of ATF6A upon transcriptional engagement apparently serves as a mechanism to rapidly turn off ERSG induction33.
Figure 6.
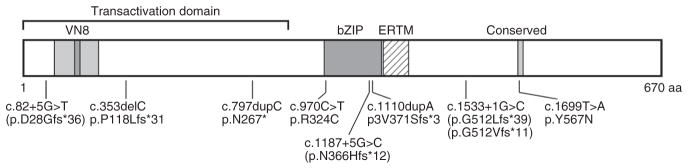
Topography of ATF6A and the location of the disease-causing variants identified in patients with ACHM. The diagram depicts the transactivation domain, including the VN8 domain, the bZIP domain and an ERTM domain39. In the C-terminal region, a domain of unknown function that is conserved between ATF6A and ATF6B is also indicated. VN8, eight-residue domain homologous to herpes simplex viral protein VP16 necessary for optimal transcriptional activity and degradation31; bZIP, basic leucine-zipper domain; ERTM,
Another member of the ATF/CREB family of transcription factors highly homologous to ATF6A—ATF6B—is also an ER membrane protein that is cleaved during ER stress to generate an N-terminal fragment of ~400 residues34. N-ATF6A and N-ATF6B possess highly conserved bZIP domains and DNA-binding domains. This conservation allows N-ATF6A and N-ATF6B to bind to ERSEs as homo- or heterodimers35; however, the functional relevance of the binding of N-ATF6B to ERSEs, either alone or as a heterodimer with N-ATF6A, is currently unknown. In addition, these proteins share an 18-residue motif of unknown function in the C terminus (Supplementary Fig. 2). Whole-body deletion of either Atf6 or Atf6b in mouse does not result in markedly aberrant phenotypes in the absence of ER stress, but deletion of both genes results in an early embryonic lethal phenotype, indicating that these genes provide overlapping functions early in development28,36. In the presence of ER stress, Atf6−/− mice cannot activate adaptive UPR transcription, whereas Atf6b−/− mice can adapt to ER stress28,36–38. Thus, ATF6A and not ATF6B is the major mechanism through which UPR gene expression is activated and promotes the survival of cells with ER stress. Functional and morphological analysis in vivo of the corresponding gene-deletion mouse model showed a phenotype affecting both the rod and cone photoreceptor systems at older ages (Fig. 5).
The missense substitution p.Arg324Cys found in this study localizes to the basic region of the bZIP domain, affecting an arginine residue that is not only conserved among transcription factors of the ATF family but also in those of the AP-1 family (Fig. 6 and Supplementary Fig. 2). The impact on UPR in cells expressing this disease-causing variant was tested in fibroblasts from family CHRO628 (Fig. 4). Fibroblasts homozygous for the ATF6 mutation encoding p.Arg324Cys showed substantially reduced HSPA5 mRNA and GRP78 (BiP) protein induction after tunicamycin exposure in comparison to the fibroblasts heterozygous for the mutation. The heterologous expression of ATF6A Arg324Cys fused to DHFR protein in HEK293 cells resulted in complete loss of HSPA5 mRNA and GRP78 (BiP) protein, whereas there was significant induction by wild-type ATF6A fused to DHFR in the control experiment. Both results indicate that the ATF6 disease-causing variant encoding p.Arg324Cys severely impairs ATF6 transcriptional activity.
The second missense substitution, p.Tyr567Asn, alters a tyrosine residue in the C-terminal part of ATF6A. This residue is completely conserved among all known ATF6 homologs down to zebrafish and is located and conserved in an 18-residue sequence motif of unknown function that is present in both ATF6A and ATF6B (Fig. 6 and Supplementary Fig. 2).
The disease-causing variants affecting consensus splice-site sequences, as well as the 1-bp deletion c.353delC and the 1-bp duplications c.797dupC and c.1110dupA, were all shown to result in frameshift and PTC (Figs. 1b and 6, and Table 1). Most likely, the corresponding aberrant transcripts will undergo nonsense-mediated decay. If translated, all transcripts would lead to considerably truncated ATF6 polypeptides. However, cDNA analysis of the disease-causing splice-site variants also showed that, in the analyzed whole-blood samples derived from patients, small amounts of correctly spliced ATF6 transcripts could be amplified, suggesting that these patients may also produce small amounts of wild-type ATF6A polypeptide (Fig. 1b).
