Abstract
The liver plays a central role in iron metabolism. It is the major storage site for iron and also expresses a complex range of molecules which are involved in iron transport and regulation of iron homeostasis. An increasing number of genes associated with hepatic iron transport or regulation have been identified. These include transferrin receptors (TFR1 and 2), a ferrireductase (STEAP3), the transporters divalent metal transporter-1 (DMT1) and ferroportin (FPN) as well as the haemochromatosis protein, HFE and haemojuvelin (HJV), which are signalling molecules. Many of these genes also participate in iron regulatory pathways which focus on the hepatic peptide hepcidin. However, we are still only beginning to understand the complex interactions between liver iron transport and iron homeostasis. This review outlines our current knowledge of molecules of iron metabolism and their roles in iron transport and regulation of iron homeostasis.
Keywords: Liver, Iron homeostasis, Iron uptake, Iron release, Iron transporters, Hereditary haemochromatosis
INTRODUCTION
Iron is an essential trace element for almost all forms of life. However, under physiological conditions, the free form of iron is practically insoluble and potentially toxic. Thus, iron is always found bound to specific ligands in such a way as to render it both soluble and non-toxic. The toxicity of iron stems from its ability to redox cycle. The release of an electron from ferrous iron, if uncontrolled, may result in the formation of highly reactive oxygen species capable of oxidising lipids, proteins and DNA[1] causing damage to the structures and processes in which they are involved. However, many catalytic and other biological processes rely on the redox properties of iron; hence, iron must be available in a form which allows it to donate and accept electrons without causing non-specific damage.
In mammals, iron is transported around the plasma bound mainly to the glycoprotein transferrin, although other forms are also present in small amounts. In normal human plasma, transferrin has a concentration of between 25 and 50 μmol/L, and is usually about one-third saturated with iron. The remaining, unoccupied, binding sites on transferrin provide a large buffering capacity in case of an acute increase in plasma iron levels, an important consideration given the toxicity of free iron. Following uptake by the tissues, iron is transferred into a cytosolic pool (the “transit pool”) from where it is distributed to ferritin for storage or to iron-requiring moieties, such as haem or iron-sulphur clusters. The majority of hepatocellular iron is contained in ferritin (80%) with 2%-3% present as haem; the remainder is either bound to transferrin or present in the transit pool[2].
The liver plays a central role in iron metabolism. It is responsible for approximately 8% of plasma iron turnover in humans[3], most of which is mediated by hepatocytes[4,5] and it has long been known that it is the major site for storage of iron. Histologically, iron is distributed around the periportal regions of the liver with a decreasing gradient towards the centrilobular regions. In iron overload disorders, this gradient becomes more pronounced[6], involving mainly hepatocytes with the resident liver macrophages, Kupffer cells, loading to a much lesser extent[7] .
More recently, it has been shown that the liver expresses a complex range of molecules which regulate iron homeostasis. The liver and, more specifically, hepatocytes, also express the vast majority of genes that have been associated with hereditary iron disorders. Our understanding of these disorders as well as the normal function of the liver in iron homeostasis is, as yet, incomplete. This review focuses on iron metabolism in hepatocytes, with a specific section on the role of Kupffer cells, and on the molecules known to be involved in iron metabolism in the liver and their role in our current understanding of liver iron transport.
TRANSFERRIN RECEPTOR 1 (TFR1)- MEDIATED UPTAKE OF TRANSFERRIN
TFR1-mediated uptake of diferric transferrin is, perhaps, the best described process of iron uptake (Figure 1, pathway 1). Briefly, transferrin binds to TFR1 and is endocytosed[2,5]. At the pH of the extracellular fluid, diferric transferrin is bound preferentially, the affinity of the receptor being higher for diferric transferrin than for either monoferric- or apo- transferrin[7-10]. Following formation of the endosome, it is acidified which results in a decrease in the affinity of transferrin for iron and subsequent detachment of the metal[11,12]. The affinity of the receptor for the (now) apotransferrin is increased by the acidic environment[7] and apotransferrin remains bound to the receptor as the endosome returns to and fuses with the plasma membrane. At the higher extracellular pH, the affinity of the receptor for apotransferrin decreases[7] and apotransferrin is released back into the circulation[2]. The entire process takes between 3.8 and 15 min[13-15].
Figure 1.
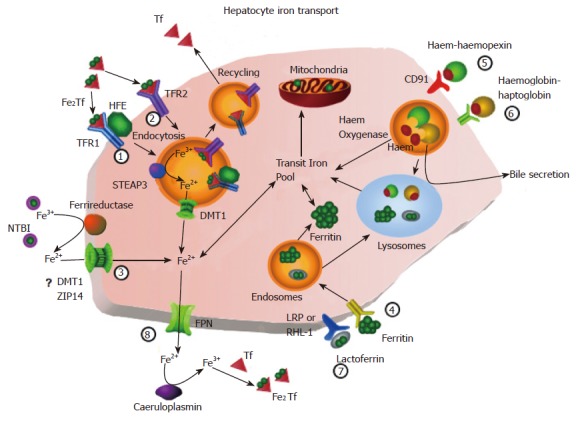
Hepatocyte iron transport. (1) TFR1-mediated uptake of diferric transferrin. Diferric transferrin binds to its specific receptor and is endocytosed. The endosome is acidified and Fe3+ is reduced by STEAP3. The iron is released and transported out of the endosome via DMT1 and apotransferrin is exocytosed. (2) TFR2-mediated uptake of transferrin. This mechanism is similar to the TFR1-specific mechanism except that transferrin binds to TFR2. (3) Uptake of NTBI. Iron is reduced and is transported into the cell via a carrier-mediated process. (4) Uptake of ferritin. Ferritin binds to its specific receptor and is endocytosed. The endosome is directed to lysosomes and the iron is transferred to the transit pool or endogenous ferritin. (5) Uptake of haem-haemopexin. The haem-haemopexin complex binds to its specific receptor CD91 and is endocytosed. Haem is removed and is degraded by haem oxygenase. (6) Uptake of haemoglobin-haptoglobin. The haemoglobin-haptoglobin complex binds to a specific receptor. Following endocytosis, the complex may be directed to the canalicular membrane for release into the bile or to the lysosomes for degradation. (7) Uptake of lactoferrin. Lactoferrin binds to LRP or RHL-1 and is endocytosed and targeted to the lysosomes for degradation. (8) Iron release Iron is released by FPN and oxidised by caeruloplasmin and binds to apotransferrin. TFR1, transferrin receptor 1; TFR2, transferrin receptor 2; STEAP3, six-transmembrane epithelial antigen of the prostate 3; DMT1, divalent metal transporter 1; NTBI, non-transferrin bound iron; ZIP14, zinc-regulated transporter and iron-regulated transporter-like protein 14; LRP, low-density lipoprotein receptor-related protein; FPN, ferroportin.
TFR1
TFR1 expression is regulated by iron primarily by a post-transcriptional mechanism. The transcript contains five iron-responsive elements (IRE) in its 3’ untranslated region (UTR) along with a number of instability elements that facilitate breakdown of the message[16-19]. Under low-iron conditions, iron regulatory proteins (IRP) bind to the IREs, placing an inhibition on the instability elements[18,19], increasing the half-life of the mRNA and, hence, increasing translation. When iron is abundant, IRPs do not bind IREs, resulting in a decrease of the stability of the transferrin receptor message. Two isoforms of IRP have been identified. The first (IRP1) is an iron-free form of cytosolic aconitase[20-22]. The second (IRP2) does not exhibit any aconitase activity[23] and appears to be the physiologically active IRP since it can respond to iron under conditions of low oxygen tension[24], a situation which occurs in the liver in vivo.
TFR1 is also regulated by other mechanisms. The gene contains an hypoxia response element in its promoter region which mediates up-regulation of transcription in the presence of hypoxia-inducible factor 1[25-27]. Transcription is also up-regulated by cytokines, such as interleukin-2, mitogens and growth factors[28-30]. Furthermore, TFR1 expression is increased in proliferating cells and reduced in quiescent cells[31-33], consistent with cellular demand for iron during periods of growth.
HFE
A mutation in HFE was the first to be shown to be causative for the iron overload disorder, haemochromatosis[34]. HFE is a major histocompatibility complex-like protein and was originally designated HLA-H. The gene is widely expressed, with highest expression in the liver and small intestine. HFE requires the protein β2-microglobulin for its correct localisation to the cell surface[35].
Despite this knowledge, the normal function of the protein has been difficult to elucidate. The crystal structure of the transferrin-TFR1 complex[36] indicates that the C-lobe of transferrin interacts with the helical domain of one of the TFR1 monomers. In contrast, the N-lobe of transferrin appears to interact partially with the helical domain, partially with the protease-like domain and, unusually, with the stalk connecting the extracellular region of TFR1 to its transmembrane region. HFE also interacts with the helical domain of TFR1[37,38], competing with transferrin for its binding site[39,40]. The resulting inhibition causes a reduction in transferrin-bound iron uptake in a variety of cell types[41-44], suggesting that HFE is involved in the regulation of iron uptake by TFR1, possibly by limiting the amount of iron released from transferrin[44]. HFE cycles with TFR1[44], but its effect on cycling is controversial, with different groups reporting no effect[44], a reduction in endocytosis[45] or a reduction in exocytosis[43].
The physiological consequences of the HFE-TFR1 interaction are difficult to ascertain given that the affinity of TFR1 for HFE is one to two orders of magnitude lower than for diferric transferrin[46], implying that, at normal transferrin concentrations, almost no HFE would be associated with the receptor. However, these measurements were conducted on isolated proteins, so it is possible that, in vivo, the local environment of these proteins changes their interactions leading to a shift in the balance of competition between HFE and transferrin.
STEAP3
Following endocytosis and vesicle acidification, iron is reduced to its ferrous form prior to being transferred across the endosomal membrane. It was suggested some years ago that the transferrin receptor appeared to facilitate detachment of iron from transferrin in the endosome[47]. More recently, it was shown that, at endosomal pH, the reduction potential of ferric iron co-ordinated by transferrin is increased when diferric transferrin is complexed to TFR1[48], confirming the earlier observation and suggesting that reduction of iron occurs prior to release from transferrin. However, purely chemical reduction is unlikely to result in the highly efficient process of iron uptake seen in biological systems.
