ABSTRACT
Type III secretion systems (T3SSs) are multiprotein machines employed by many Gram-negative bacteria to inject bacterial effector proteins into eukaryotic host cells to promote bacterial survival and colonization. The core unit of T3SSs is the needle complex, a supramolecular structure that mediates the passage of the secreted proteins through the bacterial envelope. A distinct feature of the T3SS is that protein export occurs in a strictly hierarchical manner in which proteins destined to form the needle complex filament and associated structures are secreted first, followed by the secretion of effectors and the proteins that will facilitate their translocation through the target host cell membrane. The secretion hierarchy is established by complex mechanisms that involve several T3SS-associated components, including the “switch protein,” a highly conserved, inner membrane protease that undergoes autocatalytic cleavage. It has been proposed that the autocleavage of the switch protein is the trigger for substrate switching. We show here that autocleavage of the Salmonella enterica serovar Typhimurium switch protein SpaS is an unregulated process that occurs after its folding and before its incorporation into the needle complex. Needle complexes assembled with a precleaved form of SpaS function in a manner indistinguishable from that of the wild-type form. Furthermore, an engineered mutant of SpaS that is processed by an external protease also displays wild-type function. These results demonstrate that the cleavage event per se does not provide a signal for substrate switching but support the hypothesis that cleavage allows the proper conformation of SpaS to render it competent for its switching function.
IMPORTANCE
Bacterial interaction with eukaryotic hosts often involves complex molecular machines for targeted delivery of bacterial effector proteins. One such machine, the type III secretion system of some Gram-negative bacteria, serves to inject a multitude of structurally diverse bacterial proteins into the host cell. Critical to the function of these systems is their ability to secrete proteins in a strict hierarchical order, but it is unclear how the mechanism of switching works. Central to the switching mechanism is a highly conserved inner membrane protease that undergoes autocatalytic cleavage. Although it has been suggested previously that the autocleavage event is the trigger for substrate switching, we show here that this is not the case. Rather, our results show that cleavage allows the proper conformation of the protein to render it competent for its switching function. These findings may help develop inhibitors of type III secretion machines that offer novel therapeutic avenues to treat various infectious diseases.
INTRODUCTION
Many bacteria pathogenic or symbiotic for mammals, insects, or plants utilize specialized protein-delivering nanomachines known as type III secretion systems (T3SSs) (1). These systems evolved to “inject” bacterial effector proteins into host cells to modulate cellular functions and facilitate bacterial colonization and survival (2). The core component of T3SSs is the needle complex (Fig. 1A), a multiprotein nanostructure composed of a membrane-anchored multiring base, an inner membrane-embedded export apparatus, a filamentous needle-like structure that protrudes from the bacterial surface, the inner rod, which connects the needle filament to the base substructure, and a cytoplasmic apparatus involved in targeting and preparation of substrates prior to their secretion (3). The entire needle complex is traversed by a channel, which allows passage of the secreted proteins through the bacterial envelope (4). The needle complex is evolutionarily related to flagella and exhibits an architecture that resembles that of the hook/basal body complex of the flagellar structure (5). Consistent with this evolutionary relationship, several components of the T3SS exhibit amino acid sequence similarity to flagellar proteins (6) (Fig. 1C). During needle complex assembly and subsequently during the delivery of effector proteins, type III secretion occurs in a highly hierarchical fashion (7, 8). Assembly of this complex machine is a multistep and sequential process, which starts by the assembly of the base and associated cytoplasmic substructures (9–12). This substructure can function as a type III secretion machine but with very limited specificity restricted to the proteins necessary to form the inner rod and needle (early substrates) (Fig. 1A). Once a fully functional needle complex is assembled, the secretion machine switches substrate specificity and becomes competent for the secretion of the protein translocases (middle substrates) and effectors (late substrates) (13–15) (Fig. 1A). Needle complexes assembled from mutants unable to switch substrate specificity exhibit much longer needles, as these secretion machines are locked in a secretion mode that is compatible only with the secretion of early substrates (13, 14, 16, 17). The process by which the T3SS switches specificity, however, is poorly understood, and it involves several components. One of these proteins is a highly conserved membrane-associated protease known as the “switch protein,” which undergoes autocatalytic cleavage (15, 18–25) (Fig. 1A and B). It has been proposed that autocleavage of the switch protein determines the timing of substrate switching from early to middle and late substrates (26, 27). However, we show here that a Salmonella enterica serovar Typhimurium strain expressing a catalytic, autocleavage-deficient mutant of SpaS, the switch protein of the Salmonella pathogenicity island 1 (SPI-1)-encoded T3SS, assembles needle complexes with wild-type needle length. Furthermore, we show that needle complexes assembled with preprocessed SpaS function in a manner indistinguishable from that of the wild-type SpaS. These results indicate that the cleavage of the switch protein cannot be the regulatory signal for substrate switching. Rather, we show that SpaS autocleavage is an unregulated process that occurs after its folding and before its incorporation into the needle complex. Our results support the hypothesis that cleavage is necessary for SpaS to achieve a conformation compatible with its secretion function.
