Abstract
Objectives
To evaluate the influence of IGFBP-3 methylation on recurrence in patients with stage II colorectal cancer (CRC) from 2 independent cohorts.
Background
The relationship between IGFBP-3 methylation in primary tumors (PTs) or lymph nodes (LNs) and risk of recurrence in patients with stage II CRC treated with surgery alone is unknown.
Methods
IGFBP-3 methylation of DNA from 115 PTs and 1641 LNs in patients with stage II CRC from 2 independent cohorts was analyzed. Forty patients developed recurrence, whereas 75 matched patients remained recurrence free for more than 2 years after surgery. Cox proportional hazard models were used to calculate hazard ratios (HRs) of recurrence, adjusted for patient and tumor characteristics.
Results
Methylation of IGFBP-3 in PTs was identified to be significantly associated with risk of recurrence in the training set. The signature was tested in a validation set and classified 40.7% of patients as high risk. Five-year recurrence-free survival rates were 76.4% and 58.3% for low- and high-risk patients, respectively, with an HR of 2.21 (95% confidence interval, 1.04–4.68; P = 0.039). In multivariate analysis, the signature remained the most significant prognostic factor, with an HR of 2.40 (95% confidence interval, 1.10–5.25; P = 0.029). A combined analysis of 1641 LNs from the 2 sets identified IGFBP-3 methylation in LNs was not associated with risk of recurrence.
Conclusions
Detection of IGFBP-3 methylation in PTs, but not in LNs, provides a powerful tool for the identification of patients with stage II CRC at high risk of recurrence.
Keywords: colorectal cancer, IGFBP-3, methylation, recurrence
Colorectal cancer (CRC) is the fourth most common cancer in the United States.1 Worldwide, the incidence is increasing, with an annual estimate of 1,233,700 new cases diagnosed and 608,700 deaths.2 The 5-year survival of patients with stage II is approximately 75% to 82%.3,4 There are still about 20% of patients with this stage of tumors who die of recurrent disease. There is clearly a need to identify prognostic factors to guide the identification of stage II patients who are likely to experience recurrence. This information would allow more informed planning for patients who are more likely to require and possibly benefit from intensive surveillance or adjuvant therapy.
In current practice, a limited set of clinical and pathologic markers (ie, T4 tumors, poor histologic grade, lymphovascular invasion, perineural invasion, bowel obstruction, lesions with localized perforation or close, indeterminate, or positive margins, less than 12 nodes examined) can identify small groups of patients with stage II disease who have higher recurrence risk. The majority of patients do not have a marker that categorizes them as higher risk. In light of the importance of this issue, there have been many attempts to find novel molecular markers, such as microsatellite instability (MSI)/mismatch repair,5,6 LOH 18q,7 expression/mutation/methylation of individual genes or groups of genes,8–13 to identify patients with the potential for CRC recurrence. However, the clinical utilities of these markers are still under study.
Dissemination to locoregional lymph nodes (LNs) is also an important prognostic factor in CRC. Current clinical detection of micrometastases by standard immunohistochemistry techniques is limited to those with a minimal number of cells. Technical advances now permit the detection of micrometastases at the molecular level. For example, somatic gene mutations or methylation and amplification of cancer-specific RNA that occur in the primary tumor (PT) are detectable in LNs.14–19
We have recently correlated DNA methylation of 6 extracellular matrix genes with outcome of patients with CRC. Among all genes analyzed, DNA methylation of the insulin-like growth factor binding protein 3 (IGFBP-3) gene showed the strongest association with poor survival.20 In experimental models, insulin-like growth factor-I (IGF-I) promotes the growth and metastasis of CRC cells,21–24 whereas IGFBP-3 inhibits growth through ligand sequestration and may also have antiproliferative and proapoptotic activities through actions independent of the IGF-I/IGF-I receptor.25 Several clinical studies have shown that circulating IGF-I is elevated and IGFBP-3 levels reduced in patients before the diagnosis of CRC and that increased plasma levels of IGFBP-3 are associated with a decreased risk26,27 and better prognosis of CRC.28 Importantly, IGFBP-3 promoter methylation is observed in many cancers and has been associated with poor clinical outcome. However, the possible prognostic value of IGFBP-3 methylation in PTs or LNs for tumor recurrence after surgical resection of early-stage CRC is unknown. Therefore, we assessed the influence of IGFBP-3 methylation on recurrence in patients with stage II CRC in 2 independent set studies.
