Abstract
West Nile virus (WNV) is an arbovirus that was first reported in North America in New York in 1999 and, by 2003, had spread more than 4,000 km to California. However, variation in viral genetics associated with spread are not well understood. Herein, we report sequences for more than 100 WNV isolates made from mosquito pools that were collected from 2003 – 2011 as part of routine surveillance by the California Mosquito-borne Virus Surveillance System. We performed phylogeographic analyses and demonstrated that 5 independent introductions of WNV (1 WN02 genotype strain and 4 SW03 genotype strains) occurred in California. The SW03 genotype of WNV was constrained to the southwestern U.S. and had a more rapid rate of spread. In addition, geographic constraint of WNV strains within a single region for up to 6 years suggest viral maintenance has been driven by resident, rather than migratory, birds and overwintering in mosquitoes.
Keywords: West Nile virus, arbovirus, phylogenetics, evolution
INTRODUCTION
West Nile virus (WNV) is an arbovirus of the family Flaviviridae that was first identified in North America in 1999. In the U.S., Culex mosquitoes are the primary vectors for WNV (1–4), and passerine birds are the most common avian host (1, 5). As incidental hosts, humans, equines, other mammals, and many bird species do not contribute to the enzootic cycle or evolution of WNV.
WNV was first reported in New York and reached the West Coast within 4 years. Beginning in July 2003, mosquito pools from California tested positive for WNV (6). In addition, three autochthonous human cases of WNV in southern California were reported to ArboNET in 2003. Human WNV disease cases in northern and southern California were reported to ArboNET the following year, and, by the conclusion of the 2004 transmission season, WNV had been detected in every county in the state of California (7).
The ecology of California has presented WNV with physical and climatic barriers as well as an assortment of vector and host communities. Although much of California has a Mediterranean climate with cool, rainy winters and dry summers, it is a very long state extending 1,240 km from the dry SE deserts at the Mexican border to the NW coastal rain forests at the Oregon border. The state is divided longitudinally by the Coast Range and the Sierra Nevadas, which form the east and west boundaries of the intense Central Valley agroecosystem, respectively, and by the Tehachapi and San Gabriel mountains, which enclose the southern end of the Central Valley and isolate the highly urbanized Los Angeles basin and San Diego areas from the remainder of the state. Natural physical divisions have altered mosquito and avian host communities. Culex tarsalis Coq., the primary rural vector throughout most of the state, is divided into distinct clades found in the SE Desert biome and the northern areas of the state (8). Culex pipiens L complex populations vary longitudinally, and recent genetic studies have detected four major groups based on structure analysis: Cx. p. quinquefasciatus south of the Tehachapi Mountains, Cx. pipiens form pipiens and Cx. pipiens form molestus in urban areas near San Francisco and Sacramento, and admixtures of all three forms in the Central Valley (9).
By mapping the sampling location and date of WNV-positive mosquito pools, a single introduction of WNV into southern California, followed by northward expansion, was proposed as the route for WNV emergence in California (6). The large-scale spatial dynamics of WNV emergence in the U.S. also was determined using sequence data from isolates collected across the country between 1999 and 2006 (10). In this analysis, the virus was shown to have spread from the East Coast to the West Coast at an average diffusion rate of 1,200 km/year, which includes a phase of increasing rate from 1999 to 2003 followed by a reduced rate of diffusion from 2004 to 2006. The high rate of WNV dispersal in the U.S. could be explained by WNV infection of migratory birds that travel long distances annually (11); however, evidence for infection in vernal northbound migrants has been limited for WNV (12, 13) as well as other North American encephalitides (14, 15). In contrast, movements by resident birds have also been shown to be important for WNV enzootic maintenance and movement (16). Furthermore, the phylodynamics of two emergent WNV genotypes, WN02 and SW03, have not been studied. These genotypes are characterized by the amino acid substitutions E-V159A and NS4A-A85T, respectively (17–19). WN02 genotype isolates have a shorter extrinsic incubation period in Culex mosquitoes than the founding NY99 strain (17, 20, 21), and WN02 and SW03 genotype isolates replicate to higher peak titers in house sparrows compared to the original NY99 genotype (22), suggesting that WNV transmission dynamics may differ by genotype.
In the current study, we used a consistent sampling method of collecting WNV isolates from mosquitoes, followed by sequence analysis, to evaluate WNV dynamics during its emergence in California from 2003 to 2011. We show evidence for multiple introductions of WNV into California. Next, we compared the rate of dispersal of WN02 and SW03 genotypes to infer adaptive phenotypes. Finally, using isolates collected during winter months, we provide evidence for overwintering of WNV in mosquitoes in southern California.
MATERIALS AND METHODS
Mosquito pools
Mosquitoes were collected and pooled by species during routine surveillance by California mosquito control agencies from 2003 to 2011. Pools were triturated, and viral RNA was extracted at the UC Davis Center for Vectorborne Diseases. Samples were screened for WNV using quantitative real time RT-PCR, as recently described (23). Pools were selected for further analysis based on Ct score and date and location of WNV RNA detection. Isolates were stratified by year and month of collection, genotype, or mosquito species. Significance was determined using a chi-square test.
