Abstract
Crustose coralline algae (CCA) are important components of many marine ecosystems. They aid in reef accretion and stabilization, create habitat for other organisms, contribute to carbon sequestration and are important settlement substrata for a number of marine invertebrates. Despite their ecological importance, little is known about the bacterial communities associated with CCA or whether differences in bacterial assemblages may have ecological implications. This study examined the bacterial communities on four different species of CCA collected in Belize using bacterial tag-encoded FLX amplicon pyrosequencing of the V1–V3 region of the 16S rDNA. CCA were dominated by Alphaproteobacteria, Gammaproteobacteria and Actinomycetes. At the operational taxonomic unit (OTU) level, each CCA species had a unique bacterial community that was significantly different from all other CCA species. Hydrolithon boergesenii and Titanoderma prototypum, CCA species that facilitate larval settlement in multiple corals, had higher abundances of OTUs related to bacteria that inhibit the growth and/or biofilm formation of coral pathogens. Fewer coral larvae settle on the surfaces of Paragoniolithon solubile and Porolithon pachydermum. These CCA species had higher abundances of OTUs related to known coral pathogens and cyanobacteria. Coral larvae may be able to use the observed differences in bacterial community composition on CCA species to assess the suitability of these substrata for settlement and selectively settle on CCA species that contain beneficial bacteria.
Introduction
Crustose coralline algae (CCA) are widely distributed and can be found in virtually all marine habitats. They are important calcifying species and contribute greatly to both reef accretion and stabilization (Littler and Littler, 2013). CCA have long been recognized as important settlement substrata for a variety of marine invertebrate larvae including commercially important species of abalone and sea urchins (Pawlik, 1992; Hadfield and Paul, 2001). CCA also induce larval settlement in many coral species, and this interaction has received much attention in light of the ecological importance of coral reefs and their drastic declines in recent years. Although several studies have demonstrated the ability of CCA to induce the settlement of coral larvae, the effects are not ubiquitous among CCA species. Important reef-building acroporid species, in both the Indo-Pacific and Caribbean, exhibit settlement preferences for certain CCA species (Harrington et al., 2004; Ritson-Williams et al., 2010, 2014). On the basis of these preferences, it is clear that some coral larvae are capable of recognizing and discriminating among CCA species, however, the mechanism by which they are able to differentiate among CCA is unknown. There is growing evidence that bacteria associated with potential settlement substrata induce settlement activity in many invertebrates including corals (Webster et al., 2004; reviewed in Hadfield, 2011), and that in some cases, they respond to specific bacterial strains within the bacterial community (Negri et al., 2001; Tebben et al., 2011; Tran and Hadfield, 2011; Sneed et al., 2014). It is therefore likely that larvae may use bacteria to recognize appropriate settlement substrata.
Several studies have demonstrated that different species of macroalgae harbor unique bacterial communities (Longford et al., 2007; Lachnit et al., 2009, 2011; Sneed and Pohnert, 2011). Within a given geographical area, bacterial communities are often more similar on individuals from the same algal species than they are to individuals from other algal species (Sneed and Pohnert, 2011). In some cases, algal species host specific bacterial assemblages across large geographic distances (Lachnit et al., 2009). To date, most studies on algal associated bacterial communities have focused on temperate, fleshy algal species (Lachnit et al., 2011) with the exception of a few studies of tropical fleshy algae (Barott et al., 2011; Sneed and Pohnert, 2011). Although CCA are known to be important settlement substrata for a number of invertebrate species and bacteria are known to produce chemical cues that induce invertebrate larval settlement, little is known about the bacterial communities associated with CCA. A limited number of studies have characterized CCA-associated bacteria using molecular techniques; however, these studies did not compare bacterial communities among different CCA species (Barott et al., 2011; Webster et al., 2011, 2013). In the early 1990s, Johnson et al. (1991) characterized the culturable bacteria associated with two species of South African CCA based on morphological and physiological characteristics and suggested that some bacterial strains may have a specialized association with their CCA host. Although this work suggests that, like other macroalgae, CCA may also harbor specific bacterial communities, culture-independent techniques are necessary to more fully characterize these microbial communities.
In this study, we examined the bacterial communities on four different species of CCA collected in Belize using bacterial tag-encoded FLX amplicon pyrosequencing of the V1–V3 region of the 16S rDNA. We chose CCA species that had previously been tested for their ability to facilitate coral recruitment. The CCA Titanoderma prototypum and Hydrolithon boergesenii facilitate larval settlement and post-settlement survival of two important reef-building acroporid species in the Caribbean (Acropora palmata and Acropora cervicornis) (Ritson-Williams et al., 2010, 2014). These corals are of special interest because they have declined so drastically that they are now listed as threatened on the endangered species list. Two co-occurring CCA species, Paragoniolithon solubile and Porolithon pachydermum, are common members of the reef community but do not directly facilitate recruitment in these acroporid corals (Ritson-Williams et al., 2010, 2014). We compared the bacterial communities associated with these four CCA species to determine whether these algae harbor species-specific bacterial communities that may have implications for coral larval settlement preferences.
Materials and methods
Sample collection
CCA were collected on small pieces of coral rubble from a variety of locations along the Meso-American Barrier Reef at Carrie Bow Cay (CBC), Belize in July 2011. P. solubile (n=5) and H. boergesenii (n=3) were collected at 1 m depth in coral rubble found on the reef flat directly adjacent to the Carrie Bow Cay Field Station (Site 1). T. prototypum (n=5) was collected from 5 m depth from the bottom of the spur and groove habitat in front of South Water Cay (Site 2). P. pachydermum (n=5) was collected at 0.5 m depth from the top of spurs located just over the reef crest at CBC (Site 3). Additional H. beorgesenii (n=2) samples were collected from 10 to 15 m depth in a rubble field along the barrier reef (Site 4). CCA were identified to species based on morphological characteristics (Ritson-Williams et al., 2014).
