Abstract
4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR), also known as isoprenoid synthesis H (IspH) or lysis-tolerant B (LytB), catalyzes the last step of the methylerythritol phosphate pathway to synthesize isopentenyl diphosphate and dimethylallyl diphosphate. The structure and reaction mechanism of IspH have been actively investigated in Escherichia coli but little is known in plants. Compared with the bacterial IspH, cyanobacterial and plant HDRs all contain an extra N-terminal conserved domain (NCD) that is essential for their function. Tyr72 in the NCD and several plant-specific residues around the central active site are critical for Arabidopsis HDR function. These results suggest that the structure and reaction mechanism of HDR/IspH may be different between plants and bacteria. The E. coli IspH is an iron-sulfur protein that is sensitive to oxygen. It is possible that the cyanobacterial HDR may independently evolve from the common ancestor of prokaryotes to obtain the NCD, which may protect the enzyme from high concentration of oxygen during photosynthesis.
Keywords: Arabidopsis, cyanobacterium, dimethylallyl diphosphate, HDR, iron-sulfur protein, isopentenyl diphosphate, isoprenoid, IspH, methylerythritol phosphate, oxygen evolving
Isoprenoids, also known as terpenoids, are a large and diverse group of natural products found in all living organisms. Some isoprenoids, for instance, steroid hormones in mammals, photosynthetic pigments in plants, and ubiquinone or menaquinone in bacteria, are essential or have important physiological functions. Still, some other isoprenoids including vitamins, hormones, and anticancer agents such as Taxol are medically important for human health.1 All isoprenoids are synthesized from the common 5-carbon precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP).
There are 2 metabolic pathways that use completely different precursors to synthesize IPP and DMAPP: acetyl-CoA for the mevalonate (MVA) pathway, and pyruvate and glyceraldehyde 3-phosphate for the methylerythritol phosphate (MEP) pathway. In animals, IPP and DMAPP are synthesized via the MVA pathway, whereas in most eubacteria, including many pathogenic bacteria, isoprenoids are derived from the MEP pathway.2 Interestingly, plants use both MVA and MEP pathways to synthesize IPP and DMAPP. The MVA pathway is localized to the cytosol, whereas the MEP pathway is compartmentalized in the plastid (Fig. 1). It is known that IPP and DMAPP synthesized in distinct subcellular locations are used for the biosynthesis of different isoprenoids in plants. For instance, sesquiterpenes, triterpenes and polyterpenes are derived from the cytosolic MVA pathway, whereas isoprene, monoterpenes, phytol, plastoquinones, tocopherols, carotenoids, and plant hormones gibberellin and abscisic acid are mainly synthesized in the plastids via the MEP pathway.3 Because the MEP pathway, not existing in humans, is essential for plants and many pathogenic bacteria, so enzymes of the MEP pathway are attractive targets for herbicide and drug discovery.
Figure 1.