Materials and Methods
Study Population
Evidence of recurrent disease was confirmed in 40 patients with pathologically verified stage II (T3, 4N0M0) cancer who received a diagnosis of CRC and underwent radical surgery at the Johns Hopkins Bayview Hospital (JHBH) and the Johns Hopkins Hospital (JHH) between 1995 and 2009. Cases included 12 patients from the JHBH and 28 patients from the JHH in whom the tumor recurred after surgery. On the basis of age, date of surgery (±5 years), and sex, we matched the case patients with 75 controls with stage II CRC in whom there was no recurrence with at least 24-month follow-up, by which time most of the CRC recurrences occur.29 Patients with neoadjuvant chemotherapy were excluded from the current study. Thus, formalin-fixed and paraffin-embedded (FFPE) CRC tissue and adjacent nonneoplastic colorectal tissue samples from 115 patients with coded stage II CRC were obtained from the JHBH and the JHH with approval by the Institutional Review Board and deemed in accordance with the Health Insurance Portability and Accountability Act regulations. The histopathology of each specimen was reviewed to confirm diagnosis. Uniform follow-up information was available from electronic health databases at Johns Hopkins University.
The JHBH training set consisted of 34 tissue samples from patients with stage II CRC (median follow-up of 61.4 months). The JHH validation set consisted of 81 tissue samples from patients with stage II CRC (median follow-up of 65.2 months). Patients in both cohorts were similar with respect to age, sex, location, tumor size, differentiation, LNs examined, proportion of cases with pT4, lymphovascular invasion, mucin production, proportion of cases with recurrence, recurrence type, death, and recurrence-free survival (RFS; Table 1). Clinicopathologic features of the patients and their recurrence status are listed in Supplementary Table S1 (available at http://links.lww.com/SLA/A765). The patient and tumor characteristics did not differ significantly between the recurrence and no-recurrence patients.
Table 1. Patient Demographics and Clinicopathologic Characteristics for the Training and Validation Sets.
| Demographic or Clinical Characteristic | Traning Set (n = 34) | Validation Set (n = 81) | P | ||
|---|---|---|---|---|---|
|
|
|
||||
| No. Patients | % | No. Patients | % | ||
| Hospital | |||||
| JHH | 81 | ||||
| JHBH | 34 | ||||
| Age, mean ± SD | 65.3 ± 12.8 | 66.8 ± 11.6 | 0.573 | ||
| Sex | |||||
| Male | 14 | 41.2 | 38 | 46.9 | |
| Female | 20 | 58.8 | 43 | 53.1 | |
| Localization | 0.665 | ||||
| Left colon | 14 | 41.2 | 27 | 33.3 | |
| Right colon | 15 | 44.1 | 43 | 53.1 | |
| Rectum | 5 | 14.7 | 11 | 13.6 | |
| Tumor size, cm, mean + SD | 4.7 ± 2.0 | 5.1 ± 2.5 | 0.402 | ||
| Differentiation | 0.261 | ||||
| Well and moderate | 26 | 76.5 | 69 | 85.2 | |
| Poor | 8 | 23.5 | 12 | 14.8 | |
| LNs examined, mean + SD | 14.3 ± 6.2 | 14.3 ± 5.5 | 0.995 | ||
| T4 tumors | 0.101 | ||||
| Yes | 6 | 17.6 | 6 | 7.4 | |
| No | 28 | 82.4 | 75 | 92.6 | |
| Lymphovascular invasion | 0.261 | ||||
| Yes | 8 | 23.5 | 12 | 14.8 | |
| No | 26 | 76.5 | 69 | 85.2 | |
| Mucin production | 0.523 | ||||
| Yes | 5 | 14.7 | 16 | 19.8 | |
| No | 29 | 85.3 | 65 | 80.2 | |
| Recurrence | 0.941 | ||||
| Yes | 12 | 35.3 | 28 | 34.6 | |
| No | 22 | 64.7 | 53 | 65.4 | |
| Recurrent type | 1.000* | ||||
| Local | 2 | 16.7 | 5 | 17.9 | |
| Distant | 10 | 83.3 | 23 | 82.1 | |
| Death | 0.899 | ||||
| Yes | 13 | 38.2 | 32 | 39.5 | |
| No | 21 | 61.8 | 49 | 60.5 | |
| RFS, mo, mean + SD | 61.0 ±41.2 | 59.0 ± 39.9 | 0.809 | ||
Fisher exact test.
SD indicates standard deviation.
From the training set, a total of 462 LNs were harvested (mean 14; range, 2–26 nodes). The LNs were embedded in a total of 117 paraffin blocks. The validation set included 466 blocks and 1179 LNs. On average 15 LNs (range, 6–43 nodes) were dissected per patient. Each block contained a variable number of nodes (1–6 nodes).
