Abstract
We previously demonstrated the clonal complex 81 (CC81) subtype 1 lineage is the major staphylococcal food poisoning (SFP)-associated lineage in Japan (Y. Sato'o et al., J Clin Microbiol 52:2637–2640, 2014, http://dx.doi.org/10.1128/JCM.00661-14). Strains of this lineage produce staphylococcal enterotoxin H (SEH) in addition to SEA. However, an evaluation of the risk for the recently reported SEH has not been sufficiently conducted. We first searched for staphylococcal enterotoxin (SE) genes and SE proteins in milk samples that caused a large SFP outbreak in Japan. Only SEA and SEH were detected, while there were several SE genes detected in the samples. We next designed an experimental model using a meat product to assess the productivity of SEs and found that only SEA and SEH were detectably produced in situ. Therefore, we investigated the regulation of SEH production using a CC81 subtype 1 isolate. Through mutant analysis of global regulators, we found the repressor of toxin (Rot) functioned oppositely as a stimulator of SEH production. SEA production was not affected by Rot. seh mRNA expression correlated with rot both in media and on the meat product, and the Rot protein was shown to directly bind to the seh promoter. The seh promoter sequence was predicted to form a loop structure and to hide the RNA polymerase binding sequences. We propose Rot binds to the promoter sequence of seh and unfolds the secondary structure that may lead the RNA polymerase to bind the promoter, and then seh mRNA transcription begins. This alternative Rot regulation for SEH may contribute to sufficient toxin production by the CC81 subtype 1 lineage in foods to induce SFP.
INTRODUCTION
Staphylococcus aureus produces several virulence factors causing human and animal diseases. Among virulence factors produced by S. aureus, staphylococcal enterotoxins (SEs) show emetic and superantigen activities and are the causative agents for staphylococcal food poisoning (SFP) and toxic shock syndrome (1, 2). Presently, 23 SEs and SE-like toxins (SEls) have been reported. SEA-SEE are classical SEs, while SEG-SElX are newly described SEs/SEls (1, 2). Of these, SEA is shown to be the most important in SFP outbreaks (2). Conversely, reports suggest the newly described SEs also contribute to SFP outbreaks (3–5), but contributions for these SEs remain unknown. Thus, it is necessary to evaluate the importance of the newly described SEs in SFP outbreaks.
Our previous report described the clonal complex 81 (CC81) subtype 1 lineage as the major SFP-associated lineage in Japan. Almost all of the CC81 subtype 1 isolates carried sea and showed high SEA production (6). Moreover, another unique genetic characteristic is the presence of seh. All CC81 subtype 1 isolates carried seh (all 30 strains), while other isolates rarely carried it (1 strain in 341 strains) (6).
Though limited in numbers, previous epidemiological reports found SEH (seh) in SFP cases. In 2000, a large outbreak involving 13,420 patients consuming contaminated low-fat milk occurred in Japan (7, 8). SEH and SEA were detected in the causative low-fat milk. A recent study in Germany showed almost all of the isolates from food poisoning by ice cream were positive for seh as well as sea (9). Further, an epidemiology study in South Korea showed that S. aureus strains positive for seh formed one of the dominant groups causing SFP, similar to findings of our previous study (6, 10). In the Netherlands, there was a report of an outbreak with S. aureus positive for seh only (3).
In addition to epidemiology, the emetic activity of SEs/SEls is an important characteristic as an etiological agent of SFP. Several studies concerning vomiting activities caused by SEs are reported (5, 11–14) where the emetic activity of SEH in primates was similar to that of SEA (2). Like SEA, SEH may play an important role in SFP, but there is little information about the production of SEH. Therefore, we explored the unique production mechanism of SEH in clone no. 10, which was classified into CC81 subtype 1.
MATERIALS AND METHODS
Detection of se genes and SE proteins in SFP-causing low-fat milk from a factory.
To demonstrate the involvement of SEH in food poisoning, we first retrospectively determined the repertoires of SE/SEl genes and proteins in food samples. We used daily product samples from the historically largest outbreak in Japan (7, 8). Six milk samples processed during different periods in the factory that caused a large outbreak in Japan were used as samples 1 to 6. Multiplex PCR was carried out to detect the genes for SE/SEl in the samples as described previously (15). SEs/SEls in the samples were purified and concentrated for enzyme-linked immunosorbent assay (ELISA). The excess protein, e.g., casein, was removed by adding HCl (to pH 3.8) at room temperature for 10 min. After centrifugation (4,000 × g, 20 min, 4°C), the supernatant was filtered using a Millex-GP 0.22-μm filter (Millipore, MA). After neutralizing the supernatant with NaOH (pH 6.8), 10% (wt/vol, final concentration) chloroform (Wako Pure Chemical Industries, Osaka, Japan) was added to remove lipids from the supernatant. After centrifugation (4,000 × g, 20 min, 4°C), a 20% volume of 30% (wt/vol) trichloroacetic acid (Wako Pure Chemical Industries) was added and incubated at 4°C for 30 min. The precipitated protein was collected using centrifugation (4,000 × g, 20 min, 4°C). After discarding the supernatant and drying, the protein was resuspended in 0.1 M Tris-HCl (pH 8.0; Wako Pure Chemical Industries) and neutralized with NaOH (Wako Pure Chemical Industries) (pH 7.0 to 8.0). The sample was concentrated 25 times. Sandwich ELISA was performed to detect SEA, SEC, SEG-SEI, SEK, and SEM-SEQ as described by Omoe et al. and Sato'o et al. (6, 13, 16), with modifications. For detection, SuperSignal ELISA Femto maximum sensitivity substrate (Thermo Fisher Scientific, Waltham, MA) was used as the substrate.
Staphylococcus aureus strains and assay for toxin production.
