ABSTRACT
HIV-1 is typically CCR5 using (R5) and T cell tropic (T-tropic), targeting memory CD4+ T cells throughout acute and chronic infections. However, viruses can expand into alternative cells types. Macrophage-tropic (M-tropic) HIV-1 variants have evolved to infect macrophages, which have only low levels of surface CD4. Most M-tropic variants have been isolated from the central nervous system during late-stage chronic infection. We used the HIV-1 env genes of well-defined, subject-matched M-tropic and T-tropic viruses to characterize the phenotypic features of the M-tropic Env protein. We found that, compared to T-tropic viruses, M-tropic viruses infect monocyte-derived macrophages (MDMs) on average 28-fold more efficiently, use low-density CD4 more efficiently, have increased sensitivity to soluble CD4 (sCD4), and show trends toward sensitivity to some CD4 binding site antibodies but no difference in sensitivity to antibodies targeting the CD4-bound conformation. M-tropic viruses also displayed a trend toward resistance to neutralization by monoclonal antibodies targeting the V1/V2 region of Env, suggesting subtle changes in Env protein conformation. The paired M- and T-tropic viruses did not differ in autologous serum neutralization, temperature sensitivity, entry kinetics, intrinsic infectivity, or Env protein incorporation. We also examined viruses with modestly increased CD4 usage. These variants have significant sensitivity to sCD4 and may represent evolutionary intermediates. CD4 usage is strongly correlated with infectivity of MDMs over a wide range of CD4 entry phenotypes. These data suggest that emergence of M-tropic HIV-1 includes multiple steps in which a phenotype of increased sensitivity to sCD4 and enhanced CD4 usage accompany subtle changes in Env conformation.
IMPORTANCE HIV-1 typically replicates in CD4+ T cells. However, HIV-1 can evolve to infect macrophages, especially within the brain. Understanding how CCR5-using macrophage-tropic viruses evolve and differ from CCR5-using T cell-tropic viruses may provide insights into viral evolution and pathogenesis within the central nervous system. We characterized the HIV-1 env viral entry gene from subject-matched macrophage-tropic and T cell-tropic viruses to identify entry features of macrophage-tropic viruses. We observed several differences between T cell-tropic and macrophage-tropic Env proteins, including functional differences with host CD4 receptor engagement and possible changes in the CD4 binding site and V1/V2 region. We also identified viruses with phenotypes between that of “true” macrophage-tropic and T cell-tropic viruses, which may represent evolutionary intermediates in a multistep process to macrophage tropism.
INTRODUCTION
HIV-1 host cell entry is determined solely by the virion surface protein Env. The Env protein precursor gp160 is cleaved into two proteins: the external gp120 protein and the membrane-spanning gp41 protein, which remain associated as a heterodimer and form trimers of these heterodimers. Attachment of gp120 to the host CD4 receptor induces conformational changes in gp120 that allow a secondary interaction with the host CCR5 coreceptor. CCR5 binding induces conformational changes in gp41, which promotes fusion of the viral and cellular membranes. Because the Env protein is the sole determinant of target cell entry specificity, any change in the cell types targeted must reflect a change in the properties of this protein.
The vast majority of HIV-1 isolates sampled during acute and chronic infections are CCR5-using T cell-tropic (R5 T-tropic) viruses, which are adapted to (1–3), and replicating in (4–6), CD4+ memory T cells. R5 T-tropic viruses require the high densities of the CD4 receptor found on CD4+ T cells for efficient entry and use the CCR5 coreceptor, which is most abundant on the memory subset of CD4+ T cells. In approximately one-half of late-stage HIV-1 infections, a viral population evolves the ability to use CXCR4 as a coreceptor (7–9). These CXCR4-using T cell-tropic (X4 T-tropic) viruses use CXCR4 to target CD4+ naive T cells (10, 11), which express lower densities of CCR5 and higher densities of CXCR4 than do CD4+ memory T cells (12, 13). Alternatively, viral populations can evolve to use lower densities of the CD4 receptor, enabling more-efficient entry into macrophages, which express CD4 at densities 20-fold less than is found on CD4+ memory T cells but express similar levels of the CCR5 coreceptor (14). Other studies have also observed that macrophages express lower levels of CD4 than CD4+ T cells (13, 15). Most M-tropic variants use the CCR5 coreceptor (R5 M-tropic), but X4 M-tropic viruses have been reported (16). Because M-tropic variants are detected so rarely (3, 17), the true frequency and characteristics of M-tropic viruses are only beginning to be explored.
Historically, M-tropic variants have been identified by detecting infection of monocyte-derived macrophages (MDMs) in cell culture. However, different preparations of MDMs can vary widely in their capacity to be infected—varying both between different donors and from the same donor at different times (13, 14). Because MDMs have a lower surface density of CD4 than CD4+ T cells, which is a significant impediment to entry by T-tropic viruses (14, 18, 19), it has been possible to use entry efficiency as a function of CD4 density to identify viruses that have adapted to entering macrophages. Initially, this was done using cells engineered to have either high or low levels of CD4 (20). The dependence on receptor level for viral entry can now be demonstrated most convincingly using the Affinofile cell line, in which the surface density of CD4 and/or CCR5 can be experimentally manipulated (21). Using this approach, it has been possible to identify M-tropic viruses, most often isolated from the cerebrospinal fluid (CSF) of subjects late in disease, by their ability to efficiently enter cells with low CD4 densities (CD4low cells) as a surrogate marker for macrophage tropism (14, 22). Although it is possible to observe differences in entry phenotype using MDMs, it is difficult to account for the inherent variability among both the viruses and the cells in unequivocally assigning cellular tropism. Differences in how macrophage tropism is defined can lead to substantially different observations about M-tropic viruses. Although it is widely observed that M-tropic viruses are able to enter more efficiently at low CD4 densities (14, 19, 22–26) and are more sensitive to neutralization by soluble CD4 (sCD4) (27–29) than T-tropic viruses, few other characteristics are widely agreed upon. Similarly, several amino acid changes in the HIV-1 Env protein have been associated with macrophage tropism (24, 25, 27, 30–39), but the changes are not consistent across different subjects when macrophage tropism is defined as a distinct set of evolutionary variants within that subject (40). Furthermore, viruses isolated from the brain are sometimes referred to as neurotropic but without a clear definition of what this means phenotypically other than being located in the central nervous system (CNS) at the time of isolation.
In this study, we sought to gain insight into the mechanisms and consequences of evolving macrophage tropism by investigating characteristics that are unique to M-tropic and T-tropic viruses. We generated pseudotyped viruses using HIV-1 env clones from rigorously defined M-tropic viruses and subject-matched R5 (and in one case X4) T-tropic viruses, which reduced the effect of natural variation observed in viruses between subjects and allowed us to evaluate the phenotypic consequences specific to the evolution of macrophage tropism. We first sought to clarify the magnitude of improved entry efficiency of M-tropic variants for MDMs by pooling data from a panel of MDM donors infected by our virus pairs. Next, we carried out neutralization assays using reagents that target the CD4 binding site (CD4bs) to evaluate the accessibility of epitopes within the CD4bs after the evolution of macrophage tropism. To evaluate the hypothesis that M-tropic Env proteins adopt an “open” conformation (similar to viruses adapted to tissue culture) to enhance the interaction with CD4, we examined the overall stability of the Env protein at different temperatures and the ability of neutralizing antibodies to access epitopes that are normally hidden on primary isolates. We also tested Env proteins for neutralization by autologous sera to reveal differential contributions of the antibody-mediated immune pressure to the evolution of macrophage tropism in the CNS compared to T cell tropism in the blood. We assessed the relative incorporation of Env protein into the virus particle as a potential mechanism for increasing interactions with target cell CD4 molecules. Finally, we used a large panel of subject-matched pairs of Env-pseudotyped viruses with a range of CD4 usage phenotypes to evaluate CD4 usage as a predictor of MDM infection. By comparing CD4 usage and MDM infectivity, we identified a subset of viruses with an intermediate entry phenotype that have increased sensitivity to sCD4 and may represent an evolutionary intermediate on the way to evolving the full macrophage tropism phenotype. Collectively, this work provides new information about the nature of HIV-1 Env proteins that have evolved to become macrophage tropic.
MATERIALS AND METHODS
Cells.
293T cells and TZM-bl cells (41) were maintained in Dulbecco's modified Eagle medium (DMEM) with 4.5 g/liter glucose (Cellgro) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (Sigma). Affinofile cells (21) were maintained in DMEM with 4.5 g/liter glucose (Cellgro) supplemented with 10% dialyzed fetal bovine serum (dFBS) and 50 mg/ml blasticidin (Invitrogen). Affinofile cells can be induced to express a range of CD4 densities on the cell surface that at maximum induction is similar to CD4 densities on TZM-bl cells (E. N. Dukhovlinova and K. T. Arrildt, unpublished data) and approaches but is still lower than that on CD4+ T cells (14).
Monocyte-derived macrophages were prepared as previously described (14). Briefly, blood was collected from four healthy donors. Buffy coats were prepared from whole blood by centrifugation into a Ficoll gradient (Ficoll-Paque Plus; GE Healthcare). Monocytes were isolated from the buffy coats by negative selection (EasySep human enrichment kit without CD16 depletion; StemCell Technologies). Purified monocytes were maintained in RPMI 1640 medium (Cellgro) supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin (Sigma), and 10 ng/ml recombinant human macrophage colony-stimulating factor (M-CSF; Gibco).
Study subjects and sources of env gene clones.
We examined env gene clones generated in several studies, including previously described clones generated from the blood and CSF of five adult subjects infected with HIV-1 subtype B and diagnosed with HIV-associated neurological disease (22) and from four pediatric subjects infected with HIV-1 subtype C and diagnosed with delays in neurological development (42). We also examined new env clones that were generated from the blood and subject-matched CSF, semen, or cervicovaginal fluid (CVF) of five subjects infected with HIV-1 subtype B or C (Table 1). In all cases, env gene amplicons were generated from virion RNA by synthesizing cDNA and using endpoint dilution PCR. Selected amplicons were cloned into an expression vector (pcDNA3.1), and the env clones were used in a transfection procedure to generate pseudotyped viruses.
TABLE 1.
Source material and characteristics of HIV-1 env genes
| Designation | Subject ID | env clone | Tissuea | Tropismb | Age class | Reference or sourcec |
|---|---|---|---|---|---|---|
| 4013 T | 4013 | P9 | Blood | R5 T | Adult | 22 |
| 4013 M | 4013 | C7 | CSF | R5 M | Adult | 22 |
| 4051 T | 4051 | P25 | Blood | R5 T | Adult | 22 |
| 4051 M | 4051 | C3 | CSF | R5 M | Adult | 22 |
| 4059 T | 4059 | P26 | Blood | R5 T | Adult | 22 |
| 4059 M | 4059 | C19 | CSF | R5 M | Adult | 22 |
| 5002 T | 5002 | P10 | Blood | X4 T | Adult | 22 |
| 5002 M | 5002 | C1 | CSF | R5 M | Adult | 22 |
| 7115 T | 7115 | P6 | Blood | R5 T | Adult | 22 |
| 7115 M | 7115 | C21 | CSF | R5 M | Adult | 22 |
| 27569 T | 27569 | P-B10 | Blood | R5 T | Adult | Joseph and Kincer |
| 27569 M | 27569 | C-A11 | CSF | R5 M | Adult | Joseph and Kincer |
| 30005 T | 30005 | P-F10 | Blood | R5 T | Adult | Joseph and Kincer |
| 30005 M | 30005 | C-C12 | CSF | R5 M | Adult | Joseph and Kincer |
| S4007 T | 4007 | P13 | Blood | R5 T | Pediatric | 42 |
| S4007 Int. | 4007 | C02 | CSF | R5 Int. | Pediatric | 42 |
| S4013 T | 4013 | P14 | Blood | R5 T | Pediatric | 42 |
| S4013 Int. | 4013 | C14 | CSF | R5 Int. | Pediatric | 42 |
| S4049 T | 4049 | P23 | Blood | R5 T | Pediatric | 42 |
| S4049 Int. | 4049 | C03 | CSF | R5 Int. | Pediatric | 42 |
| S4058 T | 4058 | P05 | Blood | R5 T | Pediatric | 42 |
| S4058 Int. | 4058 | C12 | CSF | R5 Int. | Pediatric | 42 |
| D929 T | 929 | PD11 | Blood | R5 T | Adult | Dukhovlinova |
| D929 Int. | 929 | VB7 | CVF | R5 Int. | Adult | Dukhovlinova |
| D1361 T | 1361 | P6G1 | Blood | R5 T | Adult | Dukhovlinova |
| D1361 Int. | 1361 | VE9 | CVF | R5 Int. | Adult | Dukhovlinova |
| PC018 T | C018 | PG11 | Blood | R5 T | Adult | Ping |
| PC018 Int. | C018 | S1B2 | Semen | R5 Int. | Adult | Ping |
CSF, cerebrospinal fluid; CVF, cervicovaginal fluid.
R5, CCR5 using; X4, CXCR4 using; T, T cell-tropic; M, macrophage-tropic; Int., intermediate.
Unpublished sources: B. Joseph and L. P. Kincer, unpublished data; E. N. Dukhovlinova, unpublished data; L. Ping, unpublished data.
Generation of pseudotyped viruses.
293T cells were plated at a density of 2.5 × 106 cells in a 100-mm dish and incubated at 37°C. After 18 to 20 h, the 293T cells were transfected with 2 μg of an env expression vector, 5 μg of pNL4-3.LucR-E- plasmid (obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH), and 30 μl FuGENE 6 (Promega) in serum-free DMEM. After 5 h, the medium was fully changed. After 48 h, the supernatant was collected, filtered through a 0.45-μm syringe filter (Millipore), and stored at −80°C in small, single-use aliquots. Virus stocks were not subjected to multiple freeze-thaw cycles.
