Abstract
Histone deacetylases (HDAC) control gene expression through their ability to acetylate proteins, thereby influencing a diverse range of cellular functions. Class I HDAC (HDAC1–3 and 8) and HDAC6 are predominantly upregulated in malignancies and their altered expression in some cancers has a significant prognostic implication. The expression and prognostic consequence of dysregulated Class I HDAC and HDAC6, key players in multiple myeloma (MM), are unknown. This study hypothesized that HDAC are dysregulated in MM and patients with high expression have significantly poorer prognostic outcomes. Quantitative PCR for 11 HDAC (Class I, II, and IV) was performed in genetically heterogeneous human myeloma cell lines (HMCL) and primary MM and compared to normal plasma cells (PC). In HMCL, HDAC1–3 and 8 (Class I), and HDAC5 and HDAC10 (Class II) were significantly upregulated compared to normal PC. In primary MM, the median expression level of all of the HDAC, except HDAC1 and HDAC11, were elevated when compared to normal PC. Patients with higher levels of HDAC1–3, HDAC4, HDAC6, and HDAC11 transcripts demonstrated a significantly shorter progression-free survival (PFS). Immunohistochemical staining for HDAC1 and HDAC6 on bone marrow trephines from a uniformly treated cohort of transplant eligible MM patients revealed that HDAC1 protein was detectable in most patients and that higher levels of MM cell HDAC1 protein expression (≥90 % versus ≤20 % MM cell positivity) correlated with both shorter PFS (P = 0 .07) and shorter overall survival (P = 0 .003). Conversely, while the majority of patients expressed HDAC6, there was no correlation between HDAC6 levels and patient outcome. Together, these results indicate that overexpression of Class I HDAC, particularly HDAC1, is associated with poor prognosis in MM.
Keywords: Class I HDAC, HDAC expression, hematological malignancy, histone deacetylases, multiple myeloma, prognosis
Abbreviations
Introduction
Multiple myeloma (MM) is an incurable clonal B-cell malignant neoplasm characterized by de novo drug resistance and complex cytogenetic abnormalities that are associated with unique clinical and prognostic implications.1-5 In contrast to the genomic abnormalities, limited information is known about the role of the epigenome in MM pathogenesis and maintenance. Epigenetic modifications, such as DNA methylation and histone acetylation of structurally intact genes have been recognized as critical facets of cancer pathogenesis and maintenance.6 Acetylation is modulated by the dynamic and antagonistic action of 2 classes of enzymes, histone deacetylases (HDAC) and histone acetyltransferases (HAT), wherein HDAC catalyze the removal of acetyl groups and HAT acetylate the N-terminal lysine residues. HDACs are a highly conserved group of enzymes currently consisting of 18 genes grouped into 4 classes, based on their homology to yeast orthologues. Class I (HDAC1–3 and 8), Class IIA (HDAC4, 5, 7, and 9), Class IIB (HDAC6 and 10) and Class IV (HDAC11) require zinc for catalyzing deacetylase activity, and Class III (Sirtuins 1–7) utilize nicotine adenine dinucleotide (NAD+) for their catalytic mechanisms.7,8 HDACs orchestrate a myriad of cellular functions, including proliferation, differentiation, and apoptosis, through the deacetylation of histones and non-histone proteins.
Dysregulation of HDAC expression, predominantly overexpression, has been observed in a number of malignancies.9 Class I HDAC expression, in particular, is known to be increased in a number of cancers, including gastric, prostate, colon, breast, renal, and cervical.10-16 Specific to hematological malignancies, dysregulated HDAC expression has been reported in peripheral T-cell lymphomas (PTCL), cutaneous T-cell lymphomas (CTCL), diffuse large B-cell lymphomas (DLBCL), pediatric acute lymphoblastic leukemia (ALL), and myeloproliferative neoplasms.17-20 In all instances, the expression of one or more of Class I HDACs was increased. Prognostic correlates of upregulated HDAC expression are, however, more complex and appear to be highly context-dependent, with the majority of studies demonstrating a worse prognosis with higher levels of HDAC1 and/or HDAC2 expression.10-14,21 The opposite effect was seen in breast cancer (HDAC1), ALL, and chronic lymphocytic leukemia (CLL; HDAC3), in which overexpression was a favorable prognostic indicator.16,22,23
HDAC6 has also been studied in cancers because of its to ability to orchestrate a variety of cellular processes that are crucial for cancer pathogenesis.24 Overexpression of HDAC6 has been demonstrated in hepatocellular carcinomas,25 CTCL,17 ALL, 22 and breast cancers.26,27 Moreover, in contrast to increased Class I HDAC expression, which for the most part is an indicator of inferior survival, overexpression of HDAC6 has been largely associated with both improved overall (OS) and progression-free survival (PFS), including in studies of CTCL, 17 breast cancer,26,27 lung cancer,28 DLBCL, 20 and CLL.23 Conversely, a small number of studies have demonstrated that increased HDAC6 expression (breast carcinomas and PTCL) is a negative prognostic factor.15,20
In MM, neither the pattern of expression of HDAC nor any potential association with prognosis has been systematically studied. In vitro HDAC6 appears to be a key modulator of MM cell survival, with studies showing the efficacy of HDAC6-selective inhibitors to induce cell death in MM cells.29-31 Similarly, Class I HDAC also play a critical role in MM cell survival, with evidence showing that inhibition of Class I HDAC induces MM cell death and that inhibition of Class I HDAC is superior to HDAC6 inhibition alone in promoting MM cell apoptosis.32,33 Dysregulation of HDAC may also to contribute to MM drug resistance, with overexpression of HDAC1 possibly conferring resistance to bortezomib-induced apoptosis. This resistance is reversed by the simultaneous addition of FK228, a Class I HDAC inhibitor (HDACi).33 Given the role HDACs play in tumor cell survival and drug resistance and the emergence of HDACi as a potentially available class of anti-MM therapeutics, characterizing the patterns of HDAC expression in MM, and any correlation with patient outcome, is of interest.
Results
HDAC levels in HMCL are significantly elevated
The levels of expression of HDAC1–11 were determined by qRT-PCR in a genetically heterogeneous panel (n = 14 ) of HMCL and compared to the levels of expression in a panel of normal PC (n = 9 ). All the HDAC transcripts were detected in the tested HMCL, with HDAC11 showing the highest expression and the most variability (mean ± SEM: 21.29 ± 13.66), followed by HDAC1 (5.89 ± 1.03), and HDAC5 (4.96 ± 0.56) (Fig. 1). The mean values represent the relative expression levels of the particular gene in HMCL compared to normal PC, which have been normalized to one. Moreover, all Class I HDAC isoforms (HDAC1, 2, 3, and 8), HDAC5, and HDAC10 were significantly elevated in HMCL when compared to normal PC (Fig. 1).