Despite evidence of endosomal ferrireductase activity[49], it wasn’t until recently that a candidate ferrireductase was identified[50]. The gene STEAP3 (“six-transmembrane epithelial antigen of the prostate 3”) is one of four genes wholly or partially deleted in the nm1054 iron deficiency anaemia mouse. Under normal conditions, it is highly expressed in the liver, and its product is a protein which co-localises in endosomes with TFR1 and DMT1 (divalent metal transporter 1; see below). It is predicted to be a haemoprotein containing an N-terminal flavin-NADH binding domain. Most importantly, ferrireductase activity and iron uptake were lower in reticulocytes obtained from nm1054 and steap3 knockout mice and overexpression in HEK293T cells resulted in increased ferrireductase activity. A follow-up paper from the same group[51] showed that the remaining three members of the Steap family (STEAP1, 2 and 4) were also ferri- and cupric- reductases. The four genes are ubiquitously expressed; however, different members are expressed more highly in some tissues than others. Foetal liver expresses all four transcripts, but adult liver expresses predominantly STEAP3 with a small amount of STEAP1[50,51]. Like STEAP3, the other Steap proteins co-localise, at least partially, in an endosomal compartment with transferrin and TFR1.
Divalent metal transporter 1 (DMT1)
The released ferrous iron is transported from the interior of the endosome to the cytosol by DMT1 (also known as “natural resistance-associated macrophage protein 2”, NRAMP2, “divalent cation transporter 1”, DCT1, or “solute carrier family 11 member 2”, SLC11A2). This protein is a transmembrane glycoprotein with 12 predicted transmembrane helices[52,53] although there is no structure currently available to confirm this.
There are four known isoforms of DMT1, resulting from splice variation at the mRNA level. Alternative first exons (1 A or 1 B) give rise to the first level of variation[54]. Secondly, each of the 5′ splice variants may contain one of two 3′ splice variations[55]. The first of these contains an IRE in its 3′UTR. The second results in replacement of the final 18 codons of the open reading frame with a different sequence of 25 codons and a different 3′UTR, which, importantly, does not contain an IRE[55]. The predominant form in the liver is the exon 1B + IRE form, although a small amount of the 1B-IRE form may also be present[54].
Studies comparing the variants of DMT1 have indicated that the +IRE isoform is localised predominantly to the plasma membrane, exhibits slower internalisation kinetics than the -IRE isoform, and is targeted to lysosomes. In contrast, the C-terminal region of the -IRE isoform contains peptide signals which are required for efficient endocytosis and subsequent targeting to recycling endosomes[56,57]. Thus, it is possible that the +IRE isoform is predominantly involved in iron transport across the plasma membrane whereas the -IRE isoform is involved in endosomal transport.
Evidence that DMT1 is the endosomal transporter is supported by the finding that DMT1 co-localises with TFR1[58-60] and cycles through the endosomal compartment, appearing in acidic endosomes[61]. DMT1 transports iron optimally at pH 5.5[62], consistent with its presence in acidic endosomes and suggesting the energy for iron transport may be provided by a proton gradient. However, considerable transport also occurs at pH 7.4, and a model for metal transport by DMT1 has been proposed which is consistent with symport of Fe2+ and H+ from acidic endosomes and uniport of Fe2+ from a neutral environment[63].
DMT1 appears to be regulated by iron levels with protein expression increased in iron loaded liver, lower in control liver, and not detected in iron deficient livers[64]. Similar results have been obtained with the HepG2 hepatoma cell line[65]. These findings are inconsistent with an IRE located in the 3′UTR of the transcript, which would be expected to result in a decrease in mRNA stability in iron loading with a concomitant decrease in protein expression. However, regulation of DMT1 is complex, and it is possible that the 5′UTR of the transcript or the N-terminal domain of the protein may modify the regulatory effects of the IRE in a tissue-specific manner[54]. Additionally, regulation based around the stability of the protein cannot be ruled out.
LOW AFFINITY TRANSFERRIN UPTAKE
A second transferrin-mediated route of iron uptake (Figure 1, pathway 2) has been recognised in hepatocytes for many years[14,66,67] and is probably responsible for the bulk of iron uptake by hepatocytes since, at the concentrations of transferrin present in the plasma, TFR1 would be saturated[15,66,68]. The mechanism of uptake is similar to that of the TFR1-mediated pathway[5,14,69]. After binding to the low affinity binding site, transferrin is endocytosed and iron is removed following acidification of the vesicle. Iron is sequestered away from the vesicle and apotransferrin is exocytosed[69].
Transferrin receptor 2 (TFR2)
In 1999, Kawabata and colleagues[70] reported the cloning of transferrin receptor 2 (TFR2), a type II transmembrane protein which shared significant sequence similarity to TFR1. It is currently the best candidate gene to code for the low-affinity binding site, with which it shares many similarities. TFR2 binds diferric transferrin specifically in a pH-dependent manner with an affinity 25-30 times lower than TFR1[38,71]. In the liver, it is expressed predominantly in hepatocytes[70,72,73] and mediates cellular transferrin and iron uptake[70,73].
Regulation of TFR2 is different from regulation of TFR1. TFR2 mRNA does not contain any iron-responsive elements and cellular iron levels do not appear to change TFR2 mRNA or protein expression. Dietary and pathological iron loading do not result in decreased hepatic expression of TFR2 mRNA and neither does iron deficiency result in increased hepatic expression[74]. Instead, TFR2 appears to be regulated at the protein level by cell cycle, with proliferating cells expressing approximately twice as many receptors as stationary cells[75] and by the presence of diferric transferrin. Diferric transferrin causes an upregulation of receptor number and a redistribution of the protein to the cell surface in liver and hepatoma cells[72,76]. The upregulation is caused by an increase in the half-life of the receptor conferred by its binding diferric transferrin. Removal of diferric transferrin results in a return to baseline expression[77]. Consistent with these findings, TFR2 protein levels were decreased with iron deficiency, and increased with iron loading in genetic models of iron overload, such as haemochromatosis, but not in the atransferrinaemic mouse which has impaired transferrin synthesis[76]. Evidence suggests that TFR2 is a sensor of transferrin saturation and controls iron metabolism by regulating hepcidin expression[78,79]. However, it is still not known whether changes in levels of TFR2 expression are correlated with changes in transferrin-bound iron uptake by the liver.
Given the similarities between TFR1 and TFR2, it was thought that, like TFR1, TFR2 may bind HFE. Co-localisation studies suggested an interaction in the duodenum. TFR2 was shown to co-localise with wild-type HFE in an early endosomal compartment whereas, in the presence of HFEC282Y, the mutation predominantly associated with haemochromatosis type 1, TFR2 was distributed mainly to basolateral membrane[80]. Despite this, initial in vitro binding studies indicated that there was no detectable interaction between soluble HFE and the soluble TFR2 ectodomain[38]. More recently, an interaction has been demonstrated between the full-length, membrane-anchored HFE and TFR2 proteins[81] suggesting that HFE may, indeed, be involved in TFR2-mediated iron uptake and TFR2-dependent regulation of hepcidin.
It has also been shown that TFR2, unlike TFR1, is present in lipid rafts and binding of diferric transferrin to TFR2 can activate the ERK1/2 and p38-MAPK signalling pathways[82]. However, any connection of this to the hepcidin signalling pathway has yet to be demonstrated.
NON-TRANSFERRIN-BOUND IRON UPTAKE
The liver is one of the major sites of accumulation of iron delivered as low molecular weight chelates (Figure 1, pathway 3)[83,84]. The pathophysiological relevance of such a process is apparent in diseases of iron overload such as hereditary haemochromatosis[85]. The form of this low molecular weight plasma pool is likely to comprise several species; however, citrate appears to be the major component in both normal[86,87] and haemochromatotic[88] sera. In experimental situations, hepatocytes and their derivatives have been shown to take up iron from a variety of chelators[89-105]. Iron taken up from these chelators has been shown to be distributed to haem and ferritin in hepatocytes[90,101,103].
NTBI uptake by hepatocytes is linear for at least the first 15 to 60 min of incubation[91,94,101,104]. It is also concentration dependent, with both ferrous and ferric iron, delivered as a variety of low molecular weight chelates, showing saturation kinetics[95,97,106-108], indicating that this process is carrier-mediated. Uptake of iron as ferric citrate has been shown to be most efficient in normal rat hepatocytes at neutral pH[92].
DMT1 as a major transporter of NTBI
NTBI uptake is increased in cells in which DMT1 mRNA and protein expression are upregulated[109,110]. Furthermore, NTBI uptake appears to share at least one common pathway with TBI uptake since diferric transferrin has consistently been shown to competitively inhibit uptake of NTBI[93,102,111]. These observations, together with findings that DMT1 is active at neutral pH[62], are consistent with DMT1 being a major transporter of NTBI in hepatocytes.
The specificity of NTBI uptake has been investigated by a number of groups and it has been generally observed that Cd, Co, Cu, Mn and Zn decrease iron uptake by normal and transformed hepatocytes[97,104,108,109,112,113]. These observations match the range of divalent metals transported by DMT1[53,114], adding further credence to the suggestion that DMT1 is a transporter of NTBI. Indeed, Mn and Cd appear to be transported by DMT1 with higher affinity than Fe[114]. Although this observation is probably not relevant under normal physiological conditions, where the concentration of Fe is considerably higher than either Mn or Cd, it may become important in pathological conditions such as heavy metal poisoning in which competition for the transporter may result in a reduction in iron uptake. The alternative N and C termini conferred by the splice variants do not appear to affect the metal transport abilities[62].
Certain inconsistencies in the data showing that some metals cause inhibition in some cell types, but not others have led to the suggestion of a family of transporters for iron and other transition metals[101,111,115]. Several candidate transporters have been identified including calcium channels and specific transporters of other metals such as the zinc transporter, ZIP14 (zinc-regulated transporter and iron-regulated transporter-like protein 14).
ZIP14
ZIP14 (SLC39A14) is a transmembrane protein with eight predicted transmembrane helices[112,113]. It is highly expressed in the liver, and is localised to the plasma membrane[112]. There are two splice variants of the transcript; however, the biological functions of these two forms are yet to be determined. Originally shown to transport zinc, ZIP14 has also been shown to transport non-transferrin bound iron[112,113,116]. But, it is not currently known whether ZIP14 is involved in hepatic iron loading. Importantly, ZIP14 has also been shown to be upregulated by interleukin-6[112], which also upregulates hepcidin during inflammation[117].
