Subcellular localization of mRNAs contributes to the generation of cellular asymmetries and cell fate determination. A comparative global analysis is given of animally and vegetally enriched RNAs in oocytes from two closely related Xenopus species.
Abstract
RNAs that localize to the vegetal cortex during Xenopus laevis oogenesis have been reported to function in germ layer patterning, axis determination, and development of the primordial germ cells. Here we report on the genome-wide, comparative analysis of differentially localizing RNAs in Xenopus laevis and Xenopus tropicalis oocytes, revealing a surprisingly weak degree of conservation in respect to the identity of animally as well as vegetally enriched transcripts in these closely related species. Heterologous RNA injections and protein binding studies indicate that the different RNA localization patterns in these two species are due to gain/loss of cis-acting localization signals rather than to differences in the RNA-localizing machinery.
INTRODUCTION
The closely related Pipid species Xenopus laevis (African clawed frog) and Xenopus (Silurana) tropicalis (western clawed frog) are members of two different clades of the Xenopus genus, originating from a common ancestor ∼50 million years ago (Bewick et al., 2012). Whereas X. laevis is allotetraploid, with 18 chromosome pairs and a genome size of 3.1 GB, X. tropicalis has a diploid karyotype, with 10 chromosome pairs and a genome size of 1.7 GB (Sater, 2012). A high degree of conservation has been described for the coding regions of orthologous genes in X. laevis and X. tropicalis (90% sequence identity; Yanai et al., 2011). Comparative transcriptome analysis revealed strongly conserved gene expression for the majority of orthologues, with changes affecting mainly the transcript levels rather than differences in the temporal profile of expression. Largest differences in expression levels were observed for maternally supplied transcripts (Yanai et al., 2011).
mRNA localization is understood to play an important role in early embryonic patterning and cell fate determination. In X. laevis, vegetally localized maternal mRNAs were found to be crucial for germ cell development and germ layer formation in the early embryo (King et al., 2005). Vegetal RNA localization in Xenopus oocytes is achieved by two major pathways. Early-pathway localization is initiated during the earliest stages of oogenesis by the entrapment of a subpopulation of mRNAs in the germplasm containing mitochondrial cloud, also referred to as Balbiani body; such RNAs become restricted to a relatively narrow region at the tip of the vegetal cortex, overlapping with the germplasm, and many such early pathway mRNAs have been found to be critical for proper germ cell development and migration (Houston, 2013). Late-pathway RNAs translocate toward the vegetal cortex at stages III–IV of oogenesis. In contrast to early-pathway RNAs, late-pathway transcripts localize to a much broader region of the vegetal cortex and function mainly during germ layer formation and patterning in the early embryo (White and Heasman, 2008). These two main localization pathways differ in the underlying mechanisms that drive vegetal enrichment. Whereas association of germplasm RNAs with the mitochondrial cloud is achieved by passive diffusion and entrapment, late-pathway RNAs are actively transported to the vegetal cortex and require dynein as well as kinesin motor proteins for proper localization (Betley et al., 2004; Chang et al., 2004; Messitt et al., 2008; Gagnon et al., 2013). In addition to the RNAs that associate with the vegetal cortex, a separate group of RNAs are enriched in the animal hemisphere of the oocyte, revealing a more diffuse distribution pattern (reviewed in King et al., 2005). The molecular mechanisms by which animal enrichment is achieved are largely unknown (Imbrie et al., 2012; Snedden et al., 2013).
RNA localization during anuran oogenesis has been most extensively studied in the Pipid X. laevis but has also been described to occur in Rana pipiens and Eleutherodactylus coqui, members of the Ranidae and Eleutherodactylidae families, respectively, which separated from the Pipidae ∼200 million years ago (Bossuyt and Roelants, 2009). However, vegetal localization of germ layer determinants such as vegt got lost in E. coqui after separation from the Ranidae ∼150 million years ago (Beckham et al., 2003; Bossuyt and Roelants, 2009). The mechanism by which germplasm RNAs such as dazl and ddx25 translocate to the vegetal cortex will differ in these species, since R. pipiens and E. coqui oocytes appear to be devoid of a mitochondrial cloud (Nath et al., 2005; Elinson et al., 2011).
RNA localization has also been reported to occur in the oocytes of the teleost Danio rerio. As in Xenopus, individual germplasm RNAs are transported via the Balbiani body to the oocyte vegetal cortex (Maegawa et al., 1999; Howley and Ho, 2000), and zebrafish RNAs localizing to the animal pole during late stages of oogenesis have also been described (Bally-Cuif et al., 1998; Houston, 2013). However, RNA localization does not seem to be strongly conserved between zebrafish and Xenopus, since several RNAs have been identified that localize in the zebrafish but not in the frog and vice versa (Bally-Cuif et al., 1998; Suzuki et al., 2000; Kosaka et al., 2007).
In the context of this study, we report on a comparative, genome-wide analysis for differentially localizing RNAs in X. laevis and X. tropicalis oocytes with RNAs isolated from vegetal or animal oocyte halves, respectively. Although we were able to identify a large group of novel vegetally localizing and animally enriched RNAs, there is only a very low degree of conservation with respect to the identity of individual such RNAs in a comparison between the two closely related Xenopus species. Furthermore, heterologous RNA localization assays and protein binding studies indicate that this is due to alterations in the RNA signal sequences rather than to differences in the RNA localization machinery.
RESULTS
Global RNA sequencing analysis identifies a large number of novel vegetally localizing transcripts in X. laevis oocytes
To achieve a global analysis of differentially localizing RNAs in X. laevis and X. tropicalis oocytes, we analyzed RNA preparations from animal and vegetal oocyte halves by next-generation sequencing. Sequences obtained were aligned to the transcript reference sequence collection of X. tropicalis and analyzed for differential enrichment in either hemisphere. With the exception of the noncoding xlsirts and gptl1 RNAs, which were not detected in the analysis, as well as exd2 and ldlrap1, all other X. laevis transcripts that were previously reported and proven to localize to the vegetal cortex by whole-mount in situ hybridization were also found to be significantly enriched in the vegetal hemisphere (Kloc et al., 1993; Zhou et al., 2004; Kataoka et al., 2005; Cuykendall and Houston, 2010; Supplemental Table S1). The degree of enrichment varies between a log2 fold change (FC) of 8.3 and 1.3. Selected transcripts (gdf1/vg1, dnd1, and vegt) were also analyzed by quantitative PCR (qPCR) and showed excellent quantitative correlation with the deep sequencing data (Supplemental Table S1). Depending on the stringency of the threshold applied, an additional 114 (log2FC ≥ 2) or 324 (log2FC ≥ 1) vegetally enriched RNAs were identified (Supplemental Table S2). We noticed considerable overlap of candidate genes identified in this study with those from two previously performed microarray-based screens for vegetally localizing RNAs in X. laevis oocytes (Horvay et al., 2006; Cuykendall and Houston, 2010; our unpublished results). An arbitrary selection of 34 novel candidate genes was assayed for vegetal localization by means of whole-mount in situ hybridization (Figure 1, Figure 4 later in this article, and Supplemental Figure S1). Of these, 14 were found to localize via the early and 12 via the late pathway. Although they were significantly enriched (log2FC between 2.25 and 5.02), the remaining eight clones were not found to localize or gave only weak signals. It could be that these RNAs are significantly enriched in the vegetal hemisphere but not associated with the vegetal cortex or germplasm. In summary, this RNA sequencing analysis confirms vegetal enrichment of previously published transcripts and yields a high number of novel, vegetally localizing RNAs that travel either the early or the late pathway in Xenopus oocytes.