The titers of virus stocks were determined on Affinofile cells induced to maximum expression of CD4 and CCR5. Infectivity was measured using a luciferase assay system kit (Promega) and a luminometer (Veritas), which measures in relative light units (RLU). Titration curves within the linear range of a virus dilution series were used to calculate the volume of viral stock resulting in 800,000 RLU, which is near the upper limit of the linear range, for each individual virus. This virus-specific titration volume was used for infection of MDMs and Affinofile cells. Alternatively, the volume of viral stock resulting in 150,000 RLU was used for infection of TZM-bl cells in neutralization assays (43). The p24-Gag concentration of viral stocks was determined using the p24 AlphaLISA assay (PerkinElmer).
MDM assay.
Monocyte-derived macrophages were plated at a density of 5.0 × 104 cells per well of a 48-well plate. Five days after plating, a 50% medium change was made. After an additional 2 days, viruses were added to the well and the plates were centrifuged at 2,000 rpm for 2 h at 37°C. The plates were washed once with phosphate-buffered saline (PBS) and washed once with medium, and then a full medium change was done to remove unbound virus. After 48 h, the cells were washed twice with PBS and lysed with 50 μl of Reporter lysis buffer (Promega), and the lysate was stored at −80°C. Virus entry was then assessed by thawing the lysates and quantifying luciferase expression by using a luciferase assay system (Promega).
Affinofile cell assay.
Assays were performed as described previously (44). Briefly, Affinofile cells were plated at a density of 2.0 × 104 cells per well of a 96-well black plate (Costar) that had been pretreated with poly-l-lysine (75 μl of 0.1 g/liter poly-l-lysine in PBS was added to each well, plates were incubated at 37°C, and then the solution was removed before adding the cells). After 18 to 20 h, ponasterone A (Invitrogen) was added to the medium at a final concentration of 5 nM to induce maximum expression of CCR5, and doxycycline (Sigma) was added to the medium at 10 different final concentrations ranging from no drug (minimal induction) to 5 ng/ml (maximum induction) to induce different levels of CD4 expression. After 20 additional hours, the medium with the chemical inducers was replaced with normal medium (i.e., lacking ponasterone A and doxycycline) and virus was added to the cells. The plates were centrifuged at 2,000 rpm for 2 h at 37°C and then incubated at 37°C. After 48 h, the medium was removed, and the cells were washed once with PBS and then lysed with 50 μl of Reporter lysis buffer (Promega). The lysate was stored at −80°C. Virus entry was then assessed by thawing the lysates and quantifying luciferase expression by using the luciferase assay system for firefly luciferase (Promega) or the Renilla luciferase assay system (Promega).
Flow cytometry.
Monocyte-derived macrophages were plated at a density of 1.1 × 106 cells in a 60-mm dish. Affinofile cells were plated at a density of 1.8 × 105 cells in a well of a 24-well plate. Cells were harvested for flow cytometry simultaneously with the infection of identically treated cells for experiments that incorporate both measurements. Cells were removed from culture dishes using nonenzymatic methods to avoid disruption of surface molecules. MDMs were removed using Cell Dissociation Buffer (Gibco), and Affinofile cells were removed using chilled PBS (CellGro). All cells were stained with Fixable Aqua dead-cell stain (Invitrogen) and saturating concentrations of phycoerythrin (PE)-conjugated anti-human CD4 antibody (clone RPA-T4; BD Biosciences). We used QuantiBRITE beads (BD Biosciences), which are conjugated with PE, as a standard to translate mean fluorescence per cell to the number of CD4 antibody binding sites (ABS) per cell. Flow cytometry was performed using a Cyan flow cytometer (Beckman Coulter) and analyzed using FlowJo software (version 9.3.1).
Neutralization assays.
Neutralization assays using TZM-bl cells have been previously described (43). Briefly, a neutralizing antibody, heat-inactivated polyclonal serum, soluble CD4 (sCD4), or T20 was serially diluted across a 96-well black plate (Costar). Virus was added at a single concentration (approximately 150,000 RLU per well) and incubated with the antibody, serum, or sCD4 for 60 min at 37°C. TZM-bl cells were added to a density of 1.0 × 104 cells per well and incubated for 48 h at 37°C. For the T20 time course, a single inhibitory concentration (50 μg/ml) of T20 was added at different times postinfection. The cells were lysed 48 h postinfection with BriteLite (PerkinElmer), and luciferase activity was assessed by measuring luminescence (Victor-Wallac luminometer). Alternatively, the cells were washed once with PBS and lysed with 50 μl of Reporter Lysis Buffer (Promega) for firefly luciferase reporter viruses or Renilla luciferase assay lysis buffer for Renilla luciferase reporter viruses, and the lysate was stored at −80°C. Virus entry was then assessed by thawing the lysates and quantifying luciferase expression by using the luciferase assay system (Promega) for firefly luciferase reporter viruses or the Renilla luciferase assay system (Promega) for Renilla luciferase reporter viruses.
To assay the stability of infectivity at different temperatures, virus was incubated at −80, 0, 25, 37, 49, or 60°C for 1 h before adding TZM-bl cells and infecting them as described above. To test the rate of temperature inactivation as a function of time, viruses were incubated at 49°C for up to 60 min or were incubated at 0°C for up to 53 h before adding TZM-bl cells and infecting as described above.
Western blotting.
To evaluate the relative quantities of Env protein on the surface of a virion, viruses were made as described above, except that they were never frozen. After filtering, 8.0 ml of the cell medium was centrifuged at 20,500 × g for 90 min at 4°C to pellet the virus. The supernatant was removed, and the pellet was resuspended in PBS at 1/50 of the original volume. To denature and inactivate the resuspended virus, 5× Laemmli buffer (312.5 mM Tris-Cl pH 6.8, 10% SDS, 25% 2-mercaptoethanol, 0.01% bromophenol blue, 50% glycerol) was added to a final concentration of 1× and incubated at 95°C for 10 min.
The denatured virus was diluted 1:5 to detect Env and 1:50 to detect p24. Using a 4 to 20% polyacrylamide Tris-glycine gel (Invitrogen), the denatured virus was loaded at seven different volumes from 1 μl to 10 μl to create a titration curve. The gels were electrophoresed at 100 V for 2 h and then transferred to polyvinylidene difluoride (PVDF) membranes by electrophoresing at 150 mA per gel for 2 h. The PVDF membrane was blocked in 5% nonfat dry milk (Bio-Rad) in Tris-buffered saline (TBS) for 1 h at ambient temperature. The blocking solution was decanted, and fresh blocking solution was added. The primary antibody, either a purified rabbit anti-gp140 IgG (generously provided by Nancy Haigwood, Oregon National Primate Research Center, Oregon Health and Science University) or a purified rabbit anti-p24 antibody (NIH AIDS Research and Reference Reagent Program), was added to the blocking solution at a final dilution of 1:10,000 and incubated 16 h at 4°C. The primary antibody and blocking solution were decanted. The PVDF was washed three times for 10 min with 0.03% Tween 20 in TBS. Fresh blocking solution with the secondary antibody, horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Bio-Rad), at a final dilution of 1:10,000 was added to the PVDF and incubated at ambient temperature for 1.5 h. The PVDF was washed six times for 10 min with 0.03% Tween 20 in TBS. The HRP was detected using the ECL Prime kit (GE Healthcare) and recorded by exposure to film (GE Healthcare) for 10 s, 20 s, 30 s, 60 s, or 5 min. After the film was developed, it was analyzed using ImageQuant TL (GE Healthcare). Titration curves were made for each exposure time, and the inference of relative amount was made within the dynamic range of exposures for subsequent analyses.
Statistical analyses.
All statistical analyses were calculated using Prism GraphPad software. A four-parameter logistic equation (4PL; also called a sigmoidal dose-response curve with a variable slope) was used for all nonlinear regression analysis, except for cold sensitivity, which followed an exponential decay. Log-transformed MDM infectivity values were analyzed by paired t test. Antibody and heat sensitivity 50% inhibitory concentrations (IC50s) and cold sensitivity half-life values were analyzed by Wilcoxon signed-rank test, and values outside the limit of detection of each assay were reported at the limit of detection for the purposes of nonparametric comparison. CD4 usage descriptive statistics and log-transformed IC50s for sCD4 were analyzed by paired t test, except when comparing non-subject-matched intermediate viruses to M-tropic viruses, where the unpaired t test was used.
RESULTS
Identification of macrophage-tropic and paired T cell-tropic HIV-1 env isolates.
We used env gene expression vectors derived from one macrophage-tropic (M-tropic) and one T cell-tropic (T-tropic) virus isolated from each of seven subjects (Table 1). Five of these subject-matched pairs (from subjects 4013, 4051, 4059, 5002, and 7115) were previously described, with the M-tropic viruses being classified using the following 3-fold definition (22, 45). First, the viruses were isolated from the cerebrospinal fluid of subjects who had slow viral load (VL) decay after initiation of antiretroviral therapy (ART), which implies that the viruses were being produced from long-lived infected cells (e.g., macrophages/microglia) that continued to produce viruses longer after new infections were blocked by ART, compared to the rapid VL decay seen in the blood, consistent with virus produced from infected T cells. Second, phylogenetic analysis of the env genes showed that these CSF viruses were genetically diverse and distinct from the T-tropic viruses found in the blood of the same subjects (i.e., compartmentalized), implying that the CSF viruses were replicating actively and independently from the viruses found in the blood. Third, reporter viruses pseudotyped with these CSF-derived Env proteins were able to infect Affinofile cells expressing the minimum density of CD4 (CD4low Affinofile cells) more efficiently than reporter viruses with the subject-matched blood-derived T-tropic Env proteins, indicative of viral evolution to infect cells with a low surface density of CD4, such as macrophages within the central nervous system (CNS) compartment. We have shown that infectivity in CD4low Affinofile cells is an assay that can reliably identify M-tropic HIV-1 (14, 22) and provides a method amenable to high-throughput screening. Posttherapy initiation samples were not available for the remaining two subjects (27569 and 30005), and thus their tropism was assessed by phylogenetic analyses (compartmentalization) and infection of CD4low cells.
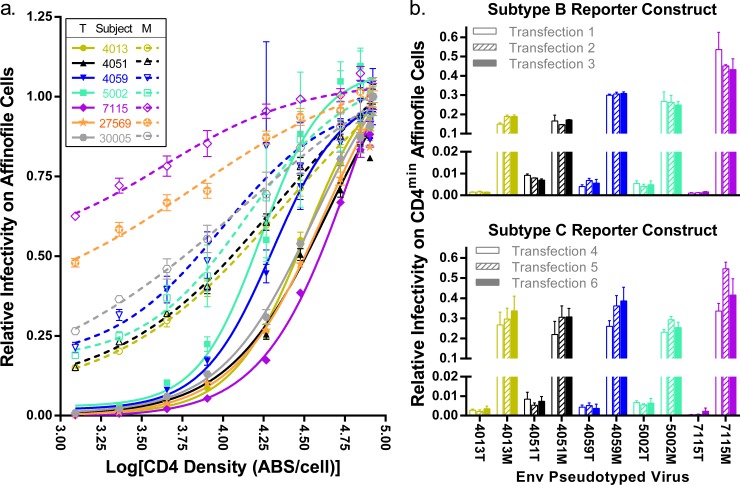
Although Env proteins derived from some of these subjects were previously assessed in separate studies (14, 22), we reanalyzed Env-pseudotyped viruses for entry phenotypes over a wide range of cell surface CD4 densities, which are shown in Fig. 1a using a semi-log plot to display the data. Consistent with previous studies, the Env proteins from the M-tropic clones were significantly better at mediating infection of cells expressing the lowest densities of CD4 (P value = 0.006). In addition, there are differences over the entire CD4 usage curves, which also clearly distinguish the CSF-derived M-tropic viruses from the blood plasma-derived T-tropic viruses. We found a 1.8-fold change in average Hill slope (P value < 0.0001) and a 2.7-fold change in the average CD4 50% effective concentration (EC50) (P value = 0.02), which indicates that the M-tropic Env proteins are able to mediate entry with significantly fewer CD4 molecules than their paired T-tropic Env proteins and confirms that CD4 receptor density is a potent restriction factor in Env-mediated entry.
FIG 1.
