Abstract
Vertebrate photoreceptor cells are exquisite light detectors operating under very dim and bright illumination. The photoexcitation and adaptation machinery in photoreceptor cells consists of protein complexes that can form highly ordered supramolecular structures and control the homeostasis and mutual dependence of the secondary messengers cyclic guanosine monophosphate (cGMP) and Ca2+. The visual pigment in rod photoreceptors, the G protein-coupled receptor rhodopsin is organized in tracks of dimers thereby providing a signaling platform for the dynamic scaffolding of the G protein transducin. Illuminated rhodopsin is turned off by phosphorylation catalyzed by rhodopsin kinase (GRK1) under control of Ca2+-recoverin. The GRK1 protein complex partly assembles in lipid raft structures, where shutting off rhodopsin seems to be more effective. Re-synthesis of cGMP is another crucial step in the recovery of the photoresponse after illumination. It is catalyzed by membrane bound sensory guanylate cyclases (GCs) and is regulated by specific neuronal Ca2+-sensor proteins called guanylate cyclase-activating proteins (GCAPs). At least one GC (ROS-GC1) was shown to be part of a multiprotein complex having strong interactions with the cytoskeleton and being controlled in a multimodal Ca2+-dependent fashion. The final target of the cGMP signaling cascade is a cyclic nucleotide-gated (CNG) channel that is a hetero-oligomeric protein located in the plasma membrane and interacting with accessory proteins in highly organized microdomains. We summarize results and interpretations of findings related to the inhomogeneous organization of signaling units in photoreceptor outer segments.
Keywords: multi-protein complexes, second messenger signaling, phototransduction, cGMP, calcium-binding proteins
Introduction
Vertebrate photoreceptor cells are neurosensory cells of unique morphology and specialized function. They are divided into two general types, rods and cones, which mediate vision at night and daylight, respectively. Absorption of photons by visual pigments, rhodopsin in rods and cone opsins in cones, triggers a well understood signaling cascade that has been thoroughly investigated in the past decades. Numerous articles in the last decades have therefore summarized the basic features of the phototransduction process (e.g., Stryer, 1991; Kaupp and Koch, 1992; Koch, 1994; Pugh and Lamb, 2000; Luo et al., 2008; Wensel, 2008; Arshavsky and Burns, 2012; Korenbrot, 2012; Palczewski, 2012): coupling of visual pigments to the heterotrimeric G protein transducin, activation of the effector phosphodiesterase PDE6 by the G protein and efficient hydrolysis of the second messenger cyclic nucleotide guanosine 3′,5′-cyclic monophosphate (cGMP) with high turnover rates, regulation of the cyclic nucleotide-gated (CNG)-channel by cGMP and re-synthesis of cGMP by a guanylate cyclase (GC) complex that is controlled by a powerful Ca2+-dependent feedback loop (Dizhoor et al., 2010; Koch et al., 2010). The cytoplasmic Ca2+-concentration in the outer segments of photoreceptor cells is maintained by two transport routes: Ca2+-influx through the CNG-channel and Ca2+-extrusion by Na+/Ca2+, K+-exchanger. Due to the light-dependent closure of the CNG-channel (Kaupp and Seifert, 2002), Ca2+ cannot enter the cell, but is expelled via the exchanger leading to a net decrease in cytoplasmic Ca2+. Termination of the signaling cascade is an equally critical step for the precise formation of light responses and photoresponse recovery. All activation steps of the excitation pathway need efficient shut-off mechanisms (Burns, 2010). Thus, rhodopsin is phosphorylated by rhodopsin kinase (GRK1) under control of Ca2+/recoverin (Senin et al., 2002b) and competitive binding of arrestin to phospho-rhodopsin prevents further interaction of transducin with rhodopsin (Sommer et al., 2014). As regards the effector, the acceleration of the intrinsic GTPase activity of transducin by RGS9-1 and accessory proteins Gβ5L and R9AP leads to a sub-second deactivation of transducin (Wensel, 2008), which in turns dissociates from PDE6, thus restoring the low basal activity controlled by its small inhibitory γ subunits. Figure 1 summarizes the basic features of the phototransduction cascade.
Figure 1.
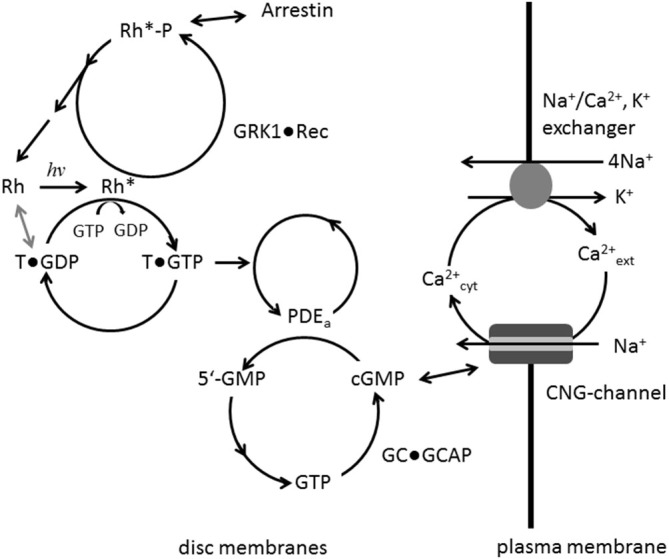
Main signaling steps in phototransduction. Photo-activation of rhodopsin (Rh to Rh*) leads to GDP/GTP exchange at the G protein transducin (T) which in turn activates its effector PDE. Hydrolysis of cGMP is catalyzed by activated PDE; resynthesis of cGMP by guanylate cyclase (GC) is under control of a negative Ca2+-feedback involving the GC-activating proteins (GCAP) Ca2+-sensor proteins. Ca2+ enters the cell via the cyclic nucleotide-gated (CNG)-channel and is extruded by the exchanger. Rh* is phosphorylated by GRK1, when inhibition by Ca2+-bound recoverin is relieved. Arrestin can bind to phosphorylated Rh* preventing further activation of transducin.
The detailed knowledge about the physiology and biochemistry of phototransduction has been covered in numerous in-depth reviews (for a small selection see references above). The present article, however will address issues, which had been investigated in more recent years showing that the photoreceptor outer segment is not a well-stirred compartment, but appears rather inhomogeneous. Publications indicating different aspects of heterogeneity date back several decades, but have been mainly interpreted as signs of the natural ageing and renewal process in outer segments. For example, gradients along the longitudinal axis of outer segments have been described for second messenger molecules (Leibovic and Bandarchi, 1997a,b; Gray-Keller et al., 1999), disc membrane composition (Boesze-Battaglia et al., 1989, 1990; Boesze-Battaglia and Albert, 1990), light response variation (Baylor and Lamb, 1982; Schnapf, 1983; Mazzolini et al., 2015) and enzymatic reactions (Shichi and Williams, 1979). While the ageing process is out of question the physiological consequences of gradients of intracellular components and the heterogeneous formation of multi-protein units come into focus. Proteins of the photoexcitation and adaptation machinery are assembled in complexes that can form highly ordered supramolecular structures by interacting with the supporting disc membrane vesicles (Wensel, 2008). Moreover, some of the main molecular components of the phototransduction cascade have been found to be involved in other non-visual related signaling networks (Kiel et al., 2011). Thus, we will discuss in the present review how these signal transducing modules contribute to the efficient and precise processing of light signals. We have restricted our review to rod biochemistry and physiology, since knowledge about cones and about protein and signaling networks in cones is less advanced. Moreover, it is worth noting that some aspects concerning for example the supramolecular organization of rhodopsin (see below) have not been investigated yet for the cone system so far.
Photoresponse Gradient Along the Longitudinal Axis of Photoreceptor Outer Segments
Suction electrode recordings from amphibian rods showed already in the 1980s that single flash response kinetics are slower when the cell is illuminated at the tip of the rod outer segment than at the base. Response amplitudes become smaller the farther away from the base the flash is delivered (Schnapf, 1983). Outer segments are renewed every 10 days in mammalians and 6–7 weeks in amphibians by a process called disc shedding, in which the disc components at the tip of the outer segments are phagocytized by the retinal pigment epithelium (Young, 1967). This process keeps the length of the outer segment constant under physiological conditions and the heterogeneity of flash response kinetics and amplitudes were interpreted as the results of an ageing process. Mazzolini et al. (2015) have recently confirmed and extended these earlier observations using dissociated rods from adult male Xenopus laevis frogs. However, their analysis showed that the amplitude of saturating and single photon responses decreased by 5–10 times, when illumination of the tip is compared with that of the base. Previous recordings showed only a twofold difference in sensitivity. Mazzolini et al. (2015) excluded a lower probability of photon capture by rhodopsin, but instead suggested that a reduced amplification of the transduction cascade leads to a reduction in efficacy due to a progressive depletion of PDE6 along the longitudinal axis of rod outer segment. Further, the kinetic variability of the light responses at the base, middle and tip of the outer segment is not contemplated by the existing quantitative models, which operate under the assumption that the biochemical components of the transduction machinery are more or less uniformly distributed (Hamer et al., 2005; Bisegna et al., 2008; Dell’Orco et al., 2009; Shen et al., 2010). Instead, the authors suggest a “series of interconnected compartments” that might have different local concentrations of key factors and therefore vary in their responsiveness to light.
Supramolecular Organization of Rhodopsin
Efficient photon capture by photoreceptor cells is due to the high density of the visual pigment rhodopsin in stacks of disc membranes reaching ≥25,000 μm-2. Work from the early 1970s came to the conclusion that rhodopsin is homogenously distributed in disc membranes and can laterally diffuse without much restriction (Cone, 1972; Liebman and Entine, 1974; Poo and Cone, 1974). This concept of random distribution and free diffusion in the membrane was further supported by results obtained by neutron diffraction and electron microscopy (Saibil et al., 1976; Roof and Heuser, 1982). After being accepted in the field for 30 years this “classical” view was challenged by an atomic force microscopy (AFM) study of disc membranes, which showed in 2003 that rhodopsin dimers are found in a paracrystalline arrangement suggesting a lower degree of lateral diffusion (Fotiadis et al., 2003a, 2004). Subsequent studies were in agreement with a more inhomogeneous distribution of immobile rhodopsin fractions (Govardovskii et al., 2009). Another very recent independent AFM study revealed that dimers of rhodopsin are organized in nanodomains, whose supramolecular features seem to be conserved in humans and mice. For example, the discs diameter was found to depend on the number of rhodopsin molecules embedded in the membrane, however it was found to be independent of rhodopsin’s spatial density (Whited and Park, 2015). A different study further reported highly concentrated patches of rhodopsin in the central region of a disc. No rhodopsin was located near the rim region that was occupied by peripherin and Rom proteins, which are well established marker proteins for this region of the outer segment (Buzhynskyy et al., 2011).
Very recently, using cryosections of dark-adapted intact rod photoreceptors from mice Gunkel et al. (2015) demonstrated by cryoelectron tomography the presence of tracks of rhodopsin dimers. In a scenario only partly similar to previous AFM determinations, the authors observed that at least ten rhodopsin dimers form a row, rows form pairs (tracks), and tracks are aligned parallel to the disc incisures, with profound implications for the kinetics of phototransduction.
Soon after the classical view of randomly distributed and freely diffusing rhodopsin monomers was challenged, a controversial discussion on this topic started and had not ended so far (Chabre et al., 2003; and reply by Fotiadis et al., 2003b). Previous biochemical and biophysical studies demonstrated that monomeric rhodopsin is sufficient to activate transducin, leading to the conclusion that no mechanistic need for a rhodopsin dimer exists (Chabre and le Maire, 2005; Ernst et al., 2007). Biochemical studies of detergent solubilized rhodopsin however provided evidence for the existence of rhodopsin dimers or oligomers depending on the detergent and solubilization conditions (Jastrzebska et al., 2004). But the use of detergents is prone to affect the quaternary structure of proteins and was therefore considered as a weak argument to support the existence of higher order organization rhodopsin in disc membranes (Chabre and le Marie, 2004). The paracrystalline arrangement of rhodopsin dimers observed in the early AFM studies was interpreted as the results of separation of the lipid phase from proteins at low temperatures (Chabre et al., 2003), but the higher order topography was also observed at room temperature (Fotiadis et al., 2003b), moreover recent AFM determinations confirmed that higher order nano-domains of rhodopsin are found both in mice and humans (Whited and Park, 2015).
A very recent study investigated the nature of rhodopsin dimers by showing that peptides encompassing the transmembrane domains (TMs) of rhodopsin block its dimerization both in vitro and in living cells, without however affecting the rates of transducin activation (Jastrzebska et al., 2015). This suggests that the supramolecular organization of rhodopsin could be essential for the stabilization of rod outer segments and receptor trafficking rather than for activating the G protein.
