Abstract
In mammals, DNA methylation profiles vary substantially between tissues. Recent genome-scale studies report that blood displays a highly distinctive methylomic profile from other somatic tissues. In this study, we sought to understand why blood DNA methylation state is so different to the one found in other tissues. We found that whole blood contains approximately twice as many tissue-specific differentially methylated positions (tDMPs) than any other somatic tissue examined. Furthermore, a large subset of blood tDMPs showed much lower levels of methylation than tDMPs for other tissues. Surprisingly, these regions of low methylation in blood show no difference regarding genomic location, genomic content, evolutionary rates, or histone marks when compared to other tDMPs. Our results reveal why blood displays a distinctive methylation profile relative to other somatic tissues. In the future, it will be important to study how these blood specific tDMPs are mechanistically involved in blood-specific functions.
Keywords: blood, Illumina 450K Array, emthylation, tissue specific
Abbreviations
- RRBS
reduced representation bisulfite sequencing
- Illumina 450K
Illumina Infinium HumanMethylation450 array
- tDMPs
tissue specific differentially methylated position
- tDMRs
tissue specific differentially methylated region
Introduction
DNA methylation, the addition of a methyl group to a cytosine, is one of the most widely studied epigenetic modifications. It plays an important role in transcriptional regulation, genomic imprinting, and X-inactivation,1 and is perturbed in complex diseases such as cancer2 as well as during aging.3 It is well known that DNA methylation profiles are tissue specific4-6 but, recently, we reported7 that the global methylation pattern of blood cells, as measured by the Illumina Infinium HumanMethylation450 array (Illumina 450K), was significantly different to that of a large number of other tissues (using unsupervised hierarchical clustering of 1,052 healthy samples). Varley et al.8 also reported a similar observation using reduced representation bisulfite sequencing (RRBS), in which blood is separated from primary cell lines and tissues, but, not surprisingly, they found that the DNA methylation state in blood is more similar to these samples than cancer cell lines. It is therefore of great interest to understand why the methylation state of blood is so different to that of other tissues.
To investigate this distinct clustering of blood compared to other tissues, we extracted a set of samples for a variety of different tissues from Marmal-aid,9 a large publicly available database of Illumina 450K experiments. We found that blood cells contained twice as many tissue specific differentially methylated positions (tDMPs) and a large number of these regions showed low DNA methylation in blood. In comparison, tDMPs for other tissues were mostly located in regions of fractional methylation, a β value of 0.3–0.7, for the respective tissue. Interestingly, these lowly methylated regions in blood show no difference in genome location, sequence content, evolutionary rates, or histone marks, suggesting that the methylation state alone may play an important functional role in blood.
Results
Blood shows distinct patterns of methylation at tissue differentially methylated positions (tDMPs)
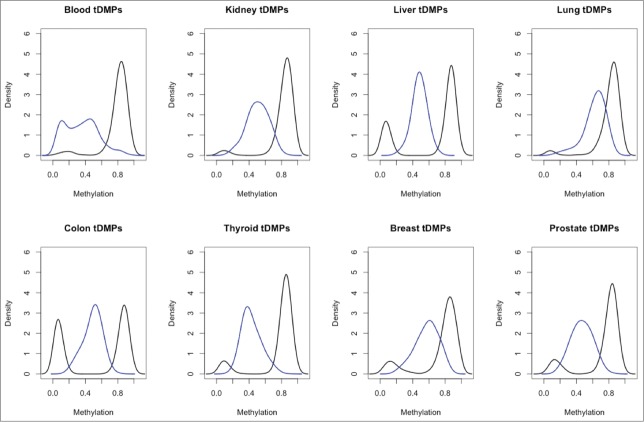