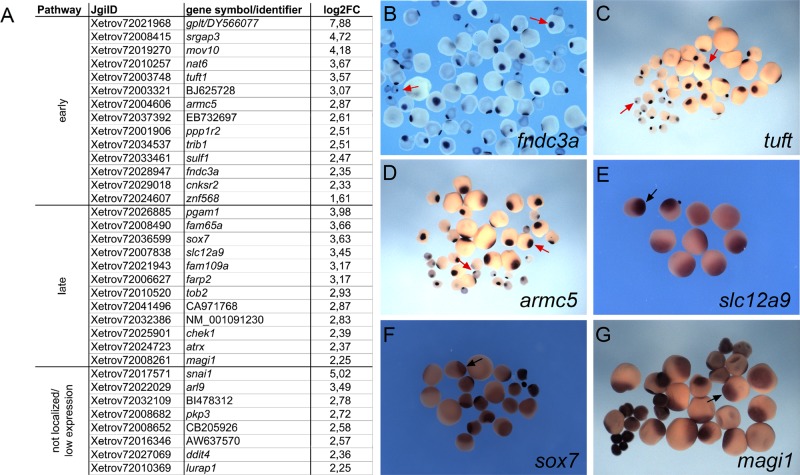
FIGURE 1:
Identification of novel vegetally localizing RNAs in X. laevis oocytes. (A) Candidate RNAs were tested for vegetal localization by in situ hybridization with X. laevis oocytes and listed according to their localization pattern (early and late). JgiID and gene symbol/GenBank accession number, as well as relative enrichment in the vegetal hemisphere as revealed by deep sequencing analysis (expressed as log2FC). RNAs for which no vegetal localization was detectable by in situ hybridization, as well as RNAs with very low expression levels that did not allow for the determination of localization patterns, are also listed. (B–D) Early-pathway localization pattern with characteristic mitochondrial cloud staining and spatially restricted localization at the vegetal pole (red arrows) for fnd3ca, tuft, and armc5 in stage I/II oocytes. (E–G) Late-pathway localization with typical broader vegetal cortex staining (black arrows) for slc12a9, sox7, and magi1 in stage III/IV oocytes.
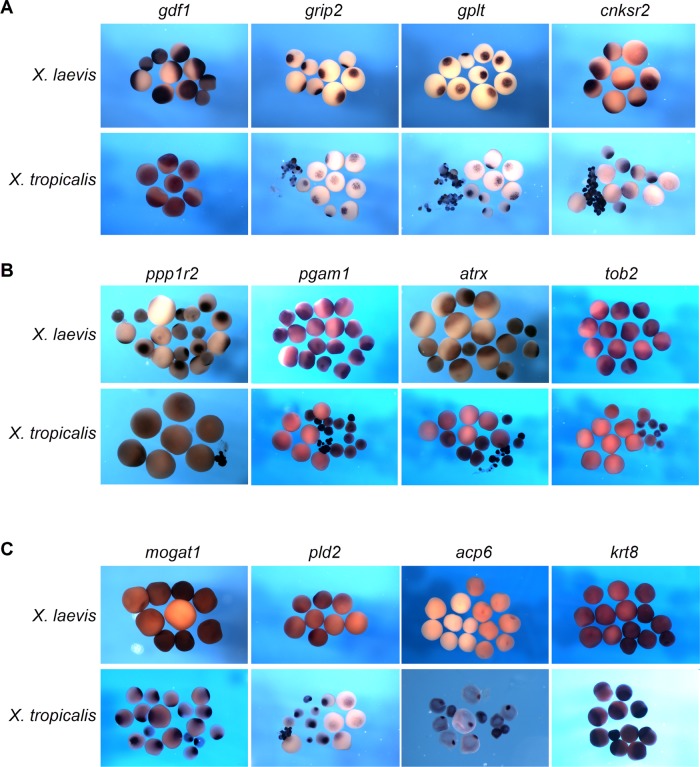
FIGURE 4:
Comparative in situ hybridization analysis confirms species-specific localization in X. laevis and X. tropicalis oocytes. (A–C) In situ hybridization with species-specific antisense RNA probes was performed with stage I–IV oocytes from X. laevis and X. tropicalis. (A) Gdf1, grip2, gplt, and cnksr2 localize to the vegetal cortex in both X. laevis and X. tropicalis oocytes. (B) Ppp1r2, pgam1, atrx, and tob2 vegetally localize in X. laevis only. (C) Mogat1, pld2, acp6, and krt8 transcripts localize to the vegetal cortex in X. tropicalis but not X. laevis oocytes.
RNA sequencing identifies many novel but not the previously known animally enriched RNAs in X. laevis oocytes
Surprisingly, fewer than one-third of the previously described animally localizing RNAs were found to be enriched in the animal hemisphere above a threshold of twofold (log2FC ≤ −1), and none of them had more than fourfold enrichment according to the RNA sequencing data (Supplemental Table S3). Lack of animal enrichment was further confirmed by quantitative reverse transcription PCR (qPCR) analysis for zfand4 and ddx3x (previously known as An1 and An3; Rebagliati et al., 1985; Gururajan et al., 1991; Linnen et al., 1993; Supplemental Table S3). On the other hand, deep sequencing analysis revealed a list of novel candidate RNAs with enrichment in the animal hemisphere: 338 with at least twofold and 51 with at least fourfold enrichment (Supplemental Table S4). Four of the novel animally enriched RNAs from X. laevis oocytes were selected for qPCR analysis and revealed very similar log2FC values as obtained by deep sequencing analysis (Figure 2A). To further verify animal enrichment, we cloned cDNA fragments of randomly selected candidate RNAs and performed in situ hybridization with staged X. laevis oocytes. Eight of 12 RNAs analyzed were enriched in the animal hemisphere (Figure 2 and Supplemental Figure S2). The remaining four RNAs tested had only very weak signals in the in situ hybridization, precluding classification as animally enriched. The animally enriched RNAs were either homogenously distributed within the animal cytoplasm or enriched within the perinuclear region rather than being restricted to the cortex. In summary, this study allows for the definition of a substantial group of novel, animally enriched RNAs, but it fails to verify many of the previously identified ones.
FIGURE 2:
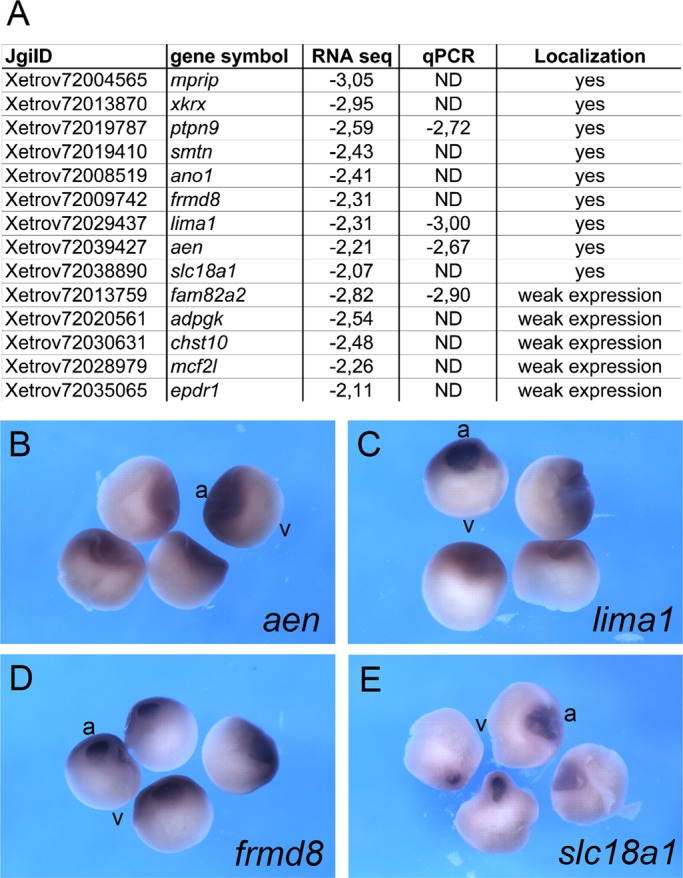
Identification of animally enriched RNAs in X. laevis oocytes. (A) Fourteen candidate transcripts with at least fourfold animal enrichment were selected for in situ hybridization analysis with X. laevis oocytes. JgiID, gene symbol, and relative enrichment in the animal hemisphere (expressed as log2FC) as revealed by deep sequencing and qPCR analysis. Detection of animal localization by in situ hybridization is indicated. For some of the transcripts, the localization pattern could not be determined due to very low expression levels. (B–E) Animal enrichment as revealed by in situ hybridization for aen, lima, frmd8, and scl18a1 transcripts. Bisected X. laevis stage VI oocytes. Animal (a) and vegetal (v) poles.