Dynamics of Transient Protein Complexes Involving Rhodopsin
The supramolecular organization of nearly immobile rhodopsin imposes a conceptual problem on the mechanistic understanding of the phototransduction cascade, which operates on a millisecond time base with high sensitivity. Intuitively one might predict that arrays of paracrystalline rhodopsin would slow down the activation kinetics. The investigation of how the diffusion properties of rhodopsin and transducin are affected by specific supramolecular assemblies is not trivial, and has so far been investigated only by computational analyses that incorporate available experimental data. Results were not always intuitive, because rhodopsin supramolecular assemblies and molecular crowding in discs imply anomalous and anisotropic diffusion paths for peripheral membrane proteins such as transducin, which significantly differ from the classical free diffusion case. In a first approach to the problem, Monte Carlo simulations of rhodopsin-transducin encounters in both the classical free diffusion and emerging paracrystalline rafts scenarios suggested that an unexpected favorable effect on the temporal response of early phototransduction reactions may occur, if rhodopsin molecules were packed in highly ordered assemblies (Dell’Orco and Schmidt, 2008). Moreover, recent surface plasmon resonance studies (Dell’Orco and Koch, 2011) performed with detergent solubilized native rhodopsin demonstrated the existence of a protein-protein interaction between dark-adapted rhodopsin and transducin, which was postulated earlier based on the analysis of their structural complementarity (Fanelli and Dell’Orco, 2005; Dell’Orco et al., 2007). Dark rhodopsin-transducin binding occurs with submicromolar affinity and is characterized by very fast association and dissociation rates in a “dynamic scaffolding” frame, where concerted diffusion/binding phenomena give rise to a dynamic hopping of transducin on rhodopsin supramolecular assemblies (Dell’Orco and Koch, 2011). The transient precoupling step was integrated into the framework of phototransduction models of both amphibian and murine rods, and was found to be compatible with the overall cascade kinetics (Invergo et al., 2014; Dell’Orco, 2015). The physiological presence of rhodopsin-transducin transient complexes has been somewhat questioned and debated (Schöneberg et al., 2014, 2015; Dell’Orco and Koch, 2015), however it appears now quite clear that it may have deep implications for the capability of rods to detect single photons (Cangiano and Dell’Orco, 2013; Dell’Orco, 2013), and seems to be an essential mechanistic step in the recently emerged picture of rhodopsin tracks observed by cryoelectron tomography, in order to create the “kinetic traps”: owing to the frequent, rapid formation and breakup of precomplexes, transducin molecules could scan a rhodopsin track by discrete hopping events, resulting in an activation rate that, in the single-photon regime, would be determined by the number of the preassembled transducin molecules per track rather than the photoactivated rhodopsin lifetime. The number of transducin molecules activated per photoactivated rhodopsin would be therefore of the same order as the number of preassembled transducin molecules per unit track (Gunkel et al., 2015). The concepts developed in the last years in opposition to the “fluid mosaic” classical organization of the disc membrane have built up a novel structural picture of the early mechanisms triggering phototransduction, whose supramolecular features are summarized in Figure 2.
Figure 2.
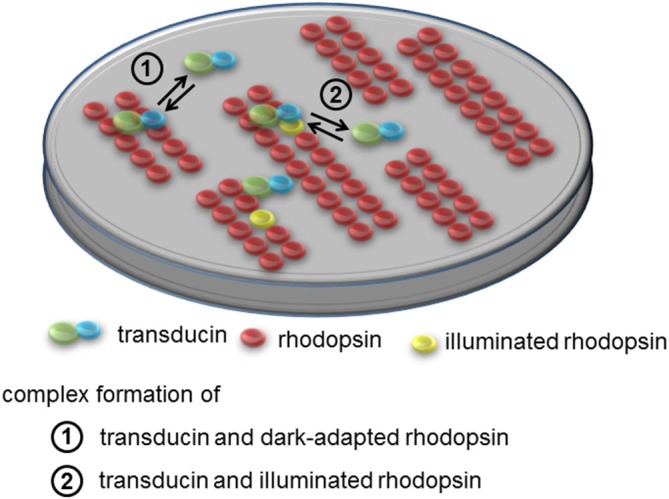
Supramolecular organization of rhodopsin and interaction with transducin. Rhodopsin is present in tracks of dimers in the disc membrane. In the dark rhodopsin-transducin complexes form with submicromolar affinity that is characterized by very fast association and dissociation rates. Movements of transducin can be described as dynamic hopping on rhodopsin supramolecular assemblies thus constituting “dynamic scaffolding”. Apparent dissociation rates of transducin from dark-adapted rhodopsin are >300-fold faster than corresponding rates from light-activated rhodopsin.
Proteomic profiling and protein network analysis of outer segments led to the prediction of signaling and/or trafficking pathways in addition to the activation and deactivation pathways that govern photoreceptor excitation and recovery. An important level of regulation of such alternative pathways seems to be played by small GTPases (Kiel et al., 2011). The monomeric G-protein Rac1 is among the putative binding partners of rhodopsin (Balasubramanian and Slepak, 2003), but its lower cellular concentration (~100-fold excess of rhodopsin) and its medium affinity for rhodopsin (apparent KD = 1.3 μM) would not allow a significant competition with the binding of transducin (Köster et al., 2014). However, under strong illumination, when transducin is depleted from the outer segment by transport to the inner segment (Pulvermüller et al., 2002; Sokolov et al., 2002; Lobanova et al., 2007), only about 10% of all rhodopsin molecules could form a complex with Rac1. Therefore, it is more likely that Rac1 binds to rhodopsin during transport after protein biosynthesis. Intracellular trafficking, in particular under conditions of changing illumination has attained increasing interest in the photoreceptor research community. In order to keep this review focused we will not discuss this field in depth, but will refer to some reviews on this topic (Calvert et al., 2006; Karan et al., 2010; Pearring et al., 2013; Wang and Deretic, 2014).
Deactivation of Rhodopsin
The efficient shut-off of the phototransduction cascade requires as initial step the deactivation of photoactivated rhodopsin. This crucial step is performed by the interplay of several proteins and binding events: GRK-1 phosphorylates illuminated rhodopsin at its C-terminus (Maeda et al., 2003), which allows subsequent binding of arrestin (p48) or the arrestin splice variant p44 (Granzin et al., 2012; Kim et al., 2013). Binding of arrestin to phosphorylated rhodopsin prevents further activation of transducin (Pulvermüller et al., 1993).
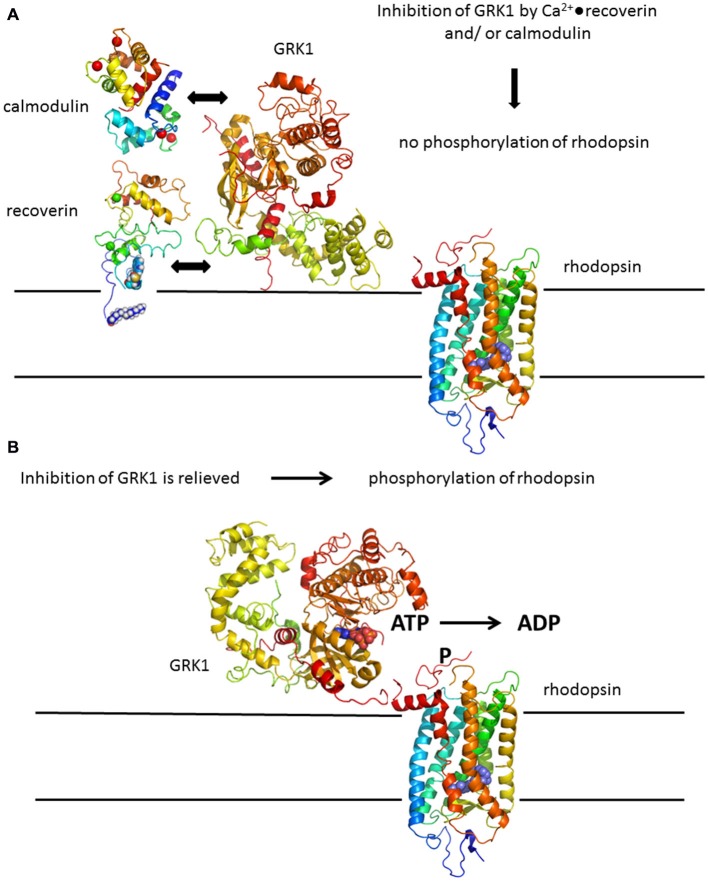
Serine and threonine residues present in the C-terminus of rhodopsin within the amino acid positions 324–348 are potential sites for phosphorylation by GRK1. Like other members of the GRK family, GRK1 phosphorylates only the light-stimulated (bleached) form of the receptor and does not act upon the unbleached receptor (for reviews, see Senin et al., 2002b; Maeda et al., 2003; Premont and Gainetdinov, 2007). Activity of GRK1 is under control of a Ca2+-dependent negative feedback loop, but GRK1 itself is not sensitive to Ca2+. Changes in intracellular [Ca2+] are sensed by retina specific neuronal Ca2+-sensor proteins including the Ca2+-binding protein recoverin (Senin et al., 2002b). By interacting with GRK1 in a Ca2+-dependent manner it controls GRK1 enzyme activity (Kawamura, 1993; Gorodovikova et al., 1994; Klenchin et al., 1995). Recoverin is heterogeneously acylated at its N-terminus by an N-myristoyl-transferase like activity (Dizhoor et al., 1992). When acylated recombinant forms of recoverin are investigated, a myristoyl group is routinely attached to the protein by co-expression of a N-myristoyl-transferase. The myristoyl moiety has a strong impact on the structural and functional properties of recoverin. For example, in the absence of Ca2+, the myristoyl group is buried within a hydrophobic pocket of the protein (Tanaka et al., 1995). Binding of Ca2+ to recoverin triggers a conformational change that leads to exposure of the myristoyl group and the hydrophobic pocket (Zozulya and Stryer, 1992; Ames et al., 1997), which facilitates its association with biological membranes and inhibition of GRK1 (Chen et al., 1995; Senin et al., 1995). Since the cytoplasmic Ca2+-concentration is high in the resting dark state of the photoreceptor cell, Ca2+-loaded recoverin associates with the disc membranes, interacts with GRK1 and inhibits its activity. Decrease of intracellular Ca2+ after illumination triggers dissociation of recoverin from the membranes and from GRK1 thereby stopping the inhibition of GRK1 and allowing rhodopsin phosphorylation (Figures 3A,B).
Figure 3.
Rhodopsin phosphorylation by GRK1. (A) Deactivation of rhodopsin is under control of a Ca2+-feedback loop involving Ca2+-sensor proteins recoverin and calmodulin. Both Ca2+-binding proteins have non-overlapping binding sites in GRK1 and act in a synergetic mode, for example by increasing the Ca2+-sensitivity of GRK1 regulation. (B) Inhibition of GRK1 is relieved at decreasing Ca2+-concentration after illumination leading to phosphorylation of rhodopsin. Structures were prepared with pymol; corresponding PDB codes are: 1F88 for rhodopsin (Palczewski et al., 2000); 3C51 for GRK1 (Singh et al., 2008); 1JSA for recoverin (Ames et al., 1997); 1CDM for calmodulin (Meador et al., 1993).
The Ca2+-myristoyl switch of recoverin (Zozulya and Stryer, 1992) is prototypical for a variety of myristoylated proteins and has served as a benchmark for studying related switching mechanisms in other Ca2+-sensors (O‘Callaghan et al., 2003; Li et al., 2011; Lim et al., 2011, 2014; Burgoyne and Haynes, 2015; Marino et al., 2015; Sulmann et al., 2015) or by characterizing the biocompatibility of nanoparticles (Marino et al., 2014). The Ca2+-binding and switching process in recoverin had been investigated in more detail by comparing WT recoverin with mutants containing disabled EF-hands or a truncated C-terminus (Senin et al., 2002a, 2003; Weiergräber et al., 2003, 2006). These studies revealed critical steps in sequential Ca2+-binding and defined crucial regions that control the Ca2+-sensitive regulation of GRK1. Several studies were also focused on regulatory aspects of GRK1 using site-directed mutagenesis, pull down methods, SPR spectroscopy and rhodopsin phosphorylation assays (Huang et al., 2009, 2011; Komolov et al., 2009; Zernii et al., 2011; Orban et al., 2012). The N-terminus of GRK1 forms an amphipathic α–helix, of which the first 25 amino acid residues interact with an exposed hydrophobic groove in recoverin (Ames et al., 2006; Higgins et al., 2006). A combination of structural analysis and computational modeling of the recoverin-kinase complex revealed that the protein-protein interface involves also the C-terminus of recoverin by forming a cation-π interaction pair, which is essential for GRK1 target recognition by recoverin (Zernii et al., 2011). Fine-tuning of Ca2+-dependent regulation of GRK1 is further achieved by synergetic action of calmodulin (Figure 3A), which binds to a region in GRK1 distant from the recoverin binding site (Grigoriev et al., 2012).