Figure 1.
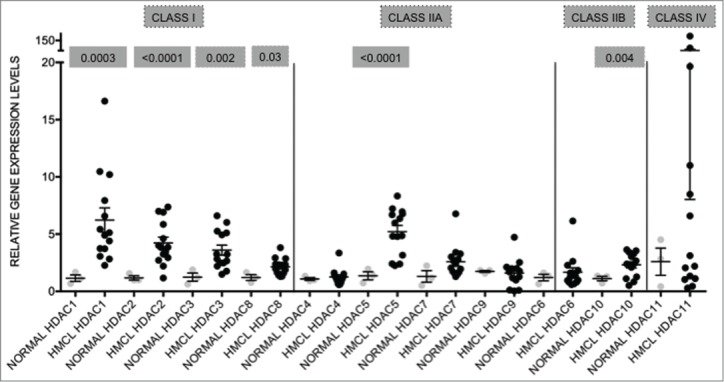
HDAC genes are overexpressed in human myeloma cell lines when compared to normal plasma cells Expression levels of HDAC in 14 human myeloma cell lines (HMCL) were compared to normal plasma cells (normal; n = 9 ). Class I HDAC (HDAC1, HDAC2, HDAC3, and HDAC8) were all significantly overexpressed. Among the class II HDAC, HDAC5 and HDAC10 were significantly overexpressed. Data are presented as mean±SEM. Significant differences were calculated with t-test utilizing GraphPad Prism 5.0d. p-values are indicated.
To determine if HDAC expression showed any correlation with recognized recurring cytogenetic abnormalities, the HMCL were grouped based on their translocation status and expression levels of HDAC in these groups were compared. Primary IgH translocations in MM are represented predominantly by 4 recurring abnormalities: 4p16 [t(4;14); FGFR3 and MMSET], 11q13 [t(11;14); CYCLIN D1], 6p21 [t(6;14); CYCLIN D3] and 16q23 [t(14;16); C-MAF]. Among the HMCL chosen for this study, 5/14 harbor t(4;14) (OPM-2, NCI-H929, LP-1, KMS-18, and JIM-1), 4/14 have t(11;14) (KMS-12BM, KMS-12PE, XG-1, and U266), 3/14 have t(14;16) (OCI-MY1, RPMI-8226, and ANBL-6), and 2/14 have both t(4;14) and t(14;16) (KMS-11 and KMS-26).34-38 KMS-11 and KMS-26, which have more than one primary translocation, were not used for analysis. The t(4;14) HMCL had the highest number of HDAC that were overexpressed (8/11; 72.72%) when compared to normal PC, with all of Class I HDAC, HDAC5, HDAC7, HDAC10, and HDAC11 significantly elevated (Table 1). Class I HDAC, HDAC5, and HDAC7 were also significantly elevated in t(11;14) HMCL (6/11; 54.54%), while t(14;16) HMCL showed overexpression of HDAC1, HDAC2, HDAC3, HDAC5, and HDAC10 and downregulation of HDAC6 (6/11; 54.54%). Secondary translocations, which are more complex and associated with tumor progression, are also prevalent in HMCL, most notably involving 8q24 (c-myc). Eleven of the HMCL used in this study have a translocation involving the c-myc locus (the exceptions being U266, ANBL-6, and JIM-1).39,40 The expression of the majority of the HDAC tested (7/11; 63.63%) was also significantly elevated in these HMCL (Table 1). Notably, HDAC1–3 were elevated in all HMCL irrespective of the translocations. These results demonstrate that HDAC are broadly dysregulated in MM with the dysregulation of the Class I HDACs being most prominent, but with no apparent correlation between the pattern of HDAC dysregulation and any particular karyotypic subsets of MM.
Table 1.
HDAC1-2 (Class I HDAC) are overexpressed in all HMCL
| Primary translocations |
Secondary translocations |
||||
|---|---|---|---|---|---|
| Gene | Normal | t(4;14) (FGFR3 and MMSET) | t(11;14) (CYCLIN D1) | t(14;16) (C-MAF) | 8q24 (c-MYC) |
| HDAC1 | 1.15±0.25 | 5.75±0.80**** | 7.73±2.04**** | 3.17±0.41*** | 6.67±0.95**** |
| HDAC2 | 1.18±0.26 | 4.83±0.48**** | 5.13±0.88**** | 2.21±0.31** | 4.33±0.39**** |
| HDAC3 | 1.24±0.29 | 3.44±0.76** | 4.00±0.71*** | 2.65±0.23** | 3.68±0.44**** |
| HDAC8 | 1.21±0.24 | 2.03±0.12** | 2.29±0.36* | 1.66±0.17 | 2.29±0.15*** |
| HDAC5 | 1.36±0.32 | 5.41±0.70**** | 6.23±1.24*** | 3.15±0.54** | 5.32±0.57**** |
| HDAC7 | 1.30±0.33 | 2.57±0.31** | 2.53±0.20* | 1.61±0.17 | 2.68±0.31** |
| HDAC6 | 1.21±0.81 | 1.57±0.26 | 2.70±0.93 | 0.69±0.08* | 1.82±0.37 |
| HDAC10 | 1.12±0.17 | 2.76±0.26**** | 1.47±0.34 | 2.25±0.36* | 2.53±0.22** |
| HDAC11 | 2.59±1.06 | 54.10±26.85* | 7.85±2.73 | 1.77±0/36 | 9.93±3.37 |
HMCLs were grouped based on their translocation status [t(4;14), t(11;14), t(14;16) and translocations involving c-myc locus – 8q24] and expression levels of HDAC were compared to those in normal plasma cells. HMCL with t(4;14) had significantly higher levels of HDAC (8/11) followed by translocations involving c-myc locus (7/11). HMCLs with t(11;14) and t(14;16) had 6/11 HDACs significantly dysregulated. Significant differences were observed using t-test and GraphPad Prism Version 5.0d. Gray-filled boxes represent significant upregulation in expression. Asterisks represent p-values (* <0.05, **<0.01, ***<0.001, ****<0.0001).