Viral genome sequencing and assembly
Mosquito homogenates were passaged once on Vero or C6/36 cells. Viral RNA was extracted from clarified primary cultures using an ABI MagMax kit. RNA amplification was performed as previously described (24). Illumina library construction was performed using NexteraXT (Illumina) by following the manufacturer’s protocol. Sequencing was performed on the Illumina HiSeq2500 platform, generating paired-end 101 bp reads. The reads were assembled using the VICUNA assembly program (25). Sequences for 112 isolates were deposited into GenBank with accession numbers KR348915–KR349026 (http://www.ncbi.nlm.nih.gov/Genbank/).
Phylogenetic analyses
143 previously sequenced full-length WNV genomes were selected from GenBank based on date and location of sampling and added to the dataset of 112 newly sequenced isolates, creating a final dataset of 255 sequences. These 143 previously-sequenced isolates were collected between 1999 and 2012. 23 out of the 143 isolates were collected in CA, and the remaining 120 isolates were collected across North America outside of CA. Latitude, longitude, and date were taken from GenBank entries or estimated as previously described (10). Dates were converted into decimals.
The coding regions of all 255 sequences were aligned using Clustal Omega (26) and edited manually. jModelTest2 was used to determine the most appropriate nucleotide substitution model, which was a generalized time reversible (GTR) model with a gamma (Γ) distribution of rate variation among sites (27, 28). Phylogenies were constructed using the Bayesian MCMC method in BEAST v1.8 (29) with a GTR + Γ substitution model, a lognormal relaxed molecular clock, and a time-aware smoothed GMRF Bayesian Skyride coalescent model (30, 31). Distances were calculated based on great circle distances. Spatial parameters were estimated using relaxed random walks (RRW), a heterogeneous probability distribution of diffusion in order to allow for variation in dispersal rate, as previously determined to be most appropriate for WNV in North America (10). However, selection for the dispersal model was also performed by using the harmonic mean estimator (HME) to calculate log Bayes Factors for gamma, lognormal, and cauchy heterogeneous models and a homogenous Brownian model. The homogeneous model was rejected. A lognormal distribution of heterogeneous dispersal rates was used for further analyses because the cauchy RRW produced results that were very similar, and MCMC convergence was not reached with the analysis using a gamma RRW. MCMC chains were run for 200 million states, sampling every 20,000 states. Convergence was evaluated using Tracer v1.6. The maximum clade credibility (MCC) tree was determined using TreeAnnotator and visualized using FigTree v1.4.2 and SPREAD (32). Computing resources were used from the Cipres Science Gateway (33).
RESULTS
Sampling and sequencing of WNV from mosquito pools
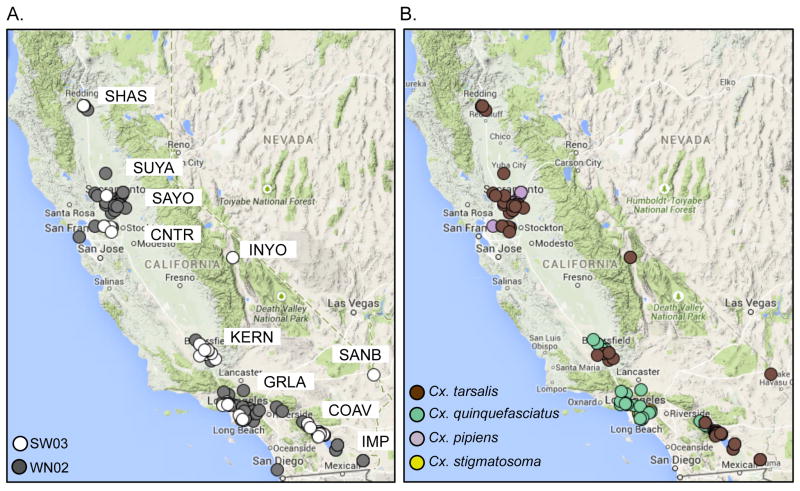
A total of 192 mosquito pools that were collected between 2003 and 2011 in California and that were RT-PCR positive for WNV RNA were inoculated onto Vero cells. Of these, infectious virus was collected from primary passages of 136 pools, from which viral RNA was extracted and the consensus sequence determined. Full-length WNV genomes were successfully assembled from 112 mosquito pools from 10 mosquito control districts in California: Sacramento-Yolo (SAYO), Shasta (SHAS), Coachella Valley (COAV), Greater Los Angeles County (GRLA), Sutter-Yuba (SUYA), Kern (KERN), Contra Costa (CNTR), Imperial County (IMPR), Owens Valley (INYO), and San Bernardino (SANB).