Our study identified ATF6 as an additional gene implicated in ACHM and associated with foveal hypoplasia. It links ATF6 to human disease and highlights the finding that the ubiquitously expressed ATF6, known for its function in UPR, has an important role in the development and function of the fovea and cone photoreceptors.
URLs
HomozygosityMapper, http://www.homozygositymapper.org/; dbSNP, http://www.ncbi.nlm.nih.gov/SNP/; National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/; Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/; SIFT, http://sift.jcvi.org/; PolyPhen-2, http://genetics.bwh. harvard.edu/pph2/; MutationTaster, http://www.mutationtaster.org/; ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/; HomoloGene, http://www.ncbi.nlm.nih.gov/homologene.
ONLINE METHODS
Subjects and clinical examination
Study participants were recruited over 10 years from international collaborating centers specialized in inherited retinal diseases. Blood or DNA samples were sent to Tübingen, Germany, for genetic investigation. This and all our studies were performed according to the tenets of the Declaration of Helsinki, and all participants gave written consent, approved by the respective local research and ethical review boards.
Patients underwent comprehensive ophthalmological examination including, depending on the center, psychophysical testing (best-corrected visual acuity, color vision, visual field testing (n = 2) and dark adaptation (n = 2)), electrophysiological assessment (including full-field and multifocal ERG) and retinal imaging (including color fundus photography, FAF and SD-OCT). Color vision was assessed with various tests including Ishihara plates, Lanthony’s Tritan album (LTA), American Optical Hand-Hardy-Rittler (AOHRR), the saturated Roth 28-Hue test, Farnsworth Munsell 100-Hue tests and the Nagel anomaloscope. Dark-adaptation thresholds, full-field ERG and multifocal ERG recordings were measured in accordance with ISCEV recommendations40,41. AOSLO was performed in patient CHRO628-II:2 as described previously42–45.
Molecular genetic analysis
DNA was isolated from peripheral blood according to standard procedures in the different centers and banked at the Institute for Ophthalmic Research (Tübingen, Germany) according to standard protocol.
Linkage analysis and homozygosity mapping
For one family (CHRO628), DNA for all family members was genotyped using Affymetrix GeneChip Human Mapping 250K NspI SNP arrays. Linkage analysis was performed by applying the easyLINKAGE software package46, using Allegro v1.2c and assuming autosomal recessive inheritance in the family (Fig. 1a). Homozygosity mapping was performed by applying Homozygosity mapper software47.
Exome sequencing
Whole-exome sequencing of DNA from CHRO628-II:4 was performed by a commercial provider (CeGaT), applying the Agilent insolution SureSelect All Exon kit (Agilent Technologies) run on a SOLiD 4 system (Applied Biosystems). All reads were mapped to the human reference genome (GRCh37/hg19), applying the Bioscope and Lifescope packages (Applied Biosystems). SNV calling was implemented with a Frequentist algorithm on high-coverage positions or with a Bayesian algorithm. In addition, the Lifescope and Bioscope programs both supported indel calling. Diseasecausing variants will be submitted to ClinVar. Exome data can be provided upon request within a scientific cooperation.
Candidate gene screening by Sanger sequencing
All coding exons and flanking intronic and UTR sequences of ATF6 (RefSeq, NM_007348.3) were PCR amplified from genomic DNA (purified by treatment with ExoSAP-IT (GE Healthcare)) and sequenced with dye-termination chemistry (BigDye Terminator version 1.1, Applied Biosystems). For primer sequences and PCR conditions, see Supplementary Table 2. Sequences were run on a capillary sequencer (ABI 3100, Applied Biosystems) and analyzed with Sequence Analysis software (version 5.1, Applied Biosystems) and sequence trace alignment software (SeqMan, DNASTAR). Segregation analysis was performed by Sanger sequencing.