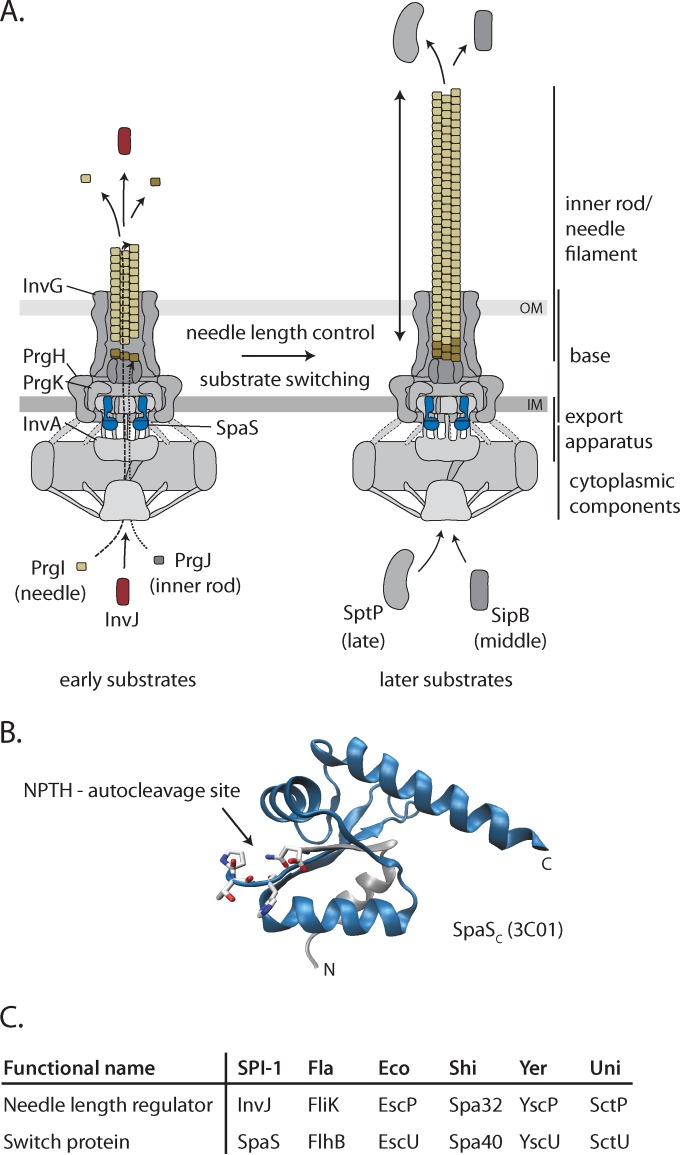
FIG 1 .
The needle complex of T3SSs and components involved in needle length control and substrate specificity switching. (A) Model of the needle complex of T3SSs highlighting components involved in needle length control and substrate specificity switching (3). Needle and inner rod assembly is facilitated by early secretion of the substrates PrgI, PrgJ, and InvJ. In response to the needle reaching a sufficient length, the system switches substrate specificity to the secretion of middle substrates (e.g., SipB) and late substrates (e.g., SptP). Evidence suggests that the InvJ proteins (shown in red) and SpaS proteins (shown in blue) are involved in these processes. The InvG, PrgH, PrgK, and InvA proteins are indicated because they are used throughout this study in mutants and as expression and loading controls. (B) Structure of the C terminus of SpaS indicating the conserved N|PTH autocleavage motif (PDB accession no. 3C01). The C-terminal fragment resulting from autocleavage (SpaScc) is shown in blue. (C) List of homologs of the needle length regulator InvJ and the switch protein SpaS. Abbreviations: OM, outer membrane; IM, inner membrane; SPI-1, Salmonella pathogenicity island 1; Fla, flagella; Eco, Escherichia coli; Shi, Shigella; Yer, Yersinia; Uni, unified nomenclature.
RESULTS
An S. Typhimurium strain expressing a catalytic mutant of the SpaS switch protein exhibits altered type III secretion hierarchy but normal needle length.
To clarify the potential role of the autocatalytic processing of SpaS in determining the timing of substrate switching, we examined the phenotype of S. Typhimurium strains expressing C-terminally FLAG-tagged SpaS mutants containing alanine substitutions at the conserved catalytic site. We found that the S. Typhimurium strain expressing SpaS with a N258A substitution (SpaSN258A) was unable to secrete middle substrates (translocases [e.g., SipB]) and late substrates (effectors [e.g., SptP]), although it was competent for the secretion of early substrates (Fig. 2A). This SpaS mutant exhibited no autocatalytic processing. In contrast, S. Typhimurium strains expressing SpaST260A or SpaSH261A were unaffected in type III secretion, and these mutants exhibit normal autocleavage (Fig. 2A). The S. Typhimurium strain expressing SpaSP259A showed an intermediate phenotype with partial autocleavage and greatly diminished secretion of intermediate and late substrates (Fig. 2A). These results confirmed that, as in other homologous systems, SpaS autocleavage is required for the efficient secretion of middle and late substrates. Accordingly, the capability of host cell invasion of the autocleavage-deficient S. Typhimurium SpaSN258A mutant was drastically reduced (see Fig. S1 in the supplemental material). The absence of secretion of middle and late substrates in the presence of secretion of early substrates suggests that the autocatalytic mutants of SpaS may fail to undergo substrate switching. If this were the case, the SpaS autocatalytic mutant should behave similarly to a mutant lacking the regulatory protein InvJ, which is locked in early substrate secretion mode and therefore exhibits needle complexes with much longer needles (13). However, we found that in stark contrast to the phenotype of the ΔinvJ mutant, the length of the needles of complexes obtained from an S. Typhimurium strain expressing the autocatalytic SpaSN258A mutant was indistinguishable from the length of the needles of complexes obtained from an S. Typhimurium strain expressing the wild-type SpaS. Needle complexes obtained from the S. Typhimurium spaSN258A (spaS gene with N-to-A change at position 258) mutant exhibited needles with an average length of 36 nm, in close agreement with the needle length observed in complexes isolated from S. Typhimurium expressing the wild-type SpaS, which exhibited an average length of 37 nm (Fig. 2B and C). Furthermore, and also in contrast to the phenotype of the ΔinvJ mutant (28), the S. Typhimurium spaSN258A mutant exhibited needle complexes with wild-type levels of the inner rod protein PrgJ (Fig. 2D). We also observed that the loss of needle length control in a ΔinvJ mutant occurred independent of SpaS autocleavage (Fig. S2). These results indicate that, contrary to what has been proposed previously for the homologous flagellar system (29), SpaS autocleavage is not involved in needle length determination.