Calcium channels
The role of calcium channels in uptake of NTBI by the liver remains unclear. It has been suggested that L-type calcium channels are responsible for a significant component of ferrous iron uptake by cardiomyocytes, particularly under iron loaded conditions[118,119]. However, information about any role for calcium channels in liver iron uptake is scant. Available evidence indicates that the transcripts coding for calcium channel subunits are expressed at low levels in the liver[120], suggesting their participation in iron uptake by the liver is likely to be minor. However, levels of mRNA do not take into account any post-transcriptional modifications or functional regulation such as gating. Hence, the contribution of calcium channels to iron uptake by the liver requires further investigation.
It has also been suggested that calcium itself plays a functional role in NTBI uptake; however, this, too, needs further clarification. Some studies have reported stimulation of NTBI uptake in cell types including hepatocytes[99,101,104,121],
whilst other studies have reported inhibition[122] or no effect[92]. It is possible that this spectrum of observations is due to variable chelation of calcium by the variety of chelators used to solubilise the iron[123].
TRANSPORT OF OTHER IRON COMPLEXES
A number other forms of iron are recognised as being cleared from the circulation by the liver; however, these are likely to be mechanisms of clearance for their respective ligands rather than for uptake of iron, per se. Specifically, these are ferritin, lactoferrin, the haem-haemopexin complex and the haemoglobin-haptoglobin complex. Circulating ferritin contains very small amounts of iron[124-126] and, as such, it is not a major source of iron in the normal human. Nevertheless, the liver clears ferritin by a method involving binding to a specific ferritin receptor[127-129] followed by endocytosis (Figure 1, pathway 4). There are several possible fates for endocytosed ferritin including catabolism of the protein in lysosomes[130-132], excretion in the bile or inclusion in the endogenous ferritin pool[133]. Any iron released is distributed to the mitochondria and endogenous ferritin[130,132].
The uptake of the haem-haemopexin complex is mediated by its specific receptor, CD91 (Figure 1, pathway 5)[134]. Following endocytosis, haem is degraded by haem oxygenase. Like transferrin, haemopexin was thought to be recycled back to the circulation[135-137]; however, this fate has recently been questioned with evidence suggesting that it is substantially degraded in lysosomes[134].
The haemoglobin-haptoglobin complex also binds to a high-affinity specific receptor and is endocytosed (Figure 1, pathway 6)[138]. However, from this point, two possibilities exist for the fate of the complex. Both haemoglobin and haptoglobin may be directed to lysosomes for degradation[139] or transported to the canalicular membrane of hepatocytes where haemoglobin is released into the bile and the receptor is recycled to the sinusoidal membrane[140]. Clearance of the haemopexin and haptoglobin complexes by the liver is of importance in haemolytic states, especially those associated with intravascular haemolysis.
Lactoferrin is an iron-binding protein similar to transferrin which is present mainly in milk. Two lactoferrin binding sites have been reported on hepatocytes, although neither is specific for lactoferrin. The first is low-density lipoprotein receptor-related protein (LRP)[141] and the second is the major (RHL-1) subunit of the asialoglycoprotein receptor[142]. Lactoferrin appears to be cleared via receptor-mediated endocytosis regardless of its binding site (Figure 1, pathway 7)[141,142]. Most of the internalised lactoferrin is directed to lysosomes for degradation[143].
IRON RELEASE
Ferroportin (FPN)
The transporter, ferroportin (FPN; “solute carrier family 40 member 1”, SLC40A1; IREG1 or “metal transporter protein-1”, MTP1) was reported independently by three groups in 2000[144-147] and appears to be the sole mediator of iron release from hepatocytes (Figure 1, pathway 8)[148]. Although it has not been shown directly, FPN appears to transport ferrous iron. Evidence comes from the apparent requirement of transport for ferroxidase activity. Caeruloplasmin knockout mice exhibit impaired hepatocellular and reticuloendothelial iron efflux which can be rescued by injection of caeruloplasmin[149]. Similarly, mice with mutations in the membrane-bound ferroxidase, hephaestin, also exhibit impaired iron efflux[150]. Further, iron efflux was stimulated in Xenopus oocytes over-expressing FPN in the presence of caeruloplasmin[147]. There have been no reports to date indicating whether FPN-mediated iron transport is linked to transport of any other ion or whether there is any energy requirement for the process.
The structure and membrane topology of FPN is currently unclear with various models predicting between nine and twelve transmembrane helices[151-153]. However, both the N- and C-termini appear to be located intracellularly[153,154], which precludes an odd number of transmembrane segments. Similarly, the quaternary structure of FPN has been the subject of debate. Initial reports suggested that FPN was oligomeric[155,156], but later reports cast doubt on this, suggesting a monomer[152,157]. Recently, a comprehensive study by de Domenico et al[154], demonstrated that FPN was most likely a dimer.
The quaternary structure of FPN may have important implications for regulation of iron homeostasis. Under the oligomeric model, the dominant negative phenotype of FPN-associated haemochromatosis (type 4) can be interpreted as interaction between wild-type and mutant forms of the protein interfering with its normal function[155,156]. The alternative interpretation, haploinsufficiency, is less likely given that mice heterozygotic for a FPN knockout demonstrated a very mild phenotype and homozygotic knockout mice died in utero[148]. Also, the majority of reports of human FPN-associated haemochromatosis with demonstrable iron loading involve heterozygotic point mutations which are at least partially functional[156,158,159].
Like ferritin, FPN mRNA contains a functional IRE in its 5′-UTR[144,147,160] indicating that translation should be augmented when iron is abundant. This has been shown to be true in HepG2 and Kupffer cells but not in the duodenum[144,160,161] suggesting that regulation of FPN is cell-specific and one or more other regulatory mechanisms may be involved.
OTHER MOLECULES INVOLVED IN IRON HOMEOSTASIS
Hepcidin
Hepcidin is a 25 residue peptide containing four internal disulphide bonds which is produced in hepatocytes under conditions of iron sufficiency[162-164]. It is created as a pre-pro-peptide which undergoes post-translational cleavage[163], and its expression is regulated by inflammation and hypoxia as well as iron levels[164,165]. It appears to be the focal point of an iron-regulatory pathway involving HFE, TFR2 and HJV, since disruption of these genes in haemochromatosis results in decreased hepcidin expression[166-168]. Its expression is enhanced by cytokines such as interleukin-6 (IL-6)[117].
In 2004, it was shown that FPN was a receptor for hepcidin[169]. In HEK293 cells, the peptide was shown to bind to FPN and induce its internalisation in a dose-dependent manner. The complex was targeted to lysosomes for degradation[169]. This is consistent with results that show increased FPN in the duodenum under conditions of iron deficiency, when hepcidin levels would be low[144], and offers a mechanism for hepcidin-mediated anaemia of inflammation[117,170] in which FPN levels are decreased resulting in a reduction of iron efflux to the plasma. The N-terminus of hepcidin is necessary for binding and internalisation of FPN, and the disulphide bonds appear to be necessary for its stability in the plasma[171]. It is unclear whether hepcidin acts in vivo as an autocrine hormone, signalling to FPN in hepatocytes or as a paracrine hormone, signalling to FPN in Kupffer cells.
Haemojuvelin (HJV)
HJV is a protein known to play a very important role in hepatic iron homeostasis although its exact function and whether it plays a role in iron transport have yet to be ascertained. It is expressed in adult skeletal muscle, and foetal and adult liver in the periportal hepatocytes[168,172]. Both soluble and membrane anchored forms have been demonstrated in Hep3B cells[173]. Identified as the protein mutated in many cases of juvenile haemochromatosis[168], human HJV shares 48% sequence identity with repulsive guidance molecules which are important in retinal development[174]. Absence of functional HJV results in increased plasma transferrin saturation and ferritin in humans[175,176] and studies in HJV knockout mice demonstrate decreased hepatic hepcidin expression and increased liver iron loading[172,177]. The major function of HJV appears to be regulation of hepcidin levels, and it has been shown that HJV can bind bone morphogenic protein-2 (BMP-2), a member of the TGF-β superfamily of cytokines and activate hepcidin transcription via SMAD-4[178,179]. This pathway is independent of HFE, TFR2 and IL-6[180].
KUPFFER CELLS
Kupffer cells are the resident macrophages of the liver. Their main function in iron metabolism appears to be as a clearing house for iron from phagocytosed red blood cells[181]. Haem breakdown is catalysed by haem oxygenase, and the products are ultimately excreted in the bile[182]. Iron can be stored in Kupffer cells as ferritin. But, much of it is released back into the circulation[183]. Consistent with this, Kupffer cells have been shown to strongly express both FPN transcript and protein[184,185]; indeed, FPN is more highly expressed in Kupffer cells than hepatocytes[144,184,186].
No functional studies on the role of FPN in iron release by Kupffer cells have been carried out. However, a number of studies have been undertaken using bone marrow-derived macrophages or macrophage cell lines. Following erythrophagocytosis or experimentally induced iron loading, the expression of many genes involved in iron metabolism, including FPN and haem oxygenase 1, are upregulated[185,187]. In these cells, FPN is localised to intracellular vesicles, redistributing to the cell surface following erythrophagocytosis[188]. The upregulation of FPN results in an increase in iron release and its down-regulation results in a decrease in iron release[189], consistent with involvement in iron recycling by Kupffer cells.
That FPN has been observed localised to intracellular vesicles in the absence of hepcidin suggests that FPN may play a role in intracellular redistribution of iron within Kupffer cells[190] as well as in iron export. Addition of hepcidin resulted in rapid disappearance of FPN from the cell membrane, and subsequent degradation of the protein[188], suggesting that in the absence of hepcidin, FPN may be able to cycle to intracellular compartments as necessary.
Kupffer cells also express TFR1[191] indicating that they can obtain iron from transferrin if necessary. Interestingly, Kupffer cells also express high levels of HFE[192]; however, they appear to be spared the level of iron loading associated with hepatocytes in HFE-associated haemochromatosis[7]. This may indicate a difference in regulation of HFE in macrophages compared to hepatocytes or simply that iron loading of Kupffer cells is partially negated by the high level of iron exported from these cells[183]. A recent report has suggested that GAPDH functions as a transferrin receptor in macrophages[193]. However, the affinity of the interaction was extremely low and its importance is yet to be determined.