Individual pieces of CCA were collected in individual sterile Whirl-Pak bags or 50 ml Falcon tubes. Samples were kept in the shade in a 5-gallon bucket with seawater until processing (<3 h). CCA were rinsed under a stream of 30 ml 0.22 μm-filtered seawater and the surfaces (3–14 cm2) were swabbed vigorously with a sterile swab. Swabs were placed in 1 ml filtered seawater and vortexed for 1 min on high. Swabs were removed with sterile forceps, 200 μl was removed for use in other assays and the remaining 800 μl suspension was centrifuged for 25 min at 13 000 r.c.f. The supernatant was removed and the resulting pellet was dissolved in 175 μl of lysis buffer (400 mM NaCl, 750 mM sucrose, 20 mM ethylenediaminetetraacetic acid, 50 mM Tris-HCl pH 9.0) and stored at −20 °C.
Water samples (n=5) were taken from each CCA collection site within Belize in 2 ml sterile microcentrifuge tubes by opening the tubes underwater, allowing them to fill, and then closing them before removing them from the water column. Using aseptic technique, 100 μl were removed from each sample for other assays and the remaining volume was centrifuged at 13 000 r.c.f. for 25 min. The supernatant was removed and 175 μl of lysis buffer was added. Samples were stored at −20 °C.
Sequencing
Samples were sent to Molecular Research LP (MR DNA, Shallowater, TX, USA) for DNA extraction and Bacterial Tag Encoded FLX Amplicon Pyrosequencing of the V1–V3 region of the 16S rDNA (Dowd et al., 2008). 16S rDNA was amplified using universal bacterial primers 27Fmod (AGRGTTTGATCMTGGCTCAG) and 519Rmodbio (GWATTACCGCGGCKGCTG). These primers span the V1 to V3 regions of the 16S rRNA gene, which allows for coverage of multiple hypervariable regions that have been commonly used as taxonomic marker regions for bacteria in marine environments (for example, Barott et al., 2011; Morrow et al., 2012). Amplification was performed in a single step 30 cycle PCR using HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA, USA) starting with an initial denaturation step (3 min at 94 °C), followed by 28 cycles (30 s at 94 °C, 40 s at 53 °C, 1 min at 72 °C) and a final elongation step (5 min at 72 °C). Amplicon products were mixed in equal concentrations, purified using Agencourt Ampure beads (Beckman Coulter, Inc., Danvers, MA, USA), and sequenced with Roche 454 FLX titanium instruments (Roche Diagnostics, Basel, Switzerland) and reagents following the manufacturer's guidelines. The resulting sequence data were processed using a proprietary analysis pipeline (MR DNA, Shallowater, TX, USA). Barcodes and primers were removed and sequences that had less than 200 bp remaining, contained ambiguous base calls or had homopolymer runs exceeding 6 bp were excluded from analysis. The read length of the remaining sequences ranged from 200 to 597 bp with an average read length of 373 bp. These sequences were denoised and chimeras were removed using UCLUST and UCHIME. Sequences that clustered together (UCLUST) with a 97% similarity were defined as an operational taxonomic unit (OTU) and the centroid of each cluster was assigned as the representative sequence. OTUs were taxonomically classified using the curated Greengenes database (DeSantis et al., 2006). Sequences of OTUs that were identified as contributing >0.5% to the dissimilarity among CCA species were also subjected to a BLAST search of the NCBI database. Sequences were submitted to the NCBI SRA database under the study accession number SRP056487. Rarefaction curves for CCA samples (Supplementary Figure S1) and seawater samples (Supplementary Figure S2) can be found in the supplementary information.
Statistical analysis
All statistical analyses were performed based on relative abundances of OTUs within samples. Bacterial community composition of CCA surface and seawater samples were compared using a two-way analysis of similarity (ANOSIM, 9999 permutations) based on the Bray-Curtis distance measure with collection site and sample source (CCA or seawater) as factors. Subsequently, one-way ANOSIMs (9999 permutations) were performed to determine differences in bacterial communities among CCA species and in seawater among collection sites. Similarity percentage analyses based on the Bray-Curtis distance measure were performed to determine which taxa contributed the most to the differentiation among CCA-associated bacterial communities. Data were also subjected to nonmetric multidimensional scaling using the Bray-Curtis measure of similarity for graphical representation. The Chao1 index was used to estimate richness and the Buzas and Gibson evenness index (eH/S, where H=Shannon Diversity Index and S=total number of OTUs) was used to measure evenness. Evenness was compared among CCA species using a one-way analysis of variance (ANOVA). Chao 1 richness estimates failed to meet the assumptions of the one-way ANOVA, and therefore, bacterial richness among CCA species was compared using the Kruskal–Wallis one-way ANOVA followed by the Dunn's Method for multiple comparisons. Statistical analyses were performed using PAST v. 2.17b (Paleontological Statistics) (Hammer et al., 2001) and SigmaPlot 11. Percent abundance data of taxonomic groups within the CCA-associated bacterial communities are presented as the mean±s.d. among replicate individuals within each CCA species unless otherwise stated.