The identity of differentially localizing mRNAs is only weakly conserved in a comparison of the two closely related species X. laevis and X. tropicalis
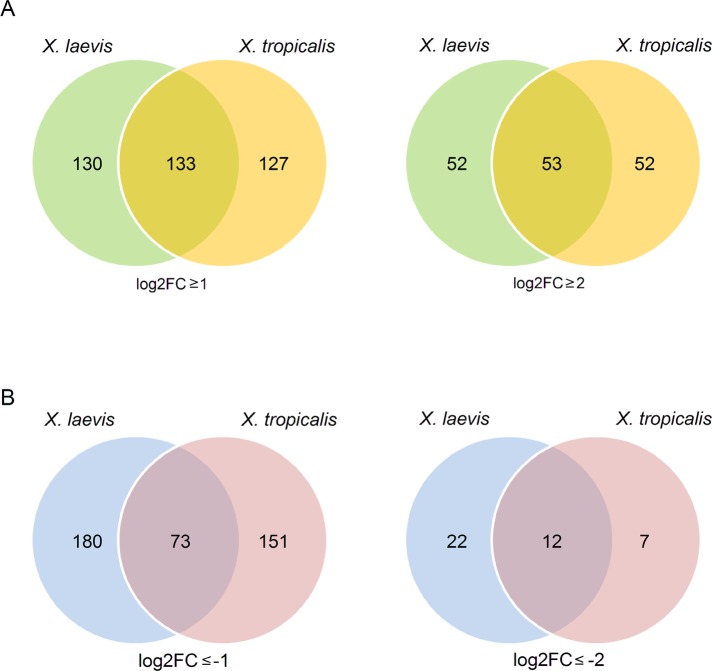
Similar to what we described for X. laevis, we also searched for vegetally localizing and animally enriched RNAs in oocytes from X. tropicalis. This analysis resulted in the definition of groups of RNAs with numbers very similar to what was observed for X. laevis (Supplemental Tables S5 and S6). In general, only about half of the transcripts that were classified to be vegetally localizing were shared between the two species, regardless of whether a relatively low threshold (log2FC ≥ 1) or a more stringent one (log2FC ≥ 2) was applied (Figure 3A and Supplemental Table S7). Similarly, the analysis of animally enriched RNAs in X. tropicalis also found an equally small overlap with animally enriched RNAs from X. laevis (Figure 3B and Supplemental Table S8). Scatterplot analysis of mean normalized counts per million for vegetal and animal samples indicate comparable general distributions of data points for X. laevis and X. tropicalis, with an enrichment of genes exhibiting higher expression levels in the group of vegetally enriched transcripts (Supplemental Figure S3).
FIGURE 3:
Differential RNA distribution is only weakly conserved in a comparison between X. laevis and X. tropicalis. (A) Numbers of vegetally enriched RNAs from X. laevis (green) and X. tropicalis (orange) as identified by deep sequencing analysis in the form of Venn diagrams. Thresholds for vegetal localization were set to either log2FC ≥ 1 (left) or ≥ 2 (right). (B) Numbers of RNAs with animal enrichment in X. laevis (blue) and X. tropicalis (pink) oocytes in the form of Venn diagrams with thresholds set to log2FC ≤ 1 (left) and ≤ 2 (right). This comparative analysis was restricted to transcripts with expression in oocytes from both species.
Conservation of vegetal RNA localization was verified for several transcripts by in situ hybridization, using species-specific probes on X. laevis and X. tropicalis oocytes, respectively. Transcripts classified to localize to the vegetal cortex in both species were verified for two novel and two previously known genes in both X. laevis and X. tropicalis oocytes (Figure 4A). Four different RNAs found to be vegetally enriched in X. laevis but not in X. tropicalis were verified to localize to the vegetal cortex in X. laevis oocytes only (Figure 4B). Finally, four RNAs vegetally enriched only in X. tropicalis indeed localize in X. tropicalis but not X. laevis oocytes (Figure 4C). Species-specific vegetal enrichment was also analyzed and confirmed by qPCR for a subset of the foregoing transcripts (Table 1). In summary, we conclude that vegetal localization and animal enrichment of maternal mRNAs are only weakly conserved in a comparison of X. laevis and X. tropicalis.
TABLE 1:
Summary of next-generation sequencing, qPCR, and in situ hybridization results for selected known and novel candidate RNAs.
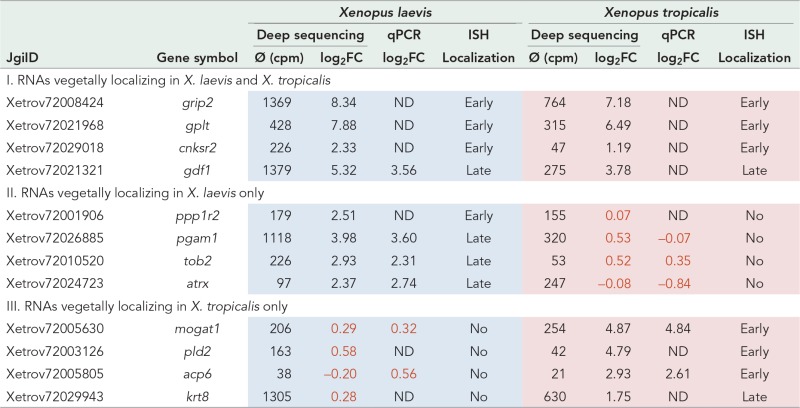 |
Vegetally enriched RNAs can be grouped into RNAs identified to localize in oocytes from both X. laevis and X. tropicalis (I), in X. laevis only (II), and in oocytes from X. tropicalis but not X. laevis (III). Candidate vegetally localizing transcripts that were selected for in situ hybridization (ISH) with X. laevis and X. tropicalis oocytes are listed with JgiID and gene symbol. Average Ø (cpm) per oocyte and relative enrichment in the vegetal hemisphere (expressed as log2FC) are as shown by deep sequencing analysis and qPCR, respectively. Values below the threshold for vegetal localization of log2FC ≥ 1 are indicated in red. Detection of vegetal localization by in situ hybridization and presumptive localization pathway is indicated.
Furthermore, gene ontology (GO) analysis of genes with conserved animal or vegetal enrichment, as well as species-specific enrichment in either hemisphere, was also performed (Supplemental Figures S4 and S5). Genes with conserved vegetal localization were enriched in categories associated with germ cell development, regulation of gene expression, and vesicular and lipid transport processes (Supplemental Figure S4). An enrichment of genes in categories associated with reproductive processes was also observed for transcripts with X. laevis–specific vegetal localization behavior. In addition, we also noticed enrichment in categories associated with cellular signaling, catabolic processes, and others. Gene ontology analysis for transcripts with X. tropicalis–specific vegetal enrichment revealed an enrichment of categories associated with cytoskeleton organization, cell cycle processes, skeletal system development, and protein catabolism. For transcripts with conserved animal enrichment, only one category of biological processes stood out, namely “response to drug,” which contains genes encoding for cortactin binding protein 2, a sorting nexin family member, acetyl-CoA carboxylase, and ABC superfamily members (Supplemental Figure S5). X. laevis–specific, animally enriched transcripts were enriched in categories associated with protein localization and Ras GTPase signal transduction, as well as nuclear export of mRNAs, whereas animally enriched transcripts from X. tropicalis were be enriched in categories associated with transcription of rRNA, mitotic cell cycle, and programmed cell death. Thus we observe a low conservation of GO term categories associated with species-specific animally and vegetally enriched transcripts, respectively.