Recent electrophysiological recordings on transgenic mice lacking recoverin (Rv−/−) showed shortened light responses indicating a reduced lifetime of light-activated rhodopsin (Makino et al., 2004; Bush and Makino, 2006; Chen et al., 2010), which is consistent with an inhibition of GRK1 by recoverin at high [Ca2+]. The same effect was recently observed by intra ocular and ex-vivo retinal delivery of liposomes loaded with recombinant recoverin or its antibody, which showed effects comparable to recoverin overexpression and downregulation, respectively (Asteriti et al., 2015). Similarly, overexpression of GRK1 in transgenic mice (12-fold higher than in wildtype (WT) mice) also leads to shortened light responses (Chen et al., 2012). By investigating mice harboring different genetic manipulations Chen et al. (2012) challenged previous investigations on mice expressing GRK1 at lower and higher levels than WT (Sakurai et al., 2011) and postulated a time constant of rhodopsin deactivation of about 50 ms. In a different approach using bottom-up modeling, the rate limiting steps in the recovery of rods after illumination were investigated by simulating conditions, in which the expression levels of GRK1 and recoverin were altered individually or in combination (Invergo et al., 2013). The analysis provided a mechanistic explanation for the puzzling evidence that GRK1 over expression does not influence the saturation time of rods under bright light stimulation (Krispel et al., 2006; Sakurai et al., 2011), indeed attributing a compensating effect to arrestin oligomerization (Invergo et al., 2013). The recoverin-GRK1 complex in cones is operating under dim light, but not in bright light demonstrating significant differences in the response recovery between rod and cone physiology (Sakurai et al., 2015). Very recently, Chen et al. (2015) suggested that GRK1 and recoverin participate in the activity regulation of PDE6, but biochemical evidence to support this hypothesis is lacking so far.
Signaling Modules and Lipid Rafts
Patches of rhodopsin seen in AFM images (see above) were interpreted as lipid raft structures and biochemical fractionation studies pointed to the existence of lipid rafts in rod outer segments. Detergent resistant membranes or lipid rafts can be isolated from sucrose step gradients of cell suspensions that are solubilized with a low concentration of the nonionic detergent Triton X-100. Main features of lipid rafts are high cholesterol content, saturated fatty acids, glycolipids and the marker protein caveolin (Martin et al., 2005; Elliott et al., 2008) that was shown to co-localize with signaling proteins like transducin in rod outer segments (Elliott et al., 2003). In fact, whole signaling units have been identified in isolated lipid rafts from preparations of rod outer segments. For example, rhodopsin, transducin, its effector cGMP-phosphodiesterase, the shorter splice variant of arrestin p44 and the RGS9-Gβ5L complex translocate to raft structures in a light-dependent manner (Seno et al., 2001; Nair et al., 2002; Balasubramanian and Slepak, 2003; Liu et al., 2003). Main conclusions from these studies were that the rhodopsin-transducin coupling is reduced and that phototransduction is less efficient in lipid micro-domains. Raft-specific protein complexes including rhodopsin were also suggested to have a role in the formation of outer segments (Berta et al., 2011).
Interestingly, relative protein composition of the raft fraction from rod outer segments was not only dependent on illumination, but also on the free Ca2+-concentration (Senin et al., 2004). Changing free Ca2+ causes proteins to translocate between the soluble fraction and the detergent resistant membrane fraction. Manipulation of the cholesterol content by the reagent methyl-β-cyclodextrin showed a clear dependence of recoverin controlled GRK1 activity: the Ca2+-dependent activity profile of GRK1 is shifted to higher Ca2+, when cholesterol is low and to lower Ca2+, when cholesterol is high (Senin et al., 2004). In native cells cholesterol is not homogeneously distributed in outer segment disc membranes, but instead rod outer segments contain a cholesterol gradient (high to low) from the base to the tip (Boesze-Battaglia et al., 1989, 1990; Boesze-Battaglia and Albert, 1990). This means, in terms of the cell’s physiology, that rhodopsin shut-off by phosphorylation would be more effective at the base than at the tip leading to faster deactivation kinetics at the base. It remains an open question, whether the photoresponse gradient along the longitudinal axis (see above) results from a lipid raft gradient. If this were the case, the photoresponse gradient would in fact depend on the local supramolecular assembly of phototransduction components.
GC Protein Complex
In addition to signaling proteins that translocate to raft structures in a light—dependent manner, the membrane bound GC in photoreceptor cells is associated with rafts independent of the illumination conditions. This membrane bound sensory GC is expressed in two forms in vertebrate rod and cone cells (Dizhoor et al., 1994; Goraczniak et al., 1994; Lowe et al., 1995) and association of one form, presumably ROS-GC1 (synonymously named GC-E or RetGC1), had been detected by immunoblotting in detergent resistant membrane fractions (Nair et al., 2002; Senin et al., 2004). It is known, however from previous studies that GC activities co-fractionate with axonemes (Fleischmann and Denisevich, 1979) of rod cells and purification of the enzyme to an apparent homogeneity of a protein band at 110–112 kDa is achieved by detergent/high salt extraction from Triton X-100 insoluble pellets (Hayashi and Yamazaki, 1991; Koch, 1991; Margulis et al., 1993). Association with cytoskeletal structures was also demonstrated by direct interaction studies showing co-immunoprecipitation of ROS-GC1 and tubulin (Schrem et al., 1999) and binding of actin (Hallett et al., 1996) to the cyclase (Figure 4 of GC signaling complex). Based on translocation studies on mice lacking ROS-GC1 (GC-E−/−) it was suggested that ROS-GC1 is crucial for the intracellular trafficking of peripheral membrane proteins (Baehr et al., 2007; Karan et al., 2010). A co-immunoprecipitation study showed further that ROS-GC1 and the α–subunit of transducin form a complex, which might regulate the light-dependent translocation of transducin (Rosenzweig et al., 2009).
Figure 4.
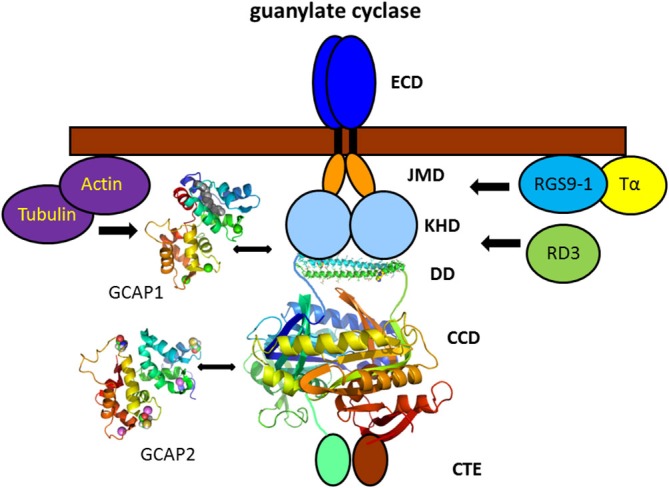
Vertebrate photoreceptor GC and interacting proteins. Photoreceptor GCs contain several domains denoted as extracellular domain (ECD, which in rod outer segment is present in the intradiscal lumen; TM, transmembrane domain; JMD, juxtamembrane domain; KHD, kinase homology domain; DD, dimerization domain; CCD, cyclase catalytic domain; and CTE, a C-terminal extension). The tertiary structure of the DD and CCD is adapted from the solved three-dimensional structure of soluble GCs (Ma et al., 2010; Allerston et al., 2013) that display a high sequence homology with membrane bound GCs in these domains (PDB codes: 3HLS and 3UVJ). Assembly and topography of GC domains, in particular of the DD and CCD is arbitrarily chosen. GCAP1 and GCAP2 activate the target GC at low cytoplasmic Ca2+-concentration bringing the cell back to the dark state. The exact regions of interaction and/or regulation by GCAPs are a matter of debate (see main text). Interaction with other proteins was shown by biochemical procedures, but physiological meaning is lacking so far. Structures of GCAP1 and GCAP2 are based on the published x-ray and NMR-structure, respectively (GCAP1: 2R2I, Stephen et al., 2007; GCAP2: 1JBA, Ames et al., 1999).
Both GCs are regulated by Ca2+-sensor proteins (Koch and Stryer, 1988) named GC-activating proteins (GCAP) that detect changes in cytoplasmic Ca2+ via their EF-hand Ca2+-binding motifs (Palczewski et al., 1994; Dizhoor et al., 1995; Frins et al., 1996). Guanylate cyclase-activating proteins (GCAPs) interact with the target GC at low and high Ca2+-concentration, although the binding affinity is not very high. Apparent affinity constants are in the lower micromolar to submicromolar range, which would allow transitory and flexible complex formation (Hwang and Koch, 2002; Peshenko et al., 2004, 2011a). Binding or dissociation of Ca2+ triggers conformational changes in GCAPs causing probably a rearrangement of the whole complex to increase or decrease GC activities (Sokal et al., 1999a; Hwang et al., 2001; Lim et al., 2009; Sulmann et al., 2014; Marino et al., 2015), which are high at low Ca2+ (and having Mg2+-bound in exchange for Ca2+; Peshenko and Dizhoor, 2006) and are low or even suppressed at saturating Ca2+-concentration. GCAPs are expressed in different isoforms from two or three in mammals to six or eight in teleost fish (Imanishi et al., 2004; Rätscho et al., 2009; Scholten and Koch, 2011; Fries et al., 2012). Expression of two or more isoforms in one cell type has raised questions as to the physiological meaning. Detailed biochemical characterization of GCAP properties in combination with the analysis of transgenic mice showed that GCAPs differ in their Ca2+-sensitivity, Ca2+-binding properties and target regulatory features (Mendez et al., 2001; Hwang et al., 2003; Peshenko et al., 2011b). These results led to a concept of GCAPs activating the target in a sequential order depending on the actual cytoplasmic Ca2+-concentration in the rod or cone cell (Hwang et al., 2003; Koch, 2006; Makino et al., 2012; Koch and Dell’Orco, 2013). For example, mammalian GCAP1 is active at higher Ca2+-concentration than GCAP2. This means in physiological terms that after a single flash of light, when Ca2+-decreases in the cell, GCAP1 will first loose its bound Ca2+ and turn into an activator before GCAP2 would step into this Ca2+-feedback loop. Electrophysiological recordings and computational modeling of photoresponses have further supported a Ca2+-relay or recruitment model of GCAP action (Koch and Dell’Orco, 2013; Wen et al., 2014). Based on differences in spatial-temporal expression profiles (Rätscho et al., 2009), Ca2+-sensitive target regulation and Ca2+-binding of multiple GCAP isoforms (Scholten and Koch, 2011; Sulmann et al., 2015) a similar concept has been proposed for the operation of these Ca2+-sensors in zebrafish rod and cone cells (Koch, 2013).
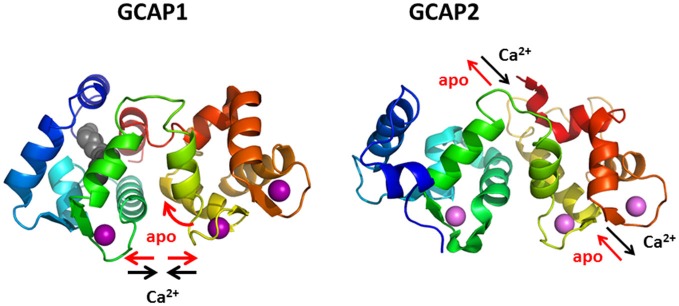
GCAPs share a high degree of amino acid sequence identity and structural homology in cases where the three-dimensional folding had been determined (Ames et al., 1999; Stephen et al., 2007). However, the protein dynamics of Ca2+-triggered conformational changes show significant differences between mammalian GCAP1 and GCAP2 on a nanosecond time scale. Fluorescence lifetime and anisotropy measurements of GCAP1 and GCAP2 that were site specifically labeled with the dye Alexa647 revealed different movements of secondary structural elements (Kollmann et al., 2012; Robin et al., 2015). In the case of GCAP1 these findings were further supported by molecular dynamics simulation showing that GCAP1 undergoes a twisted accordion-like movement upon changing Ca2+-concentration (Robin et al., 2015). GCAP2 in contrast responds to a change in Ca2+ by an up-and-down or piston-like movement of an α-helix between amino acid positions 111 and 131 (Kollmann et al., 2012; Figure 5). A main conclusion from these studies was that two structurally similar proteins can differ significantly in their conformational structural dynamics thereby providing structural framework for their different regulatory responses.