Results and discussion
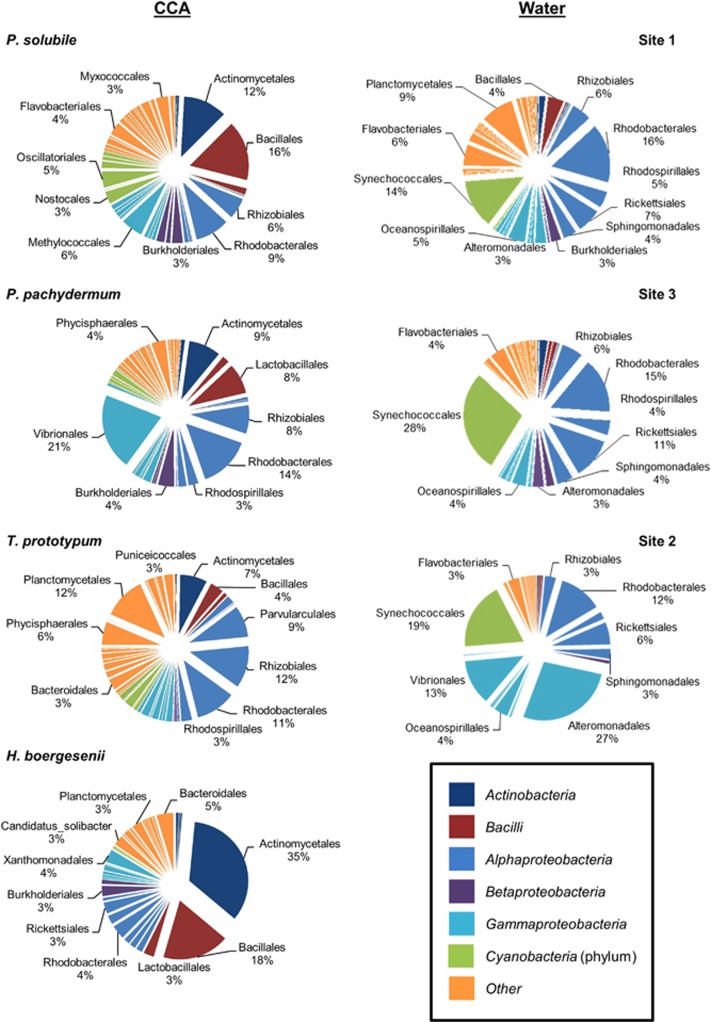
Surfaces of CCA collected in Belize were dominated by Proteobacteria, which made up an average of 27±14% to 61±12% of the total bacterial abundance (Figure 1). Of these, most OTUs belonged to the Alphaproteobacteria with average relative abundances ranging from 15±16% on samples of H. boergesenii to 37±23% on samples of T. prototypum (Figure 1). Gammaproteobacteria were also present in high abundances and accounted for 7±7% (H. boergesenii) to 27±21% (P. pachydermum) of the total relative abundance (Figure 1). Firmicutes were abundant on H. boergesenii (23±11%) and on P. solubile (20±27%), but were not major contributors to the bacterial communities on other samples. These results were similar to those found associated with two species of CCA (Neogoniolithon fosliei and Hydrolithon onkodes) from Australia and unspecified CCA species from Curacao. CCA species from Australia were dominated by Alphaproteobacteria, Gammaproteobacteria and Bacteriodetes (Webster et al., 2011; Webster et al., 2013), and whole tissues of unspecified CCA from Curacao were dominated by Proteobacteria, Firmicutes and unknown bacteria (Barott et al., 2011). Contrary to the results of studies of N. fosliei and H. onkodes from Australia, Bacteriodetes were not among the most dominant bacterial phyla found on the surfaces of CCA from Belize. The highest average relative abundance of Bacteroidetes was found on H. boergesenii (7±6%, Figure 1). In addition to the major phyla found associated with CCA in previous studies (Barott et al., 2011; Webster et al., 2011; Webster et al., 2013), CCA samples from Belize had relatively high numbers of Actinomycetes, ranging from 8±17% on T. prototypum to 37±14% on H. boergesenii (Figure 1).
Figure 1.
Relative abundances of OTUs found on the surfaces of four CCA species and in seawater samples from their corresponding collection sites. H. boergesenii was collected at sites 1 and 4 (no seawater samples). Pie wedges represent order level classification and colors indicate class. Orders making up ⩾3% of the relative abundance are labeled.
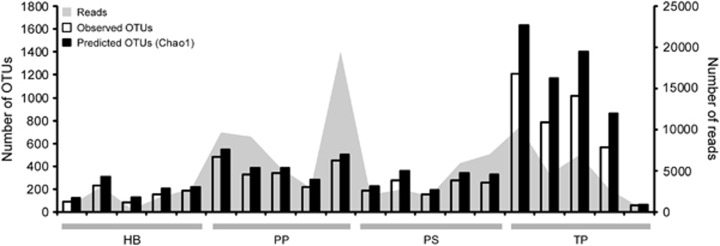
Interspecies variability: diversity
There was a significant difference in the species richness of bacteria associated with different species of CCA as estimated by the Chao1 richness estimator (Kruskal–Wallis one-way ANOVA, P=0.034, Figure 2). Pairwise comparisons revealed that T. prototypum samples had significantly higher estimated species richness (1026.58±610.00 OTUs) compared with H. boergesenii (196.08±74.96 OTUs, Dunn's Method, P<0.05, Figure 2). Bacterial species richness increases with algal age for some algal species (Bengtsson et al., 2012). The high level of species richness found on the surface of T. prototypum may be related to the fact that this CCA species, unlike the others tested here, does not slough its surface cells (see supplementary material in Ritson-Williams et al., 2014) allowing for the development of a more mature bacterial community. Estimated species richness of neither P. solubile (288.06±76.87 OTUs) nor P. pachydermum (419.04±104.90 OTUs) was significantly different from any of the other CCA species tested. There was no difference in the evenness of bacterial communities among CCA species (one-way ANOVA, P=0.852). There was no significant difference in species richness among water samples (777.24±217.82–822.48±261.67 OTUs) collected from the three collection sites (one-way ANOVA, P=0.945) suggesting that variations in richness among CCA species is not determined by the available pool of bacteria in the water column.
Figure 2.
Number of observed and predicted OTUs and number of total reads in samples of CCA surface-associated bacteria. PS, P. solubile; HB, H. boergesenii; TP, T. prototypum; PP, P. pachydermum. Predicted OTUs were calculated using the Chao1 index.