In order to investigate the differences in methylation between blood and all other tissues it was necessary to obtain a robust set of samples for a number of different tissues. At present, while whole genome bisulfite sequencing is the gold standard, there is a very limited number of samples available. In addition, for samples that are available, the data is of very low coverage. In contrast, a large number of studies have been performed using the Illumina 450K array, which has good coverage. Therefore, we extracted Illumina 450K arrays from Marmal-aid,9 a database of publicly available Illumina 450K arrays, for tissues that contained a minimum of 50 healthy samples, including whole blood, breast, colon, kidney, liver, lung, prostate, and thyroid. Due to the public nature of the data, it was necessary to perform an initial quality control step to remove samples that were outliers (Materials and Methods). We then randomly selected 50 samples for each of the tissues, and all further analysis was performed on these samples (Supplementary File 1). We then called tDMPs for each tissue between the tissue of interest and all the other tissues (Materials and Methods). We found that for each set of tDMPs, a large number of probes (25,035–171,070) were significantly differently methylated (P-value < 2 × 10−8) (Table 1). Interestingly, blood tDMPs showed more than twice as many significant differences as those found in any of the other tissues, excluding colon, for which blood showed 1.78 times as many significant differences. A greater than 1.7-fold increase in the number of tDMPs was also maintained, even when filtering for those methylation differences >0.2-fold in absolute β value. Furthermore, there was a >6-fold increase of blood tDMPs that had methylation differences >0.5-fold compared to all other tissues (blood: 2624, thyroid: 413, prostate: 231, kidney: 148, liver: 103, colon: 66, breast: 9, and lung: 0). This is particularly striking when looking at the top 1000 tDMPs as ranked by P-value, in which blood tDMPs show a bimodality of methylation with one peak at β value ≅ 0.1 and the second peak at around 0.4. All the other tissues contain a single peak located at intermediate levels of methylation (β value = 0.3–0.7) (Fig. 1). This low methylation peak in blood explains why it separates out from the other tissues when performing hierarchical clustering. To investigate this peak further we selected the top 100 tDMPs from blood with average methylation of β value <0.2 [unmethylated-tDMP (u-tDMP)], top 100 tDMPs from blood with average methylation value β value >0.3 [fractionally methylated tDMP (f-tDMP)] and the top 100 f-tDMPs from liver, kidney, and lung, also with β value >0.3.
Table 1.
Number of significant differences called between tissue of interest and all the other tissues. Second and third column show the number of those differences after being filtered for β value differences >0.2 and 0.5, respectively
| Tissue | Number of significant differences | Number of significant differences (>0.2) | Number of significant differences (>0.5) |
|---|---|---|---|
| Blood | 171070 | 44940 | 2624 |
| Breast | 59707 | 12330 | 9 |
| Colon | 95869 | 21640 | 66 |
| Kidney | 76734 | 17482 | 148 |
| Liver | 81077 | 21643 | 103 |
| Lung | 25035 | 3210 | 0 |
| Prostate | 77328 | 26348 | 231 |
| Thyroid | 78500 | 24755 | 413 |
Figure 1.
A paneled plot of the different distribution of β values for the 8 tDMP calls. In blue in each panel is the density of β values for the top 1000 tDMPs in the specific tissue and in black is the density of average β values of all the other tissues for these same tDMPs. Blood tDMPs show a bimodal distribution in blue with 2 peaks: one with low methylation and one with intermediate level of methylation. In all the other tissues profiled, there is a single peak with intermediate level of methylation.