LIVER IRON TRANSPORT IN DISORDERS OF IRON METABOLISM
In the absence of any relevant genetic defects, an increase in plasma iron would result in an increase in transferrin saturation followed by a rise in the concentration of NTBI. Gene expression in the liver would change to sequester the iron in a non-toxic form, and signal to the duodenum to reduce iron absorption. In primary iron overload disorders, such as hereditary haemochromatosis, mutations in HFE, HJV or TFR2 result in a decrease in hepcidin production, and subsequent misregulation of iron absorption by, the duodenum (as, of course, does a lack of functional hepcidin)[194]. The mechanistic consequences of this for the liver are difficult, if not impossible, to dissect out from the resulting iron overload. This leads to the paradox of TFR2, an iron transporter which is sub-functional in type 3 haemochromatosis, resulting in hepatic iron overload[71,195] rather than hepatic iron deficiency. This apparent paradox is probably the most telling demonstration of the liver’s repertoire of iron transport and regulatory mechanisms.
Secondary iron overload is often a consequence of blood transfusions required for the treatment of certain types of anaemia such as β-thalassaemia or sideroblastic anaemia. The source of the excess iron is haem from transfused erythrocytes which are broken down in the normal way with the haem being catabolised, inter alia, by Kupffer cells. As with primary iron overload disorders, gene expression in secondary iron overload will change to reflect the cellular iron loading and the increase in plasma transferrin saturation and NTBI concentration despite the underlying anaemia.
Iron deficiency may be caused by a number of factors including genetic disorders, pregnancy, an increased requirement for iron during growth, or simply by lack of dietary iron intake. The initial stage of iron deficiency corresponds to mobilisation of storage iron from the liver with decreases in hepatocyte ferritin, and an increase in iron uptake proteins such as TFR1[196]. Plasma NTBI is generally considered to be non-existent or at very low levels in iron deficiency[85].
CONCLUSION
Iron transport by the liver is, of necessity, tightly regulated because of the liver’s myriad of transport pathways, and its role in iron homeostasis. The explosion of information in the past ten years describing many of the genes involved in liver iron transport has not only provided insight into the mechanisms involved, but also confirmed the complexities evident from the literature from previous decades. Nevertheless, much work remains to be done in piecing together this information in order to fully understand how the pathways of iron transport, the distribution of iron in the liver and the regulatory pathways interact and how they contribute to iron homeostasis.
ACKNOWLEDGMENTS
We are grateful to the National Health and Medical Research Council (Australia) and the Fremantle Medical Research Foundation for funding and to Miss Nicole McCoy for assistance with preparation of the manuscript.
Footnotes
Supported by The National Health and Medical Research Council of Australia
S- Editor Liu Y L- Editor Alpini GD E- Editor Li JL
References
- 1.Papanikolaou G, Pantopoulos K. Iron metabolism and toxicity. Toxicol Appl Pharmacol. 2005;202:199–211. doi: 10.1016/j.taap.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 2.Young SP, Roberts S, Bomford A. Intracellular processing of transferrin and iron by isolated rat hepatocytes. Biochem J. 1985;232:819–823. doi: 10.1042/bj2320819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan EH. Iron metabolism and transport. In: Zakim D, Boyer TD, eds , editors. Hepatology A textbook of liver disease. 3rd edition. Sydney: W.B. Saunders Company; 1996. pp. 526–554. [Google Scholar]
- 4.Hershko C, Cook JD, Finch DA. Storage iron kinetics. 3. Study of desferrioxamine action by selective radioiron labels of RE and parenchymal cells. J Lab Clin Med. 1973;81:876–886. [PubMed] [Google Scholar]
- 5.Morgan EH, Smith GD, Peters TJ. Uptake and subcellular processing of 59Fe-125I-labelled transferrin by rat liver. Biochem J. 1986;237:163–173. doi: 10.1042/bj2370163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halliday JW, Searle J. Hepatic iron deposition in human disease and animal models. Biometals. 1996;9:205–209. doi: 10.1007/BF00144626. [DOI] [PubMed] [Google Scholar]
- 7.Dautry-Varsat A, Ciechanover A, Lodish HF. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc Natl Acad Sci USA. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornfeld S. The effect of metal attachment to human apotransferrin on its binding to reticulocytes. Biochim Biophys Acta. 1969;194:25–33. doi: 10.1016/0005-2795(69)90175-5. [DOI] [PubMed] [Google Scholar]
- 9.Morgan EH. Transferrin, biochemistry, physiology and clinical significance. Molec Aspects Med. 1981;4:1–123. [Google Scholar]
- 10.Young SP, Bomford A, Williams R. The effect of the iron saturation of transferrin on its binding and uptake by rabbit reticulocytes. Biochem J. 1984;219:505–510. doi: 10.1042/bj2190505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldwin DA, De Sousa DM, Von Wandruszka RM. The effect of pH on the kinetics of iron release from human transferrin. Biochim Biophys Acta. 1982;719:140–146. doi: 10.1016/0304-4165(82)90317-8. [DOI] [PubMed] [Google Scholar]
- 12.Bali PK, Aisen P. Receptor-modulated iron release from transferrin: differential effects on N- and C-terminal sites. Biochemistry. 1991;30:9947–9952. doi: 10.1021/bi00105a019. [DOI] [PubMed] [Google Scholar]
- 13.Bacon BR, Tavill AS. Role of the liver in normal iron metabolism. Semin Liver Dis. 1984;4:181–192. doi: 10.1055/s-2008-1041769. [DOI] [PubMed] [Google Scholar]
- 14.Morgan EH, Baker E. Iron uptake and metabolism by hepatocytes. Fed Proc. 1986;45:2810–2816. [PubMed] [Google Scholar]
- 15.Trinder D, Morgan E, Baker E. The mechanisms of iron uptake by fetal rat hepatocytes in culture. Hepatology. 1986;6:852–858. doi: 10.1002/hep.1840060508. [DOI] [PubMed] [Google Scholar]
- 16.Casey JL, Di Jeso B, Rao K, Klausner RD, Harford JB. Two genetic loci participate in the regulation by iron of the gene for the human transferrin receptor. Proc Natl Acad Sci USA. 1988;85:1787–1791. doi: 10.1073/pnas.85.6.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casey JL, Hentze MW, Koeller DM, Caughman SW, Rouault TA, Klausner RD, Harford JB. Iron-responsive elements: regulatory RNA sequences that control mRNA levels and translation. Science. 1988;240:924–928. doi: 10.1126/science.2452485. [DOI] [PubMed] [Google Scholar]
- 18.Casey JL, Koeller DM, Ramin VC, Klausner RD, Harford JB. Iron regulation of transferrin receptor mRNA levels requires iron-responsive elements and a rapid turnover determinant in the 3' untranslated region of the mRNA. EMBO J. 1989;8:3693–3699. doi: 10.1002/j.1460-2075.1989.tb08544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harford JB, Klausner RD. Coordinate post-transcriptional regulation of ferritin and transferrin receptor expression: the role of regulated RNA-protein interaction. Enzyme. 1990;44:28–41. doi: 10.1159/000468745. [DOI] [PubMed] [Google Scholar]
- 20.Hentze MW, Argos P. Homology between IRE-BP, a regulatory RNA-binding protein, aconitase, and isopropylmalate isomerase. Nucleic Acids Res. 1991;19:1739–1740. doi: 10.1093/nar/19.8.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy MC, Mende-Mueller L, Blondin GA, Beinert H. Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. Proc Natl Acad Sci USA. 1992;89:11730–11734. doi: 10.1073/pnas.89.24.11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klausner RD, Rouault TA, Harford JB. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 23.Guo B, Yu Y, Leibold EA. Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J Biol Chem. 1994;269:24252–24260. [PubMed] [Google Scholar]
- 24.Meyron-Holtz EG, Ghosh MC, Rouault TA. Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science. 2004;306:2087–2090. doi: 10.1126/science.1103786. [DOI] [PubMed] [Google Scholar]
- 25.Bianchi L, Tacchini L, Cairo G. HIF-1-mediated activation of transferrin receptor gene transcription by iron chelation. Nucleic Acids Res. 1999;27:4223–4227. doi: 10.1093/nar/27.21.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lok CN, Ponka P. Identification of a hypoxia response element in the transferrin receptor gene. J Biol Chem. 1999;274:24147–24152. doi: 10.1074/jbc.274.34.24147. [DOI] [PubMed] [Google Scholar]
- 27.Tacchini L, Bianchi L, Bernelli-Zazzera A, Cairo G. Transferrin receptor induction by hypoxia. HIF-1-mediated transcriptional activation and cell-specific post-transcriptional regulation. J Biol Chem. 1999;274:24142–24146. doi: 10.1074/jbc.274.34.24142. [DOI] [PubMed] [Google Scholar]
- 28.Miskimins WK, McClelland A, Roberts MP, Ruddle FH. Cell proliferation and expression of the transferrin receptor gene: promoter sequence homologies and protein interactions. J Cell Biol. 1986;103:1781–1788. doi: 10.1083/jcb.103.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouyang Q, Bommakanti M, Miskimins WK. A mitogen-responsive promoter region that is synergistically activated through multiple signalling pathways. Mol Cell Biol. 1993;13:1796–1804. doi: 10.1128/mcb.13.3.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seiser C, Teixeira S, Kühn LC. Interleukin-2-dependent transcriptional and post-transcriptional regulation of transferrin receptor mRNA. J Biol Chem. 1993;268:13074–13080. [PubMed] [Google Scholar]