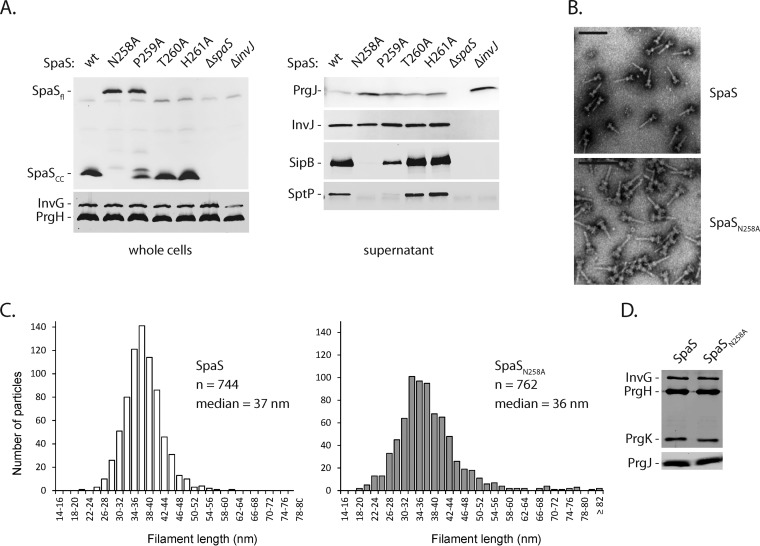
FIG 2 .
Finishing of needle elongation and substrate specificity switching are subsequent and separate events. (A) SpaS expression levels and autocleavage in the wild-type (wt) strain and indicated mutant strains with alanine substitutions in the conserved NPTH autocleavage motif were assayed by SDS-PAGE of whole-cell lysates, subsequent Western blotting, and immunodetection of the C-terminal FLAG tag of SpaS. Likewise, type III secretion into culture supernatants was profiled by immunodetection of the indicated substrate proteins. SpaSfl, full-length SpaS. (B) Purified needle complexes of Salmonella containing wt SpaS or its autocleavage-deficient allele N258A were negatively stained on copper grids and visualized by electron microscopy. Bar, 100 nm. (C) Needle lengths of about 750 isolated needle complexes were measured for each strain, and the length distribution was illustrated in a histogram. (D) The protein content of isolated needle complexes was analyzed by SDS-PAGE, Western blotting, and immunodetection of the needle complex base proteins InvG, PrgH, and PrgK and of the inner rod protein PrgJ.
SpaS autocleavage occurs before needle complex assembly.
It has been suggested previously that autocleavage of SpaS may dictate the timing of switching of the T3SS from early to middle and late substrates (26, 27). We reasoned that if this were the case, SpaS autocleavage must be regulated to ensure that it occurs prior or concomitant to substrate switching. Therefore, to investigate whether autocleavage is regulated by the state of needle complex assembly or by the state of secretion, we compared SpaS autocleavage after the arabinose-induced expression of the SPI-1 T3SS in wild-type, ΔprgHIJK (assembly-deficient), ΔinvA (secretion-deficient), ΔprgIJ (needle assembly-deficient), or ΔinvJ (switching-deficient) S. Typhimurium strains. We observed that SpaS autocleavage occurred rapidly and was complete within 15 min of induction (Fig. 3A). We estimated the half-life of full-length SpaS to be <5 min. More importantly, the cleavage and cleavage kinetics of SpaS were indistinguishable in all the strains tested, indicating that neither secretion nor the state of assembly of the needle complex influences the kinetics of its autoproteolytic processing. Blue native PAGE (BN-PAGE) analysis showed that SpaS was observed within needle complexes as early as 15 min after induction of its expression (Fig. 3B). These results suggest that SpaS autocleavage may be an unregulated process that occurs just after folding of its C-terminal domain and before incorporation into the needle complex. Taken together, these observations indicate that SpaS autocleavage is unlikely to be the triggering event that determines the timing of substrate switching.
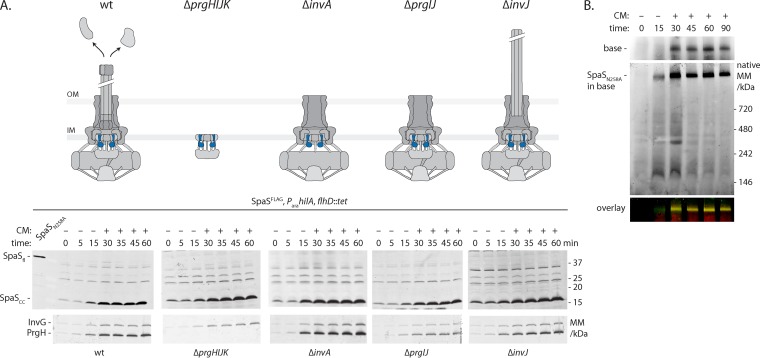
FIG 3 .
SpaS autocleavage is not regulated and occurs before needle complex assembly. All experiments shown were done with S. Typhimurium strains containing C-terminally FLAG-tagged SpaS (SpaSFLAG) and an arabinose-inducible SPI-1 master regulator HilA on the chromosome. (A) To assay the kinetics of SpaS autocleavage in wt and indicated mutant strains, bacteria were grown for 3 h, after which SPI-1 was induced by the addition of arabinose. Thirty minutes after induction, further protein synthesis was inhibited by the addition (+) of chloramphenicol (CM). Samples were taken at the indicated time points, and SpaS autocleavage was analyzed by SDS-PAGE, Western blotting, and immunodetection of its C-terminal FLAG tag. As a reference for the position of full-length SpaS, a sample of the autocleavage mutant was run in the leftmost lane. Induction of the needle complex base components InvG and PrgH is shown as a reference. The state of needle complex assembly and secretion in a given mutant is illustrated. The positions of molecular mass markers (in kilodaltons) are indicated to the right of the gel. SpaS is highlighted in blue. Abbreviations: OM, outer membrane; IM, inner membrane; SpaSfl, full-length SpaS. (B) To assay the kinetics of needle complex assembly, crude membranes of bacteria harvested at the indicated time points after SPI-1 induction were solubilized by DMNG, after which protein complexes were separated by blue native PAGE. Complex association of needle complex base components and SpaS was analyzed by Western blotting and immunodetection using an Li-Cor Odyssey system. Base components were detected in the 700-nm channel, while SpaSFLAG was detected in the 800-nm channel. The overlay of both channels is shown.
