Abstract
Chromatin boundary elements (CBEs) are widely distributed in the genome and mediate formation of chromatin loops, but their roles in gene regulation remain poorly understood. The complex expression pattern of the Drosophila homeotic gene Sex combs reduced (Scr) is directed by an unusually long regulatory sequence harboring diverse cis elements and an intervening neighbor gene fushi tarazu (ftz). Here we report the presence of a multitude of CBEs in the Scr regulatory region. Selective and dynamic pairing among these CBEs mediates developmentally regulated chromatin loops. In particular, the SF1 boundary plays a central role in organizing two subsets of chromatin loops: one subset encloses ftz, limiting its access by the surrounding Scr enhancers and compartmentalizing distinct histone modifications, and the other subset subdivides the Scr regulatory sequences into independent enhancer access domains. We show that these CBEs exhibit diverse enhancer-blocking activities that vary in strength and tissue distribution. Tandem pairing of SF1 and SF2, two strong CBEs that flank the ftz domain, allows the distal enhancers to bypass their block in transgenic Drosophila, providing a mechanism for the endogenous Scr enhancer to circumvent the ftz domain. Our study demonstrates how an endogenous CBE network, centrally orchestrated by SF1, could remodel the genomic environment to facilitate gene regulation during development.
INTRODUCTION
Organization of the eukaryotic genome is highly complex. Recent studies revealed that extensive loop structures exist in mammalian and Drosophila genomes, separating genes and gene families into distinct functional domains (1–5). Many of these loops are anchored by specialized DNAs called chromatin boundary elements (CBEs) or insulators. These elements have two well-characterized molecular functions: they block the regulatory effects of enhancers and silencers as well as limit the spread of organized chromatin (for recent reviews, see references 1 and 5 to 11). These properties, in addition to the finding that CBEs are frequently positioned between differentially expressed genes, suggest that they play important function in insulating neighboring genes to ensure their independent regulation (2, 12–17).
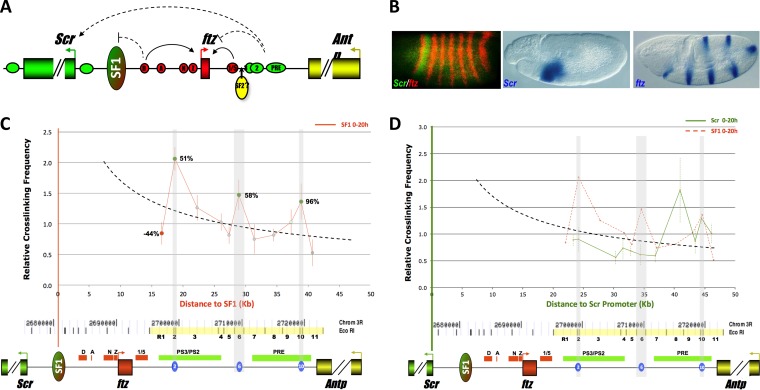
It is increasingly recognized that CBEs are also present within gene regulatory regions, often between genes and distal enhancers, raising questions about their functional significance. Examples of these CBEs include the Drosophila Fab-7, Fab-8, and SF1 elements (18–22). It has been postulated that the Fab boundaries, located within the regulatory sequences of the Abdominal B (Abd-B) gene, may divide distinct enhancer domains and mediate transcriptional regulation in a tissue-specific fashion (20, 23–25). We have previously identified a CBE named SF1 within the regulatory sequences of the homeotic gene Sex comb reduced (Scr). SF1 is located between the promoter and several distal regulatory elements of Scr, including the PS2 and PS3 enhancers, which activate Scr during early embryogenesis and a Polycomb response element (PRE) that maintains the Scr expression in late development (Fig. 1A) (21, 26–29). In addition, the Scr upstream region is further interrupted by a nested neighboring gene, fushi tarazu (ftz) (Fig. 1A). Despite such close proximity and juxtaposition of the ftz and Scr promoters and enhancers, the two neighboring genes are expressed in distinct patterns (Fig. 1B).
FIG 1.
Identification of SF1-tethering elements (STEs) in the Drosophila ftz-Antp interval. (A) Diagram of genomic region containing the Scr (green), ftz (red), and Antp (yellow) genes. Filled boxes, arrowheads, and horizontal ovals represent genes, promoters, and enhancers, respectively. Red enhancers: D, ftz distal; A, ftz AE1; N, ftz neurogenic; Z, ftz zebra; and 1/5, ftz stripe 1/5 enhancer. Green enhancers: 3, Scr PS3; 2, Scr PS2; and PRE, Scr PRE. Vertical oval, SF1; curved arrows, enhancer-promoter interactions; dashed arrows, interactions that could be modulated by SF1. (B) Scr and ftz are expressed in distinct patterns. Shown are images from whole-mount in situ hybridization showing patterns of Scr and ftz in stage 5 (left) and stage 11 (center and right) embryos. Probes used in hybridization are indicated at the bottoms of the photos. (C) SF1 boundary interacts with multiple DNA elements in the ftz-Antp (FA) interval. 3C capture frequencies between SF1 and R1 to R11 elements were quantitated by conventional PCR and plotted over distance. Dashed curve, a distance-frequency power trend line (see Materials and Methods). Elements captured at a frequency above, near, or below the expected value are in green, gray, and red, respectively. The difference between observed and expected capture frequencies is indicated as a percentage for R1, R2, R6, and R10. The gray-shaded vertical regions correspond to the three STEs. A genomic map of the Scr-Antp region, drawn to scale, is shown below with EcoRI sites. R1 to R11 elements are marked in yellow and labeled numerically. Genes and their regulatory elements are shown as horizontal bars; red, ftz; green, Scr, see panel A. Three STEs are shown as blue ovals. (D) Distinct FA elements interact with the Scr promoter. 3C capture frequencies between the Scr promoter and R1 to R11 elements were quantitated by conventional PCR and plotted over distance. Dashed curve, a distance-frequency power trend line; red dashed line, capture profile of SF1.
We used the Scr-ftz region as a model to study how loops tethered by these CBEs might configure regulatory DNA and modulate enhancer traffic in complex genetic loci. We have shown previously that SF1 could insulate Scr from ftz in the upstream direction by preventing an intergenic ftz enhancer from activating an Scr-like promoter (D in Fig. 1A) (21, 30). However, it remains unclear how ftz is insulated in the downstream direction and how Scr distal enhancers overcome the block of SF1 (dashed arrow in Fig. 1A). Further, ftz is active in tissues where Scr may be repressed. How is ftz protected from the encroachment of silent chromatin, which is known to spread along the chromosome? We hypothesize that since CBEs are known to associate with one another to form chromatin loops, SF1 may interact with a yet unknown boundary downstream of ftz, designated SF2, to form a chromatin loop (Fig. 1A) (30–34). This putative loop would insulate ftz from influences by the Scr regulatory elements and the repressive chromatin. It could also facilitate the distal Scr enhancers to the Scr promoter.