Interspecies variability: composition
The two CCA species known to facilitate coral larval settlement represent the extremes in bacterial species richness, which suggests that the composition may be more important than diversity in terms of providing a bacterial community conducive to larval settlement. What is known to date about the role of bacteria in the induction of coral larval settlement has focused on the role of individual bacterial strains. Several strains have been shown to induce settlement, suggesting that the presence of specific inductive species is more important than the overall diversity of the bacterial community (Negri et al., 2001; Tebben et al., 2011; Tran and Hadfield, 2011; Sneed et al., 2014). However, little work has been carried out to examine the importance of multi-species conglomerations of bacteria on larval settlement preferences.
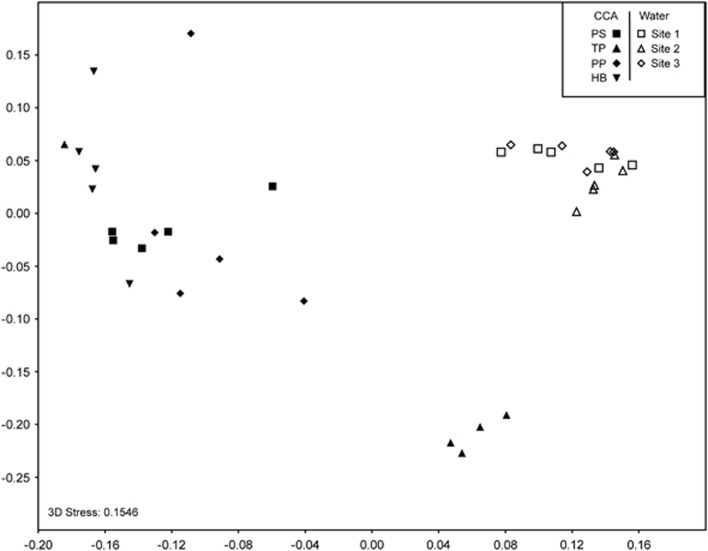
Bacterial community composition on CCA surfaces was significantly different among different CCA species (one-way ANOSIM, R=0.5308, P<0.0001, Figure 3). Pairwise comparisons showed that each CCA species had a significantly different bacterial community composition compared with every other CCA species (Figure 3, Table 1). According to similarity percentage analyses, 14 OTUs contributed >0.5% to the dissimilarity among CCA species (Table 2). The percent similarity for many of these sequences to their closest match is low (<96%) even within the NCBI database. This highlights that the diversity of marine bacteria has much still to be discovered.
Figure 3.
Nonmetric multidimensional scaling plot of bacterial communities on CCA surfaces and in seawater using the Bray-Curtis measure of similarity. Water was collected from the same sites that the CCA were collected. PS and HB were collected from Site 1. TP was collected from Site 2. PP was collected from Site 3. PS, P. solubile; HB, H. boergesenii; TP, T. prototypum; PP, P. pachydermum.
Table 1. Pairwise comparisons of bacterial communities associated with CCA species Hydrolithon boergesenii (HB), Titanoderma prototypum (TP), Paragoniolithon solubile (PS) and Porolithon pachydermum (PP) following one-way ANOSIM.
| R-values |
||||
|---|---|---|---|---|
| HB | TP | PS | PP | |
| P-values | ||||
| HB | — | 0.458 | 0.384 | 0.528 |
| TP | 0.0492 | — | 0.532 | 0.492 |
| PS | 0.0086 | 0.0459 | — | 0.508 |
| PP | 0.0063 | 0.0468 | 0.0078 | — |
Abbreviations: ANOSIM, analysis of similarity; CCA, crustose coralline algae.
R values are given above the diagonal and P values beneath the diagonal. P values <0.05 are indicated in bold.
Table 2. SIMPER analyses, OTUs contributing >0.5% to the dissimilarity among CCA species Hydrolithon boergesenii (HB), Titanoderma prototypum (TP), Paragoniolithon solubile (PS) and Porolithon pachydermum (PP).
| OTU # |
Mean % abundance (# of samples containing OTU) |
% contr.to dissim. | Order | Closest taxonomic match—Greengenes | % ID | Closest match source—NCBI (accession #) | % ID | |||
|---|---|---|---|---|---|---|---|---|---|---|
| HB | TP | PS | PP | |||||||
| 1504 | 12.10 (5) | 4.06 (1) | 3.64 (5) | 2.61 (5) | 3.90 | Actinomycetales | Propionibacterium acnes | 96.5 | Deep-sea sediment (JN914883.1) | 98 |
| 4606 | 0.00 (0) | 0.26 (4) | 0.05 (1) | 5.87 (4) | 1.59 | Vibrionales | Vibrio harveyi | 97.5 | Coral-associated (KC545338.1) | 97 |
| 4092 | 0.90 (4) | 0.00 (0) | 3.89 (3) | 0.23 (5) | 1.22 | Bacillales | Staphylococcus epidermidis | 96.1 | Deep-sea sediment (JN913945.1) | 96 |
| 1778 | 3.64 (5) | 0.98 (1) | 1.48 (5) | 1.29 (5) | 1.17 | Actinomycetales | Propionibacterium acnes | 99.2 | Ocean drilling core (AB803950.1) | 99 |
| 198 | 2.20 (3) | 0.53 (1) | 0.00 (0) | 0.00 (0) | 0.69 | Bacillales | Planomicrobium spp. | 95.7 | River sediment (HF545332.1) | 98 |
| 2743 | 0.93 (4) | 0.00 (0) | 1.82 (2) | 0.11 (5) | 0.68 | Bacillales | Staphylococcus epidermidis | 97.3 | Coral-associated (FJ809136.1) | 99 |
| 911 | 0.00 (0) | 0.00 (0) | 2.41 (2) | 0.00 (0) | 0.64 | Oscillatoriales | Oscillatoria spongeliae | 85.5 | Coral-associated (JQ347397.1) | 94 |
| 5734 | 0.00 (0) | 0.00 (0) | 2.31 (1) | 0.00 (0) | 0.61 | Nostocales | Anabaena cylindrica | 85.8 | Coral-associated (JQ347397.2) | 93 |