Eleven Staphylococcus aureus strains were used for toxin production. The SE/SEl genotypes of each strain are described in Table 1. We used nine clinical isolates from SFP, one human nasal swab isolate (IVM50), and MW2 as a control. Staphylococcus aureus was cultured in medium and on the meat product. Medium culture was performed as described previously (6), with some modifications. Staphylococcus aureus was precultured in 3 ml brain heart infusion (BHI) broth (Becton Dickinson, Sparks, MD) supplemented with 1% (wt/vol) yeast extract (Becton Dickinson) at 37°C overnight. About 6 × 109 precultured cells were inoculated into 60 ml fresh BHI broth supplemented with 1% yeast extract and cultured at 37°C for 24 h with constant agitation. If necessary, antibiotics (chloramphenicol final concentration, 10 μg/ml; tetracycline final concentration, 5 μg/ml) and/or xylose (final concentration, 1%, wt/vol) were added before inoculation of bacterial cells. After culture and centrifugation (15,600 × g, 20 min, 4°C), filtration was performed using a 0.2-μm-pore-size Minisart membrane filter (Sartorious, Göttingen, Germany) to prepare supernatant samples for sandwich ELISA. To evaluate SE production in foods we used a meat product, salted ham, because it is commonly associated with SFP (1) and it is relatively easy to measure SEs produced on the surface. For culture on meat product, about 108 S. aureus cells precultured overnight were spread on the surface of a 5-g salted ham slice. The bacterium-contaminated salted ham slices were incubated at 37°C for 24 h. After incubation, the slices were washed with 2 ml of Dulbecco's phosphate-buffered saline (pH 7.4) supplemented with 0.1% (wt/vol) bovine serum albumin (Sigma, St. Louis, MO). The washed buffer was centrifuged (15,600 × g, 20 min, 4°C) and filtered with a 0.2-μm-pore-size Minisart membrane filter to eliminate meat debris and bacteria, and the filtrates were used as ELISA samples. Sandwich ELISA was performed to detect the seven SE products (SEA to SED and SEG to SEI) as described above.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Staphylococcus aureus | ||
| No. 10 | CC81 subtype 1, sea, seb, seh, sek, seq | 42 |
| 01240 | CC81 subtype 1, sea, seh, sek, seq | 6 |
| Nagasaki | CC81 subtype 1, sea, seb, seh, sek, seq | 16 |
| Hiroshima3 | CC5, seg, sei, selj, sem, sen, seo, ser | 6 |
| IVM50 | CC508, sec, seg, sei, sel, sem, sen, seo | 15 |
| 01235 | CC96, sea, sec, sel | 6 |
| Oita1 | CC508, seg, sei, sem, sen, seo | 6 |
| 11727 | CC8, sea, sed, selj, ser | 15 |
| MW2 | CC1, sea, sec, seh, sek, sel, seq | 43 |
| 196E | CC not assigned, sea, sed, selj, ser | 44 |
| S6 | CC not assigned, sea, seb, sek, seq | 45 |
| RN4220 | Genetic manipulation strain | 46 |
| No. 10 ΔsarA | Deletion mutant | This study |
| No. 10 ΔsarA pKAT | Vector control mutant | This study |
| No. 10 ΔsarA pKAT::sarA | Complemented mutant | This study |
| No. 10 ΔsarS | Deletion mutant | This study |
| No. 10 ΔsarS pWH1520 | Vector control mutant | This study |
| No. 10 ΔsarS pWH1520::sarS | Complemented mutant | This study |
| No. 10 ΔsarT | Deletion mutant | This study |
| No. 10 ΔsarT pKAT | Vector control mutant | This study |
| No. 10 ΔsarT pKAT::sarT | Complemented mutant | This study |
| No. 10 ΔsarU | Deletion mutant | This study |
| No. 10 ΔsarU pKAT | Vector control mutant | This study |
| No. 10 ΔsarU pKAT::sarU | Complemented mutant | This study |
| No. 10 Δrot | Deletion mutant | This study |
| No. 10 Δrot pKAT | Vector control mutant | This study |
| No. 10 Δrot pKAT::rot | Complemented mutant | This study |
| No. 10 ΔsaeR | Deletion mutant | This study |
| No. 10 ΔsaeR pWH1520 | Vector control mutant | This study |
| No. 10 ΔsaeR pWH1520::saeR | Complemented mutant | This study |
| Escherichia coli | ||
| DH5α | Cloning strain | TaKaRa |
| BL21(DE3) | Expression strain | Novagen |
Genetic manipulation.
We prepared gene deletion mutants, deletion mutants containing empty vector, and mutants complemented with the deleted sarA, sarS, sarT, sarU, saeR, and rot genes from the wild type of the CC81 subtype 1 lineage, no. 10. Allelic replacement and gene complementation were performed as described previously (17, 18). Plasmids and primers used in this study are listed in Tables 2 and 3. The constructed mutants in this study are listed in Table 1.
TABLE 2.
Plasmids used in this study
| Plasmid name | Purpose | Source or reference |
|---|---|---|
| pKFT | Genetic manipulation | 17 |
| pKFT (sarA FR) | Genetic manipulation | This study |
| pFK25 (pKFT (sarS FR)) | Genetic manipulation | 17 |
| pKFT (sarT FR) | Genetic manipulation | Sugai laboratory |
| pKFT (sarU FR) | Genetic manipulation | Sugai laboratory |
| pKFT (rot FR) | Genetic manipulation | In this study |
| pKFT (saeR FR) | Genetic manipulation | Sugai laboratory |
| pKAT | Genetic manipulation | Sugai laboratory |
| pKAT::sarA | Genetic manipulation | Sugai laboratory |
| pKAT::sarT | Genetic manipulation | Sugai laboratory |
| pKAT::sarU | Genetic manipulation | Sugai laboratory |
| pKAT::rot | Genetic manipulation | This study |
| pWH1520 | Genetic manipulation | MoBiTec |
| pWH1520::sarS | Genetic manipulation | 18 |
| pWH1520::saeR | Genetic manipulation | Sugai laboratory |
| pGEM-T Easy | EMSA | Promega |
| pGEM-T Easy/SEH promoter | EMSA | This study |
| pET28a | EMSA | Novagen |
| pGrota | EMSA | 20 |
Reconstructed in this study.
TABLE 3.