Increased CD4 usage differentiates M-tropic viruses from T-tropic viruses. (a) Paired T-tropic and M-tropic env genes were used to pseudotype luciferase reporter viruses. These paired viruses were then used to infect Affinofile cells expressing various levels of CD4, and infectivity was measured by the relative light units (RLU) produced by luciferase. CD4 densities were measured by flow cytometry and reported as the log10 value of antibody binding sites (ABS) per cell. Infectivity is normalized to infectivity at the maximum induction of CD4 and fitted to a dose-response curve, which represents the CD4 usage of each virus and can be described using the Hill slopes and EC50s. Viruses expressing T-tropic Env proteins (T) are represented with closed symbols and solid lines. Viruses expressing M-tropic Env proteins (M) are represented with open symbols and broken lines. Unique color and shape combinations (as specified in the legend within the figure) identify the subjects from which the env genes were isolated, and these identifiers are maintained for all of the figures. M-tropic Env proteins effected a 62-fold increase in average entry at the lowest density of CD4 over subject-matched T-tropic Env proteins (tpaired = 4.2, df = 6, P value = 0.006). M-tropic Env proteins also had enhanced CD4 usage over the entire range of CD4 densities as described by a 2.7-fold-lower EC50 (tpaired = 3.1, df = 6, P value = 0.02) and a 1.8-fold-lower Hill slope (tpaired = 10, df = 6, P value < 0.0001) compared to subject-matched T-tropic Env proteins. (b) Five pairs of T-tropic and M-tropic env genes were pseudotyped in triplicate with either a subtype B (top, transfections 1 to 3) or subtype C (bottom, transfections 4 to 6) env-deficient HIV-1 genome containing a firefly (subtype B) or Renilla (subtype C) luciferase reporter gene. The resulting 60 pseudoviruses (10 env vectors × 2 reporter vectors × 3 replicates) were used to infect CD4low Affinofiles, and infectivity was reported as a fraction of infectivity on CD4high Affinofiles (infected concurrently). The average standard deviation (SD) across transfections for viruses pseudotyped with the subtype B construct is 1.1% (a range of 0.012% to 5.5%) and with the subtype C construct is 2.9% (a range of 0.061% to 11%). There were minor differences in the relative infectivity of viruses produced with the subtype B reporter versus that produced with the subtype C reporter (SD of differences = 5.4%), but these differences were not statistically significant (tpaired = 1.4, df = 9, P value = 0.21).
Because most of the variation in CD4 usage between viruses is represented by the ability to enter using the minimum CD4 levels expressed on Affinofile cells, we can use a simplified “CD4low usage” assay in which virus infection of CD4low Affinofiles is normalized to infection of CD4high Affinofiles (14, 22) to enable screening larger panels of viruses for CD4 usage. To assess the variation in pseudotyped virus preparations and evaluate the effects of the env-deficient genome reporter construct, we pseudotyped five subject-matched pairs of env expression plasmids in triplicate with either an env-deficient subtype B HIV-1 genome expression plasmid encoding a firefly luciferase reporter (transfections 1 to 3) or an env-deficient subtype C HIV-1 genome expression plasmid encoding a Renilla luciferase reporter (transfections 4 to 6) and performed a CD4low usage assay (Fig. 1b). Overall, the results between transfections and with the different reporter constructs were similar, with some detectable variation in CD4low usage when the same env gene was pseudotyped with the different reporter constructs (i.e., 4013M and 4051M had somewhat higher CD4low usage values with the subtype C reporter construct), which could be due to differences in the interactions between the Env proteins and the structural proteins in the pseudoviruses or to variation in the Affinofile cells between experiments (i.e., infection using the subtype B or subtype C reporter construct). For each reporter construct, there was very little variation in CD4low usage between transfections of the same env plasmid. This assay emphasizes the substantial difference in the ability of M-tropic viruses to use low-density CD4 compared with T-tropic viruses and reveals the high fidelity of the CD4 usage assays.
Macrophage-tropic Env proteins increase the infectivity of monocyte-derived macrophages 28-fold over that of T cell-tropic Env proteins.
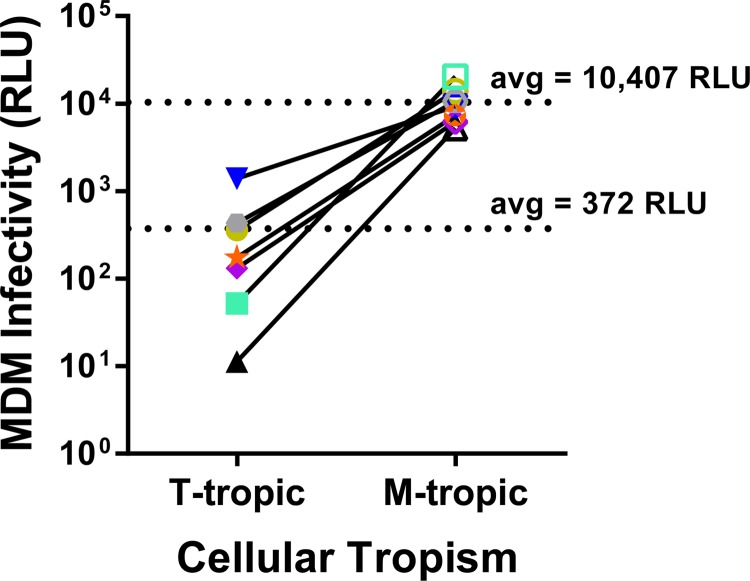
To evaluate the extent to which infectivity of monocyte-derived macrophages (MDMs) can distinguish M-tropic viruses from T-tropic viruses, we used this panel of seven paired pseudotyped viruses to infect MDMs derived from four donors. Similar to previous reports (14), we found that virus infectivity varied up to 5-fold across MDM donors (data not shown). However, by pooling data from multiple donors we were able to estimate the magnitude of the differential infectivity on MDMs for these two groups of viruses. When the relative infectivity data were averaged across the four MDM donors and the seven virus pairs, we observed that the ability of M-tropic viruses to infect MDMs was 28-fold higher than that of T-tropic viruses (P value = 0.0004; Fig. 2).
FIG 2.
M-tropic viruses are better adapted than T-tropic viruses to infection of MDMs. Paired T-tropic and M-tropic pseudotyped reporter viruses were used to infect monocyte-derived macrophages (MDMs) isolated from four donors. Infectivity on MDMs was normalized to infectivity on Affinofile cells expressing maximum CD4 levels. Normalized infectivity values were averaged across donors for each virus and plotted. Closed symbols represent T-tropic Env proteins (T), and open symbols represent M-tropic Env proteins (M) with links between subject-matched pairs. Colors and symbol shapes identify the originating subject as detailed in Fig. 1. Comparing the mean infectivities of T-tropic and M-tropic viruses revealed that M-tropic Env proteins confer a 28-fold increase in the average MDM infectivity over subject-matched T-tropic Env proteins. Mean values are listed and marked with broken lines. The log normalized values were compared between T-tropic and M-tropic infectivities by paired t test (tpaired = 7.0, df = 6, P value = 0.0004).
Macrophage-tropic Env proteins are potently inhibited by soluble CD4 and show a trend toward enhanced sensitivity to CD4 binding site antibodies but are not differentially inhibited by a small-molecule drug targeting the CD4 binding site.
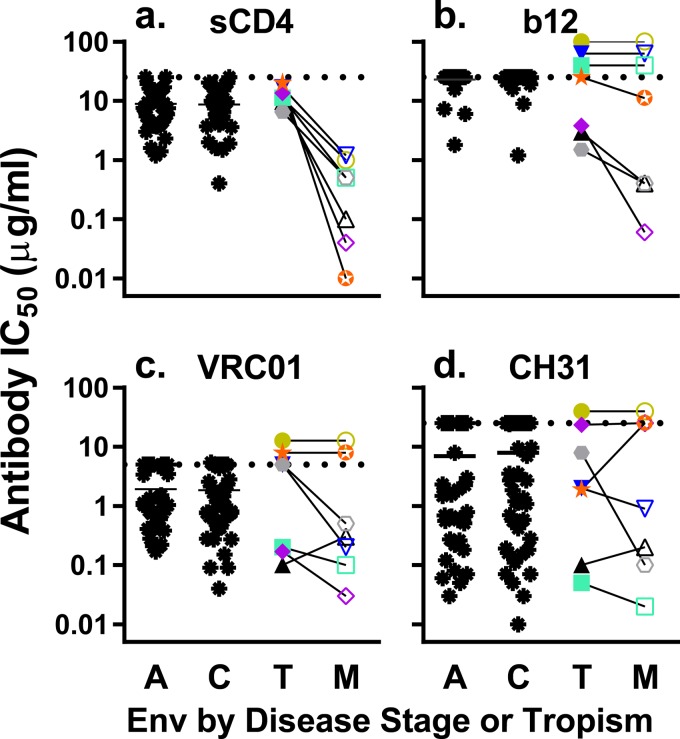
In order to better understand how the M-tropic Env proteins are able to mediate entry using lower densities of CD4, we performed a competitive inhibition assay by incubating sCD4 with the pseudotyped viruses prior to infection and then measured the relative infectivity (normalized to the viruses incubated without sCD4) on TZM-bl cells (Fig. 3a). The viruses pseudotyped with M-tropic Env proteins were, on average, 27-fold more sensitive to sCD4 than those pseudotyped with the paired T-tropic Env proteins (P value = 0.002), confirming that there is a statistically significant difference in how M-tropic Env proteins interact with CD4 compared to T-tropic Env proteins. We then compared the values for our panel of 14 pseudotyped viruses (7 subject-matched pairs of M- and T-tropic Env proteins) with our previously published data on a large panel of acute and chronic infection subtype C Env-pseudotyped viruses (3). The subtype B T-tropic pseudoviruses did not differ significantly in their sensitivity to sCD4 relative to the acute or chronic subtype C pseudoviruses, suggesting that the T-tropic pseudoviruses display a sensitivity to sCD4 that is typical of plasma-derived viruses.
FIG 3.
M-tropic viruses are significantly more sensitive to neutralization by sCD4 and show trends toward increased sensitivity to some CD4bs-targeting antibodies compared to paired T-tropic viruses. Pseudotyped reporter viruses were exposed to various concentrations of sCD4 (a) or an antibody with an epitope overlapping the CD4 binding site (CD4bs) b12 (b), VRCO1 (c), or CH31 (d) in a TZM-bl neutralization assay. IC50s were calculated from dose-response curves and plotted. Dashed lines represent the limits of detection. IC50s above the limit of detection (LOD) were plotted at the LOD, except for pairs where both viruses had IC50s that exceeded the LOD, in which case the symbols were stacked above the LOD line. The same Env-pseudotyped viruses were used in each panel: a control group of T-tropic acute (A) and chronic (C) infection viruses (black asterisks) and our panel of matched T-tropic viruses (closed symbols) and M-tropic viruses (open symbols) from seven subjects. The data for the acute and chronic subtype C viruses are reproduced from Ping et al. (3) to allow a comparison to a large data set of typical viral Env proteins. Viruses expressing M-tropic Env proteins had a statistically significant 27-fold increase in sensitivity to neutralization by sCD4 over subject-matched T-tropic Env proteins (statistical analysis performed on log normalized EC50s; tpaired = 5.5, df = 6, P value = 0.002). M-tropic Env proteins showed trends toward sensitivity to neutralization by b12 (Wpaired = 10, P value = 0.1) and VRC01 (Wpaired = 10, P value = 0.3) over subject-matched T-tropic Env proteins that did not reach statistical significance using a Wilcoxon matched-pair test. No difference in sensitivity to neutralization by CH31 was observed (Wpaired = −3, P value = 0.8).
The enhanced interaction between M-tropic Env proteins and CD4 could be due to having a more exposed CD4 binding site (CD4bs), to an increased affinity for CD4, or to prematurely shifting into the CD4-induced (CD4i) state (a conformational shift that occurs following CD4 binding and results in the conformation that can bind the CCR5 coreceptor). If sensitivity to sCD4 is due to a more exposed CD4bs, then sensitivity to sCD4 would predict sensitivity to neutralizing antibodies targeting the CD4bs. To evaluate exposure of the CD4bs, we performed infectivity assays in the presence of antibodies targeting the CD4bs, specifically b12 (46), VRCO1 (47), and CH31 (48) (Fig. 3b to d). We found that four of the seven M-tropic clones were more sensitive to neutralization by b12 than were the paired T-tropic clones, but the remaining three pairs were completely resistant, making the overall difference between M-tropic and T-tropic neutralization not statistically different. Similarly, four of seven M-tropic clones were more sensitive than the paired T-tropic clones to VRC01 (though one pair was reversed), but again the overall differences in neutralization were not significant. There was no detectable difference in CH31 neutralization between the two groups. Two additional CD4bs antibodies, F105 and CH103, were tested on a smaller panel of 10 pseudotyped viruses (five pairs: 4013, 4051, 4059, 5002, and 7115), but none of the viruses showed any neutralization at the maximum level of F105 tested (14 μg/ml) and all but one pair of viruses (4051) had IC50s above the 10-μg/ml limit of detection for CH103 (data not shown). The paired viruses that were susceptible to neutralization by CH103 had similar IC50s (4051T, 0.14 μg/ml; 4051M, 0.16 μg/ml). Taken together, there is a trend toward increased sensitivity to neutralization by some anti-CD4bs antibodies, but this conclusion is limited by sample size, which is due to the limited number of independent M-tropic viruses isolated. Larger panels of paired Env proteins will be needed to definitively determine whether there are changes in the CD4bs that can be probed by antibody neutralization.
We also probed the CD4bs of these viruses using the inhibitor BMS-626529 (Table 2), which is thought to interact with a target overlapping the CD4bs, though there are conflicting reports about whether BMS-626529 competes with CD4 for binding to the CD4bs or whether it binds outside the CD4bs and prevents assumption of the CD4i Env protein conformation (49, 50). In general, the IC50s were indistinguishable between paired M-tropic and T-tropic viruses and ranged from 0.34 nM to 4.2 nM, which is typical for primary isolates. Two exceptions were the 5002T Env protein, which was highly resistant (IC50 = 310 nM) and contained the known resistance mutation S375M (50), and the 4013T Env protein, which was moderately resistant (IC50 = 42.0 nM) and had no known resistance mutations. A tight range of IC50s across Env proteins suggests that the BMS-626529 inhibitor is able to access and interact similarly with its target in the CD4bs on both M-tropic and T-tropic Env proteins.
TABLE 2.
