Abstract
The magnitude, duration and oscillation of cellular signalling pathway responses are often limited by negative feedback loops, defined as an ‘activator-induced inhibitor’ regulatory motif. Within the NFκB signalling pathway, a key negative feedback regulator is IκBα. We show here that, contrary to current understanding, NFκB-inducible expression is not sufficient for providing effective negative feedback. We then employ computational simulations of NFκB signalling to identify IκBα molecular properties that are critical for proper negative feedback control and test the resulting predictions in biochemical and single-cell live-imaging studies. We identified nuclear import and nuclear export of IκBα and the IκBα–NFκB complex, as well as the free IκBα half-life, as key determinants of post-induction repression of NFκB and the potential for subsequent reactivation. Our work emphasizes that negative feedback is an emergent systems property determined by multiple molecular and biophysical properties in addition to the required ‘activator-induced inhibitor’ relationship.
Keywords: negative feedback, gene regulation, NFκB, IκBα, oscillation
1. Introduction
Negative feedback control is a ubiquitous regulatory motif in many biological systems, critical to the maintenance of proper homeostasis, dynamic control in response to perturbations, or oscillatory patterns [1]. The defining feature of a negative feedback motif is an activator–inhibitor pair in which the activator induces expression or activity of the inhibitor. Indeed, many studies focus on the molecular mechanism(s) that provide(s) inducibility, often characterized by the fold change and any intrinsic delay. However, actual molecular circuits within cells are incompletely described by the activator-inducible inhibitor paradigm, as they may need to contend with physical realities within the cell such as the biochemistry of molecular interactions, sub-cellular compartmentalization or protein half-life. Thus, proper functioning of a negative feedback circuit may depend on biochemical properties other than the activator-responsive control of the inhibitor.
IκBα is a prominent negative feedback regulator in the NFκB signalling system [2,3]. IκBα directly controls the dynamics of the transcription factor NFκB, a central regulator of inflammatory and immune response gene expression [4,5]. Through its reversible sequestration of NFκB in the cytoplasm, IκBα not only controls the duration of NFκB activity [4,6] but also enables reactivation that can result in oscillatory dynamics observed both in population studies [4] and in single cells [7]. Mathematical models were shown to recapitulate these dynamic features [8,9], and reduced models have identified NFκB-responsive expression of IκBα as a key determinant of oscillatory dynamics [10–12]. The dynamics of NFκB signalling are stimulus-specific, and a critical determinant of inflammatory and immune gene expression programmes [13,14], prompting pioneering work to focus drug-targeting strategies on dynamical features to achieve superior specificity [15].
Here, we examine the molecular properties that confer IκBα's ability to control NFκB dynamics. We find that while inducible expression of IκBα is required for proper NFκB dynamics [16], inducible expression is not sufficient as inducible expression of another IκB family member, IκBβ, is unable to support normal dynamical control of NFκB. This finding prompts us to characterize other IκBα properties that are required for proper negative feedback control of NFκB. Our study delineates how several molecular properties combine to produce the emergent systems property of dynamic negative feedback control of NFκB.
2. Results
2.1. NFκB-responsive transcriptional control is necessary but not sufficient for IκBα negative feedback
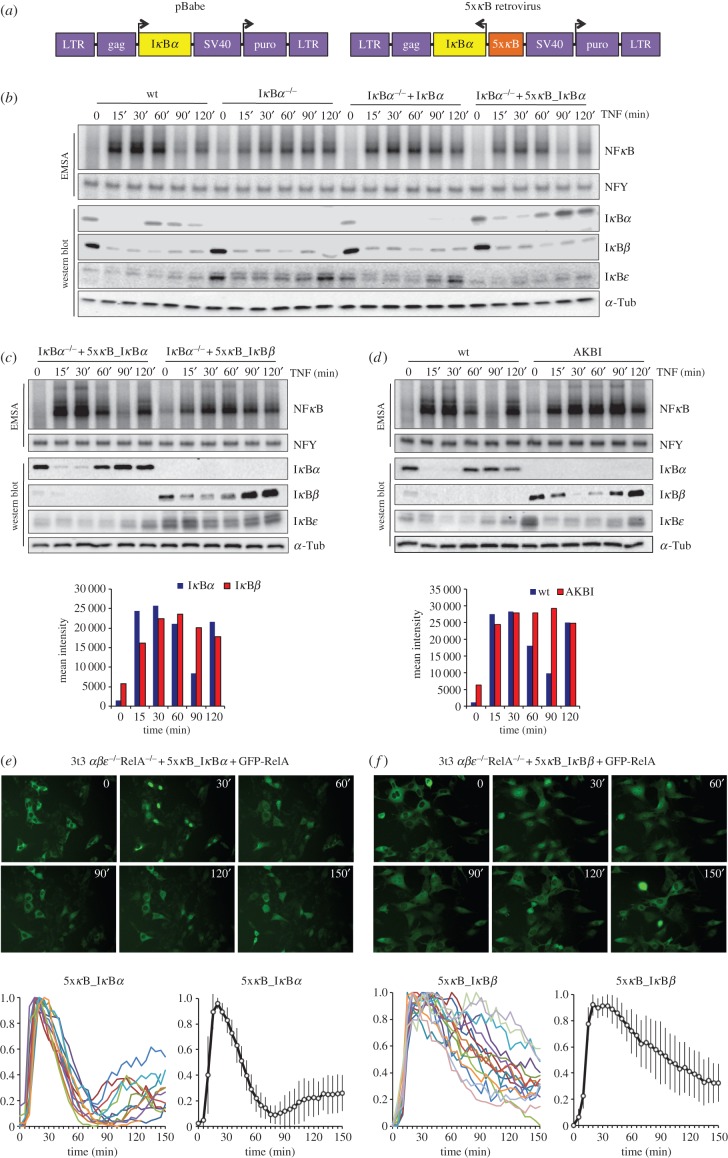