Procedures
Genomic DNA from FFPE tissue was extracted by phenol-chloroform, and polymerase chain reaction (PCR) targeted for KRAS codons 12 and 13 was performed as previously described.30 PCR products were sequenced in both directions by use of an M13F primer (5′-GTAAAACGACGGCCAGT-3′) and an M13R primer (5′-CAGGAAACAGCTATGACC-3′) that were incorporated into the forward and reverse primers of each primer pair, respectively (Agencourt Bioscience Corporation). Sequence data were analyzed with Sequencher 4.8 software (Gene Codes). Verification of all mutations was accomplished by bidirectional sequencing of a second PCR product derived independently from the original template.
MSI status was determined using D2S123, D5S346, D17S250, BAT25, and BAT26.31 Microsatellite sizes were compared with those of normal adjacent tissue, and tumors with 2 or more of the markers exhibiting instability were classified as high MSI (MSI-H). Tumors with only 1 marker exhibiting instability or no markers with instability were classified as low MSI (MSI-L) or microsatellite stable (MSS), respectively. In this study, MSI-L and MSS tumors were grouped together and henceforth are referred to as MSS, and MSI-H is referred to as MSI.
Quantitative real-time methylation-specific PCR (Q-MSP) was conducted to assess IGFBP-3 methylation in PTs. Quantitative multiplex methylation-specific PCR (QM-MSP)32 was conducted to assess IGFBP-3 methylation in LNs. Primer sequences for analysis were designed using MSPprimer33 and are listed in Supplementary Table S2 (available at http://links.lww.com/SLA/A765). For analysis, DNA was extracted and bisulfite modification was carried out using the EZ DNA methylation Kit (Zymo Research).
Q-MSP was performed using Applied Biosystems 7500 One-Step Plus Real-Time PCR System. Each reaction contained 10.0 μL of 2× Power SYBR Green PCR Master Mix (Applied Biosystems), 2.5 pmol each of forward and reverse primers, and 2 μL of DNA template in a total reaction volume of 20 μ L. To confirm specificity of amplicons from PCR, we performed dissociation curve analysis. The PCR conditions were 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds, 60°C for 60 seconds.
The QM-MSP procedure required 2 stages of PCR reactions. In the first-stage PCR reaction, 2 μL of bisulfite-treated DNA was added to 23 μL of reaction buffer [100 pmol of deoxynucleotide triphosphates, 2.5 μL of 10× PCR buffer, and 1 unit of JumpStart Red Taq DNA Polymerase (Sigma)] containing 5 pmol each of the forward and reverse primers. Conditions were 95°C for 5 minutes, followed by 25 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, with a final extension cycles of 72°C for 5 minutes. The first-stage PCR products were diluted 1:50, and 2 μL was introduced to the second-stage PCR. The second-stage PCR conditions were same as Q-MSP procedure but 30 cycles of 95°C for 15 seconds.
Threshold cycles (Ct) were used to calculate methylation index (MI): MI = 100/[1 + 2ˆ(Ctm − Ctu)]; Ctm and Ctu denote threshold cycles of the probes specific for the methylated and unmethylated states, respectively. The mean of at least 2 replicate measurements was calculated for each sample and used for statistical analysis.
Statistical Analysis
Statistical analysis was performed by using the SPSS software (version 18.0; SPSS, Chicago, IL). Determination of cutoff values was made by the receiver-operator characteristics (ROC) curve. The area under the curve and the best sensitivity and specificity were then computed. The comparison of clinicopathologic factors was analyzed using the t test, χ2 tests, and analysis of variance. Results were considered significant at P < 0.05. Survival was estimated by using the Kaplan-Meier method and log-rank test. Cox proportional hazard regression models were used to determine univariate and multivariate hazard ratios (HRs).
Results
The sensitivity of Q-MSP and QM-MSP assays for IGFBP3 was tested by assessing dilutions of RKO DNA into normal control DNA. Dilution experiments showed linearity of amplification down to a dilution of 1:104 for methylated DNA in Q-MSP assay and 1:106 in QM-MSP assay.
Cutoff value of Q-MSP to distinguish cases from normal controls was determined in the training data set (n = 34) and 38 normal controls. Cutoff value of QM-MSP was determined in 41 LNs from 38 patients with stage III CRC, which were all histologically positive, and 43 normal LN controls. Optimal cutoffs were determined by maximizing the sensitivity and specificity for detection of PTs or LNs for all values of Q-MSP or QM-MSP, respectively. Optimal Q-MSP value distinguishing cases from controls was found to be 4, and QM-MSP value to be 0.8.