Cellular heterogeneity does not explain the differences in tDMPs
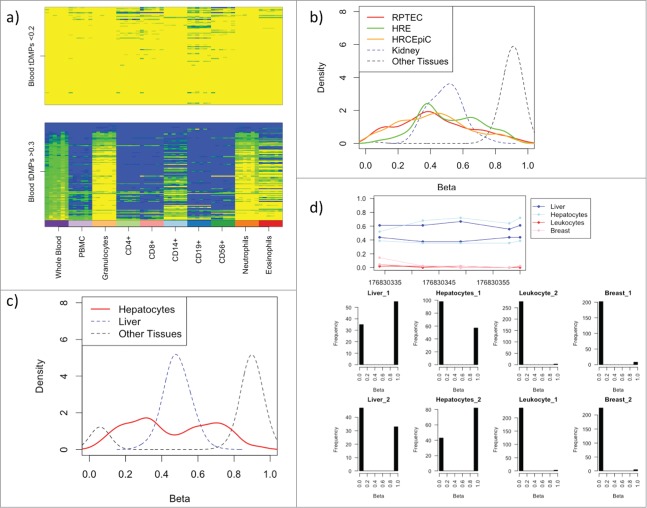
Whole blood consists of a number of distinct cell types including neutrophils (∼60%), CD4+ (∼13%), CD8+ (∼6%), CD14+ (∼5%), eosinophils (∼4%), CD19+ (∼3%), and CD56+ (∼2%). To investigate the effect of this cellular heterogeneity on the tDMPs we looked at the top 100 whole blood u-tDMPs and f-tDMPs in a series of blood subtypes (GSE35069).10 The whole blood u-tDMPs show a consistent unmethylated state in all of the different blood subtypes, while a large proportion of whole blood f-tDMPs are actually unmethylated, specifically in granulocytes (42%), neutrophils (32%), and eosinophils (28%), and, hence, the f-tDMPs found in whole blood are due to the mixture of cell types present (Fig. 2A). This suggests that the f-tDMPs in liver, kidney, and lung may also be due to cell subtypes of the specific tissue. To explore this further we downloaded Illumina 450K data for different kidney cell lines from ENCODE11 [renal proximal tubule epithelial cells (RPTEC), human renal epithelial cells (HRE) and human renal cortical epithelial cells (HRCEpiC)]. While a small number of the probes (RPTEC: 18%, HRE: 4%, HRCEpiC: 17%) are u-tDMPs in the appropriate cell line, the majority are maintained as f-tDMPs (RPTEC: 62%, HRE: 80%, HRCEpiC: 58%) (Fig. 2B). Additionally, we also downloaded hepatocyte data from ENCODE11 and found a small number of the liver f-tDMPs are u-tDMPs in hepatocytes (17%), but the majority were still f-tDMPs (47%) (Fig. 2C). While it is possible that the hepatocyte and kidney cell lines may themselves be a mixture of different cell types, it seems more likely that the tDMPs in these tissues are not fractionally methylated due to cellular heterogeneity.
Figure 2.
(A) The blood t-DMPs split into u-tDMPs (top panel) and f-tDMPs (bottom panel) with the methylation state recorded from low (yellow) to high (blue). The u-tDMPs found in whole blood are shared among all the blood subsets profiled while the majority of f-tDMPs in whole blood are specific to granulocytes. (B) Renal proximal tubule epithelial cells (RPTEC; red), human renal epithelial cells (HRE; green) and human renal cortical epithelial cells (HRCEpiC; orange) were downloaded from ENCODE and the methylation state of kidney tDMPs is shown here as a density plot. In blue is the average methylation state of the kidney tissues used in our calls and in black is the average methylation of the other tissues. The majority of these kidney cells show similar intermediate levels of methylation for these sites. (C) A hepatocyte sample was downloaded from ENCODE and the methylation state is shown in red for the liver tDMPs. In blue is the average methylation state of the liver samples originally used to call the tDMPs and in black the average methylation of the other tissues. Hepatocytes show similar intermediate levels of methylation to that of the liver tissue. (D) RRBS data taken from ENCODE for liver, hepatocytes, leukocytes, and breast. The top panel shows the average methylation across a region of liver tDMPs with 5 CpGs covered by a single read. The liver and hepatocytes show an increase in methylation in this region over that of the leukocytes and breast tissue. The bottom panel shows a histogram of the average methylation state of each read across the 5 CpGs. Each read can have one of 5 states (0, 0.2, 0.4, 0.6, 0.8, or 1). This clearly shows that this liver tDMR is maintained by approximately half the reads having 0 state (or all the CpGs in the read unmethylated), while the other half of the reads are in state 1.