Studies of NFκB dynamic control by IκB family members have identified IκBα as the key negative feedback regulator due to its highly inducible NFκB-responsive promoter [2–4]. To characterize the role of NFκB-inducible expression, we complemented IκBα-deficient murine embryo fibroblasts (MEFs) with retroviral plasmids that express IκBα from either a constitutive (pBabe) or an NFκB-inducible (5xκB) promoter (figure 1a). Unlike pBabe-reconstituted cells, 5xκB_IκBα reconstituted cells showed dynamic resynthesis profiles similar to endogenous IκBα in wild-type cells following stimulation with tumour necrosis factor (TNF) (figure 1b). Importantly, when we examined the control of NFκB activity by electrophoretic mobility shift assay (EMSA), we found that 5xκB cells showed post-induction repression and the transient trough of NFκB activity characteristic of wild-type cells, correcting the misregulation in IκBα-deficient cells, whereas cells constitutively expressing IκBα were unable to capture this response (figure 1b).
Figure 1.
NFκB-dependent transcriptional control is not sufficient for IκB negative functions. (a) Schematic diagram of pBabe and 5xκB retroviral expression constructs consisting of a tandem repeat of 5xκB sites driving the expression of IκBα. (b) Electrophoretic mobility shift assay (EMSA) and immunoblot analysis of wt, IκBα−/−, IκBα−/−+ pBabe_IκBα, and IκBα−/−+ 5xκB_ IκBα murine embryo fibroblast (MEF) cell lines. EMSA indicates NFκB activity over a 120 min time course after stimulation with 1 ng ml−1 of TNF; NFY binding was used as an EMSA control. Western blot shows protein abundances for IκBα, IκBβ and IκBε with α-tubulin as a loading control. (c) IκBα−/− MEFs reconstituted with NFκB-inducible IκBα or IκBβ were treated with 1 ng ml−1 of TNF and nuclear extracts analysed by EMSA for NFκB binding activity; NFY binding was used as an EMSA control. Immunoblots of corresponding cytoplasmic fractions were probed with antibodies specific for IκBα, IκBβ and IκBε; α-tubulin was used as a loading control. Densitometric quantification of NFκB is presented as a bar graph below. (d) EMSA and immunoblot analysis of wild-type MEFs and MEFs that have the endogenous coding region for IκBα replaced by IκBβ knock-in (AKBI). Cells were treated with 1 ng ml−1 of TNF and nuclear extracts analysed by EMSA for NFκB-binding activity; NFY binding was used as an EMSA control. Immunoblots of corresponding cytoplasmic fractions were probed with antibodies specific for IκBα, IκBβ and IκBε; α-tubulin was used as a loading control. Densitometric quantification of NFκB is presented as a bar graph below. (e,f) NFκB nuclear localization at the single-cell level. IκBα−/−β−/−ε−/−RelA−/− MEFs reconstituted with AcGFP1-RelA and NFκB-inducible IκBs. (e) IκBα cells were treated with 10 ng of TNF and fluorescent images were captured every 5 min and cellular localization of AcGFP1-RelA was measured and plotted as a normalized nuclear to cytoplasmic ratio individually (bottom left colour traces) and as a combined average (bottom right black trace). (f) IκBβ cells were treated with 10 ng of TNF and fluorescent images were captured every 5 min and cellular localization of AcGFP1-RelA was measured and plotted as a normalized nuclear to cytoplasmic ratio individually (bottom left colour traces) and as a combined average (bottom right black trace).
To test whether NFκB-inducible control was not only required but also sufficient for NFκB dynamic control, we complemented IκBα-deficient cells with a 5xκB retrovirus expressing IκBβ, a highly homologous IκB family member capable of inhibiting NFκB but not normally providing negative feedback. Interestingly, these cells did not show proper dynamic control of NFκB even though IκBβ expression was under NFκB control similar to IκBα (figure 1c; electronic supplementary material, figure S1A). However, when another known IκB negative feedback regulator, IκBε [17], was linked to this promoter, NFκB activity did show post-induction repression (electronic supplementary material, figure S1B). These results indicate that, despite the high degree of sequence homology, IκBα and IκBβ have distinct molecular properties that, along with differential gene expression control, render IκBα an effective negative feedback regulator but not IκBβ. In order to confirm the validity of this conclusion, we obtained fibroblasts from a genetic knock-in mouse in which the IκBβ coding region was engineered to replace the IκBα open reading frame such that IκBβ expression was under the control of the endogenous IκBα promoter [18]. Remarkably, these so-called AKBI cells also failed to show proper NFκB post-induction attenuation despite highly inducible IκBβ expression (figure 1d; electronic supplementary material, figure S1c).