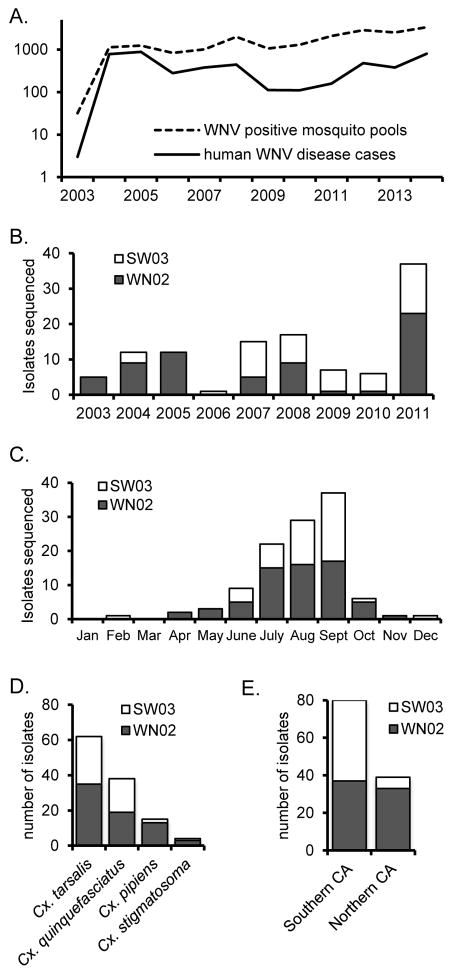
WNV positive mosquito pools were first detected in California in 2003, and the number detected varied annually (Fig. 1A). Therefore, the number of WNV genomes sequenced from mosquito pools in this study also varied by year (Fig. 1B). In addition, the total number of sequenced isolates varied by month (Fig. 1C), mosquito species (Fig. 1D), and location of collection (Fig. 1E and Fig. 2A). Across the years sampled, the number of isolates that were sequenced was generally proportional to the total number of WNV positive mosquito pools detected in California. The number of sequenced isolates were underrepresented in 2006, likely due degradation of samples, and overrepresented in 2003 due to an attempt to understand the genetic composition of isolates at the presumed time of WNV introduction into California. The majority of sequenced WNV RNA-positive mosquito pools were collected during July, August, and September (Fig. 1C) in a manner proportional to the number of positive mosquito pools and human WNV disease cases reported to ArboNET from California. Most isolates were collected in 5 mosquito control districts: GRLA, KERN, COAV, SAYO, and CNTR (Fig. 2A), which together accounted for the locations of 50% of human WNV disease cases in CA. In total, isolates from ca. 1% of all WNV RNA-positive mosquito pools collected in CA between 2003 and 2011 were sequenced in this study.
Figure 1.
Temporal distribution of WNV isolates sequenced in this study. (A) The number of WNV positive mosquito pools identified in California (dashed line) and number of human WNV disease cases per year in California (solid line). (B) The number of sequenced WN02 (filled bars) and SW03 isolates (open bars) that were collected per year. (C) The number of sequenced WN02 (filled bars) and SW03 (open bars) isolates that were collected each month. (D) The number of WN02 (filled bars) and SW03 (open bars) isolates stratified by mosquito species. (E) The number of WN02 (filled bars) and SW03 (open bars) isolates collected in northern or southern CA.
Figure 2.
Spatial distribution of WNV isolates sequenced in this study. (A) A map showing the location of WN02 (filled circles) and SW03 (open circles) isolates sampled in this study, with locations of mosquito control districts indicated. (B) A map showing the location of isolates sampled in this study stratified by mosquito species.
Isolates were stratified by genotype (WN02 or SW03). Approximately 60% of WNV isolates were WN02 genotype, and 40% were SW03 genotype. Analyses of isolates based on genotype showed significant differences based on year of collection (p<0.001; Fig. 1C). There were more WN02 genotype viruses than SW03 genotype viruses collected from 2003–2005, indicating that the WN02 genotype was likely responsible for human outbreaks during these years. The distribution of SW03 genotype viruses was also restricted based on location, such that the proportion of WN02 and SW03 genotype viruses from southern CA was significantly different compared to northern CA (p<0.001; Fig. 1D). The majority of viruses collected in northern CA were WN02 genotype. The mosquito species represented by the isolates included Cx. pipiens and Cx. tarsalis in the north and Cx. quinquefasciatus, Cx. tarsalis, and Cx. stigmatosoma in the south (Fig. 2B). However, there was not a significant difference in the proportion of WNV genotypes by mosquito species.
Phylodynamics of WNV in California
The coding region of the 112 newly sequenced time-stamped WNV genomes from CA were aligned with 143 publicly available WNV genomes from GenBank. A Bayesian MCMC analysis was performed to infer their phylodynamic relationship. On average, WNV in the U.S. exhibited a mutation rate of 5.3×10−4 substitutions/site/year [95% highest posterior density (HPD) interval: 4.7×10−4 to 5.8×10−4 substitutions/site/year] from 1999 to 2012. The origin of the WNV epidemic in the U.S. was estimated to be 1998.4 (95% HPD interval: 1997.9–1998.9) at 40.47°N, −72.59°W, which is near Long Island, NY. This estimate fits well with the onset of the first known case of human WNV disease in the U.S., which was August 5, 1999 (1999.6) in New York (34).