Primers used in this study
| Primer name | Sequence (5′-3′) | Purpose |
|---|---|---|
| gyrB forward | AGGTCTTGGAGAAATGAATG | qPCR |
| gyrB reverse | CAAATGTTTGGTCCGCTT | qPCR |
| seh forward | TCAAGGTGATAGTGGCAAT | qPCR |
| seh reverse | CCAATCACCCTTTCCTGT | qPCR |
| rot forward | TGCAGTATTTCAACCACACAC | qPCR |
| rot reverse | GTATCGTTAATGCGCCAGT | qPCR |
| M13 forward | GTAAAACGACGGCCAGT | Genetic manipulation |
| M13 reverse | CAGGAAACAGCTATGAC | Genetic manipulation |
| SarA1 | AGGATCCAAAAATTCTAATGGA | Genetic manipulation |
| SarA2 | GGATCCTCAACTCATTCTTAAG | Genetic manipulation |
| SarA3 | GGTTTTGCACTAATGGCACT | Genetic manipulation |
| SarA4 | TTGCTCGATACATTTGCCCGA | Genetic manipulation |
| sarS1 | GCCAAAGCTTATACATGGCTAGTCGG | Genetic manipulation |
| sarS4 | TCAAGGATCCATAGAAGGCGCTTTG | Genetic manipulation |
| sarT F-F | GGCTGCAGGATAGTGACTTAATGTTCTT | Genetic manipulation |
| sarT F-R | CCGGATCCGTCTTCATCCCTTACTTTTAA | Genetic manipulation |
| sarT R-F | GGGGATCCAGTATGTTTCGAGATTTTAAT | Genetic manipulation |
| sarT R-R | CCGAGCTCTACACCCTGTGGTGCAGTGTC | Genetic manipulation |
| sarT F | CGGGATCCCGTTATGTTTCATTAATATTTATTTC | Genetic manipulation |
| sarT R | CCCAAGCTTGGGCCTTACATTCTCCTACTA | Genetic manipulation |
| sarU F-F | CGGGATCCCGTTATGTTTCATTAATATTTATTTC | Genetic manipulation |
| sarU F-R | GGCTGCAGCTTACATTCTCCTACTACTATTTTC | Genetic manipulation |
| sarU R-F | GGCTGCAGATTTAACAGATTTACCTCTTG | Genetic manipulation |
| sarU R-R | CCCAAGCTTGTGACGATATTGTTGAATCTG | Genetic manipulation |
| sarU F | GGGTTCGAAGTCTTCATCCCTTACTTTTAA | Genetic manipulation |
| sarU R | CCCAAGCTTTTAAAAGAAAAATTTTCTTGG | Genetic manipulation |
| rot1 | AATGGATCCACAAGGTATTA | Genetic manipulation |
| rot2 | GGGGATCCTGTTAATTTCTCCT | Genetic manipulation |
| rot3 | TCAAATGTTGACTACTCAAT | Genetic manipulation |
| rot4 | ATAAACTTGCTTTCTATTCA | Genetic manipulation |
| rot comp S | GCCGAAGCTTTAAGGTTGAGAATGTATATC | Genetic manipulation |
| rot comp AS | GCCGAAGCTTTTACACAGCAATAATTGCGT | Genetic manipulation |
| SaeF | TGGAAAGCTTATGATTTCACAGCACCC | Genetic manipulation |
| SaeSR | TTGCAAGCTTGATACAAGTAATTGGTC | Genetic manipulation |
| T7 promoter-1 | TAATACGACTCACTATAGGG | EMSA |
| T7 promoter-1 cy3a | TAATACGACTCACTATAGGG | EMSA |
| SP6 promoter-1 | CAAGCTATTTAGGTGACACTATAG | EMSA |
| SEH upstream S | TCTAACTACTATAGCAACTG | EMSA |
| SEH upstream AS | TTTAAAACTCCTCAATATAT | EMSA |
| g-rot-f-NcoI | GCGCCATGGTGAAAAAAGTAAATAACGACACT | EMSA |
| g-rot-r-XhoI | GCGCTCGAGCACAGCAATAATTGCGTTTA | EMSA |
cy3 labeled at the 5′ end.
RNA extraction, reverse transcription, and real-time PCR.
Bacterial culture was performed as described above. From medium culture, cells were collected from broth by centrifugation (2,800 × g, 10 min, 4°C). For meat culture, bacterium-contaminated salted ham incubated for 24 h was washed with 2 ml Dulbecco's phosphate-buffered saline (pH 7.4), and then the cells were pelleted by centrifugation (2,800 × g, 10 min, 4°C). The bacterial cells were subjected to RNA extraction.
RNA extraction was performed with the FastRNA pro blue kit (MP Biomedicals, Irvine, CA). The remnant DNA was treated using RQ1 RNase-free DNase (Promega, Madison, WI). Subsequently, reverse transcription from purified RNA was performed with a Transcriptor first-strand cDNA synthesis kit (Roche, Basel, Switzerland). All cDNA preparation procedures were performed according to the manufacturer's directions. All cDNA solutions were 10-fold diluted with TE buffer (containing 10 mM Tris-HCl and 1 mM EDTA, pH 8.0), and 4 μl of the diluted cDNA solution was subjected to real-time PCR assay (20-μl [total volume] reaction mixture containing 0.5 μM each primer set in each tube). Real-time PCR was performed with SsoAdvanced SYBR green supermix (Bio-Rad, Hercules, CA) and a Bio-Rad CFX96 system (Bio-Rad). The real-time PCR primers are listed in Table 3. Real-time PCR conditions included denaturing at 95°C for 1 min, 40 cycles of 95°C for 15 s and 60°C (rot and seh) or 62°C (gyrB) for 15 s, and then 72°C for 30 s. Fluorescence was measured at the end of every extension step. After cycling, melt curves analysis was performed between 70°C and 90°C. All quantitative PCR (qPCR) data were analyzed using CFX Manager software version 3.0 (Bio-Rad) according to the manufacturer's instructions. The relative expression level of rot and seh was calculated by relating the target gene expression to the constant expression of the reference gene, gyrB. To determine the PCR amplification efficiency of each qPCR, the cDNA solution was serially diluted with 10 mM Tris-HCl (pH 7.5) containing 0.5% Tween 20 (Wako Pure Chemical Industries). No inhibition of PCR amplification was observed, and PCR amplification efficiency of all samples was between 90 and 110%.
Electrophoretic mobility shift assay (EMSA).
We adapted a previous method with modification to perform the mobility shift assay (19). The SEH promoter sequence was amplified with PrimeSTAR GXL DNA polymerase (TaKaRa, Shiga, Japan) with the primers listed in Table 3. The PCR product was purified using gel extraction with a FastGene gel/PCR extraction kit (Nippon Genetics, Tokyo, Japan), and then a 3′ A was added to the purified DNA with a Mighty TA cloning reagent set for PrimeSTAR (TaKaRa). TA cloning was performed with the DNA fragment with the added A by using pGEM-T Easy (Promega) and the Ligation high Ver. 2 kit (Toyobo, Osaka, Japan). Blue-white selection used DH5α and LB agar supplemented with ampicillin (Wako Pure Chemical Industries), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Wako Pure Chemical Industries), and isopropyl-β-d-thiogalactopyranoside (IPTG; Wako Pure Chemical Industries). The inserted sequence was confirmed by sequencing with a BigDye Terminator v 3.1 kit (Life Technologies Corporation, Carlsbad, CA), T7 promoter primer, SP-6 promoter primer (Table 3), and Hi-Di formamide (Life Technologies Corporation) using a 3130 genetic analyzer (Life Technologies Corporation). Promoter cloning (construction of pGEM-T Easy/SEH promoter; Table 2) was performed according to the manufacturer's directions.
The DNA probe was amplified with the pGEM-T Easy/SEH promoter, PrimeSTAR GXL DNA polymerase, T7 promoter primer (5′-cy3 labeled or nonlabeled), and SEH upstream AS primer (Table 3) and purified using gel extraction with a FastGene gel/PCR extraction kit. DNA concentration was measured using a NanoDrop 1000 (NanoDrop Technologies, Inc., Wilmington, DE). Rot protein was prepared according to a previously described method (20) using the vectors and primers listed in Tables 2 and 3, respectively. SEH promoter probe and Rot protein were incubated under previously described conditions (20). The reaction mixture was electrophoresed with a 6% polyacrylamide gel and 0.5× Tris-borate-EDTA (TBE) buffer under cold conditions. After electrophoresis, the gel was imaged using a Molecular Imager FX system (Bio-Rad).