| 6908 | 0.00 (0) | 2.26 (4) | 0.00 (0) | 0.00 (0) | 0.60 | Rhodobacterales | Rhodobacteraceae spp. | 88.6 | Coral-associated (GU119607.1) | 94 |
| 635 | 0.00 (0) | 0.06 (2) | 0.01 (1) | 2.06 (4) | 0.56 | Vibrionales | Vibrio harveyi | 97.1 | V. harveyi/V. coralliilyticus genome (CP009467.2/CP009617.1) | 97/97 |
| 1555 | 1.19 (2) | 0.98 (1) | 0.00 (0) | 0.00 (0) | 0.54 | Bacillales | Planomicrobium spp. | 96.8 | Bovine rumen (EU845583.1) | 98 |
| 1706 | 0.49 (4) | 0.04 (1) | 1.66 (3) | 0.13 (5) | 0.53 | Bacillales | Staphylococcus epidermidis | 96.1 | Human skin (JF030251.1) | 96 |
| 8276 | 0.00 (0) | 1.93 (4) | 0.00 (0) | 0.00 (0) | 0.51 | Planctomycetales | Blastopirellula spp. | 87.7 | Nuclear plant biofilm/marine sponge (HM596372.1/FJ652502.1) | 99/96 |
| 580 | 0.00 (0) | 0.00 (0) | 0.07 (1) | 1.85 (1) | 0.51 | Rhodobacterales | Roseovarius spp. | 94.0 | Coral reef microbial mat (AB294940.1) | 95 |
Abbreviations: CCA, crustose coralline algae; OTU, operational taxonomic unit; SIMPER, similarity percentage.
Closest taxonomic match based on percent similarity to sequences in the Greengenes database. Closest match source based on a BLAST search of the NCBI database. The highest mean % abundances are highlighted in bold for each OTU.
Surfaces of H. boergesenii had higher abundances of Actinomycetes compared with the other three CCA species (Figure 1, Table 2). The OTU (1504) that contributed the most to the dissimilarity between CCA species aligns most closely (96.5%) with an Actinomycete in the genus Propionibacterium that was broadly present across CCA species, but made up a large proportion (12.10±8.25%) of the sequences found on the surfaces of H. boergesenii and was less common (2.61±1.69% to 4.06±9.09%) on the other three species (Table 2). Another OTU (1778) aligning most closely (99.2%) to Propionibacterium acne was also present in higher abundances on H. boergesenii (3.64±3.33%) compared with the other CCA species (0.98±2.19–1.48±0.48%, Table 2). Propionibacteria are known mostly for their role as commensal microbes or opportunistic pathogens on human skin and as fermenters in food making (Grice and Segre, 2011; Thierry et al., 2011). They have been found in marine samples (La Rivière et al., 2013; Li et al., 2014), and a BLAST search of these two OTUs reveals close matches (98% and 99%, respectively) to uncultured bacteria from deep-sea sediments and ocean drilling cores (Table 2). Although their ecology in this environment remains largely unknown, two strains of Propionibacterium isolated from the coral Acropora digitifera exhibit broad-spectrum antibiotic activity (Nithyanand et al., 2011). Two OTUs (198 and 1555) aligning most closely (95.7% and 96.8%, respectively) to Planomicrobium spp. (order Bacillales) were found associated with H. boergesenii (2.20±3.32% and 1.19±2.35%). The same two OTUs were found in high abundances on one of the five T. prototypum samples (4.90% and 2.63%) but were not found on either of the other two CCA species (Table 2). Planomicrobium spp. have been isolated from diverse marine environments and exhibit algicidal and antimicrobial activity (Skerratt et al., 2002). Members of this genus isolated from coral mucus reduced growth and biofilm formation of coral pathogens (Skerratt et al., 2002; Shnit-Orland and Kushmaro, 2009; Alagely et al., 2011). T. prototypum also had higher abundances than other CCA of an OTU (6908) in the order Rhodobacterales (2.26±3.67%) that aligned weakly (88.6%) to species of Rhodobacteraceae and an OTU (8276) in the order Planctomycetales (1.93±1.47%) that aligned weakly (87.7%) to Blastopirellula spp. Rhodobacterales have often been associated with coral disease (Mouchka et al., 2010); however, the low percent similarity of OTU 6908 to its closest match in the curated Greengenes database makes it difficult to attribute taxonomic identification. A BLAST search reveals that this OTU is most closely related (92–94% similarity) to uncultured bacteria associated with a variety of corals including early life history stages of Porites astreoides. The closest match (99%) for OTU 8276 within the NCBI database is that of an uncultured bacterium from a biofilm within a nuclear plant (Table 2). The closest match to a marine source was a 96% match to an uncultured bacterium associated with a marine sponge (Table 2).