To examine this further, we obtained RRBS data from ENCODE11 for breast, hepatocyte, leukocyte, and liver samples (IDs in Supplementary File 2). By examining this sequencing data it is possible to get a measure of the methylation state on a per molecule basis by looking at the methylation state of the CpGs on individual reads. Unfortunately, we found that only 1 of the liver f-tDMPs was covered with enough coverage (minimum of 10 reads in each sample) (Fig. 2D). Interestingly, in this single liver f-tDMP, the approximately 50% average methylation is maintained by bimodality of the methylation state of the reads, e.g., the average methylation across the CpGs for each read is either 100% or 0%. This implies that at the single cell level using this single f-tDMP (or region) it would not be possible to determine whether each individual cell was a liver cell or a blood cell.
Whole blood u-tDMPs do not show differences in genomic location or sequence
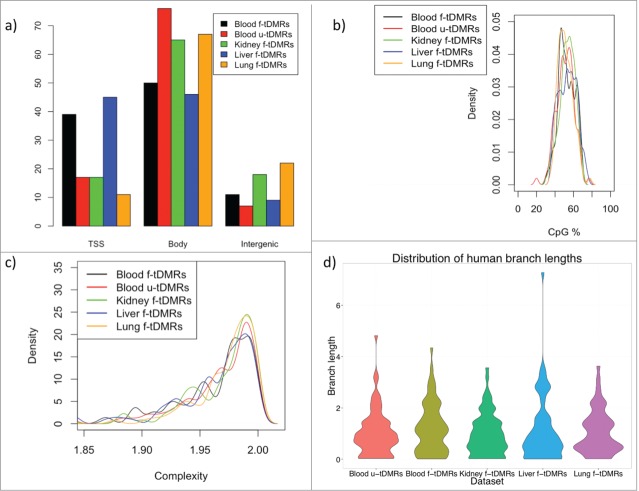
We next wondered whether these f-tDMPs in the other tissues showed any difference to that of the blood u-tDMPs in terms of their genomic location or sequence content. For further analysis it was necessary to define tissue specific differentially methylated regions (tDMRs) and this was done by combining the top tDMPs into regions for each of the 4 tissues to create a list of the top 100 tDMRs for u-tDMRs (blood) and f-tDMRs (blood, kidney, liver and lung) (Materials and Methods). We first investigated the location of these tDMRs in relation to annotated genes and found that blood u-tDMRs showed no significant enrichment or depletion for being near the transcriptional start site (TSS), within the gene body or in an intergenic region (P-value > 0.1 for all tests). Blood f-tDMRs did, however, show significant enrichment for being located within the gene body (1.7-fold enrichment; P-value < 0.001) as did kidney f-tDMRs (1.4-fold enrichment; P-value < 0.001) and lung f-tDMRs (1.5-fold enrichment; P-value < 0.001), while liver tDMRs showed enrichment for the TSS (1.3-fold enrichment; P-value = 0.01) (Fig. 3A).
Figure 3.
(A) The percentage of tDMRs that are located within the promoter, defined as being 2 kb upstream of the TSS to the TSS (labeled as TSS), the gene body or intergenic region. A tDMR is associated with a promoter if it overlaps any part of the promoter, while a gene body tDMR must overlap the gene body but not the promoter. (B) CpG% for each of the different tDMRs. No significant difference in CpG% is found between any of the tDMRs. (C) Complexity of the sequence for the tDMRs (Methods). No significant difference is found between the tDMRs. (D) Distribution of the branch lengths from the human-chimp ancestor to the human for the different tDMRs as a measure of the evolutionary rate. Again, no difference is seen between the different tDMRs.