- 31.Chitambar CR, Massey EJ, Seligman PA. Regulation of transferrin receptor expression on human leukemic cells during proliferation and induction of differentiation. Effects of gallium and dimethylsulfoxide. J Clin Invest. 1983;72:1314–1325. doi: 10.1172/JCI111087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trowbridge IS, Omary MB. Human cell surface glycoprotein related to cell proliferation is the receptor for transferrin. Proc Natl Acad Sci USA. 1981;78:3039–3043. doi: 10.1073/pnas.78.5.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Chen G, Pantopoulos K. Inhibition of transferrin receptor 1 transcription by a cell density response element. Biochem J. 2005;392:383–388. doi: 10.1042/BJ20050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Ellis MC, Fullan A, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 35.Feder JN, Tsuchihashi Z, Irrinki A, Lee VK, Mapa FA, Morikang E, Prass CE, Starnes SM, Wolff RK, Parkkila S, et al. The hemochromatosis founder mutation in HLA-H disrupts beta2-microglobulin interaction and cell surface expression. J Biol Chem. 1997;272:14025–14028. doi: 10.1074/jbc.272.22.14025. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- 37.Bennett MJ, Lebrón JA, Bjorkman PJ. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature. 2000;403:46–53. doi: 10.1038/47417. [DOI] [PubMed] [Google Scholar]
- 38.West AP, Bennett MJ, Sellers VM, Andrews NC, Enns CA, Bjorkman PJ. Comparison of the interactions of transferrin receptor and transferrin receptor 2 with transferrin and the hereditary hemochromatosis protein HFE. J Biol Chem. 2000;275:38135–38138. doi: 10.1074/jbc.C000664200. [DOI] [PubMed] [Google Scholar]
- 39.Feder JN, Penny DM, Irrinki A, Lee VK, Lebrón JA, Watson N, Tsuchihashi Z, Sigal E, Bjorkman PJ, Schatzman RC. The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci USA. 1998;95:1472–1477. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giannetti AM, Björkman PJ. HFE and transferrin directly compete for transferrin receptor in solution and at the cell surface. J Biol Chem. 2004;279:25866–25875. doi: 10.1074/jbc.M401467200. [DOI] [PubMed] [Google Scholar]
- 41.Arredondo M, Tapia V, Rojas A, Aguirre P, Reyes F, Marzolo MP, Núñez MT. Apical distribution of HFE-beta2-microglobulin is associated with inhibition of apical iron uptake in intestinal epithelia cells. Biometals. 2006;19:379–388. doi: 10.1007/s10534-005-6687-x. [DOI] [PubMed] [Google Scholar]
- 42.Carlson H, Zhang AS, Fleming WH, Enns CA. The hereditary hemochromatosis protein, HFE, lowers intracellular iron levels independently of transferrin receptor 1 in TRVb cells. Blood. 2005;105:2564–2570. doi: 10.1182/blood-2004-03-1204. [DOI] [PubMed] [Google Scholar]
- 43.Ikuta K, Fujimoto Y, Suzuki Y, Tanaka K, Saito H, Ohhira M, Sasaki K, Kohgo Y. Overexpression of hemochromatosis protein, HFE, alters transferrin recycling process in human hepatoma cells. Biochim Biophys Acta. 2000;1496:221–231. doi: 10.1016/s0167-4889(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 44.Roy CN, Penny DM, Feder JN, Enns CA. The hereditary hemochromatosis protein, HFE, specifically regulates transferrin-mediated iron uptake in HeLa cells. J Biol Chem. 1999;274:9022–9028. doi: 10.1074/jbc.274.13.9022. [DOI] [PubMed] [Google Scholar]
- 45.Salter-Cid L, Brunmark A, Li Y, Leturcq D, Peterson PA, Jackson MR, Yang Y. Transferrin receptor is negatively modulated by the hemochromatosis protein HFE: implications for cellular iron homeostasis. Proc Natl Acad Sci USA. 1999;96:5434–5439. doi: 10.1073/pnas.96.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.West AP, Giannetti AM, Herr AB, Bennett MJ, Nangiana JS, Pierce JR, Weiner LP, Snow PM, Bjorkman PJ. Mutational analysis of the transferrin receptor reveals overlapping HFE and transferrin binding sites. J Mol Biol. 2001;313:385–397. doi: 10.1006/jmbi.2001.5048. [DOI] [PubMed] [Google Scholar]
- 47.Sipe DM, Murphy RF. Binding to cellular receptors results in increased iron release from transferrin at mildly acidic pH. J Biol Chem. 1991;266:8002–8007. [PubMed] [Google Scholar]
- 48.Dhungana S, Taboy CH, Zak O, Larvie M, Crumbliss AL, Aisen P. Redox properties of human transferrin bound to its receptor. Biochemistry. 2004;43:205–209. doi: 10.1021/bi0353631. [DOI] [PubMed] [Google Scholar]
- 49.Scheiber B, Goldenberg H. NAD(P)H: ferric iron reductase in endosomal membranes from rat liver. Arch Biochem Biophys. 1993;305:225–230. doi: 10.1006/abbi.1993.1415. [DOI] [PubMed] [Google Scholar]
- 50.Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37:1264–1269. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohgami RS, Campagna DR, McDonald A, Fleming MD. The Steap proteins are metalloreductases. Blood. 2006;108:1388–1394. doi: 10.1182/blood-2006-02-003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gruenheid S, Cellier M, Vidal S, Gros P. Identification and characterization of a second mouse Nramp gene. Genomics. 1995;25:514–525. doi: 10.1016/0888-7543(95)80053-o. [DOI] [PubMed] [Google Scholar]
- 53.Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 54.Hubert N, Hentze MW. Previously uncharacterized isoforms of divalent metal transporter (DMT)-1: implications for regulation and cellular function. Proc Natl Acad Sci USA. 2002;99:12345–12350. doi: 10.1073/pnas.192423399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee PL, Gelbart T, West C, Halloran C, Beutler E. The human Nramp2 gene: characterization of the gene structure, alternative splicing, promoter region and polymorphisms. Blood Cells Mol Dis. 1998;24:199–215. doi: 10.1006/bcmd.1998.0186. [DOI] [PubMed] [Google Scholar]
- 56.Lam-Yuk-Tseung S, Gros P. Distinct targeting and recycling properties of two isoforms of the iron transporter DMT1 (NRAMP2, Slc11A2) Biochemistry. 2006;45:2294–2301. doi: 10.1021/bi052307m. [DOI] [PubMed] [Google Scholar]
- 57.Lam-Yuk-Tseung S, Touret N, Grinstein S, Gros P. Carboxyl-terminus determinants of the iron transporter DMT1/SLC11A2 isoform II (-IRE/1B) mediate internalization from the plasma membrane into recycling endosomes. Biochemistry. 2005;44:12149–12159. doi: 10.1021/bi050911r. [DOI] [PubMed] [Google Scholar]
- 58.Canonne-Hergaux F, Levy JE, Fleming MD, Montross LK, Andrews NC, Gros P. Expression of the DMT1 (NRAMP2/DCT1) iron transporter in mice with genetic iron overload disorders. Blood. 2001;97:1138–1140. doi: 10.1182/blood.v97.4.1138. [DOI] [PubMed] [Google Scholar]
- 59.Su MA, Trenor CC, Fleming JC, Fleming MD, Andrews NC. The G185R mutation disrupts function of the iron transporter Nramp2. Blood. 1998;92:2157–2163. [PubMed] [Google Scholar]
- 60.Tabuchi M, Tanaka N, Nishida-Kitayama J, Ohno H, Kishi F. Alternative splicing regulates the subcellular localization of divalent metal transporter 1 isoforms. Mol Biol Cell. 2002;13:4371–4387. doi: 10.1091/mbc.E02-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Touret N, Furuya W, Forbes J, Gros P, Grinstein S. Dynamic traffic through the recycling compartment couples the metal transporter Nramp2 (DMT1) with the transferrin receptor. J Biol Chem. 2003;278:25548–25557. doi: 10.1074/jbc.M212374200. [DOI] [PubMed] [Google Scholar]
- 62.Garrick MD, Kuo HC, Vargas F, Singleton S, Zhao L, Smith JJ, Paradkar P, Roth JA, Garrick LM. Comparison of mammalian cell lines expressing distinct isoforms of divalent metal transporter 1 in a tetracycline-regulated fashion. Biochem J. 2006;398:539–546. doi: 10.1042/BJ20051987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mackenzie B, Ujwal ML, Chang MH, Romero MF, Hediger MA. Divalent metal-ion transporter DMT1 mediates both H+ -coupled Fe2+ transport and uncoupled fluxes. Pflugers Arch. 2006;451:544–558. doi: 10.1007/s00424-005-1494-3. [DOI] [PubMed] [Google Scholar]
- 64.Trinder D, Oates PS, Thomas C, Sadleir J, Morgan EH. Localisation of divalent metal transporter 1 (DMT1) to the microvillus membrane of rat duodenal enterocytes in iron deficiency, but to hepatocytes in iron overload. Gut. 2000;46:270–276. doi: 10.1136/gut.46.2.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scheiber-Mojdehkar B, Sturm B, Plank L, Kryzer I, Goldenberg H. Influence of parenteral iron preparations on non-transferrin bound iron uptake, the iron regulatory protein and the expression of ferritin and the divalent metal transporter DMT-1 in HepG2 human hepatoma cells. Biochem Pharmacol. 2003;65:1973–1978. doi: 10.1016/s0006-2952(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 66.Cole ES, Glass J. Transferrin binding and iron uptake in mouse hepatocytes. Biochim Biophys Acta. 1983;762:102–110. doi: 10.1016/0167-4889(83)90122-2. [DOI] [PubMed] [Google Scholar]
- 67.Thorstensen K, Romslo I. Albumin prevents nonspecific transferrin binding and iron uptake by isolated hepatocytes. Biochim Biophys Acta. 1984;804:393–397. doi: 10.1016/0167-4889(84)90065-x. [DOI] [PubMed] [Google Scholar]
- 68.Thorstensen K, Romslo I. Uptake of iron from transferrin by isolated hepatocytes. Biochim Biophys Acta. 1984;804:200–208. doi: 10.1016/0167-4889(84)90150-2. [DOI] [PubMed] [Google Scholar]
- 69.Trinder D, Zak O, Aisen P. Transferrin receptor-independent uptake of differic transferrin by human hepatoma cells with antisense inhibition of receptor expression. Hepatology. 1996;23:1512–1520. doi: 10.1053/jhep.1996.v23.pm0008675172. [DOI] [PubMed] [Google Scholar]
- 70.Kawabata H, Yang R, Hirama T, Vuong PT, Kawano S, Gombart AF, Koeffler HP. Molecular cloning of transferrin receptor 2. A new member of the transferrin receptor-like family. J Biol Chem. 1999;274:20826–20832. doi: 10.1074/jbc.274.30.20826. [DOI] [PubMed] [Google Scholar]