RESULTS
Detection of SE genes and SE proteins in SFP-causing samples of a large SFP outbreak.
We first attempted to detect SE/SEl genes and proteins in milk samples reported to contain SEA and SEH (7, 8). Including these two genes, nine toxin genes (sec, seg, sek, sei, sem, sen, seo, sep, and seq) were detected in four of the six milk samples (see Fig. S1 in the supplemental material). To confirm toxin proteins, we used sandwich ELISA, which detects >0.2 ng/ml SE protein, and detected only SEA and SEH (0.46 ng/ml and 0.63 ng/ml, respectively) but not the other serotype SEs. This suggested the samples associated with SFP contain inactivated S. aureus cells carrying a variety of SE genes besides sea and seh or their DNA, but only limited types of enterotoxins were detectably produced. We questioned if there is a preference in the production of enterotoxin serotypes by SFP-causing S. aureus, especially in foods, and this led us to investigate SE production in food samples.
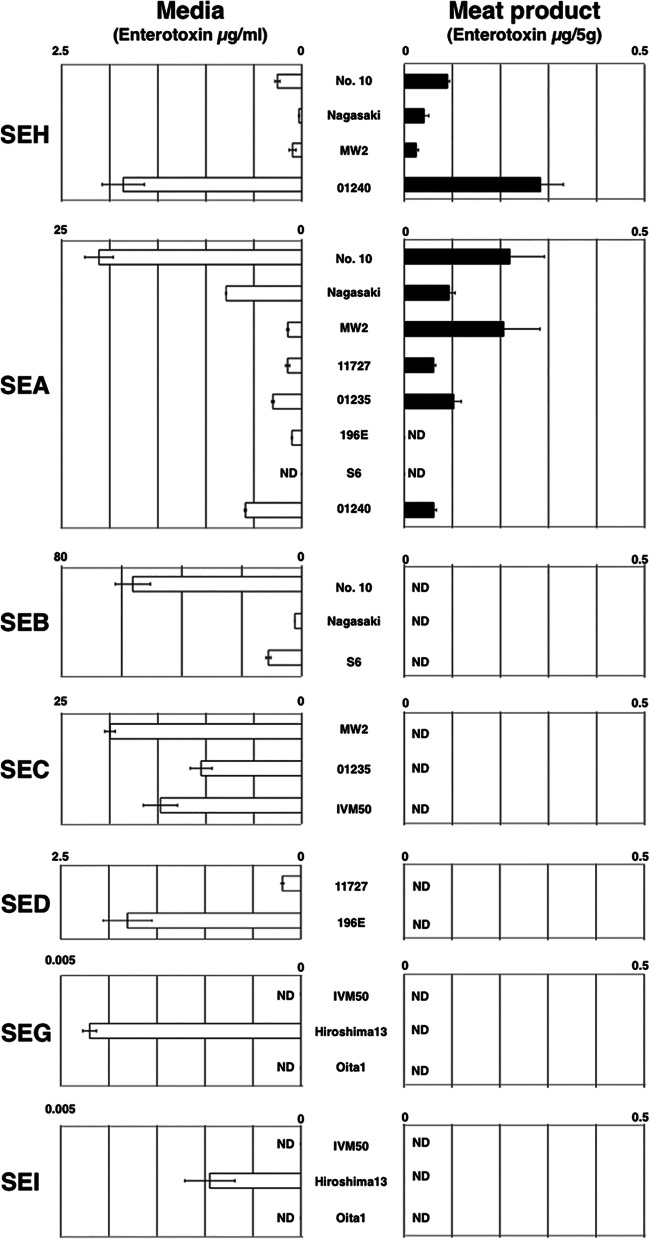
Comparison of SE production in media and on salted ham.
We compared the production of SEs by S. aureus grown on salted ham and in media. The data are shown in Fig. 1. Only SEA and SEH, but no other serotype SEs, were detectably produced on meat product after incubation (Fig. 1). SEH was detected in all samples, and its amounts ranged from 0.025 to 0.28 μg/5 g meat product. Also, SEA was detected in six of eight strains, and its amounts ranged from 0.061 to 0.22 μg/5 g meat product. These results indicated not all SEs were produced at detectable levels on meat products under this condition. Of note, the amount of SEH was similar to that of SEA, the most important SE in SFP (Fig. 1). Conversely, various SEs were detected in the media (Fig. 1). Among SEs, the level of SEB was the highest, followed by SEC, SEA, SEH, and SED. These data further support the idea that SEH, as well as SEA, is preferentially produced in (on) foods and is therefore an etiologically important SE in SFP.
FIG 1.
Comparison of SE production in media and on meat product. Briefly, various S. aureus strains were cultured in media and on salted ham slices, and the filtrates of culture broth and wash from the slices were used for ELISA. The averages and standard errors from each sample are shown. All cultures and all ELISAs were repeated two times (n = 4 per sample). White bar, SEs in media (μg/ml); black bar, SEs on meat product (μg/5 g). ND, not detectable (<0.2 ng/ml or 5 g salted ham). (A portion of the data for SEB, SEH, SEG, and SEI were a reexamination of the studies by Omoe et al. and Sato'o et al. using improved ELISA [16, 42]).
Identification of the regulatory pathway of SEH.
We determined the factors affecting SEH production in media using a series of regulatory gene mutants. The data from the ELISA for SEH are shown in Fig. 2. The deletion of sarA, sarT, sarU, rot, and saeR decreased SEH production, while complementation of mutants recovered production. In contrast, sarS deletion increased production while its complementation decreased production. The schematic image of the regulatory pathway of SEH expression, based on our data and previous studies, is illustrated in Fig. 3. Rot was named after the function “repressor of toxins” in S. aureus (21). However, our results clearly demonstrated that Rot is an enhancer of SEH production. In addition, SEA production was not affected by Rot (see Fig. S2 in the supplemental material).
FIG 2.