HDAC are dysregulated in primary MM, with higher expression correlating to significantly shorter survival
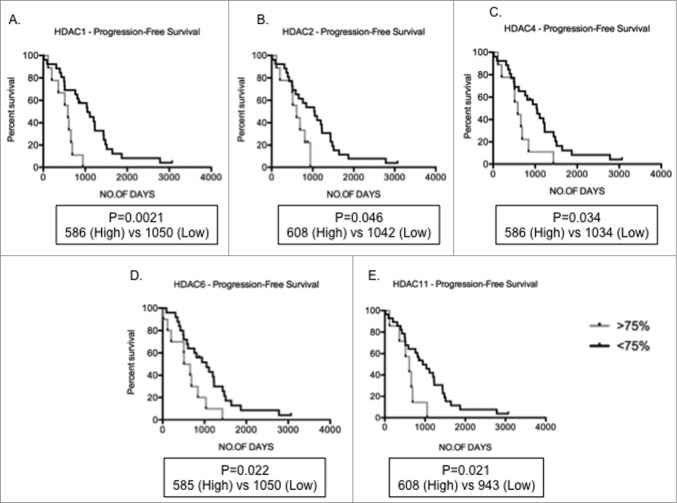
To determine if HDAC expression is abnormal in primary MM compared to normal plasma cells cohorts of purified MM tumor cells from both newly diagnosed and relapsed MM patients were evaluated. Table 2 illustrates the patient characteristics of the studied MM population. Analyses of gene expression levels of HDAC in primary MM cells (n = 55 ; clinical details available for n = 49 ) and normal PC (n = 9 ) revealed that the median levels of expression of all HDAC, with the exception of HDAC1, HDAC4, and HDAC11, were significantly elevated in MM, compared to normal PC, in which expression levels were normalized to one for that particular gene (Fig. 2). Median HDAC4 expression levels were reduced in comparison to normal PC. HDAC9 had the highest fold change in median expression levels when compared to normal PC (median 3.31) followed by HDAC3 (median 2.23) and HDAC7 (median: 2.20). The potential prognostic significance of dysregulated HDAC expression was then analyzed. The overall survival (OS – from diagnosis to time of death) and progression-free survival (PFS – from diagnosis to time of progression or death) in MM patients with high expression levels (≥ 75th centile) compared to low levels (≤ 75th centile) were derived utilizing Kaplan-Meier survival plots. Of the n = 55 MM samples used for qRT-PCR, 37 had information available to determine OS and PFS. Although the median level of expression of HDAC1 in MM patients was not significantly altered compared to normal PC, those patients with elevated HDAC1 expression had shorter PFS compared to patients with low expression (median PFS; High 586 vs Low 1050; P = 0 .0021; Fig. 3A). Similarly, MM patients with elevated HDAC2, HDAC4, HDAC6, and HDAC11 also showed shorter PFS compared to patients with lower expression levels (Fig. 3B-E) indicating that overexpression of HDAC is a predictor for poor prognosis.
Table 2.
Patient characteristics-Gene expression cohort
| MEDIAN AGE AT DIAGNOSIS | 57 YEARS (31-91) |
|---|---|
| MALE : FEMALE | 32 : 17 |
| EARLY : ADVANCED | 26 : 23 |
| ISS STAGE AT DIAGNOSIS I : II : III | 10 : 15 : 12 |
Table indicates characteristics of primary MM patient cohort utilized for assessment of HDAC1 -11 expression levels by quantitative RT-PCR.
Figure 2.
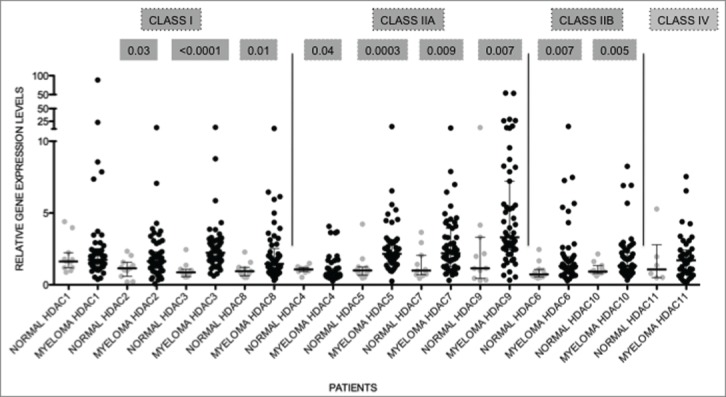
HDAC levels are dysregulated in primary MM Expression levels of HDAC in primary MM (n = 55 ) were compared to normal plasma cells (normal; n = 9 ). All genes, except HDAC1 and HDAC11, were significantly altered. Data are presented as median with range. Significant differences were calculated utilizing Mann-Whitney test in GraphPad Prism 5.0d. P-values are indicated.
Figure 3.
Increased HDAC levels correlate to poor prognosis Kaplan-Meir plots showing progression-free survival (PFS) in MM patients with high (>75th centile of MM patient cohort) compared to low (<75th centile) expression profiles. (A-E) Patients with high HDAC1, HDAC2, HDAC4, HDAC6, and HDAC11 expression depicted significantly shorter PFS days than patients with low levels of these genes. Significant differences and median survival days were calculated utilizing GraphPad Prism 5.0d. P-values are indicated.
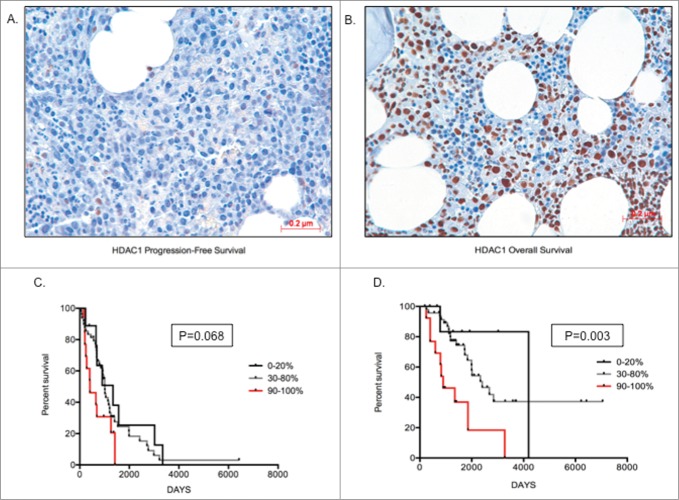
Increased HDAC1 protein expression correlates with poor overall and progression-free survival
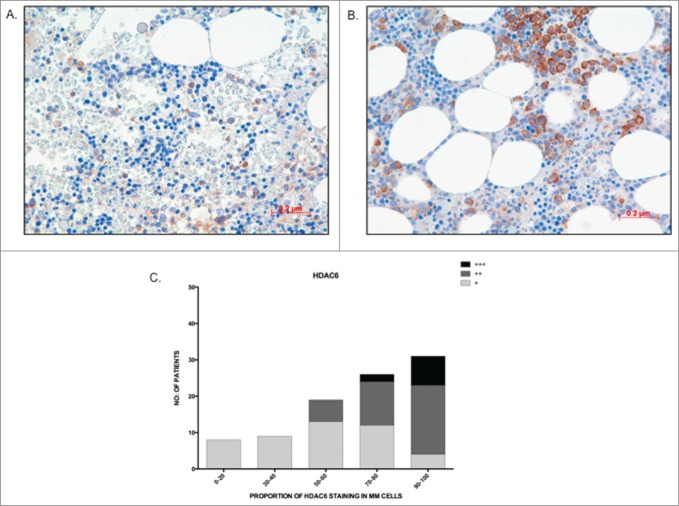
The qRT-PCR data from the primary MM cohort demonstrated that overexpression of Class I HDAC genes is significantly correlated with a poorer prognosis. To validate our findings at the protein level, a preliminary analysis of HDAC1–3 and HDAC6 protein expression in BM core biopsies from MM patients was first performed to determine the feasibility of such an approach. HDAC2 and HDAC3 protein expression was ubiquitous, being seen in all MM cells and in a majority of non-MM cells (data not shown); therefore, it was considered that a correlative analysis of HDAC2 and HDAC3 protein expression would be uninterpretable. In contrast, HDAC1 and HDAC6 expression was more MM