Type III secretion machines assembled with precleaved SpaS are fully functional.
If cleavage of SpaS were not involved in the regulation of type III secretion, it follows that needle complexes assembled with precleaved forms of the switch protein should be fully functional and display no hierarchical secretion defects. To test this hypothesis, we designed an experiment in which SpaS was expressed and cleaved before expression of all other T3SS components (Fig. 4A). SpaS expression was then halted, and the expression of the rest of the SPI-1 T3SS was subsequently induced. Under these experimental conditions, no unprocessed SpaS was detected prior to the induction of expression of the rest of the T3SS components (Fig. 4B). Type III secretion and needle complex assembly under these conditions were compared in samples obtained from the same strains subjected to conditions that led to the simultaneous expression of SpaS and the rest of the T3SS components. We found no difference in needle complex assembly or type III secretion in samples obtained from T3SS machines assembled under both experimental conditions, and the needle complexes exhibit similar needle length and architecture (Fig. 4C and D). Furthermore, no differences were found in the secretion of middle and late substrates (Fig. 4B). These results indicate that the SpaS autocleavage event does not play a role in needle complex assembly, needle length regulation, or the establishment of a hierarchy in the secretion process.
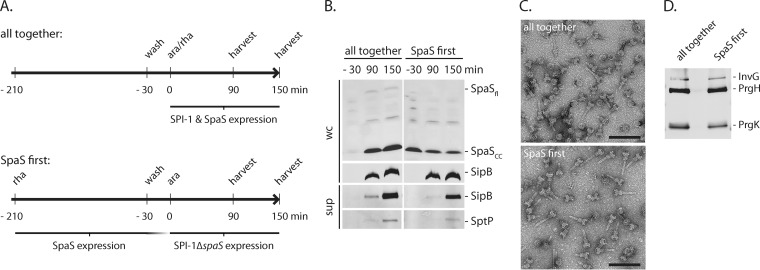
FIG 4 .
Fully functional secretion systems can be obtained from previously provided cleaved SpaS. (A) Illustration of the experimental set up. The experiments were done with Salmonella ΔspaS mutant containing an arabinose-inducible SPI-1 master regulator HilA on the chromosome. C-terminally FLAG-tagged SpaS was complemented from a rhamnose-inducible low-copy-number plasmid. For the assembly control, bacteria were grown for 3 h in the absence of inducing sugars and washed, and expression of SPI-1 and SpaSFLAG was simultaneously induced by the addition of arabinose and rhamnose (ara/rha). To express SpaSFLAG prior to the other needle complex components, bacteria were grown for 3 h in the presence of rhamnose. Then, SpaS expression was switched off by rhamnose removal, and SPI-1 expression (without SpaS) was induced by the addition of arabinose after 30 min, 90 min, and 150 min after SPI-1 induction. SpaS expression and secretion was profiled, and needle complexes were isolated. (B) SpaS expression levels and autocleavage was assayed by SDS-PAGE of whole-cell lysates, subsequent Western blotting, and immunodetection of the FLAG epitope. Likewise, type III secretion into culture supernatants was profiled by immunodetection of the indicated substrate proteins. Abbreviations: wc, whole cells; sup, culture supernatant. (C) Purified needle complexes were negatively stained on copper grids and visualized by electron microscopy. Bars, 100 nm. (D) The protein content of isolated needle complexes was analyzed by SDS-PAGE, Western blotting, and immunodetection of the needle complex base proteins InvG, PrgH, and PrgK.
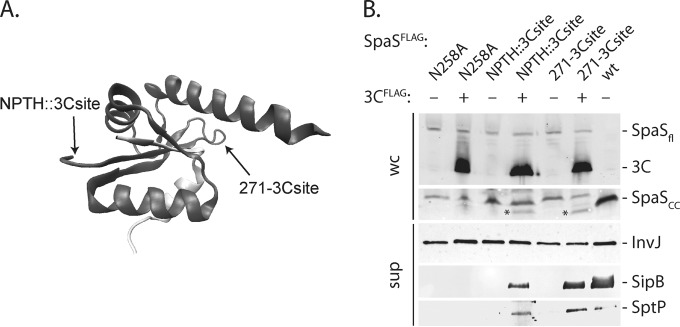
Exogenous cleavage of SpaS results in a fully functional protein.
The observation that SpaS cleavage occurs rapidly and can occur prior to its insertion into the secretion apparatus suggests that autoprocessing may not be a regulatory event but rather a requirement to achieve its proper functional conformation. We reasoned that if this were the case, it should be possible to obtain a functional SpaS protein even if its processing is mediated by an external protease. We replaced the NPTH processing site of SpaS with a cleavage site for the human rhinovirus 3C protease. We also introduced a human rhinovirus 3C protease cleavage site at a different position in the SpaS C terminus (E271) in the context of an autocleavage-deficient mutant (Fig. 5A). The resulting mutants exhibited the same InvJ secretion phenotype as that shown by a strain expressing a catalytic mutant of SpaS, indicating that introduction of the novel processing site did not result in gross conformational changes. We examined the effect of the expression of the human rhinovirus 3C protease in trans in the context of S. Typhimurium strains expressing the engineered SpaS mutants. We found that expression of the viral protease resulted in the specific cleavage of SpaS at the predicted sites. Even though the efficiency of exogenous SpaS cleavage was low, possibly for SpaS being inaccessible due to quick assembly into the needle complex, the rhinovirus 3C protease-mediated cleavage yielded functional SpaS competent for mediating the secretion of the middle substrate SipB and the late substrate SptP in both cases (Fig. 5B). These results indicate that external SpaS cleavage, even if not at the site of the conserved NPTH motif, results in a fully functional protein and further support the hypothesis that the cleavage of SpaS per se does not play a regulatory role. Rather, these results support the hypothesis that cleavage allows the proper conformational flexibility of SpaS to render it competent for substrate specificity switching and secretion of middle and late substrates.