In order to examine translocation dynamics in single cells, and without the confounding contributions of other IκB family members, we generated IκBα−/−β−/−ε−/−RelA−/−3T3 cells that lack all three classical NFκB inhibitors and RelA, and reconstituted them with a constitutively expressed fluorescent GFP-RelA and NFκB-responsively expressed IκBα or IκBβ. Whereas reconstitution with IκBα resulted in transient NFκB activation in response to TNF treatment, defined by a trough at about 60 min, followed by a second phase in some cells (figure 1e), reconstitution with NFκB-inducible IκBβ resulted in sustained NFκB activation showing only slow and incomplete post-induction repression (figure 1f). Of note, in both conditions, the mean RelA nuclear localization profile of the collection of individual cells (figure 1e,f, black trace) closely resembled the population level in biochemical studies (figure 1c,d). These data clearly indicate that when expression of IκBα or IκBβ is driven by the same NFκB-responsive promoter, resulting in ostensibly similar expression profiles, only IκBα can provide effective dynamic negative feedback control on NFκB. Thus, inducible inhibitor expression in and of itself is not sufficient for proper negative feedback control of NFκB.
2.2. Mathematical modelling identifies multiple molecular properties of IκBα contributing to the negative feedback control of NFκB
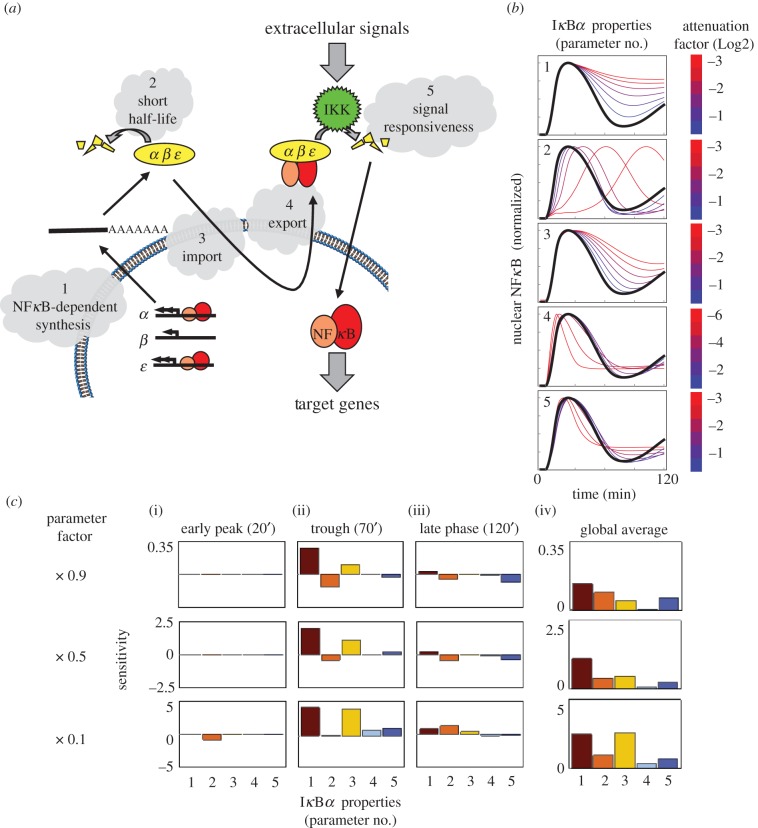
IκBα has several characteristics—other than NFκB-dependent synthesis—that in principle may contribute to its negative feedback function, e.g. its nuclear import and export properties, as well as constitutive and signal-induced degradation of free and NFκB-bound IκBα (figure 2a). Here, we use a previously established in silico model of NFκB regulation to investigate the contributions of each of these processes to the control of dynamic NFκB signals. When normalized for maximum activity, we confirmed that partial inhibition of the NFκB-dependent synthesis of IκBα potently impaired post-induction attenuation, with 10% inhibition resulting in a 30% increase in the signalling level at 70 min (figure 2b, row 1, and 2c). However, we also found that partial inhibition of IκBα nuclear import had a similar effect with a 10% inhibition causing an 11% increase in signalling at 70 min (figure 2b, row 3). Weak inhibition of the degradation of free IκBα had little effect on post-attenuation induction, whereas stronger inhibition shifted the peak of NFκB to later times resulting in a modest increase in late activity (90% inhibition caused 14% increase in signalling at 120 min, figure 2b, row 2). Partial inhibition of nuclear export or of signal-induced degradation of NFκB-bound IκBα also reduced the post-attenuation reactivation, with a 10% inhibition causing 1% and 9.5% decreased signalling at 120 min, respectively (figure 2b, rows 4 and 5).