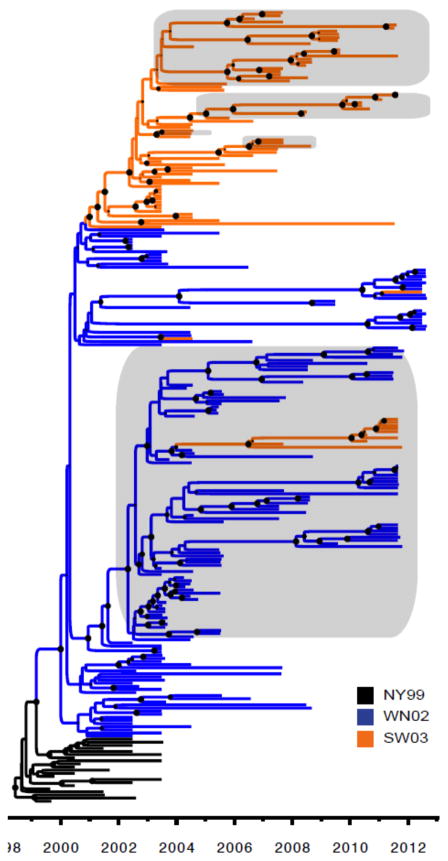
WNV was first identified in Imperial Valley, CA in July 2003 in mosquitoes, and then in sentinel chickens, dead birds, and humans at multiple locations throughout southern CA. WNV was contained south of the Tehachapi Mountains during 2003 but then spread rapidly throughout CA. The first detection of WNV outside of southern CA was in July 2004 near Fresno (6, 7). If a single introduction of WNV into CA was assumed, the time to most recent common ancestor (TMRCA) of CA WNV would indicate an undetected introduction of WNV into CA in 2000.3 (95% HPD interval: 1999.7–2000.9). However, surveillance programs had been actively testing for WNV in mosquitoes and sentinel chickens since 2000, and WNV was not detected in CA until July 16, 2003 (2003.5) (6), making a single, undetected introduction of WNV into CA in 2000 very unlikely. Instead, if multiple introductions of WNV into CA were allowed, a more reasonable time of introduction could be estimated. In this scenario, there was genetic evidence of 1 introduction of WNV genotype WN02 in 2002.3 between the GRLA and COAV districts (95% HPD interval: 2001.9–2002.7) and 2 introductions of WNV genotype SW03 in the GRLA district approximately 1 year later in 2003.3 (95% HPD interval: 2002.8–2003.8) and 2003.5 (95% HPD interval: 2003.0–2003.9), followed by at least 2 more introductions of SW03 in 2005 and 2006 in the COAV district (Fig. 3). The WN02 genotype virus spread to northern CA at least three times during the year after its initial introduction into CA. However, only the first of the four SW03 introductions spread to northern CA, and this occurred several years later in 2006.5. The other three introduced SW03 viruses remained locally near the site of introduction in southern CA (COAV or GRLA). Co-circulation of WN02 and SW03 genotypes was evident in most locations from 2006 through 2008, although the SW03 genotype appeared to have displaced the WN02 genotype in COAV and GRLA by 2009. This apparent displacement is unlikely to be due to sampling bias, as nearly twice as many isolates were sequenced per year in the COAV and GRLA districts after 2008 as compared to before 2008.
Figure 3.
Phylogenetic relationship between WNV isolates. A MCC phylogeny constructed from the open reading frame of 196 WNV isolates is shown. Nodes with >0.9 posterior probability are indicated by filled circles. Isolates from California are highlighted in gray. The two emergent genotypes of WNV are indicated on the phylogeny by color: WN02 in blue lines and SW03 in orange lines.
Phylogeography of WNV in California
During 2003, WNV was only detected in the 6 most southern counties of California. The most likely sources of the WNV strains introduced into CA were the western and southern states, primarily Arizona and Texas (Fig. 4A and B). Isolates from Mexico that were included in this analyses were also derived from Arizona (SW03 genotype) or CA (WN02 genotype) and were introduced in 2002–2003 (Fig. 4A and B).
Figure 4.
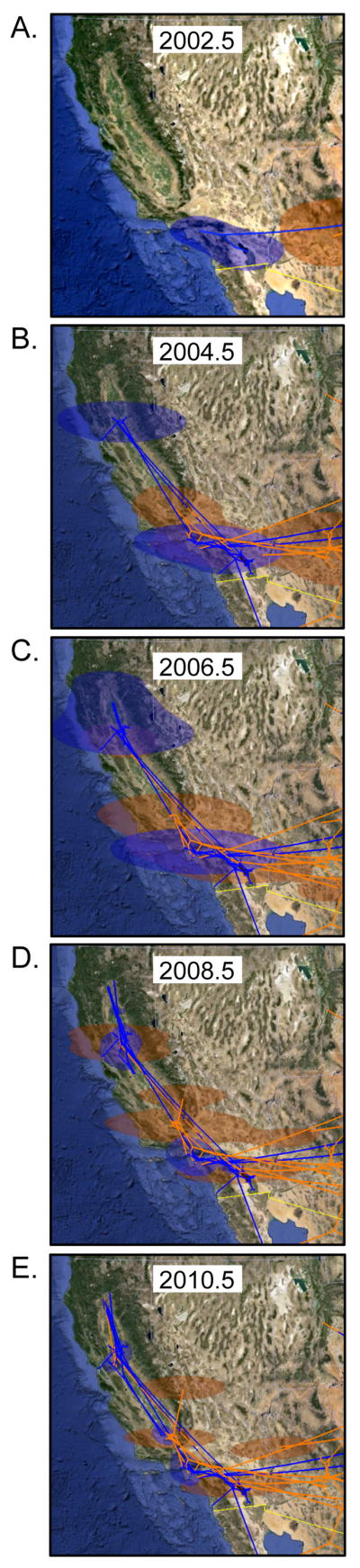
Dispersal of WNV isolates in California from 2002 – 2010 (A through E). Solid lines represent the MCC tree of WNV isolates, with 80% HPD regions for the location of uncertain nodes shown by clouds. The two emergent genotypes of WNV are indicated by color: WN02 in blue and SW03 in orange.