We next investigated whether there was any underlying difference in the sequence of these regions but found little difference in either the CpG content or complexity of the sequence for the blood u-tDMRs when compared to the f-tDMRs (Fig. 3B and C). To investigate this further we wondered whether the blood u-tDMRs showed any differences in evolutionary rates. We estimated these rates as the length of the tree branch from the human-chimp ancestor to the human using a normalization approach to correct for differences in local mutation rates (Materials and Methods). The normalization adds variance to the estimates of the branch length and, hence, it was necessary to remove some extreme outliers that obscure the bulk of the distribution. The mean rates for blood u-tDMRs, blood f-tDMRs, kidney f-tDMRs, liver f-tDMRs, and lung f-tDMRs were 1.04 ± 0.11, 1.21 ± 0.12, 0.98 ± 0.1, 1.15 ± 0.15, 1.06 ± 0.12, respectively. The evolutionary rates in regions for blood u-tDMRs appeared to be no different than in the other regions (P = 0.7, 0.44, 0.16, and 0.9, respectively, for the comparisons of blood u-tDMRS vs. each other region; Welch's t-test) (Fig. 3D).
tDMRs are associated with H3K4me1 and H3K4me3 marks but show lack of correlation with steady state gene expression
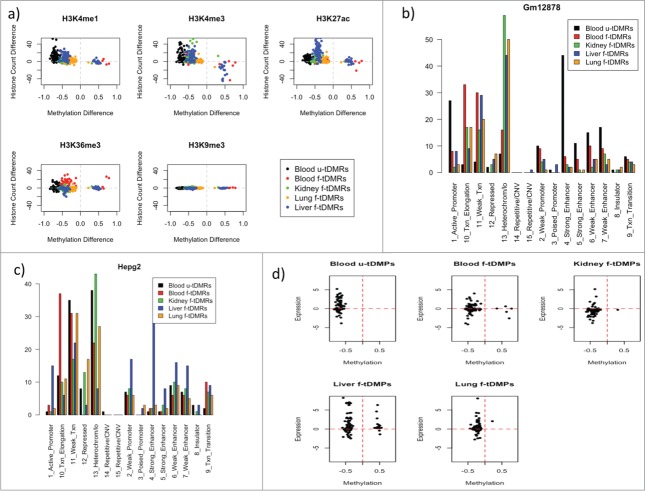
We extracted histone (H3K4me1, H3K4me3, H3K27ac, H3K36me3, and H3K9me3) read counts from the roadmap epigenomics project12 for each of the 4 tissues. For each of the 5 sets of tDMRs, we calculated the read counts for the relevant tissue and subtracted these from the average read counts of the other 3 tissues for that region. We found, for H3K4me1, a highly significant enrichment for blood u-tDMRs (mean: 9.7; P-value = 1.2 × 10−15 t-test) and liver f-tDMRs (mean: 9.3; P-value <2.2 × 10−16; t-test for regions with methylation difference <0). For H3K4me3, we see highly significant enrichment for blood u-tDMRs (mean: 11.0; P-value = 6.3 x 10−9), kidney f-tDMRs (mean: 5.9; P-value = 1.1 × 10−6), lung f-tDMRs (mean: 2.0; P-value = 3.9 × 10−10), and liver f-tDMRs (mean: 8.4; P-value = 1.9 × 10−12), but no significant enrichment or depletion for blood f-tDMRs. This is somewhat surprising as H3K4me3 is often associated with promoters and, yet, kidney f-tDMRs and lung f-tDMRs are highly enriched for regions associated with gene bodies. It has been previously reported, however, that DNA methylation within intragenic regions regulated intragenic promoter activity and, that this occurs tissue- and cell type-specific manner.13 Blood f-tDMRs do however show enrichment for H3K36me3 (mean: 4.6; P-value = 2.5 × 10−7). For H3K27ac, we see small but significant enrichment for blood u-tDMRs (mean: 5.0; P-value < 2.2 × 10−16), blood f-tDMRs (mean: 1.18; P-value = 2.0 × 10−7), kidney f-tDMRs (mean: 2.6; P-value = 1.8 × 10−8) and a large enrichment for liver f-tDMRs (mean: 25; P-value < 2.2 × 10−16) (Fig. 4A). To investigate this further we downloaded the chromHMM states from the UCSC genome browser.14 We found for Gm12878 (a lymphoblastoid cell line) a strong association of the blood u-tDMRs with state 4 - strong enhancer (blood u-tDMRs: 44%, blood f-tDMRs: 6%, kidney f-tDMRs: 3%; liver f-tDMRs: 2%; lung f-tDMRs: 2%) (Fig. 4B). Meanwhile, for Hepg2, a cell line derived from a male patient with liver carcinoma we found liver f-tDMRs to be similarly enriched at state 4 (blood u-tDMRs: 1%, blood f-tDMRs: 2%, kidney f-tDMRs: 2%, liver tDMRs: 28%; lung tDMRs: 3%) (Fig. 4C). Due to the limited number of samples available with chromHMM states, these were the only 2 relevant cell lines for comparison with our results. Therefore, the combination of both the chromHMM and histone data shows that these tDMRs show similar tissue specificity in histone marks for these regions but there is no distinct difference in blood tDMRs to that of other tissues.