cell-restricted, allowing MM cell specific determination to be undertaken. Consequently, validation via analysis of HDAC1 and HDAC6 MM cell protein expression was undertaken utilizing a second independent cohort of 97 uniformly treated transplant-eligible MM patients. Of the 97 tested patients, 79 patients showed assessable HDAC1 nuclear staining of MM cells and, within this cohort, 73.4% (58/79) of patients showed ≥50 % staining (Table 3). Representative images showing 20% HDAC1 staining and a second specimen with 90% HDAC1 staining are shown (Fig. 4A, B). Comparing patients with the lowest (0–20%) and highest (90–100%) HDAC1 expression demonstrated that MM patients with a higher proportion of MM cell HDAC1 expression had both shorter OS (P = 0 .003) and PFS (P = 0 .068) than MM patients with lower levels of expression (Fig. 4C, D). Similarly, for HDAC6, among the 97 tested patients, 93 showed assessable cytoplasmic staining. HDAC6 staining was categorized both quantitatively (number of positive MM cells), with 82% (76/93) of patients showing positivity in ≥50 % of MM cells (Table 3), and qualitatively (relative intensity of staining in positive MM cells), with MM cells graded as weak (+), moderate (++), or strong (+++) (Fig. 5B). Representative images of weak and strong HDAC6 staining in MM cells are shown in Fig. 5A, B. Patients with ≤40% MM cell positivity (17/93 patients; 18%) manifested only weak staining of HDAC6, whereas moderate staining intensity was present in the majority of the patients with ≥50% MM cell positivity (38% weak, 49% moderate, and 13% strong) (Fig. 5C). However, unlike HDAC1, there was no correlation between the level of HDAC6 expression and patient outcome (PFS and OS; data not shown). Furthermore, there was no correlation between the levels of HDAC1 and HDAC6 expression (data not shown).
Table 3.
Immunohistochemistry for HDAC1 and HDAC6 on bone marrow trephines of MM patients
| HDAC1 | HDAC6 |
|---|---|
| Total no. of samples: 97 | Total no. of samples: 97 |
| Assessable no. of samples: 79 | Assessable no. of samples: 93 |
| Proportions (no. of samples) | Proportions (no. of samples) |
| 0 (1) | 0 (2) |
| 10 (1) | 10 (3) |
| 20 (10) | 20 (3) |
| 30 (5) | 30 (4) |
| 40 (4) | 40 (5) |
| 50 (19) | 50 (9) |
| 60 (5) | 60 (10) |
| 70 (6) | 70 (9) |
| 80 (15) | 80 (17) |
| 90 (12) | 90 (18) |
| 100 (1) | 100 (13) |
Table representing the split in the proportions of MM cells positive for HDAC1 and HDAC6 in the samples assessed.
Figure 4.
Higher HDAC1 levels correlate with poor prognosis (A) Image depicting low staining of HDAC1 (20%) in MM cells; (B) Image depicting high proportion of HDAC1 staining (90%) in MM cells; (C) Kaplan-Meir survival plots for patients with 0–20%, 30–80%, and 90–100% proportion of HDAC1 staining, indicating patients with higher proportion of HDAC1 have shorter progression-free survival days; (D) Kaplan-Meir survival plots for patients with 0–20%, 30–80% and 90–100% proportion of HDAC1 staining indicating patients with higher proportion of HDAC1 have shorter overall survival days. Patients with >70% infiltration of MM cells was chosen for image capture. Significant differences and median survival days were calculated utilizing GraphPad Prism 5.0d and p-values are indicated.
Figure 5.
The majority of patients show high HDAC6 staining in MM cells (A) Image depicting low proportion (30%) and weak staining intensity (+) of HDAC6 in MM cells; (B) Image depicting high proportion (90%) and strong staining intensity of HDAC6 in MM cells; (C) Graph representing the distribution of HDAC6 staining proportion and intensity in bone marrow trephines tested. Higher proportion of HDAC6 staining correlated to stronger staining intensity.
Discussion
This comprehensive analysis of HDAC expression in MM has demonstrated that HDACs are dysregulated in MM and that overexpression of Class I HDACs, particularly HDAC1, are adverse prognostic factors. Although a number of studies have studied the expression and prognostic implications of altered levels of HDAC in cancer, this is the first study to characterize the expression pattern of HDACs in MM and correlate expression with patient outcomes.