We report here the identification of an array of CBE-like elements in the ftz downstream region through their association with SF1. These SF1-tethering elements (STEs) organize topologically independent chromatin loops that separate ftz from Scr and subdivide the Scr regulatory sequences into early and late enhancer access domains. SF1-STE associations are both selective and regulated during development, leading to stage-specific chromatin loops. We show that these dynamic loops restrict the range of the ftz promoter and Scr enhancers. They also demarcate domains of different histone modification in a temporally regulated fashion. We show that these STEs exhibit diverse enhancer-blocking activities that vary in strength and tissue distribution. Significantly, tandem pairing of SF1 and an STE we named SF2 allows distal enhancers to bypass the block of both boundaries in transgenic Drosophila. This provides a potential mechanism for the endogenous Scr distal enhancers to circumvent the ftz domain and multiple CBEs. This unique enhancer bypass is mediated by two endogenous CBEs in the Drosophila Antennapedia complex, suggesting that CBEs within gene regulatory sequences can play multifaceted roles essential for proper gene regulation. Taken together, the results of our study suggest that diverse CBEs may be integral components of complex genetic loci that organize dynamic and/or tissue-specific chromatin loops to modulate enhancer access and the local chromatin environment during development.
MATERIALS AND METHODS
3C.
Chromosome conformation capture (3C) experiments were performed according to published protocols, with modifications (35). All captures were repeated minimally three times (biological replicates). Approximately 3 × 107 nuclei were used in the chromatin preparation according to existing protocols (36). The optimal quantity of template DNA used in PCRs was determined empirically by serial dilutions. Quantitative PCR (qPCR) was used for the all 3C experiments except those whose results are shown in Fig. 1, which used conventional PCR. Briefly, 3C samples were amplified for 20 to 22 cycles with the outer primer pair. Five to 10 percent of the outer PCR product was amplified with nested inner primers. For conventional PCR, capture products were fractionated on an agarose gel and digitally imaged. Quantitation and analysis were done using ImageJ software. qPCR was performed on an Applied Biosystems 7500 real-time PCR system with SYBR green PCR master mix (Applied Biosystems) or a Bio-Rad CFX-Connect using SYBR green Sso mix (Bio-Rad). The specificity of the signal was validated by agarose gel and melt-curve analyses. Sequences of 3C primers are listed in Table S1 to S4 in the supplemental material. All primers were designed to be ∼100 to 150 bp from the restriction site so that all capture products would be comparable in size. To generate the control template, purified fly genomic DNA was digested with EcoRI or other restriction enzymes and ligated at a concentration of ∼500 ng/μl. The frequencies of capture expressed as relative cross-linking, PCRE/PCRC, is generally plotted over distance (37). All statistical analyses and charts were made using the Microsoft Excel program. The distance-capture frequency curves were generated using data from ∼500 captures between sites with a linear distance up to 150 kb apart, of which the relevant distance ranges are shown in the figures below. We also generated separate curves for conventional and quantitative PCRs to control for the data range. As a result, 46 or 32 pairs of sites were within the range for conventional or qPCR, respectively.
Enhancer-blocking assays with transgenic Drosophila embryos.
Enhancer-blocking assays including spacer- and SF1-containing transgenic Drosophila lines were described previously (21, 38, 39). Individual STEs were cloned by PCR (see Table S5 for primers), purified after further digestions, and inserted into the NotI site between the Neuroectoderm (NEE) and Hairy stripe 1 (H1) enhancers in pWNHZ vector or pFWNHZ, which contains a copy of SF1 in the Nsi site downstream of the miniwhite gene. For transgene containing tandem SF1 and SF2B, the two insulators were inserted in the same relative position and orientation as in their native genomic loci. For transgenes containing SF2B, genomic integration at the VK33 attP site was mediated by phiC-31 site-specific insertion. For these transgenes, a phiC31 attB site was inserted at the NsiI site downstream of the miniwhite gene. For control, an independent pWNHZ-SF1 genomic insertion at the VK33 attP was also generated and the enhancer-blocking profile of this line is comparable to that shown in Fig. 6C and K. Microinjections were performed in the Cai laboratory or by Rainbow Transgene (Camarillo, CA).
FIG 6.
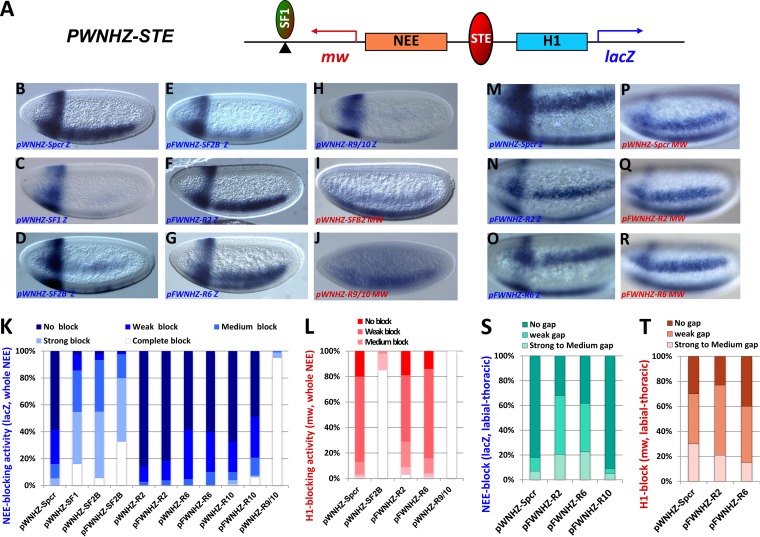
STEs exhibit diverse enhancer-blocking activity in transgenic Drosophila. (A) Diagram of the pWNHZ-STE transgene vector containing divergently transcribed lacZ (blue arrow) and miniwhite (mw; red arrow) reporters flanking the NEE and H1 enhancers (see Materials and Methods). Red oval, STEs or spacer control (Spcr) between NEE and H1; green oval, SF1 inserted in a subset of transgenes (pFWNHZ). (B to J) Representative images of transgenic embryos after whole-mount in situ hybridization with the anti-lacZ (B to H) or the antiwhite (I and J) RNA probes. Transgenes contained in these embryos are labeled at the bottom of each photo, with probes indicated (Z, lacZ; mw, miniwhite). Embryos are shown in sagittal views with anterior to the left and dorsal up. (K) Quantitation of NEE blocking in the whole neuroectoderm in transgenic embryos stained with the lacZ probe (see Materials and Methods for details). (L) Quantitation of H1 blocking in the head stripe in transgenic embryos stained with the miniwhite probe. (M to R) Representative images of embryos after whole-mount in situ hybridization with the anti-lacZ (M to O) or the antiwhite (P-R) RNA probes. Transgenes in these embryos are labeled at the bottom of the photo, with probes indicated. Anterior region of embryos are shown in ventral lateral views, anterior to the left and dorsal up. (S) Quantitation of NEE blocking in the labial thoracic tissues (gap) in embryos stained with the lacZ probe (see Materials and Methods for details). (T) Quantitation of the labial thoracic gap in embryos stained with the miniwhite probe.
In situ hybridizations with lacZ, white, and Scr antisense RNA probes were performed as previously described (30). Whole-mount in situ hybridization and visual assessments of reporter expression were performed according to previously published procedures (21, 38). For each pCANH transgene, 50 to 100 embryos were scored double-blindly from at least three independent insertion lines. Briefly, blastoderm stage embryos were scored for lacZ level in the NEE domain against the H1 domain (see Fig. 6B and C). Based on the ratio of lacZ in the H1 and NEE domains (H/N ratio), embryos were ranked into five categories as follows: no block, H/N ratio ≤ 1; weak block, H/N ratio = 2; medium block, H/N ratio = 4; strong block, H/N ratio ≥ 5; and complete block, H/N ratio ≥ 10. For lacZ expression in the thoracic neuroectoderm (gap in Fig. 6D and E), the staining level was compared to that of the control NEE-driven lacZ and ranked into three categories: no gap, weak gap, and medium to strong gap.