- 71.Kawabata H, Germain RS, Vuong PT, Nakamaki T, Said JW, Koeffler HP. Transferrin receptor 2-alpha supports cell growth both in iron-chelated cultured cells and in vivo. J Biol Chem. 2000;275:16618–16625. doi: 10.1074/jbc.M908846199. [DOI] [PubMed] [Google Scholar]
- 72.Deaglio S, Capobianco A, Calì A, Bellora F, Alberti F, Righi L, Sapino A, Camaschella C, Malavasi F. Structural, functional, and tissue distribution analysis of human transferrin receptor-2 by murine monoclonal antibodies and a polyclonal antiserum. Blood. 2002;100:3782–3789. doi: 10.1182/blood-2002-01-0076. [DOI] [PubMed] [Google Scholar]
- 73.Robb AD, Ericsson M, Wessling-Resnick M. Transferrin receptor 2 mediates a biphasic pattern of transferrin uptake associated with ligand delivery to multivesicular bodies. Am J Physiol Cell Physiol. 2004;287:C1769–C1775. doi: 10.1152/ajpcell.00337.2004. [DOI] [PubMed] [Google Scholar]
- 74.Fleming RE, Migas MC, Holden CC, Waheed A, Britton RS, Tomatsu S, Bacon BR, Sly WS. Transferrin receptor 2: continued expression in mouse liver in the face of iron overload and in hereditary hemochromatosis. Proc Natl Acad Sci USA. 2000;97:2214–2219. doi: 10.1073/pnas.040548097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee AW, Oates PS, Trinder D. Effects of cell proliferation on the uptake of transferrin-bound iron by human hepatoma cells. Hepatology. 2003;38:967–977. doi: 10.1053/jhep.2003.50422. [DOI] [PubMed] [Google Scholar]
- 76.Robb A, Wessling-Resnick M. Regulation of transferrin receptor 2 protein levels by transferrin. Blood. 2004;104:4294–4299. doi: 10.1182/blood-2004-06-2481. [DOI] [PubMed] [Google Scholar]
- 77.Johnson MB, Enns CA. Diferric transferrin regulates transferrin receptor 2 protein stability. Blood. 2004;104:4287–4293. doi: 10.1182/blood-2004-06-2477. [DOI] [PubMed] [Google Scholar]
- 78.Camaschella C. Why do humans need two types of transferrin receptor? Lessons from a rare genetic disorder. Haematologica. 2005;90:296. [PubMed] [Google Scholar]
- 79.Le Gac G, Mons F, Jacolot S, Scotet V, Férec C, Frébourg T. Early onset hereditary hemochromatosis resulting from a novel TFR2 gene nonsense mutation (R105X) in two siblings of north French descent. Br J Haematol. 2004;125:674–678. doi: 10.1111/j.1365-2141.2004.04950.x. [DOI] [PubMed] [Google Scholar]
- 80.Griffiths WJ, Cox TM. Co-localization of the mammalian hemochromatosis gene product (HFE) and a newly identified transferrin receptor (TfR2) in intestinal tissue and cells. J Histochem Cytochem. 2003;51:613–624. doi: 10.1177/002215540305100507. [DOI] [PubMed] [Google Scholar]
- 81.Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281:28494–28498. doi: 10.1074/jbc.C600197200. [DOI] [PubMed] [Google Scholar]
- 82.Calzolari A, Raggi C, Deaglio S, Sposi NM, Stafsnes M, Fecchi K, Parolini I, Malavasi F, Peschle C, Sargiacomo M, et al. TfR2 localizes in lipid raft domains and is released in exosomes to activate signal transduction along the MAPK pathway. J Cell Sci. 2006;119:4486–4498. doi: 10.1242/jcs.03228. [DOI] [PubMed] [Google Scholar]
- 83.Anghileri LJ, Cordova Martinez A, Maincent P, Robert J. In vivo behaviour of low molecular weight iron complexes. Eur J Drug Metab Pharmacokinet. 1991;16:203–206. doi: 10.1007/BF03189960. [DOI] [PubMed] [Google Scholar]
- 84.Craven CM, Alexander J, Eldridge M, Kushner JP, Bernstein S, Kaplan J. Tissue distribution and clearance kinetics of non-transferrin-bound iron in the hypotransferrinemic mouse: a rodent model for hemochromatosis. Proc Natl Acad Sci USA. 1987;84:3457–3461. doi: 10.1073/pnas.84.10.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Batey RG, Lai Chung Fong P, Shamir S, Sherlock S. A non-transferrin-bound serum iron in idiopathic hemochromatosis. Dig Dis Sci. 1980;25:340–346. doi: 10.1007/BF01308057. [DOI] [PubMed] [Google Scholar]
- 86.Sarkar B. State of iron(3) in normal human serum: low molecular weight and protein ligands besides transferrin. Can J Biochem. 1970;48:1339–1350. doi: 10.1139/o70-208. [DOI] [PubMed] [Google Scholar]
- 87.Stojkovski S, Goumakos W, Sarkar B. Iron(III)-binding polypeptide in human cord and adult serum: isolation, purification and partial characterization. Biochim Biophys Acta. 1992;1137:155–161. doi: 10.1016/0167-4889(92)90197-j. [DOI] [PubMed] [Google Scholar]
- 88.Grootveld M, Bell JD, Halliwell B, Aruoma OI, Bomford A, Sadler PJ. Non-transferrin-bound iron in plasma or serum from patients with idiopathic hemochromatosis. Characterization by high performance liquid chromatography and nuclear magnetic resonance spectroscopy. J Biol Chem. 1989;264:4417–4422. [PubMed] [Google Scholar]
- 89.BASS RL, BERNICK S, SALTMAN P. The nucleus in the accumulation of iron by liver cell suspensions. Exp Cell Res. 1957;13:395–397. doi: 10.1016/0014-4827(57)90018-6. [DOI] [PubMed] [Google Scholar]
- 90.Batey RG, Shamir S, Wilms J. Properties and hepatic metabolism of non-transferrin-bound iron. Dig Dis Sci. 1981;26:1084–1088. doi: 10.1007/BF01295972. [DOI] [PubMed] [Google Scholar]
- 91.Brissot P, Wright TL, Ma WL, Weisiger RA. Efficient clearance of non-transferrin-bound iron by rat liver. Implications for hepatic iron loading in iron overload states. J Clin Invest. 1985;76:1463–1470. doi: 10.1172/JCI112125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Graham RM, Morgan EH, Baker E. Characterisation of citrate and iron citrate uptake by cultured rat hepatocytes. J Hepatol. 1998;29:603–613. doi: 10.1016/s0168-8278(98)80156-6. [DOI] [PubMed] [Google Scholar]
- 93.Graham RM, Morgan EH, Baker E. Ferric citrate uptake by cultured rat hepatocytes is inhibited in the presence of transferrin. Eur J Biochem. 1998;253:139–145. doi: 10.1046/j.1432-1327.1998.2530139.x. [DOI] [PubMed] [Google Scholar]
- 94.Grohlich D, Morley CG, Bezkorovainy A. Some aspects of iron uptake by rat hepatocytes in suspension. Int J Biochem. 1979;10:797–802. doi: 10.1016/0020-711x(79)90051-x. [DOI] [PubMed] [Google Scholar]
- 95.Kaplan J, Jordan I, Sturrock A. Regulation of the transferrin-independent iron transport system in cultured cells. J Biol Chem. 1991;266:2997–3004. [PubMed] [Google Scholar]
- 96.Planas-Bohne F, Jung W, Neu-Müller M. Uptake of 59Fe and 239Pu by rat liver cells and human hepatoma cells. Int J Radiat Biol Relat Stud Phys Chem Med. 1985;48:797–805. doi: 10.1080/09553008514551891. [DOI] [PubMed] [Google Scholar]
- 97.Randell EW, Parkes JG, Olivieri NF, Templeton DM. Uptake of non-transferrin-bound iron by both reductive and nonreductive processes is modulated by intracellular iron. J Biol Chem. 1994;269:16046–16053. [PubMed] [Google Scholar]
- 98.Saltman P, Fiskin RD, Bellnger SB. The metabolism of iron by rat liver slices; the effect of physical environment and iron concentration. J Biol Chem. 1956;220:741–750. [PubMed] [Google Scholar]
- 99.Saltman P, Fiskin RD, Bellinger SB, Alex T. The metabolism of iron by rat liver slices; the effect of chemical agents. J Biol Chem. 1956;220:751–757. [PubMed] [Google Scholar]
- 100.Alex T, Fiskin RD, Frisch HL, Saltman P. The kinetics of iron metabolism in rat liver slices. J Biol Chem. 1956;221:777–780. [PubMed] [Google Scholar]
- 101.Sturrock A, Alexander J, Lamb J, Craven CM, Kaplan J. Characterization of a transferrin-independent uptake system for iron in HeLa cells. J Biol Chem. 1990;265:3139–3145. [PubMed] [Google Scholar]
- 102.Trinder D, Morgan E. Inhibition of uptake of transferrin-bound iron by human hepatoma cells by nontransferrin-bound iron. Hepatology. 1997;26:691–698. doi: 10.1053/jhep.1997.v26.pm0009303500. [DOI] [PubMed] [Google Scholar]
- 103.White GP, Jacobs A. Iron uptake by Chang cells from transferrin, nitriloacetate and citrate complexes: the effects of iron-loading and chelation with desferrioxamine. Biochim Biophys Acta. 1978;543:217–225. doi: 10.1016/0304-4165(78)90066-1. [DOI] [PubMed] [Google Scholar]
- 104.Wright TL, Brissot P, Ma WL, Weisiger RA. Characterization of non-transferrin-bound iron clearance by rat liver. J Biol Chem. 1986;261:10909–10914. [PubMed] [Google Scholar]
- 105.Wright TL, Fitz JG, Weisiger RA. Non-transferrin-bound iron uptake by rat liver. Role of membrane potential difference. J Biol Chem. 1988;263:1842–1847. [PubMed] [Google Scholar]
- 106.Basset P, Quesneau Y, Zwiller J. Iron-induced L1210 cell growth: evidence of a transferrin-independent iron transport. Cancer Res. 1986;46:1644–1647. [PubMed] [Google Scholar]
- 107.Brissot P, Zanninelli G, Guyader D, Zeind J, Gollan J. Biliary excretion of plasma non-transferrin-bound iron in rats: pathogenetic importance in iron-overload disorders. Am J Physiol. 1994;267:G135–G142. doi: 10.1152/ajpgi.1994.267.1.G135. [DOI] [PubMed] [Google Scholar]
- 108.Baker E, Baker SM, Morgan EH. Characterisation of non-transferrin-bound iron (ferric citrate) uptake by rat hepatocytes in culture. Biochim Biophys Acta. 1998;1380:21–30. doi: 10.1016/s0304-4165(97)00120-7. [DOI] [PubMed] [Google Scholar]
- 109.Chua AC, Olynyk JK, Leedman PJ, Trinder D. Nontransferrin-bound iron uptake by hepatocytes is increased in the Hfe knockout mouse model of hereditary hemochromatosis. Blood. 2004;104:1519–1525. doi: 10.1182/blood-2003-11-3872. [DOI] [PubMed] [Google Scholar]