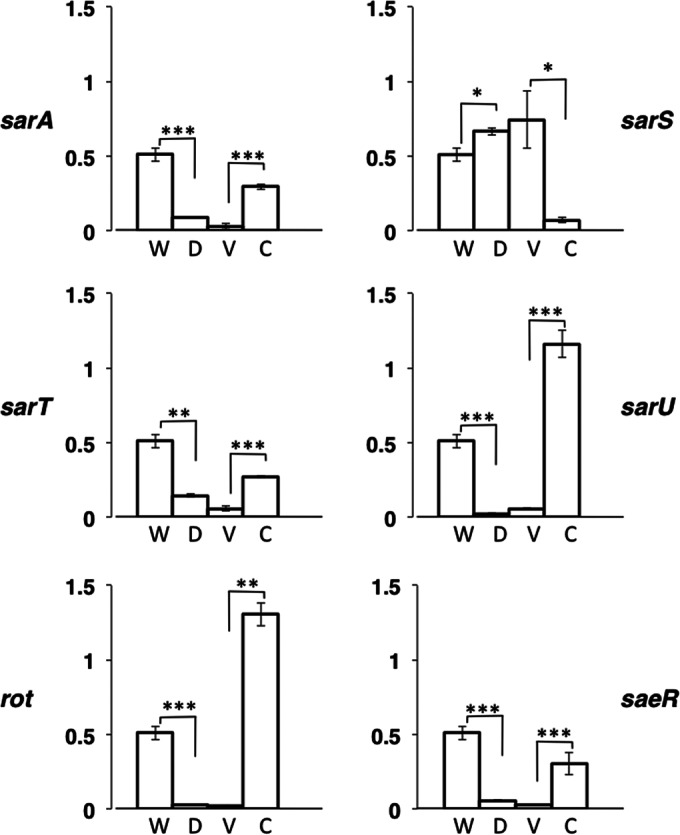
Effects of global regulators on SEH production. SEH production of global regulator mutants in medium was compared with that of the wild type. S. aureus SFP clone no. 10 and its global regulator mutants were cultured in medium, and the filtrates of the cultures were used for ELISA. The averages and standard errors from each sample are shown. All cultures and all ELISAs were repeated two times (n = 4 per sample). W, no. 10 wild type (the value in this figure is the same as that in Fig. 1); D, global regulator deletion mutants; V, vector control mutants (deletion mutants containing pKAT or pWH1520); C, complemented mutants (deletion mutants containing complementing vector). Significance was determined by Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 3.
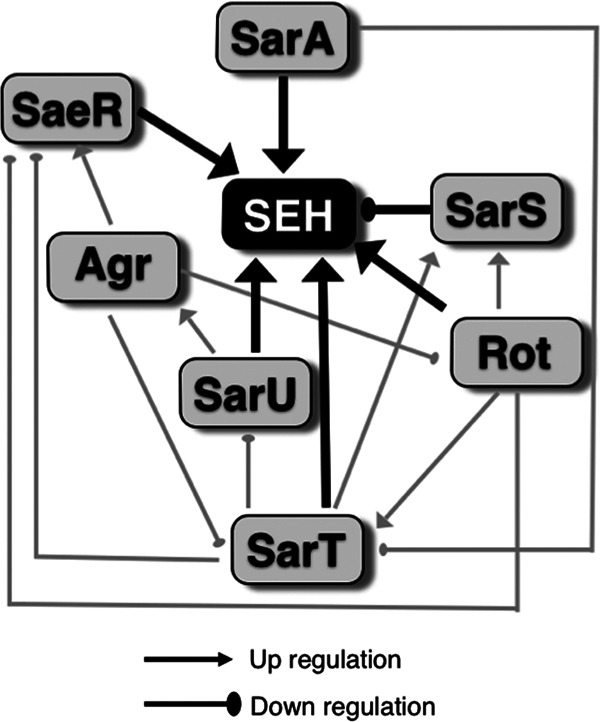
Regulatory pathway of SEH. Arrowheads, upregulation; circle heads, downregulation; black lines, found in this study; gray lines, found in previous studies (22, 25, 28–32, 47).
mRNAs of rot and seh were expressed in both media and meat product.
Subsequently, we performed a temporal sampling of S. aureus from media and meat product and conducted qPCR to investigate the expression of rot and seh genes. As shown in Fig. 4, mRNA expression of rot and seh correlated (correlation coefficient [R2] of 0.99 in media and 0.44 on meat product) in S. aureus both in media and on meat product.
FIG 4.
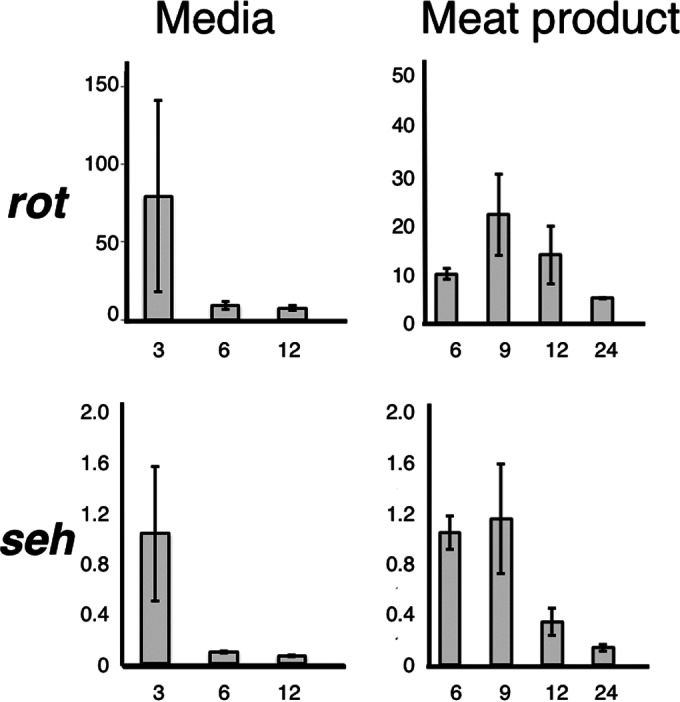
Expression time course of seh and rot in S. aureus SFP clone no. 10. The bacterium was cultured at the indicated times and recovered from the broth culture (or the wash from ham slices) by centrifugation. RNA purification, cDNA synthesis, and subsequent qPCR were performed as described in Materials and Methods. Vertical lines, relative expression (rot or seh and gyrB); horizontal lines, incubation time (hours). Two independent cultures and two independent assays were performed, and a total of four pieces of data per sample were averaged. Standard errors are shown. Correlation coefficients of relative rates (seh and gyrB as well as rot and gyrB) were R2 = 0.99 (in media) and R2 = 0.44 (on meat product).
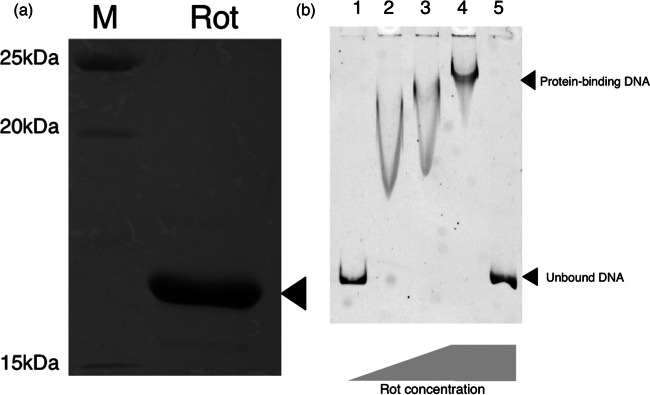
Rot protein-seh promoter sequence interaction.