RESULTS
Identification of multiple SF1-tethering elements in the Scr regulatory region.
To search for potential CBEs that loop with SF1, we used chromosomal conformation capture (3C) to survey the entire ftz-Antennapedia (Antp) (FA) interval (Fig. 1C, yellow bar) for DNA sequences that capture SF1 in 0- to 20-h fly embryos (Fig. 1C) (35, 40–43). Since genomic sites located within 100 kb are known to capture each other at relatively high frequencies, we generated a distance-frequency reference curve using 46 pairs of Drosophila genomic sites (dashed curve) (see Materials and Methods). Among the 11 EcoRI fragments (R1 to R11) from the ftz-Antp region, R2, R6, and R10 captured SF1 at a frequency of 50% or higher above the expected value (Fig. 1C). In contrast, other regions captured SF1 at near or below expected values. The negative controls without ligation or without cross-linking produced no capture products, as expected (see Fig. S1 in the supplemental material). These results indicate that SF1 interacts with multiple DNA elements, termed SF1-tethering elements (STEs), in the ftz downstream regions in the developing fly embryos. It is worth noting that R2 is located near the distal end of the ftz gene, between the ftz stripe 1/5 enhancer and the Scr distal elements (Fig. 1C) (27, 44). Interaction between R2 and SF1 could loop out ftz, bringing the distal enhancers closer to the Scr promoter. Similarly, SF1-R6 pairing could facilitate interactions between the Scr promoter and its late Polycomb response element (PRE). R10 is located at the end of the Scr distal region. This region is bound by multiple insulator proteins and has been shown to contain tethering activities (14–16, 40, 45–47). An SF1-R10 loop could separate Scr enhancers from the neighboring Antp (27).
The Scr promoter interacts with distinct elements in the ftz-Antp interval.
The location of the three STEs near the Scr regulatory sequences raises the question of whether their capture with SF1 might be influenced by the Scr enhancer-promoter interactions. To address this, we performed a parallel 3C assay to examine how the Scr promoter interacts with the elements in the FA interval (Fig. 1D). Our results show that the region interacts with the Scr promoter in a distinct profile from that with SF1 (Fig. 1D). The Scr promoter failed to capture R2 and R6. However, it captured R8, which is an Scr PRE that maintains Scr expression in late development (26). Another element that was effectively captured by the Scr promoter is R10. It overlaps with a known tether element that facilitates the Scr distal enhancers to the Scr promoter (44). Therefore, our results indicate that SF1-tethered chromatin loops are largely distinct from the loops resulting from Scr enhancer-promoter interactions.
Selective pairing between SF1 and STEs.
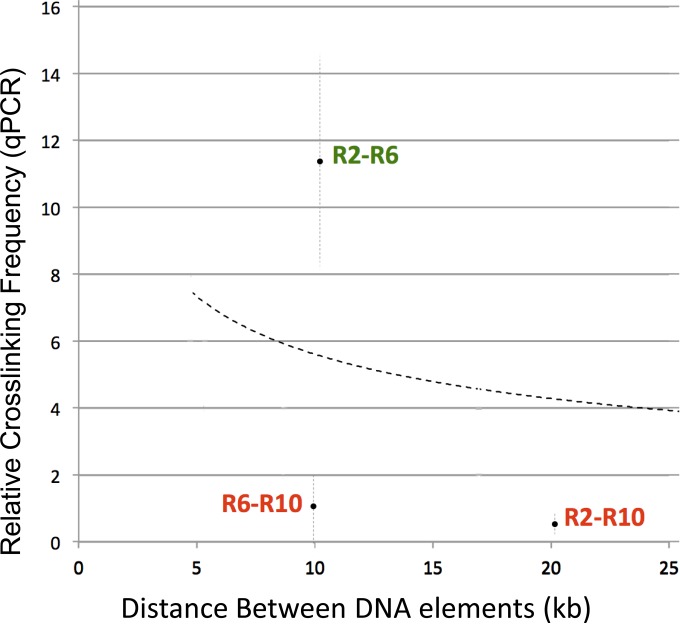
Given that multiple DNA elements were captured with SF1, we wondered whether they all interact with SF1 simultaneously, a scenario that would predict high capture among all STEs. Alternatively, individual STEs may contact SF1 selectively, forming unique loops in different tissues and/or different developmental stages. To distinguish between these possibilities, we used 3C qPCR to examine the association between pairs of STEs in 0- to 20-h embryos (Fig. 2). For this experiment, we generated a new distance-frequency curve by qPCR (dashed curve) (see Materials and Methods). Our results show that all STEs did not capture with one another at high frequencies (Fig. 2). For example, R10 failed to capture R2 or R6, even though high capture was observed between R2 and R6. These results suggest that SF1 may be capable of simultaneously pairing with some, but not all, partners and that such selective pairings may contribute to the alternative formation of distinct chromatin loops in various tissues and/or developmental stages.
FIG 2.
SF1 does not contact all STEs simultaneously. 3C capture frequencies between pairs of STEs were quantitated by qPCR and plotted over distance. Pairs captured at a frequency above or below the expected value are in green or red, respectively. Dashed curve, a new distance-frequency power trend line by qPCR (see Materials and Methods).
STEs delimit domains of genomic access for the Scr early and late enhancers.
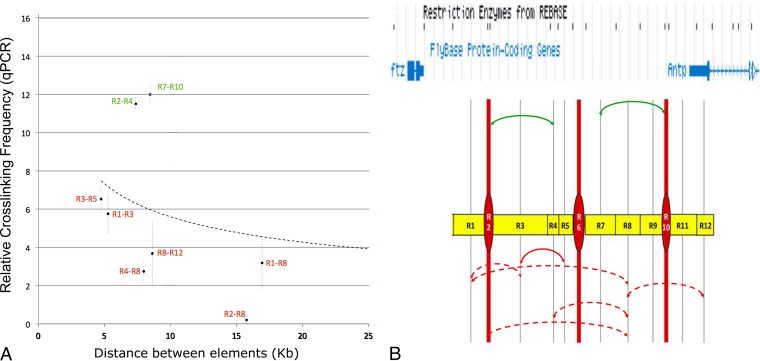
Boundary-tethered chromatin loops are believed to restrict enhancer access (31, 32, 48, 49). The three STEs are located at or near the ends of the Scr early and late regulatory elements and may be involved in organizing these enhancer domains. To evaluate whether SF1-STE loops physically constrain genomic access in the native chromatin environment, we randomly picked eight pairs of nonjoining EcoRI fragments from the ftz-Antp interval and measured their capture frequencies by 3C qPCR (Fig. 3A). Our results show that the capture between these genomic fragments cannot be predicted based on the distances that separate them. Rather, it appears to depend on whether the two elements are separated by STEs. For example, all five pairs of sites separated by at least one STE were captured at frequencies below expected values (Fig. 3, dashed arrows). In contrast, two of the three pairs of sites not separated by an STE were captured at frequencies above expected values (Fig. 3, solid arrows). These results suggest that SF1-tethered loops constrain genomic access in the native chromatin environment, and they may underlie the structural as well as functional divisions in the Scr upstream regulatory sequences. These results also suggest that physical restriction of enhancer access may contribute to enhancer-blocking mechanisms by CBEs.
FIG 3.