Previous studies have shown that WNV followed a heterogeneous dispersal pattern during emergence in the U.S., in which rapid dispersal from 1999–2003 was followed by a much slower rate of dispersal from 2003–2006, with an average rate of 1200 km/year (10). Using a heterogeneous dispersal model, in this study WNV had an average rate of dispersal of 130 km/year (95% HPD: 118–142 km/year) from 1999–2012, which was much slower than the previous estimate for WNV spread during emergence. This suggests that the long-range movements that drove high WNV dispersal rates early in the epidemic were no longer characteristic of WNV once it became endemic. However, a homogeneous model of dispersal was still rejected. Excluding the CA WN02 genotype isolates from the analysis, WNV had an average rate of dispersal of 167 km/year (95% HPD interval: 150–185 km/year), suggesting that CA WN02 genotype viruses had a rate of dispersal that is lower than the average for North American isolates. Excluding the SW03 clade from the analysis, WNV had an average rate of dispersal of 113 km/year (95% HPD interval: 100–124 km/year). This suggests that SW03 genotype viruses had a rate of dispersal that is higher than average for North American isolates. The SW03 clade spread at a dispersal rate that was 10% higher than the relative rate of dispersal of the CA WN02 clade.
The SW03 genotype was estimated to have first arisen in New Mexico in 2001, whereas the WN02 genotype was estimated to have first arisen in Connecticut in 2000. Interestingly, the SW03 genotype evolved independently on at least 3 other occasions in the southwest: in GRLA between 2005.8 and 2007.3, in Dallas, Texas between 2010.5 and 2011.9, and in Colorado between 2003.5 and 2004.6. Based on the 80% HPD estimates for the location of ancestral nodes, the SW03 genotype thrived in New Mexico, Arizona, southern CA, and northern Mexico (Fig. 4B). In contrast, the WN02 genotype was not found among the 10 isolates from Arizona included in the analysis and was restricted in its diffusion in the southwest (Fig. 4A). However, it spread within northern CA to a much greater extent than the SW03 genotype (Fig. 4A). This suggests that the observation in Fig. 1E that the SW03 and WN02 genotypes were found in different proportions based on location may be due to genetic determinants of viral spread within the different climates, mosquito subspecies, or avian hosts in the southwestern U.S.
Overwintering of WNV in mosquitoes
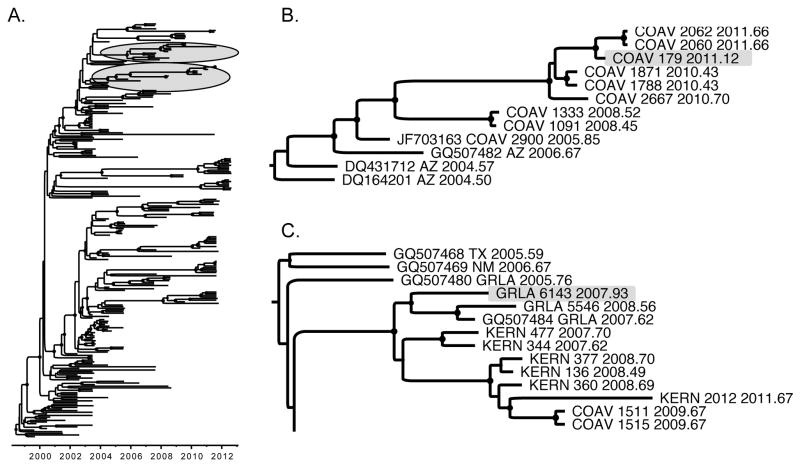
Despite the collection of most mosquito pools during summer and fall, the seasons of greatest WNV transmission, two mosquito pools containing WNV were collected during December 2007 and February 2011 in the GRLA and COAV districts, respectively. If WNV had been maintained in southern California through the winter by mosquitoes, these winter isolates should phylogenetically cluster with viruses collected at the end of one transmission season and viruses collected at the beginning of the next transmission season. In addition, spatial clustering of these WNV variants would be expected.