From the correlation between clinicopathologic features and postoperative recurrence of patients with stage II CRC using univariate analyses, none of the factors were statistically significant in the training set and only lymphovascular invasion (P = 0.031) was statistically significant in the validation set (Table 2).
Table 2. Univariate Analysis for RFS in Training Set and Validation Set.
| Characteristic | Training Set (n = 34) | Validation Set (n = 81) | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| n | HR (95%CI) | P | n | HR (95%CI) | P | |
| Age,yr | ||||||
| <60 | 14 | 24 | ||||
| ≥60 | 20 | 0.99 (0.31–3.12) | 0.981 | 57 | 1.41 (0.60–3.32) | 0.430 |
| Sex | ||||||
| Male | 14 | 38 | ||||
| Female | 20 | 1.46 (0.44–4.85) | 0.538 | 43 | 0.85 (0.41–1.79) | 0.672 |
| LNs examined | ||||||
| ≥12 | 23 | 57 | ||||
| <12 | 11 | 2.44 (0.79–7.59) | 0.123 | 24 | 1.44 (0.68–3.08) | 0.345 |
| pT4 | ||||||
| No | 28 | 75 | ||||
| Yes | 6 | 0.34 (0.04–2.64) | 0.302 | 6 | 0.99 (0.24–4.19) | 0.994 |
| Lymphovascular invasion | ||||||
| No | 26 | 69 | ||||
| Yes | 8 | 0.69 (0.15–3.14) | 0.627 | 12 | 2.58 (1.09–6.12) | 0.031 |
| Mucin production | ||||||
| No | 29 | 65 | ||||
| Yes | 5 | 0.50 (0.06–3.86) | 0.504 | 16 | 1.40 (0.59–3.29) | 0.446 |
| Differentiation | ||||||
| Well and moderate | 26 | 69 | ||||
| Poor | 8 | 1.14 (0.31–4.20) | 0.848 | 12 | 0.42 (0.10–1.77) | 0.238 |
| Location | ||||||
| Right colon | 15 | 43 | ||||
| Left colon and rectum | 19 | 2.62 (0.71–9.68) | 0.150 | 38 | 0.92 (0.44–1.93) | 0.817 |
| KRAS mutations | ||||||
| No | 19 | 56 | ||||
| Yes | 15 | 1.83 (0.59–5.73) | 0.298 | 25 | 1.11 (0.50–2.45) | 0.802 |
| MSI status | ||||||
| MSS | 19 | 40 | ||||
| MSI | 15 | 0.66 (0.20–2.19) | 0.495 | 41 | 0.71 (0.33–1.49) | 0.705 |
| Group by cutoff 4 | ||||||
| U group | 22 | 42 | ||||
| M group | 12 | 2.02 (0.65–6.26) | 0.226 | 39 | 1.50 (0.71–3.18) | 0.288 |
| Group by cutoff 10 | ||||||
| Low-risk group | 26 | 48 | ||||
| High-risk group | 8 | 4.60 (1.46–14.44) | 0.009 | 33 | 2.21 (1.04–4.68) | 0.039 |
M indicates IGFBP-3 methylated; U, IGFBP-3 unmethylated.
In the training set, KRAS mutations, MSI, and IGFBP-3 methylation (MI, ≥4) were shown in 44.1%, 44.1%, and 32.4% PTs, respectively. None of the 3 markers was significantly associated with RFS (P = 0.298, 0.495, and 0.226 in univariate analysis, respectively, Table 2). Figure 1A shows scatterplots of the MI values of IGFBP-3 in the tumors and normal controls. When ROC analysis was used to separate all cases into 2 groups, a cutoff value of 10 for MI level recurrence was noted with 50.0% sensitivity and 90.9% specificity (Fig. 1B). Therefore, tumors were dichotomized into both a methylation-negative (MI, <4) or -low (MI, ≥4 but < 10) group (low-risk group) and a methylation-high (MI ≥ 10) group (high-risk group). Patients with IGFBP-3 methylation-high tumors (high-risk group) had significantly reduced RFS (log-rank P = 0.004). The 5-year RFS rate was 3-fold greater for patients in the low-risk group than for those in the high-risk group (75.7% vs 25.0%, respectively; Fig. 2A). The signature predicted recurrence with an HR of 4.60 (95% confidence interval (CI), 1.46–14.44; P = 0.009; Table 2) in the high-risk group.