STEs demarcate domains of enhancer access in vivo. (A) 3C capture frequencies of 8 pairs of FA elements were quantitated by qPCR and plotted over distance. Pairs captured at frequency above or below the expected frequency are in green or red, respectively. Dashed curve, a distance-frequency power trend line by qPCR. (B) Diagram of genomic access domains based on panel A. Yellow boxes represent R1 to R11 elements, except STEs, which are represented by red ovals. The gray vertical lines indicate the midpoint of each fragment. Curved arrows indicate captures between pairs of elements, with those across an STE drawn as dashed curves and those not across any STEs as solid curves. Captures above or below the expected frequency are green or red, respectively. A map of the FA region is shown at the top with EcoRI sites.
Dynamic pairing between SF1 and partner STEs restrict access to the ftz promoter during development.
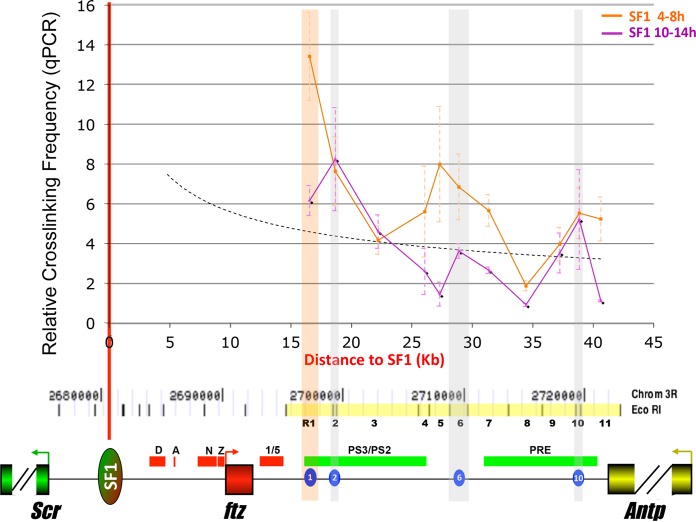
The evidence for selective pairing between SF1 and STEs led us to postulate that their interactions may be developmentally regulated. To test this, we examined captures between SF1 and the FA elements in more synchronized embryos 4 to 8 or 10 to 14 h of age. These two developmental stages were selected to enrich chromatin conformations corresponding to the activation (4 to 8 h) or maintenance (10 to 14 h) phase of homeotic genes. Surprisingly, in 4- to 8-h embryos, SF1 was captured strongly with R1, which is located immediately downstream of ftz (Fig. 4). However, such a strong association between SF1 and R1 was not observed at 10 to 14 h (Fig. 4). In addition, SF1 also showed dynamic capture with regions surrounding R6, including R4 to R7 elements, during the 4- to 8-h stage. In contrast, SF1 captured R2 and R10 at comparably high levels in both 4- to 8-h and 10- to 14-h embryos. Taken together, these results show that the chromatin loops tethered by SF1 and STEs are highly regulated during development.
FIG 4.
SF1-STE associations are developmentally regulated. 3C capture frequencies between SF1 and R1-R11 in 4-h to 8-h (orange line) and 10-h to 14-h (purple line) embryos were quantitated by qPCR and plotted over distance. Dashed curve, a distance-frequency trend line generated by qPCR. Vertical shaded bars indicate regions of STEs (orange, R1; gray, R2, R6, and R10). At the bottom, a genomic map of the Scr-Antp region, drawn to scale, is shown with EcoRI sites. R1 to R11 elements are marked in yellow and labeled numerically (see Fig. 1C). STEs are shown as blue ovals.
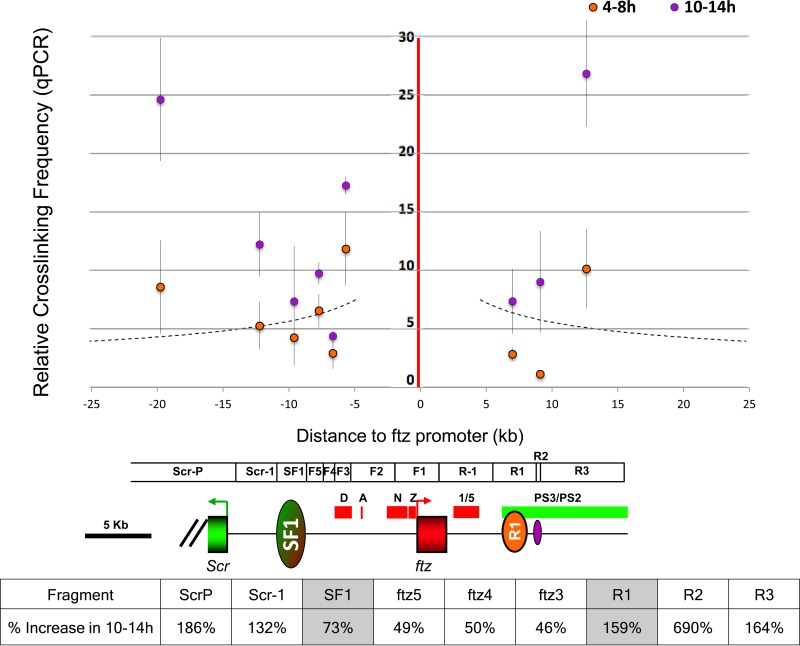
The strong association between SF1 and R1 occurs at a time when both Scr and ftz genes are activated by their long-range enhancers. The timing of formation and the extent of the SF1-R1 loop, which coincides precisely with the limit of the ftz gene, suggest that it may serve to prevent these long-range enhancers from interfering with the inappropriate promoters. To test this hypothesis, we probed the accessibility of the ftz promoter by the surrounding Scr enhancers in 4- to 8-h as well as 10- to 14-h embryos. Nine nonjoining EcoRI fragments surrounding the ftz promoter were tested, including three within the SF1-R1 loop (F3 to F5), SF1 and R1, and four outside the loop (Scr-P, Scr-1, R2, and R3,) (Fig. 5). Our results show that DNA elements located within 10 kb of the ftz promoter were captured at a reduced frequency, possibly due to the constraint by the loop (Fig. 5) (35). The only exception is F3, which contains a scaffold/matrix attachment region (S/MAR) known to anchor chromatin (50–52). In addition, we noticed that there was a general upshift in the capture frequency in the 10- to 14-h embryos (Fig. 5). However, all of the fragments located outside the SF1-R1 loop showed greater increases in their capture with the ftz promoter at 10 to 14 h than the fragments located inside the SF1-R1 loop (Fig. 5). This is consistent with the SF1-R1 loop restricting access to the ftz promoter by the surrounding Scr enhancers in early embryos. We also compared the interactions between SF1 and R1 with that with R-1, which is also located within the loop, and observed a similar increase in the capture during the late stage, suggesting that SF1-R1 interactions may suppress other SF1 interactions (see Fig. S2 in the supplemental material).
FIG 5.
SF1 and R1 restrict access to the ftz promoter in early embryos. (Top) 3C capture frequencies between the ftz promoter and nine surrounding nonjoining EcoRI elements in 4-h to 8-h and 10-h to 14-h embryos were quantitated by qPCR and plotted over distance. EcoRI fragments tested in the capture drawn as boxes with an Scr-Antp genomic map, both to scale with the plot above (see also Fig. 1C). (Bottom) Percentage increase of capture frequency between 4- to 8-h and 10- to 14-h stages. Data for SF1 and R1 are shown in gray.
The SF1-R1 loop defines the active ftz chromatin domain in Drosophila embryos.