Figure 5.
Protein dynamics of GCAP1 and GCAP2. Binding and dissociation of Ca2+ (violet spheres) triggers different conformational changes and movements of secondary structural elements, a twisted accordion-like movement in GCAP1 and a piston-like movement of one α-helix (yellow) in GCAP2.
Mutations in the retinal degeneration 3 protein (RD3) correlate with type 12 of Leber congenital amaurosis causing a severe form of blindness at early age (Friedman et al., 2006; Preising et al., 2012). The RD3 protein binds to photoreceptor specific GCs 1 and 2 (ROS-GC1 and ROS-GC2) and both proteins are not detectable in rods and cones of RD3 deficient mice (Azadi et al., 2010). Studies using RD3 and ROS-GC1 transfected COS-7 cells and by delivering the RD3 gene into an RD3 deficient mice strain by subretinal injection (Molday et al., 2013) revealed that RD3 is necessary for the correct translocation and cellular localization of ROS-GC1 (Azadi et al., 2010; Zulliger et al., 2015). RD3 does not only bind to GC1, but can act as an allosteric modulator inhibiting ROS-GC1 activity (Peshenko et al., 2011b; Figure 4).
The multiple binding and regulatory processes of the GC signaling unit leave several issues unresolved. For example, published work concerning the binding sites in the GC are inconclusive, since direct interaction sites for GCAP1 and GCAP2 have been mapped to different intracellular regions in ROS-GC1 (Figure 4) including the juxtamembrane domain (JMD; Duda et al., 1999a; Lange et al., 1999), the kinase homology domain (KHD; Krylov and Hurley, 2001) and the cyclase catalytic domain (CCD; Sokal et al., 1999b; Duda et al., 2005). Findings were based on different experimental approaches like peptide competition using peptide libraries, site-directed mutagenesis, and crosslinking in combination with mass spectrometry and might reflect different protein complexes due to the transitory nature of the GCAP-GC interaction mode (see above). However, a more recent paper claims that GCAP1 and GCAP2 bind to overlapping binding sites in a mutually exclusive manner (Peshenko et al., 2015a). A further major unresolved issue in understanding the GC signaling complex is how conformational changes in allosteric regulators (GCAPs) are transmitted to the catalytic site to increase the GC enzymatic turnover rate at least by a factor of 10. A critical role in sensing these conformational changes seems to play the dimerization domain (DD) of the cyclase. This domain forms a coiled—coil structure that is disrupted or in some cases even tightened in a number of retinal disease related mutations causing defects in the Ca2+-dependent control of GC activity (Duda et al., 1999b, 2000; Ramamurthy et al., 2001; Zägel et al., 2013). Very recently experimental evidence was presented showing that the DDis part of the GCAP binding interface (Peshenko et al., 2015b).
The differential regulation of GCs is not limited to GCAPs, but includes other Ca2+-sensor proteins as well. Neurocalcin δ (Kumar et al., 1999; Venkataraman et al., 2008) and S100B (Duda et al., 2002) target to binding sites in ROS-GC1 that are localized in the CCD or the C-terminal extension (CTE). Both Ca2+-sensors regulate the activity of ROS-GC1 in a Ca2+-dependent manner, but opposite to the GCAP mode indicating that these operation modes do not participate in outer segment physiology (Venkataraman et al., 2003), but are rather localized in other photoreceptor cell compartments, for example in modulating the signal transmission to cone ON-bipolar cells (Wen et al., 2012).
Protein Assembly Providing a Structural Link Between Plasma and Disc Membrane
CNG channels are heterotetrameric proteins consisting, in rod and cone cells, of three α–subunits and one β–subunit (Weitz et al., 2002; Zheng et al., 2002). The subunits however are specific for each cell type and are designated CNGA1 and CNGB1 for the rod channel and CNGA3 and CNGB3 for the cone channel. The β–subunit CNGB1 of the rod channel has a unique structure, since it has an extended NH2-terminal part containing four proline-rich repeats and a glutamic acid rich part, which is therefore named glutamic acid rich protein (GARP) part (Sugimoto et al., 1991; Körschen et al., 1995; Colville and Molday, 1996). GARP is also expressed as two soluble protein variants (GARP1 and GARP2). Interaction of the GARP part of CNGB1 with disc membrane proteins at the rim region of the discs has been shown for the retina specific ABC-binding cassette (ABCR) transporter (Körschen et al., 1999) and for the peripherin-2/ROM-1 complex (Poetsch et al., 2001; Becirovic et al., 2014). The CNG channel interacts further in the plasma membrane with the Na+/Ca2+, K+-exchanger (Schwarzer et al., 2000; Kang et al., 2003). The oligomeric protein assembly around the CNG channel constitutes a highly organized microdomain in the rod cell consistent with a role of GARPs in maintaining structural integrity (Körschen et al., 1999; Poetsch et al., 2001; Zhang et al., 2009). The physiological role of different GARP variants however has not been fully explored and is still under discussion. For example, the soluble GARP variant GARP2 binds PDE6 with high affinity thereby inhibiting its basal activity (Pentia et al., 2006); it further controls the open probability of the CNG channel by acting as a gate keeper (Michalakis et al., 2011). These two mechanisms would lead to a reduction in dark noise (Pentia et al., 2006; Michalakis et al., 2011). Overexpressing GARP2 has an effect on the phototransduction gain as derived from fitting the rising phase of the electroretinogram a-wave response (Sarfare et al., 2014). The same study showed also a slower shutoff of the photoresponse. Reconciliation of physiological recordings with functional studies of isolated GARP proteins is hampered by their “intrinsically unfolded nature”, which is known from biochemical and structural analysis of GARP forms (Batra-Safferling et al., 2006). This property however, would make them suitable as flexible tethering proteins linking the CNG channel/exchanger complex with the rim region of the disc membrane.
Concluding Remarks
Signaling proteins in photoreceptor cells form building blocks and signaling modules involving the disc membranes as platforms. These units might preferably exist of transitory protein complexes that are determined by the affinities of their binding partners and their lateral and longitudinal diffusion. This flexible regulatory network of proteins therefore creates the heterogeneous distribution of signaling units in the outer segment under conditions of a crowded intracellular environment. It will be a future challenge to investigate how the exchange, flow and mutual interaction of key factors in signaling units determines the preciseness of visual phototransduction and adaptation.
Author Contributions
KWK developed the concept of the manuscript, wrote the first draft and approved the final version. DDO added to all parts of the manuscript and approved the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The theoretical and experimental work performed in the laboratories of the authors has been funded by different grants from the Deutsche Forschungsgemeinschaft (DFG) to KWK and by the Italian Ministry for Education and Research via departmental grants to DDO.
References
- Allerston C. K., von Delft F., Gileadi O. (2013). Crystal structures of the catalytic domain of human soluble guanylate cyclase. PLoS One 8:e57644. 10.3410/f.718029832.793480138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames J. B., Dizhoor A. M., Ikura M., Palczewski K., Stryer L. (1999). Three-dimensional structure of guanylyl cyclase activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases. J. Biol. Chem. 274, 19329–19337. 10.1074/jbc.274.27.19329 [DOI] [PubMed] [Google Scholar]
- Ames J. B., Ishima R., Tanaka T., Gordon J. I., Stryer L., Ikura M. (1997). Molecular mechanics of calcium-myristoyl switches. Nature 389, 198–202. [DOI] [PubMed] [Google Scholar]
- Ames J. B., Levay K., Wingard J. N., Lusin J. D., Slepak V. Z. (2006). Structural basis for calcium-induced inhibition of rhodopsin kinase by recoverin. J. Biol. Chem. 281, 37237–37245. 10.1074/jbc.m606913200 [DOI] [PubMed] [Google Scholar]
- Arshavsky V. Y., Burns M. E. (2012). Photoreceptor signaling: supporting vision across a wide range of light intensities. J. Biol. Chem. 287, 1620–1626. 10.1074/jbc.r111.305243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asteriti S., Dal Cortivo G., Pontelli V., Cangiano L., Buffelli M., Dell’Orco D. (2015). Effective delivery of recombinant proteins to rod photoreceptors via lipid nanovesicles. Biochem. Biophys. Res. Commun. 461, 665–670. 10.1016/j.bbrc.2015.04.088 [DOI] [PubMed] [Google Scholar]
- Azadi S., Molday L. L., Molday R. S. (2010). RD3, the protein associated with Leber congenital amaurosis type 12, is required for guanylate cyclase trafficking in photoreceptor cells. Proc Natl. Acad. Sci. U S A 107, 21158–21163. 10.3410/f.6564958.6698056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehr W., Karan S., Maeda T., Luo D. G., Li S., Bronson J. D., et al. (2007). The function of guanylate cyclase 1 and guanylate cyclase 2 in rod and cone photoreceptors. J. Biol. Chem. 282, 8837–8847. 10.1074/jbc.m610369200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian N., Slepak V. Z. (2003). Light-mediated activation of Rac-1 in photoreceptor outer segments. Curr. Biol. 13, 1306–1310. 10.1016/s0960-9822(03)00511-6 [DOI] [PubMed] [Google Scholar]
- Batra-Safferling R., Abarca-Heidemann K., Körschen H. G., Tziatzios C., Stoldt M., Budyak I., et al. (2006). Glutamic acid-rich proteins of rod photoreceptors are natively unfolded. J. Biol. Chem. 281, 1449–1460. 10.1074/jbc.m505012200 [DOI] [PubMed] [Google Scholar]
- Baylor D. A., Lamb T. D. (1982). Local effects of bleaching in retinal rods of the toad. J. Physiol 328, 49–71. 10.1113/jphysiol.1982.sp014252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becirovic E., Nguyen O. N., Paparizos C., Butz E. S., Stern-Schneider G., Wolfrum U., et al. (2014). Peripherin-2 couples rhodopsin to the CNG channel in outer segments of rod photoreceptors. Hum. Mol. Genet. 23, 5989–5997. 10.1093/hmg/ddu323 [DOI] [PubMed] [Google Scholar]
- Berta A. I., Boesze-Battaglia K., Magyar A., Szél A., Kiss A. L. (2011). Localization of caveolin-1 and c-src in mature and differentiating photoreceptors: raft proteins co-distribute with rhodopsin during development. J. Mol. Histol. 42, 523–533. 10.1007/s10735-011-9360-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisegna P., Caruso G., Andreucci D., Shen L., Gurevich V. V., Hamm H. E., et al. (2008). Diffusion of the second messengers in the cytoplasm acts as a variability suppressor of the single photon response in vertebrate phototransduction. Biophys J. 94, 3363–3383. 10.1529/biophysj.107.114058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesze-Battaglia K., Albert A. D. (1990). Cholesterol modulation of photoreceptor function in bovine retinal rod outer segments. J. Biol. Chem. 265, 20727–20730. [PubMed] [Google Scholar]
- Boesze-Battaglia K., Fliesler S. J., Albert A. D. (1990). Relationship of cholesterol content to spatial distribution and age of disc membranes in retinal rod outer segments. J. Biol. Chem. 265, 18867–18870. [PMC free article] [PubMed] [Google Scholar]
- Boesze-Battaglia K., Hennessey T., Albert A. D. (1989). Cholesterol heterogeneity in bovine rod outer segment disk membranes. J. Biol. Chem. 264, 8151–8155. [PMC free article] [PubMed] [Google Scholar]