Evaluation of variants in consensus splice-site sequences
Total RNA was extracted from PaxGene blood tubes (Qiagen) for patients CHRO282, CHRO593 and CHRO709 (GC4040) according to the manufacturer’s recommendations and reverse transcribed to test for missplicing caused by the following variants in canonical splice-site sequences: c.82+5G>T, c.1187+5G>C and c.1533+1G>C. Exonic ATF6-specific primers located in exon 1 and exon 4, exon 8 and exon 10, and exon 11 and exon 13, respectively, were used for amplification of wild-type versus mutant cDNA. PCR fragments were electrophoretically separated on 2% agarose gels, visualized by ethidium bromide staining and evaluated. PCR products were then purified, Sanger sequenced and analyzed as described above.
Atf6 mouse model
In this study, male and female Atf6+/+, Atf6+/− and Atf6−/− mice on a pure C57BL/6J background were used as described previously28. All mouse care and procedures were conducted according to protocols and guidelines approved by the Sanford-Burnham Medical Research Institute Institutional Animal Care and Use Committee as well as the Tübingen Institutional Animal Care and Use Committee. Sample size was not predetermined.
Morphological and functional assessment in Atf6−/− mice
Electroretinography in Atf6−/− knockout mice
ERGs were recorded binocularly from Atf6−/− mice (n = 3) in comparison to control littermates (n = 3) at the ages of 3, 5 and 18 months postnatally as described previously21. Mice were anaesthetized using a combination of ketamine (66.7 mg/kg body weight) and xylazine (11.7 mg/kg body weight). Their pupils were dilated, and single-flash ERG responses were obtained under scotopic (dark adapted overnight) and photopic (light adapted with a background illumination of 30 cd/m2 starting 10 min before recording) conditions. Single white-flash stimuli ranged from −4 to 1.5 log cd s m−2 under scotopic conditions and from −2 to 1.5 log cd s m−2 under photopic conditions. Ten responses were averaged with interstimulus intervals of 5 s (for −4 to 0.5 log cd s m−2) or 17 s (for 0 to 1.5 log cd s m−2).
Confocal scanning-laser ophthalmoscopy
The retinal structures of the anesthetized mice were visualized via cSLO imaging with HRA1 and HRA2 systems (Heidelberg Engineering) according to previously described procedures22. Briefly, HRA1 and HRA2 systems feature lasers in the short (visible) wavelength range (488 nm for both and 514 nm for HRA1 only) and also in the long (infrared) wavelength range (795/830 nm and 785/815 nm, respectively). The 488- and 795-nm lasers are used for fluorescein (FLA) and indocyanine green (ICG) angiography, respectively.
Spectral domain–optical coherence tomography
SD-OCT imaging was performed in the same session as cSLO, and it was carried out with a Spectralis HRA+OCT instrument (Heidelberg Engineering). This device features a superluminescent diode at 870 nm as a low-coherence light source. Scans are acquired at a speed of 40,000 scans per second, and each two-dimensional B scan contains up to 1,536 A scans48. Images were acquired with the equipment set at a 30° field of view and with Heidelberg Eye Explorer software (HEYEX version 5.3.3.0).
Mouse retina whole-mount analyses
All mice were maintained in accordance with Institutional Animal Care and Use Committee guidelines. Heterozygous Atf6+/− (n = 4) and Atf6−/− (n = 4) mice were sacrificed at 12 months of age, and retinal tissues were collected for retinal whole-mount imaging and molecular analyses. For retinal whole-mount studies, mouse eyes were fixed in 4% paraformaldehyde for 15 min. Eyes were dissected to isolate the retinas. Retinas were fixed in 4% paraformaldehyde for an additional 10 min, washed three times with PBS and incubated with FITC-conjugated PNA (1:50 dilution; Vector Laboratories) overnight at 4 °C. After several washes in PBS, the retinas were flat mounted and covered by a coverslip after the application of several drops of Antifade solution (Prolong, Invitrogen). Flat-mounted retinas were imaged using a Keyence BZ-9000 Fluorescence Microscope. PNA-positive cone cells were quantified with Keyence BZ image analysis software. The average number of cone cells per mm2 with standard deviation was based on measurements from eight retinas. At least four fields were quantified per retina.