Localization activity of endogenous RNAs can be recapitulated for synthetic RNAs injected into X. laevis and X. tropicalis oocytes
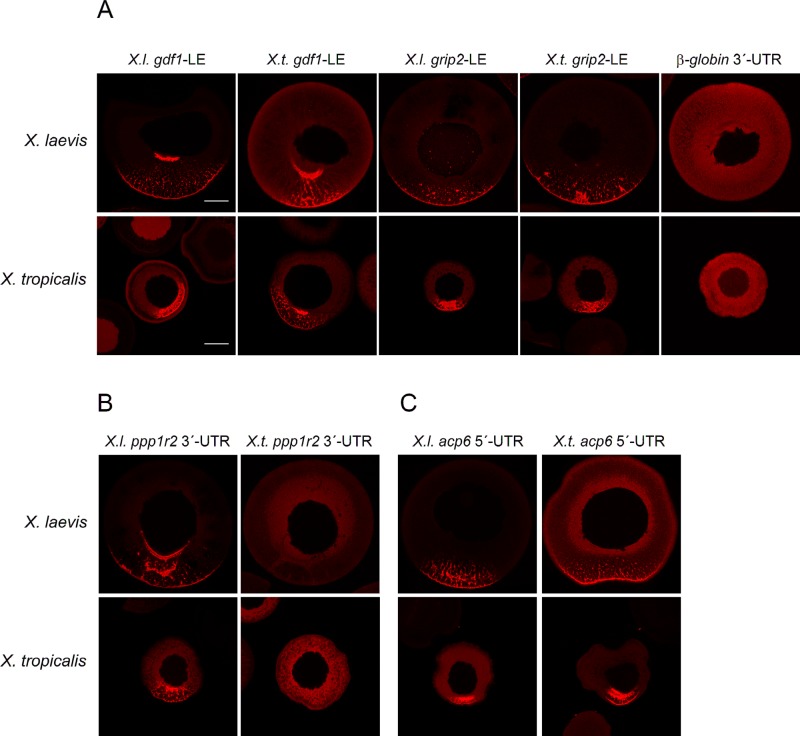
Differential localization behavior of transcripts in the two species analyzed here could be explained by either gain/loss of cis-acting RNA localization signals or differences in the trans-acting RNA localization machinery. These two possibilities were addressed by microinjection of synthetic RNA fragments from either X. laevis or X. tropicalis into oocytes from either the same or the other species. The conserved vegetal enrichment of gdf1 and grip2 could imply that both the functionality of cis-acting RNA signals and trans-acting localization proteins have been conserved between species. To test this hypothesis, we injected X. tropicalis RNA fragments corresponding to the localization elements (LEs) as identified in X. laevis (Mowry and Melton, 1992; Claussen et al., 2011) into early to mid-stage oocytes from X. laevis and X. tropicalis females and analyzed them for localization by confocal imaging. RNA localization elements from one species also localized in oocytes from the other species, whereas a control RNA (β-globin 3′-untranslated region [UTR]) did not localize in oocytes from both species (Figure 5A and Supplemental Figure S6C). RNA localization was quantified as described previously (Bauermeister et al., 2015) and given as average ratio of mean pixel intensity at the vegetal cortex versus mean pixel intensity in the animal cytoplasm (Supplemental Table S9).
FIGURE 5:
Differential localization behavior of orthologous RNAs appears to rely on the RNA signal sequence but not on differences in the RNA localization machinery. (A–C) Isolated localization elements, as well as 5′-UTR, ORF, and 3′-UTRs from different transcripts and species as indicated, were labeled with cyanine-3 and injected into X. laevis and X. tropicalis oocytes. Representative confocal images of fixed oocytes. Average vegetal/animal ratios of injected RNA are listed in Supplemental Table S9. Vegetal poles are oriented toward the bottom (if assignable). Scale bars, 100 μm. (A) Injection of gdf1 and grip2 localization elements from X. laevis and X. tropicalis, as well as nonlocalizing β-globin 3′-UTR (negative control), into oocytes from both species. (B) Injection of X. laevis and X. tropicalis ppp1r2 3′-UTRs. (C) Injection of X. laevis and X. tropicalis acp6-5′-UTRs.
Ppp1r2 belongs to the group of RNAs that were vegetally enriched in oocytes from X. laevis but not X. tropicalis (as described earlier). Because the localization element in ppp1r2 has not yet been characterized, the complete 3′-UTR was injected, since the vast majority of RNA localization elements have been mapped to this region. The X. laevis ppp1r2 3′-UTR indeed mediated vegetal localization after injection into X. laevis oocytes (Figure 5B). Vegetal localization was also achieved upon injection into X. tropicalis oocytes, showing that the localization machinery in X. tropicalis oocytes is able to recognize and mediate transport of the heterologous X. laevis RNA. Consistent with the ubiquitous distribution of ppp1r2 in X. tropicalis, as observed by in situ hybridization, the X. tropicalis 3′-UTR does not localize upon injection into X. tropicalis oocytes or into X. laevis oocytes, indicating that it is devoid of a functional localization element. Early-pathway localization activities, with characteristic enrichment in the mitochondrial cloud, were observed upon injection of X. laevis ppp1r2-3′-UTR into stage I oocytes, whereas X. tropicalis ppp1r2-3′-UTR remains evenly distributed in the cytoplasm or is even excluded from the mitochondrial cloud (Supplemental Figure S6, A and B).
Vegetal localization of acp6 transcripts has been detected in X. tropicalis but not in X. laevis oocytes (as described earlier). Injection of acp6 RNA fragments from X. laevis and X. tropicalis identified the vegetal localization activity in the 5′-UTR but not in the open reading frame (ORF) or 3′-UTR of the corresponding transcripts (Figure 5C and unpublished data). Of interest, the localization activities found in the injection assay are not consistent with the distribution of the endogenous RNAs in oocytes from the two different species; whereas endogenous acp6 RNA localizes in X. tropicalis oocytes only, injected 5′-UTRs from both species efficiently localized after injection into both X. laevis and X. tropicalis oocytes (Figure 5C). This finding indicates that the vegetal localization signal of the X. tropicalis acp6 mRNA could be masked by an unknown mechanism in the context of the full-length transcript.
Assuming that differences in the primary sequence of cis-acting vegetal localization elements are responsible for differential localization behavior in the two closely related Xenopus species, we compared 3′-UTRs from RNAs with conserved or species-specific localization activity. Indeed, we observe slightly higher degrees of average overall identities with 3′-UTRs from transcripts with conserved vegetal localization than with 3′-UTRs from transcripts with species-specific vegetal enrichment (Supplemental Table S10). However, because sequence identity values scatter widely within the groups of RNAs with conserved and nonconserved localization behavior, this correlation does not appear to provide a predictive value.
The proper localization of orthologous RNAs after injection into oocytes from a different species, as shown for gdf1-LE, grip2-LE, and ppp1r2-3′-UTR, indicates that the trans-acting localization machineries acting during vegetal localization seem to be conserved between X. laevis and X. tropicalis. To test whether similar vegetal localization complexes are assembled in X. tropicalis oocytes as described previously for X. laevis (Bauermeister et al., 2015), we used a ribonucleoprotein reconstitution approach, using X. tropicalis oocyte extracts and in vitro–transcribed early- and late-pathway LEs from X. laevis (gdf1-LE and grip2-LE). With the exception of Stau1 (Allison et al., 2004; Yoon and Mowry, 2004), X. tropicalis orthologues of known localization proteins, such as Igf2bp3 (Deshler et al., 1997; Havin et al., 1998), Ptbp1 (Cote et al., 1999), Elavl1 and 2 (Arthur et al., 2009), and Hnrnpab (Czaplinski et al., 2005), coeluted with gdf1- and grip2-LE RNAs but not or to a much lesser extent with the β-globin control RNA (Figure 6). Comparable protein binding to late-pathways LEs has been observed with X. laevis oocyte extracts (Bauermeister et al., 2015). The relatively strong interaction of Stau1 with the control RNA may be explained by a non–sequence-specific interaction of this double-stranded RNA binding domain–containing protein with the stem-loop structures present in the RNA-affinity tag.