- Burgoyne R. D., Haynes L. P. (2015). Sense and specificity in neuronal calcium signalling. Biochim. Biophys. Acta 1853, 1921–1932. 10.1016/j.bbamcr.2014.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M. E. (2010). Deactivation mechanisms of rod phototransduction. Invest. Ophthalmol. Vis. Sci. 51, 1282–1288. 10.1167/iovs.09-4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush R. A., Makino C. (2006). “Recoverin shapes the photoresponse of retinal rods,” in Neuronal Calcium Sensor Proteins eds Philippov P. P., Koch K.-W. (Hauppauge, NY: Nova Publishers; ), 153–180. [Google Scholar]
- Buzhynskyy N., Salesse C., Scheuring S. (2011). Rhodopsin is spatially heterogeneously distributed in rod outer segment disk membranes. J. Mol. Recognit. 24, 483–489. 10.1002/jmr.1086 [DOI] [PubMed] [Google Scholar]
- Calvert P. D., Strissel K. J., Schiesser W. E., Pugh E. N., Arshavsky V. Y. (2006). Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 16, 560–568. 10.1016/j.tcb.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Cangiano L., Dell’Orco D. (2013). Detecting single photons: a supramolecular matter? FEBS Lett. 587, 1–4. 10.1016/j.febslet.2012.11.015 [DOI] [PubMed] [Google Scholar]
- Chabre M., Cone R., Saibil H. (2003). Biophysics: is rhodopsin dimeric in native retinal rods? Nature 426, 30–31. 10.1038/426030b [DOI] [PubMed] [Google Scholar]
- Chabre M., le Maire M. (2005). Monomeric G-protein-coupled receptor as a functional unit. Biochemistry 44, 9395–9403. 10.1021/bi050720o [DOI] [PubMed] [Google Scholar]
- Chen C. K., Inglese J., Lefkowitz R. J., Hurley J. B. (1995). Ca2+-dependent interaction of recoverin with rhodopsin kinase. J. Biol. Chem. 270, 18060–18066. 10.1074/jbc.270.30.18060 [DOI] [PubMed] [Google Scholar]
- Chen C. K., Woodruff M. L., Fain G. L. (2015). Rhodopsin kinase and recoverin modulate phosphodiesterase during mouse photoreceptor light adaptation. J. Gen. Physiol. 145, 213–224. 10.1085/jgp.201411273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. K., Woodruff M. L., Chen F. S., Chen Y., Ciluffo M. C., Tranchina D., et al. (2012). Modulation of mouse rod response decay by rhodopsin kinase and recoverin. J. Neurosci. 32, 15998–16006. 10.1523/jneurosci.1639-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. K., Woodruff M. L., Chen F. S., Chen D., Fain G. L. (2010). Background light produces a recoverin-dependent modulation of activated-rhodopsin lifetime in mouse rods. J. Neurosci. 30, 1213–1220. 10.1523/jneurosci.4353-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colville C. A., Molday R. S. (1996). Primary structure and expression of the human beta-subunit and related proteins of the rod photoreceptor cGMP-gated channel. J. Biol. Chem. 271, 32968–32974. 10.1074/jbc.271.51.32968 [DOI] [PubMed] [Google Scholar]
- Cone R. A. (1972). Rotational diffusion of rhodopsin in the visual receptor membrane. Nat. New Biol. 236, 39–43. 10.1038/newbio236039a0 [DOI] [PubMed] [Google Scholar]
- Dell’Orco D. (2013). A physiological role for the supramolecular organization of rhodopsin and transducin in rod photoreceptors. FEBS Lett. 587, 2060–2066. 10.1016/j.febslet.2013.05.017 [DOI] [PubMed] [Google Scholar]
- Dell’Orco D. (2015). Rhodopsin transient complexes investigated by systems biology approaches. Methods Mol. Biol. 1271, 251–263. 10.1007/978-1-4939-2330-4_17 [DOI] [PubMed] [Google Scholar]
- Dell’Orco D., Koch K. W. (2011). A dynamic scaffolding mechanism for rhodopsin and transducin interaction in vertebrate vision. Biochem. J. 440, 263–271. 10.1042/bj20110871 [DOI] [PubMed] [Google Scholar]
- Dell’Orco D., Koch K. W. (2015). Transient complexes between dark rhodopsin and transducin: circumstantial evidence or physiological necessity? Biophys. J. 108, 775–777. 10.1016/j.bpj.2014.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Orco D., Schmidt H. (2008). Mesoscopic Monte Carlo simulations of stochastic encounters between photoactivated rhodopsin and transducin in disc membranes. J. Phys. Chem. B. 112, 4419–4426. 10.1021/jp709963f [DOI] [PubMed] [Google Scholar]
- Dell’Orco D., Schmidt H., Mariani S., Fanelli F. (2009). Network-level analysis of light adaptation in rod cells under normal and altered conditions. Mol. Biosyst. 5, 1232–1246. 10.1039/b908123b [DOI] [PubMed] [Google Scholar]
- Dell’Orco D., Seeber M., Fanelli F. (2007). Monomeric dark rhodopsin holds the molecular determinants for transducin recognition: insights from computational analysis. FEBS Lett. 581, 944–948. 10.1016/j.febslet.2007.01.074 [DOI] [PubMed] [Google Scholar]
- Dizhoor A. M., Ericsson L. H., Johnson R. S., Kumar S., Olshevskaya E., Zozulya S., et al. (1992). The NH2 terminus of retinal recoverin is acylated by a small family of fatty acids. J. Biol. Chem. 267, 16033–16036. [PubMed] [Google Scholar]
- Dizhoor A. M., Lowe D. G., Olshevskaya E. V., Laura R. P., Hurley J. B. (1994). The human photoreceptor membrane guanylyl cyclase, RetGC, is present in outer segments and is regulated by calcium and a soluble activator. Neuron 12, 1345–1352. 10.1016/0896-6273(94)90449-9 [DOI] [PubMed] [Google Scholar]
- Dizhoor A. M., Olshevskaya E. V., Peshenko I. V. (2010). Mg2+/Ca2+cation binding cycle of guanylyl cyclase activating proteins (GCAPs): role in regulation of photoreceptor guanylyl cyclase. Mol. Cell. Biochem. 334, 117–124. 10.1007/s11010-009-0328-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizhoor A. M., Olshevskaya E. V., Henzel W. J., Wong S. C., Stults J. I., Ankoudinova I., et al. (1995). Cloning, sequencing and expression of a 24-kDa Ca2+-binding protein activating photoreceptor guanylyl cyclase. J. Biol. Chem. 270, 25200–25206. 10.1074/jbc.270.42.25200 [DOI] [PubMed] [Google Scholar]
- Duda T., Fik-Rymarkiewicz E., Venkataraman V., Krishnan R., Koch K. W., Sharma R. K. (2005). The calcium-sensor guanylate cyclase activating protein type 2-specific site in rod outer segment membrane guanylate cyclase type 1. Biochemistry 44, 7336–7345. 10.1021/bi050068x [DOI] [PubMed] [Google Scholar]
- Duda T., Koch K. W., Venkataraman V., Lange C., Beyermann M., Sharma R. K. (2002). Ca2+-sensor S100β modulated sites of membrane guanylate cyclase in the photoreceptor-bipolar synapse. EMBO J. 21, 2547–2556. 10.1093/emboj/21.11.2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda T., Krishnan A., Venkataraman V., Lange C., Koch K. W., Sharma R. K. (1999a). Mutations in the rod outer segment membrane guanylate cyclase (ROS-GC1) in a cone-rod dystrophy cause defects in calcium signaling. Biochemistry 38, 13912–13919. 10.1021/bi9915972 [DOI] [PubMed] [Google Scholar]
- Duda T., Venkataraman V., Goraczniak R., Lange C., Koch K.-W., Sharma R. K. (1999b). Functional consequences of a rod outer segment membrane guanylate cyclase (ROS-GC1) gene mutation linked with leber’s congenital amaurosis. Biochemistry 38, 509–515. 10.1021/bi9824137 [DOI] [PubMed] [Google Scholar]
- Duda T., Venkataraman V., Jankowska A., Lange C., Koch K. W., Sharma R. K. (2000). Impairmment of the rod outer segment membrane guanylate cyclase dimerization in a cone-rod dystrophy results in defective calcium signaling. Biochemistry 39, 12522–12533. 10.1021/bi001514d [DOI] [PubMed] [Google Scholar]
- Elliott M. H., Fliesler S. J., Ghalayini A. J. (2003). Cholesterol-dependent association of caveolin-1 with the transducin alpha subunit in bovine photoreceptor rod outer segments: disruption by cyclodextrin and guanosine 5’-O-(3-thiotriphosphate). Biochemistry 42, 7892–7903. 10.1021/bi027162n [DOI] [PubMed] [Google Scholar]
- Elliott M. H., Nash Z. A., Takemori N., Fliesler S. J., McClellan M. E., Naash M. I. (2008). Differential distribution of proteins and lipids in detergent-resistant and detergent-soluble domains in rod outer segment plasma membranes and disks. J. Neurochem. 104, 336–352. 10.1111/j.1471-4159.2007.04971.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst O. P., Gramse V., Kolbe M., Hofmann K. P., Heck M. (2007). Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc. Natl. Acad. Sci. U S A 104, 10859–10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanelli F., Dell’Orco D. (2005). Rhodopsin activation follows precoupling with transducin: inferences from computational analysis. Biochemistry 44, 14695–14700. 10.1021/bi051537y [DOI] [PubMed] [Google Scholar]
- Fleischmann D., Denisevich M. (1979). Guanylate cyclase of isolated bovine retinal rod axonemes. Biochemistry 18, 5060–5066. 10.1021/bi00590a006 [DOI] [PubMed] [Google Scholar]
- Fotiadis D., Liang Y., Filipek S., Saperstein D. A., Engel A., Palczewski K. (2003a). Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature 421, 127–128. 10.1038/421127a [DOI] [PubMed] [Google Scholar]
- Fotiadis D., Liang Y., Filipek S., Saperstein D. A., Engel A., Palczewski K. (2003b). Reply to Chabre et al., biophysics: is rhodopsin dimeric in native retinal rods? Nature 426, 31 10.1038/426030b [DOI] [PubMed] [Google Scholar]
- Fotiadis D., Liang Y., Filipek S., Saperstein D. A., Engel A., Palczewski K. (2004). The G protein-coupled receptor rhodopsin in the native membrane. FEBS Lett. 564, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. S., Chang B., Kannabiran C., Chakarova C., Singh H. P., Jalali S., et al. (2006). Premature truncation of a novel protein, RD3, exhibiting subnuclear localization is associated with retinal degeneration. Am. J. Hum. Genet. 79, 1059–1070. 10.1086/510021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries R., Scholten A., Säftel W., Koch K. W. (2012). Operation profile of zebrafish guanylate cyclase-activating protein 3. J. Neurochem. 121, 54–65. 10.1111/j.1471-4159.2011.07643.x [DOI] [PubMed] [Google Scholar]
- Frins S., Bönigk W., Müller F., Kellner R., Koch K. W. (1996). Functional characterization of a guanylyl cyclase-activating protein from vertebrate rods. J. Biol. Chem. 271, 8022–8027. 10.1074/jbc.271.14.8022 [DOI] [PubMed] [Google Scholar]
- Goraczniak R. M., Duda T., Sitaramayya A., Sharma R. K. (1994). Structural and functional characterization of the rod outer segment membrane guanylate cyclase. Biochem J. 302, 455–461. 10.1042/bj3020455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodovikova E. N., Senin I. I., Philippov P. P. (1994). Calcium-sensitive control of rhodopsin phosphorylation in the reconstituted system consisting of photoreceptor membranes, rhodopsin kinase and recoverin. FEBS Lett 353, 171–172. 10.1016/0014-5793(94)01030-7 [DOI] [PubMed] [Google Scholar]
- Govardovskii V. I., Korenyak D. A., Shukolyukov S. A., Zueva L. V. (2009). Lateral diffusion of rhodopsin in photoreceptor membrane: a reappraisal. Mol. Vis. 15, 1717–1729. [PMC free article] [PubMed] [Google Scholar]
- Granzin J., Cousin A., Weirauch M., Schlesinger R., Büldt G., Batra-Safferling R. (2012). Crystal structure of p44, a constitutively active splice variant of visual arrestin. J. Mol. Biol. 416, 611–618. 10.1016/j.jmb.2012.01.028 [DOI] [PubMed] [Google Scholar]
- Gray-Keller M., Denk W., Shraiman B., Detwiler P. B. (1999). Longitudinal spread of second messenger signals in isolated rod outer segments of lizards. J. Physiol. 519, 679–692. 10.1111/j.1469-7793.1999.0679n.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I. I., Senin I. I., Tikhomirova N. K., Komolov K. E., Permyakov S. E., Zernii E. Y., et al. (2012). Synergetic effect of recoverin and calmodulin on regulation of rhodopsin kinase. Front. Mol. Neurosci. 5:28. 10.3389/fnmol.2012.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunkel M., Schöneberg J., Alkhaldi W., Irsen S., Noé F., Kaupp U. B., et al. (2015). Higher-order architecture of rhodopsin in intact photoreceptors and its implication for phototransduction kinetics. Structure 23, 628–638. 10.1016/j.str.2015.01.015 [DOI] [PubMed] [Google Scholar]