During embryogenesis, ftz is highly expressed in many tissues in which Scr is inactive or repressed due to the assembly of silent chromatin. To assess whether SF1-tethered loops play any roles in organizing distinct chromatin structures, we examined the histone modification profiles of the Scr-Antp interval in the modENCODE data set (http://gbrowse.modencode.org/fgb2/gbrowse/fly/) (14). As shown in Fig. S3 in the supplemental material, the SF1-R1 loop detected in 4- to 8-h embryos coincides precisely with a domain of low H3K9me3 repressive chromatin marks (red bracket) at this stage. The H3K27me3 repressive chromatin marks also show a similar, albeit more moderate, depletion in this domain (see http://gbrowse.modencode.org/fgb2/gbrowse/fly/). The highly active ftz transcription in multiple body segments at the 4- to 8-h stage is likely responsible for the strong depletion of the repressive marks. Furthermore, the disruption of the SF1-R1 loop in 10- to 14-h embryos is accompanied by an increase of repressive marks in the ftz domain and a slight decrease of these marks in the surrounding Scr regions. In 16- to 24-h embryos, the boundary at R1 in the H3K9me3 profile gradually disappeared. In contrast to the strong but transient border at R1, a weaker border at R2 persists through late development in the H3K9me3 profile. This demarcation could reflect the more stable SF1-R2 association (see Fig. S3). The H3K4me1 marks are enriched around regulatory elements in both the ftz and Scr domains during early development, consistent with enhancer-mediated activation of both genes (53–56). These marks appear to peak earlier in the ftz domain (0 to 4 h) than in the Scr domain (4 to 8 h), echoing the different transcriptional onset of the two genes (57). Finally, the histone modification signatures around R10 are persistent throughout embryogenesis, reflecting its stable genomic association revealed by 3C (Fig. 1C and 4). Taken together, the histone modification profiles in the Scr-Antp region suggest that the dynamic interactions between SF1 and STEs could underlie the chromatin domains in this region. The R1 element, positioned at the distal end of the ftz domain, loops with SF1 to insulate ftz and Scr genes by blocking their long-range enhancers and delimiting their distinct chromatin structures in vivo. It displays a strong fit to the defining criteria of the hypothetical SF2 and is therefore named so from here on.
STEs contain diverse enhancer-blocking activity.
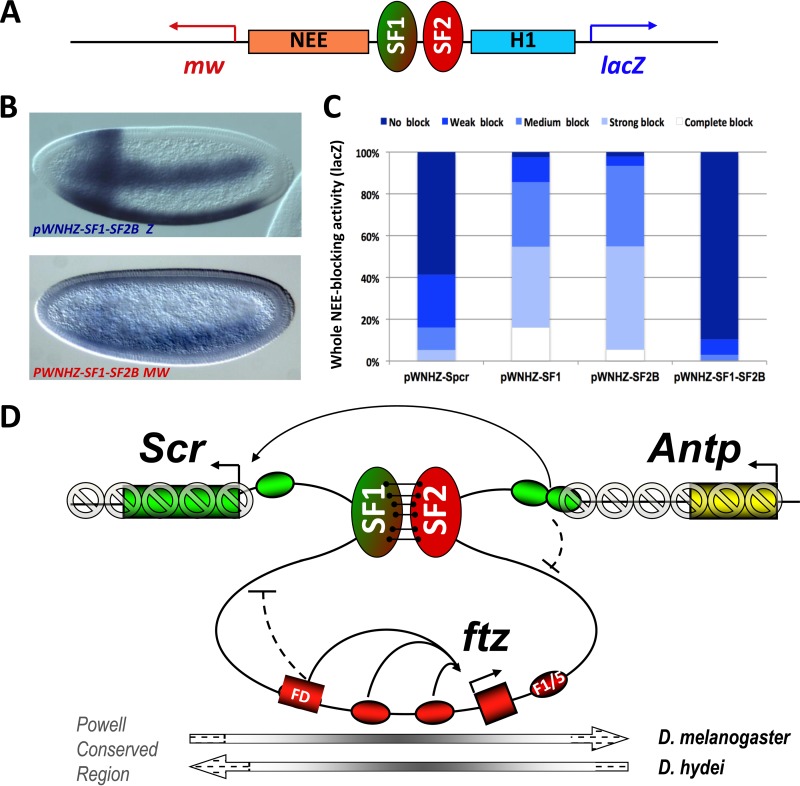
The abilities of STEs to tether loops as well as to constrain promoter and enhancer access reflect key characteristics of insulators (31, 32). A survey of the genome-wide chromatin immunoprecipitation (ChIP) data reveals that STEs also bind to distinct sets of insulator proteins (see Fig. S4 in the supplemental material) (15, 40, 45–47). We tested STEs for enhancer-blocking activity using an established insulator assay in transgenic fly embryos (21, 38). The assay transgene contains two tissue-specific enhancers, Neuroectoderm (NEE) and Hairy stripe 1 (H1), between divergently transcribed lacZ and miniwhite reporters (pWNHZ) (Fig. 6A). Previous studies have shown that insulators such as SF1 can block the distal NEE enhancer and reduce lacZ expression in the horizontal stripes when inserted between NEE and H1 (Fig. 6B, C, and K; see also Materials and Methods) (21, 38, 58, 59). We first tested SF2B, a 2-kb subfragment of SF2 that spans the major insulator binding peaks. When inserted between NEE and H1, SF2B significantly reduced NEE-driven lacZ expression without affecting the anterior vertical stripe driven by the H1 enhancer (Fig. 6D and K). We further showed that SF2B can block the H1 enhancer without affecting NEE function on the miniwhite reporter (Fig. 6I and L). These results indicate that SF2B contains strong insulator activity. We have previously shown that two Gypsy insulators flanking an enhancer increases the block of the enclosed enhancer, possibly due to improved pairing from the proximity of the two insulators (31). To test whether SF1 can augment the insulator function of SF2B, we inserted a copy of SF1 (green oval) distal to NEE in the downstream region of miniwhite (pFWNHZ) (Fig. 6A). Indeed, increased blocking of NEE was observed in these transgenic embryos (Fig. 6E and K). This result suggests that SF1 and SF2B can loop with each other to better restrict the NEE enhancer. It also suggests that pairing between SF1 and SF2B can occur independently of their endogenous genomic contexts and in multiple body segments. We also tested a 2-kb element consisting of R10 and part of R9 (R9/10) that includes major peaks of insulator proteins. We found that R9/10 also displayed a strong enhancer-blocking activity (Fig. 6H and J to L). Together, these findings suggest that both SF2B and R9/10 exhibit ubiquitous insulator activities in early embryos.
However, little ubiquitous insulator activity was observed for R2, R6, or R10 alone (Fig. 6K). Insertion of SF1 distal to NEE failed to significantly improve enhancer-blocking function by these elements (Fig. 6F and G and K). Intriguingly, we observed reduced lacZ expression in a small anterior region of the NEE stripe in pFWNHZ-R2 and pFWNHZ-R6 embryos but not in pFWNHZ-R10 or control embryos (Fig. 6F and G, N to O, and S). Such a gap was consistently observed in multiple transgenic lines (see Fig. S5E to M in the supplemental material). The endogenous Scr expression extends from posterior maxillary to anterior thoracic tissues in early fly embryos (27, 28, 60–62). Double staining using lacZ and Scr probes revealed that cells in the gap are positive for Scr transcripts, indicating that they are a part of the Scr-expressing labial and first thoracic segments (see Fig. S5N to Q). Furthermore, staining with a miniwhite probe showed little difference between the experimental and control embryos, consistent with a localized NEE-blocking activity in these transgenes (Fig. 6P to R and T). These results suggest that R2 and R6 may depend on tissue-specific protein factors to tether loops with SF1.