In this study, the expression of HDAC at a transcriptional level was evaluated utilizing both genetically heterogeneous HMCL and primary MM and then compared to normal PC. This showed overexpression of class I HDAC (HDAC1, HDAC2, HDAC3, and HDAC8) in all HMCL tested, while only 2 of the Class II (HDAC5 and HDAC10) were overexpressed. Importantly, the analysis of primary MM tumors was performed on purified patient MM cells, thus eliminating any relevant contamination by non-MM bone marrow cells. While the median level of MM cell HDAC1 and HDAC11 gene transcription was not increased when compared to normal PC, patients with the highest levels of HDAC1, HDAC2, HDAC4, HDAC6, and HDAC11 all experienced shorter PFS when compared to patients with lower levels of expression. When we analyzed patient outcome based on MM cell HDAC1 protein expression in a second independent cohort of patients, overexpressors similarly demonstrated inferior outcomes. Our findings are consistent with studies in other malignancies demonstrating a correlation between dysregulation of Class I HDAC expression and prognosis. Immunohistochemistry on bone marrow core biopsies indicated that HDAC1, HDAC2, and HDAC3 were also present in limited populations of non-MM cells consistent with previously published result.41
Class I HDAC are the most studied class of HDAC and are known to be predominantly overexpressed, both in solid and hematological malignancies, with higher levels usually, but not always, correlating with a worse prognosis (reviewed in 9,42). HDAC1 and HDAC2 have significant functional redundancy in relation to proliferation, cell cycle, and apoptosis. 43 In addition, while the impact of specifically inhibiting HDAC1 or HDAC2 in MM cells is not yet known, we have previously clearly demonstrated that inhibition of Class I HDAC is superior to inhibition of HDAC6 in killing both HMCL and primary MM cells.32 Available evidence also suggests that MM resistance to proteasome inhibitors may be conferred through overexpression of HDAC1 but not overexpression of HDAC2 or HDAC3.33 HDAC1 overexpression could result in several MM-related physiological impacts including proliferation, through repression of tumor suppressor cyclin-dependent kinase inhibitor p21WAF1, and angiogenesis, by repression of p53 and induction of hypoxia-responsive genes hypoxia inducible factor (HIF-1) and vascular endothelial growth factor (VEGF).44 HDAC1 and HDAC2 are also recruited to the E-cadherin promoter by the SNAIL/SLUG family of transcription factors to downregulate E-cadherin, a gene vital for cell adhesion. Loss of E-cadherin promotes metastasis; therefore, overexpression of HDAC could potentially increase metastatic potential of MM cells.44 Collectively, these data and our findings described here suggest that dysregulation of Class I HDAC is a feature of MM that likely has a significant impact on disease biology. Dysregulation of HDACs and their potential to regulate multiple physiological processes justifies the ongoing efforts to better define the utilization of both pan- or Class I specific-HDAC inhibitors as potential anti-MM therapeutics. HDAC inhibitors have been utilized in MM therapy, with common toxicity related to off-target effects (gastrointestinal, fatigue, and reversible thrombocytopenia) due to the non-selectivity of pan-inhibitors.45 Based on our observations that HDAC1 overexpression has significant impact on the prognosis of MM patients, using HDAC1 selective inhibitors may help abrogate the common toxicities of pan-inhibitors.
While class I HDACs have been extensively studied, fewer studies have examined the expression of Class II and IV HDACs in cancers. The exception to this is Class IIB HDAC6, which has been previously evaluated as a potential therapeutic target in MM.29,30 Our data show that HDAC6 was dysregulated in primary MM, with higher levels correlating to poor prognosis. Moreover, on IHC assessment of BM core biopsies, it was evident that the majority of MM cells express HDAC6 protein; however, in contrast to transcriptional expression, we found no correlation between the level of HDAC6 protein expression and patient outcome. Whether overexpression of HDAC6 modulates the aggresome and proteasome pathways in MM and whether the impact o this is uniform across all MM patients, is yet to be determined. Furthermore, it is unclear if patients with higher levels of HDAC6 expression may be more responsive to HDAC6-specific inhibition when compared to patients with lower levels of expression.
The median level of expression of Class IIA HDAC4 was found to be significantly lower in primary MM when compared to normal PC; however, higher HDAC4 expression was associated with shorter PFS, suggesting that downregulated HDAC4 expression may be a favorable prognostic factor in MM. HDAC4 is known to act either as a tumor-suppressor or an oncogene in a highly context-dependent manner46 and is known to be involved in both the DNA repair process after ionizing radiation47 and the modulation of cellular proliferation through repression of p21.48,49 HDAC11 showed a remarkable degree of variability of expression, both in HMCL and in primary MM (Figs. 1 and 2). HDAC11 is functionally pleiotropic known to regulate immune response, interact with a number of chromatin modifying genes and to modulate RNA editing and processing.50,51 HDAC11 depletion is also known to reduce metabolic activity and the viability of cancer cells,51,52 suggesting that increased HDAC11 expression may be beneficial to the tumor cells. The functional consequence of overexpression in MM is uncertain and the further exploration of HDAC11 in MM would be of interest. Finally, given the known role of HDAC11 in modulating immune response, HDAC11 inhibition could perhaps be explored to enhance the immune response against MM, particularly in the context of present and emerging immunomodulatory approaches to MM.
In conclusion, it is evident from this study that HDAC are broadly dysregulated in MM, with Class I HDAC expression, particularly HDAC1, being the most aberrantly expressed. Importantly, we have shown that overexpression of HDAC is associated with inferior patient outcomes. Further studies exploring the protein expression pattern of other Class I and II HDACs (specifically HDAC8, HDAC5, and HDAC10) and the functional consequences of manipulating HDAC expression will help in determining the specific roles of these proteins in MM pathogenesis. Moreover, as a range of HDAC appear to be dysregulated in MM, the utilization of pan-HDAC inhibitors may represent a more rational approach for MM therapy than the use of isoform-specific inhibitors.
Materials and methods
Human myeloma cell lines
The human myeloma cell lines (HMCL) U266, NCI-H929, LP-1, and RPMI-8226 were obtained from the American Type Culture Collection (Rockville, MD, USA). OPM-2 was obtained from Deutshe SammLung von Mikro-orgaanismen und Zellculturen (Braunshwieig, Germany). KMS-11, KMS-12PE, KMS-12BM, KMS-18, and KMS-26 were kind gifts from Dr. Takemi Otsuki, Kawasaki Medical School, Japan. ANBL-6, XG-1 and OCI-MY1 were kind gifts from Winthrop P Rockefeller Cancer Institute (Little Rock, AR, USA). JIM-1 was from I. MacLennan from Birmingham, United Kingdom. HMCL were maintained in RPMI-1640 media supplemented with 10% heat inactivated fetal calf serum (FCS, Lonza, Victoria, Australia) and 2 mM L-glutamine (Life Technologies, Mulgrave, VIC, Australia).
Ficoll isolation of bone marrow mononuclear cells, determination of MM cell proportion, and isolation of CD138+ MM cells
Bone marrow (BM) from MM patients (newly diagnosed, relapsed/refractory) or non-myeloma patients (patients with hematological disorders and no known BM disease established through long term follow-up) was obtained following written informed consent as per Alfred Hospital Human Ethics Committee-approved protocol. Ficoll Paque Plus (GE Healthcare, Rydalmere, NSW, Australia) was utilized to isolate buffy layers containing bone marrow mononuclear cells (BMMNC) as per manufacturer's guidelines. Red blood cells were removed using red blood cell lysis buffer (10 mmol/L KHCO3, 150 mmol/L NH4Cl and 0.1 mmol/L EDTA, pH 8.0) for 5 minutes at 37°C followed by removal of any lysis buffer by washing with sterile phosphate buffered saline (PBS). Cells were then cultured overnight in RPMI-1640 media supplemented with 10% FCS and 2 mM L-glutamine. Proportion of MM or normal plasma cells (PC) in BMMNC isolated from each patient was determined through flow cytometric enumeration of CD138, CD38, and CD45 staining. Briefly, cells were stained with CD45-FITC (Becton Dickinson (BD) Biosciences, North Ryde, NSW, Australia), CD38-PerCP (BD Biosciences), and CD138-PE (Miltenyi Biotec, Macquarie Park, NSW, Australia) for 20 minutes at room temperature, washed, and resuspended in 300 μL of buffer (0.5% FCS/PBS). Samples were acquired on a BD FACSCalibur Flow Cytometer and proportion of CD38+/CD45-/CD138+ cells from MM BM was determined. To isolate MM cells, CD138 microbeads was employed using manufacturers guidelines (Miltenyi Biotec). Cells were washed in beads buffer (PBS/ 2mM EDTA / 0.5% BSA) and stained with microbeads for 20 mins. CD138+ cells were selected through magnetic isolation using an MS-column (Miltenyi Biotec). For normal BM, flow cytometry was performed to ensure that proportion of PC fell within the normal range of plasma cells (<3%). Normal plasma cells were isolated through the utilization of Plasma cell isolation kit II (Miltenyi biotec) according to manufacturer's instructions. The resultant population of cells was enriched for the proportion of PC. Purified cells were stored in TRIzol (Life Technologies).