Mouse retina outer nuclear layer analysis
Homozygous Atf6−/− (3 mice, 4 retinas, 14 sections) and wild-type littermates (Atf6+/+; 2 mice, 4 retinas, 9 sections) mice at 18 months of age were used to evaluate the effect of Atf6 depletion on the ONL. Briefly, mice were sacrificed, and the oriented eyes were fixed for 15 min in 4% paraformaldehyde at room temperature. The cornea and lens were dissected, and the eyeballs were cryoprotected in graded sucrose solutions (10% for 1 h at room temperature, 20% for 2 h at room temperature and 30% overnight at 4 °C). Sagital sections (12 μm) were obtained and mounted with Vectashield with DAPI (Vectorlabs) for imaging. Retinal mosaics on the central retina were obtained on an Axio Imager Z.1 ApoTome Microscope equipped with a Zeiss Axiocam MRm digital camera. The number of photoreceptor nuclei rows was counted every 500 μm (with the optic nerve being point 0). Several sections per eye and a minimum of six measurements per point were performed (technical replicates).
Functional analysis
Cell culture
Human primary fibroblast cells or HEK293 cells were maintained at 37 °C under 5% CO2 in DMEM (Mediatech) supplemented with 10% FCS (Mediatech) and 1% penicillin-streptomycin (Invitrogen). For human primary fibroblast cells, 1× GlutaMAX (Invitrogen) was also added to the medium. Mycoplasma-free HEK293 cells were obtained from Invitrogen.
Chemicals
Tunicamycin was obtained from Calbiochem EMD Bioscience. Trimethoprim was obtained from Teknova.
Plasmid construction and transfection
DHFR.YFP.pcDNA-DEST40 and DHFR.ATF6(1–373).pcDNA-DEST40 plasmids were generous gifts from R.L. Wiseman (Scripps Research Institute). The mutation encoding p.Arg324Cys was introduced into the DHFR.ATF6(1–373).pcDNA-DEST40 plasmid by site-direct mutagenesis using overlapping PCR. To express YFP, ATF6 (1–373) or the Arg324Cys ATF6 mutant in the cell culture system, plasmids containing the corresponding cDNA were transiently transfected into HEK293 cells using Lipofectamine 2000 (Invitrogen). Trimethoprim was added 4 h after transfection to induce protein expression.
Molecular biology
Cells were lysed, and total RNA was collected using the RNeasy mini kit according to the manufacturer’s instructions (Qiagen). mRNA was reverse transcribed using the iScript cDNA Synthesis kit (Bio-Rad). For qPCR analysis, cDNA was used as template in SYBR Green qPCR SuperMix (Bio-Rad). The primers used are listed in Supplementary Table 3. GAPDH or Rpl19 mRNA levels served as an internal normalization standard. qPCR was carried out at 95 °C for 5 min followed by 40 cycles of amplification with 95 °C for 10 s, 60 °C for 10 s and 72 °C for 10 s.
Protein biochemistry
Human fibroblasts or HEK293 cells were lysed in SDS lysis buffer (2% SDS and 62.5 mM Tris-HCl, pH 6.8, containing protease inhibitors (Sigma-Aldrich) and phosphatase inhibitor (Thermo Scientific)). Retinas from wild-type Atf6+/+, heterozygous Atf6+/− and knockout Atf6−/− mice were lysed in 300 μl of lysis buffer (0.5 g/ml n-dodecyl-β-D-maltoside (Calbiochem EMD Bioscience) in PBS with protease inhibitors and phosphatase inhibitor) and sonicated three times for 5 s. Protein concentrations for the whole-cell lysates were determined by BCA protein assay (Pierce). Equal amounts of protein were loaded onto 4–15% Mini-PROTEAN TGX precast gels (Bio-Rad) and analyzed by immunoblotting. The following antibodies and dilutions were used: antibody to Gαt1 (sc-389) and 1D4 antibody to rod opsin (sc-57432) at a 1:1,000 dilution (Santa Cruz Biotechnology); antibody to human ATF6A (ab122897) at a 1:1,000 dilution (Abcam); antibody to calreticulin (SPA-600) and antibody to PDI (SPA-890) at a 1:1,000 dilution (Stressgen); antibody to BiP (GTX113340) at a 1:1,000 dilution, antibody to GAPDH (GTX627408) at a 1:5,000 dilution, antibody to HSP90 (GTX101448) at a 1:5,000 dilution and antibody to β-tubulin (GTX101279) at a 1:5,000 dilution (GeneTex); antibody to M-opsin (AB5405) and antibody to S-opsin (AB5407) at a 1:1,000 dilution (EMD Millipore); and antibody to glycogen phosphorylase at a 1:1,000 dilution (a kind gift from S. Tsang (Columbia University)). After overnight incubation with primary antibody, membranes were washed in TBS with 0.1% Tween-20 (TBST) and then incubated with a horseradish peroxidase–coupled secondary antibody (Cell Signaling Technology). Immunoreactivity was detected using SuperSignal West chemiluminescent substrate (Pierce).