Env sensitivity to BMS-626529
| Subject ID | IC50 for BMS-626529 (nM) |
|
|---|---|---|
| T-tropic Env | M-tropic Env | |
| 4013 | 42 | 2.0 |
| 4051 | 0.87 | 0.5 |
| 4059 | 0.34 | 0.45 |
| 5002 | 310 | 1.3 |
| 7115 | 1.4 | 4.2 |
Macrophage-tropic Env proteins do not have enhanced sensitivity to HIV-1-neutralizing antibodies.
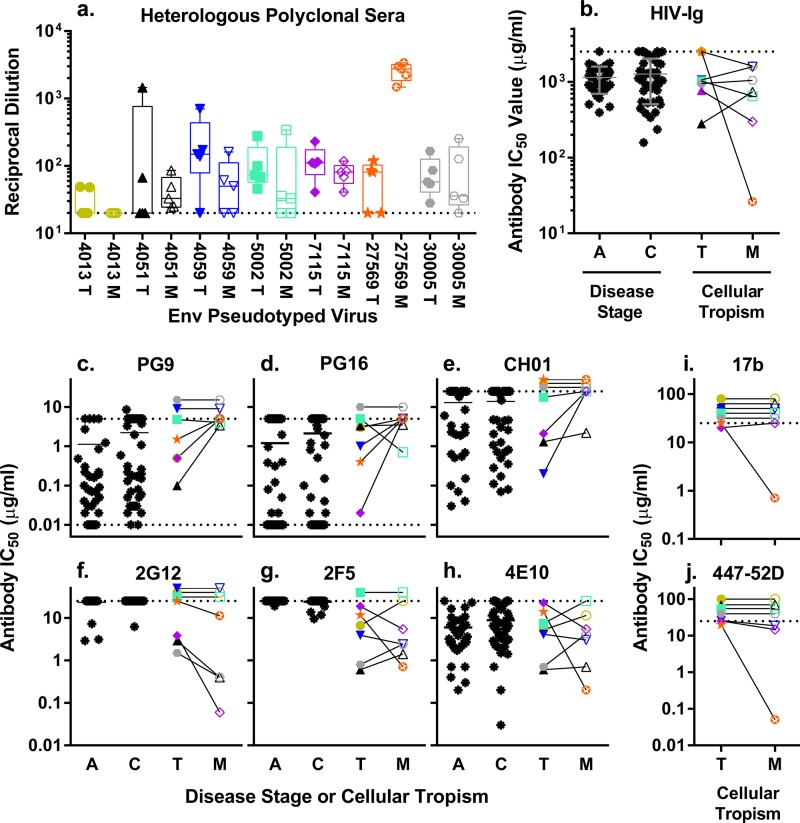
To evaluate whether macrophage tropism is associated with changes in Env protein conformation in regions outside the CD4bs, we tested the neutralization sensitivity of our well-defined panel of 14 pseudotyped viruses against a large panel of neutralizing HIV-1 antibodies. We used five polyclonal sera and HIV-Ig to detect gross changes in Env protein conformation (Fig. 4a and b) and six monoclonal antibodies (MAbs) to examine changes in specific epitopes in the V1/V2 region (Fig. 4c to e), the N-linked glycosylation coat (Fig. 4f), or the membrane-proximal external region of gp41 (MPER; Fig. 4g to h). There were no significant differences in sensitivity between the M-tropic and T-tropic Env proteins to any of the polyclonal antibodies (Fig. 4a and b), implying that there are no significant changes in neutralization sensitivity as a whole. However, analysis of the sensitivity to MAbs again suggested possible differences.
FIG 4.
Similar to T-tropic viruses, M-tropic viruses are generally resistant to neutralization by non-CD4bs-targeting antibodies with subtle trends toward increased resistance to V1/V2-targeting antibodies and decreased resistance to a glycan-targeting antibody. Pseudotyped reporter viruses were exposed to various concentrations of the following in a TZM-bl neutralization assay: polyclonal sera from five HIV-infected subjects (a); purified polyclonal HIV-Ig (b); anti-V1/V2 MAbs PG9 (c), PG16 (d), and CH01 (e); antiglycosylation MAb 2G12 (f); anti-MPER MAbs 2F5 (g), and 4E10 (h); and anti-CD4i MAbs 17b (i) and 447-52D (j). The data for the acute and chronic infection subtype C viruses are reproduced from Ping et al. (3) to allow a comparison to a large data set of typical viral Env proteins. IC50s were calculated from dose-response curves and plotted. Dashed lines represent the limits of detection. IC50s above the limit of detection (LOD) were plotted at the LOD, except for pairs where both viruses had IC50s that exceeded the LOD, in which case the symbols were stacked above the LOD line. Data for the same Env-pseudotyped viruses were used in each panel: T-tropic acute (A) and chronic (C) infection viruses (black asterisks; panels b to h only), T-tropic viruses (T; closed symbols), and M-tropic viruses (M; open symbols). Subject-matched viruses are linked, and the IC50s of T-tropic and M-tropic viruses were compared using a Wilcoxon matched-pair test. M-tropic and subject-matched T-tropic Env proteins were not significantly different in neutralization by polyclonal sera (FANOVA = 0.8 [where ANOVA is analysis of variance], r2 = 0.1, P value = 0.6) (a) or HIV-Ig (Wpaired = 12, P value = 0.5) (b) or by MPER-targeting MAbs 2F5 (Wpaired = 1, P value = 1) (g) and 4E10 (Wpaired = −2, P value = 0.9) (h). M-tropic Env proteins trended toward increased resistance to neutralization by V1/V2-targeting MAbs PG9 (Wpaired = −13, P value = 0.1) (c), PG16 (Wpaired = −13, P value = 0.2) (d), and CH01 (Wpaired = −10, P value = 0.1) (e) and increased sensitivity to neutralization by glycosylation-targeting MAb 2G12 (Wpaired = 10, P value = 0.1) (h) compared to subject-matched T-tropic Env proteins. Thirteen of the 14 viruses were resistant to neutralization by MAbs 17b (i) and 447-52D (j), which target epitopes typically present only in the CD4-bound conformation of the viral Env protein from primary isolates (for 17b, Wpaired = 1, P value = 1; for 447-52D, Wpaired = 6, P value = 0.3).
The epitopes of the PG9 and PG16 antibodies (51) are in the V1/V2 region of HIV-1 Env protein and require interactions with the N-linked glycosylation sites at amino acids 156 and 160 (52). Loss of glycosylation at either of these sites confers resistance to neutralization by PG9 and PG16. Within our panel of seven paired Env proteins, only one (4013M) has a known resistance mutation (N160Y), and this Env protein was resistant to all three V1/V2-antibodies tested. The remaining six M-tropic viruses were almost uniformly resistant to neutralization by both PG9 and PG16 (Fig. 4c and d), which is in contrast to the paired T-tropic viruses that showed the wide variation in neutralization sensitivity that was also observed in the larger panel of acute and chronic infection Env proteins. This pattern of neutralization resistance in M-tropic viruses and variable neutralization in T-tropic viruses was continued in the analysis of V1/V2 antibody CH01 (Fig. 4e), which has a more complex and conformation-dependent epitope (53). However, due to the high variation in the control groups and the small sample size, we were unable to show a statistically significant difference in neutralization between M-tropic and T-tropic viruses. These data suggest that the conformation of Env that defines the surface epitopes, perhaps through the orientation of the carbohydrate side chains, may be altered as part of the evolution to be able to enter cells with a low density of CD4.
The epitope of 2G12 is dependent on glycans at amino acid positions 295, 332, and 392 with some interactions with glycans at 386 and 448 (54). In our panel of seven pairs, four of the seven pairs (eight viruses) were resistant to neutralization by 2G12 (Fig. 4f), which is partially explained by loss of glycosylation sites in seven of eight viruses. The remaining three pairs showed a pattern of increased sensitivity in the M-tropic viruses compared to the paired T-tropic viruses. However, the small sample size compounded by resistance mutations that obscured comparisons of four of seven pairs again limited our ability to assign statistical significance to this observation. In contrast, antibodies targeting the MPER showed no difference in neutralization between M-tropic and T-tropic viruses either in directionality or variability (Fig. 4g to h). The sensitivity of M-tropic and T-tropic viruses to 2F5 and 4E10 (55) covered similar ranges and contained similar variance, which does not seem to be correlated by subject or cellular tropism.
Primary isolates from the blood are typically resistant to antibodies targeting the CD4-induced epitopes on the Env protein. However, most M-tropic viruses were isolated from the relatively immune-privileged CNS, which may release these viruses from the selective pressure that is thought to restrict exposure of CD4i epitopes. In an effort to probe the exposure of CD4i epitopes, we used two additional MAbs directed at regions of the Env protein that interact with the coreceptor and are exposed after CD4 binding. Specifically, we examined MAbs 17b (56), which has an epitope overlapping the CCR5 binding domain, and 447-52D (57), which targets the Env protein V3 loop (Fig. 4i to j). Viruses pseudotyped with either the M-tropic or T-tropic Env proteins were highly resistant to both 17b and 447-52D with IC50s near or above the detection threshold (>20 μg/ml). Only one virus, 27569 M, showed substantial neutralization sensitivity to 17b or 447-52D, and it was also the virus most sensitive to neutralization by polyclonal antibodies.
Resistance to CD4i antibodies could reflect that M-tropic viruses, like most primary isolates, do not expose CD4i epitopes, or it could reflect an absence of the specific antibody epitopes used to probe these conformations, which could mask any potential exposure of CD4i epitopes. To determine whether these Env proteins contain epitopes recognizable by the b12, 17b, and 447-52D antibodies, we have carried out a preliminary analysis by inserting single-point mutations to encode alanine at position 155 or 177 in the V1/V2 loop, which is physically distal from the targeted epitopes. These mutations individually induce an open conformation in the Env protein of the JR-CSF isolate as measured by increased sensitivity to b12, 17b, and 447-52D (W. D. Graham and K. T. Arrildt, unpublished data). Neutralizations assays using the b12, 17b, and 447-52D antibodies were repeated for viruses pseudotyped with the mutant Env proteins that maintained viability. We observed that at least one of the Env proteins for four of the five pairs of Env proteins showed increased sensitivity to either one or two of the antibodies, indicative of the presence of these epitopes. Because high neutralization sensitivity to antibodies directed at CD4i epitopes is typically associated with tissue culture-adapted strains in which the Env protein has adopted an overall open conformation (2, 17, 58, 59), the lack of sensitivity to these antibodies is consistent with M-tropic Env proteins that, despite high sensitivity to sCD4, are not in an open conformation.
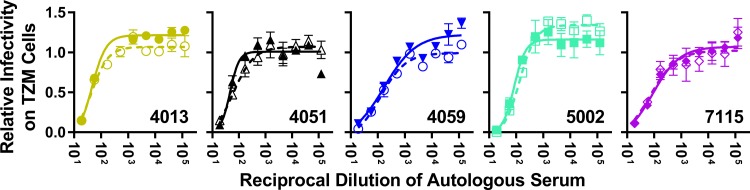
Macrophage-tropic Env proteins do not have enhanced sensitivity to autologous serum antibodies.
Most viruses currently identified as being M-tropic were isolated from the relatively immune-privileged CNS, which may have protected these viral populations from antibody-mediated selective pressure. The CNS typically maintains IgG at concentrations far lower than that in the plasma (approximately 400 times lower) (60), though levels may increase in subjects with HIV-associated neurocognitive disease (61–63). To better understand the antibody environment in which M-tropic viral populations evolve, we tested the sensitivity of five pairs of viruses to neutralization by autologous serum (Fig. 5) that was contemporaneous with the collection of the viruses. The neutralization levels of paired T-tropic and M-tropic Env-pseudotyped viruses by autologous serum were nearly identical in each case, with no distinguishable differences in the IC50s (P value = 1.0). Furthermore, the IC50s for all viruses fell within the normal range of sensitivity for primary isolates from chronic infection, with an average IC50 of 1:100. (serum dilution) and a range of 1:48 to 1:200 (64–74). These observations suggest that differences in antibody-mediated immune pressure between anatomical compartments are not a major factor in the evolution of macrophage tropism.
FIG 5.
M-tropic viruses have not evolved an increased sensitivity to autologous serum. Five paired T-tropic (closed symbols, solid lines) and M-tropic (open symbols, broken lines) pseudoviruses were exposed to heat-inactivated autologous serum (from the same subject and sampling time as the viruses isolated) in a TZM-bl neutralization assay. The relative infectivity was analyzed as a function of the reciprocal dilution of the autologous serum. No detectable differences in sensitivity to autologous serum were detected between paired T-tropic and M-tropic viruses (Wpaired = −1.0, P value = 1.0). The average IC50 across subjects and tropism is 1:100, with a range from 1:48 to 1:200 (SD = 1:50, standard error of the mean [SEM] = 1:16).
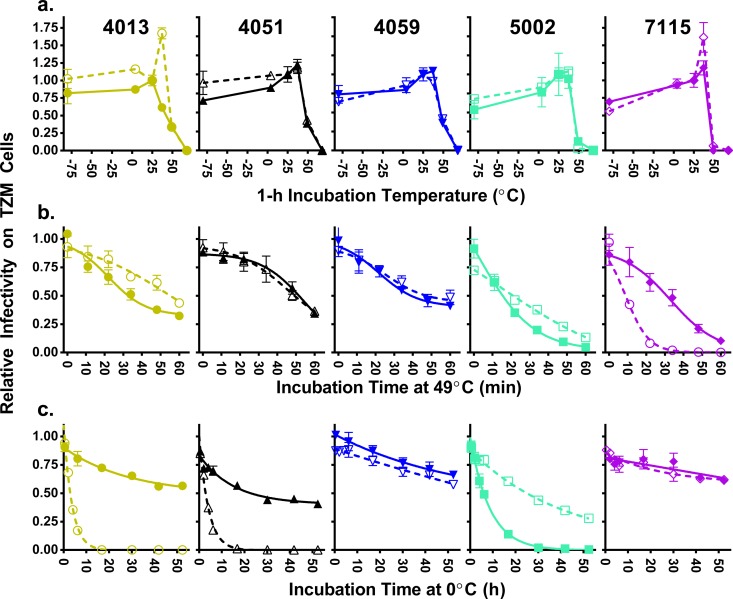
There are no gross stability differences between M-tropic Env proteins and paired T-tropic Env proteins.