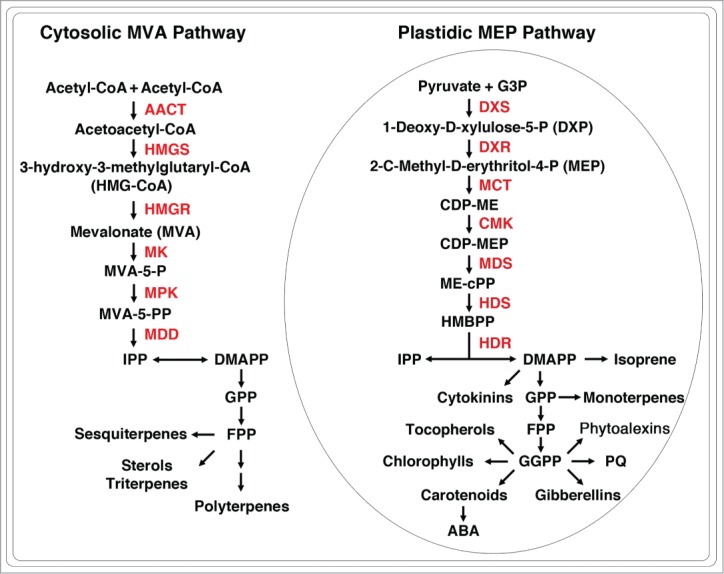
The mevalonate (MVA) and methylerythritol phosphate (MEP) pathways of isoprenoid biosynthesis in plants. MVA-5-P, mevalonate-5-phosphate; MVA-5-PP, mevalonate-5-pyrophosphate; Enzymes of the MVA pathway: AACT, acetoacetyl-CoA transferase; HMGS, HMG-CoA synthase; HMGR, HMG-CoA reductase; MK, MVA kinase; MPK, MVA-5-P kinase; MDD; MVA-5-PP decarboxylase. CDP-ME, 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol; CDP-MEP, 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol-2-phosphate; ME-cPP, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate; HMBPP, 4-hydroxy-3-methylbut-2-enyl diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl diphosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate; PQ, plastoquinone; ABA, abscisic acid. Enzymes of the MEP pathway: DXS, DXP synthase; DXR, DXP reductoisomerase; MCT, MEP cytidylyltransferase; CMK, CDP-ME kinase; MDS, ME-cPP synthase; HDS, HMBPP synthase; HDR, HMBPP reductase.
There are 7 steps in the MEP pathway. In the first step, pyruvate and glyceraldehyde 3-phosphate are converted into 1-deoxy-D-xylulose 5-phosphate (DXP) by DXP synthase.4,5 DXP is also a precursor for the biosynthesis of vitamins B1 and B6 in bacteria.4,6 In the second step of the MEP pathway, DXP reductoisomerase transforms DXP into 2C-methyl-D-erythritol 4-phosphate (MEP) that is the first committed metabolite in the pathway.7 MEP is then converted to IPP and DMAPP in consecutive steps catalyzed by the other MEP pathway enzymes.8-13 In Arabidopsis, all the MEP pathway enzymes are predicted to contain transit peptides, which are able to target the plant enzymes to the plastids.14 Because the photosynthetic pigments are derived from the MEP pathway, mutants defective in any of the MEP pathway genes are albino lethal.14-20
The 4-hydroxy-3-methylbut-2-enyl diphosphate (HMBPP) reductase, commonly known as HDR, isoprenoid synthesis H (IspH), or lysis-tolerant B (LytB), catalyzes the last step of the MEP pathway that converts HMBPP into a 5:1 mixture of IPP and DMAPP in Escherichia coli.12,13 In Arabidopsis thaliana, the hdr-1 knockout mutant is albino lethal and the HDR transgene-induced gene-silencing lines are albino, pale green, or variegated.18,21 The Arabidopsis HDR was able to complement the E. coli ispH mutant indicating that the function of this enzyme is conserved from bacteria to plants.18,22 The structure and reaction mechanisms of IspH have been extensively studied in E. coli and Aquifex aeolicus.23-29 By contrast, little is known about the structure and reaction mechanisms of HDR in plants. Recently, we used the E. coli IspH (Protein Data Bank code 3F7T) as a template to predict the structure of Arabidopsis HDR. The prediction revealed that the IspH domain of the Arabidopsis HDR (amino acids 111-466) had a trefoil-like structure similar to that of E. coli IspH (Fig. 2A).21 Unexpectedly, the IspH domain of the Arabidopsis HDR corresponding to the full-length IspH of E. coli failed to complement the E. coli ispH mutant.21 Nonetheless, several key amino acids involved in iron-sulfur formation, substrate binding or catalytic reaction in E. coli IspH were conserved in Arabidopsis HDR. Homology modeling and substrate docking of the IspH domain of the Arabidopsis HDR revealed that these key residues were also located in the central active site.21 In addition to the conserved key amino acids, the Arabidopsis HDR contains several critical residues that are absent in the bacterial enzyme.21 These results implicate that the reaction mechanism of plant HDR may be different from that of E. coli IspH.
Figure 2.