Statistical analysis
All results are presented as means ± s.d. from at least three independent experiments or mice per experimental condition. Student’s two-tailed t tests (for paired samples) were performed to determine P values. A value of P < 0.05 was considered significant: *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Acknowledgments
We want to thank C.W. Seok for data analysis. These studies were supported by various grants to the different authors and institutions: Bundesministerium für Bildung und Forschung (BMBF) grant 01GM1108A to B.W. and S.K.; US National Institutes of Health grants EY001919 and EY020846 to J.H.L. and DK042394, DK088227 and HL052173 to R.J.K. and a post-doctoral Foundation Fighting Blindness fellowship to W.-C.C.; National Institute for Health Research, Biomedical Research Centre at Moorfields Eye Hospital, National Health Service (NHS) Foundation Trust and University College London Institute of Ophthalmology, Fight For Sight, Moorfields Eye Hospital Special Trustees, Retinitis Pigmentosa Fighting Blindness and the Foundation Fighting Blindness (US) all to A.T.M., M.M. and A.R.W.; and the Wellcome Trust (099173/Z/12/Z) to M.M. and A.R.W. M.M. is supported by a Foundation Fighting Blindness Career Development Award; Mira Godard Research fund to E.H.; the imaging facilities at the Barbara and Donald Jonas Laboratory of Stem Cells and Regenerative Medicine and the Bernard and Shirlee Brown Glaucoma Laboratory are supported by Cannon, US National Institutes of Health Core grant 5P30EY019007, National Cancer Institute Core grant 5P30CA013696 and unrestricted funds from Research to Prevent Blindness (RPB), a Columbia University, New York RPB Physician-Scientist Award, the Schneeweiss Stem Cell Fund, New York State (N09G-302 and N13G-275) and the Gebroe Family Foundation, grant R01EY018213 to S.H.T.; Foundation Fighting Blindness (US) grants BR-GE-0510-0489-RAD to A.I.d.H. and C-GE-0811-0545-RAD01 to F.P.M.C., the Prof. Dr. H.J. Flieringa Foundation Stichting Wetenschappelijk Onderzoek het Oogziekenhuis (SWOO) and the Rotterdam Eye Hospital to F.P.M.C. and A.I.d.H. E.Z. is supported by Center for Integrative Neuroscience–DFG Center of Excellence EXC 307, University of Tübingen, Germany. R.K.K. is supported by the Foundation Fighting Blindness (Canada) and the CIHR (Canadian Institutes for Health Research).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
S.K., B.W., J.H.L. and R.J.K. conceived and designed the project and analyzed and interpreted data. D.Z., F.S., F.B., F.I., E.H., A.V., J.B., G.R., A.T.M., A.W., M.M., R.K.K., E.Z. and S.H.T. provided clinical data collection and interpretation. N.W., J.S., W.-C.C., S.R., A.I.d.H., F.P.M.C., I.L. and H.R. designed and performed experiments and analyzed and interpreted data. Specifically, N.W. performed cDNA analysis and haplotyping. J.S. performed all candidate gene sequencing. I.G.M. performed mouse retinal histology. T.M.S. was responsible for exome sequencing. S.C. and S.H.T. provided the AOSLO data. S.C.B., M.G.G., V.S. and M.W.S. provided the in vivo morphological and functional analyses of the mouse model, data generation and analysis, and writing of the manuscript. S.K., J.H.L. and D.Z. drafted the manuscript. M.M., R.K.K., E.H., A.V., A.T.M., A.W., M.M. and R.K.K. critically revised the manuscript for intellectual content. All authors discussed the results and commented on the manuscript. All authors read and approved the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Thiadens AA, et al. Progressive loss of cones in achromatopsia: an imaging study using spectral-domain optical coherence tomography. Invest Ophthalmol Vis Sci. 2010;51:5952–5957. doi: 10.1167/iovs.10-5680. [DOI] [PubMed] [Google Scholar]