Antibodies can access only the outer surface of the Env spike and cannot reveal changes in the core structure or stability of the protein. To assess whether there were differences in the stability of the Env proteins, we incubated a smaller panel of 10 (representative) pseudotyped viruses (5 pairs) at different temperatures to infer the stability of the Env trimers as measured by the loss of infectivity. We first incubated each of the viruses for 60 min at six different temperatures (−80°C, 0°C, 25°C, 37°C, 49°C, and 68°C) prior to infection of TZM-bl cells and normalized the data to infectivity after incubation at 25°C (Fig. 6a). Overall, the pseudotyped viruses were most stable at either 25°C or 37°C and were fully inactivated at 68°C. Although different temperatures produced variable levels of inactivation for individual viruses, neither sensitivity nor resistance to any temperature was consistently associated with either M-tropic or T-tropic viruses. Our second assay measured the loss of infectivity over time at 49°C, which has been previously shown to result in a moderate rate of virus inactivation (75), to provide a more quantitative assessment of the level of heat sensitivity for each Env protein (Fig. 6b). Four of the five pairs showed similar viral decay curves when the T-tropic and M-tropic viruses were compared. One pair showed a significant difference in temperature sensitivity over time, with the M-tropic virus being the most sensitive virus tested (subject 7115). However, this sensitivity was not recapitulated for other M-tropic and T-tropic clones generated from the same subject (data not shown). Finally, we measured the decay of infectivity of the panel of pseudotyped viruses at 0°C over the course of 53 h (Fig. 6c). Sensitivity to cold was highly variable between viruses, but neither sensitivity nor resistance to cold was consistent with cellular tropism. Taken together, these results show that the ability of M-tropic viruses to enter cells using a low density of CD4 and the enhanced sensitivity to sCD4 are not the result of a less stable Env protein.
FIG 6.
M-tropic viruses do not differ from T-tropic viruses in sensitivity to temperature. The effect of temperature on infectivity was assessed for five pairs of subject-matched T-tropic (closed symbols, solid lines) and M-tropic (open symbols, broken lines) Env-pseudotyped reporter viruses. (a) Viruses were incubated at various temperatures for 1 h prior to infecting TZM-bl cells, and the remaining infectivity was normalized to infectivity after incubation at 25°C. Viruses with M-tropic Env proteins were compared with subject-matched T-tropic Env proteins at five temperatures (−80°C, 4°C, 37°C, 49°C and 68°C) but revealed no significant differences between tropism groups (at −80°C, Wpaired = −3, P value = 0.8; at 4°C, Wpaired = −5, P value = 0.6; at 37°C, Wpaired = −7, P value = 0.4; at 49°C, Wpaired = −1, P value = 1; at 68°C, Wpaired = −1, P value = 1). Viruses were incubated at 49°C (b) or 0°C (c) for various lengths of time prior to infecting TZM-bl cells. The remaining infectivity was normalized to an untreated aliquot of each virus. M-tropic Env proteins differed in some cases from paired T-tropic Env proteins, but these differences were not correlated with tropism (panel b, Wpaired = 7, P value = 0.4; panel c, Wpaired = 3, P value = 0.8).
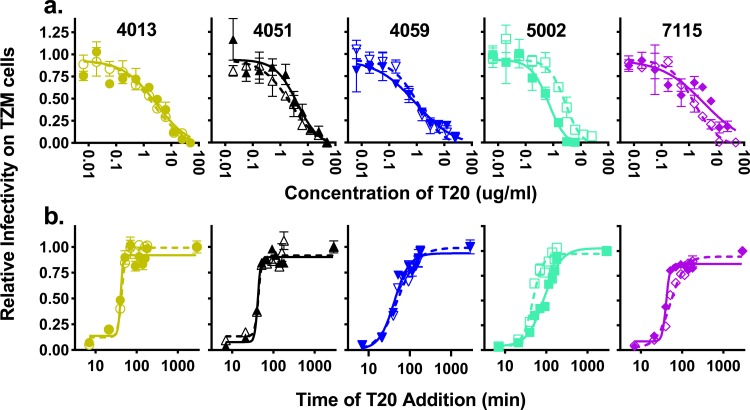
Env-mediated fusion rates do not differ between macrophage-tropic and T cell-tropic Env proteins.
We next considered the possibility that enhanced entry could be due to an enhancement in viral fusion with the cellular membrane, which may compensate for low attachment by making each attachment event more likely to result in entry (76). T20 is a fusion inhibitor that mimics the C-terminal heptad repeat (CHR) region of gp41, which normally binds to the N-terminal heptad repeat (NHR) to form a six-helix bundle that induces virus-host membrane fusion. However, when T20 binds the NHR, it stabilizes gp41 in a prefusion conformation and blocks binding by the CHR, effectively blocking fusion. To determine whether there were any significant differences in sensitivity to T20 between the five pairs of viruses, we titrated T20 to find the concentration required to inhibit fusion with TZM-bl cells for each of the 10 pseudotyped viruses (Fig. 7a). We did not detect any consistent differences in T20 sensitivity between the T-tropic and M-tropic Env proteins. We then used a fully inhibitory T20 concentration (50 μg/ml) to compare the kinetics of fusion between the two groups of viruses as they became resistant to T20, i.e., the time to formation of the six-helix bundle and fusion with the target cell (Fig. 7b). There was no difference in the rate of fusion between any of the pairs, suggesting that the rate of fusion is not a factor in macrophage tropism.
FIG 7.
M-tropic viruses have fusion kinetics similar to those of T-tropic viruses. Inhibition assays with T20 were performed on five pairs of subject-matched T-tropic (closed symbols, solid lines) and M-tropic (open symbols, broken lines) Env-pseudotyped reporter viruses. (a) Viruses were exposed to different concentrations of T20 prior to infecting TZM-bl cells, and the remaining infectivity was normalized to untreated virus. T20 dose-response curves did not differ detectably between matched pairs (Wpaired = 9, P value = 0.3) and were similar across subjects. (b) A saturating concentration (50 μg/ml) of T20 was added to viruses at various times after the addition of cells. Resistance to T20 over time was plotted as a normalized value of remaining infectivity. The resulting measures of fusion kinetics did not differ detectably between matched pairs (Wpaired = −5, P value = 0.6) and were similar across subjects.
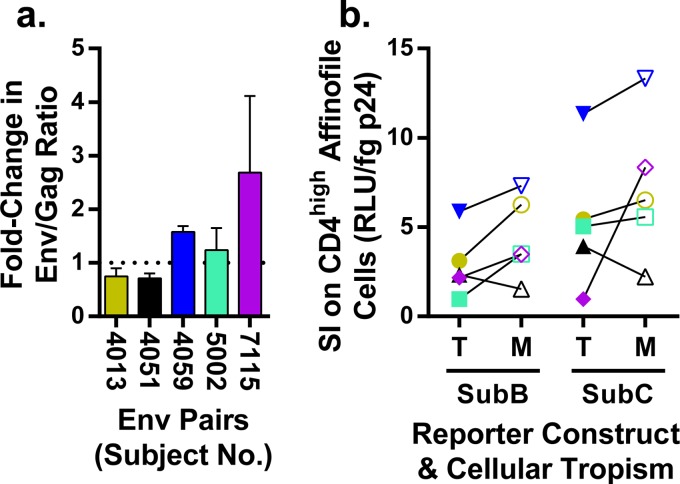
Increased Env protein incorporation does not explain increased CD4 usage by macrophage-tropic Env-pseudotyped viruses.
A simple way for a virus particle to increase the probability of interacting with CD4 would be to increase the number of Env trimers on the surface of the virion. To evaluate whether the M-tropic Env proteins had increased expression on the surface of the virion, we used Western blotting to quantify the ratio of gp120 (to represent Env) to p24 (to represent the number of virions). We then calculated the relative ratio of M-tropic Env proteins per virion as a fold change over the paired T-tropic Env proteins per virion (Fig. 8a). For these experiments, the estimation of the amount of each protein was conducted using a dilution series to work within the dynamic (although nonlinear) range of Western blot analysis (data not shown). The fold change values did not significantly differ from 1. Based on these results, we conclude that differential Env protein incorporation does not explain the enhanced CD4 usage observed for the M-tropic pseudotyped viruses.
FIG 8.
M-tropic Env proteins are incorporated at levels similar to those of T-tropic Env proteins. (a) Western blot analysis was used to evaluate the relative incorporation of Env proteins into virions using five pairs of T-tropic and M-tropic Env-pseudotyped viruses. For each virus, the abundance of Env protein was detected by Western blotting normalized to the abundance of Gag. The normalized values of Env proteins in M-tropic viruses were compared to those of subject-matched T-tropic viruses and reported as the fold change value of M-tropic over T-tropic Env protein incorporation. The fold change values (mean = 1.4, SD = 0.8) were not found to deviate significantly from a hypothetical mean of 1 (tpaired = 1, df = 4, P value = 0.3), which represents the null hypothesis of an equal abundance of Env proteins on the surface of viruses expressing T-tropic and M-tropic Env proteins. (b) Five paired T-tropic (T, closed symbols) and M-tropic (M, open symbols) env genes were pseudotyped with an env-deficient HIV-1 genome that was either subtype B with a firefly luciferase reporter gene (SubB) or subtype C with a Renilla luciferase reporter gene (SubC). Virion density was represented by p24 concentration and the titers of infectivity were determined on CD4high Affinofile cells. The specific infectivity (SI) values reported are the linear relationship of infectivity (RLU) per virion as represented by a structural protein product, p24-Gag (fg). Although the subtype C construct gives overall higher SI values, the effects of HIV-1 subtypes cannot be directly compared, because different reporters are used. Despite a small apparent trend of increased SI of M-tropic viruses in both reporter constructs, any differences in SI between paired T-tropic and M-tropic viruses are not significant when taken together (Wpaired = 13, P value = 0.13) or separated by reporter construct (for SubB, Wpaired = 13, P value = 0.13; for SubC, Wpaired = 9, P value = 0.31). Also, because the CD4 density on CD4high Affinofiles is saturating for M-tropic viruses and not saturating for T-tropic viruses, the SI values for T-tropic viruses are likely an underestimate, which may account for a slight reduction in the observed SI for T-tropic viruses compared to that for M-tropic viruses.
Env protein incorporation can be affected by interactions been Env and structural proteins (reviewed in reference 77). We examined whether such interactions alter specific infectivity (SI) in our experimental system, where SI is defined as the infectivity per viral particle (Fig. 8b). SI was assessed by measuring the infectivity of pseudoviruses produced from the same 10 pairs of env clones carrying either the structural proteins encoded by the env-deficient HIV-1 subtype B reporter genome or subtype C reporter genome and using p24 concentration to infer the per virion relative infectivity. There were similar trends within each reporter construct in that there was an approximately 10-fold range of SI values when comparing a single reporter construct (subtype B or C) across the different Env proteins. Also, there was a trend toward a small increase in infectivity of the M-tropic viruses; however, CD4 densities on CD4high Affinofile cells are not saturating for infection by T-tropic viruses, making it possible that their SI values are underestimated. Also, while the subtype C reporter virus appears to confer a slight enhancement in SI overall, the two reporter constructs cannot be directly compared because the different luciferase reporter enzymes (firefly versus Renilla) may exhibit different amounts of luminescence, resulting from the same number of infection events. Thus, although we see variation between Env proteins, the variation between different subjects is greater than between the paired M- and T-tropic Env proteins, indicating that macrophage tropism, defined by the Env protein, is not achieved by increasing the per-particle infectivity.
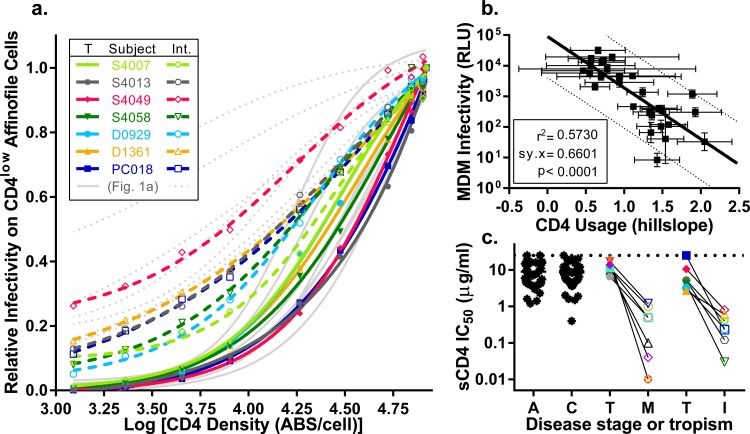
CD4 usage is predictive of MDM infectivity and, along with sensitivity to sCD4, reveals viruses with intermediate phenotypes.