FIGURE 6:
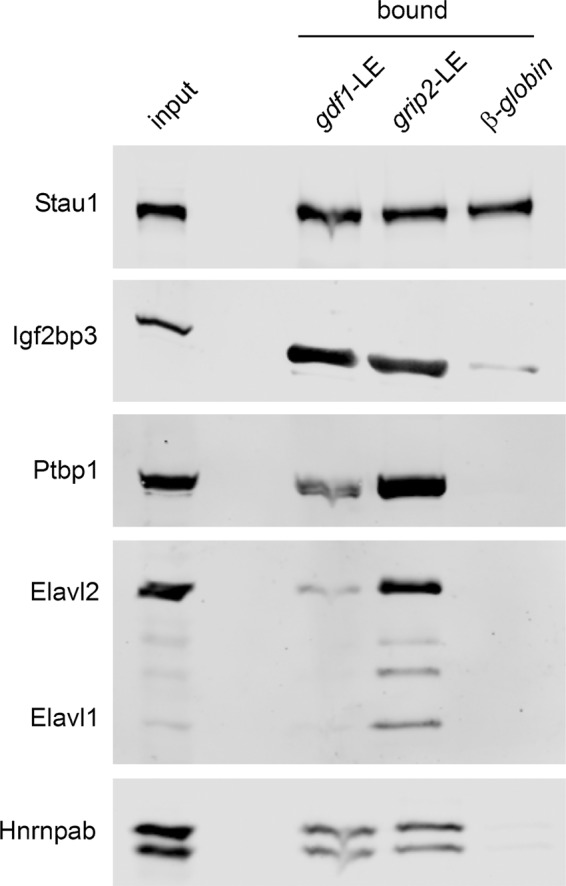
Interaction of localization proteins with LE-RNAs is conserved in X. tropicalis. Assembly of localization complexes with X. tropicalis oocyte extracts and tagged X. laevis gdf1-LE, grip2-LE, and β-globin-3′-UTR control RNA was performed in vitro. Copurifying localization proteins were detected by Western blot analysis.
In summary, data obtained here suggest that the differential localization behavior of orthologous RNAs is most likely due to differences in the cis-acting RNA signal sequences rather than differences in the trans-acting RNA localization machineries.
DISCUSSION
In this study, next-generation sequencing techniques were used for the identification of known and novel differentially localized transcripts in oocytes from X. laevis and the closely related X. tropicalis. Of interest, the overlap of differentially localizing RNAs in a comparison of the two species was found to be much lower than would have been expected from their close evolutionary relationship. Injection analysis suggests that this differential localization behavior is most likely caused by species-specific differences in the cis-acting RNA signals rather than in the trans-acting localization machineries.
Vegetally localizing RNAs function as important regulators during early Xenopus development, and numerous screens have been performed to isolate such transcripts from Xenopus oocyte RNA preparations (Rebagliati et al., 1985; Mosquera et al., 1993; Chan et al., 1999; Claussen and Pieler, 2004; Kataoka et al., 2005; Horvay et al., 2006; Cuykendall and Houston, 2010). Here we successfully used next-generation sequencing analysis for the identification of differentially enriched transcripts in Xenopus oocytes. Depending on the selected threshold for vegetal enrichment, 140 (at least fourfold enriched) to 350 (at least twofold enriched) different transcripts were identified from X. laevis oocytes. This is in good agreement with microarray analysis data, which led to an estimated minimal number of ∼275 vegetally localized RNAs (Cuykendall and Houston, 2010). About half of the top 120 enriched sequences identified in the same study were also vegetally enriched in the study described here. However, other RNAs were not enriched by a factor of more than twofold (e.g., nrf1, igf1r, and nucks1) or were found to be even animally enriched (e.g., exd2, sesn1, and trim69.1); thus additional analyses are required to prove vegetal enrichment of candidate genes identified in these various screens, including ours.
About 15–20% of the vegetally enriched transcripts with expression in both Xenopus species are encoded by unknown or nonannotated genes; when subjected to gene ontology analysis, the remaining genes were found to group into biological processes such as germ cell development, regulation of gene expression, and vesicular and lipid transport processes. Cellular signaling, catabolic, and other processes were found for transcripts with X. laevis unique vegetal enrichment, and cytoskeleton organization, cell cycle processes, skeletal system development, and protein catabolism were identified for transcripts with X. tropicalis–specific vegetal enrichment. However, a function of novel localizing transcripts in the given processes remains to be determined.
Although we were able to reidentify almost all of the previously published vegetally localizing RNAs in this study, this was not the case for RNAs with described animal enrichment. In general, the number of highly enriched RNAs (at least eightfold) is much lower for the group of RNAs with animal enrichment than for the group of vegetally enriched RNAs, indicating that animal enrichment in general may not result in a similarly steep gradient of animal–vegetal RNA distribution as shown for vegetally localizing RNAs (King et al., 2005; Sindelka et al., 2010). Because, for a subset of previously described animally localizing RNAs, animal enrichment has been identified in eggs or early embryos (Supplemental Table S3), it cannot be ruled out that differential RNA distribution might also be a result of differential RNA stability that might occur at egg maturation and thus not be detectable in the immature oocytes used in this study. However, animal enrichment has been verified by in situ hybridization as well as qPCR analysis for a group of selected novel candidate RNAs in the context of the present study. Knowledge about the effect of animally enriched RNAs on early Xenopus development is limited. The animally enriched ubiquitin ligase trim33 was shown to be essential for ectoderm specification by antagonizing mesoderm-inducing activities, thus allowing for the “default” neural induction (Dupont et al., 2005). Detailed in situ hybridization analysis with newly identified RNAs that exhibit enrichment in animal blastomeres of eight-cell-stage embryos revealed preferential expression of these RNAs in neural ectoderm during later developmental stages, which may indicate a similar function of these RNAs in the establishment of neural fate in ectodermal derivatives (Grant et al., 2014). Gene ontology analysis with animally enriched RNAs identified in this study revealed an enrichment of genes involved in biological processes, such as response to drug for transcripts with conserved animal enrichment, as well as protein and RNA transport, cell cycle processes, and regulation of small GTPase–mediated signal transduction for X. laevis–specific, animally enriched RNAs and transcription of rRNA, mitotic cell cycle, and programmed cell death for transcripts with X. tropicalis–specific animal enrichment. However, the effect of animally enriched transcripts associated with the aforementioned biological processes on neural ectodermal development in Xenopus remains to be determined.
Of interest, our analysis revealed a surprisingly low overlap of differentially enriched RNAs between X. laevis and X. tropicalis oocytes. These differences in localization behavior could be due to either loss or gain of localization activity in one of the species during evolution. However, to address this question, additional information about differential RNA distribution in oocytes from further related frog species is required. Comparative transcriptome analysis revealed strong conservation of temporal gene expression profiles during early development of X. laevis and X. tropicalis, with the largest differences in expression levels occurring in maternally supplied transcripts. This was consistent with the hourglass model of embryonic patterning (Yanai et al., 2011). According to this model, developmental constraints are highest during mid-embryogenesis, allowing for larger variations in gene expression before and after this critical phase (Kalinka and Tomancak, 2012). The weak conservation of spatial transcript enrichment found in this study might similarly reflect one aspect of higher divergence during early development of the two closely related Xenopus species.
Conserved localization behavior is likely to indicate a conserved developmental function of the respective genes in the two Xenopus species. However, the absence of conserved spatial RNA enrichment between the relatively closely related species may be due to a certain extent of functional redundancy of localized transcripts, which would have lowered the evolutionary pressure to maintain asymmetric localization of a particular transcript. Alternatively, additional fail-safe mechanisms may ensure restriction of transcripts to specific regions in the developing embryo even if vegetal localization during oogenesis is lost. For example, restriction of dnd1 and ddx25 transcripts to primordial germ cells be achieved not only by initial localization to the germplasm during oogenesis, but also by microRNA-mediated clearance of these transcripts in somatic cells and stabilization in the primordial germ cells in the developing embryo (Koebernick et al., 2010; Yamaguchi et al., 2014).