P. solubile had higher numbers of Staphylococcus epidermidis and cyanobacteria (Oscillatoriales and Nostocales). Three OTUs (4092, 2743 and 1706) aligning with S. epidermidis contributed greater than 0.5% to the differences among CCA species and were found in higher relative abundances on P. solubile (3.89±7.41%, 1.82±3.36% and 1.66±3.12%) compared with the other three CCA species. Additionally, two OTUs that align with cyanobacteria in the orders Oscillatoriales and Nostocales were found only on P. solubile (2.41±5.36% and 2.31±5.16%, respectively) and not on any of the other species. Although these OTUs were not found on every P. solubile sample, they constituted a relatively high percentage of the total reads among P. solubile samples. Staphylococcus epidermidis is most often associated with human skin; however two of the three S. epidermidis OTUs found on P. solubile had close matches within the NCBI database to uncultured bacteria from marine environments (Table 2). Staphylococcus spp. have been isolated from algal biofilms and coral mucus (de Castro et al., 2010; Horta et al., 2014), and there is some evidence linking Staphylococcus strains to coral disease (Kellogg et al., 2013). Cyanobacteria, especially those in the order Oscillatoriales, inhibit settlement of coral larvae and have been implicated as causative agents of black band disease (Frias-Lopez et al., 2003; Kuffner and Paul, 2004; Kuffner et al., 2006; Casamatta et al., 2012). Larvae of multiple coral species avoid settling on the surface of P. solubile (Ritson-Williams et al., 2010; Ritson-Williams et al., 2014), and this may be due to inhibitory microbial OTUs that cause larval avoidance of this species of CCA. Interestingly, two methanotrophic OTUs (1796 and 378) were found exclusively on P. solubile and were found on every P. solubile sample. A BLAST search of both OTUs revealed that the closest matches (93%) are uncultured bacteria associated with the coral Montastraea (=Orbicella) faveolata (Sunagawa et al., 2009). The presence of methanotrophs within marine algal biofilms has rarely been reported and is more common on freshwater plants (Yoshida et al., 2014). The presence of these bacteria on the surfaces of P. solubile and their relatedness to OTUs found in coral mucus makes further exploration of these bacteria and their ecological roles an interesting direction for future research.
Porolithon pachydermum samples had high abundances of Vibrios with sequences in the order Vibrionales making up 0–63.1% (21.0±25.0%) of the total sequences found on the surfaces of individuals (Figure 1). No other CCA species had an average abundance of Vibrionales greater than 3% (Figure 1). Of these sequences, two OTUs (4606 and 635) aligning with V. harveyi were found in high abundances on P. pachydermum (5.87±7.60% and 2.06±2.76%) and contributed greater than 0.5% to the differences between CCA species. Additionally, an OTU (580) aligning (94%) with Roseovarius spp. in the family Rhodobacteraceae was found in higher abundances on P. pachydermum (1.85±4.14%) compared with other CCA species. Both V. harveyi and members of the Rhodobacteraceae have been associated with invertebrate diseases including coral disease (Godwin et al., 2012; Roder et al., 2014).
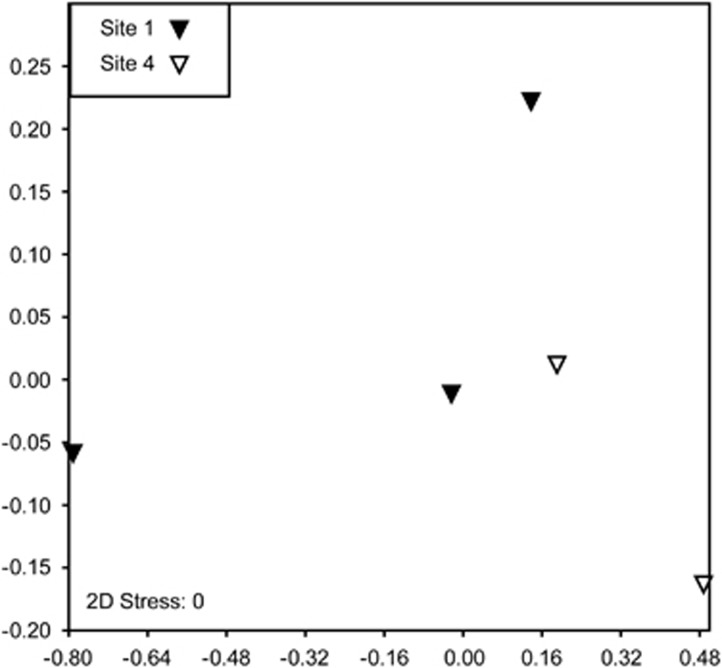
Water samples collected from the CCA collection sites had significantly different bacterial communities compared with CCA surfaces (two-way ANOSIM, R=0.7506, P<0.001, Figure 3). Bacterial communities (including those associated with CCA and seawater) also differed by collection site (two-way ANOSIM, R=0.4214, P<0.001), indicating that collection site may be impacting the bacteria associated with CCA. However, H. boergesenii samples were collected from two different sites and showed no significant difference in bacterial communities across sites (one-way ANOSIM, R=0, P=0.5034, Figure 4). Coral larvae will settle on individuals of H. boergesenii collected from both sites (Ritson-Williams, personal observation) providing further support that properties specific to the species and not the collection site are responsible for the settlement inductive activity of this CCA.
Figure 4.
Nonmetric multidimensional scaling plot of bacterial communities on the surface H. boergesenii samples collected from two different sites based on the Bray-Curtis measure of similarity.