The N-terminal conserved domain of HDR only occurs in the oxygenic photosynthesis organisms. (A) The trefoil-like structure of active E. coli IspH (Protein Data Bank: 3F7T) contains a [3Fe-4S] cluster in the catalytic center. (B) Schematic diagrams of Arabidopsis thaliana, Synechocystis sp. PCC 6803 and E. coli HDR/IspH. TP, transit peptide; NCD, N-terminal conserved domain. (C) Alignment of HDR/IspH proteins from plants and bacteria showing the N-terminal conserved domain among the oxygenic photosynthesis organisms. The E. coli IspH does not contain the NCD, and only part of the alignment is shown here. Sm, Selaginella moellendorffii; Pp, Physcomitrella patens; At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Os, Oryza sativa; Zm, Zea mays; Pd, Pinus densiflora; Sy, Synechocystis sp PCC 6803; Cr, Chlamydomonas reinhardtii; Ec, Escherichia coli. The arrow indicates the conserved Tyr residue that is critical for Arabidopsis HDR function.
The Arabidopsis MEP pathway enzymes are predicted to localize to the plastid. Indeed, transit peptides of the Arabidopsis MEP pathway enzymes are able to target the green fluorescent protein (GFP) to the plastid.14 Amino acid sequence alignment revealed that all plant HDRs had an extra N-terminal conserved domain (NCD) following the transit peptides (Fig. 2B and C).21 The NCD of Arabidopsis HDR includes amino acids 56 to 110, which is followed by the IspH domain. We used protein BLAST (http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi) to search the database and did not find any domain or motif that was similar to the NCD. Because the NCD immediately follows the transit peptide, it is reasonable to suspect that the NCD may play a role in targeting the plant enzyme to the chloroplast. However, it has been shown that the first 52 amino acids encompassing the predicted transit peptide of Arabidopsis HDR were sufficient to target the GFP reporter protein to the chloroplast.14 In a recent report, we further demonstrated that the Arabidopsis HDRΔNCD-GFP fusion protein was localized to the chloroplast.21 Thus, the NCD is not required for targeting the Arabidopsis HDR enzyme to the chloroplast.
The NCD is highly conserved in cyanobacteria, green algae, and land plants. The Arabidopsis and cyanobacterial HDRs would fail to complement the E. coli ispH mutant without the NCD.21 In addition to the critical amino acids involved in substrate binding or catalysis located in the central active site of the IspH domain, we further uncovered that the absolutely conserved tyrosine-72 residue in the NCD was critical for Arabidopsis HDR function.21 These results suggest that the NCD is essential for Arabidopsis and cyanobacterial HDR function. The exact functions of the NCD in plant and cyanobacterial HDRs have yet to be established. We proposed that the NCD might be involved in protein stability, substrate-binding, or catalytic reaction in Arabidopsis HDR.21 Further studies are required to define the function of the NCD in plant HDRs.
Although the involvement of HDR in the last step of the MEP pathway is conserved from bacteria to plants, the NCD is specific to oxygen-evolving photosynthesis organisms.21 The occurrence of the NCD and some specific amino acids in cyanobacterial HDR indicate that the structure and reaction mechanism of HDR have evolved independently in cyanobacteria versus other prokaryotes. The E. coli HDR is an iron-sulfur protein that is sensitive to oxygen.30 The critical Cys residues involved in iron-sulfur cluster formation are conserved in plant and cyanobacterial HDRs.21 It is likely that the HDR enzyme is also an iron-sulfur protein in these oxygen-evolving photosynthetic organisms. This raises an obvious question whether the HDR enzymes in cyanobacteria and plants are sensitive to oxygen. In non-oxygen evolving photosynthetic bacteria, their HDR amino acid sequences, and presumably their structure and reaction mechanism, are highly similar to those of E. coli and A. aeolicus.21 It is likely that the production of oxygen is the driving force for the early divergence of HDR between cyanobacteria and the other prokaryotes. The occurrence of the NCD, and possibly the other conserved motifs in cyanobacteria and plants, may enable the HDR enzyme to survive the high oxygen environment during photosynthesis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the Ministry of Science and Technology and Academia Sinica (98-CDA-L04).