RNA localization assays with heterologous ppp1r2 3′-UTRs indicate that differences in the localization behavior are most likely due to alterations in the RNA signal sequence rather than differences in the RNA localization machinery in a comparison of the two Xenopus species. Sequence comparisons reveal an overall identity for the X. laevis and X. tropicalis ppp1r2 3′-UTRs of ∼70%, which is above the average sequence identity of ∼50% for 3′-UTRs from selected transcripts with differential behavior in the two species. However, even lower sequence identities were revealed for 3′-UTRs or isolated localization elements from RNAs with similar localization behavior in X. laevis and X. tropicalis (e.g., gdf1, nanos1). This indicates that differential localization behavior is not necessarily reflected by a lower degree of primary sequence conservation and vice versa. Similarly, it has been shown that even relatively small changes in primary sequence can alter the functional properties of RNA localization elements, either by changing protein binding activities or secondary structure of critical RNA regions (Claussen and Pieler, 2004).
The proper localization of orthologous RNAs after injection into oocytes from a different frog species indicates that the trans-acting vegetal localization machineries are conserved. In this context, a detailed comparative analysis of the gdf1-LEs from X. laevis and the closely related Kenyan clawed frog, Xenopus borealis, revealed that X. borealis and X. laevis gdf1-LE are recognized by the same localization proteins from X. laevis oocyte extracts, and both direct vegetal localization after injection into X. laevis oocytes (Lewis et al., 2004). Here we demonstrated that a similar set of orthologous localization proteins from X. tropicalis oocyte extracts, including Igf2bp3, Ptbp1, Elavl1 and 2, and Hnrnpab, assemble with X. laevis gdf1- and grip2-LEs, strongly indicating that vegetal localization machineries are conserved between different Xenopus species.
Of interest, microinjected acp6 5′-UTR did not recapitulate the localization behavior of the endogenous transcript; even though acp6 does not localize to the vegetal cortex in X. laevis oocytes, injected 5′-UTR mediates vegetal localization in oocytes from both species. This could indicate that the localization element of the X. tropicalis acp6 transcript is masked in the context of the full-length transcript. However, it has not been possible to identify this “masking activity” in the adjacent coding region (unpublished data), and it will be interesting to examine why such an inhibiting activity would be present in the 3′-UTR. Alternatively, vegetal localization of endogenous X. laevis acp6 transcripts may be inhibited by competition with other localizing RNAs for limiting components of the trans-acting localization machinery, as injection of high amounts of synthetic RNA may have uncovered this cryptic localization activity.
MATERIALS AND METHODS
RNA preparation from X. laevis and X. tropicalis oocytes
X. laevis and X. tropicalis ovaries were obtained by surgery from adult females and subjected to collagenase treatment (Liberase; Roche Diagnostics, Mannheim, Germany). Animal and vegetal oocyte halves were prepared by manual dissection using a scalpel blade and snap frozen in liquid nitrogen. RNA was prepared from either 10 or 30 animal and vegetal halves as described (Claussen and Pieler, 2004). Briefly, animal and vegetal halves were homogenized in RNA extraction buffer (50 mM Tris/HCl, pH 7.5, 5 mM EDTA, pH 8.0, 40 mM NaCl, 0.5% SDS) containing proteinase K (Merck, Darmstadt, Germany) and incubated for 1 h at 45°C, followed by several extraction steps with acidic phenol/chloroform/isoamyl alcohol and chloroform/isoamyl alcohol. Afterward, the RNA was precipitated by addition of an equal volume of 8 M LiCl. After being dissolve and reprecipitated with ammonium acetate/ethanol, the RNA was treated with DNase (Thermo Scientific, Waltham, MA).
RNA sequencing, data processing, and statistical analysis
For sequencing, the RNA samples were prepared with the TruSeq RNA Sample Prep Kit, version 2, according to the manufacturer’s instructions (Illumina, San Diego,CA) and sequenced using a HiSeq 2000 (Illumina). For X. laevis samples, 95–base pair paired-end runs were performed, whereas X. tropicalis samples were analyzed by 50–base pair single-end runs. Sequencing quality was checked via FastQC software (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Sequence images were transformed to BCL files with the Illumina BaseCaller software and demultiplexed to FASTQ files with CASAVA, version 1.8.2. Duplicated reads were not removed. To allow for direct comparison, sequences from both experimental setups (X. laevis and X. tropicalis) were aligned to the transcript reference sequence of X. tropicalis (by courtesy of Michael J. Gilchrist: http://genomics.nimr.mrc.ac.uk/resources/xenopus/Xt-transcripts-Xenbase-v72b-MJG-27oct11.fasta), taking unique gene identifiers only. Read mapping was performed with Bowtie2, version 2.1.0, using the “very sensitive” local alignment mode and standard parameters for alignment scoring (Langmead and Salzberg, 2012). Roughly, this allows up to six mismatches (12%) in reads of 50 base pairs and 39 mismatches (∼21%) in 2× 95–base pair paired-end reads. Subsequently conversion of resulting .sam files to sorted .bam files, indexing, and filtering of unique hits (MAPQ value >20, “-q20”) was conducted with Samtools, version 0.1.19. Counting of hits to transcript was performed using the Samtools idxstat function (with subsequent halving of counts for paired-end reads). Data were preprocessed and analyzed in the R/Bioconductor environment (www.bioconductor.org), loading edgeR and biomaRt packages (Robinson et al., 2010). After filtering, genes with >50 counts for at least half of the samples were selected and normalized to trimmed mean of M values and dispersions estimated. Furthermore, genes were filtered for group (animal and vegetal) means >50 counts and tested for differentially expressed genes based on a generalized linear model likelihood ratio test, assuming negative binomial data distribution via edgeR. For comparability between the two different data sets (X. laevis and X. tropicalis), counts per million (cpm) were calculated, and thresholds were set to mean cpm for at least one group (animal or vegetal) in each data set ≥5. Fold changes (ratios) between animal and vegetal hemispheres were calculated as log2 values (log2FC). Gene annotation was enriched by data from Xenbase. The assignment to previously identified vegetally enriched candidate genes was performed by BLAST searches (Altschul et al., 1990) of the X. tropicalis reference sequence collection with UniGeneID sequences from Cuykendall and Houston (2010), as well as candidate sequences from our own microarray screen (unpublished results). The data discussed in this article have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus and are accessible through GEO Series accession number GSE58420 (Edgar et al., 2002).
Gene ontology term analysis
GO term analysis was performed using the functional annotation tool of DAVID, version 6.7 (Huang da et al., 2009a, b). Homo sapiens, Mus musculus, X. laevis, X. tropicalis, D. rerio, and Drosophila melanogaster were selected to limit annotations of the gene list, and H. sapiens was selected as population background. GO term analysis was performed on gene lists of transcripts with conserved and species-specific animal or vegetal enrichment.
Pairwise sequence alignments
Pairwise global nucleotide sequence alignments were performed with EMBOSS Needle, applying the default alignment settings (Rice et al., 2000).
Cloning procedures
For verification of differential RNA localization, in situ hybridization with X. laevis and X. tropicalis oocytes was performed. Gene- and species-specific in situ hybridization probes were generated by cloning of the corresponding cDNA fragments into pGEMTeasy (Promega, Madison, WI). Primers were designed to match the corresponding nucleotide reference sequence listed in Xenbase (Supplemental Table S11). In cases in which no gene had been assigned to the corresponding JgiID sequence, BLAST searches (Altschul et al., 1990) with these sequences were performed and primers designed for the best-matching expressed sequence tag (EST) sequence from X. laevis/X. tropicalis. GenBank accession numbers of these transcripts are given in the respective tables and figures. The sequence with the JgiID Xetrov72021968 best matched to ESTs DY566077 (X. laevis) and BX772720 (X. tropicalis) and was named germ plasm localized transcript (gplt) according to the localization observed in oocytes from both Xenopus species.
For the generation of Cy3-labeled and lacZ-tagged injection constructs, cDNA fragments corresponding to the 5′-UTR, ORF, 3′-UTR, or localization sequence were amplified by PCR and cloned into BamHI/XhoI or EcoRI sites of pBK-CMV-lacZ (Claussen and Pieler, 2004). Oligonucleotides used for cloning of cDNA subfragments into pBK-CMV-lacZ are listed in Supplemental Table S12.