FIG 5 .
NPTH-dependent autocleavage is not necessary to achieve substrate specificity switching. (A) Illustration of the positions of the introduction of human rhinovirus 3C protease cleavage sites on the crystal structure of the C-terminal domain of SpaS (SpaSc) (PDB accession no. 3C01). (B) The expression and cleavage of SpaSFLAG and 3C protease in the indicated strains were analyzed in whole-cell lysates by SDS-PAGE, Western blotting, and immunodetection of the FLAG epitope. Functionality of substrate specificity switching was profiled by SDS-PAGE of TCA-precipitated culture supernatants, Western blotting, and immunodetection of InvJ, SipB, and SptP. Asterisks indicate the cleaved C-terminal fragments of the engineered SpaS variants. Abbreviations: 3C, human rhinovirus 3C protease; wc, whole cells; sup, culture supernatant.
DISCUSSION
During needle complex assembly and later, during secretion of its substrates, the type III secretion machine functions in a hierarchical fashion. After completion of the assembly of the base substructure, the T3SS becomes competent for the engagement of the early substrate proteins that form the inner rod and needle substructures as well as the regulatory protein InvJ (13, 14, 17). Subsequent to completion of the assembly of the needle complex, the T3SS changes substrate specificity, losing the ability to secrete early substrates and becoming competent for the secretion of the effector proteins and the proteins necessary for their translocation through the target host cell membrane (translocases) (15). The mechanism of substrate reprogramming is poorly understood and the subject of some controversy. It is clear that the regulatory protein InvJ (and its homologs in other bacteria) plays an important role in this process, since mutants lacking this regulator are “locked” in the “early secretion mode,” therefore exhibiting abnormally long needles (13, 14, 16, 17). Mutant strains lacking InvJ are also unable to assemble the inner rod protein, which suggests that the role of this regulatory protein in substrate switching may be indirect, through its involvement in needle complex assembly (28, 30). It has been suggested previously that the process of substrate switching also involves the export apparatus component SpaS (or its homologs in other bacteria [Fig. 1C]), a highly conserved membrane protease, which undergoes autocatalytic cleavage (15, 18–25). It has been proposed that SpaS works in concert with InvJ to mediate substrate switching and that its autocleavage (potentially triggered by its interaction with InvJ) is the signal that determines substrate switching from early to middle and late substrates (15, 25–27, 31). In this study, we present evidence that is incompatible with this model.
We have confirmed that the autocatalytic mutant of SpaS exhibits altered secretion. While competent for the secretion of early substrates, this mutant is unable to secrete middle and late substrates, a phenotype that originally led to the proposed implication of SpaS in substrate switching (18–20, 32–34). However, we found that an S. Typhimurium strain expressing the autocatalytic mutant SpaSN258A assembled wild-type needle complexes with normal needle lengths, a phenotype drastically different from that of a strain lacking the regulatory protein InvJ. We also found that the kinetics of SpaS processing were very fast and unaffected by the state of assembly of the needle complex or by its secretion status. More importantly, we found that exogenous processing of SpaS did not alter its function and that needle complexes assembled with a precleaved form of SpaS exhibited normal needle length and a normal secretion profile. These findings indicate that the cleavage event per se has no role in substrate switching. Rather, we propose that cleavage may be necessary for SpaS to achieve a structural configuration compatible with its role in secretion.
The autocatalytic mutant of SpaS exhibits normal needle length, although it is unable to secrete middle and late substrates. This observation indicates that in the case of this mutant, the secretion machine can become incompetent for the secretion of early substrates without automatically gaining the ability to secrete middle and late substrates. This phenotype is in sharp contrast with that of a mutant lacking the regulatory protein InvJ, which is “locked” in an early substrate secretion mode and exhibits abnormally long needles (13). These observations suggest that the substrate switching process may involve two steps: losing competency for secretion of early substrates (i.e., needle and inner rod proteins) and gaining the ability to secrete middle and late substrates (effectors and protein translocases). While it is clear that the latter cannot occur with an unprocessed form of SpaS, our data indicate that the cleavage event is not involved in switching. Rather, our results favor the notion that the cleavage of SpaS may confer this protein the conformational flexibility that is compatible with the secretion of all substrates. More experiments will be required to demonstrate that a two-step process is involved under physiological conditions.
It had been shown in the flagellar system that loss of autocleavage of the SpaS homolog FlhB results in an impairment of hook length control (29, 35). Based on these results, it was hypothesized that FlhB autocleavage per se plays a role in hook length control. Also in Yersinia, an impaired needle length control was observed in an autocleavage mutant of the switch protein YscU (33). However, this defect could be compensated for by overexpression of the needle length regulator YscP, indicating that a general secretion defect of the autocleavage mutant might be responsible for the observed phenotype. FliK secretion had not been tested in the FlhB autocleavage mutant in the study of the flagellar system so that reduced FliK secretion remains a possible explanation for impaired hook length control in the FlhB autocleavage mutant. In our study, secretion of the needle length regulator InvJ was indistinguishable in the wild type and SpaSN258A autocleavage mutant (Fig. 2), which strengthens the ground for our hypothesis that autocleavage plays no role in needle length control.
Based on the structures of uncleaved and cleaved versions of the C-terminal domain of SpaS and its homologs, it had been suggested that changes in surface features after autocleavage are the critical facilitators of substrate specificity switching (21). Our results with SpaS cleaved by an exogenous protease suggest that the specific surface of SpaS created by the autocleaved N|PTH motif is not critical for substrate specificity switching, as it can be replaced by the dissimilar human rhinovirus 3C protease LEVLFQ|GP cleavage motif. More importantly, since SpaS capable of substrate specificity switching also results from cleavage on the site of the C-terminal domain opposite the N|PTH motif, it is likely that conformational flexibility allowed by the separated polypeptide chains is the critical facilitator of substrate specificity switching. This notion is supported by the observation that a decreased thermal stability—indicating an increased conformational flexibility—correlates with a higher probability of YscP-independent switching in mutants of the Yersinia SpaS homolog YscU (36).