We next determined if Rot directly binds to the seh promoter sequence. The nucleotide sequences of rot and seh promoters in no. 10 were the same as those of MW2 (data not shown). Recombinant Rot was purified with a His tag (Fig. 5a) to show binding of His-tagged Rot to the promoter sequence of seh. Using EMSA, the binding of Rot caused an apparent mobility shift of the band (Fig. 5b). Also, the addition of redundant nonlabeled DNA totally cancelled the shift. Taken together with mutation/complementation experiments and qRNA analysis, the data strongly suggested Rot protein directly binds to the seh promoter and positively enhances seh mRNA transcription.
FIG 5.
Gel shift assay of seh promoter in the presence and absence of Rot. (a) Purified Rot protein. (b) Gel shift assay. EMSA was performed with seh promoter sequence DNA probe (50 ng/reaction) incubated with various concentrations of Rot (ng/reaction). Lane 1, 0 ng; lane 2, 1 ng; lane 3, 2 ng; lanes 4 and 5, 10 ng. Nonlabeled DNA probe (with 50-fold more nonlabeled than labeled DNA) was added to lane 5 in the reaction sample. Electrophoresis was carried out with 6% polyacrylamide gel and 0.5× TBE buffer at a 100-V constant voltage for 2.5 h under cold conditions. Subsequently, the gel was imaged by the Molecular Imager FX system (Bio-Rad).
DISCUSSION
Gene regulation and virulence factor production are closely related to the virulence of S. aureus (22, 23). Among regulatory proteins, Rot was first identified in 2000, and several reports showed Rot represses many secretory proteins but enhances cell wall proteins (21, 24–27). The origin of the name of Rot came from its function as a repressor of the expression of toxin genes. Until now, the Rot-dependent inhibitory expression of toxins, including SEB, SED, α-hemolysin, β-hemolysin (truncated), and Panton-Valentine leukocidin, is demonstrated.
Other global regulators, such as the two-component system and the SarA family, interact with Rot (28–32). Among these, the interaction between the Agr system and Rot was characterized. The Agr system is one of the two-component regulatory mechanisms in S. aureus that sense cell density. RNAIII, functional RNA of the Agr system, prevents the translation of rot and indirectly changes gene expression (30, 32). This interactive mechanism also affects some SE expression. The repression of SEB and SED by Rot is inhibited by RNAIII (26, 27). In addition, the upstream sequence of sec has imperfect 10-bp inverted repeats (in MW2 [GenBank accession number BA000033], positions 851652 to 851660 and 851689 to 851698), closely similar to the inverted repeats important for Rot binding to the seb promoter (27). Similar to SEB and SED, the expression of SEC seems to be repressed by Rot, and this repression may be cancelled by RNAIII (33). Effects of the Agr system on the expression of egc-related SEs, such as SEG and SEI, remains to be investigated. However, the expression of these genes changes when the cell density changes (34). Therefore, egc-related SEs also may be affected by RNAIII (and Rot). Of note, these levels of SE production were much lower than those of others in media (Fig. 1). As a possible reason for this, it may be that these SEs could not be detected on meat product, on which the production of other SEs also decreased (Fig. 1). In contrast, we found sarA, sarT, sarU, rot, and saeR are upregulators and sarS is a downregulator for SEH production (Fig. 2 and 3). In addition, we observed SEA was not repressed by Rot. The expression of SEA and SEH was reported to be unaffected by agr (35, 36). This suggests Rot-Agr interaction does not directly interfere with these cases of expression, unlike the other SEs mentioned above. We constructed the mutants from a single strain, no. 10, classified into CC81 subtype 1. We likewise observed similar SEH production in media and meat product with both of the other CC81 subtype 1 strains (Nagasaki and 01240) and the closely related CC1 lineage (MW2) (Fig. 1). Further, the nucleotide sequences of the Rot and seh promoter in no. 10 was the same as those of MW2. Although further investigations are needed, we speculate that an alternative control mechanism for SEH production by Rot is common among these closely related lineages.
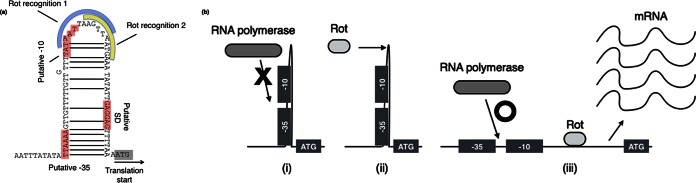
Recently, Benson et al. reported Rot binds the promoters of superantigen-like protein (SSL) and activates SSL expression (37). They also suggested Rot contributes to the stabilization of promoters and then may aid the binding of SaeR to the promoters. In this study, a similar phenomenon was observed. There was a positive correlation of mRNA expression between SEH and Rot under both conditions (Fig. 4), and the data suggest Rot regulates SEH production at the transcriptional level and was essential for expression of seh. Additionally, Rot and SaeR both activated SEH production, similar to a previous study (37). We analyzed the secondary structure of the seh promoter sequence. As shown in Fig. 6a, part of the sequence includes putative Shine-Dalgarno (SD), −10, and −35 sequences that were predicted to form a possible loop-like structure, indicating that loop formation interferes with the access of the RNA polymerase. There are two sequences important for binding of Rot, similar to a previous study (27), at the loop region in this structure, depicted as Rot recognition 1 and 2. Based on the analysis of the seh promoter structure, we hypothesized the following mechanism, as shown in Fig. 6b: (i) RNA polymerase cannot bind to the promoter, (ii) Rot unwinds and prevents the formation of the loop structure, and (iii) this leads RNA polymerase to bind to the promoter sequence, and then the transcription begins. Rot may rewind the SEH promoter to stabilize the structure of the promoter, aiding binding of RNA polymerase to −10 and −35 sequences (Fig. 6). The data suggest alternative Rot regulation is crucial for SEH production on (in) foods and in causing SFP.
FIG 6.
Structure of upstream sequence of seh and schematic image of seh transcription. (a) Prediction of secondary structure of seh promoter region. GENETYX-MAC v.15 (Software Development Co., Ltd., Tokyo, Japan) was used for analysis. The loop-like structure of the promoter sequence is shown. The sequence of no. 10 was identical to that of MW2. −35, −10, and SD sequences and transcription start codons (ATG) are boxed. Two nucleotide sequences important for Rot binding are represented by curved bars. The sequences are similar to those in a previous study (27), but orientation and order differed. (b) Hypothetical interaction scheme of Rot and the promoter sequence of seh. Topological images are shown. (i) The hairpin-like structure inhibits binding of RNA polymerase. (ii) Rot binds to free non-base-pairing regions. (iii) Structure rewinds. RNA polymerase is able to bind the promoter sequence.