References
- 1. Sacchettini JC, Poulter CD. Creating isoprenoid diversity. Science 1997; 277:1788-9; PMID:9324768; http://dx.doi.org/ 10.1126/science.277.5333.1788 [DOI] [PubMed] [Google Scholar]
- 2. Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J 1993; 295:517-24; PMID:8240251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 2013; 64: 665-700; PMID: 23451776; http://dx.doi.org/ 10.1146/annurev-arplant-050312-120116 [DOI] [PubMed] [Google Scholar]
- 4. Sprenger GA, Schorken U, Wiegert T, Grolle S, de Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H. Identification of a thiamin dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc Natl Acad Sci USA 1997; 94:12857-62; PMID:9371765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lois LM, Campos N, Putra SR, Danielsen K, Rohmer M, Boronat A. Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of D-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc Natl Acad Sci USA 1998; 95:2105-10; PMID:9482846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. White RH. Stable isotope studies on the biosynthesis of the thiazole moiety of thiamin in Escherichia coli. Biochemistry 1978; 17:3833-40; PMID:359046 [DOI] [PubMed] [Google Scholar]
- 7. Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci USA 1998; 95:9879-84; PMID: 9707569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk MH. Cytidine 5’-triphosphate dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc Natl Acad Sci USA 1999; 96:11758-63; PMID:10518523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luttgen H, Rohdich F, Herz S, Wungsintaweekul J, Hecht S, Schuhr CA, Fellermeier M, Sagner S, Zenk MH, Bacher A, et al. Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-D-erythritol. Proc Natl Acad Sci USA 2000; 97:1062-67; PMID:10655484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herz S, Wungsintaweekul J, Schuhr CA, Hecht S, Luttgen H, Sagner S, Fellermeier M, Eisenreich W, Zenk MH, Bacher A, et al. Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate to 2C-methyl-D-erythritol 2,4-cyclodiphosphate. Proc Natl Acad Sci USA 2000; 97: 2486-90; PMID:10694574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F. Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc Natl Acad Sci USA 2001; 98: 14837-42; PMID:11752431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rohdich F, Hecht S, Gartner K, Adam P, Krieger C, Amslinger S, Arigoni D, Bacher A, Eisenreich W. Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc Natl Acad Sci USA 2002; 99:1158-63; PMID:11818558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adam P, Hecht S, Eisenreich W, Kaiser J, Grawert T, Arigoni D, Bacher A, Rohdich F. Biosynthesis of terpenes: studies on 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase. Proc Natl Acad Sci USA 2002; 99:12108-13; PMID:12198182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsieh MH, Chang CY, Hsu SJ, Chen JJ. Chloroplast localization of methylerythritol 4-phosphate pathway enzymes and regulation of mitochondrial genes in ispD and ispE albino mutants in Arabidopsis. Plant Mol Biol 2008; 66:663-73; PMID:18236010; http://dx.doi.org/ 10.1007/s11103-008-9297-5 [DOI] [PubMed] [Google Scholar]
- 15. Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, Leon P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J 1996; 9:649-58; PMID:8653115 [DOI] [PubMed] [Google Scholar]