In summary, we have presented evidence indicating that the cleavage of SpaS is not involved in determining the timing for substrate switching. We thus propose that cleavage per se is necessary for the structural flexibility that allows the switch protein to acquire a conformation compatible with its function in substrate specificity switching and secretion of middle and late substrates.
MATERIALS AND METHODS
Materials.
Chemicals were from Sigma-Aldrich unless otherwise specified. The detergents n-dodecyl-N,N-dimethylamine-N-oxide (LDAO) and decyl-maltose-neopentyl-glycol (DMNG) were from Affymetrix-Anatrace. Primers are listed in Table S1 in the supplemental material and were synthetized by Eurofins and Integrated DNA Technologies.
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table S2 in the supplemental material. All strains were derived from Salmonella enterica serovar Typhimurium strain SL1344 (37). Strains expressing chromosomally FLAG-tagged SpaS and mutants were constructed by allelic exchange as previously described (38). FLAG-tagged SpaS functions indistinguishably from the wild-type protein with respect to the secretion of early, intermediate, and late substrates (see Fig. S3 in the supplemental material) and the invasion of host cells (Fig. S1). Salmonella Typhimurium strains were grown at 37°C in LB broth supplemented with 0.3 M NaCl with low aeration to enhance expression of genes of Salmonella pathogenicity island 1 (SPI-1). Cultures were supplemented as required with streptomycin (50 µg/ml), tetracycline (12.5 µg/ml), ampicillin (100 µg/ml), kanamycin (25 µg/ml), or chloramphenicol (50 µg/ml).
Secretion assay.
S. Typhimurium strains were grown at 37°C in LB broth supplemented with 0.3 M NaCl for 5 h (or as indicated). Whole cells and culture supernatants were separated by centrifugation. Whole cells were resuspended in an appropriate volume of SDS-PAGE loading buffer. Supernatants were passed through a 0.2-µm filter, supplemented with 0.1% Na-deoxycholic acid, and precipitated with 10% trichloroacetic acid (TCA) for a period of time ranging from 30 min to overnight at 4°C. Precipitated culture supernatant samples were washed with acetone after centrifugation and resuspended in SDS-PAGE loading buffer. Whole-cell and culture supernatant samples were subjected to SDS-PAGE, followed by immunoblotting.
Immunoblotting.
For protein detection, samples were subjected to SDS-PAGE using SERVAGel TG PRiME 8 to 16% precast gels, transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad), and probed with primary antibodies anti-SipB, anti-InvJ, anti-PrgJ, anti-SptP, anti-needle complex base (anti-NC base), and M2 anti-FLAG. Secondary antibodies were goat anti-mouse IgG DyLight 800 conjugate and goat anti-rabbit IgG DyLight 680 conjugate (Thermo Scientific Pierce). Detection was performed using the Odyssey imaging system (Li-Cor).
Cell fractionation or crude membrane preparation.
Ten optical density (OD) units of bacterial lysates were resuspended in 750 µl buffer K (50 mM triethanolamine [pH 7.5], 250 mM sucrose, 1 mM EDTA, 1 mM MgCl2, 10 µg/ml DNase, 2 mg/ml lysozyme, 1:100 protease inhibitor cocktail) and incubated for 30 min on ice. Samples were bead milled, and beads, unbroken cells, and debris were removed by centrifugation for 10 min at 10,000 × g and 4°C. Crude membranes contained in the supernatant were precipitated by centrifugation for 45 min at 55,000 rpm and 4°C in a Beckman TLA 55 rotor. Pellets containing crude membranes were frozen until use.
BN-PAGE.
Blue native PAGE (BN-PAGE) was carried out as previously described (9) with some modifications. Crude membranes were resuspended in 1× phosphate-buffered saline (PBS), and protein concentration was determined using the bicinchoninic acid (BCA) protein assay kit (Thermo Scientific Pierce). The membranes (10 μg) were solubilized with a final concentration of 1% DMNG for 1 h at 4°C. BN loading dye (5% [wt/vol] Serva Blue G [Serva Electrophoresis GmbH], 250 mM aminocaproic acid, 50% glycerol) (0.5%) was added to the samples, mixed, and run on a NativePAGE Novex Bis-Tris gel system (Thermo Life Technologies), followed by immunoblotting. NC bases were visualized using anti-NC base antibodies and secondary anti-rabbit IgG DyLight 680 conjugate secondary antibodies. FLAG-tagged SpaS was visualized using M2 anti-FLAG antibodies and secondary anti-mouse IgG DyLight 800 conjugate antibodies. Detection was performed using the Odyssey imaging system (Li-Cor).
SpaS cleavage before T3SS induction.
The coordinated expression of T3SS components was carried out as previously described (9) by using S. Typhimurium strains in which expression of examined components was uncoupled by the employment of two different compatible promoter systems. Chromosomal expression of the master regulator of the SPI-1 T3SS (HilA) was under control of an arabinose-inducible promoter. SpaS was deleted from the chromosome and complemented in trans from a low-copy-number plasmid under control of a rhamnose-inducible promoter.
To achieve complete autocleavage of SpaS before expression of all other T3SS components, SpaS was expressed by the addition of rhamnose for 3 h. Then, the inducer was washed out. After a 30-min period to allow depletion of residual SpaS expression, arabinose was added for induction of all other T3SS components. In control samples, expression of SpaS and all other T3SS components was induced by the simultaneous addition of rhamnose and arabinose, respectively, after 3 h of bacterial growth. Samples were taken at different time points after induction of the T3SS, and secretion assays were performed.
SpaS exogenous cleavage.