Using well-characterized, subject-matched M-tropic and T-tropic env genes, we have demonstrated that MDM infectivity (when averaged over multiple donors), low CD4 usage, and sensitivity to sCD4 are clear phenotypes that can be used to distinguish M-tropic viruses from paired T-tropic viruses. Based on these defined characteristics, we identified seven additional HIV-1 env genes cloned from compartmentalized virus in the CSF early after HIV-1 infection of infants (42) or in the genital tract of adults (E. N. Dukhovlinova and L. Ping, unpublished data) that showed a modest enhancement of entry on CD4low Affinofile cells (i.e., “intermediate” between the values of T-tropic and M-tropic viruses) and seven subject-matched T-tropic Env proteins isolated from the blood. Viruses with intermediate CD4 usage phenotypes are rare and may provide information about the evolution of macrophage tropism. We analyzed the viral entry phenotypes of these additional seven intermediate pairs using Affinofile cells expressing CD4 across the full range of densities. We found that viruses with an intermediate entry phenotype on CD4low cells also showed differences in entry phenotype compared to both M-tropic and T-tropic viruses over the entire range of CD4 densities (Fig. 9a). When we used our entire 14-pair panel of 28 viruses (14 T-tropic, 7 M-tropic, and 7 intermediate) to infect MDMs, we found that differences in the ability to use CD4 (as captured by the Hill slope of the CD4 titration curve) was predictive for infectivity on MDMs (r2 = 0.57) (Fig. 9b). This result demonstrates that a substantial fraction of MDM infectivity can be accounted for by the ability of the Env protein to use low CD4 densities for entry.
FIG 9.
Viruses of intermediate CD4 usage reveal a correlation between CD4 usage and MDM infectivity. Seven pairs of subject-matched Env-pseudotyped viruses in which one virus had a modest enhancement in entry of CD4low Affinofile cells (intermediate, Int.) and the other virus was T-tropic (T) were analyzed and plotted, and the data were overlapped for the original seven pairs of M-tropic (M) and T-tropic (T) viruses shown in Fig. 1. (a) CD4 usage of intermediate (open symbols, broken lines) and T-tropic (closed symbols, solid lines) viruses was evaluated by the Affinofile cell assay as previously described and plotted against previously shown data for subject-matched T-tropic (solid light gray lines) and M-tropic (dotted light gray lines) viruses from Fig. 1. Unique colors are used to identify each subject as specified in the figure legend (the overlap in symbols with those from Fig. 1 is not meaningful). The intermediate viruses differ from paired T-tropic viruses in Hill slope (tpaired = 2.9, df = 6, P value = 0.03) but not in EC50 (tpaired = 1.0, df = 6, P value = 0.34). Similarly, intermediate viruses also differ from unpaired M-tropic viruses, but only in EC50 (tpaired = 2.5, df = 12, P value = 0.03) and not Hill slope (tpaired = 0.09, df = 12, P value = 0.93). (b) CD4 usage was represented by Hill slope values for each virus, which was plotted against log normalized values of infectivity on MDMs. We evaluated the correlation of CD4 usage and MDM infection and plotted the linear regression (solid line) with the 95% confidence band (broken lines). Variation in CD4 usage could explain 57% of the variation in MDM infectivity (r2 = 0.57, Sy.x = 0.66 [where Sy.x is residual standard deviation], P value < 0.0001). (c) Neutralization sensitivity to sCD4 was evaluated in a TZM-bl neutralization assay as previously described and reported as IC50s. Subject-matched pairs of intermediate (Int., open symbols) and T-tropic (T, closed symbols) viruses were plotted against data from Fig. 3a for subject-matched pairs of T-tropic (T, closed symbols) and M-tropic (M, open symbols) viruses and the panel of viruses from acute (A) and chronic (C) infections (black asterisks). The colors and symbols are the same as those identified in Fig. 1a and 9a. Subject-matched viruses are linked. Similar to M-tropic Env proteins, Int. Env proteins are 23-fold more sensitive to neutralization by sCD4 than subject-matched T-tropic Env proteins (tpaired = 6.8, df = 6, P value = 0.0005).
Given that the CD4 entry phenotype appears to be continuous within the dynamic range that was tested, we asked whether this was also the case for sCD4 sensitivity. As shown in Fig. 3, M-tropic Env proteins are significantly more sensitive to sCD4 than the paired T cell-tropic Env proteins. Using the same approach, we were also able to show that intermediate Env proteins are significantly more sensitive to sCD4 than subject-matched T-tropic Env proteins (P value = 0.0005) (Fig. 9c). Furthermore, the sCD4 IC50s of the intermediate viruses were similar to those of the well-characterized M-tropic viruses. Taken together, this suggests that evolution of the M-tropic entry phenotype may be a multistep process marked by increased sensitivity to sCD4 and enhanced entry at low CD4 densities.
DISCUSSION
HIV-1 evolution to enter and replicate in macrophages results in Env proteins that can more efficiently utilize the low densities of CD4 found on macrophages for viral entry. This ability to use a low density of CD4 is a key feature that distinguishes M-tropic viruses from the more typical R5 T-tropic viruses, which require high densities of CD4 for entry into target T cells. We found that low-density CD4 usage and sensitivity to soluble CD4 (sCD4) are distinct criteria for macrophage tropism. Furthermore, CD4 entry phenotypes are more reproducible than the relative infection of monocyte-derived macrophages (MDMs). We also found suggestive evidence that the evolution of macrophage tropism includes multiple stages. Although we observed that the enhanced CD4 usage of all the M-tropic viruses was accompanied by an increased sensitivity to sCD4, we also observed multiple “intermediate” viral lineages that had significantly increased sensitivity to sCD4 but, in some cases, had only a slight increase in the ability to use low CD4 densities. Based on these results, we hypothesize that macrophage tropism evolves through multiple stages that can be marked by an increase in sCD4 sensitivity, enhancement of CD4 usage, and other subtle changes in the Env protein conformation.
In order to make a more accurate assessment of the phenotypic changes that emerge during the evolution of macrophage tropism, we designed our analysis to contain three vital attributes. First, we examined pairs of M-tropic and T-tropic env genes isolated from the same subjects without ex vivo passaging of their virus. This minimizes the effect of strain-to-strain variation and focuses our analyses on the evolutionary event(s) that took place within each subject. Also, because we used a single pair of env genes from each subject, each comparison was an independent observation. Second, we analyzed the phenotypes of Env proteins expressed from env genes that had been generated by endpoint dilution PCR, which avoids artificial recombinants during amplification. Third, we made phenotypic comparisons across a group of env gene pairs from different subjects to identify generalizable differences between T-tropic and M-tropic viruses. These key features allowed us to further define the properties of Env proteins that result in low-density CD4 usage and an M-tropic phenotype.
Infection of MDMs ex vivo has long been used as a measure of macrophage tropism (17, 23, 26, 78–81). However, different preparations of MDMs vary in their ability to be infected in ways that have not been controlled experimentally (13, 14, 80). The typical approach to this substantial experimental problem has been to compare several donors and infer a result. An additional complication of this approach is that R5 T-tropic viruses also vary widely in their ability to infect MDMs (14). This two-dimensional variability (virus and cells) makes it difficult to unequivocally assign cellular tropism in isolation. However, by pooling MDM infection data from four donors and multiple subject-matched M-tropic and T-tropic pairs of viruses, we found a 28-fold increase in average MDM infection by M-tropic over T-tropic viruses (Fig. 2). These data provide a quantitative estimate of the typical gain in efficiency of MDM infectivity achieved in transitioning to macrophage tropism.
The difference in CD4 density on the surface of macrophages and CD4+ T cells inspired the use of cell lines with either high or low levels of CD4 as a method to assess the relationship between efficiency of CD4 usage and macrophage tropism (26, 80, 82, 83). The efficiency of CD4 usage as a surrogate phenotype for macrophage tropism is most easily demonstrated using Affinofile cells, in which CD4 expression can be varied (21) over a wide range and can approximate the surface densities of CD4 on T cells and on MDMs (14). This more-sensitive measure of CD4 usage allowed us to identify HIV-1 variants that are characterized by both an intermediate entry phenotype on CD4low cells and an intermediate entry phenotype on MDMs (Fig. 9b). Based on this analysis, we were able to demonstrate that HIV-1 infection efficiency on CD4low cells predicts macrophage tropism and also predicts intermediate entry phenotypes.
We have focused our study on the role of HIV-1 Env in mediating cell entry and so have not considered other potential viral proteins or sequences that may evolve as part of the adaptation to macrophages. We have ruled out that the differences observed between M-tropic and T-tropic viruses result from differences in the specific infectivity as conferred by their Env proteins (Fig. 8b). We also evaluated whether increased Env protein incorporation, which is mediated by interactions between Env and Gag proteins, could contribute to enhanced CD4 usage but found comparable levels of Env proteins per viral particle between subject-matched T-tropic and M-tropic virions (Fig. 8a). Thus, the M-tropic and T-tropic viruses are equivalent in how they contribute to particle formation and confer infectivity in the presence of high levels of CD4, with the differences in viruses being much greater in viruses isolated between subjects than between the M-tropic and T-tropic viruses present within a subject.
The enhanced ability of M-tropic viruses to use low CD4 densities for entry suggests an altered interaction between Env and CD4. This suggestion is supported by the observation that the Hill slope determined by CD4 titration is significantly different between these two groups of viruses (Fig. 1a). Previous studies have reported that M-tropic variants are also more sensitive to sCD4 than T-tropic variants (27–29). We confirmed this observation by showing that M-tropic variants were on average 27-fold more sensitive to sCD4 than their T-tropic counterparts (Fig. 3a). However, we found no difference in the ability of M-tropic or T-tropic viruses to interact with the CD4 binding site (CD4bs) small-molecule agonist BMS-529626 (Table 2). A previous report found differences in sensitivity to BMS-663068 (the prodrug form of BMS-626529) when comparing M-tropic and T-tropic viruses (84); however, that study included multiple and unequal numbers of clones from each compartment of several subjects, which complicates the statistical analysis, which assumes independence of observations. When we probed with a series of CD4bs antibodies, we found suggestive patterns in two of the three CD4bs antibodies tested that hinted at an increased sensitivity of M-tropic viruses in the pairs of viruses that were not completely resistant (Fig. 3b to d). Two additional CD4bs antibodies, F105 and CH103, are typically nonneutralizing in primary isolates, which has been suggested to result from steric restriction related to the angle of access between the antibodies and the Env trimer (85–87). The five pairs of viruses tested against F105 and CH103 were almost uniformly resistant to neutralization (data not shown), suggesting that the angle of CD4 access is not dramatically altered in the M-tropic viruses. The variability in antibody sensitivity between Env proteins to these antibodies limits the statistical power of these analyses, given the limited number of subject-matched M-tropic and T-tropic Env pairs available. However, the patterns of increased sensitivity to the relatively bulky CD4bs antibodies, when coupled with the significant increases in sensitivity to sCD4, suggest that conformational differences around the CD4 binding site may exist in Env proteins from M-tropic viruses.
We also examined whether conformational differences around the CD4 binding site create a more open conformation. Early observations revealed that tissue culture-adapted viruses, in the absence of antibody selection, result in the premature exposure of CD4-induced epitopes (88–91), indicating that the Env protein for these viruses is in a more open conformation that exposes the coreceptor binding site. Furthermore, studies have shown that tissue culture-adapted viruses evolved to use low CD4 for entry (92, 93) were more sensitive to sCD4 (94–96) and had greater sensitivity to antibody neutralization than typical primary isolates (58, 89, 92, 97). Together, these observations raised the possibility that these phenotypes covary and are generated by the same mechanism. To determine whether selection to use low CD4 densities for cell entry results in an open Env protein conformation, we examined Env protein sensitivity to polyclonal sera and MAbs directed against the V3 and CD4i epitopes as a general probe for these two conformational states. We found that there was no difference in antibody sensitivity to polyclonal sera or to most MAbs when comparing the M-tropic and T-tropic viruses (Fig. 4), indicating that M-tropic viruses have not assumed the open conformation of tissue culture-adapted viruses. Furthermore, there was no difference in T-tropic and M-tropic viruses in sensitivity to autologous, contemporaneous serum (Fig. 5), and the IC50s from all viruses tested were similar to those typically reported for chronic infection (64–74). However, M-tropic viruses appeared to be more resistant to the V1/V2-targeted MAb PG9 (Fig. 4c), suggesting that M-tropic viruses may differ in the Env protein conformation of the V1/V2 region, which is now understood to be highly ordered (52, 98) and is thought to contribute to the structure and function of the Env trimer (99–103). Although our ability to assign significance to V1/V2 antibody resistance as a feature of macrophage tropism is limited by sample size (n = 7) and the inherent variability to the neutralization of primary isolates, this observation is consistent with another obtained using a larger panel of unpaired T-tropic and M-tropic viruses (29). Overall, the neutralizing antibody sensitivity properties of M-tropic Env proteins reveals them to be more like R5 T-tropic Env proteins than tissue culture-adapted variants, but with intriguing differences in the CD4bs and perhaps the V1/V2 loop region.
Dissociation of attached virions prior to successful entry is common (76) and is likely to be a more substantial problem when CD4 is at a low density. Increased efficiency of postattachment entry steps may be able to compensate when attachment conditions are poor, which led us to examine the properties of fusion (the final step) and the properties that may affect the conformational changes that occur between attachment and fusion. Differences in Env protein stability affect the conformational changes required for entry (104), and differences in stability can be probed by sensitivity to inactivation at different temperatures. Env proteins display variation in sensitivity to heat (75, 104, 105) and cold (105–107). Heat lability is a feature of the open conformation of tissue culture-adapted viruses but not of comparatively stable primary isolates (75, 104). Although there was some variation in heat sensitivity between Env proteins from different subjects, the subject-matched M-tropic and T-tropic virus pairs were generally indistinguishable (Fig. 6b). There were significant differences in cold sensitivity within pairs, but sensitivity was inconsistent across T-tropic or M-tropic Env protein pairs (Fig. 6c). Therefore, thermal sensitivity is also unlikely to be a coevolving feature of HIV-1 cellular tropism or CD4 usage. In contrast, fusion was consistent within a subject-pair, despite some variation between viruses from different subjects (Fig. 7). Overall, we found no evidence that fusion efficiency or Env protein instability is linked to cellular tropism or CD4 usage. From these observations, features of macrophage tropism appear to be limited to the Env-CD4 interaction.