- 2.Thomas MG, et al. Structural grading of foveal hypoplasia using spectral-domain optical coherence tomography a predictor of visual acuity? Ophthalmology. 2011;118:1653–1660. doi: 10.1016/j.ophtha.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genead MA, et al. Photoreceptor structure and function in patients with congenital achromatopsia. Invest Ophthalmol Vis Sci. 2011;52:7298–7308. doi: 10.1167/iovs.11-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sundaram V, et al. Retinal structure and function in achromatopsia: implications for gene therapy. Ophthalmology. 2014;121:234–245. doi: 10.1016/j.ophtha.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aboshiha J, et al. A prospective longitudinal study of retinal structure and function in achromatopsia. Invest Ophthalmol Vis Sci. 2014;55:5733–5743. doi: 10.1167/iovs.14-14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohl S, et al. Mutations in the cone photoreceptor G-protein α–subunit gene GNAT2 in patients with achromatopsia. Am J Hum Genet. 2002;71:422–425. doi: 10.1086/341835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aligianis IA, et al. Mapping of a novel locus for achromatopsia (ACHM4) to 1p and identification of a germline mutation in the αsubunit of cone transducin (GNAT2) J Med Genet. 2002;39:656–660. doi: 10.1136/jmg.39.9.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang B, et al. A homologous genetic basis of the murine Cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc Natl Acad Sci USA. 2009;106:19581–19586. doi: 10.1073/pnas.0907720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiadens AA, et al. Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am J Hum Genet. 2009;85:240–247. doi: 10.1016/j.ajhg.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohl S, et al. A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am J Hum Genet. 2012;91:527–532. doi: 10.1016/j.ajhg.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohl S, et al. Total colourblindness is caused by mutations in the gene encoding the α-subunit of the cone photoreceptor cGMP-gated cation channel. Nat Genet. 1998;19:257–259. doi: 10.1038/935. [DOI] [PubMed] [Google Scholar]
- 12.Sundin OH, et al. Genetic basis of total colourblindness among the Pingelapese islanders. Nat Genet. 2000;25:289–293. doi: 10.1038/77162. [DOI] [PubMed] [Google Scholar]
- 13.Kohl S, et al. Mutations in the CNGB3 gene encoding the β-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum Mol Genet. 2000;9:2107–2116. doi: 10.1093/hmg/9.14.2107. [DOI] [PubMed] [Google Scholar]
- 14.Zhu C, Johansen FE, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 16.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hai TW, Liu F, Coukos WJ, Green MR. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 20.Shoulders MD, et al. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanimoto N, Sothilingam V, Seeliger MW. Functional phenotyping of mouse models with ERG. Methods Mol Biol. 2013;935:69–78. doi: 10.1007/978-1-62703-080-9_4. [DOI] [PubMed] [Google Scholar]
- 22.Seeliger MW, et al. In vivo confocal imaging of the retina in animal models using scanning laser ophthalmoscopy. Vision Res. 2005;45:3512–3519. doi: 10.1016/j.visres.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 23.Joly S, et al. Cooperative phagocytes: resident microglia and bone marrow immigrants remove dead photoreceptors in retinal lesions. Am J Pathol. 2009;174:2310–2323. doi: 10.2353/ajpath.2009.090023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander JJ, et al. Restoration of cone vision in a mouse model of achromatopsia. Nat Med. 2007;13:685–687. doi: 10.1038/nm1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michalakis S, et al. Restoration of cone vision in the CNGA3−/− mouse model of congenital complete lack of cone photoreceptor function. Mol Ther. 2010;18:2057–2063. doi: 10.1038/mt.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komáromy AM, et al. Gene therapy rescues cone function in congenital achromatopsia. Hum Mol Genet. 2010;19:2581–2593. doi: 10.1093/hmg/ddq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho LS, et al. Long-term and age-dependent restoration of visual function in a mouse model of CNGB3-associated achromatopsia following gene therapy. Hum Mol Genet. 2011;20:3161–3175. doi: 10.1093/hmg/ddr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J, et al. ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, et al. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 30.Roy B, Lee AS. The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic Acids Res. 1999;27:1437–1443. doi: 10.1093/nar/27.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thuerauf DJ, Morrison LE, Hoover H, Glembotski CC. Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J Biol Chem. 2002;277:20734–20739. doi: 10.1074/jbc.M201749200. [DOI] [PubMed] [Google Scholar]