The human rhinovirus 3C protease cleavage site was introduced into the chromosome at two different positions of SpaS: first, replacing the NPTH motif and second, after residue E271 in a SpaSN258A background. A rhamnose-inducible plasmid expressing 3C protease with an N-terminal InvJ signal sequence (residues 1 to 15) and a C-terminal FLAG tag were constructed and introduced in the strains containing SpaS with the engineered cleavage site. Bacteria were grown in the presence of 500 µM rhamnose for expression of the 3C protease and 0.05% arabinose for expression of the SPI-1 T3SS, including FLAG-tagged SpaS. Secretion assays were performed in the presence (expression) or absence of the 3C protease, and whole-cell and culture supernatant samples were subjected to SDS-PAGE, followed by immunoblotting.
Purification of Salmonella needle complexes.
Purification of needle complexes was carried out with slight modification of a previously published protocol (39). The cultures were grown at 37°C in LB supplemented with 0.3 M NaCl and appropriate antibiotics. The bacteria were harvested and resuspended in 150 mM Tris (pH 7.4), 0.5 M sucrose, 1 mg/ml hen egg lysozyme, and 7 mM EDTA and incubated on ice while stirring for 45 min followed by 20 min at 37°C. The cells were lysed with 0.5% LDAO, and then 450 mM NaCl and 12 mM MgCl2 were added to the lysate. Cell debris was removed by low-speed centrifugation at 15,000 × g for 30 min, and NCs were pelleted by high-speed centrifugation at 264,900 × g for 2.5 h. The pellet was resuspended in a buffer composed of 0.5% LDAO, 1× PBS (pH 7.4), 375 mM NaCl, and 5 mM EDTA and adjusted to a final concentration of 27.5% (wt/vol) of CsCl. Samples were centrifuged for 14 to 16 h at 286,000 × g. Aliquots (0.5 ml) were collected by using a Biocomp gradient station. Diluted fractions were pelleted at 121,139 × g for 2 h. Needle complexes were resuspended in buffer composed of 0.1% LDAO, 1× PBS (pH 7.4), 375 mM NaCl, and 5 mM EDTA. The composition of NC preparations was assessed by SDS-PAGE, followed by immunoblotting.
Electron microscopy and image analysis.
Purified needle complexes were negatively stained with 2% phosphotungstic acid (pH 4.5) or 1% aqueous uranyl acetate on carbon-coated copper grids. Micrographs were recorded either using an FEI Tecnai Biotwin microscope at 80 kV or using a JEM-1400Plus (JEOL) microscope at 120 kV, and needle lengths were measured as published previously (28).
SUPPLEMENTAL MATERIAL
Host cell invasion of the autocleavage-deficient SpaSN258A mutant. Host cell invasion of indicated S. Typhimurium strains was assessed by a gentamicin protection assay as described previously (C. M. Collazo and J. E. Galan, Mol Microbiol 24:747–756, 1997). The results for three independent replicate samples are shown. Error bars indicate standard deviations (Student’s t test). Download
Loss of needle length control in invJ deletion mutant is independent of autocleavage of SpaS. Purified needle complexes of a SpaSN258AFLAG-expressing S. Typhimurium ΔinvJ mutant strain were negatively stained on copper grids and visualized by electron microscopy. The image on the right shows a magnified view of the boxed area on the image on the left. Download
C-terminally FLAG-tagged SpaS functions indistinguishable from wt SpaS. Expression levels and type III secretion into culture supernatants by the S. Typhimurium strains indicated was profiled by SDS-PAGE, Western blotting, and immunodetection of InvJ (early substrate), SipB (intermediate substrate), and SptP (late substrate). Abbreviations: wc, whole cells; sup, culture supernatant. Download
Strains and plasmids used in this study.
Oligonucleotides used in this study.
ACKNOWLEDGMENTS
Funding of this work was provided by the Alexander von Humboldt Foundation in the framework of the Sofja Kovalevskaja Award endowed by the Federal Ministry of Education and Research (to S.W.), and in the framework of the Georg Forster Research Fellowship (to J.V.M.F.). J.V.M.F. was also supported by CONACyT (238898). Work in J. E. Galán’s laboratory was supported by National Institute of Allergy and Infectious Diseases grant AI030492 (to J.E.G.).
Thomas Marlovits is acknowledged for providing antisera against SptP.
Footnotes
Citation Monjarás Feria JV, Lefebre MD, Stierhof Y-D, Galán JE, Wagner S. 2015. Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar Typhimurium. mBio 6(5):e01459-15. doi:10.1128/mBio.01459-15.