Figure 4.
(A) A series of scatter plots of the difference in read count for the 5 different tDMRs for 5 different histone marks. Each point represents a different tDMR and on the x-axis is the methylation difference between the tissue of interest and the other tissues and on the y-axis is the difference in read count of the particular histone and the other tissues. (B) Percentage overlap of the various different tDMRs with the chromHMM states of GM12878. (C) Percentage overlap of the various different tDMRs with the chromHMM states of Hepg2. (D) Correlation of the methylation difference of each of the tDMRs with the log fold difference in expression between the tissue of interest and the all the other tissues.
Finally, we extracted gene expression data from BioGPS [Human U133A/GNF1H Gene Atlas (GSE1133)] for each of the 4 different tissues. For each of the tDMRs we calculated the expression difference for the relevant tissue against the other 3 tissues for the gene nearest to the tDMR. For all tDMRs, we found very little correlation between the difference in methylation and a difference in expression. Some genes did seem to be more expressed when the tDMR was less methylated, but this was not true generally and there was a distinct lack of correlation (Fig. 4D).
Discussion
Using Marmal-aid, a publicly available database of Illumina 450k arrays, it was possible to define tissue-specific differentially methylated probes that can be used to define various tissues. Interestingly, there was double the amount of probes that showed differential methylation in blood to that of other tissues used in this study. In particular, we found that, for blood, the probes were unmethylated (or u-tDMPs), while for all the other tissues the probes were fractionally methylated in the specific tissue (or f-tDMP). Using three kidney cell lines and a hepatocyte sample we were able to show that the fractional methylated positions were unlikely to be caused by cellular heterogeneity.
Why do blood tDMPs in particular have such relatively low levels of methylation, and what is the functional consequence of this? Although standard bisulfite conversion methods cannot distinguish between methylated and hydroxymethylated cytosines, the latter is present at very low levels in blood and most somatic tissues (not including brain)15 and, hence, hydroxymethylation cannot account for this. We were unable to find specific differences in genomic location, sequence content, sequence conservation, or histone marks, in comparison to tDMPs for other tissues. Therefore, could it be that the cytosine methylation state per se, as opposed to these other molecular correlates, plays an important role at these tDMPs? In future studies, it could be interesting to investigate whether the dynamic interplay between TET and DNMT enzymes16 is different in blood compared with other tissues. With regards to how the preponderance of u-tDMPs in blood is involved in blood-specific functions, we can only speculate. First, blood is not a solid tissue. Secondly, blood subsets are critical components of the immune system that has to react to invading pathogens with varying degrees of response time. The potential role of blood u-tDMPs in either of these blood-specific properties is a key avenue of research for the future.