SF1-SF2 pairing facilitates enhancer bypass in transgenic embryos.
A unique behavior called “enhancer bypass” has been reported for certain CBEs, in which a block of distal enhancers is neutralized when two such CBEs pair in cis or in trans (31–33, 63–67). We have hypothesized that SF1 and SF2, when paired in vivo, could allow the Scr distal enhancers to overcome both boundaries to interact with the Scr promoter (Fig. 7D). To test this hypothesis, we inserted both SF1 and SF2B in tandem between the NEE and the H1 enhancers in the enhancer-blocking transgene (Fig. 7A). Indeed, we observed a strong recovery of NEE-driven lacZ expression and H1-driven miniwhite expression in these transgenic embryos (Fig. 7B and C). This result indicates that pairing of SF1 and SF2B can neutralize their enhancer-blocking activity. This is the first enhancer bypass reported for two endogenous pairing CBEs in the Drosophila Antennapedia complex.
FIG 7.
The SF1-SF2 loop remodels enhancer traffic and modulates chromatin structure in the Scr-Antp region. (A) Diagram of the enhancer bypass transgene containing tandemly paired SF1 and SF2B insulators between NEE and H1 in the pWNHZ vector (see Fig. 6A and Materials and Methods). (B) Representative images of transgenic embryos containing the bypass transgene after hybridization with anti-lacZ (top) or antiwhite (bottom) RNA probes. (C) Quantitation of NEE blocking in pWNHZ-SF1SF2B bypass transgenic embryos stained with the lacZ probe. Quantitation of spacer and single insulator controls are from Fig. 6K. (D) Model showing SF1 attached to SF2, leading to the formation of a chromatin loop that contains the entire the ftz transcription unit. The loop may restrict access between enhancers and promoters of neighboring genes (blocked arrows). It may allow the Scr distal enhancers and PRE element to bypass the SF1 block (long curved arrow). The loop may also insulate the ftz domain from the encroachment of repressive chromatin assembled on Scr and Antp genes in the anterior tissues (shaded block signs). Arrows below the map show the evolutionarily conserved Powell genomic region (see Discussion).
DISCUSSION
The genomes of insects and mammals are widely populated with CBEs that may serve as anchoring sites for chromatin loops. An increasing body of evidence suggests that in addition to the CBEs that reside between genetic loci and insulate genes, some CBEs can also be found in the introns and in regulatory sequences of a single gene (15). Using the Drosophila Scr locus as a model, we have attempted to elucidate the roles of this new class of CBEs in gene regulation. We have found that within a 50-kb Scr regulatory region, there are at least four CBE-like elements that interact with SF1. We provide evidence that SF1 tethers multiple developmentally regulated chromatin loops through selective and dynamic pairing with these STEs during development. One subset of the loops functionally isolates the ftz gene embedded in the Scr regulatory sequences, while others subdivide and possibly facilitate the Scr early and late regulatory elements. In particular, an STE we call SF2 loops with SF1 to enclose and insulate ftz from Scr by blocking the Scr long-range enhancers and repressive chromatin structures. Importantly, association of SF1-SF2 facilitates enhancer bypass in transgenic embryos, suggesting a mechanism that could assist the Scr distal enhancers in circumventing the ftz domain in vivo. These findings validate a mechanism that not only allows CBE-like elements to be tolerated within gene regions but also may provide diverse utility in other genomic functions. Our study provides a comprehensive analysis of how an endogenous CBE network, centrally orchestrated by SF1, might provide multilayered control of Scr and ftz gene activities by coordinating dynamic and selective formation of chromatin loops in rapidly developing embryos.
Potential regulatory roles of chromatin loops in the Scr-ftz-Antp region.
The chromatin loops tethered by SF1 and STEs may address several major challenges to proper gene regulation in the Scr-ftz-Antp gene region (Fig. 7C).
(i) Regulation of enhancer access.
The Scr regulatory region contains a nested pair rule gene, ftz. The Scr and ftz promoters are located close to each other, and their enhancers are scattered on both sides of ftz (Fig. 1A and 7D, green and red ovals). How are enhancer-promoter interactions specified for these two genes? We have shown that SF1 may play a role by blocking an intergenic enhancer from a Scr-like promoter (D in Fig. 1A) (21). However, it remains unclear how ftz is insulated from Scr in the downstream direction. In this work, we showed that SF1 and SF2 pairs transiently to enclose the ftz gene domain, including all its enhancers. The timing and extent of the loop coincide with a reduced access of the ftz promoter to the outside Scr enhancers in vivo. In transgenic embryos, SF1 inserted distal to NEE can also augment the block of the enhancer by SF2B, supporting the notion that an SF1-SF2 loop restricts enhancer access.
Our study further indicates that the SF1-STE loops correlate with domains of enhancer access for the Scr distal regulatory elements. By pairing individually with SF1, these loops could facilitate selected access of these elements to the Scr promoter. Such delineation of enhancer domains by CBE-like elements is reminiscent of the Fab boundaries subdividing the iab enhancer domains in the Abd-B regulatory region (16, 18, 20, 22, 25, 68). Compared to Fab-7, which was shown to restrict enhancer domains in a tissue-specific fashion, the STEs appear to separate the Scr distal regulatory sequences into early and late regulatory domains (18). Our data further show that the R9/10 region contains a constitutive boundary that may separate as well as insulate neighboring Scr and Antp genes (14–16, 40, 45–47). This region was also known to tether to both Scr and Antp promoters, possibly regulating the access or activity of the Scr distal enhancers (23, 29, 44).
(ii) Separation of distinct chromatin structure.
The ftz gene is transcribed in many tissues in which Scr is inactive during early development, and the two genes continue to be expressed in distinct tissues in later stages. How does ftz remain active amid the repressive chromatin assembled in the surrounding Scr regions (Fig. 7D)? Among the STEs, both SF2 and R2 are located at the end of the ftz domain (44, 50, 69–72). We show that the transient SF1-SF2 loop in 4- to 8-h embryos indeed defines the active ftz domain marked by low H3K9me3 and low H3K27me3 at this stage. The stable SF1-R2 loop also correlates with a small but visible border of distinct chromatin structures between the two genes during late development, possibly protecting ftz from the encroachment of PRE-mediated silencing.
(iii) Facilitation of the Scr distal enhancers.
The Scr regulatory sequences are interrupted by the ftz gene domain and multiple CBEs, among which SF1 and SF2 contain strong and ubiquitous enhancer-blocking activity. These could pose impediments to the Scr distal enhancers. Previous studies have shown that tandem arrangement of CBEs may lead to reduction or cancellation of their enhancer-blocking function due to changes in chromatin loop configurations (31, 33, 63, 64). Based on this, we have postulated that pairing between SF1 and SF2 would loop out the ftz domain and allow the Scr distal enhancers to “bypass” the block of both boundaries (Fig. 7D, long curved arrow). In this study, we have shown that tandem arrangement of SF1 and SF2 indeed neutralizes the block of the distal enhancers in a transgenic setting. This provides a potential mechanism for the Scr distal regulatory elements to overcome multiple CBEs to interact with the Scr promoter.
Our study suggests that the unique SF1-SF2 loop may fulfill multiple functional roles as listed above. Interestingly, the SF1-SF2 interval corresponds to an evolutionarily conserved genomic block (Powell conserved region) that contains the entire ftz gene and is found in a “flipped” orientation in several Drosophila species (Fig. 7D, horizontal arrows at bottom) (73). These observations suggest that chromatin loops may shield gene regulation from local chromosome rearrangements, resulting in intermingling as well as interdependence of genes and their regulatory environment during evolution.