In Fig. 5A, isolate COAV 179_2011, which was collected on Feb. 15, 2011 from Culex tarsalis, is highlighted on an MCC Bayesian phylogeny. This winter isolate, which clustered with the SW03 genotype viruses, clustered temporally between isolates collected during the 2010 and 2011 WNV transmission seasons (Fig. 5B). COAV isolates were also geographically clustered north of the Salton Sea for 6 years. Similar results were found for a second winter isolate (GRLA 6143_2007, SW03 genotype, collected Dec. 5, 2007 from Cx. quinquefasciatus), also highlighted on the MCC Bayesian phylogeny (Fig. 5A). This isolate was phylogenetically and geographically clustered in the same location for 2 years (Fig. 5C). These data indicate the overwintering of WNV amongst winter Culex mosquitoes in southern CA.
Figure 5.
Overwintering of WNV in southern CA. (A) The MCC phylogeny of WNV, with the areas of interest highlighted. (B) A close-up view of one branch from the MCC phylogeny, with winter isolate COAV179_2011 highlighted. (B) A close-up view of another branch from the MCC phylogeny, with winter isolate GRLA6143_2007 highlighted. Isolate names include location of collection, isolate number, and date of collection in decimal format.
WNV was introduced into California at least 5 times.
The SW03 genotype of WNV was constrained to southern California.
The SW03 genotype spread with a more rapid dispersal rate than the WN02 genotype.
The overall dispersal rate of WNV has decreased dramatically since its emergence.
WNV overwintered in California in Culex mosquitoes.
While no winter isolates were collected from northern CA, there was phylogenetic clustering of WN02 isolates from SAYO, SUYA, SHAS, and CNTR districts from 2005 to 2011. This indicates that WNV was also likely maintained in northern CA, rather than reintroduced annually from southern CA.
DISCUSSION
By sampling mosquitoes from the same locations throughout the emergence and maintenance of WNV in California (Fig. 1 and 2), a unique dataset of viral isolates was created to evaluate the spatial and temporal evolution of WNV. With this dense sampling strategy, we were able to determine that WNV was introduced into California at least 5 times via southern CA, with the majority of introductions occurring around 2003 and by SW03 genotype viruses (Fig. 3). The first three of the introduced WNV strains (one WN02 and two SW03 genotype viruses) were maintained in California through the most recent sampling dates in 2011, and the first two of the introduced strains also spread to northern California (Fig. 3). These studies build upon previous reports of the variable rate of diffusion of WNV (10) by adding a genetic component to this heterogeneous dispersal pattern. SW03 genotype viruses had a higher dispersal rate than the average of all WNV isolates, and SW03 and WN02 genotype viruses occupy different spatial niches in the U.S. (Fig. 4). Horizontal transmission of WNV during winter months has previously been shown to occur in southern California in birds (35) and Culex tarsalis mosquitoes (36). The phylogeographic continuity of WNV evolution (Fig. 5) confirmed that overwintering contributes to enzootic maintenance. Together, these analyses suggest that the spread and maintenance of WNV is dependent on viral genetics and climate.
Since the introduction of WNV to the U.S., only two non-synonymous mutations in WNV have risen to high frequencies. The E-V159A substitution (WN02 genotype) has been found in all sequenced WNV isolates for several years. The NS4A-A85T substitution (SW03 genotype) has arisen independently at least four times in the southwestern U.S. While WNV lacked strong spatial clustering during its emergence in the U.S., likely due to a complex enzootic cycle involving birds, the isolates from CA showed evidence of spatial restriction based on genotype. Although there were no differences between WN02 and SW03 genotypes in the location of their introductions into California (Fig. 4), SW03 genotype viruses were more frequently located in southern CA than WN02 genotype viruses (Fig. 1), were introduced more frequently to CA than WN02 genotype viruses (Fig. 3), and were estimated to have a higher dispersal rate than WN02 genotype viruses (Fig. 4). These seemingly contradictory characteristics of a spatially restricted yet more diffuse virus suggest that the amino acid substitution that defines the SW03 genotype, NS4A-A85T, may be a temperature-sensitive adaptive mutation that drives a higher dispersal rate only in warmer climates. The mechanism by which SW03 viruses could diffuse faster than WN02 viruses is unknown but may be due to faster kinetics of viral replication or higher competence in mosquitoes (17, 21) or peridomestic passerine birds such as house sparrows (22) compared to WN02 genotype viruses. In addition, the warmer climate in southern CA may naturally decrease the extrinsinic incubation period of WNV in mosquitoes.
Other WNV mutations that have been previously characterized include several that are known to increase temperature sensitivity of WNV (37). These mutations were only found in CA isolates from 2003 through 2005. The decreased ability of WNV isolates containing these temperature-sensitive mutations to replicate at the high temperatures that are generated during corvid infections (38) may have contributed to the disappearance of these mutations. Therefore, temperature sensitivity is a critical phenotype to consider in the analysis of WNV spread.
In this dataset, there is evidence of few long-distance WNV dispersal events after the virus was introduced to CA. While the introduction of WNV into a naïve location may be more visibly dependent on migrating birds, the maintenance of WNV in an endemic area is likely dependent on resident birds, which contribute only small-scale movements. In order to understand whether the differential dispersal of emergent WNV genotypes in the U.S. is due to differences in host or vector competence, the replication dynamics of the WNV genotypes will need to be compared in multiple avian and mosquito species.