Methods
Extracting data and calling tDMPs
Normalized samples were extracted from Marmal-aid9 for tissues that contained a minimum of 50 samples. An initial call of tissue specific methylation differences was made using dmpFinder function in minfi.17 This was performed for each tissue by defining 2 categories: the tissue of interest was assigned to Group 1, while the remaining tissues were assigned as Group 2. The top 2 probes for each of the tissue specific calls were used to visually inspect the data and any samples that were found to be closer to the mean of Group 2 than of Group 1 were removed. The remaining samples were then used to produce the final calls. For each tissue, we randomly selected 50 samples and, following a similar procedure to above, we called differences using dmpFinder. The IDs of the samples used in the experiment can be found in Supplementary file 1. The calls for each of the tissues for all probes can be found at http://webspace.qmul.ac.uk/rlowe/tdmps/.
RRBS analysis
Raw sequence files were downloaded from the Sequence Read Archive (SRA) for breast, hepatocyte, leukocyte and liver samples. Sample IDs are available in Supplementary file 2. These were mapped to the human genome (hg19) using BISMARK18 and the methylation state for each read covering the top 100 tDMPs was extracted using custom scripts from the aligned BAM file.
Combining tDMPs into tDMRs
For certain analyses, it was necessary to define regions of differential methylation and, hence, we combined the tDMPs into regions.
We combined 2 or more tDMPs into regions if the tDMPs were within 500 bp of each other and the methylation difference was consistent. If no probes were located within 500 bp of the tDMP then we set a default width to the region if 500 bp centered on the called tDMP.
Sequence complexity
To produce a measure for the sequence complexity of each DMR we calculated the Shannon Entropy using the following equation:
where is the frequency of base within the sequence.
Evolutionary analysis
For each of the regions identified, EPO alignments19,20 for 15 mammalian species were retrieved using Ensembl REST API.21 Alignments that did not include sequences for human, chimpanzee and at least one out-group species were filtered out. For each region with an alignment retained, the first intron of the closest gene was selected as a control region. Intron coordinates were obtained using the BioMart21 R23 package. Alignments for these intron sequences were obtained with Ensembl API.21 Again, only alignments where sequences for human, chimpanzee and at least one out-group species were present in the control region were retained for further analysis. Evolutionary rates were estimated using the baseml program of the PAML package24 (options: model = REV plus Gamma-distributed rate variation over sites, ncatG = 4). Alignments for regions of interest were concatenated with alignments for the intronic regions and analyzed as site partitions (option Mgene = 1, permitting different substitution dynamics and different evolutionary rates for each lineage and for the regions of interest and introns).
Histone analysis
Histone marks (H3K4me1, H3K4me3, H3K27ac, H3K36me3, and H3K9me3) for each tissue were downloaded from the Roadmap Epigenomics Project12 as wig files. These were converted to BigWig using the wigToBigWig program from UCSC. Read counts were calculated for each of the tDMRs using the bigWigSummary program.