- 16. Gutierrez-Nava M de L, Gillmor CS, Jimenez LF, Guevara-Garcia A, Leon P. CHLOROPLAST BIOGENESIS genes act cell and noncell autonomously in early chloroplast development. Plant Physiol 2004; 135: 471-82; PMID:15133149; http://dx.doi.org/ 10.1104/pp.103.036996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guevara-Garcia A, San Roman C, Arroyo A, Cortes ME, de la Luz Gutierrez-Nava M, Leon P. Characterization of the Arabidopsis clb6mutant illustrates the importance of posttranscriptional regulation of the methyl-D-erythritol 4-phosphate pathway. Plant Cell 2005; 17:628-43; PMID:15659625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsieh MH, Goodman HM. The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol 2005; 138:641-53; PMID:15863698; http://dx.doi.org/ 10.1104/pp.104.058735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsieh MH, Goodman HM. Functional evidence for the involvement of Arabidopsis IspF homolog in the nonmevalonate pathway of plastid isoprenoid biosynthesis. Planta 2006; 223:779-84; PMID:16231155; http://dx.doi.org/ 10.1007/s00425-005-0140-9 [DOI] [PubMed] [Google Scholar]
- 20. Tseng CC, Lee CJ, Chung YT, Sung TY, Hsieh MH. Differential regulation of Arabidopsis plastid gene expression and RNA editing in non-photosynthetic tissues. Plant Mol Biol 2013; 82:375-92; PMID:23645360; http://dx.doi.org/ 10.1007/s11103-013-0069-5 [DOI] [PubMed] [Google Scholar]
- 21. Hsieh WY, Sung TY, Wang HT, Hsieh MH. Functional evidence for the critical amino-terminal conserved domain and key amino acids of Arabidopsis 4-hydroxy-3-methylbut-2-enyl diphosphate reductase. Plant Physiology 2014; 166:57-69; PMID:25037211; http://dx.doi.org/ 10.1104/pp.114.243642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McAteer S, Coulson A, McLennan N, Masters M. The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J Bacteriol 2001; 183:7403-7; PMID:11717301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gräwert T, Kaiser J, Zepeck F, Laupitz R, Hecht S, Amslinger S, Schramek N, Schleicher E, Weber S, Haslbeck M, et al. IspH protein of Escherichia coli: studies on iron-sulfur cluster implementation and catalysis. J Am Chem Soc 2004; 126:12847-55; PMID:15469281 [DOI] [PubMed] [Google Scholar]
- 24. Rekittke I, Wiesner J, Röhrich R, Demmer U, Warkentin E, Xu W, Troschke K, Hintz M, No JH, Duin EC, et al. Structure of (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase, the terminal enzyme of the non-mevalonate pathway. J Am Chem Soc 2008; 130: 17206-7; PMID:19035630; http://dx.doi.org/ 10.1021/ja806668q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gräwert T, Rohdich F, Span I, Bacher A, Eisenreich W, Eppinger J, Groll M. Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction. Angew Chem Int Ed Engl 2009; 48:5756-9; PMID:19569147 [DOI] [PubMed] [Google Scholar]
- 26. Gräwert T, Span I, Eisenreich W, Rohdich F, Eppinger J, Bacher A, Groll M. Probing the reaction mechanism of IspH protein by x-ray structure analysis. Proc Natl Acad Sci USA 2010; 107:1077-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W, Wang K, Liu YL, No JH, Li J, Nilges MJ, Oldfield E. Bioorganometallic mechanism of action, and inhibition, of IspH. Proc Natl Acad Sci USA 2010; 107:4522-7; PMID:20173096; http://dx.doi.org/ 10.1073/pnas.0911087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Span I, Gräwert T, Bacher A, Eisenreich W, Groll M. Crystal structures of mutant IspH proteins reveal a rotation of the substrate's hydroxymethyl group during catalysis. J Mol Biol 2012; 416:1-9; PMID:22137895; http://dx.doi.org/ 10.1016/j.jmb.2011.11.033 [DOI] [PubMed] [Google Scholar]
- 29. Wang W, Wang K, Span I, Jauch J, Bacher A, Groll M, Oldfield E. Are free radicals involved in IspH catalysis? An EPR and crystallographic investigation. J Am Chem Soc 2012; 134: 11225-34;PMID:22687151; http://dx.doi.org/ 10.1021/ja303445z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wolff M, Seemann M, Tse BSB, Frapart Y, Tritsch D, Estrabot AG, Rodriguez-Concepcion M, Boronat A, Marquet A, Rohmer M. Isoprenoid biosynthesis via the methylerythritol phosphate pathway: the (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase (LytB/IspH) from Escherichia coli is a [4Fe-4S] protein. FEBS Lett 2003; 541:115-20; PMID:12706830; http://dx.doi.org/ 10.1016/S0014-5793(03)00317-X [DOI] [PubMed] [Google Scholar]