- 110.Shindo M, Torimoto Y, Saito H, Motomura W, Ikuta K, Sato K, Fujimoto Y, Kohgo Y. Functional role of DMT1 in transferrin-independent iron uptake by human hepatocyte and hepatocellular carcinoma cell, HLF. Hepatol Res. 2006;35:152–162. doi: 10.1016/j.hepres.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 111.Scheiber-Mojdehkar B, Zimmermann I, Dresow B, Goldenberg H. Differential response of non-transferrin bound iron uptake in rat liver cells on long-term and short-term treatment with iron. J Hepatol. 1999;31:61–70. doi: 10.1016/s0168-8278(99)80164-0. [DOI] [PubMed] [Google Scholar]
- 112.Liuzzi JP, Lichten LA, Rivera S, Blanchard RK, Aydemir TB, Knutson MD, Ganz T, Cousins RJ. Interleukin-6 regulates the zinc transporter Zip14 in liver and contributes to the hypozincemia of the acute-phase response. Proc Natl Acad Sci USA. 2005;102:6843–6848. doi: 10.1073/pnas.0502257102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taylor KM, Morgan HE, Johnson A, Nicholson RI. Structure-function analysis of a novel member of the LIV-1 subfamily of zinc transporters, ZIP14. FEBS Lett. 2005;579:427–432. doi: 10.1016/j.febslet.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 114.Garrick MD, Singleton ST, Vargas F, Kuo HC, Zhao L, Knöpfel M, Davidson T, Costa M, Paradkar P, Roth JA, et al. DMT1: which metals does it transport? Biol Res. 2006;39:79–85. doi: 10.4067/s0716-97602006000100009. [DOI] [PubMed] [Google Scholar]
- 115.Garrick LM, Dolan KG, Romano MA, Garrick MD. Non-transferrin-bound iron uptake in Belgrade and normal rat erythroid cells. J Cell Physiol. 1999;178:349–358. doi: 10.1002/(SICI)1097-4652(199903)178:3<349::AID-JCP9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 116.Liuzzi JP, Aydemir F, Nam H, Knutson MD, Cousins RJ. Zip14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc Natl Acad Sci USA. 2006;103:13612–13617. doi: 10.1073/pnas.0606424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, Ganz T. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271–1276. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Oudit GY, Sun H, Trivieri MG, Koch SE, Dawood F, Ackerley C, Yazdanpanah M, Wilson GJ, Schwartz A, Liu PP, et al. L-type Ca2+ channels provide a major pathway for iron entry into cardiomyocytes in iron-overload cardiomyopathy. Nat Med. 2003;9:1187–1194. doi: 10.1038/nm920. [DOI] [PubMed] [Google Scholar]
- 119.Tsushima RG, Wickenden AD, Bouchard RA, Oudit GY, Liu PP, Backx PH. Modulation of iron uptake in heart by L-type Ca2+ channel modifiers: possible implications in iron overload. Circ Res. 1999;84:1302–1309. doi: 10.1161/01.res.84.11.1302. [DOI] [PubMed] [Google Scholar]
- 120.Graf EM, Bock M, Heubach JF, Zahanich I, Boxberger S, Richter W, Schultz JH, Ravens U. Tissue distribution of a human Ca v 1.2 alpha1 subunit splice variant with a 75 bp insertion. Cell Calcium. 2005;38:11–21. doi: 10.1016/j.ceca.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 121.Nilsen T. Effects of calcium on hepatocyte iron uptake from transferrin, iron-pyrophosphate and iron-ascorbate. Biochim Biophys Acta. 1991;1095:39–45. doi: 10.1016/0167-4889(91)90042-v. [DOI] [PubMed] [Google Scholar]
- 122.Morgan EH. Membrane transport of non-transferrin-bound iron by reticulocytes. Biochim Biophys Acta. 1988;943:428–439. doi: 10.1016/0005-2736(88)90374-4. [DOI] [PubMed] [Google Scholar]
- 123.Mwanjewe J, Martinez R, Agrawal P, Samson SE, Coughlin MD, Brassard P, Grover AK. On the Ca2+ dependence of non-transferrin-bound iron uptake in PC12 cells. J Biol Chem. 2000;275:33512–33515. doi: 10.1074/jbc.M005435200. [DOI] [PubMed] [Google Scholar]
- 124.Arosio P, Yokota M, Drysdale JW. Characterization of serum ferritin in iron overload: possible identity to natural apoferritin. Br J Haematol. 1977;36:199–207. doi: 10.1111/j.1365-2141.1977.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 125.Pootrakul P, Josephson B, Huebers HA, Finch CA. Quantitation of ferritin iron in plasma, an explanation for non-transferrin iron. Blood. 1988;71:1120–1123. [PubMed] [Google Scholar]
- 126.Worwood M, Dawkins S, Wagstaff M, Jacobs A. The purification and properties of ferritin from human serum. Biochem J. 1976;157:97–103. doi: 10.1042/bj1570097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Adams PC, Powell LW, Halliday JW. Isolation of a human hepatic ferritin receptor. Hepatology. 1988;8:719–721. doi: 10.1002/hep.1840080402. [DOI] [PubMed] [Google Scholar]
- 128.Mack U, Powell LW, Halliday JW. Detection and isolation of a hepatic membrane receptor for ferritin. J Biol Chem. 1983;258:4672–4675. [PubMed] [Google Scholar]
- 129.Osterloh K, Aisen P. Pathways in the binding and uptake of ferritin by hepatocytes. Biochim Biophys Acta. 1989;1011:40–45. doi: 10.1016/0167-4889(89)90075-x. [DOI] [PubMed] [Google Scholar]
- 130.Sibille JC, Ciriolo M, Kondo H, Crichton RR, Aisen P. Subcellular localization of ferritin and iron taken up by rat hepatocytes. Biochem J. 1989;262:685–688. doi: 10.1042/bj2620685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Trump BF, Valigorsky JM, Arstila AU, Mergner WJ, Kinney TD. The relationship of intracellular pathways of iron metabolism to cellular iron overload and the iron storage diseases. Cell sap and cytocavitary network pathways in relation to lysosomal storage and turnover of iron macromolecules. Am J Pathol. 1973;72:295–336. [PMC free article] [PubMed] [Google Scholar]
- 132.Unger A, Hershko C. Hepatocellular uptake of ferritin in the rat. Br J Haematol. 1974;28:169–179. doi: 10.1111/j.1365-2141.1974.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 133.Anderson GJ, Frazer DM. Hepatic iron metabolism. Semin Liver Dis. 2005;25:420–432. doi: 10.1055/s-2005-923314. [DOI] [PubMed] [Google Scholar]
- 134.Hvidberg V, Maniecki MB, Jacobsen C, Højrup P, Møller HJ, Moestrup SK. Identification of the receptor scavenging hemopexin-heme complexes. Blood. 2005;106:2572–2579. doi: 10.1182/blood-2005-03-1185. [DOI] [PubMed] [Google Scholar]
- 135.Smith A, Hunt RC. Hemopexin joins transferrin as representative members of a distinct class of receptor-mediated endocytic transport systems. Eur J Cell Biol. 1990;53:234–245. [PubMed] [Google Scholar]
- 136.Smith A, Morgan WT. Transport of heme by hemopexin to the liver: evidence for receptor-mediated uptake. Biochem Biophys Res Commun. 1978;84:151–157. doi: 10.1016/0006-291x(78)90276-0. [DOI] [PubMed] [Google Scholar]
- 137.Smith A, Morgan WT. Haem transport to the liver by haemopexin. Receptor-mediated uptake with recycling of the protein. Biochem J. 1979;182:47–54. doi: 10.1042/bj1820047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, Moestrup SK. Identification of the haemoglobin scavenger receptor. Nature. 2001;409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 139.Higa Y, Oshiro S, Kino K, Tsunoo H, Nakajima H. Catabolism of globin-haptoglobin in liver cells after intravenous administration of hemoglobin-haptoglobin to rats. J Biol Chem. 1981;256:12322–12328. [PubMed] [Google Scholar]
- 140.Hinton RH, Dobrota M, Mullock BM. Haptoglobin-mediated transfer of haemoglobin from serum into bile. FEBS Lett. 1980;112:247–250. doi: 10.1016/0014-5793(80)80190-6. [DOI] [PubMed] [Google Scholar]
- 141.Meilinger M, Haumer M, Szakmary KA, Steinböck F, Scheiber B, Goldenberg H, Huettinger M. Removal of lactoferrin from plasma is mediated by binding to low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor and transport to endosomes. FEBS Lett. 1995;360:70–74. doi: 10.1016/0014-5793(95)00082-k. [DOI] [PubMed] [Google Scholar]
- 142.Bennatt DJ, Ling YY, McAbee DD. Isolated rat hepatocytes bind lactoferrins by the RHL-1 subunit of the asialoglycoprotein receptor in a galactose-independent manner. Biochemistry. 1997;36:8367–8376. doi: 10.1021/bi963079m. [DOI] [PubMed] [Google Scholar]
- 143.Ziere GJ, van Dijk MC, Bijsterbosch MK, van Berkel TJ. Lactoferrin uptake by the rat liver. Characterization of the recognition site and effect of selective modification of arginine residues. J Biol Chem. 1992;267:11229–11235. [PubMed] [Google Scholar]
- 144.Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem. 2000;275:19906–19912. doi: 10.1074/jbc.M000713200. [DOI] [PubMed] [Google Scholar]
- 145.Dadone MM, Kushner JP, Edwards CQ, Bishop DT, Skolnick MH. Hereditary hemochromatosis. Analysis of laboratory expression of the disease by genotype in 18 pedigrees. Am J Clin Pathol. 1982;78:196–207. doi: 10.1093/ajcp/78.2.196. [DOI] [PubMed] [Google Scholar]
- 146.Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt SJ, Moynihan J, Paw BH, Drejer A, Barut B, Zapata A, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403:776–781. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- 147.McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, et al. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000;5:299–309. doi: 10.1016/s1097-2765(00)80425-6. [DOI] [PubMed] [Google Scholar]
- 148.Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, Andrews NC. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 149.Harris ZL, Durley AP, Man TK, Gitlin JD. Targeted gene disruption reveals an essential role for ceruloplasmin in cellular iron efflux. Proc Natl Acad Sci USA. 1999;96:10812–10817. doi: 10.1073/pnas.96.19.10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vulpe CD, Kuo YM, Murphy TL, Cowley L, Askwith C, Libina N, Gitschier J, Anderson GJ. Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat Genet. 1999;21:195–199. doi: 10.1038/5979. [DOI] [PubMed] [Google Scholar]
- 151.Devalia V, Carter K, Walker AP, Perkins SJ, Worwood M, May A, Dooley JS. Autosomal dominant reticuloendothelial iron overload associated with a 3-base pair deletion in the ferroportin 1 gene (SLC11A3) Blood. 2002;100:695–697. doi: 10.1182/blood-2001-11-0132. [DOI] [PubMed] [Google Scholar]