Figure 2.
Modelling IκBα properties contributing to negative feedback control of NFκB. (a) Schematic illustrates negative feedback control of NFκB by IκBs. Numbers indicate potential reactions that may contribute to dynamic regulation of NFκB. (b) Normalized nuclear NFκB concentration is shown for unperturbed models (black) and models in which the indicated reactions are partially inhibited (blue-red lines reflect various degrees of inhibition). Reaction numbers as in (a). (c) Sensitivity analysis of three temporal phases of the NFκB with respect to changes in IκBα regulation. ((i)–(iii)) Sensitivity ratio (nucNFκBperturbed − nucNFκBunperturbed)/nucNFκBunperturbed in per cent units for each reaction (numbers as in (a)). (iv) Global sensitivity (RMSD) integrated over 120 min. Results are shown for three levels of inhibition (10%, twofold and 10-fold).
To compare the contribution of these processes, we determined the sensitivity of NFκB activity at three specific times representing early, post-induction attenuation and late parts of the signal to various perturbations (figure 2c). We also quantified the global sensitivity to each perturbation as the root mean square deviation (RMSD) of the perturbed and unperturbed signals over 120 min, sampled at 1 min intervals (figure 2c(iv)). This analysis posits that IκBα-mediated post-induction attenuation of NFκB activity (figure 2c(ii)) is a function not only of the NFκB-dependent synthesis rate but also of IκBα's nuclear import, as well as its constitutive degradation. It also predicts that nuclear export and IKK-dependent degradation of NFκB-bound IκBα are important for late post-attenuation signalling (figure 2c(iii)).
2.3. Experimental testing of model predictions: multiple IκBα properties contribute distinct characteristics to NFκB dynamic control
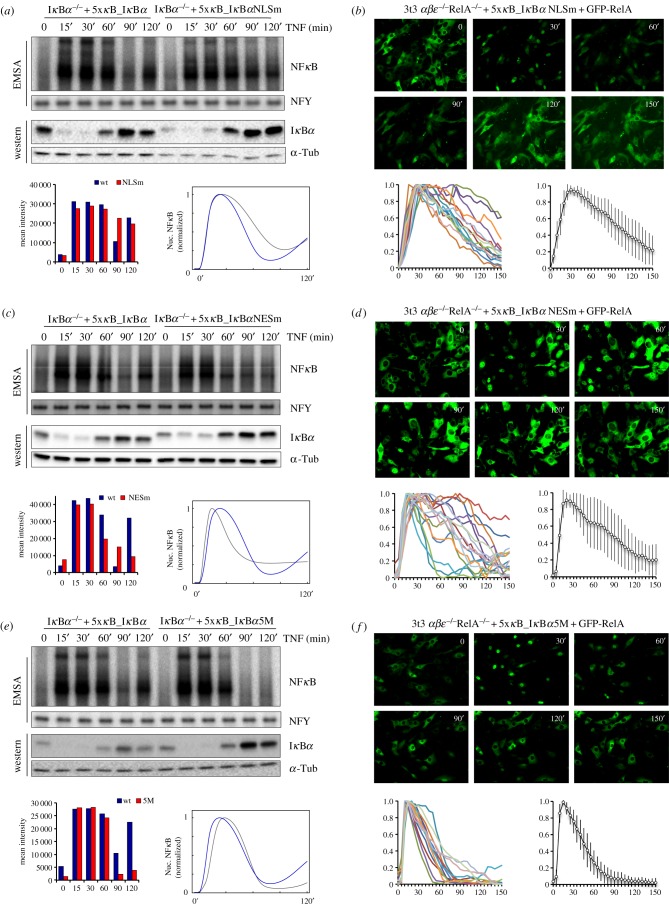
To test the computational predictions, we pursued a genetic perturbation approach. Previous work showed nuclear import of IκBα to be mediated by an unconventional NLS sequence [19,20]. Using this information, we reconstituted IκBα-deficient cells with an IκBα NLS mutant (IκBαNLSm: L110A,L115A,L117A,L120A). In DNA binding studies of IκBαNLSm cells, TNF induced NFκB activation comparable to that of wild-type IκBα cells (figure 3a), and although the resynthesis of IκBαNLSm protein was effectively induced by NFκB, the IκBαNLSm cells were defective for the rapid post-induction repression of NFκB. At 70 min, the signal in IκBαNLSm cells was 2.9-fold higher than in wild-type IκBα cells (considering the different basal levels). This is consistent with the threefold increase predicted by the model when the corresponding parameter is reduced to 35% of its wild-type value (figure 3a; electronic supplementary material, figure S2a). Consistent with population-level biochemical studies, IκBα−/−β−/−ε−/−RelA−/− cells expressing GFP-RelA showed that IκBαNLSm was defective in post-induction repression and cytoplasmic relocalization of NFκB in single cells (figure 3b). Despite the substantial heterogeneity in RelA cytoplasmic re-localization, the population mean closely resembles the population-level results obtained by EMSA. These data clearly show that the nuclear localization of IκBα is indispensable for proper termination of NFκB activity.