The evolutionary path to HIV-1 macrophage tropism remains poorly understood. A number of Env substitutions have been suggested to contribute to macrophage tropism (24, 25, 27, 30–39), but none of these suggested mutations can distinguish the paired M-tropic and T-tropic viruses used in our study. By including additional HIV-1 variants identified by intermediate CD4 usage, we observed that both MDM infection and low CD4 usage vary across a wide range. This suggests sequential adaptation to infecting macrophages that may require the accumulation of several mutations. Direct comparison revealed that variation in CD4 usage could explain more than one-half of the observed variation in MDM infection (Fig. 9b). We do not know if other factors also impact entry into macrophages or if the remaining variation is due to experimental variation.
We previously showed that T-tropic and M-tropic viruses differ in CD4 usage but not in CCR5 usage, as measured by sensitivity to maraviroc (14). We also observed that M-tropic and intermediate variants have similar increases in sensitivity to sCD4 compared to T cell-tropic variants (Fig. 9c). Other groups have also identified viruses from the CNS that, like our intermediate viruses, are sensitive to sCD4 but unable to efficiently infect MDMs (for examples, see references 28 and 108). The lack of a positive relationship between sensitivity to sCD4 and the ability to infect MDMs made it difficult for previous studies to assign tropism for these viruses. The increased sensitivity of our CD4 usage assay allowed us to detect a group of viruses with intermediate enhancement of CD4 usage, sensitivity to sCD4, and an intermediate ability to infect MDM in vivo. Together, these findings imply that sCD4 sensitivity is a phenotype that can be detected early in the enhancements in low CD4 usage. Also, the general resistance to neutralizing antibodies combined with a pattern of sensitivity to sCD4 indicates that M-tropic Env proteins exist in a state that is distinct from both R5 T cell-tropic Env proteins (which are resistant to sCD4) and tissue culture-adapted Env proteins (which are globally sensitive to neutralization).
Because our viruses represent a cross-section of viral populations rather than a longitudinal study of evolving populations, it is difficult to determine whether a continuous range of values (e.g., CD4 usage or MDM infectivity) represents a continuous phenotype or a large variation around a bimodal phenotype. However, if CD4 usage and MDM infectivity are on a multistep evolutionary path to macrophage tropism (as suggested by our detection of intermediates), then defining the mutations on this pathway and understanding the nature of the change in the interaction between the viral Env protein and the host CD4 receptor remain as important questions to inform our understanding of the evolution of macrophage tropism.
ACKNOWLEDGMENTS
We are grateful to the subjects that provided samples and those who contributed to their collection. Reagents were generously provided by Nancy Haigwood, Mark Krystal at Bristol-Myers Squibb, and through the NIH AIDS Research Reference and Reagent Program. We also thank Sook-Kyung Lee for providing her invaluable time and advice.
This work was funded by an award from the National Institutes of Health R01 MH101024 and R37 AI44667 to R.S. and P01 MH094177. K.T.A. and C.B.S. were supported by T32 AI07419. Much of the neutralization sensitivity work was supported by the NIAID-NIH contract number HHSN27201100016C (to D.C.M). R.S.H. and M.M. were supported by the MLW Core Programme Grant from the Wellcome Trust UK. We also received support from the UNC Center for AIDS Research (P30 AI50410) and the Lineberger Comprehensive Cancer Center (P30 CA16086).
Tissue samples were provided by the National NeuroAIDS Tissue Consortium (NNTC), which is funded by the NIH through the NIMH and NINDS Institutes by the following grants: Texas NeuroAIDS Research Center (U24MH100930), California NeuroAIDS Tissue Network (U24MH100928), National Neurological AIDS Bank (U24MH100929), Manhattan HIV Brain Bank (U24MH100931), and Data Coordinating Center (U24MH100925). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NNTC or NIH.
Conflict of interests statement: G.S. is an employee of AbbVie. M.S.C. served on an advisory board for Roche Molecular Systems, Inc. R.W.P. and R.S. have each been awarded an honorarium and travel support to speak at a scientific meeting by AbbVie. R.W.P. also served as a one-time consultant with Merck & Co and has received an honorarium and travel expenses from Gilead to provide a lecture at a scientific meeting.
REFERENCES
- 1.Isaacman-Beck J, Hermann EA, Yi Y, Ratcliffe SJ, Mulenga J, Allen S, Hunter E, Derdeyn CA, Collman RG. 2009. Heterosexual transmission of human immunodeficiency virus type 1 subtype C: macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J Virol 83:8208–8220. doi: 10.1128/JVI.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BTM, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol 80:11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ping L-H, Joseph SB, Anderson JA, Abrahams M-R, Salazar-Gonzalez JF, Kincer LP, Treurnicht FK, Arney L, Ojeda S, Zhang M, Keys J, Potter EL, Chu H, Moore P, Salazar MG, Iyer S, Jabara C, Kirchherr J, Mapanje C, Ngandu N, Seoighe C, Hoffman I, Gao F, Tang Y, Labranche C, Lee B, Saville A, Vermeulen M, Fiscus S, Morris L, Karim SA, Haynes BF, Shaw GM, Korber BT, Hahn BH, Cohen MS, Montefiori D, Williamson C, Swanstrom R, CAPRISA Acute Infection Study and the Center for HIV-AIDS Vaccine Immunology Consortium. 2013. Comparison of viral Env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol 87:7218–7233. doi: 10.1128/JVI.03577-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sleasman JW, Aleixo LF, Morton A, Skoda-Smith S, Goodenow MM. 1996. CD4+ memory T cells are the predominant population of HIV-1-infected lymphocytes in neonates and children. AIDS 10:1477–1484. doi: 10.1097/00002030-199611000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 6.Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol 78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koot M, Keet IP, Vos AH, de Goede RE, Roos MT, Coutinho RA, Miedema F, Schellekens PT, Tersmette M. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med 118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 8.Tscherning C, Alaeus A, Fredriksson R, Björndal A, Deng H, Littman DR, Fenyö EM, Albert J. 1998. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology 241:181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 9.Huang W, Eshleman SH, Toma J, Fransen S, Stawiski E, Paxinos EE, Whitcomb JM, Young AM, Donnell D, Mmiro F, Musoke P, Guay LA, Jackson JB, Parkin NT, Petropoulos CJ. 2007. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol 81:7885–7893. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho SH, Shek L, Gettie A, Blanchard J, Cheng-Mayer C. 2005. V3 loop-determined coreceptor preference dictates the dynamics of CD4+-T-cell loss in simian-human immunodeficiency virus-infected macaques. J Virol 79:12296–12303. doi: 10.1128/JVI.79.19.12296-12303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimura Y, Igarashi T, Donau OK, Buckler-White A, Buckler C, Lafont BAP, Goeken RM, Goldstein S, Hirsch VM, Martin MA. 2004. Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc Natl Acad Sci U S A 101:12324–12329. doi: 10.1073/pnas.0404620101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci U S A 94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci U S A 96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joseph SB, Arrildt KT, Swanstrom AE, Schnell G, Lee B, Hoxie JA, Swanstrom R. 2014. Quantification of entry phenotypes of macrophage-tropic HIV-1 across a wide range of CD4 densities. J Virol 88:1858–1869. doi: 10.1128/JVI.02477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewin S, Sonza S, Irving L, McDonald C, Mills J, Crowe S. 1996. Surface CD4 is critical to in vitro HIV infection of human alveolar macrophages. AIDS Res Hum Retroviruses 12:877–883. doi: 10.1089/aid.1996.12.877. [DOI] [PubMed] [Google Scholar]
- 16.Yi Y, Chen W, Frank I, Cutilli J, Singh A, Starr-Spires L, Sulcove J, Kolson DL, Collman RG. 2003. An unusual syncytia-inducing human immunodeficiency virus type 1 primary isolate from the central nervous system that is restricted to CXCR4, replicates efficiently in macrophages, and induces neuronal apoptosis. J Neurovirol 9:432–441. doi: 10.1080/13550280390218706. [DOI] [PubMed] [Google Scholar]
- 17.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander M, Lynch R, Mulenga J, Allen S, Derdeyn CA, Hunter E. 2010. Donor and recipient envs from heterosexual human immunodeficiency virus subtype C transmission pairs require high receptor levels for entry. J Virol 84:4100–4104. doi: 10.1128/JVI.02068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martín-García J, Cao W, Varela-Rohena A, Plassmeyer ML, González-Scarano F. 2006. HIV-1 tropism for the central nervous system: brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology 346:169–179. doi: 10.1016/j.virol.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 20.Kabat D, Kozak SL, Wehrly K, Chesebro B. 1994. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol 68:2570–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston SH, Lobritz MA, Nguyen S, Lassen K, Delair S, Posta F, Bryson YJ, Arts EJ, Chou T, Lee B. 2009. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J Virol 83:11016–11026. doi: 10.1128/JVI.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. 2011. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog 7:e1002286. doi: 10.1371/journal.ppat.1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters PJ, Sullivan WM, Duenas-Decamp MJ, Bhattacharya J, Ankghuambom C, Brown R, Luzuriaga K, Bell J, Simmonds P, Ball J, Clapham PR. 2006. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J Virol 80:6324–6332. doi: 10.1128/JVI.02328-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, Kunstman K, Wolinsky SM, Gabuzda D. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci U S A 103:15160–15165. doi: 10.1073/pnas.0605513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duenas-Decamp MJ, Peters PJ, Burton D, Clapham PR. 2009. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J Virol 83:2575–2583. doi: 10.1128/JVI.02133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, Moore JP, Wolinsky SM, Gabuzda D. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol 76:6277–6292. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musich T, Peters PJ, Duenas-Decamp MJ, Gonzalez-Perez MP, Robinson J, Zolla-Pazner S, Ball JK, Luzuriaga K, Clapham PR. 2011. A conserved determinant in the V1 loop of HIV-1 modulates the V3 loop to prime low CD4 use and macrophage infection. J Virol 85:2397–2405. doi: 10.1128/JVI.02187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Perez MP, O'Connell O, Lin R, Sullivan WM, Bell J, Simmonds P, Clapham PR. 2012. Independent evolution of macrophage-tropism and increased charge between HIV-1 R5 envelopes present in brain and immune tissue. Retrovirology 9:20. doi: 10.1186/1742-4690-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connell O, Repik A, Reeves JD, Gonzalez-Perez MP, Quitadamo B, Anton ED, Duenas-Decamp M, Peters P, Lin R, Zolla-Pazner S, Corti D, Wallace A, Wang S, Kong X-P, Lu S, Clapham PR. 2013. Efficiency of bridging-sheet recruitment explains HIV-1 R5 envelope glycoprotein sensitivity to soluble CD4 and macrophage tropism. J Virol 87:187–198. doi: 10.1128/JVI.01834-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cashin K, Roche M, Sterjovski J, Ellett A, Gray LR, Cunningham AL, Ramsland PA, Churchill MJ, Gorry PR. 2011. Alternative coreceptor requirements for efficient CCR5- and CXCR4-mediated HIV-1 entry into macrophages. J Virol 85:10699–10709. doi: 10.1128/JVI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K, Bell JE, Wolinsky SM, Gabuzda D. 2007. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 360:105–119. doi: 10.1016/j.virol.2006.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duenas-Decamp MJ, Peters P, Burton D, Clapham PR. 2008. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J Virol 82:5807–5814. doi: 10.1128/JVI.02585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunfee RL, Thomas ER, Wang J, Kunstman K, Wolinsky SM, Gabuzda D. 2007. Loss of the N-linked glycosylation site at position 386 in the HIV envelope V4 region enhances macrophage tropism and is associated with dementia. Virology 367:222–234. doi: 10.1016/j.virol.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouyang Y, Liu L, Zhang Y, Yuan L, Liu Z, Yang S, Wei F, Qiao L, Chen D. 2014. Discordant patterns of tissue-specific genetic characteristics in the HIV-1 env gene from HIV-associated neurocognitive disorder (HAND) and non-HAND patients. J Neurovirol 20:332–340. doi: 10.1007/s13365-014-0247-5. [DOI] [PubMed] [Google Scholar]
- 35.Harrington PR, Connell MJ, Meeker RB, Johnson PR, Swanstrom R. 2007. Dynamics of simian immunodeficiency virus populations in blood and cerebrospinal fluid over the full course of infection. J Infect Dis 196:1058–1067. doi: 10.1086/520819. [DOI] [PubMed] [Google Scholar]