REFERENCES
- 1.Büttner D. 2012. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev 76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galán JE. 2009. Common themes in the design and function of bacterial effectors. Cell Host Microbe 5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radics J, Königsmaier L, Marlovits TC. 2014. Structure of a pathogenic type 3 secretion system in action. Nat Struct Mol Biol 21:82–87. doi: 10.1038/nsmb.2722. [DOI] [PubMed] [Google Scholar]
- 5.Kawamoto A, Morimoto YV, Miyata T, Minamino T, Hughes KT, Kato T, Namba K. 2013. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci Rep 3:3369. doi: 10.1038/srep03369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abby SS, Rocha EPC. 2012. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet 8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collazo CM, Galan JE. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect Immun 64:3524–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lara-Tejero M, Kato J, Wagner S, Liu X, Galán JE. 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner S, Königsmaier L, Lara-Tejero M, Lefebre M, Marlovits TC, Galán JE. 2010. Organization and coordinated assembly of the type III secretion export apparatus. Proc Natl Acad Sci U S A 107:17745–17750. doi: 10.1073/pnas.1008053107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diepold A, Amstutz M, Abel S, Sorg I, Jenal U, Cornelis GR. 2010. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J 29:1928–1940. doi: 10.1038/emboj.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diepold A, Wiesand U, Cornelis GR. 2011. The assembly of the export apparatus (YscR,S,T,U,V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol Microbiol 82:502–514. doi: 10.1111/j.1365-2958.2011.07830.x. [DOI] [PubMed] [Google Scholar]
- 12.Diepold A, Wagner S. 2014. Assembly of the bacterial type III secretion machinery. FEMS Microbiol Rev 38:802–822. doi: 10.1111/1574-6976.12061. [DOI] [PubMed] [Google Scholar]
- 13.Kubori T, Sukhan A, Aizawa SI, Galán JE. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci U S A 97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamano K, Katayama E, Toyotome T, Sasakawa C. 2002. Shigella Spa32 is an essential secretory protein for functional type III secretion machinery and uniformity of its needle length. J Bacteriol 184:1244–1252. doi: 10.1128/JB.184.5.1244-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edqvist PJ, Olsson J, Lavander M, Sundberg L, Forsberg A, Wolf-Watz H, Lloyd SA. 2003. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol 185:2259–2266. doi: 10.1128/JB.185.7.2259-2266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Journet L, Agrain C, Broz P, Cornelis GR. 2003. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 17.Monjarás Feria J, García-Gómez E, Espinosa N, Minamino T, Namba K, González-Pedrajo B. 2012. Role of EscP (Orf16) in injectisome biogenesis and regulation of type III protein secretion in enteropathogenic Escherichia coli. J Bacteriol 194:6029–6045. doi: 10.1128/JB.01215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavander M, Sundberg L, Edqvist PJ, Lloyd SA, Wolf-Watz H, Forsberg A. 2002. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol 184:4500–4509. doi: 10.1128/JB.184.16.4500-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, Macnab RM. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol 48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- 20.Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM. 2005. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem 280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- 21.Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, Miller SI, Finlay BB, Strynadka NCJ. 2008. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453:124–127. doi: 10.1038/nature06832. [DOI] [PubMed] [Google Scholar]
- 22.Deane JE, Graham SC, Mitchell EP, Flot D, Johnson S, Lea SM. 2008. Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol Microbiol 69:267–276. doi: 10.1111/j.1365-2958.2008.06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiesand U, Sorg I, Amstutz M, Wagner S, van den Heuvel J, Lührs T, Cornelis GR, Heinz DW. 2009. Structure of the type III secretion recognition protein YscU from Yersinia enterocolitica. J Mol Biol 385:854–866. [DOI] [PubMed] [Google Scholar]
- 24.Lountos GT, Austin BP, Nallamsetty S, Waugh DS. 2009. Atomic resolution structure of the cytoplasmic domain of Yersinia pestis YscU, a regulatory switch involved in type III secretion. Protein Sci 18:467–474. doi: 10.1002/pro.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Björnfot AC, Lavander M, Forsberg A, Wolf-Watz H. 2009. Autoproteolysis of YscU of Yersinia pseudotuberculosis is important for regulation of expression and secretion of Yop proteins. J Bacteriol 191:4259–4267. doi: 10.1128/JB.01730-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferris HU, Minamino T. 2006. Flipping the switch: bringing order to flagellar assembly. Trends Microbiol 14:519–526. doi: 10.1016/j.tim.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Riordan KE, Schneewind O. 2008. YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol Microbiol 68:1485–1501. doi: 10.1111/j.1365-2958.2008.06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM, Galán JE. 2006. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441:637–640. doi: 10.1038/nature04822. [DOI] [PubMed] [Google Scholar]
- 29.Erhardt M, Hirano T, Su Y, Paul K, Wee DH, Mizuno S, Aizawa S-I, Hughes KT. 2010. The role of the FliK molecular ruler in hook-length control in Salmonella enterica. Mol Microbiol 75:1272–1284. doi: 10.1111/j.1365-2958.2010.07050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefebre MD, Galán JE. 2014. The inner rod protein controls substrate switching and needle length in a Salmonella type III secretion system. Proc Natl Acad Sci U S A 111:817–822. doi: 10.1073/pnas.1319698111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams AW, Yamaguchi S, Togashi F, Aizawa SI, Kawagishi I, Macnab RM. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol 178:2960–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minamino T, Macnab RM. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J Bacteriol 182:4906–4914. doi: 10.1128/JB.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorg I, Wagner S, Amstutz M, Müller SA, Broz P, Lussi Y, Engel A, Cornelis GR. 2007. YscU recognizes translocators as export substrates of the Yersinia injectisome. EMBO J 26:3015–3024. doi: 10.1038/sj.emboj.7601731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen D-K, Moriya N, Martinez-Argudo I, Blocker AJ. 2012. Needle length control and the secretion substrate specificity switch are only loosely coupled in the type III secretion apparatus of Shigella. Microbiology (Reading, Engl) 158:1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. 2011. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J 30:2948–2961. doi: 10.1038/emboj.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frost S, Ho O, Login FH, Weise CF, Wolf-Watz H, Wolf-Watz M. 2012. Autoproteolysis and intramolecular dissociation of Yersinia YscU precedes secretion of its C-terminal polypeptide YscUCC. PLoS One 7:e49349. doi: 10.1371/journal.pone.0049349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoiseth SK, Stocker BAD. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 38.Kaniga K, Bossio JC, Galán JE. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol 13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 39.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Host cell invasion of the autocleavage-deficient SpaSN258A mutant. Host cell invasion of indicated S. Typhimurium strains was assessed by a gentamicin protection assay as described previously (C. M. Collazo and J. E. Galan, Mol Microbiol 24:747–756, 1997). The results for three independent replicate samples are shown. Error bars indicate standard deviations (Student’s t test). Download
Loss of needle length control in invJ deletion mutant is independent of autocleavage of SpaS. Purified needle complexes of a SpaSN258AFLAG-expressing S. Typhimurium ΔinvJ mutant strain were negatively stained on copper grids and visualized by electron microscopy. The image on the right shows a magnified view of the boxed area on the image on the left. Download
C-terminally FLAG-tagged SpaS functions indistinguishable from wt SpaS. Expression levels and type III secretion into culture supernatants by the S. Typhimurium strains indicated was profiled by SDS-PAGE, Western blotting, and immunodetection of InvJ (early substrate), SipB (intermediate substrate), and SptP (late substrate). Abbreviations: wc, whole cells; sup, culture supernatant. Download
Strains and plasmids used in this study.
Oligonucleotides used in this study.