Figure 3.
Multiple IκBα properties contribute distinct characteristics to NFκB control. Nuclear localization of IκBα is required for the termination of NFκB activity (a,b). Nuclear export function of IκBα is required for post-repression activation of NFκB activity (c,d). IκBα protein half-life control is critical for sustained NFκB dynamics (e,f). IκBα−/− MEFs were reconstituted with NFκB-inducible wild-type or NLS mutant (a,b), NES mutant (c,d) or the 5M mutant (e,f) form of IκBα. The cells were treated with 1 ng ml−1 of TNF and nuclear extracts were analysed by EMSA for NFκB activity and corresponding cytoplasmic extracts subjected to western blotting with indicated antibodies (a,c,e). Bar graphs show quantification of EMSA; curves are modelling the result of NFκB activity in single cells upon stimulation with 10 ng ml−1 of TNF. Real-time fluorescent images of IκBα−/−β−/−ε−/−RelA−/− MEFs reconstituted with AcGFP1-RelA and NFκB-inducible IκBα NLS mutant (b), NES mutant (d) or the 5M mutant (f) IκBα (showing cellular localization of RelA at indicated time points). Below the fluorescent images, single-cell traces show the ratio of nuclear to cytoplasmic localization of AcGFP1-RelA in fluorescent images (left) as well as the average curve and standard deviation of the single-cell traces (right).
IκBα also relies on a nuclear export sequence (NES) for the efficient nuclear export of NFκB [21,22]. Mathematical modelling suggested strong inhibition of nuclear export would result in reduced late NFκB activity. To assess the role of IκBα nuclear export on the sub-cellular localization control of NFκB, we generated an NES mutant (IκBαNESm; L45A,L49A,I52A). Reconstituted cells expressing IκBαNESm were unable to produce the post-repression reactivation of NFκB characteristic of wild-type protein-controlled NFκB signalling (figure 3c; electronic supplementary material, figure S2b). The activity is qualitatively similar to the model prediction for a fivefold attenuation in the corresponding parameter. These results indicate that the nuclear export function of IκBα is crucial for the post-repression activation of NFκB signalling. Interestingly, the single-cell studies with the NES mutant revealed seemingly contradictory results (figure 3d), as most cells displayed sustained RelA nuclear localization. This apparent discrepancy is resolved by recognizing that: (1) the mutant localizes to the nucleus causing inhibition of NFκB activity but does not allow for NFκB export and reactivation, and (2) the biochemical assay detects DNA binding activity of free NFκB, whereas the single-cell imaging is a readout of NFκB localization only.
IκBα is known to have a very short half-life that is extended approximately twofold by an IκBα5M mutant (S283A, S288,T291A,S293A,T296A) [23,24]. When expressed in IκBα−/− cells from an NFκB-responsive promoter, IκBα5M achieved effective post-induction repression of NFκB activity (figure 3e; electronic supplementary material, figure S2c). However, the re-activation of NFκB was undetectable, consistent with computational predictions that indicated a signal close to basal level at 120 min when the corresponding parameter was reduced to 35% of its wild-type value. Similarly, in single live-cell studies, IκBα5M mediated efficient relocalization of RelA to the cytoplasm with a complete absence of late-phase activity (figure 3f).
3. Discussion
Given the well-documented role of NFκB activity in vital cellular processes, a number of mechanisms have evolved to ensure precise regulation of its activity. The IκBα negative feedback loop is a prominent NFκB regulatory mechanism, allowing for both post-induction repression and repeated or oscillatory bursts of activity, and is critical for providing complex dynamic control which is thought to mediate specificity in NFκB's pleiotropic physiological functions. Prior studies have established that the NFκB-responsive IκBα promoter is critical for this negative feedback control [16], but it has remained unclear whether specific characteristics of the IκBα protein may be important as well. Indeed, biochemical studies presented here using MEFs derived from mice in which IκBβ was engineered into the IκBα locus to (AKBI MEFs) clearly demonstrate that, even when IκBβ is under NFκB-transcriptional induction, it is unable to provide proper negative feedback. These data motivated our characterization of IκBα protein properties that contribute to proper negative feedback function. Our strategy was to complement IκBα−/− cells with retroviral transgenes providing for κB-responsive expression of engineered IκBα variants defective in specific molecular characteristics.
In addition to traditional biochemical approaches to study NFκB response and regulation, we examined NFκB response dynamics in single cells. Recent studies have characterized NFκB dynamics in single cells, but, to date, no studies have employed gene knock-out cells to probe underlying molecular mechanisms. Thus, regulatory control mechanisms identified at the biochemical/population level have yet to be reconciled with single-cell microscopy tracking studies that boast high temporal resolution and individual cellular histories. In this work, we employed a cell line lacking RelA and all canonical IκB proteins (IκBα−/−IκBβ−/−IκBε−/−RelA−/− cells), which we then reconstituted with NFκB-inducible IκB variants and fluorescent RelA reporter in order to examine the contributions of specific IκB protein characteristics.