- Hallett M. A., Delaat J. L., Arikawa K., Schlamp C. L., Kong F., Williams D. S. (1996). Distribution of guanylate cyclase within photoreceptor outer segments. J. Cell Sci. 109, 1803–1812. [DOI] [PubMed] [Google Scholar]
- Hamer R. D., Nicholas S. C., Tranchina D., Lamb T. D., Jarvinen J. L. (2005). Toward a unified model of vertebrate rod phototransduction. Vis. Neurosci. 22, 417–436. 10.1017/s0952523805224045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F., Yamazaki A. (1991). Polymorphism in purified guanylate cyclase from vertebrate rod photoreceptors. Proc. Natl. Acad. Sci. U S A 88, 4746–4750. 10.1073/pnas.88.11.4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins M. K., Oprian D. D., Schertler G. F. X. (2006). Recoverin binds exclusively to an amphipathic peptide at the N terminus of rhodopsin kinase, inhibiting rhodopsin phosphorylation without affecting catalytic activity of the kinase. J. Biol. Chem. 281, 19426–19432. 10.1074/jbc.m602203200 [DOI] [PubMed] [Google Scholar]
- Huang C. C., Orban T., Jastrzebska B., Palczewski K., Tesmer J. J. (2011). Activation of G protein-coupled receptor kinase 1 involves interactions between its N-terminal region and its kinase domain. Biochemistry 50, 1940–1949. 10.1021/bi101606e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. C., Yoshino-Koh K., Tesmer J. J. (2009). A surface of the kinase domain critical for the allosteric activation of G protein-coupled receptor kinases. J. Biol. Chem. 284, 17206–17215. 10.1074/jbc.m809544200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. Y., Koch K. W. (2002). The myristoylation of the neuronal Ca2+-sensors guanylate cyclase-activating protein 1 and 2. Biochim. Biophys. Acta 1600, 111–117. 10.1016/s1570-9639(02)00451-x [DOI] [PubMed] [Google Scholar]
- Hwang J. Y., Lange C., Helten A., Höppner-Heitmann D., Duda T., Sharma R. K., et al. (2003). Regulatory modes of rod outer segment membrane guanylate cyclase differ in catalytic efficiency and Ca2+-sensitivity. Eur. J. Biochem. 270, 3814–3821. 10.1046/j.1432-1033.2003.03770.x [DOI] [PubMed] [Google Scholar]
- Hwang J. Y., Schlesinger R., Koch K. W. (2001). Calcium-dependent cysteine reactivities in the neuronal calcium sensor guanylate cyclase-activating protein 1. FEBS Lett. 508, 355–359. 10.1016/s0014-5793(01)03094-0 [DOI] [PubMed] [Google Scholar]
- Imanishi Y., Yang L., Sokal I., Filipek S., Palczewski K., Baehr W. (2004). Diversity of guanylate cyclase-activating proteins (GCAPS) in teleost fish: characterization of three novel GCAPs (GCAP4, GCAP5, GCAP7) from zebrafish (Danio rerio) and prediction of eight GCAPs (GCAP1–8) in pufferfish (Fugu rubripes). J. Mol. Evol. 59, 204–217. 10.1007/s00239-004-2614-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Invergo B. M., Dell’Orco D., Montanucci L., Koch K. W., Bertrandpetit J. (2014). A comprehensive model of the phototransduction cascade in mouse rod cells. Mol. Biosyst. 10, 1481–1489. 10.1039/c3mb70584f [DOI] [PubMed] [Google Scholar]
- Invergo B. M., Montanucci L., Koch K. W., Bertranpetit J., Dell’Orco D. (2013). Exploring the rate-limiting steps in visual phototransduction recovery by bottom-up kinetic modeling. Cell Commun. Signal 11: 36. 10.1186/1478-811x-11-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B., Chen Y., Orban T., Jin H., Hofmann L., Palczewski K. (2015). Disruption of rhodopsin dimerization with synthetic peptides targeting an interaction interface. J. Biol. Chem. 290, 25728–25744. 10.1074/jbc.m115.662684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B., Maeda T., Zhu L., Fotiadis D., Filipek S., Engel A., et al. (2004). Functional characterization of rhodopsin monomers and dimers in detergents. J. Biol. Chem. 279, 54663–54675. 10.1074/jbc.m408691200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K., Bauer P. J., Kinjo T. G., Szerencsei R. T., Bönigk W., Winkfein R. J., et al. (2003). Assembly of retinal rod or cone Na+/Ca2+-K+ exchanger oligomers with cGMP-gated channel subunits as probed with heterologously expressed cDNAs. Biochemistry 42, 4593–4600. 10.1021/bi027276z [DOI] [PubMed] [Google Scholar]
- Karan S., Frederick J. M., Baehr W. (2010). Novel functions of photoreceptor guanylate cyclases revealed by targeted deletion. Mol. Cell. Biochem. 334, 141–155. 10.1007/s11010-009-0322-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp U. B., Koch K. W. (1992). Role of cGMP and Ca2+ in vertebrate photoreceptor excitation and adaptation. Ann. Rev. Physiol. 54, 153–175. 10.1146/annurev.physiol.54.1.153 [DOI] [PubMed] [Google Scholar]
- Kaupp U. B., Seifert R. (2002). Cyclic nucleotide-gated ion channels. Physiol. Rev. 82, 769–824. 10.1152/physrev.00008.2002 [DOI] [PubMed] [Google Scholar]
- Kawamura S. (1993). Rhodopsin phosphorylation as a mechanism of cyclic GMP phosphodiesterase regulation by S-modulin. Nature 362, 855–857. 10.1038/362855a0 [DOI] [PubMed] [Google Scholar]
- Kiel C., Vogt A., Campagna A., Chatr-aryamontri A., Swiatek-de Lange M., Beer M., et al. (2011). Structural and fuctional protein network analyses predict novel signalling functions for rhodopsin. Mol. Syst. Biol. 7: 551. 10.1038/msb.2011.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Hofmann K. P., Ernst O. P., Scheerer P., Choe H. W., Sommer M. E. (2013). Crystal structure of pre-activated arrestin p44. Nature 497, 142–146. 10.1038/nature12133 [DOI] [PubMed] [Google Scholar]
- Klenchin V. A., Calvert P. D., Bownds M. D. (1995). Inhibition of rhodopsin kinase by recoverin. Further evidence for a negative feedback system in phototransduction. J. Biol. Chem. 270, 16147–16152. 10.1074/jbc.270.27.16147 [DOI] [PubMed] [Google Scholar]
- Koch K. W. (1991). Purification and identification of photoreceptor guanylate cyclase. J. Biol. Chem. 266, 8634–8637. [PubMed] [Google Scholar]
- Koch K. W. (1994). Calcium as modulator of phototransduction in vertebrate photoreceptor cells. Rev. Physiol. Biochem. Pharmacol. 125, 149–192. 10.1007/bfb0030910 [DOI] [PubMed] [Google Scholar]
- Koch K. W. (2006). GCAPs, the classical neuronal calcium sensors in the retina - a Ca2+-relay model of guanylate cyclase activation. Calcium Binding Proteins 1, 3–6. [Google Scholar]
- Koch K. W. (2013). The guanylate cyclase signaling system in zebrafish photoreceptors. FEBS Lett. 587, 2055–2059. 10.1016/j.febslet.2013.04.023 [DOI] [PubMed] [Google Scholar]
- Koch K. W., Dell’Orco D. (2013). A Calcium-Relay Mechanism in Vertebrate Phototransduction. ACS Chem. Neurosci. 4, 909–917. 10.1021/cn400027z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K. W., Stryer L. (1988). Highly cooperative feedback control of retinal rod guanylate cyclase by calcium ions. Nature 334, 64–66. 10.1038/334064a0 [DOI] [PubMed] [Google Scholar]
- Koch K.-W., Duda T., Sharma R. K. (2010). Ca2+-modulated vision-linked ROS-GC guanylate cyclase transduction machinery. Mol. Cell. Biochem. 334, 105–115. 10.1007/s11010-009-0330-z [DOI] [PubMed] [Google Scholar]
- Kollmann H., Becker S. F., Shirdel J., Scholten A., Ostendorp A., Lienau C., et al. (2012). Probing the Ca2+ switch of the neuronal Ca2+ sensor GCAP2 by time-resolved fluorescence spectroscopy. ACS Chem. Biol. 7, 1006–1014. 10.1021/cb3000748 [DOI] [PubMed] [Google Scholar]
- Komolov K. E., Senin I. I., Kovaleva N. A., Christoph M. P., Churumova V. A., Grogoriev I. I., et al. (2009). Mechanism of rhodopsin kinase regulation by recoverin. J. Neurochem. 110, 72–79. 10.1111/j.1471-4159.2009.06118.x [DOI] [PubMed] [Google Scholar]
- Korenbrot J. I. (2012). Speed, sensitivity and stability of the light response in rod and cone photoreceptors: facts and models. Prog. Ret. Eye Res. 31, 442–466. 10.1016/j.preteyeres.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körschen H. G., Beyermann M., Müller F., Heck M., Vantler M., Koch K. W., et al. (1999). Interaction of glutamic acid-rich proteins with components of the cGMP-signaling pathway in rod photoreceptors. Nature 400, 761–766. 10.1038/23468 [DOI] [PubMed] [Google Scholar]
- Körschen H. G., Illing M., Seifert R., Sesti F., Williams A., Gotzes S., et al. (1995). A 240 kDa protein represents the complete beta subunit of the cyclic nucleotide-gated channel from rod photoreceptor. Neuron 15, 627–636. 10.1016/0896-6273(95)90151-5 [DOI] [PubMed] [Google Scholar]
- Köster M., Dell’Orco D., Koch K. W. (2014). The interaction network of rhodopsin involving the heterotrimeric G-protein transducin and the monomeric GTPase Rac1 is determined by distinct binding processes. FEBS J. 281, 5175–5185. 10.1111/febs.13064 [DOI] [PubMed] [Google Scholar]
- Krispel C. M., Chen D., Melling N., Chen Y. J., Martemyanov K. A., Quillinan N., et al. (2006). RGS expression rate-limits recovery of rod photoresponses. Neuron. 51, 409–416. 10.3410/f.1040096.489013 [DOI] [PubMed] [Google Scholar]
- Krylov D. M., Hurley J. B. (2001). Identification of proximate regions in a complex of retinal guanylyl cyclase 1 and guanylyl cyclase-activating protein-1 by a novel mass spectrometry-based method. J. Biol. Chem. 276, 30648–30654. 10.1074/jbc.m104121200 [DOI] [PubMed] [Google Scholar]
- Kumar V. D., Vijay-Kumar S., Krishnan A., Duda T., Sharma R. K. (1999). A second calcium regulator of rod outer segment membrane guanylate cyclase, ROS-GC1: neurocalcin. Biochemistry 38, 12614–12620. 10.1021/bi990851n [DOI] [PubMed] [Google Scholar]
- Lange C., Duda T., Beyermann M., Sharma R. K., Koch K. W. (1999). Regions in vertebrate photoreceptor guanylyl cyclase ROS-GC1 involved in Ca2+-dependent regulation by guanyly cyclase-activating protein GCAP-1. FEBS Lett. 460, 27–31. 10.1016/s0014-5793(99)01312-5 [DOI] [PubMed] [Google Scholar]
- Leibovic K. N., Bandarchi J. (1997a). Phototransduction and calcium exchange along the length of the retinal rod outer segment. Neuroreport 8, 1295–1300. [DOI] [PubMed] [Google Scholar]
- Leibovic K. N., Bandarchi J. (1997b). Effects of light and temperature on the response gradient of retinal rod outer segments. Brain Res. 750, 321–324. 10.1016/S0006-8993(96)01357-1 [DOI] [PubMed] [Google Scholar]
- Li C., Pan W., Braunewell K. H., Ames J. B. (2011). Structural analysis of Mg2+ and Ca2+ binding, myristoylation and dimerization of the neuronal calcium sensor and visinin-like protein 1 (VILIP-1). J. Biol. Chem. 286, 6354–6366. 10.1074/jbc.M110.173724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman P. A., Entine G. (1974). Lateral diffusion of visual pigment in photoreceptor disk membranes. Science 185, 457–459. 10.1126/science.185.4149.457 [DOI] [PubMed] [Google Scholar]
- Lim S., Dizhoor A. M., Ames J. B. (2014). Structural diversity of neuronal calcium sensor proteins and insights for activation of retinal guanylyl cyclase by GCAP1. Front. Mol. Neurosci. 7:19. 10.3389/fnmol.2014.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Peshenko I., Dizhoor A., Ames J. B. (2009). Effects of Ca2+, Mg2+ and myristoylation on guanylyl cyclase activating protein 1 structure and stability. Biochemistry 48, 850–862. 10.1021/bi801897p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Strahl T., Thorner J., Ames J. B. (2011). Structure of a Ca2+-myristoyl switch protein that controls activation of a phosphatidylinositol 4-kinase in fission yeast. J. Biol. Chem. 286, 12565–12577. 10.1074/jbc.m110.208868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Seno K., Hayashi F. (2003). Active transducin alpha subunit carries PDE6 to detergent-resistant membranes in rod photoreceptor outer segments. Biochem. Biophys. Res. Commun. 303, 19–23. 10.1016/s0006-291x(03)00284-5 [DOI] [PubMed] [Google Scholar]