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
Total RNA was from primary samples or HMCL were prepared using TRIzol or QIAGEN RNeasy mini kit, respectively (QIAGEN, Germantown, MD, USA) and any residual genomic DNA was removed utilizing the Turbo-DNase I kit (AMBION, Austin, TX, USA). The quality and quantity of the RNA obtained was assessed using Nanodrop 2000 Spectrophotometer and Quant-IT (Life Technologies), respectively. Reverse transcription was performed for 200 ng (primary samples and HMCL) of total RNA with 100 U of Superscript III reverse transcriptase (Life Technologies) and random hexamers (Life Technologies, 2.5 μM final concentration), according to manufactures guidelines. Quantitative RT-PCR was performed for HDAC1–11 in normal (n = 9 ) and MM plasma cells (n = 55 ) and HMCL (n = 14 ). Each HMCL had 3 biological replicates. The reaction mixture consisted of 8 μl of Applied Biosystems Power-SYBR PCR Master Mix with 500 nM of each forward (F) and reverse (R) primers for HDAC target genes and 2 μl of diluted template cDNA. PCR was performed in the LightCycler 480 instrument (Roche Diagnostics, Castle Hill, NSW, Australia) at 95oC for 10 min, with 45 cycles of amplification 95oC for 15s, 62oC for 30s, 72oC for 30s. Each reaction was performed in triplicate. Primers are listed in supplemental Table 1. For sample loading normalization, ACTB (encoding β-actin) was used and the mRNA levels of ACTB were not significantly different between samples. Amplified products were all verified by melting curves analysis. Data were analyzed using Roche LightCycler 480 software version 1.5 and copy number of target genes was determined by the comparative threshold cycle method (ΔΔCT) using the Pfaffl method.53 Data are presented as median with range (primary samples) or mean with SEM (HMCL), as described in the relevant figures. For statistical analyses of primary samples, non-parametric tests were utilized as the D'Agostino-Pearson normality test performed with the data set indicated that HDAC expression did not fit to a Gaussian distribution (data not shown).
Immunohistochemistry on bone marrow trephine sections
Immunohistochemistry (IHC) was undertaken on an automated immunostainer (Lab Vision Autostainer 360–2D; Lab Vision, Kalamazoo, MI, USA) according to the manufacturer's instructions. Paraffin-embedded BM trephine sections were decalcified and sequentially cut (3 μm thick sections), mounted, dewaxed, and immunostained. Peroxidase staining was done using Lab Vision UltraVision LP Large Volume Detection System HRP Polymer (Ready-to-Use). The IHC was independently scored by 2 specialist pathologists (A.K and A.C) to provide a consensus score. MM cell infiltration of the entire BM was scored in 10% increments based on morphologic assessment using both H & E and CD138 staining (clone MI15; 1:250; DAKO, Campbellfield, VIC, AUS). Sections were then scored for the presence of HDAC1 (polyclonal rabbit anti-HDAC1; 1:500; ab19845, Abcam, Cambridge, MA) or HDAC6 (Polyclonal rabbit anti-HDAC6; 1:200; H-300, Santa Cruz, Dallas, TX) with each stain scored in 10% increments according to the number of positive MM cells (assessed with reference to the total number of MM cells as described above) with the number of MM cells positive for HDAC1 or HDAC6 thus represented as a proportion. The presence and subcellular localization of HDAC1 and HDAC6 were also documented. Representative images of the sections (40x) were taken to depict high and low proportions of staining utilizing the NIKON optiphot 2 microscope.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Lona Estifo from Alfred Health Pathology Department for immunohistochemical analysis.
References
- 1. Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Blade J, Boccadoro M, Child JA, Avet-Loiseau H, Kyle RA, et al. International staging system for multiple myeloma. J Clin Oncol 2005; 23:3412-20; PMID:15809451; http://dx.doi.org/ 10.1200/JCO.2005.04.242 [DOI] [PubMed] [Google Scholar]
- 2. Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011; 471:467-72; PMID:21430775; http://dx.doi.org/ 10.1038/nature09837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer 2012; 12:335-48; PMID:22495321; http://dx.doi.org/ 10.1038/nrc3257 [DOI] [PubMed] [Google Scholar]
- 4. Kuehl WM, Bergsagel PL. Early genetic events provide the basis for a clinical classification of multiple myeloma. Hematol Am Soc Hematol Educ Prog 2005:346-52; PMID:16304402; http://dx.doi.org/ 10.1182/asheducation-2005.1.346 [DOI] [PubMed] [Google Scholar]
- 5. Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol 2005; 23:6333-8; PMID:16155016; http://dx.doi.org/ 10.1200/JCO.2005.05.021 [DOI] [PubMed] [Google Scholar]
- 6. Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 2005; 37:391-400; PMID:15765097; http://dx.doi.org/ 10.1038/ng1531 [DOI] [PubMed] [Google Scholar]
- 7. Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev 2009; 10:32-42; PMID:19065135; http://dx.doi.org/ 10.1038/nrg2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett 2009; 277:8-21; PMID:18824292; http://dx.doi.org/ 10.1016/j.canlet.2008.08.016 [DOI] [PubMed] [Google Scholar]
- 9. Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett 2009; 280:168-76; PMID:19103471; http://dx.doi.org/ 10.1016/j.canlet.2008.10.047 [DOI] [PubMed] [Google Scholar]
- 10. Rikimaru T, Taketomi A, Yamashita Y, Shirabe K, Hamatsu T, Shimada M, Maehara Y. Clinical significance of histone deacetylase 1 expression in patients with hepatocellular carcinoma. Oncology 2007; 72:69-74; PMID:18004079; http://dx.doi.org/ 10.1159/000111106 [DOI] [PubMed] [Google Scholar]