Acknowledgments
We acknowledge the intense mosquito sampling, pool preparation, and fiscal support from the Mosquito and Vector Control Association of California member agencies. We especially thank Robert Chiles, Sandra Garcia, and the staff of the arbovirus research group in the Center for Vectorborne Diseases that screened the pools for WNV. We also thank Philippe Lemey for advice and technical assistance with BEAST.
This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Grant No. AI55607 (WKR), Grant No. AI067380 (GDE) and Contract No. HHSN272200900018C to the Broad Institute’s Genomic Sequencing Center for Infectious Diseases, and by an APHL/CDC Emerging Infectious Diseases postdoctoral fellowship (NKD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reisen WK, Fang Y, Martinez VM. Avian host and mosquito (Diptera: Culicidae) vector competence determine the efficiency of West Nile and St. Louis encephalitis virus transmission. Journal of medical entomology. 2005;42:367–375. doi: 10.1093/jmedent/42.3.367. [DOI] [PubMed] [Google Scholar]
- 2.Apperson CS, Hassan HK, Harrison BA, Savage HM, Aspen SE, Farajollahi A, Crans W, Daniels TJ, Falco RC, Benedict M, Anderson M, McMillen L, Unnasch TR. Host feeding patterns of established and potential mosquito vectors of West Nile virus in the eastern United States. Vector borne and zoonotic diseases. 2004;4:71–82. doi: 10.1089/153036604773083013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Godsey MS, Jr, Nasci R, Savage HM, Aspen S, King R, Powers AM, Burkhalter K, Colton L, Charnetzky D, Lasater S, Taylor V, Palmisano CT. West Nile virus-infected mosquitoes, Louisiana, 2002. Emerging infectious diseases. 2005;11:1399–1404. doi: 10.3201/eid1109.040443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turell MJ, O’Guinn M, Oliver J. Potential for New York mosquitoes to transmit West Nile virus. The American journal of tropical medicine and hygiene. 2000;62:413–414. doi: 10.4269/ajtmh.2000.62.413. [DOI] [PubMed] [Google Scholar]
- 5.Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, Davis B, Bowen R, Bunning M. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerging infectious diseases. 2003;9:311–322. doi: 10.3201/eid0903.020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reisen W, Lothrop H, Chiles R, Madon M, Cossen C, Woods L, Husted S, Kramer V, Edman J. West Nile virus in California. Emerging infectious diseases. 2004;10:1369–1378. doi: 10.3201/eid1008.040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hom AML, Kramer VL, Cahoon B, Glaser C, Cossen C, Baylis E, Jean C, Tu E, Eldridge BF, Carney R, Padgett K, Sun B, Reisen WK, Woods L, Husted S. Surveillance for mosquito-borne encephalitis virus activity and human disease, including West Nile virus, in California, 2004. Proceedings of the Mosquito and Vector Control Association of California. 2005;73:66–77. [Google Scholar]
- 8.Venkatesan M, Rasgon JL. Population genetic data suggest a role for mosquito-mediated dispersal of West Nile virus across the western United States. Molecular ecology. 2010;19:1573–1584. doi: 10.1111/j.1365-294X.2010.04577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kothera L, Nelms BM, Reisen WK, Savage HM. Population genetic and admixture analyses of Culex pipiens complex (Diptera: Culicidae) populations in California, United States. The American journal of tropical medicine and hygiene. 2013;89:1154–1167. doi: 10.4269/ajtmh.13-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pybus OG, Suchard MA, Lemey P, Bernardin FJ, Rambaut A, Crawford FW, Gray RR, Arinaminpathy N, Stramer SL, Busch MP, Delwart EL. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:15066–15071. doi: 10.1073/pnas.1206598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dusek RJ, McLean RG, Kramer LD, Ubico SR, Dupuis AP, 2nd, Ebel GD, Guptill SC. Prevalence of West Nile virus in migratory birds during spring and fall migration. The American journal of tropical medicine and hygiene. 2009;81:1151–1158. doi: 10.4269/ajtmh.2009.09-0106. [DOI] [PubMed] [Google Scholar]
- 12.Rappole JH, Hubalek Z. Migratory birds and West Nile virus. Journal of applied microbiology. 2003;94(Suppl):47S–58S. doi: 10.1046/j.1365-2672.94.s1.6.x. [DOI] [PubMed] [Google Scholar]
- 13.Reisen WK, Wheeler SS, Garcia S, Fang Y. Migratory birds and the dispersal of arboviruses in California. The American journal of tropical medicine and hygiene. 2010;83:808–815. doi: 10.4269/ajtmh.2010.10-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calisher CH, Maness KS, Lord RD, Coleman PH. Identification of two South American strains of eastern equine encephalomyelitis virus from migrant birds captured on the Mississippi delta. American journal of epidemiology. 1971;94:172–178. doi: 10.1093/oxfordjournals.aje.a121309. [DOI] [PubMed] [Google Scholar]