Although the IκB proteins were first identified as cytoplasmic inhibitors, it has become clear that they play a major role in regulating nuclear NFκB. IκBα has been shown to be efficiently transported into the nucleus where it binds active NFκB dimers on the promoters of NFκB-activated genes and facilitates the dissociation of the transcription factor from DNA. Comparing mutants with wild-type IκB proteins, we were able to show the contributions of IκB inducible synthesis, nucleo-cytoplasmic transport and degradation control to the various aspects of the prototypical NFκB response; namely, duration and amplitude of initial NFκB activation, post-activation repression and post-repression re-activation of NFκB signalling. Our biochemical assays, together with single-cell studies, demonstrated that IκBα nuclear localization is indispensable for the rapid termination of NFκB activity. Specifically, we showed that an NES-deficient form of IκBα supported efficient induction and post-induction repression of NFκB DNA binding activity, but not the characteristic re-activation and second phase NFκB activity. The deficiency in NES function prevents the protein from efficiently returning NFκB to the cytoplasm for the next round of activation, maintaining an inactive IκBαNESm-bound pool of NFκB in the nucleus. Finally, by employing an IκBα harbouring five mutations that confer stability, increasing the half-life of the normally rapidly turned over uncomplexed/free protein, we were able to show the importance of such rapid turnover in generating characteristic NFκB temporal profiles. In cells expressing this IκBα5M form, we found a complete absence of second-phase activation, indicating that a low level of free IκBα protein (ensured by a short half-life) is required for this aspect of the response.
Our results demonstrate not only that IκBα feedback is dependent on NFκB-inducible synthesis but also that several other processes dependent on the molecular characteristics of the protein itself, for example import, export and half-life control, must be tuned in a coordinated manner to generate the hallmark features of NFκB signalling, namely post-induction repression and reactivation. By contrast, the IκBβ protein does not support proper negative feedback control even when expressed from IκBα's promoter; we speculate that substantially reduced nucleo-cytoplasmic transport [25,26] may be a key underlying reason; a second characteristic that may play a role is IκBα's but not IκBβ's ability to strip NFκB off the DNA [27]. Indeed, these properties are not required for the NFκB dimer stabilization/chaperone function recently ascribed to IκBβ [28]. Our results illustrate the more general point: that negative feedback regulation in cells is a complex process that depends on multiple molecular properties beyond activator-induced expression [29,30]. These findings may well extend to other transcriptional networks as nuclear transport is a defining feature of many gene regulatory networks. Understanding the specific contributions of each process as well as their characteristic time scales is an important step for identifying effective druggable targets that may allow for correction of dynamic misregulation in cells associated with pathology [15].
4. Material and methods
4.1. Computational modelling
The response of the NFκB regulatory module was simulated using the computational ODE-based model described in [31]. In order to focus on regulatory mechanisms involving IκBα, the other IκB family members were removed. Following equilibration, TNF responses were simulated as in [31]. Time-course curves in figure 2 were generated by applying multipliers to the kinetic parameters corresponding to the reactions in figure 2a. The multiplier values were: 2−3,−2.5,−2,−1.5,−1,−0.5 (reactions 1, 2, 3 and 5) and 2−6,−5,−4,−3,−2,−1 (reaction 4), reflecting different sensitivities for reaction 4. NFκB time courses are normalized to their peak value. Sensitivity ratios sr(t) at a particular time ti are defined as: (nucNFκBperturbed − nucNFκBunperturbed)/nucNFκBunperturbed, where nucNFκBperturbed/unperturbed are the normalized nuclear concentrations of NFκB at time ti obtained with a model with/without a multiplicative factor (0.9, 0.5, 0.1) for the indicated kinetic rate parameter (values shown in per cent units). The global average sensitivity in figure 2c was calculated as the RMS of the sr(t) sampled at 1 min intervals between 1 and 120 min post-stimulation.
4.2. DNA constructs
NFκB-inducible IκBα and IκBβ constructs were generated in the self-inactivating (SIN) retrovirus backbone (HRSpuro) modified to express the IκBα or IκBβ transgene under the control of five tandem κB sites upstream of a minimal promoter. IκBα mutant forms were produced using site-directed mutagenesis. For live-cell studies, AcGFP1 was fused to the N-terminus of RelA and the resulting construct was sub-cloned into the constitutively expressing retroviral plasmid pBabe-Hygro.
4.3. Cells and cell culture
Immortalized IκBα−/− MEFs were previously described [4] and IκBα−/−β−/−ε−/−RelA−/− MEFs were produced by interbreeding of the four individual mouse knock-out strains and harvesting E13.5 embryos, subjecting primary MEFs to the 3T3 protocol of repeated passage until a stably proliferating cell culture emerged. AKBI MEFs were a generous gift from BingBing Jiang (Boston University). MEFs were cultured in Dulbecco's modified Eagle's medium supplemented with 100 U penicillin/streptomycin (10378016; Life Technologies), 0.3 mg ml−1 glutamine and 10% fetal calf serum (complete medium). Plat-E cells [32] were maintained in complete medium containing blasticidin (10 µg ml−1) and puromycin (1 µg ml−1).