- Lobanova E. S., Finkelstein S., Song H., Tsang S. H., Chen C. K., Sokolov M., et al. (2007). Transducin translocation in rods is triggered by saturation of the GTPase-activating complex. J. Neurosci. 27, 1151–1160. 10.1523/jneurosci.5010-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe D. G., Dizhoor A. M., Liu K., Gu Q., Spencer M., Laura R., et al. (1995). Cloning and expression of a second photoreceptor-specific membrane retina guanylyl cyclase (RetGC), RetGC-2. Proc. Natl. Acad. Sci. U S A 92, 5535–5539. 10.1073/pnas.92.12.5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D. G., Xue T., Yau K. W. (2008). How vision begins: an odyssey. Proc. Natl. Acad. Sci. U S A 105, 9855–9862. 10.1073/pnas.0708405105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Beuve A., van den Akker F. (2010). Crystal structure of the signaling helix coiled-coil domain of the beta1 subunit of the soluble guanylyl cyclase. BMC Struct. Biol. 10:2. 10.1186/1472-6807-10-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Imanishi Y., Palczewski K. (2003). Rhodopsin phosphorylation: 30 years later. Progr. Ret. Eye Res. 22, 417–434. 10.1016/s1350-9462(03)00017-x [DOI] [PubMed] [Google Scholar]
- Makino C. L., Dodd R. L., Chen J., Burns M. E., Roca A., Simon M. I., et al. (2004). Recoverin regulates light-dependent phosphodiesterase activity in retinal rods. J. Gen. Physiol. 123, 729–741. 10.1085/jgp.200308994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino C. L., Wen X. H., Olshevskaya E. V., Peshenko I. V., Savchenko A. B., Dizhoor A. M. (2012). Enzymatic relay mechanism stimulates cyclic GMP synthesis in rod photoresponse: biochemical and physiological study in guanyly cyclase activating protein 1 knockout mice. PLoS One. 7:e47637. 10.1371/journal.pone.0047637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis A., Goraczniak R. M., Duda T., Sharma R. K., Sitaramayya A. (1993). Structural and biochemical identity of retinal rod outer segment membrane guanylate cyclase. Biochem. Biophys. Res. Commun. 194, 855–861. 10.1006/bbrc.1993.1900 [DOI] [PubMed] [Google Scholar]
- Marino V., Astegno A., Pedroni M., Piccinelli F., Dell’Orco D. (2014). Nanodevice-induced conformational and functional changes in a prototypical calcium sensor protein. Nanoscale 6, 412–423. 10.1039/c3nr04978g [DOI] [PubMed] [Google Scholar]
- Marino V., Sulmann S., Koch K. W., Dell’Orco D. (2015). Structural effects of Mg2+ on the regulatory states of three neuronal calcium sensors operating in vertebrate phototransduction. Biochim. Biophys. Acta 1853, 2055–2065. 10.1016/j.bbamcr.2014.10.026 [DOI] [PubMed] [Google Scholar]
- Martin R. E., Elliott M. H., Brush R. S., Anderson R. E. (2005). Detailed characterization of the lipid composition of detergent-resistant membranes from photoreceptor rod outer segment membranes. Invest. Ophthalmol. Vis. Sci. 46, 1147–1154. 10.1167/iovs.04-1207 [DOI] [PubMed] [Google Scholar]
- Mazzolini M., Facchetti G., Andolfi L., Zaccaria P. R., Tuccio S., Treu J., et al. (2015). The phototransduction machinery in the rod outer segment has a strong efficacy gradient. Proc. Natl. Acad. Sci. U S A 112, E2715–E2724. 10.1073/pnas.1423162112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador W. E., Means A. R., Quiocho F. A. (1993). Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science 262, 1718–1721. 10.1126/science.8259515 [DOI] [PubMed] [Google Scholar]
- Mendez A., Burns M. E., Sokal I., Dizhoor A. M., Baehr W., Palczewski K., et al. (2001). Role of guanylate cyclase-activating proteins (GCAPs) in setting the flash sensitivity of rod photoreceptors. Proc. Natl. Acad. Sci. U S A 98, 9948–9953. 10.3410/f.1001761.6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalakis S., Zong X., Becirovic E., Hammelmann V., Wein T., Wanner K. T., et al. (2011). The glutamic acid-rich protein is a gating inhibitor of cyclic nucleotide-gated channels. J. Neurosci. 31, 133–141. 10.1523/jneurosci.4735-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molday L. L., Djajadi H., Yan P., Szczygiel L., Boye S. L., Chiodo V. A., et al. (2013). RD3 gene delivery restores guanylate cyclase localization and rescues photoreceptors in the Rd3 mouse model of Leber congenital amaurosis 12. Hum. Mol. Genet. 22, 3894–3905. 10.1093/hmg/ddt244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair K. S., Balasubramanian N., Slepak V. Z. (2002). Signal-dependent translocation of transducin, RGS9–1-Gbeta5L complex and arrestin to detergent-resistant membrane rafts in photoreceptors. Curr.Biol. 12, 421–425 10.1016/s0960-9822(02)00691-7 [DOI] [PubMed] [Google Scholar]
- O‘Callaghan D. W., Tepikin A. V., Burgoyne R. D. (2003). Dynamics and calcium sensitivity of the Ca2+/myristoyl switch protein hippocalcin in living cells. J. Cell Beiol. 163, 715–721. 10.1083/jcb.200306042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban T., Huang C. C., Homan K. T., Jastrzebska B., Tesmer J. J. G., Palczewski K. (2012). Substrate-induced changes in the dynamics of rhodopsin kinase (G protein-coupled receptor kinase 1). Biochemistry 51, 3404–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. (2012). Chemistry and biology of vision. J. Biol. Chem. 287, 1612–1619. 10.1074/jbc.R111.301150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., et al. (2000). Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745. 10.1126/science.289.5480.739 [DOI] [PubMed] [Google Scholar]
- Palczewski K., Subbaraya I., Gorczyca W. A., Helekar B. S., Ruiz C. C., Ohguro H., et al. (1994). Molecular cloning and characterization of retinal photoreceptor guanylyl cyclase-activating protein. Neuron 13, 395–404. 10.1016/0896-6273(94)90355-7 [DOI] [PubMed] [Google Scholar]
- Pearring J. N., Salinas R. Y., Baker S. A., Arshavsky V. Y. (2013). Protein sorting, targeting and trafficking in photoreceptor cells. Prog. Retin. Eye Res. 36, 24–51. 10.1016/j.preteyeres.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentia D. C., Hosier S., Cote R. H. (2006). The glutamic acid-rich protein-2 (GARP2) is a high affinity rod photoreceptor phosphodiesterase (PDE6)-binding protein that modulates its catalytic properties. J. Biol. Chem. 281, 5500–5505. 10.1074/jbc.m507488200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshenko I. V., Dizhoor A. M. (2006). Ca2+ and Mg2+ binding properties of GCAP-1. Evidence that Mg2+-bound form is the physiological activator of photoreceptor guanylyl cyclase. J. Biol. Chem. 281, 23830–23841. 10.1074/jbc.m600257200 [DOI] [PubMed] [Google Scholar]
- Peshenko I. V., Olshevskaya E. V., Dizhoor A. M. (2015a). Evaluating the role of retinal membrane guanylyl cyclase 1 (RetGC1) domains in binding guanylyl cyclase-activating proteins (GCAPs). J. Biol. Chem. 290, 6913–6924. 10.1074/jbc.M114.629642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshenko I. V., Olshevskaya E. V., Dizhoor A. M. (2015b). Dimerization domain of retinal membrane guanylyl cyclase 1 (RetGC1) is an essential part of guanylyl cyclase-activating protein (GCAP) binding interface. J. Biol. Chem. 290, 19584–19596. 10.1074/jbc.m115.661371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshenko I. V., Olshevskaya E. V., Azadi S., Molday L. L., Molday R. S., Dizhoor A. M. (2011a). Retinal degeneration 3 (RD3) protein inhibits catalytic activity of retinal membrane guanylyl cyclase (RetGC) and its stimulation by activating proteins. Biochemistry 50, 9511–9519. 10.1021/bi201342b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshenko I. V., Olshevskaya E. V., Savchenko A. B., Karan S., Palczewski K., Baehr W., et al. (2011b). Enzymatic properties and regulation of the native isozymes of retinal membrane guanylyl cyclase (RetGC) from mouse photoreceptors. Biochemistry 50, 5590–5600. 10.1021/bi200491b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshenko I. V., Moiseyev G. P., Olshevskaya E. V., Dizhoor A. M. (2004). Factors that determine Ca2+ sensitivity of photoreceptor guanylyl cyclase. Kinetic analysis of the interaction between the Ca2+-bound and the Ca2+-free guanylyl cyclase activating proteins (GCAPs) and recombinant photoreceptor guanylyl cyclase 1 (RetGC-1). Biochemistry 43, 13796–13804. [DOI] [PubMed] [Google Scholar]
- Poetsch A., Molday L. L., Molday R. S. (2001). The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J. Biol. Chem. 276, 48009–48016. 10.1074/jbc.M108941200 [DOI] [PubMed] [Google Scholar]
- Poo M., Cone R. A. (1974). Lateral diffusion of rhodopsin in the photoreceptor membrane. Nature 247, 438–441. 10.1038/247438a0 [DOI] [PubMed] [Google Scholar]
- Preising M. N., Hausotter-Will N., Solbach M. C., Friedburg C., Rüschendorf F., Lorenz B. (2012). Mutations in RD3 are associated with an extremely rare and severe form of early onset retinal dystrophy. Invest. Ophthalmol. Vis. Sci. 53, 3463–3472. 10.1167/iovs.12-9519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premont R. T., Gainetdinov R. R. (2007). Physiological roles of G protein-coupled receptor kinases and arrestins. Annu. Rev. Physiol. 69, 511–534. 10.1146/annurev.physiol.69.022405.154731 [DOI] [PubMed] [Google Scholar]
- Pugh E. N., Jr, Lamb T. D. (2000). “Phototransduction in vertebrate rods and cones: molecular mechanisms of amplification, recovery and light adaptation,” in Handbook of Biological Physics Elsevier Science BV, eds Stavenga D. G., DeGrip W. J., Pugh E. N., Jr. (New york, NY: Elsevier Science: ), 183–255. [Google Scholar]
- Pulvermüller A., Giessl A., Heck M., Wottrich R., Schmitt A., Ernst O. P., et al. (2002). Calcium-dependent assembly of centrin-G-protein complex in photoreceptor cells. Mol. Cell. Biol. 22, 2194–2203. 10.1128/mcb.22.7.2194-2203.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvermüller A., Palczewski K., Hofmann K. P. (1993). Interaction bewteen photoactivated rhodopsin and ist kinase: stability and kinetics of complex formation. Biochemistry 32, 14082–14088. 10.1021/bi00214a002 [DOI] [PubMed] [Google Scholar]
- Ramamurthy V., Tucker C., Wilkie S., Daggett V., Hunt D., Hurley J. B. (2001). Interactions within the coiled-coil domain of RetGC-1 guanlyly cyclase are optimized for regulation rather than for high affinity. J. Biol. Chem. 276, 26218–26229. 10.1074/jbc.m010495200 [DOI] [PubMed] [Google Scholar]
- Rätscho N., Scholten A., Koch K. W. (2009). Expression profiles of three novel sensory guanylate cyclases and guanylate cyclase-activating proteins in the zebrafish retina. Biochim. Biophys. Acta 1793, 1110–1114. 10.1016/j.bbamcr.2008.12.021 [DOI] [PubMed] [Google Scholar]
- Robin J., Brauer J., Sulmann S., Marino V., Dell’Orco D., Lienau C., et al. (2015). Differential nanosecond protein dynamics in homologous calcium sensors. ACS Chem. Biol. 10, 2344–2352. 10.1021/acschembio.5b00278 [DOI] [PubMed] [Google Scholar]
- Roof D. J., Heuser J. E. (1982). Surfaces of rod photoreceptor disk membranes: integral membrane components. J. Cell. Biol. 95, 487–500. 10.1083/jcb.95.2.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig D. H., Nair K. S., Levay K., Peshenko I. V., Crabb J. W., Dizhoor A. M., et al. (2009). Interaction of retinal guanylate cyclase with the alpha subunit of transducin: potential role in transducin localization. Biochem. J. 417, 803–812. 10.1042/bj20081513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil H., Chabre M., Worcester D. (1976). Neutron diffraction studies of retinal rod outer segment membranes. Nature 262, 266–270. 10.1038/262266a0 [DOI] [PubMed] [Google Scholar]