Implications
CCA are important components of marine environments, and CCA species are often grouped together in terms of their functioning in ecosystems. This study demonstrates that different CCA species host different bacterial communities, highlighting the importance of assessing the ecological roles of individual CCA species. Here, we examined CCA species known to elicit different settlement responses in coral larvae. Identifying the bacterial OTUs that contribute to the dissimilarity among CCA species provides insights into the microbial ecology of these algae and allows us to form hypotheses about the complex interactions between CCA and invertebrate larvae. Corals may have evolved to select settlement substrata that contain beneficial bacteria on their surfaces. For example, Planomicrobium spp. that were found on CCA species that facilitate coral larval settlement (T. prototypum and H. boergesenii) may enhance the survival of new recruits by inhibiting the growth of coral pathogens. Additionally, larvae may avoid certain substrata owing to the presence of pathogenic or allelopathic bacteria. Both of the CCA species that have been shown to be avoided by coral larvae, P. solubile and P. pachydermum, have high abundances of bacteria that are closely related to known coral pathogens (Vibrios and Rhodobacteraceae) or cyanobacteria that are known to produce allelopathic compounds (Oscillatoriales). Coral larvae respond to a variety of physical and chemical cues (Gleason and Hofmann, 2011) during the settlement process including the presence of bacterial biofilms and specific bacterial strains within biofilms (Negri et al., 2001; Tran and Hadfield, 2011; Sneed et al., 2014). Gleason and Hofmann (2011) proposed a ‘hierarchy of action' in which coral larvae hone in on a settlement location through a series of cues that function at increasingly smaller spatial scales. It is possible that coral larvae may be able to assess the suitability of substrata for settlement at a fine scale (within a reef) based on the bacterial communities on the surface of the preferred CCA. Corals are especially vulnerable during their early life history stages and recruiting into a favorable environment is critical for their survival and persistence (Ritson-Williams et al., 2009). Some corals acquire their internal bacterial communities only after settlement and metamorphosis (Sharp et al., 2010). It is therefore likely that the bacterial communities associated with settlement substrata are important to coral recruits not only in terms of direct impacts on their health, but also as a reservoir for the development of the coral's microbiome. The findings presented here demonstrate that different species of CCA have different assemblages of bacteria associated with them and provide a basis for future research into the role of microbial community composition on the selection of settlement substrata by invertebrate larvae.
Acknowledgments
Funding was provided by the Smithsonian Institution's Next Generation Sequencing Small Grant and the Smithsonian Competitive Grants Program for Science. Support for J Sneed's postdoctoral fellowship was provided by the Smithsonian Marine Station at Fort Pierce. We thank Dr Mary Rice for her helpful input on this project. We thank the Belize Fisheries Department for providing permits to conduct research at Carrie Bow Cay, Belize. This is contribution no. 992 from the Smithsonian Marine Station and 973 from the CCRE program.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alagely A, Krediet CJ, Ritchie KB, Teplitski M. (2011). Signaling-mediated cross-talk modulates swarming and biofilm formation in a coral pathogen Serratia marcescens. ISME J 5: 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barott KL, Rodriguez-Brito B, Janouskovec J, Marhaver KL, Smith JE, Keeling P et al. (2011). Microbial diversity associated with four functional groups of benthic reef algae and the reef-building coral Montastraea annularis. Environ Microbiol 13: 1192–1204. [DOI] [PubMed] [Google Scholar]
- Bengtsson MM, Sjotun K, Lanzen A, Ovreas L. (2012). Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J 6: 2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamatta D, Stanic D, Gantar M, Richardson LL. (2012). Characterization of Roseofilum reptotaenium (Oscillatoriales, Cyanobacteria) gen. et sp nov isolated from Caribbean black band disease. Phycologia 51: 489–499. [Google Scholar]
- de Castro AP, Araujo SD, Reis AMM, Moura RL, Francini RB, Pappas G et al. (2010). Bacterial community associated with healthy and diseased reef coral Mussismilia hispida from eastern Brazil. Microb Ecol 59: 658–667. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG et al. (2008). Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Bonheyo GT, Jin Q, Fouke BW. (2003). Cyanobacteria associated with coral black band disease in Caribbean and Indo-Pacific reefs. Appl Environ Microb 69: 2409–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason DF, Hofmann DK. (2011). Coral larvae: From gametes to recruits. J Exp Mar Biol Ecol 408: 42–57. [Google Scholar]
- Godwin S, Bent E, Borneman J, Pereg L. (2012). The role of coral-associated bacterial communities in Australian Subtropical White Syndrome of Turbinaria mesenterina. Plos One 7: e44243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. (2011). The skin microbiome. Nat Rev Micro 9: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield MG, Paul VJ. (2001). Natural chemical cues for settlement and metamorphosis of marine invertebrate larvae. In: McClintock J, Baker B (eds), Marine Chemical Ecology. CRC Press: Boca Raton, pp 431–462. [Google Scholar]
- Hadfield MG. (2011). Biofilms and marine invertebrate larvae: What bacteria produce that larvae use to choose settlement sites. Annu Rev Mar Sci 3: 453–470. [DOI] [PubMed] [Google Scholar]
- Hammer O, Harper DAT, Ryan PD. (2001). PAST: Paleontological Statistics software package for education and data analysis. Palaeontol Electron 4: 9. [Google Scholar]
- Harrington L, Fabricius K, De'ath G, Negri A. (2004). Recognition and selection of settlement substrata determine post-settlement survival in corals. Ecology 85: 3428–3437. [Google Scholar]
- Horta A, Pinteus S, Alves C, Fino N, Silva J, Fernandez S et al. (2014). Antioxidant and antimicrobial potential of the Bifurcaria bifurcata epiphytic bacteria. Mar Drugs 12: 1676–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CR, Muir CD, Reysenback A-L. (1991). Characteristic bacteria associated with the surfaces of coralline algae: a hypothesis for bacterial induction of marine invertebrate larvae. Mar Ecol Prog Ser 74: 281–294. [Google Scholar]