4.4. Retrovirus-mediated gene transduction
NFκB-inducible IκB and AcGFP1-RelA constructs were transfected into Plat-E packaging cells pre-conditioned in antibiotic-free complete medium using poly(ethylenimine). Supernatant was collected 48 h post-transfection, filtered and used to infect target cells with 4 µg ml−1 polybrene to enhance infection efficiency (Sigma). Infected cells were selected with puromycin hydrochloride (Sigma) for IκBs and/or with hygromycin B (InvivoGen) for the AcGFP1-RelA. Murine TNF (Roche) was used at 1 or 10 ng ml−1.
4.5. Biochemical analyses
Whole-cell extracts were prepared in radioimmunoprecipitation assay buffer with protease inhibitors and normalized for total protein before immunoblot analyses. Cytoplasmic and nuclear extracts for immunoblot analyses and EMSA, respectively, were prepared as previously described [4,24]. IκBα was probed with sc-371, IκBβ with sc-945 and α-tubulin with sc-5286. All antibodies were from Santa Cruz Biotechnology.
4.6. Microscopy
Cells were plated onto 35 mm glass bottom dishes (MatTek) or iBidi eight-well chambers (iBidi) 24 h prior to stimulation and immediate imaging. Images were acquired on an Axio Observer Z1 inverted microscope (Carl Zeiss Microscopy GmbH, Germany) with a 40×, 1.3 NA oil-immersion, or 20×, 0.8 NA air-immersion objective to a Coolsnap HQ2 CCD camera (Photometrics, Canada) using ZEN imaging software (Carl Zeiss Microscopy GmbH, Germany). Environmental conditions were maintained in a humidified chamber at 37°C, 5% CO2 (Pecon, Germany). Quantitative image processing was performed using the FIJI distribution of Image J (NIH). All cells of each frame in the microscope imaging experiments were measured for total fluorescence intensity. Time-course data were normalized by the minimum and maximum values to account for the varying overall intensities of different cells. The single-cell traces were averaged and error bars in the mean curves are the standard deviation from the mean.
Supplementary Material
Acknowledgements
We thank Bingbing Jiang (Boston University) for the generous gift of AKBI MEFs and acknowledge Santa Cruz Biotechnology for their support.
Authors' contributions
R.F. performed the experimental work, assisted by K.T.F., Y.E.L. and J.D.V. M.B. performed the computational modelling. R.F., J.D.V. and A.H. wrote the manuscript.
Competing interests
We have no competing interests.
Funding
The work was supported by grants to A.H. from the NIH: R01 GM071573 and P01 GM071862. R.F. was a Sigrid Juselius Foundation and Saatioiden postdoctoral fellow and M.B. was a Cancer Research Institute postdoctoral fellow.
References
- 1.Alon U. 2007. An introduction to systems biology: design principles of biological circuits. Boca Raton, FL: Chapman and Hall/CRC. [Google Scholar]
- 2.Scott ML, Fujita T, Liou HC, Nolan GP, Baltimore D. 1993. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 7, 1266–1276. ( 10.1101/gad.7.7a.1266) [DOI] [PubMed] [Google Scholar]
- 3.Chiao PJ, Miyamoto S, Verma IM. 1994. Autoregulation of I kappa B alpha activity. Proc. Natl Acad. Sci. USA 91, 28–32. ( 10.1073/pnas.91.1.28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann A, Levchenko A, Scott ML, Baltimore D. 2002. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science 298, 1241–1245. ( 10.1126/science.1071914) [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann A, Baltimore D. 2006. Circuitry of nuclear factor kappaB signaling. Immunol. Rev. 210, 171–186. ( 10.1111/j.0105-2896.2006.00375.x) [DOI] [PubMed] [Google Scholar]
- 6.Shih VF, Kearns JD, Basak S, Savinova OV, Ghosh G, Hoffmann A. 2009. Kinetic control of negative feedback regulators of NF-kappaB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proc. Natl Acad. Sci. USA 106, 9619–9624. ( 10.1073/pnas.0812367106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson DE, et al. 2004. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306, 704–708. ( 10.1126/science.1099962) [DOI] [PubMed] [Google Scholar]
- 8.O'Dea E, Hoffmann A. 2010. The regulatory logic of the NF-kappaB signaling system. Cold Spring Harb. Perspect. Biol. 2, a000216 ( 10.1101/cshperspect.a000216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basak S, Behar M, Hoffmann A. 2012. Lessons from mathematically modeling the NF-kappaB pathway. Immunol. Rev. 246, 221–238. ( 10.1111/j.1600-065X.2011.01092.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishna S, Jensen MH, Sneppen K. 2006. Minimal model of spiky oscillations in NF-kappaB signaling. Proc. Natl Acad. Sci. USA 103, 10 840–10 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayot F, Jayaprakash C. 2006. NF-kappaB oscillations and cell-to-cell variability. J. Theor. Biol. 240, 583–591. ( 10.1016/j.jtbi.2005.10.018) [DOI] [PubMed] [Google Scholar]