Expression analysis
We extracted gene expression data from BioGPS [Human U133A/GNF1H Gene Atlas (GSE1133)] for each of the 4 different tissues blood, liver, kidney and lung. For each of the tDMRs we calculated the nearest TSS and calculated for that gene the logFC of expression between the tissue of interest and the other 3 tissues for each of the 4 sets of tDMRs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
RL, VK are supported by the EU-FP7 “BLUEPRINT” program (282510). GS, NG are supported by the European Molecular Biology Laboratory. GS is a member of Wolfson College, University of Cambridge. This work was partly supported by the BBSRC (BB/H012494/1).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Bird AP. CpG-rich islands and the function of DNA methylation. Nature 1986; 321:209-213; PMID:2423876; http://dx.doi.org/ 10.1038/321209a0 [DOI] [PubMed] [Google Scholar]
- 2. Laird PW. The power and promise of DNA methylation markers. Nat Rev Cancer 2003; 3:253-266; PMID:12671664; http://dx.doi.org/ 10.1038/nrc1045 [DOI] [PubMed] [Google Scholar]
- 3. Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, et al. . Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013; 49(2):359-67; PMID:23177740; http://dx.doi.org/ 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Gräf S, Tomazou EM, Bäckdahl L, Johnson N, Herberth M, et al. . An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res 2008; 18:1518-29; PMID:18577705; http://dx.doi.org/ 10.1101/gr.077479.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Slieker RC, Bos SD, Goeman JJ, Bovée JV, Talens RP, van der Breggen R, Suchiman HE, Lameijer EW, Putter H, van den Akker EB, et al. . Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin 2013; 6(1):26; PMID:23919675; http://dx.doi.org/ 10.1186/1756-8935-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lokk K, Modhukur V, Rajashekar B, Märtens K, Mägi R, Kolde R, Kolt Ina M, Nilsson TK, Vilo J, Salumets A, et al. . DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014. 15(4); PMID:24690455; http://dx.doi.org/ 10.1186/gb-2014-15-4-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lowe R, Gemma C, Beyan H, Hawa MI, Bazeos A, Leslie RD, Montpetit A, Rakyan VK, Ramagopalan SV. Buccals are likely to be a more informative surrogate tissue than blood for epigenome-wide association studies. Epigenetics. 2013; 8(4):445-54; PMID:23538714; http://dx.doi.org/ 10.4161/epi.24362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, et al. . Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res 2013; 23(3):555-67; PMID:23325432; http://dx.doi.org/ 10.1101/gr.147942.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lowe R, Rakyan VK. Marmal-aid - a database for Infinium HumanMethylation450. BMC Bioinformatics 2013; 14:359; PMID:24330312; http://dx.doi.org/ 10.1186/1471-2105-14-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén SE, Greco D, Söderhäll C, Scheynius A, Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 2012; 7(7):e41361; PMID:22848472; http://dx.doi.org/ 10.1371/journal.pone.0041361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, et al. . ENCODE whole-genome data in the UCSC genome browser (2011 update). Nucleic Acids Res 2011; 39: D871-5; PMID: 21037257; http://dx.doi.org/ 10.1093/nar/gkq1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. . The NIH roadmap epigenomics mapping consortium. Nat Biotechnol 2010; 28(10):1045-8; PMID:20944595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010; 466(7303):253-7; PMID:20613842; http://dx.doi.org/ 10.1038/nature09165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 2012; 9(3):215-6; PMID:22373907; http://dx.doi.org/ 10.1038/nmeth.1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids 2011; 2011:870726; PMID:21772996; http://dx.doi.org/ 10.4061/2011/870726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martínez JA, Pape UJ, Jacobsen SE, Peters B, Rao A. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA 2014; 111(4):1361-6; PMID:24474761; http://dx.doi.org/ 10.1073/pnas.1322921111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014; 30(10):1363-9; PMID:24478339; http://dx.doi.org/ 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011; 27(11):1571-2; PMID:21493656; http://dx.doi.org/ 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res 2008; 18(11):1814-28; PMID:18849524; http://dx.doi.org/ 10.1101/gr.076554.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paten B, Herrero J, Fitzgerald S, Beal K, Flicek P, Holmes I, Birney E. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res 2008; 18(11):1829-43; PMID:18849525; http://dx.doi.org/ 10.1101/gr.076521.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. . Ensembl 2014. Nucleic Acids Res 2014; 42; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005; 21(16):3439-40; PMID:16082012; http://dx.doi.org/ 10.1093/bioinformatics/bti525 [DOI] [PubMed] [Google Scholar]
- 23. R Core Team 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria URL: http://www.R-project.org/. [Google Scholar]
- 24. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 2007; 24(8):1586-91; PMID:17483113; http://dx.doi.org/ 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.