- 152.Gonçalves AS, Muzeau F, Blaybel R, Hetet G, Driss F, Delaby C, Canonne-Hergaux F, Beaumont C. Wild-type and mutant ferroportins do not form oligomers in transfected cells. Biochem J. 2006;396:265–275. doi: 10.1042/BJ20051682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu XB, Yang F, Haile DJ. Functional consequences of ferroportin 1 mutations. Blood Cells Mol Dis. 2005;35:33–46. doi: 10.1016/j.bcmd.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 154.De Domenico I, Ward DM, Musci G, Kaplan J. Evidence for the multimeric structure of ferroportin. Blood. 2007;109:2205–2209. doi: 10.1182/blood-2006-06-032516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.De Domenico I, Ward DM, Nemeth E, Vaughn MB, Musci G, Ganz T, Kaplan J. The molecular basis of ferroportin-linked hemochromatosis. Proc Natl Acad Sci USA. 2005;102:8955–8960. doi: 10.1073/pnas.0503804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McGregor JA, Shayeghi M, Vulpe CD, Anderson GJ, Pietrangelo A, Simpson RJ, McKie AT. Impaired iron transport activity of ferroportin 1 in hereditary iron overload. J Membr Biol. 2005;206:3–7. doi: 10.1007/s00232-005-0768-1. [DOI] [PubMed] [Google Scholar]
- 157.Pignatti E, Mascheroni L, Sabelli M, Barelli S, Biffo S, Pietrangelo A. Ferroportin is a monomer in vivo in mice. Blood Cells Mol Dis. 2006;36:26–32. doi: 10.1016/j.bcmd.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 158.Drakesmith H, Schimanski LM, Ormerod E, Merryweather-Clarke AT, Viprakasit V, Edwards JP, Sweetland E, Bastin JM, Cowley D, Chinthammitr Y, et al. Resistance to hepcidin is conferred by hemochromatosis-associated mutations of ferroportin. Blood. 2005;106:1092–1097. doi: 10.1182/blood-2005-02-0561. [DOI] [PubMed] [Google Scholar]
- 159.Schimanski LM, Drakesmith H, Merryweather-Clarke AT, Viprakasit V, Edwards JP, Sweetland E, Bastin JM, Cowley D, Chinthammitr Y, Robson KJ, et al. In vitro functional analysis of human ferroportin (FPN) and hemochromatosis-associated FPN mutations. Blood. 2005;105:4096–4102. doi: 10.1182/blood-2004-11-4502. [DOI] [PubMed] [Google Scholar]
- 160.Lymboussaki A, Pignatti E, Montosi G, Garuti C, Haile DJ, Pietrangelo A. The role of the iron responsive element in the control of ferroportin1/IREG1/MTP1 gene expression. J Hepatol. 2003;39:710–715. doi: 10.1016/s0168-8278(03)00408-2. [DOI] [PubMed] [Google Scholar]
- 161.Chen Y, Qian ZM, Du J, Duan X, Chang Y, Wang Q, Wang C, Ma YM, Xu Y, Li L, et al. Iron loading inhibits ferroportin1 expression in PC12 cells. Neurochem Int. 2005;47:507–513. doi: 10.1016/j.neuint.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 162.Krause A, Neitz S, Mägert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, Adermann K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000;480:147–150. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 163.Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 164.Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loréal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–7819. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- 165.Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, Devaux I, Beaumont C, Kahn A, Vaulont S. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110:1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gehrke SG, Kulaksiz H, Herrmann T, Riedel HD, Bents K, Veltkamp C, Stremmel W. Expression of hepcidin in hereditary hemochromatosis: evidence for a regulation in response to the serum transferrin saturation and to non-transferrin-bound iron. Blood. 2003;102:371–376. doi: 10.1182/blood-2002-11-3610. [DOI] [PubMed] [Google Scholar]
- 167.Nemeth E, Roetto A, Garozzo G, Ganz T, Camaschella C. Hepcidin is decreased in TFR2 hemochromatosis. Blood. 2005;105:1803–1806. doi: 10.1182/blood-2004-08-3042. [DOI] [PubMed] [Google Scholar]
- 168.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dubé MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 169.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 170.Nemeth E, Valore EV, Territo M, Schiller G, Lichtenstein A, Ganz T. Hepcidin, a putative mediator of anemia of inflammation, is a type II acute-phase protein. Blood. 2003;101:2461–2463. doi: 10.1182/blood-2002-10-3235. [DOI] [PubMed] [Google Scholar]
- 171.Nemeth E, Preza GC, Jung CL, Kaplan J, Waring AJ, Ganz T. The N-terminus of hepcidin is essential for its interaction with ferroportin: structure-function study. Blood. 2006;107:328–333. doi: 10.1182/blood-2005-05-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115:2180–2186. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106:2884–2889. doi: 10.1182/blood-2005-05-1845. [DOI] [PubMed] [Google Scholar]
- 174.Monnier PP, Sierra A, Macchi P, Deitinghoff L, Andersen JS, Mann M, Flad M, Hornberger MR, Stahl B, Bonhoeffer F, et al. RGM is a repulsive guidance molecule for retinal axons. Nature. 2002;419:392–395. doi: 10.1038/nature01041. [DOI] [PubMed] [Google Scholar]
- 175.Gehrke SG, Pietrangelo A, Kascák M, Braner A, Eisold M, Kulaksiz H, Herrmann T, Hebling U, Bents K, Gugler R, et al. HJV gene mutations in European patients with juvenile hemochromatosis. Clin Genet. 2005;67:425–428. doi: 10.1111/j.1399-0004.2005.00413.x. [DOI] [PubMed] [Google Scholar]
- 176.Koyama C, Hayashi H, Wakusawa S, Ueno T, Yano M, Katano Y, Goto H, Kidokoro R. Three patients with middle-age-onset hemochromatosis caused by novel mutations in the hemojuvelin gene. J Hepatol. 2005;43:740–742. doi: 10.1016/j.jhep.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 177.Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115:2187–2191. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 179.Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 180.Truksa J, Peng H, Lee P, Beutler E. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL-6. Proc Natl Acad Sci USA. 2006;103:10289–10293. doi: 10.1073/pnas.0603124103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Terpstra V, van Berkel TJ. Scavenger receptors on liver Kupffer cells mediate the in vivo uptake of oxidatively damaged red blood cells in mice. Blood. 2000;95:2157–2163. [PubMed] [Google Scholar]
- 182.Maines MD, Gibbs PE. 30 some years of heme oxygenase: from a "molecular wrecking ball" to a "mesmerizing" trigger of cellular events. Biochem Biophys Res Commun. 2005;338:568–577. doi: 10.1016/j.bbrc.2005.08.121. [DOI] [PubMed] [Google Scholar]
- 183.Kondo H, Saito K, Grasso JP, Aisen P. Iron metabolism in the erythrophagocytosing Kupffer cell. Hepatology. 1988;8:32–38. doi: 10.1002/hep.1840080108. [DOI] [PubMed] [Google Scholar]
- 184.Bastin J, Drakesmith H, Rees M, Sargent I, Townsend A. Localisation of proteins of iron metabolism in the human placenta and liver. Br J Haematol. 2006;134:532–543. doi: 10.1111/j.1365-2141.2006.06216.x. [DOI] [PubMed] [Google Scholar]
- 185.Zhang AS, Xiong S, Tsukamoto H, Enns CA. Localization of iron metabolism-related mRNAs in rat liver indicate that HFE is expressed predominantly in hepatocytes. Blood. 2004;103:1509–1514. doi: 10.1182/blood-2003-07-2378. [DOI] [PubMed] [Google Scholar]
- 186.Canonne-Hergaux F, Donovan A, Delaby C, Wang HJ, Gros P. Comparative studies of duodenal and macrophage ferroportin proteins. Am J Physiol Gastrointest Liver Physiol. 2006;290:G156–G163. doi: 10.1152/ajpgi.00227.2005. [DOI] [PubMed] [Google Scholar]
- 187.Knutson MD, Vafa MR, Haile DJ, Wessling-Resnick M. Iron loading and erythrophagocytosis increase ferroportin 1 (FPN1) expression in J774 macrophages. Blood. 2003;102:4191–4197. doi: 10.1182/blood-2003-04-1250. [DOI] [PubMed] [Google Scholar]
- 188.Delaby C, Pilard N, Gonçalves AS, Beaumont C, Canonne-Hergaux F. Presence of the iron exporter ferroportin at the plasma membrane of macrophages is enhanced by iron loading and down-regulated by hepcidin. Blood. 2005;106:3979–3984. doi: 10.1182/blood-2005-06-2398. [DOI] [PubMed] [Google Scholar]
- 189.Knutson MD, Oukka M, Koss LM, Aydemir F, Wessling-Resnick M. Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin. Proc Natl Acad Sci USA. 2005;102:1324–1328. doi: 10.1073/pnas.0409409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Knutson M, Wessling-Resnick M. Iron metabolism in the reticuloendothelial system. Crit Rev Biochem Mol Biol. 2003;38:61–88. doi: 10.1080/713609210. [DOI] [PubMed] [Google Scholar]
- 191.van Berkel TJ, Dekker CJ, Kruijt JK, van Eijk HG. The interaction in vivo of transferrin and asialotransferrin with liver cells. Biochem J. 1987;243:715–722. doi: 10.1042/bj2430715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bastin JM, Jones M, O'Callaghan CA, Schimanski L, Mason DY, Townsend AR. Kupffer cell staining by an HFE-specific monoclonal antibody: implications for hereditary haemochromatosis. Br J Haematol. 1998;103:931–941. doi: 10.1046/j.1365-2141.1998.01102.x. [DOI] [PubMed] [Google Scholar]
- 193.Raje CI, Kumar S, Harle A, Nanda JS, Raje M. The macrophage cell surface glyceraldehyde-3-phosphate dehydrogenase is a novel transferrin receptor. J Biol Chem. 2007;282:3252–3261. doi: 10.1074/jbc.M608328200. [DOI] [PubMed] [Google Scholar]
- 194.Pietrangelo A. Hereditary hemochromatosis--a new look at an old disease. N Engl J Med. 2004;350:2383–2397. doi: 10.1056/NEJMra031573. [DOI] [PubMed] [Google Scholar]
- 195.Camaschella C, Roetto A, Calì A, De Gobbi M, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25:14–15. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- 196.Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]