Vomiting activity associated with SEs has been investigated using animal models. SEH was reported to have relatively lower vomiting-inducing activity than others against Suncus murinus but similar to that of SEA against the primates (2). Evenson et al. estimated that the minimum amount of SEA for an SFP outbreak is 144 ng/person (38). Assuming that the vomiting activity caused by SEH is comparable to that associated with SEA in humans, consumption of about 5 to 15 g meat product incubated for 24 h with all four seh-positive strains used in this study is enough to cause SFP symptoms. All strains positive for seh in this study except MW2 were classified into CC81, which is the major SFP lineage in Japan, as described previously (6, 39). We claim that this clone has significant potential to cause SFP.
As described above, the large SFP outbreak in Japan was caused by SEA and SEH. Other than Japan, articles reported there were outbreaks caused by S. aureus positive for the seh gene in France, Netherlands, China, Germany, and South Korea (3, 9, 10, 40, 41). Among these, the epidemiological study in South Korea showed S. aureus strains positive for seh were one of the dominant groups. SEH-producing strains belonging to CC81 subtype 1 or related clones may cause frequent SFP outbreaks in far-east Asia. Further epidemiological studies on this important SFP lineage may be necessary to show the spread of the clone positive for seh.
In this study, we demonstrated that a novel positive upregulation mechanism of SEH production by Rot may be one of the virulence mechanisms contributing to the highly frequent onset of SFP outbreaks caused by CC81 subtype 1. Our conclusion has some limits due to a single experimental condition mimicking food contamination using salted ham; however, our data show we can claim SEH and SEA are important SEs. The relationships between newly described SEs and SFP are not fully understood, and further studies are necessary to evaluate potential risks of the newly described SEs in SFP.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jim Nelson for editorial assistance, the Natural Science Center for Basic Research and Development (Hiroshima University) for EMSA imaging, and Yasunori Suzuki (Tokyo Metropolitan Institute of Public Health) for technical assistance for EMSA.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01936-15.
REFERENCES
- 1.Hennekinne JA, De Buyser ML, Dragacci S. 2012. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol Rev 36:815–836. doi: 10.1111/j.1574-6976.2011.00311.x. [DOI] [PubMed] [Google Scholar]
- 2.Hu DL, Nakane A. 2014. Mechanisms of staphylococcal enterotoxin-induced emesis. Eur J Pharmacol 722:95–107. doi: 10.1016/j.ejphar.2013.08.050. [DOI] [PubMed] [Google Scholar]
- 3.Jørgensen HJ, Mathisen T, Løvseth A, Omoe K, Qvale KS, Loncarevic S. 2005. An outbreak of staphylococcal food poisoning caused by enterotoxin H in mashed potato made with raw milk. FEMS Microbiol Lett 252:267–272. doi: 10.1016/j.femsle.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Omoe K, Hu DL, Takahashi-Omoe H, Nakane A, Shinagawa K. 2003. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect Immun 71:6088–6094. doi: 10.1128/IAI.71.10.6088-6094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ono HK, Omoe K, Imanishi K, Iwakabe Y, Hu DL, Kato H, Saito N, Nakane A, Uchiyama T, Shinagawa K. 2008. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect Immun 76:4999–5005. doi: 10.1128/IAI.00045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato'o Y, Omoe K, Naito I, Ono HK, Nakane A, Sugai M, Yamagishi N, Hu DL. 2014. Molecular epidemiology and identification of a Staphylococcus aureus clone causing food poisoning outbreaks in Japan. J Clin Microbiol 52:2637–2640. doi: 10.1128/JCM.00661-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asao T, Kumeda Y, Kawai T, Shibata T, Oda H, Haruki K, Nakazawa H, Kozaki S. 2003. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol Infect 130:33–40. doi: 10.1017/S0950268802007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeda T, Tamate N, Yamaguchi K, Makino S. 2005. Mass outbreak of food poisoning disease caused by small amounts of staphylococcal enterotoxins A and H. Appl Environ Microbiol 71:2793–2795. doi: 10.1128/AEM.71.5.2793-2795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fetsch A, Contzen M, Hartelt K, Kleiser A, Maassen S, Rau J, Kraushaar B, Layer F, Strommenger B. 2014. Staphylococcus aureus food-poisoning outbreak associated with the consumption of ice-cream. Int J Food Microbiol 187:1–6. doi: 10.1016/j.ijfoodmicro.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 10.Cha JO, Lee JK, Jung YH, Yoo JI, Park YK, Kim BS, Lee YS. 2006. Molecular analysis of Staphylococcus aureus isolates associated with staphylococcal food poisoning in South Korea. J Appl Microbiol 101:864–871. doi: 10.1111/j.1365-2672.2006.02957.x. [DOI] [PubMed] [Google Scholar]
- 11.Bergdoll MS. 1988. Monkey feeding test for staphylococcal enterotoxin. Methods Enzymol 165:324–333. doi: 10.1016/S0076-6879(88)65048-8. [DOI] [PubMed] [Google Scholar]
- 12.Hu DL, Omoe K, Shimoda Y, Nakane A, Shinagawa K. 2003. Induction of emetic response to staphylococcal enterotoxins in the house musk shrew (Suncus murinus). Infect Immun 71:567–570. doi: 10.1128/IAI.71.1.567-570.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omoe K, Imanishi K, Hu DL, Kato H, Fugane Y, Abe Y, Hamaoka S, Watanabe Y, Nakane A, Uchiyama T, Shinagawa K. 2005. Characterization of novel staphylococcal enterotoxin-like toxin type P. Infect Immun 73:5540–5546. doi: 10.1128/IAI.73.9.5540-5546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omoe K, Hu DL, Ono HK, Shimizu S, Takahashi-Omoe H, Nakane A, Uchiyama T, Shinagawa K, Imanishi K. 2013. Emetic potentials of newly identified staphylococcal enterotoxin-like toxins. Infect Immun 81:3627–3631. doi: 10.1128/IAI.00550-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omoe K, Hu DL, Takahashi-Omoe H, Nakane A, Shinagawa K. 2005. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiol Lett 246:191–198. doi: 10.1016/j.femsle.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Omoe K, Ishikawa M, Shimoda Y, Hu DL, Ueda S, Shinagawa K. 2002. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates harboring seg, seh, or sei genes. J Clin Microbiol 40:857–862. doi: 10.1128/JCM.40.3.857-862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato F, Sugai M. 2011. A simple method of markerless gene deletion in Staphylococcus aureus. J Microbiol Methods 87:76–81. doi: 10.1016/j.mimet.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Kato F, Kadomoto N, Iwamoto Y, Bunai K, Komatsuzawa H, Sugai M. 2011. Regulatory mechanism for exfoliative toxin production in Staphylococcus aureus. Infect Immun 79:1660–1670. doi: 10.1128/IAI.00872-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda S, Tomida Y, Ohta H, Takamiya K. 2007. The critical role of a hydrogen bond between Gln63 and Trp104 in the blue-light sensing BLUF domain that controls AppA activity. J Mol Biol 368:1223–1230. doi: 10.1016/j.jmb.2007.02.087. [DOI] [PubMed] [Google Scholar]