- Sakurai K., Chen J., Khani S. C., Kefalov V. J. (2015). Regulation of mammalian cone phototransduction by recoverin and rhodopsin kinase. J. Biol. Chem. 290, 9239–9250. 10.1074/jbc.m115.639591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K., Young J. E., Kefalov V. J., Khani S. C. (2011). Variation in rhodopsin kinase expression alters the dim flash response shut off and the light adaptation in rod photoreceptors. Invest. Ophthalmol. Vis. Sci. 29, 6793–6800. 10.1167/iovs.11-7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfare S., McKeown A. S., Messinger J., Rubin G., Wei H., Kraft T. W., et al. (2014). Overexpression of rod photoreceptor glutamic acid rich protein 2 (GARP2) increases gain and slows recovery in mouse retina. Cell Commun. Signal. 12: 67. 10.1186/s12964-014-0067-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapf J. L. (1983). Dependence of the single photon response on longitudinal position of absorption in toad rod outer segments. J. Physiol. 343, 147–159. 10.1113/jphysiol.1983.sp014886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten A., Koch K. W. (2011). Differential calcium signaling by cone specific guanylate cyclase-activating proteins from the zebrafish retina. PLoS One 6:e23117. 10.1371/journal.pone.0023117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J., Heck M., Hofmann K. P., Noé F. (2014). Explicit spatiotemporal simulation of receptor-G protein coupling in rod cell disk membranes. Biophys. J. 107, 1042–1053. 10.1016/j.bpj.2014.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J., Heck M., Hofmann K. P., Noé F. (2015). Response to comment “Transient complexes between dark rhodopsin and transducin: circumstantial evidence or physiological necessity?” by D. Dell’Orco and K.-W. Koch. Biophys. J. 108, 778–779. 10.1016/j.bpj.2014.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrem A., Lange C., Beyermann M., Koch K.-W. (1999). Identification of a domain in guanylyl cyclase-activating protein 1 that interacts with a complex of guanylyl cyclase and tubulin in photoreceptors. J. Biol. Chem. 274, 6244–6249. 10.1074/jbc.274.10.6244 [DOI] [PubMed] [Google Scholar]
- Schwarzer A., Schauf H., Bauer P. J. (2000). Binding of the cGMP-gated channel to the Na/Ca-K exchanger in rod photoreceptors. J. Biol. Chem. 275, 13448–13454. 10.1074/jbc.275.18.13448 [DOI] [PubMed] [Google Scholar]
- Senin I. I., Fischer T., Komolov K. E., Zinchenko D. V., Philippov P. P., Koch K.-W. (2002a). Ca2+-myristoyl switch in the neuronal calcium sensor recoverin requires different functions of Ca2+-binding sites. J. Biol. Chem. 277, 50365–50372. 10.1074/jbc.m204338200 [DOI] [PubMed] [Google Scholar]
- Senin I. I., Koch K. W., Akhtar M., Philippov P. P. (2002b). Ca2+-dependent control of rhodopsin phosphorylation: recoverin and rhodopsin kinase. Adv. Exp. Med. Biol. 514, 69–99. 10.1007/978-1-4615-0121-3_5 [DOI] [PubMed] [Google Scholar]
- Senin I. I., Hoeppner-Heitmann D., Polkovnikova O. O., Churumova V. A., Tikhomirova N. K., Philippov P. P., et al. (2004). Recoverin and rhodopsin kinase activity in detergent-resistant membrane rafts from rod outer segments. J. Biol. Chem. 279, 48647–48653. 10.1074/jbc.m402516200 [DOI] [PubMed] [Google Scholar]
- Senin I. I., Vaganova S. A., Weiergräber O. H., Ergonov N. S., Philippov P. P., Koch K.-W. (2003). Functional restoration of the Ca2+-myristoyl switch in a recoverin mutant. J. Mol. Biol. 330, 409–418. 10.1016/s0022-2836(03)00581-3 [DOI] [PubMed] [Google Scholar]
- Senin I. I., Zargarov A. A., Alekseev A. M., Gorodovikova E. N., Lipkin V. M., Philippov P. P. (1995). N-myristoylation of recoverin enhances its efficiency as an inhibitor of rhodopsin kinase. FEBS Lett. 376, 87–90. 10.1016/0014-5793(95)01187-2 [DOI] [PubMed] [Google Scholar]
- Seno K., Kishimoto M., Abe M., Higuchi Y., Mieda M., Owada Y., et al. (2001). Light- and guanosine 5′-3-O-(thio)triphosphate-sensitive localization of a G protein and its effector on detergent-resistant membrane rafts in rod photoreceptor outer segments. J. Biol. Chem. 276, 20813–20816. 10.1074/jbc.c100032200 [DOI] [PubMed] [Google Scholar]
- Shen L., Caruso G., Bisegna P., Andreucci D., Gurevich V. V., Hamm H. E., et al. (2010). Dynamics of mouse rod phototransduction and its sensitivity to variation of key parameters. IET Syst. Biol. 4, 12–32. 10.1049/iet-syb.2008.0154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichi H., Williams T. C. (1979). Rhodopsin phosphorylation suggests biochemical heterogeneities of retinal rod disks. J. Supramol. Struct. 12, 419–424. 10.1002/jss.400120402 [DOI] [PubMed] [Google Scholar]
- Singh P., Wang B., Maeda T., Palczewski K., Tesmer J. J. G. (2008). Structures of rhodopsin kinase in different ligand states reveal key elements involved in G proiten-coupled receptor kinase activation. J. Biol. Chem. 283, 14053–14062. 10.3410/f.1103994.561150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal I., Haeseleer F., Arendt A., Adman E. T., Hargrave P. A., Palczewski K. (1999a). Identification of a guanylyl cyclase-activating protein-binding site within the catalytic domain of retinal guanylyl cyclase 1. Biochemistry 38, 1387–1393. 10.1021/bi982512k [DOI] [PubMed] [Google Scholar]
- Sokal I., Otto-Bruc A. E., Surgucheva I., Verlinde C. L. M., Wang C. K., Baehr W., et al. (1999b). Conformational changes in guanylyl cyclase-activating protein 1 (GCAP1) and its tryptophan mutants as a function of calcium concentration. J. Biol. Chem. 274, 19829–19837. 10.1074/jbc.274.28.19829 [DOI] [PubMed] [Google Scholar]
- Sokolov M., Lyubarsky A. L., Strissel K. J., Savchenko A. B., Govardovskii V. I., Pugh E. N., et al. (2002). Massive light-driven translocation of transducin between the two major compartments of rod cells: a novel mechanism of light adaptation. Neuron 28, 95–106. 10.3410/f.1005371.64305 [DOI] [PubMed] [Google Scholar]
- Sommer M. E., Hofmann K. P., Heck M. (2014). Not just signal shutoff: the protective role of arrestin-1 in rod cells. Handb. Exp. Pharmacol. 219, 101–116. 10.1007/978-3-642-41199-1_5 [DOI] [PubMed] [Google Scholar]
- Stephen R., Bereta G., Golczak M., Palczewski K., Sousa M. C. (2007). Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1. Structure 15, 1392–1402. 10.1016/j.str.2007.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L. (1991). Visual excitation and recovery. J. Biol. Chem. 266, 10711–10714. [PubMed] [Google Scholar]
- Sugimoto Y., Yatsunami K., Tsujimoto M., Khorana H. G., Ichikawa A. (1991). The amino acid sequence of a glutamic acid-rich protein from bovine retina as deduced from the cDNA sequence. Proc. Natl. Acad. Sci. U S A 88, 3116–3119. 10.1073/pnas.88.8.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulmann S., Vocke F., Scholten A., Koch K. W. (2015). Retina specific GCAPs in zebrafish acquire functional selectivity in Ca2+-sensing by myristoylation and Mg2+-binding. Sci. Rep. 5:11228. 10.1038/srep11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulmann S., Dell’Orco D., Marino V., Behnen P., Koch K.-W. (2014). Conformational changes in calcium-sensor proteins under molecular crowding conditions. Chemistry Eur. J. 20, 6756–6762. 10.1002/chem.201402146 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Ames J. B., Harvey T. S., Stryer L., Ikura M. (1995). Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature 376, 444–447. 10.1038/376444a0 [DOI] [PubMed] [Google Scholar]
- Venkataraman V., Duda T., Ravichandran S., Sharma R. K. (2008). Neurocalcin delta Modulation of ROS-GC1, a New Model of Ca2+ Signaling. Biochemistry 47, 6590–6601. 10.1021/bi800394s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman V., Duda T., Vardi N., Koch K.-W., Sharma R. K. (2003). Calcium-modulated guanylate cyclase (ROS-GC1) transduction machinery in the photoreceptor-bipolar region. Biochemistry 42, 5640–5648. 10.1021/bi034025x [DOI] [PubMed] [Google Scholar]
- Wang J., Deretic D. (2014). Molecular complexes that direct rhodopsin transport to primary cilia. Prog. Retin. Eye Res. 38, 1–19. 10.1016/j.preteyeres.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiergräber O., Senin I. I., Zernii E. Y., Churumova V. A., Kovaleva N. A., Nazipova A. A., et al. (2006). Tuning of a neuronal calcium sensor. J. Biol. Chem. 281, 37594–37602. 10.1074/jbc.M603700200 [DOI] [PubMed] [Google Scholar]
- Weiergräber O. H., Senin I. I., Philippov P. P., Granzin J., Koch K. W. (2003). Impact of N-terminal myristoylation on the Ca2+-dependent conformational transition in recoverin. J. Biol. Chem. 278, 22972–22979. 10.1074/jbc.m300447200 [DOI] [PubMed] [Google Scholar]
- Weitz D., Ficek N., Kremmer E., Bauer P. J., Kaupp U. B. (2002). Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron 36, 881–889. 10.1016/s0896-6273(02)01098-x [DOI] [PubMed] [Google Scholar]
- Wen X. H., Dizhoor A. M., Makino C. L. (2014). Membrane guanylyl cyclase complexes shape the photoresponses of retinal rods and cones. Front. Mol. Neurosci. 7:45. 10.3389/fnmol.2014.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X. H., Duda T., Pertzev A., Venkataraman V., Makino C. L., Sharma R. K. (2012). S100B serves as a Ca2+ sensor for ROS-GC1 guanylate cyclase in cones but not in rods of the murine retina. Cell. Physiol. Biochem. 29, 417–430. 10.1159/000338496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensel T. G. (2008). Signal transducing membrane complexes of photoreceptor outer segments. Vision Res. 48, 2052–2061. 10.1016/j.visres.2008.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited A. M., Park P. S. (2015). Nanodomain organization of rhodopsin in native human and murine rod outer segment disc membranes. Biochim. Biophys. Acta. 1848, 26–34. 10.1016/j.bbamem.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. W. (1967). The renewal of photoreceptor cell outer segments. J. Cell. Biol. 33, 61–72. 10.1083/jcb.33.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zägel P., Dell’Orco D., Koch K.-W. (2013). The dimerization domain in outer segment guanylate cyclase isa Ca2+-sensitive control switch module. Biochemistry 52, 5065–5074. 10.1021/bi400288p [DOI] [PubMed] [Google Scholar]
- Zernii E., Komolov K., Permyakov S., Kolpakova T., Dell’Orco D., Poetzsch A., et al. (2011). Involvement of recoverin C-terminal segment in recognition of the target enzyme rhodopsin kinase. Biochem. J. 435, 441–450. 10.1042/bj20110013 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Molday L. L., Molday R. S., Sarfare S. S., Woodruff M. L., Fain G. L., et al. (2009). Knockout of GARPs and the β-subunit of the rod cGMP-gated channel disrupts disk morphogenesis and rod outer segment structural integrity. J. Cell Sci. 122, 1192–1200. 10.1242/jcs.042531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Trudeau M. C., Zagotta W. N. (2002). Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron 36, 891–896. 10.1016/s0896-6273(02)01099-1 [DOI] [PubMed] [Google Scholar]
- Zozulya S., Stryer L. (1992). Calcium-myristoyl protein switch. Proc. Natl. Acad. Sci. U S A 89, 11569–11573. 10.1073/pnas.89.23.11569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulliger R., Naash M. I., Rajala R. V., Molday R. S., Azadi S. (2015). Impaired association of retinal degeneration-3 with guanylate cyclase-1 and guanylate cyclase-activating protein-1 leads to leber congenital amaurosis-1. J. Biol. Chem. 290, 3488–3499. 10.1074/jbc.m114.616656 [DOI] [PMC free article] [PubMed] [Google Scholar]