Diverse enhancer-blocking behaviors by STEs.
Among the CBEs in the Scr-Antp interval, SF1, SF2, and the R9/10 element exhibit strong and ubiquitous enhancer-blocking activities in the transgenic insulator assay. Genome-wide chromatin immunoprecipitation (ChIP) studies showed that these three elements associate with distinct sets of insulator proteins. While SF1 and SF2 are bound by dCTCF, CP190, and SuHw, R9/10 exhibits strong binding to GAF and Mod(mdg4) (15, 40, 45–47). Although GAF binds only weakly to SF1, it has been shown to be critical for the enhancer-blocking activity of an SF1 subfragment (21). GAF also footprints weakly with SF2 but in a nonoverlapping pattern with other insulator proteins. Mod(mdg4) is the only insulator factor that binds significantly to all three elements. These observations suggest that although most known insulator proteins are ubiquitously expressed, selective or combinatorial recruitment of these proteins to various genomic sites by developmentally regulated factors may be involved in regulated boundary activity.
Two other STEs, R2 and R6, did not exhibit ubiquitous enhancer-blocking activity. Our data suggest that R2 and R6 may contain enhancer-blocking activity in labial and thoracic segments. These are the tissues in which Scr and Antp are expressed. In the insulator ChIPseq profile, the R6 region appears to be overdepleted for known insulator proteins compared with the surrounding genome, suggesting that another protein factor(s) may bind there and possibly facilitate interactions with SF1 and other STEs (15, 40, 45–47). Our results further indicate that although SF1-STE interactions appear to modulate the access of endogenous enhancers, they may not be sufficient to block heterologous enhancers in our insulator assays (30). It is possible that the strength of endogenous chromatin loops is adapted to neighboring regulatory interactions, rather than universally strong. A previously reported endogenous boundary, the 1A2 region in the Drosophila yellow locus, interacts with a full-length Gypsy insulator but exhibits relatively weak enhancer-blocking activity (74). Our results also suggest that major chromatin boundaries, such as SF1, may interact with diverse partners to organize local networks of chromatin loops. These loops may vary in strength, duration, or the tissues in which they form, but they are all physiologically relevant for local gene regulation.
Certain CBEs are known to allow enhancers to “bypass” when they are arranged in tandem or interacting in trans (31–33, 63–67). This was taken as evidence that CBEs block enhancers by tethering chromatin loops. An enhancer flanked by pairing CBEs is enclosed in a chromatin loop and blocked from promoters outside the loop, whereas an enhancer and a promoter separated by paired CBEs can interact with each other. Enhancer bypass was first demonstrated for the Gypsy insulator in transgenic Drosophila (31, 32, 65–67). Recent studies had shown that boundaries from the Bithorax complex, including Fab-7 and Fab-8, also interact with each other and mediate bypass of heterologous enhancers (75–77). Interestingly, our previous data showed that pairing of the full-length Fab-7 and Fab-8 elements did not lead to enhancer bypass in transgenic embryos (39). This might be due to the absence of the pairing partners in the genome vicinity, an indication of the diverse interactions that could occur between CBEs. The enhancer bypass we observed in SF1-SF2 paring is the first such example mediated by two authentic pairing CBEs from the Drosophila Antennapedia complex. It provides an explanation of why CBEs not only are tolerated within gene regions but also, indeed, could perform essential functions during gene regulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chris Rushlow for the generous gift of the attB-containing plasmid. We thank Jingwen Yue for assistance with microinjection. We thank Ping Shen, Edward Kipreos, Matt Romine, and Jeff Gardner for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00663-15.
REFERENCES
- 1.Dean A. 2011. In the loop: long range chromatin interactions and gene regulation. Brief Funct Genomics 10:3–10. doi: 10.1093/bfgp/elq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murrell A. 2011. Setting up and maintaining differential insulators and boundaries for genomic imprinting. Biochem Cell Biol 89:469–478. doi: 10.1139/o11-043. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Corces VG. 2011. Chromatin insulators: a role in nuclear organization and gene expression. Adv Cancer Res 110:43–76. doi: 10.1016/B978-0-12-386469-7.00003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holwerda SJ, de Laat W. 2013. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc Lond B Biol Sci 368:20120369. doi: 10.1098/rstb.2012.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maksimenko O, Georgiev P. 2014. Mechanisms and proteins involved in long-distance interactions. Front Genet 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celniker SE, Drewell RA. 2007. Chromatin looping mediates boundary element promoter interactions. Bioessays 29:7–10. doi: 10.1002/bies.20520. [DOI] [PubMed] [Google Scholar]
- 7.Van Bortle K, Corces VG. 2013. The role of chromatin insulators in nuclear architecture and genome function. Curr Opin Genet Dev 23:212–218. doi: 10.1016/j.gde.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. 2014. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. Bioessays 36:163–172. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holwerda S, de Laat W. 2012. Chromatin loops, gene positioning, and gene expression. Front Genet 3:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Z, Felsenfeld G. 2012. Order from chaos in the nucleus. Mol Cell 48:327–328. doi: 10.1016/j.molcel.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenfeld G, Dekker J. 2012. Genome architecture and expression. Curr Opin Genet Dev 22:59–61. doi: 10.1016/j.gde.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Sun JQ, Hatanaka A, Oki M. 2011. Boundaries of transcriptionally silent chromatin in Saccharomyces cerevisiae. Genes Genet Syst 86:73–81. [DOI] [PubMed] [Google Scholar]
- 13.Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. 2002. Large clusters of co-expressed genes in the Drosophila genome. Nature 420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- 14.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, Washietl S, Arshinoff BI, Ay F, Meyer PE, Robine N, Washington NL, Di Stefano L, Berezikov E, Brown CD, Candeias R, Carlson JW, Carr A, Jungreis I, Marbach D, Sealfon R, Tolstorukov MY, Will S, Alekseyenko AA, Artieri C, Booth BW, Brooks AN, Dai Q, Davis CA, Duff MO, Feng X, Gorchakov AA, Gu T, Henikoff JG, Kapranov P, Li R, MacAlpine HK, Malone J, Minoda A, Nordman J, Okamura K, Perry M, Powell SK, Riddle NC, Sakai A, Samsonova A, Sandler JE, Schwartz YB, Sher N, Spokony R, Sturgill D, van Baren M, Wan KH, Yang L, Yu C, Feingold E, Good P, Guyer M, Lowdon R, Ahmad K, Andrews J, Berger B, Brenner SE, Brent MR, Cherbas L, Elgin SC, Gingeras TR, Grossman R, Hoskins RA, Kaufman TC, Kent W, Kuroda MI, Orr-Weaver T, Perrimon N, Pirrotta V, Posakony JW, Ren B, Russell S, Cherbas P, Graveley BR, Lewis S, Micklem G, Oliver B, Park PJ, Celniker SE, Henikoff S, Karpen GH, Lai EC, MacAlpine DM, Stein LD, White KP, Kellis M. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nègre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, Stein L, Henikoff S, Kellis M, White KP. 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adryan B, Woerfel G, Birch-Machin I, Gao S, Quick M, Meadows L, Russell S, White R. 2007. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol 8:R167. doi: 10.1186/gb-2007-8-8-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galloni M, Gyurkovics H, Schedl P, Karch F. 1993. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J 12:1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagstrom K, Muller M, Schedl P. 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev 10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Levine M. 1999. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell 99:567–575. doi: 10.1016/S0092-8674(00)81546-9. [DOI] [PubMed] [Google Scholar]
- 21.Belozerov VE, Majumder P, Shen P, Cai HN. 2003. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J 22:3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127:779–790. [DOI] [PubMed] [Google Scholar]
- 23.Cléard F, Moshkin Y, Karch F, Maeda RK. 2006. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet 38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- 24.Gyurkovics H, Gausz J, Kummer J, Karch F. 1990. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J 9:2579–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Ashe H, Burks C, Levine M. 1999. Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development 126:3057–3065. [DOI] [PubMed] [Google Scholar]