- 20.Xue T, You Y, Shang F, Sun B. 2012. Rot and Agr system modulate fibrinogen-binding ability mainly by regulating clfB expression in Staphylococcus aureus NCTC8325. Med Microbiol Immunol 201:81–92. doi: 10.1007/s00430-011-0208-z. [DOI] [PubMed] [Google Scholar]
- 21.McNamara PJ, Milligan-Monroe KC, Khalili S, Proctor RA. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J Bacteriol 182:3197–3203. doi: 10.1128/JB.182.11.3197-3203.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fechter P, Caldelari I, Lioliou E, Romby P. 2014. Novel aspects of RNA regulation in Staphylococcus aureus. FEBS Lett 588:2523–2529. doi: 10.1016/j.febslet.2014.05.037. [DOI] [PubMed] [Google Scholar]
- 23.Purrello SM, Daum RS, Edwards GFS, Lina G, Lindsay J, Peters G, Stefani S. 2014. Meticillin-resistant Staphylococcus aureus (MRSA) update: new insights into bacterial adaptation and therapeutic targets. J Glob Antimicrob Resist 2:61–69. doi: 10.1016/j.jgar.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Dumitrescu O, Choudhury P, Boisset S, Badiou C, Bes M, Benito Y, Wolz C, Vandenesch F, Etienne J, Cheung AL, Bowden MG, Lina G. 2011. Beta-lactams interfering with PBP1 induce Panton-Valentine leukocidin expression by triggering sarA and rot global regulators of Staphylococcus aureus. Antimicrob Agents Chemother 55:3261–3271. doi: 10.1128/AAC.01401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saïd-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. 2003. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 185:610–619. doi: 10.1128/JB.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tseng CW, Zhang S, Stewart GC. 2004. Accessory gene regulator control of staphyloccoccal enterotoxin D gene expression. J Bacteriol 186:1793–1801. doi: 10.1128/JB.186.6.1793-1801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng CW, Stewart GC. 2005. Rot repression of enterotoxin B expression in Staphylococcus aureus. J Bacteriol 187:5301–5309. doi: 10.1128/JB.187.15.5301-5309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol 40:1–9. doi: 10.1016/S0928-8244(03)00309-2. [DOI] [PubMed] [Google Scholar]
- 29.Frees D, Sørensen K, Ingmer H. 2005. Global virulence regulation in Staphylococcus aureus: pinpointing the roles of ClpP and ClpX in the sar/agr regulatory network. Infect Immun 73:8100–8108. doi: 10.1128/IAI.73.12.8100-8108.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. 2006. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 31.Li D, Cheung A. 2008. Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect Immun 76:1068–1075. doi: 10.1128/IAI.01069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regassa LB, Couch JL, Betley MJ. 1991. Steady-state staphylococcal enterotoxin type C mRNA is affected by a product of the accessory gene regulator (agr) and by glucose. Infect Immun 59:955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derzelle S, Dilasser F, Duquenne M, Deperrois V. 2009. Differential temporal expression of the staphylococcal enterotoxins genes during cell growth. Food Microbiol 26:896–904. doi: 10.1016/j.fm.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Lis E, Podkowik M, Bystroń J, Stefaniak T, Bania J. 2012. Temporal expression of staphylococcal enterotoxin H in comparison with accessory gene regulator-dependent and -independent enterotoxins. J Food Prot 75:238–244. doi: 10.4315/0362-028X.JFP-11-336. [DOI] [PubMed] [Google Scholar]
- 36.Tremaine MT, Brockman DK, Betley MJ. 1993. Staphylococcal enterotoxin A gene (sea) expression is not affected by the accessory gene regulator (agr). Infect Immun 61:356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benson MA, Lilo S, Nygaard T, Voyich JM, Torres VJ. 2012. Rot and SaeRS cooperate to activate expression of the staphylococcal superantigen-like exoproteins. J Bacteriol 194:4355–4365. doi: 10.1128/JB.00706-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evenson ML, Hinds MW, Bernstein RS, Bergdoll MS. 1988. Estimation of human dose of staphylococcal enterotoxin A from a large outbreak of staphylococcal food poisoning involving chocolate milk. Int J Food Microbiol 7:311–316. doi: 10.1016/0168-1605(88)90057-8. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki Y, Omoe K, Hu DL, Sato'o Y, Ono HK, Monma C, Arai T, Konishi N, Kato R, Hirai A, Nakama A, Kai A, Kamata Y. 2014. Molecular epidemiological characterization of Staphylococcus aureus isolates originating from food poisoning outbreaks that occurred in Tokyo, Japan. Microbiol Immunol 58:570–580. doi: 10.1111/1348-0421.12188. [DOI] [PubMed] [Google Scholar]
- 40.Kérouanton A, Hennekinne JA, Letertre C, Petit L, Chesneau O, Brisabois A, De Buyser ML. 2007. Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int J Food Microbiol 115:369–375. doi: 10.1016/j.ijfoodmicro.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 41.Yan X, Wang B, Tao X, Hu Q, Cui Z, Zhang J, Lin Y, You Y, Shi X, Grundmann H. 2012. Characterization of Staphylococcus aureus strains associated with food poisoning in Shenzhen, China. Appl Environ Microbiol 78:6637–6642. doi: 10.1128/AEM.01165-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato'o Y, Omoe K, Ono HK, Nakane A, Hu DL. 2013. A novel comprehensive analysis method for Staphylococcus aureus pathogenicity islands. Microbiol Immunol 57:91–99. doi: 10.1111/1348-0421.12007. [DOI] [PubMed] [Google Scholar]
- 43.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 44.Kadan RS, Martin WH, Mickelsen R. 1963. Effect of ingredients use in condensed and frozen dairy products on thermal resistance of potentially pathogenic staphylococci. Appl Microbiol 11:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frea JI, McCoy E, Strong FM. 1963. Purification of type B staphylococcal enterotoxin. J Bacteriol 86:1308–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Traber K, Novick R. 2006. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate delta- and alpha-haemolysins. Mol Microbiol 59:1519–1530. doi: 10.1111/j.1365-2958.2006.04986.x. [DOI] [PubMed] [Google Scholar]
- 47.Cheung AL, Nishina KA, Trotonda MP, Tamber S. 2008. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol 40:355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.