- 32.Desterro JM, Rodriguez MS, Hay RT. Regulation of transcription factors by protein degradation. Cell Mol Life Sci. 2000;57:1207–1219. doi: 10.1007/PL00000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thuerauf DJ, Morrison L, Glembotski CC. Opposing roles for ATF6α and ATF6β in endoplasmic reticulum stress response gene induction. J Biol Chem. 2004;279:21078–21084. doi: 10.1074/jbc.M400713200. [DOI] [PubMed] [Google Scholar]
- 34.Haze K, et al. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem J. 2001;355:19–28. doi: 10.1042/0264-6021:3550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida H, et al. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6α and 6β that activates the mammalian unfolded protein response. Mol Cell Biol. 2001;21:1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Rutkowski DT, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arensdorf AM, Dezwaan McCabe D, Kaufman RJ, Rutkowski DT. Temporal clustering of gene expression links the metabolic transcription factor HNF4α to the ER stress-dependent gene regulatory network. Front Genet. 2013;4:188. doi: 10.3389/fgene.2013.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thuerauf DJ, Marcinko M, Belmont PJ, Glembotski CC. Effects of the isoform-specific characteristics of ATF6α and ATF6β on endoplasmic reticulum stress response gene expression and cell viability. J Biol Chem. 2007;282:22865–22878. doi: 10.1074/jbc.M701213200. [DOI] [PubMed] [Google Scholar]
- 40.Marmor MF, et al. International Society for Clinical Electrophysiology of Vision. ISCEV standard for full-field clinical electroretinography (2008 update) Doc Ophthalmol. 2009;118:69–77. doi: 10.1007/s10633-008-9155-4. [DOI] [PubMed] [Google Scholar]
- 41.Hood DC, et al. International Society For Clinical Electrophysiology of Vision. ISCEV standard for clinical multifocal electroretinography (mfERG) (2011 edition) Doc Ophthalmol. 2012;124:1–13. doi: 10.1007/s10633-011-9296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park SP, et al. Disruption of the human cone photoreceptor mosaic from a defect in NR2E3 transcription factor function in young adults. Graefes Arch Clin Exp Ophthalmol. 2013;251:2299–2309. doi: 10.1007/s00417-013-2296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pyo Park S, Hwan Hong I, Tsang SH, Chang S. Cellular imaging demonstrates genetic mosaicism in heterozygous carriers of an X-linked ciliopathy gene. Eur J Hum Genet. 2013;21:1240–1248. doi: 10.1038/ejhg.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SP, et al. Early structural anomalies observed by high-resolution imaging in two related cases of autosomal-dominant retinitis pigmentosa. Ophthalmic Surg Lasers Imaging Retina. 2014;45:469–473. doi: 10.3928/23258160-20140908-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong IH, et al. Cone photoreceptor abnormalities correlate with vision loss in a case of acute posterior multifocal placoid pigment epitheliopathy. Ophthalmic Surg Lasers Imaging Retina. 2014;45:74–78. doi: 10.3928/23258160-20131220-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann K, Lindner TH. easyLINKAGE Plus—automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21:3565–3567. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- 47.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper—an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer MD, et al. Noninvasive, in vivo assessment of mouse retinal structure using optical coherence tomography. PLoS ONE. 2009;4:e7507. doi: 10.1371/journal.pone.0007507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.