- 11. Weichert W, Denkert C, Noske A, Darb-Esfahani S, Dietel M, Kalloger SE, Huntsman DG, Kobel M. Expression of class I histone deacetylases indicates poor prognosis in endometrioid subtypes of ovarian and endometrial carcinomas. Neoplasia (New York, NY) 2008; 10:1021-7; PMID:18714364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weichert W, Roske A, Gekeler V, Beckers T, Ebert MP, Pross M, Dietel M, Denkert C, Rocken C. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol 2008; 9:139-48; PMID:18207460; http://dx.doi.org/ 10.1016/S1470-2045(08)70004-4 [DOI] [PubMed] [Google Scholar]
- 13. Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche FR, Niesporek S, Denkert C, Dietel M, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer 2008; 98:604-10; PMID:18212746; http://dx.doi.org/ 10.1038/sj.bjc.6604199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weichert W, Roske A, Niesporek S, Noske A, Buckendahl AC, Dietel M, Gekeler V, Boehm M, Beckers T, Denkert C. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res 2008; 14:1669-77; PMID:18347167; http://dx.doi.org/ 10.1158/1078-0432.CCR-07-0990 [DOI] [PubMed] [Google Scholar]
- 15. Yoshida N, Omoto Y, Inoue A, Eguchi H, Kobayashi Y, Kurosumi M, Saji S, Suemasu K, Okazaki T, Nakachi K, et al. Prediction of prognosis of estrogen receptor-positive breast cancer with combination of selected estrogen-regulated genes. Cancer Sci 2004; 95:496-502; PMID:15182430; http://dx.doi.org/ 10.1111/j.1349-7006.2004.tb03239.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast*. Breast Cancer Res Treat 2005; 94:11-6; PMID:16172792; http://dx.doi.org/ 10.1007/s10549-005-6001-1 [DOI] [PubMed] [Google Scholar]
- 17. Marquard L, Gjerdrum LM, Christensen IJ, Jensen PB, Sehested M, Ralfkiaer E. Prognostic significance of the therapeutic targets histone deacetylase 1, 2, 6 and acetylated histone H4 in cutaneous T-cell lymphoma. Histopathology 2008; 53:267-77; PMID:18671804; http://dx.doi.org/ 10.1111/j.1365-2559.2008.03109.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Skov V, Larsen TS, Thomassen M, Riley CH, Jensen MK, Bjerrum OW, Kruse TA, Hasselbalch HC. Increased gene expression of histone deacetylases in patients with Philadelphia-negative chronic myeloproliferative neoplasms. Leuk Lymphoma 2012; 53:123-9; PMID:21806350; http://dx.doi.org/ 10.3109/10428194.2011.597905 [DOI] [PubMed] [Google Scholar]
- 19. Wang JC, Chen C, Dumlao T, Naik S, Chang T, Xiao YY, Sominsky I, Burton J. Enhanced histone deacetylase enzyme activity in primary myelofibrosis. Leuk Lymphoma 2008; 49:2321-7; PMID:19052980; http://dx.doi.org/ 10.1080/10428190802527699 [DOI] [PubMed] [Google Scholar]
- 20. Marquard L, Poulsen CB, Gjerdrum LM, de Nully Brown P, Christensen IJ, Jensen PB, Sehested M, Johansen P, Ralfkiaer E. Histone deacetylase 1, 2, 6 and acetylated histone H4 in B- and T-cell lymphomas. Histopathology 2009; 54:688-98; PMID:19438744; http://dx.doi.org/ 10.1111/j.1365-2559.2009.03290.x [DOI] [PubMed] [Google Scholar]
- 21. Mimori K, Ogawa K, Okamoto M, Sudo T, Inoue H, Mori M. Clinical significance of enhancer of zeste homolog 2 expression in colorectal cancer cases. Eur J Surg Oncol 2005; 31:376-80; PMID:15837043; http://dx.doi.org/ 10.1016/j.ejso.2004.11.001 [DOI] [PubMed] [Google Scholar]
- 22. Moreno DA, Scrideli CA, Cortez MA, de Paula Queiroz R, Valera ET, da Silva Silveira V, Yunes JA, Brandalise SR, Tone LG. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br J Haematol 2010; 150:665-73; PMID:20636436; http://dx.doi.org/ 10.1111/j.1365-2141.2010.08301.x [DOI] [PubMed] [Google Scholar]
- 23. Van Damme M, Crompot E, Meuleman N, Mineur P, Bron D, Lagneaux L, Stamatopoulos B. HDAC isoenzyme expression is deregulated in chronic lymphocytic leukemia B-cells and has a complex prognostic significance. Epigenetics 2012; 7:1403-12; PMID:23108383; http://dx.doi.org/ 10.4161/epi.22674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aldana-Masangkay GI, Sakamoto KM. The role of HDAC6 in cancer. J Biomed Biotechnol 2011; 2011:875824; PMID:21076528; http://dx.doi.org/ 10.1155/2011/875824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ding G, Liu HD, Huang Q, Liang HX, Ding ZH, Liao ZJ, Huang G. HDAC6 promotes hepatocellular carcinoma progression by inhibiting P53 transcriptional activity. FEBS Lett 2013; 587:880-6; PMID:23402884; http://dx.doi.org/ 10.1016/j.febslet.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 26. Saji S, Kawakami M, Hayashi S, Yoshida N, Hirose M, Horiguchi S, Itoh A, Funata N, Schreiber SL, Yoshida M, et al. Significance of HDAC6 regulation via estrogen signaling for cell motility and prognosis in estrogen receptor-positive breast cancer. Oncogene 2005; 24:4531-9; PMID:15806142; http://dx.doi.org/ 10.1038/sj.onc.1208646 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Z, Yamashita H, Toyama T, Sugiura H, Omoto Y, Ando Y, Mita K, Hamaguchi M, Hayashi S, Iwase H. HDAC6 expression is correlated with better survival in breast cancer. Clin Cancer Res 2004; 10:6962-8; PMID:15501975; http://dx.doi.org/ 10.1158/1078-0432.CCR-04-0455 [DOI] [PubMed] [Google Scholar]
- 28. Osada H, Tatematsu Y, Saito H, Yatabe Y, Mitsudomi T, Takahashi T. Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int J Cancer 2004; 112:26-32; PMID:15305372; http://dx.doi.org/ 10.1002/ijc.20395 [DOI] [PubMed] [Google Scholar]
- 29. Hideshima T, Bradner JE, Wong J, Chauhan D, Richardson P, Schreiber SL, Anderson KC. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc Natl Acad Sci 2005; 102:8567-72; PMID:15937109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hideshima T, Richardson PG, Anderson KC. Mechanism of action of proteasome inhibitors and deacetylase inhibitors and the biological basis of synergy in multiple myeloma. Mol Cancer Ther 2011; 10:2034-42; PMID:22072815; http://dx.doi.org/ 10.1158/1535-7163.MCT-11-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Santo L, Hideshima T, Kung AL, Tseng JC, Tamang D, Yang M, Jarpe M, van Duzer JH, Mazitschek R, Ogier WC, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 2012; 119:2579-89; PMID:22262760; http://dx.doi.org/ 10.1182/blood-2011-10-387365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mithraprabhu S, Khong T, Jones SS, Spencer A. Histone deacetylase (HDAC) inhibitors as single agents induce multiple myeloma cell death principally through the inhibition of class I HDAC. Br J Haematol 2013; PMID:23692150; http://dx.doi.org/ 10.1111/bjh.12388 [DOI] [PubMed] [Google Scholar]