- 12.Mothes J, Busse D, Kofahl B, Wolf J. 2015. Sources of dynamic variability in NF-kappaB signal transduction: a mechanistic model. BioEssays 37, 452–462. ( 10.1002/bies.201400113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werner SL, Barken D, Hoffmann A. 2005. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science 309, 1857–1861. ( 10.1126/science.1113319) [DOI] [PubMed] [Google Scholar]
- 14.Behar M, Hoffmann A. 2010. Understanding the temporal codes of intra-cellular signals. Curr. Opin. Genet. Dev. 20, 684–693. ( 10.1016/j.gde.2010.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behar M, Barken D, Werner SL, Hoffmann A. 2013. The dynamics of signaling as a pharmacological target. Cell 155, 448–461. ( 10.1016/j.cell.2013.09.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werner SL, Kearns JD, Zadorozhnaya V, Lynch C, O'Dea E, Boldin MP, Ma A, Baltimore D, Hoffmann A. 2008. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 22, 2093–2101. ( 10.1101/gad.1680708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A. 2006. IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, inflammatory gene expression. J. Cell Biol. 173, 659–664. ( 10.1083/jcb.200510155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng JD, Ryseck RP, Attar RM, Dambach D, Bravo R. 1998. Functional redundancy of the nuclear factor kappa B inhibitors I kappa B alpha and I kappa B beta. J. Exp. Med. 188, 1055–1062. ( 10.1084/jem.188.6.1055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachdev S, Bagchi S, Zhang DD, Mings AC, Hannink M. 2000. Nuclear import of IkappaBalpha is accomplished by a ran-independent transport pathway. Mol. Cell Biol. 20, 1571–1582. ( 10.1128/MCB.20.5.1571-1582.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachdev S, Hoffmann A, Hannink M. 1998. Nuclear localization of IkappaB alpha is mediated by the second ankyrin repeat: the IkappaB alpha ankyrin repeats define a novel class of cis-acting nuclear import sequences. Mol. Cell Biol. 18, 2524–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang TT, Kudo N, Yoshida M, Miyamoto S. 2000. A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes. Proc. Natl Acad. Sci. USA 97, 1014–1019. ( 10.1073/pnas.97.3.1014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang TT, Miyamoto S. 2001. Postrepression activation of NF-kappaB requires the amino-terminal nuclear export signal specific to IkappaBalpha. Mol. Cell Biol. 21, 4737–4747. ( 10.1128/MCB.21.14.4737-4747.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathes E, O'Dea EL, Hoffmann A, Ghosh G. 2008. NF-kappaB dictates the degradation pathway of IkappaBalpha. EMBO J. 27, 1357–1367. ( 10.1038/emboj.2008.73) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Dea EL, Kearns JD, Hoffmann A. 2008. UV as an amplifier rather than inducer of NF-kappaB activity. Mol. Cell 30, 632–641. ( 10.1016/j.molcel.2008.03.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Wu J, Ghosh G. 2003. KappaB-Ras binds to the unique insert within the ankyrin repeat domain of IkappaBbeta and regulates cytoplasmic retention of IkappaBbeta×NF-kappaB complexes. J. Biol. Chem. 278, 23 101–23 106. ( 10.1074/jbc.M301021200) [DOI] [PubMed] [Google Scholar]
- 26.Malek S, Chen Y, Huxford T, Ghosh G. 2001. IkappaBbeta, but not IkappaBalpha, functions as a classical cytoplasmic inhibitor of NF-kappaB dimers by masking both NF-kappaB nuclear localization sequences in resting cells. J. Biol. Chem. 276, 45 225–45 235. ( 10.1074/jbc.M105865200) [DOI] [PubMed] [Google Scholar]
- 27.Bergqvist S, Alverdi V, Mengel B, Hoffmann A, Ghosh G, Komives EA. 2009. Kinetic enhancement of NF-kappaBxDNA dissociation by IkappaBalpha. Proc. Natl Acad. Sci. USA 106, 19 328–19 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsui R, Kearns JD, Lynch C, Vu D, Ngo KA, Basak S, Ghosh G, Hoffmann A. 2015. IkappaBbeta enhances the generation of the low-affinity NFkappaB/RelA homodimer. Nat. Commun. 6, 7068 ( 10.1038/ncomms8068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen LK, Kulasiri D. 2009. On the functional diversity of dynamical behaviour in genetic and metabolic feedback systems. BMC Syst. Biol. 3, 51 ( 10.1186/1752-0509-3-51) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen LK. 2012. Regulation of oscillation dynamics in biochemical systems with dual negative feedback loops. J. R. Soc. Interface 9, 1998–2010. ( 10.1098/rsif.2012.0028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee SP, Behar M, Birnbaum HA, Hoffmann A, Wright PE, Ghosh G. 2013. Analysis of the RelA:CBP/p300 interaction reveals its involvement in NF-kappaB-driven transcription. PLoS Biol. 11, e1001647 ( 10.1371/journal.pbio.1001647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita S, Kojima T, Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066. ( 10.1038/sj.gt.3301206) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.