- 26.Gindhart JG Jr, Kaufman TC. 1995. Identification of Polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics 139:797–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gindhart JG Jr, King AN, Kaufman TC. 1995. Characterization of the cis-regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics 139:781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorman MJ, Kaufman TC. 1995. Genetic analysis of embryonic cis-acting regulatory elements of the Drosophila homeotic gene sex combs reduced. Genetics 140:557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Southworth JW, Kennison JA. 2002. Transvection and Silencing of the Scr homeotic gene of Drosophila melanogaster. Genetics 161:733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai HN, Zhang Z, Adams JR, Shen P. 2001. Genomic context modulates insulator activity through promoter competition. Development 128:4339–4347. [DOI] [PubMed] [Google Scholar]
- 31.Cai HN, Shen P. 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 32.Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 33.Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. 2007. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol Cell Biol 27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai HN. 2006. Function and mechanism of chromatin boundaries, p 343–363. In Ma J. (ed), Gene expression and regulation. Higher Education Press, Beijing, China. [Google Scholar]
- 35.Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 36.Blanton J, Gaszner M, Schedl P. 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev 17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10:1453–1465. doi: 10.1016/S1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 38.Cai HN, Levine M. 1997. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J 16:1732–1741. doi: 10.1093/emboj/16.7.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majumder P, Cai HN. 2003. The functional analysis of insulator interactions in the Drosophila embryo. Proc Natl Acad Sci U S A 100:5223–5228. doi: 10.1073/pnas.0830190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mito Y, Henikoff JG, Henikoff S. 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 41.Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. 2012. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. 2006. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 44.Calhoun VC, Levine M. 2003. Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc Natl Acad Sci U S A 100:9878–9883. doi: 10.1073/pnas.1233791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bushey AM, Ramos E, Corces VG. 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith ST, Wickramasinghe P, Olson A, Loukinov D, Lin L, Deng J, Xiong Y, Rux J, Sachidanandam R, Sun H, Lobanenkov V, Zhou J. 2009. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol 328:518–528. doi: 10.1016/j.ydbio.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. 2009. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J 28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ameres SL, Drueppel L, Pfleiderer K, Schmidt A, Hillen W, Berens C. 2005. Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer. EMBO J 24:358–367. doi: 10.1038/sj.emboj.7600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bondarenko VA, Jiang YI, Studitsky VM. 2003. Rationally designed insulator-like elements can block enhancer action in vitro. EMBO J 22:4728–4737. doi: 10.1093/emboj/cdg468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dearolf CR, Topol J, Parker CS. 1989. Transcriptional control of Drosophila fushi tarazu zebra stripe expression. Genes Dev 3:384–398. doi: 10.1101/gad.3.3.384. [DOI] [PubMed] [Google Scholar]
- 51.Gasser SM, Laemmli UK. 1986. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell 46:521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- 52.Mirkovitch J, Mirault ME, Laemmli UK. 1984. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell 39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 53.Hon GC, Hawkins RD, Ren B. 2009. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet 18:R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 55.Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 56.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tautz D, Pfeifle C. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 59.Li M, Belozerov VE, Cai HN. 2010. Modulation of chromatin boundary activities by nucleosome-remodeling activities in Drosophila melanogaster. Mol Cell Biol 30:1067–1076. doi: 10.1128/MCB.00183-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahaffey JW, Kaufman TC. 1987. Distribution of the Sex combs reduced gene products in Drosophila melanogaster. Genetics 117:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pattatucci AM, Kaufman TC. 1991. The homeotic gene Sex combs reduced of Drosophila melanogaster is differentially regulated in the embryonic and imaginal stages of development. Genetics 129:443–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pattatucci AM, Otteson DC, Kaufman TC. 1991. A functional and structural analysis of the Sex combs reduced locus of Drosophila melanogaster. Genetics 129:423–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Melnikova L, Juge F, Gruzdeva N, Mazur A, Cavalli G, Georgiev P. 2004. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc Natl Acad Sci U S A 101:14806–14811. doi: 10.1073/pnas.0403959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Golovnin A, Mazur A, Kopantseva M, Kurshakova M, Gulak PV, Gilmore B, Whitfield WG, Geyer P, Pirrotta V, Georgiev P. 2007. Integrity of the Mod(mdg4)-67.2 BTB domain is critical to insulator function in Drosophila melanogaster. Mol Cell Biol 27:963–974. doi: 10.1128/MCB.00795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morris JR, Chen JL, Geyer PK, Wu CT. 1998. Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc Natl Acad Sci U S A 95:10740–10745. doi: 10.1073/pnas.95.18.10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen JL, Huisinga KL, Viering MM, Ou SA, Wu CT, Geyer PK. 2002. Enhancer action in trans is permitted throughout the Drosophila genome. Proc Natl Acad Sci U S A 99:3723–3728. doi: 10.1073/pnas.062447999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuhn EJ, Viering MM, Rhodes KM, Geyer PK. 2003. A test of insulator interactions in Drosophila. EMBO J 22:2463–2471. doi: 10.1093/emboj/cdg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. 1994. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res 22:3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hiromi Y, Kuroiwa A, Gehring WJ. 1985. Control elements of the Drosophila segmentation gene fushi tarazu. Cell 43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- 70.Hiromi Y, Gehring WJ. 1987. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell 50:963–974. doi: 10.1016/0092-8674(87)90523-X. [DOI] [PubMed] [Google Scholar]
- 71.Maier D, Preiss A, Powell JR. 1990. Regulation of the segmentation gene fushi tarazu has been functionally conserved in Drosophila. EMBO J 9:3957–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pick L, Schier A, Affolter M, Schmidt-Glenewinkel T, Gehring WJ. 1990. Analysis of the ftz upstream element: germ layer-specific enhancers are independently autoregulated. Genes Dev 4:1224–1239. doi: 10.1101/gad.4.7.1224. [DOI] [PubMed] [Google Scholar]
- 73.Maier D, Sperlich D, Powell JR. 1993. Conservation and change of the developmentally crucial fushi tarazu gene in Drosophila. J Mol Evol 36:315–326. [DOI] [PubMed] [Google Scholar]
- 74.Parnell TJ, Viering MM, Skjesol A, Helou C, Kuhn EJ, Geyer PK. 2003. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc Natl Acad Sci U S A 100:13436–13441. doi: 10.1073/pnas.2333111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kyrchanova O, Toshchakov S, Podstreshnaya Y, Parshikov A, Georgiev P. 2008. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol Cell Biol 28:4188–4195. doi: 10.1128/MCB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kyrchanova O, Ivlieva T, Toshchakov S, Parshikov A, Maksimenko O, Georgiev P. 2011. Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process. Nucleic Acids Res 39:3042–3052. doi: 10.1093/nar/gkq1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gohl D, Aoki T, Blanton J, Shanower G, Kappes G, Schedl P. 2011. Mechanism of chromosomal boundary action: roadblock, sink, or loop? Genetics 187:731–748. doi: 10.1534/genetics.110.123752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.