Quantitative RT-PCR analysis
To assess the relative enrichment of candidate RNAs in either animal or vegetal hemispheres, quantitative RT analysis using the iQ SYBR Green Super Mix and CFX 96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA) was performed on the RNA preparations as for RNA sequencing analysis. Mean relative enrichments were determined for two biological replicates and two technical replicates, respectively, and are given as log2FC. Oligonucleotides used for quantiative RT-PCR analysis are listed in Supplemental Table S13.
In situ hybridization analysis
In situ hybridization analysis on X. laevis and X. tropicalis oocytes was as described (Harland, 1991; Hollemann et al., 1999) using digoxigenin-labeled antisense RNA probes.
In vitro synthesis of cyanine-3– and Atto-633–labeled transcripts and RNA localization assays
In vitro synthesis of cyanine-3–labeled transcripts and oocyte injection was as described (Claussen and Pieler, 2010). For synthesis of Atto-633–labeled transcripts, 1 nmol of aminoallyl-UTP-Atto-633 (NU-821-633; Jena Bioscience, Jena, Germany) was added to a 25 μl of in vitro transcription reaction. In contrast to the previously used protocols, X. laevis and X. tropicalis mid-stage oocytes were incubated in vitellogenin-free oocyte culture medium after injection and analyzed by confocal microscopy using the LSM 780 (Zeiss, Oberkochen, Germany). Fixation, clearing, and imaging of injected oocytes were done as described in Gagnon and Mowry (2011). Quantification of vegetal enrichment was performed as described previously (Bauermeister et al., 2015). In brief, confocal images of injected oocytes were opened in ImageJ (National Institutes of Health, Bethesda, MD), and mean pixel intensities at the vegetal cortex and in the animal cytoplasm were measured by using the segmented line tool and used to calculate the average vegetal/animal ratio of pixel intensity. For transcripts with no vegetal localization activity, the assignment of animal and vegetal poles is hampered. In these cases, mean pixel intensities of arbitrarily defined opposite cytoplasmic regions were used for quantification.
RNA affinity purification
For in vitro assembly of localization complexes from X. tropicalis oocyte extracts, X. laevis gdf1- and grip2-LEs were cloned into BamHI/XhoI sites of the PP7 RNA-tag–containing pBluescript II derivative (Bauermeister et al., 2015). XhoI-linearized plasmids were used as templates for T7-mediated in vitro synthesis of PP7-tagged RNAs (MEGA script T7; Ambion; Thermo Scientific). Gdf1-LE, grip2-LE, and β-globin 3′-UTRs were immobilized on ZZ-tev-PP7coat protein bound immunoglobulin G–agarose, and RNA affinity purification was essentially as described previously (Bauermeister et al., 2015). For the preparation of X. tropicalis oocyte S16 extracts, ∼250 μl of early- and late-stage oocytes were homogenized in 500 μl of IPP145 buffer (50 mM Tris-HCl, pH 8, 145 mM NaCl, 5% glycerol, 0.05% NP-40 substitute, 1 mM dithiothreitol) with protease inhibitors (Complete; Roche) and centrifuged for 30 min at 16,000 × g. Reconstitution of localization complexes and washing and elution steps were all performed in IPP145 buffer. S16 input and eluates were subsequently analyzed by Western blot for copurification of X. tropicalis localization proteins using the following antibodies: anti-Igf2bp3 (Zhang et al., 1999), 1:5000; anti-Ptbp1 (4E11; BSBS Antibody Facility, Braunschweig, Germany), 1:10; anti-HuR (sc-5261; Santa Cruz Biotechnology, Dallas, TX), 1:5000; anti-Hnrnpab (Czaplinski et al., 2005), 1:10,000; and anti-Stau1 (Allison et al., 2004), 1:5000. All antibodies showed cross-reactivity with orthologous proteins from X. tropicalis. Secondary IRDye 680– or 800–labeled anti-mouse and anti-rabbit antibodies (Li-Cor, Lincoln, NE) were used at 1:20,000 dilutions and detection was performed using a Li-Cor Odyssey infrared imaging system.
Note added in proof. While the manuscript was under review, De Domenico et al. (2015) reported on the spatial distribution of maternal mRNAs in the X. tropicalis eight-cell-stage embryo as revealed by single-blastomere sequencing. The vast majority of novel transcripts with consistent differential enrichment reported in their analysis were also identified to be asymmetrically distributed in our study.
Supplementary Material
Acknowledgments
We thank Mike Gilchrist for providing access to the X. tropicalis transcript reference sequence collection and helpful advice during the initial phase of this study, as well as Gregor Bucher for discussions and Marion Dornwell for excellent technical assistance. Antibodies were kindly provided by J. Yisraeli (Igf2bp3), K. Czaplinski and I.W. Mattaj (Hnrnpab), and N. Standart (Stau1).
Abbreviations used:
- ISH
in situ hybridization
- qPCR
quantitative reverse transcription PCR
- UTR
untranslated region.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-02-0115) on September 2, 2015.
REFERENCES
- Allison R, Czaplinski K, Git A, Adegbenro E, Stennard F, Houliston E, Standart N. Two distinct Staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA. 2004;10:1751–1763. doi: 10.1261/rna.7450204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arthur PK, Claussen M, Koch S, Tarbashevich K, Jahn O, Pieler T. Participation of Xenopus Elr-type proteins in vegetal mRNA localization during oogenesis. J Biol Chem. 2009;284:19982–19992. doi: 10.1074/jbc.M109.009928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally-Cuif L, Schatz WJ, Ho RK. Characterization of the zebrafish Orb/CPEB-related RNA binding protein and localization of maternal components in the zebrafish oocyte. Mech Dev. 1998;77:31–47. doi: 10.1016/s0925-4773(98)00109-9. [DOI] [PubMed] [Google Scholar]
- Bauermeister D, Claussen M, Pieler T. A novel role for Celf1 in vegetal RNA localization during Xenopus oogenesis. Dev Biol. 2015;405:214–224. doi: 10.1016/j.ydbio.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Beckham YM, Nath K, Elinson RP. Localization of RNAs in oocytes of Eleutherodactylus coqui, a direct developing frog, differs from Xenopus laevis. Evol Dev. 2003;5:562–571. doi: 10.1046/j.1525-142x.2003.03061.x. [DOI] [PubMed] [Google Scholar]
- Betley JN, Heinrich B, Vernos I, Sardet C, Prodon F, Deshler JO. Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr Biol. 2004;14:219–224. doi: 10.1016/j.cub.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Bewick AJ, Chain FJ, Heled J, Evans BJ. The pipid root. Syst Biol. 2012;61:913–926. doi: 10.1093/sysbio/sys039. [DOI] [PubMed] [Google Scholar]
- Bossuyt F, Roelants K. Frogs and toads (Anura) In: Hedges SB, Kumar S, editors. The Timetree of Life. Oxford, UK: Oxford University Press; 2009. pp. 357–364. [Google Scholar]
- Chan AP, Kloc M, Etkin LD. fatvg encodes a new localized RNA that uses a 25-nucleotide element (FVLE1) to localize to the vegetal cortex of Xenopus oocytes. Development. 1999;126:4943–4953. doi: 10.1242/dev.126.22.4943. [DOI] [PubMed] [Google Scholar]
- Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, King ML. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell. 2004;15:4669–4681. doi: 10.1091/mbc.E04-03-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen M, Pieler T. Xvelo1 uses a novel 75-nucleotide signal sequence that drives vegetal localization along the late pathway in Xenopus oocytes. Dev Biol. 2004;266:270–284. doi: 10.1016/j.ydbio.2003.09.043. [DOI] [PubMed] [Google Scholar]
- Claussen M, Pieler T. Identification of vegetal RNA-localization elements in Xenopus oocytes. Methods. 2010;51:146–151. doi: 10.1016/j.ymeth.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Claussen M, Tarbashevich K, Pieler T. Functional dissection of the RNA signal sequence responsible for vegetal localization of XGrip2.1 mRNA in Xenopus oocytes. RNA Biol. 2011;8:873–882. doi: 10.4161/rna.8.5.16028. [DOI] [PubMed] [Google Scholar]