- 33. Kikuchi J, Wada T, Shimizu R, Izumi T, Akutsu M, Mitsunaga K, Noborio-Hatano K, Nobuyoshi M, Ozawa K, Kano Y, et al. Histone deacetylases are critical targets of bortezomib-induced cytotoxicity in multiple myeloma. Blood 2010; 116:406-17; PMID:20351311; http://dx.doi.org/ 10.1182/blood-2009-07-235663 [DOI] [PubMed] [Google Scholar]
- 34. Gazdar AF, Oie HK, Kirsch IR, Hollis GF. Establishment and characterization of a human plasma cell myeloma culture having a rearranged cellular myc proto-oncogene. Blood 1986; 67:1542-9; PMID:2423157 [PubMed] [Google Scholar]
- 35. Hitzler JK, Martinez-Valdez H, Bergsagel DB, Minden MD, Messner HA. Role of interleukin-6 in the proliferation of human multiple myeloma cell lines OCI-My 1 to 7 established from patients with advanced stage of the disease. Blood 1991; 78:1996-2004; PMID:1912582 [PubMed] [Google Scholar]
- 36. Jelinek DF, Ahmann GJ, Greipp PR, Jalal SM, Westendorf JJ, Katzmann JA, Kyle RA, Lust JA. Coexistence of aneuploid subclones within a myeloma cell line that exhibits clonal immunoglobulin gene rearrangement: clinical implications. Cancer Res 1993; 53:5320-7; PMID:8221668 [PubMed] [Google Scholar]
- 37. Kuipers J, Vaandrager JW, Weghuis DO, Pearson PL, Scheres J, Lokhorst HM, Clevers H, Bast BJ. Fluorescence in situ hybridization analysis shows the frequent occurrence of 14q32.3 rearrangements with involvement of immunoglobulin switch regions in myeloma cell lines. Cancer Genet Cytogenet 1999; 109:99-107; PMID:10087940; http://dx.doi.org/ 10.1016/S0165-4608(98)00157-5 [DOI] [PubMed] [Google Scholar]
- 38. Lombardi L, Poretti G, Mattioli M, Fabris S, Agnelli L, Bicciato S, Kwee I, Rinaldi A, Ronchetti D, Verdelli D, et al. Molecular characterization of human multiple myeloma cell lines by integrative genomics: insights into the biology of the disease. Genes Chromosomes Cancer 2007; 46:226-38; PMID:17171682; http://dx.doi.org/ 10.1002/gcc.20404 [DOI] [PubMed] [Google Scholar]
- 39. Dib A, Gabrea A, Glebov OK, Bergsagel PL, Kuehl WM. Characterization of MYC translocations in multiple myeloma cell lines. J Natl Cancer Inst Monogr 2008:25-31; PMID:18647998; http://dx.doi.org/ 10.1093/jncimonographs/lgn011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, Dewald G, Kirsch IR, Bergsagel PL, Kuehl WM. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci U S A 2000; 97:228-33; PMID:10618400; http://dx.doi.org/ 10.1073/pnas.97.1.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wada T, Kikuchi J, Nishimura N, Shimizu R, Kitamura T, Furukawa Y. Expression levels of histone deacetylases determine the cell fate of hematopoietic progenitors. J Biol Chem 2009; 284:30673-83; PMID:19736310; http://dx.doi.org/ 10.1074/jbc.M109.042242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barneda-Zahonero B, Parra M. Histone deacetylases and cancer. Mol Oncol 2012; 6:579-89; PMID:22963873; http://dx.doi.org/ 10.1016/j.molonc.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Segre CV, Chiocca S. Regulating the regulators: the post-translational code of class I HDAC1 and HDAC2. J Biomed Biotechnol 2011; 2011:690848; PMID:21197454; http://dx.doi.org/ 10.1155/2011/690848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene 2007; 26:5420-32; PMID:17694083; http://dx.doi.org/ 10.1038/sj.onc.1210610 [DOI] [PubMed] [Google Scholar]
- 45. Ocio EM, San Miguel JF. The DAC system and associations with multiple myeloma. Invest New Drugs 2010; 28 Suppl 1:S28-35; PMID:21120582; http://dx.doi.org/ 10.1007/s10637-010-9589-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clocchiatti A, Florean C, Brancolini C. Class IIa HDACs: from important roles in differentiation to possible implications in tumourigenesis. J Cell Mol Med 2011; 15:1833-46; PMID:21435179; http://dx.doi.org/ 10.1111/j.1582-4934.2011.01321.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kao GD, McKenna WG, Guenther MG, Muschel RJ, Lazar MA, Yen TJ. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J Cell Biol 2003; 160:1017-27; PMID:12668657; http://dx.doi.org/ 10.1083/jcb.200209065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilson AJ, Byun DS, Nasser S, Murray LB, Ayyanar K, Arango D, Figueroa M, Melnick A, Kao GD, Augenlicht LH, et al. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol Biol Cell 2008; 19:4062-75; PMID:18632985; http://dx.doi.org/ 10.1091/mbc.E08-02-0139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mottet D, Pirotte S, Lamour V, Hagedorn M, Javerzat S, Bikfalvi A, Bellahcene A, Verdin E, Castronovo V. HDAC4 represses p21(WAF1/Cip1) expression in human cancer cells through a Sp1-dependent, p53-independent mechanism. Oncogene 2009; 28:243-56; PMID:18850004; http://dx.doi.org/ 10.1038/onc.2008.371 [DOI] [PubMed] [Google Scholar]
- 50. Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol 2013; 9:672; PMID:23752268; http://dx.doi.org/ 10.1038/msb.2013.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, Nguyen D, Wright KL, Atadja PW, Bhalla K, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol 2009; 10:92-100; PMID:19011628; http://dx.doi.org/ 10.1038/ni.1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deubzer HE, Schier MC, Oehme I, Lodrini M, Haendler B, Sommer A, Witt O. HDAC11 is a novel drug target in carcinomas. Int J Cancer 2013; 132:2200-8; PMID:23024001; http://dx.doi.org/ 10.1002/ijc.27876 [DOI] [PubMed] [Google Scholar]
- 53. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; PMID:11328886; http://dx.doi.org/ 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.