- Kellogg CA, Piceno YM, Tom LM, DeSantis TZ, Gray MA, Zawada DG et al. (2013). Comparing bacterial community composition between healthy and White Plague-Like Disease states in Orbicella annularis using PhyloChip G3 microarrays. Plos One 8: e79801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffner IB, Paul VJ. (2004). Effects of the benthic cyanobacterium Lyngbya majuscula on the larval settlement of the reef corals Acropora surculosa and Pocillopora damicornis. Coral Reefs 23: 455–458. [Google Scholar]
- Kuffner IB, Walters LJ, Becerro MA, Paul VJ, Ritson-Williams R, Beach K. (2006). Inhibition of coral recruitment by macroalgae and cyanobacteria. Mar Ecol Prog Ser 323: 107–117. [Google Scholar]
- La Rivière M, Roumagnac M, Garrabou J, Bally M. (2013). Transient shifts in bacterial communities associated with the temperate gorgonian Paramuricea clavata in the Northwestern Mediterranean Sea. Plos One 8: e57385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachnit T, Blumel M, Imhoff JF, Wahl M. (2009). Specific epibacterial communities on macroalgae: phylogeny matters more than habitat. Aquat Biol 5: 181–186. [Google Scholar]
- Lachnit T, Meske D, Wahl M, Harder T, Schmitz R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ Microbiol 13: 655–665. [DOI] [PubMed] [Google Scholar]
- Li ZY, Wang YZ, He LM, Zheng HJ. (2014). Metabolic profiles of prokaryotic and eukaryotic communities in deep-sea sponge Lamellomorpha sp indicated by metagenomics. Sci Rep 4: 3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littler MM, Littler DS. (2013). The nature of crustose coralline algae and their interactions on reefs. Smith Contrib Mar Sci 39: 199–212. [Google Scholar]
- Longford SR, Tujula NA, Crocetti GR, Holmes AJ, Holmstrom C, Kjelleberg S et al. (2007). Comparisons of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat Microb Ecol 48: 217–229. [Google Scholar]
- Morrow KM, Moss AG, Chadwick NE, Liles MR. (2012). Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl Environ Microb 78: 6438–6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchka ME, Hewson I, Harvell CD. (2010). Coral-associated bacterial assemblages: Current knowledge and the potential for climate-driven impacts. Integr Comp Biol 50: 662–674. [DOI] [PubMed] [Google Scholar]
- Negri AP, Webster NS, Hill RT, Heyward AJ. (2001). Metamorphosis of broadcast spawning corals in response to bacteria isolated from crustose algae. Mar Ecol Prog Ser 223: 121–131. [Google Scholar]
- Nithyanand P, Manju S, Karutha Pandian S. (2011). Phylogenetic characterization of culturable actinomycetes associated with the mucus of the coral Acropora digitifera from Gulf of Mannar. Fems Microbiol Lett 314: 112–118. [DOI] [PubMed] [Google Scholar]
- Pawlik JR. (1992). Chemical ecology of the settlement of benthic marine invertebrates. Oceanogr Mar Biol 30: 273–335. [Google Scholar]
- Ritson-Williams R, Arnold SN, Fogarty N, Steneck RS, Vermeij MJ, Paul. VJ. (2009). New perspectives on ecological mechanisms affecting coral recruitment on reefs. Smith Contrib Mar Sci 38: 437–457. [Google Scholar]
- Ritson-Williams R, Paul VJ, Arnold SN, Steneck RS. (2010). Larval settlement preferences and post-settlement survival of the threatened Caribbean corals Acropora palmata and A. cervicornis. Coral Reefs 29: 71–81. [Google Scholar]
- Ritson-Williams R, Arnold SN, Paul VJ, Steneck RS. (2014). Larval settlement preferences of Acropora palmata and Montastraea faveolata in response to diverse red algae. Coral Reefs 33: 59–66. [Google Scholar]
- Roder C, Arif C, Bayer T, Aranda M, Daniels C, Shibl A et al. (2014). Bacterial profiling of White Plague Disease in a comparative coral species framework. ISME J 8: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp KH, Ritchie KB, Schupp PJ, Ritson-Williams R, Paul VJ. (2010). Bacterial acquisition in juveniles of several broadcast spawning coral species. Plos One 5: e10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnit-Orland M, Kushmaro A. (2009). Coral mucus-associated bacteria: a possible first line of defense. Fems Microbiol Ecol 67: 371–380. [DOI] [PubMed] [Google Scholar]
- Skerratt JH, Bowman JP, Hallegraeff G, James S, Nichols PD. (2002). Algicidal bacteria associated with blooms of a toxic dinoflagellate in a temperate Australian estuary. Mar Ecol Prog Ser 244: 1–15. [Google Scholar]
- Sneed JM, Pohnert G. (2011). The green alga Dicytosphaeria ocellata and its organic extracts alter natural bacterial biofilm communities. Biofouling 27: 347–356. [DOI] [PubMed] [Google Scholar]
- Sneed JM, Sharp KH, Ritchie KB, Paul VJ. (2014). The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. P Roy Soc B-Biol Sci 281: 20133086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, DeSantis TZ, Piceno YM, Brodie EL, DeSalvo MK, Voolstra CR et al. (2009). Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J 3: 512–521. [DOI] [PubMed] [Google Scholar]
- Tebben J, Tapiolas DM, Motti CA, Abrego D, Negri AP, Blackall LL et al. (2011). Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. Plos One 6: e19082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry A, Deutsch SM, Falentin H, Dalmasso M, Cousin FJ, Jan G. (2011). New insights into physiology and metabolism of Propionibacterium freudenreichii. Int J Food Microbiol 149: 19–27. [DOI] [PubMed] [Google Scholar]
- Tran C, Hadfield MG. (2011). Larvae of Pocillopora damicornis (Anthozoa) settle and metamorphose in response to surface-biofilm bacteria. Mar Ecol Prog Ser 433: 85–96. [Google Scholar]
- Webster NS, Smith LD, Heyward AJ, Watts JEM, Webb RI, Blackall LL et al. (2004). Metamorphosis of a scleractinian coral in response to microbial biofilms. Appl Environ Microb 70: 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Soo R, Cobb R, Negri AP. (2011). Elevated seawater temperature causes a microbial shift on crustose coralline algae with implications for the recruitment of coral larvae. ISME J 5: 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Uthicke S, Botte ES, Flores F, Negri AP. (2013). Ocean acidification reduces induction of coral settlement by crustose coralline algae. Global Change Biol 19: 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Iguchi H, Yurimoto H, Murakami A, Sakai Y. (2014). Aquatic plant surface as a niche for methanotrophs. Front Microbiol 5: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.