- 36.Rossi F, Querido B, Nimmagadda M, Cocklin S, Navas-Martín S, Martín-Garcia J. 2008. The V1-V3 region of a brain-derived HIV-1 envelope glycoprotein determines macrophage tropism, low CD4 dependence, increased fusogenicity and altered sensitivity to entry inhibitors. Retrovirology 5:89. doi: 10.1186/1742-4690-5-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evering TH, Kamau E, Bernard LS, Farmer CB, Kong X-P, Markowitz M. 2014. Single genome analysis reveals genetic characteristics of Neuroadaptation across HIV-1 envelope. Retrovirology 11:65. doi: 10.1186/s12977-014-0065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sterjovski J, Churchill MJ, Ellett A, Gray LR, Roche MJ, Dunfee RL, Purcell DFJ, Saksena N, Wang B, Sonza S, Wesselingh SL, Karlsson I, Fenyo E-M, Gabuzda D, Cunningham AL, Gorry PR. 2007. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology 4:89. doi: 10.1186/1742-4690-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards KH, Aasa-Chapman MM, McKnight A, Clapham PR. 2010. Modulation of HIV-1 macrophage-tropism among R5 envelopes occurs before detection of neutralizing antibodies. Retrovirology 7:48. doi: 10.1186/1742-4690-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arrildt KT, Joseph SB, Swanstrom R. 2012. The HIV-1 env protein: a coat of many colors. Current HIV/AIDS Rep 9:52–63. doi: 10.1007/s11904-011-0107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sturdevant CB, Dow A, Jabara CB, Joseph SB, Schnell G, Takamune N, Mallewa M, Heyderman RS, Van Rie A, Swanstrom R. 2012. Central nervous system compartmentalization of HIV-1 subtype C variants early and late in infection in young children. PLoS Pathog 8:e1003094. doi: 10.1371/journal.ppat.1003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montefiori DC. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol Chapter 12:Unit 12.11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 44.Joseph SB, Lee B, Swanstrom R. 2014. Affinofile assay for identifying macrophage-tropic HIV-1. Bio-protocol 4(14):e1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnell G, Spudich S, Harrington P, Price RW, Swanstrom R. 2009. Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1-associated dementia. PLoS Pathog 5:e1000395. doi: 10.1371/journal.ppat.1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burton DR, Barbas CF, Persson MA, Koenig S, Chanock RM, Lerner RA. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A 88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang Z-Y, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schön A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB, Sodroski JG. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci U S A 101:5036–5041. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nowicka-Sans B, Gong Y-F, McAuliffe B, Dicker I, Ho H-T, Zhou N, Eggers B, Lin P-F, Ray N, Wind-Rotolo M, Zhu L, Majumdar A, Stock D, Lataillade M, Hanna GJ, Matiskella JD, Ueda Y, Wang T, Kadow JF, Meanwell NA, Krystal M. 2012. In vitro antiviral characteristics of HIV-1 attachment inhibitor BMS-626529, the active component of the prodrug BMS-663068. Antimicrob Agents Chemother 56:3498–3507. doi: 10.1128/AAC.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang G-Y, Diwanji D, Georgiev I, Do Kwon Y, Lee D, Louder MK, Moquin S, Schmidt SD, Yang Z-Y, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang L-X, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonsignori M, Hwang K-K, Chen X, Tsao C-Y, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Yongping Y, Zhang B, Zhu J, Kwong PD, O'Dell S, Mascola JR, Wu L, Nabel GJ, Phogat S, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori DC, Kepler TB, Tomaras GD, Alam SM, Liao H-X, Haynes BF. 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol 85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol 76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses 10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 56.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buchbinder A, Zolla-Pazner S, Karwowska S, Gorny MK, Burda ST. 1992. Synergy between human monoclonal antibodies to HIV extends their effective biologic activity against homologous and divergent strains. AIDS Res Hum Retroviruses 8:1395. [DOI] [PubMed] [Google Scholar]
- 58.Moore JP, Cao Y, Qing L, Sattentau QJ, Pyati J, Koduri R, Robinson J, Barbas CF, Burton DR, Ho DD. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol 69:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keir G, Thompson EJ. 1986. Proteins as parameters in the discrimination between different blood-CSF barriers. J Neurol Sci 75:245–253. doi: 10.1016/0022-510X(86)90072-9. [DOI] [PubMed] [Google Scholar]
- 61.Singer EJ, Syndulko K, Fahy-Chandon BN, Shapshak P, Resnick L, Schmid P, Conrad AJ, Tourtellotte WW. 1994. Cerebrospinal fluid p24 antigen levels and intrathecal immunoglobulin G synthesis are associated with cognitive disease severity in HIV-1. AIDS 8:197–204. doi: 10.1097/00002030-199402000-00007. [DOI] [PubMed] [Google Scholar]
- 62.Anthony IC, Crawford DH, Bell JE. 2003. B lymphocytes in the normal brain: contrasts with HIV-associated lymphoid infiltrates and lymphomas. Brain 126:1058–1067. doi: 10.1093/brain/awg118. [DOI] [PubMed] [Google Scholar]
- 63.Meira CS, Vidal JE, Costa-Silva TA, Motoie G, Gava R, Hiramoto RM, Pereira-Chioccola VL. 2013. IgG4 specific to Toxoplasma gondii excretory/secretory antigens in serum and/or cerebrospinal fluid support the cerebral toxoplasmosis diagnosis in HIV-infected patients. J Immunol Methods 395:21–28. doi: 10.1016/j.jim.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 64.Mahalanabis M, Jayaraman P, Miura T, Pereyra F, Chester EM, Richardson B, Walker B, Haigwood NL. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naïve human immunodeficiency virus controllers. J Virol 83:662–672. doi: 10.1128/JVI.01328-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bunnik EM, Pisas L, Nuenen ACV, Schuitemaker H. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J Virol 82:7932–7941. doi: 10.1128/JVI.00757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, Li H, Leseka N, Treurnicht F, Mlisana K, Shaw GM, Karim SSA, Williamson C, Morris L, CAPRISA 002 Study Team. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol 81:6187–6196. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deeks SG, Schweighardt B, Wrin T, Galovich J, Hoh R, Sinclair E, Hunt P, McCune JM, Martin JN, Petropoulos CJ, Hecht FM. 2006. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J Virol 80:6155–6164. doi: 10.1128/JVI.00093-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frost SDW, Wrin T, Smith DM, Pond SLK, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, Petropoulos CJ, Little SJ, Richman DD. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A 102:18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aasa-Chapman MMI, Hayman A, Newton P, Cornforth D, Williams I, Borrow P, Balfe P, McKnight A. 2004. Development of the antibody response in acute HIV-1 infection. AIDS 18:371–381. doi: 10.1097/00002030-200402200-00002. [DOI] [PubMed] [Google Scholar]
- 70.Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A 100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 72.Pilgrim AK, Pantaleo G, Cohen OJ, Fink LM, Zhou JY, Zhou JT, Bolognesi DP, Fauci AS, Montefiori DC. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J Infect Dis 176:924–932. doi: 10.1086/516508. [DOI] [PubMed] [Google Scholar]
- 73.Moog C, Fleury HJ, Pellegrin I, Kirn A, Aubertin AM. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J Virol 71:3734–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moore PL, Gray ES, Morris L. 2009. Specificity of the autologous neutralizing antibody response. Curr Opin HIV AIDS 4:358–363. doi: 10.1097/COH.0b013e32832ea7e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agrawal N, Leaman DP, Rowcliffe E, Kinkead H, Nohria R, Akagi J, Bauer K, Du SX, Whalen RG, Burton DR, Zwick MB. 2011. Functional stability of unliganded envelope glycoprotein spikes among isolates of human immunodeficiency virus type 1 (HIV-1). PLoS One 6:e21339. doi: 10.1371/journal.pone.0021339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Platt EJ, Kozak SL, Durnin JP, Hope TJ, Kabat D. 2010. Rapid dissociation of HIV-1 from cultured cells severely limits infectivity assays, causes the inactivation ascribed to entry inhibitors, and masks the inherently high level of infectivity of virions. J Virol 84:3106–3110. doi: 10.1128/JVI.01958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tedbury PR, Freed EO. 2014. The role of matrix in HIV-1 envelope glycoprotein incorporation. Trends Microbiol 22:372–378. doi: 10.1016/j.tim.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li S, Juarez J, Alali M, Dwyer D, Collman R, Cunningham A, Naif HM. 1999. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J Virol 73:9741–9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aquino-De Jesus MJ, Anders C, Miller G, Sleasman JW, Goodenow MM, Andiman WA. 2000. Genetically and epidemiologically related “non-syncytium-inducing” isolates of HIV-1 display heterogeneous growth patterns in macrophages. J Med Virol 61:171–180. doi:. [DOI] [PubMed] [Google Scholar]
- 80.Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR. 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol 78:6915–6926. doi: 10.1128/JVI.78.13.6915-6926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping L-H, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walter BL, Wehrly K, Swanstrom R, Platt E, Kabat D, Chesebro B. 2005. Role of low CD4 levels in the influence of human immunodeficiency virus type 1 envelope V1 and V2 regions on entry and spread in macrophages. J Virol 79:4828–4837. doi: 10.1128/JVI.79.8.4828-4837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peters PJ, Duenas-Decamp MJ, Sullivan WM, Brown R, Ankghuambom C, Luzuriaga K, Robinson J, Burton DR, Bell J, Simmonds P, Ball J, Clapham PR. 2008. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology 5:5. doi: 10.1186/1742-4690-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang Z-Y, Zhang M-Y, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tran K, Poulsen C, Guenaga J, de Val N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, Hedestam GBK, Wyatt RT. 2014. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc Natl Acad Sci U S A 111:E738–E747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y, O'Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. 2011. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol 85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y-j, Fredriksson R, McKeating JA, Fenyö EM. 1997. Passage of HIV-1 molecular clones into different cell lines confers differential sensitivity to neutralization. Virology 238:254–264. doi: 10.1006/viro.1997.8812. [DOI] [PubMed] [Google Scholar]
- 89.Trkola A, Ketas T, KewalRamani VN, Endorf F, Binley JM, Katinger H, Robinson J, Littman DR, Moore JP. 1998. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J Virol 72:1876–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beddows S, Louisirirotchanakul S, Cheingsong-Popov R, Easterbrook PJ, Simmonds P, Weber J. 1998. Neutralization of primary and T-cell line adapted isolates of human immunodeficiency virus type 1: role of V3-specific antibodies. J Gen Virol 79(Part 1):77–82. doi: 10.1099/0022-1317-79-1-77. [DOI] [PubMed] [Google Scholar]
- 91.Hoffman TL, LaBranche CC, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci U S A 96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pugach P, Kuhmann SE, Taylor J, Marozsan AJ, Snyder A, Ketas T, Wolinsky SM, Korber BT, Moore JP. 2004. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology 321:8–22. doi: 10.1016/j.virol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 93.Shieh JT, Martín J, Baltuch G, Malim MH, González-Scarano F. 2000. Determinants of syncytium formation in microglia by human immunodeficiency virus type 1: role of the V1/V2 domains. J Virol 74:693–701. doi: 10.1128/JVI.74.2.693-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Daar ES, Li XL, Moudgil T, Ho DD. 1990. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc Natl Acad Sci U S A 87:6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O'Brien WA, Mao SH, Cao Y, Moore JP. 1994. Macrophage-tropic and T-cell line-adapted chimeric strains of human immunodeficiency virus type 1 differ in their susceptibilities to neutralization by soluble CD4 at different temperatures. J Virol 68:5264–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moore JP, Burkly LC, Connor RI, Cao Y, Tizard R, Ho DD, Fisher RA. 1993. Adaptation of two primary human immunodeficiency virus type 1 isolates to growth in transformed T cell lines correlates with alterations in the responses of their envelope glycoproteins to soluble CD4. AIDS Res Hum Retroviruses 9:529–539. doi: 10.1089/aid.1993.9.529. [DOI] [PubMed] [Google Scholar]
- 97.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse P-J, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp 120 trimers. Nature 455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hraber P, Korber BT, Lapedes AS, Bailer RT, Seaman MS, Gao H, Greene KM, McCutchan F, Williamson C, Kim JH, Tovanabutra S, Hahn BH, Swanstrom R, Thomson MM, Gao F, Harris L, Giorgi E, Hengartner N, Bhattacharya T, Mascola JR, Montefiori DC. 2014. Impact of clade, geography, and age of the epidemic on HIV-1 neutralization by antibodies. J Virol 88:12623–12643. doi: 10.1128/JVI.01705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rusert P, Krarup A, Magnus C, Brandenberg OF, Weber J, Ehlert A-K, Regoes RR, Günthard HF, Trkola A. 2011. Interaction of the gp120 V1V2 loop with a neighboring gp120 unit shields the HIV envelope trimer against cross-neutralizing antibodies. J Exp Med 208:1419–1433. doi: 10.1084/jem.20110196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol 78:5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cavrois M, Neidleman J, Santiago ML, Derdeyn CA, Hunter E, Greene WC. 2014. Enhanced fusion and virion incorporation for HIV-1 subtype C envelope glycoproteins with compact V1/V2 domains. J Virol 88:2083–2094. doi: 10.1128/JVI.02308-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leaman DP, Zwick MB. 2013. Increased functional stability and homogeneity of viral envelope spikes through directed evolution. PLoS Pathog 9:e1003184. doi: 10.1371/journal.ppat.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kassa A, Finzi A, Pancera M, Courter JR, Smith AB, Sodroski J. 2009. Identification of a human immunodeficiency virus type 1 envelope glycoprotein variant resistant to cold inactivation. J Virol 83:4476–4488. doi: 10.1128/JVI.02110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog 7:e1002101. doi: 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Medjahed H, Pacheco B, Désormeaux A, Sodroski J, Finzi A. 2013. The HIV-1 gp120 major variable regions modulate cold inactivation. J Virol 87:4103–4111. doi: 10.1128/JVI.03124-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dunfee RL, Thomas ER, Gabuzda D. 2009. Enhanced macrophage tropism of HIV in brain and lymphoid tissues is associated with sensitivity to the broadly neutralizing CD4 binding site antibody b12. Retrovirology 6:69. doi: 10.1186/1742-4690-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]