- Cote CA, Gautreau D, Denegre JM, Kress TL, Terry NA, Mowry KL. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- Cuykendall TN, Houston DW. Identification of germ plasm-associated transcripts by microarray analysis of Xenopus vegetal cortex RNA. Dev Dyn. 2010;239:1838–1848. doi: 10.1002/dvdy.22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Kocher T, Schelder M, Segref A, Wilm M, Mattaj IW. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-beta-related mRNA during oogenesis. Dev Cell. 2005;8:505–515. doi: 10.1016/j.devcel.2005.01.012. [DOI] [PubMed] [Google Scholar]
- De Domenico E, Owens ND, Grant IM, Gomes-Faria R, Gilchrist MJ. Molecular asymmetry in the 8-cell stage Xenopus tropicalis embryo described by single blastomere transcript sequencing. Dev Biol. 2015;2015 doi: 10.1016/j.ydbio.2015.06.010. pii: S0012-1606(15)30015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinson RP, Sabo MC, Fisher C, Yamaguchi T, Orii H, Nath K. Germ plasm in Eleutherodactylus coqui, a direct developing frog with large eggs. EvoDevo. 2011;2:20. doi: 10.1186/2041-9139-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Kreiling JA, Powrie EA, Wood TR, Mowry KL. Directional transport is mediated by a Dynein-dependent step in an RNA localization pathway. PLoS Biol. 2013;11:e1001551. doi: 10.1371/journal.pbio.1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Mowry KL. Visualization of mRNA localization in Xenopus oocytes. Methods Mol Biol. 2011;714:71–82. doi: 10.1007/978-1-61779-005-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Yan B, Johnson MA, Johnson DL, Moody SA. Novel animal pole-enriched maternal mRNAs are preferentially expressed in neural ectoderm. Dev Dyn. 2014;243:478–496. doi: 10.1002/dvdy.24082. [DOI] [PubMed] [Google Scholar]
- Gururajan R, Perry-O’Keefe H, Melton DA, Weeks DL. The Xenopus localized messenger RNA An3 may encode an ATP-dependent RNA helicase. Nature. 1991;349:717–719. doi: 10.1038/349717a0. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 1998;12:1593–1598. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollemann T, Panitz F, Pieler T. In situ hybridization techniques with Xenopus embryos. In: Richter JD, editor. A Comparative Methods Approach to the Study of Oocytes and Embryos. Oxford, UK: Oxford University Press; 1999. pp. 279–290. [Google Scholar]
- Horvay K, Claussen M, Katzer M, Landgrebe J, Pieler T. Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev Biol. 2006;291:1–11. doi: 10.1016/j.ydbio.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Houston DW. Regulation of cell polarity and RNA localization in vertebrate oocytes. Int Rev Cell Mol Biol. 2013;306:127–185. doi: 10.1016/B978-0-12-407694-5.00004-3. [DOI] [PubMed] [Google Scholar]
- Howley C, Ho RK. mRNA localization patterns in zebrafish oocytes. Mech Dev. 2000;92:305–309. doi: 10.1016/s0925-4773(00)00247-1. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Imbrie GA, Wu H, Seldin DC, Dominguez I. Asymmetric localization of Ck2α during Xenopus oogenesis. Hum Genet Embryol. 2012;Suppl 4:11328. doi: 10.4172/2161-0436.S4-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka AT, Tomancak P. The evolution of early animal embryos: conservation or divergence? Trends Ecol Evol. 2012;27:385–393. doi: 10.1016/j.tree.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Tazaki A, Kitayama A, Ueno N, Watanabe K, Mochii M. Identification of asymmetrically localized transcripts along the animal-vegetal axis of the Xenopus egg. Dev Growth Differ. 2005;47:511–521. doi: 10.1111/j.1440-169X.2005.00826.x. [DOI] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- Kloc M, Spohr G, Etkin LD. Translocation of repetitive RNA sequences with the germ plasm in Xenopus oocytes. Science. 1993;262:1712–1714. doi: 10.1126/science.7505061. [DOI] [PubMed] [Google Scholar]
- Koebernick K, Loeber J, Arthur PK, Tarbashevich K, Pieler T. Elr-type proteins protect Xenopus Dead end mRNA from miR-18-mediated clearance in the soma. Proc Natl Acad Sci USA. 2010;107:16148–16153. doi: 10.1073/pnas.1004401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka K, Kawakami K, Sakamoto H, Inoue K. Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech Dev. 2007;124:279–289. doi: 10.1016/j.mod.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RA, Kress TL, Cote CA, Gautreau D, Rokop ME, Mowry KL. Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech Dev. 2004;121:101–109. doi: 10.1016/j.mod.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Linnen JM, Bailey CP, Weeks DL. Two related localized mRNAs from Xenopus laevis encode ubiquitin-like fusion proteins. Gene. 1993;128:181–188. doi: 10.1016/0378-1119(93)90561-g. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Yasuda K, Inoue K. Maternal mRNA localization of zebrafish DAZ-like gene. Mech Dev. 1999;81:223–226. doi: 10.1016/s0925-4773(98)00242-1. [DOI] [PubMed] [Google Scholar]
- Messitt TJ, Gagnon JA, Kreiling JA, Pratt CA, Yoon YJ, Mowry KL. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell. 2008;15:426–436. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera L, Forristall C, Zhou Y, King ML. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development. 1993;117:377–386. doi: 10.1242/dev.117.1.377. [DOI] [PubMed] [Google Scholar]
- Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- Nath K, Boorech JL, Beckham YM, Burns MM, Elinson RP. Status of RNAs, localized in Xenopus laevis oocytes, in the frogs Rana pipiens and Eleutherodactylus coqui. J Exp Zool B Mol Dev Evol. 2005;304:28–39. doi: 10.1002/jez.b.21020. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Weeks DL, Harvey RP, Melton DA. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell. 1985;42:769–777. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sater KA, Gilchrist MJ. Xenopus genomics and genetics: progress and prospects. In: Denny P, Kole C, editors. Genome Mapping and Genomics in Laboratory Animals. Berlin: Springer-Verlag; 2012. pp. 183–196. [Google Scholar]
- Sindelka R, Sidova M, Svec D, Kubista M. Spatial expression profiles in the Xenopus laevis oocytes measured with qPCR tomography. Methods. 2010;51:87–91. doi: 10.1016/j.ymeth.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Snedden DD, Bertke MM, Vernon D, Huber PW. RNA localization in Xenopus oocytes uses a core group of trans-acting factors irrespective of destination. RNA. 2013;19:889–895. doi: 10.1261/rna.038232.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Maegawa S, Nishibu T, Sugiyama T, Yasuda K, Inoue K. Vegetal localization of the maternal mRNA encoding an EDEN-BP/Bruno-like protein in zebrafish. Mech Dev. 2000;93:205–209. doi: 10.1016/s0925-4773(00)00270-7. [DOI] [PubMed] [Google Scholar]
- White JA, Heasman J. Maternal control of pattern formation in Xenopus laevis. J Exp Zool B Mol Dev Evol. 2008;310:73–84. doi: 10.1002/jez.b.21153. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Kataoka K, Watanabe K, Orii H. Restriction of the Xenopus DEADSouth mRNA to the primordial germ cells is ensured by multiple mechanisms. Mech Dev. 2014;131:15–23. doi: 10.1016/j.mod.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YJ, Mowry KL. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development. 2004;131:3035–3045. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Yaniv K, Oberman F, Wolke U, Git A, Fromer M, Taylor WL, Meyer D, Standart N, Raz E, Yisraeli JK. Vg1 RBP intracellular distribution and evolutionarily conserved expression at multiple stages during development. Mech Dev. 1999;88:101–106. doi: 10.1016/s0925-4773(99)00162-8. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang J, King ML. Polarized distribution of mRNAs encoding a putative LDL receptor adaptor protein, xARH (autosomal recessive hypercholesterolemia) in Xenopus oocytes. Mech Dev. 2004;121:1249–1258. doi: 10.1016/j.mod.2004.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.