- 15.Lord RD, Calisher CH. Further evidence of southward transport of arboviruses by migratory birds. American journal of epidemiology. 1970;92:73–78. doi: 10.1093/oxfordjournals.aje.a121181. [DOI] [PubMed] [Google Scholar]
- 16.Rappole JH, Compton BW, Leimgruber P, Robertson J, King DI, Renner SC. Modeling movement of West Nile virus in the Western hemisphere. Vector borne and zoonotic diseases. 2006;6:128–139. doi: 10.1089/vbz.2006.6.128. [DOI] [PubMed] [Google Scholar]
- 17.Ebel GD, Carricaburu J, Young D, Bernard KA, Kramer LD. Genetic and phenotypic variation of West Nile virus in New York, 2000–2003. The American journal of tropical medicine and hygiene. 2004;71:493–500. [PubMed] [Google Scholar]
- 18.Davis CT, Ebel GD, Lanciotti RS, Brault AC, Guzman H, Siirin M, Lambert A, Parsons RE, Beasley DW, Novak RJ, Elizondo-Quiroga D, Green EN, Young DS, Stark LM, Drebot MA, Artsob H, Tesh RB, Kramer LD, Barrett AD. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: evidence for the emergence of a dominant genotype. Virology. 2005;342:252–265. doi: 10.1016/j.virol.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 19.McMullen AR, May FJ, Li L, Guzman H, Bueno R, Jr, Dennett JA, Tesh RB, Barrett AD. Evolution of new genotype of West Nile virus in North America. Emerging infectious diseases. 2011;17:785–793. doi: 10.3201/eid1705.101707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilpatrick AM, Meola MA, Moudy RM, Kramer LD. Temperature, viral genetics, and the transmission of West Nile virus by Culex pipiens mosquitoes. PLoS Pathog. 2008;4:e1000092. doi: 10.1371/journal.ppat.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moudy RM, Meola MA, Morin LL, Ebel GD, Kramer LD. A newly emergent genotype of West Nile virus is transmitted earlier and more efficiently by Culex mosquitoes. The American journal of tropical medicine and hygiene. 2007;77:365–370. [PubMed] [Google Scholar]
- 22.Duggal NK, Bosco-Lauth A, Bowen RA, Wheeler SS, Reisen WK, Felix TA, Mann BR, Romo H, Swetnam DM, Barrett AD, Brault AC. Evidence for co-evolution of West Nile Virus and house sparrows in North America. PLoS neglected tropical diseases. 2014;8:e3262. doi: 10.1371/journal.pntd.0003262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reisen WK, Padgett K, Fang Y, Woods L, Foss L, Anderson J, Kramer V. Chronic infections of West Nile virus detected in California dead birds. Vector borne and zoonotic diseases. 2013;13:401–405. doi: 10.1089/vbz.2012.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malboeuf CM, Yang X, Charlebois P, Qu J, Berlin AM, Casali M, Pesko KN, Boutwell CL, DeVincenzo JP, Ebel GD, Allen TM, Zody MC, Henn MR, Levin JZ. Complete viral RNA genome sequencing of ultra-low copy samples by sequence-independent amplification. Nucleic acids research. 2013;41:e13. doi: 10.1093/nar/gks794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Charlebois P, Gnerre S, Coole MG, Lennon NJ, Levin JZ, Qu J, Ryan EM, Zody MC, Henn MR. De novo assembly of highly diverse viral populations. BMC genomics. 2012;13:475. doi: 10.1186/1471-2164-13-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 28.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular biology and evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Molecular biology and evolution. 2008;25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bielejec F, Rambaut A, Suchard MA, Lemey P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics. 2011;27:2910–2912. doi: 10.1093/bioinformatics/btr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller MA, Pfeiffer W, Schwartz T. Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA. 2010. pp. 1–8. [Google Scholar]
- 34.Centers for Disease C and Prevention. Outbreak of West Nile-like viral encephalitis--New York, 1999. MMWR Morbidity and mortality weekly report. 1999;48:845–849. [PubMed] [Google Scholar]
- 35.Reisen WK, Fang Y, Lothrop HD, Martinez VM, Wilson J, Oconnor P, Carney R, Cahoon-Young B, Shafii M, Brault AC. Overwintering of West Nile virus in Southern California. Journal of medical entomology. 2006;43:344–355. doi: 10.1603/0022-2585(2006)043[0344:oownvi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 36.Nelms BM, Macedo PA, Kothera L, Savage HM, Reisen WK. Overwintering biology of Culex (Diptera: Culicidae) mosquitoes in the Sacramento Valley of California. Journal of medical entomology. 2013;50:773–790. doi: 10.1603/me12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrade CC, Maharaj PD, Reisen WK, Brault AC. North American West Nile virus genotype isolates demonstrate differential replicative capacities in response to temperature. The Journal of general virology. 2011;92:2523–2533. doi: 10.1099/vir.0.032318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinney RM, Huang CY, Whiteman MC, Bowen RA, Langevin SA, Miller BR, Brault AC. Avian virulence and thermostable replication of the North American strain of West Nile virus. The Journal of general virology. 2006;87:3611–3622. doi: 10.1099/vir.0.82299-0. [DOI] [PubMed] [Google Scholar]