Figure 1.
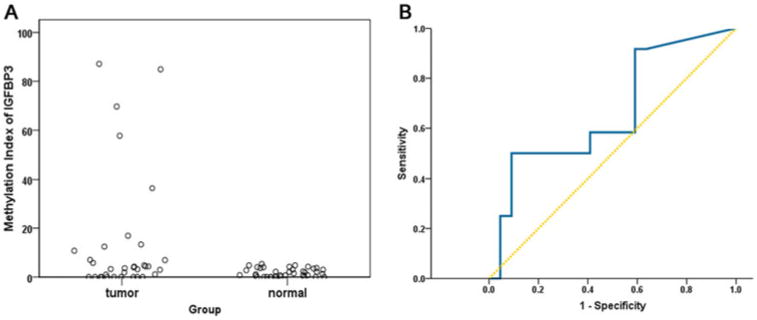
A, Distribution of methylation indices of IGFBP-3 in samples from 38 normal controls and 34 patients with CRC. B, ROC analysis for prediction of recurrence. Dividing all cases into 2 qroups (cutoff value of 10), recurrence was noted with 50.0% sensitivity and 90.9% specificity. Group 1 (low-risk group): a methylation-negative (Ml, <4) or -low (Ml, 4 to < 10) group; group 2 (high-risk group): a methylation-high (Ml, ≥10) group.
Figure 2.
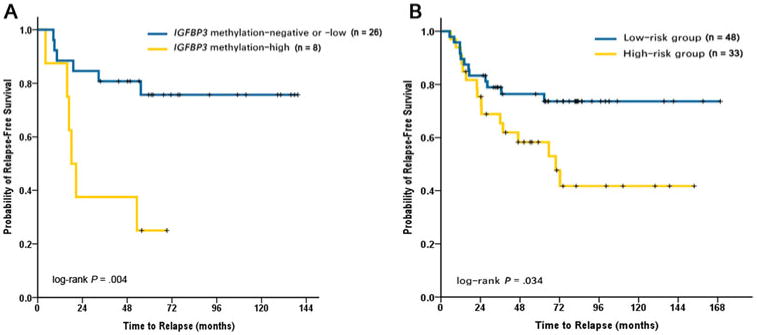
A, Kaplan-Meier analysis of RFS in the training set. Patients with ICFBP-3 methylation-high tumors (high-risk group) had significantly reduced RFS (log-rank P = 0.004). Five-year RFS rate for low-risk patients was 75.7% (95% CI, 67.0% –84.4%) and for high-risk patients was 25.0% (95% CI, 9.7%–40.3%). B, Kaplan-Meier analysis of RFS in the validation set. Patients in the high-risk group had significantly reduced RFS (log-rank P = 0.034). Five-year RFS rate for low-risk patients was 76.4% (95% CI, 70.1%–82.7%) and for high-risk patients was 58.3% (95% CI, 49.4%–67.2%).
An independent patient cohort of 81 patients was then used to evaluate the performance of the IGFBP-3 methylation in PT classifier using the cutoff value identified in the training set. In the validation set, 48 (59.3%) patients were identified as low risk, whereas 33 (40.7%) patients were high risk. The signature predicted recurrence with an HR of 2.21 (95% CI, 1.04–1.68; P = 0.039; Table 2) in the high-risk group. Patients in the high-risk group had significantly reduced RFS (log-rank P = 0.034). The low-risk group had a 5-year RFS rate of 76.4% (95% CI, 70.1%–82.7%), whereas the high-risk group had a 5-year RFS rate of only 58.3% (95% CI, 49.4%–67.2%, Fig. 2B).
Patients with colon cancer on the right side were more often classified as high risk than patients with cancer in the left colon and rectum (P < 0.001). High risk was also positively associated with mucin production (P = 0.048). Classification as low or high risk was not associated with age, sex, number of assessed LNs, pT4, lymphovascular invasion, differentiation, KRAS mutations, or MSI (Table 3).
Table 3. Clinicopathologic Characteristics of High-risk and Low-risk Groups in Validation Set.
| Characteristic | n (%) | High-risk Group (n = 33) n (%) | Low-risk Group (n = 48) n (%) | P |
|---|---|---|---|---|
| Age, yr | 0.061 | |||
| <60 | 24 (29.6) | 6 (18.2) | 18 (37.5) | |
| ≥60 | 57 (70.4) | 27 (81.8) | 30 (62.5) | |
| Sex | 0.502 | |||
| Male | 38 (46.9) | 14 (42.4) | 24 (50.0) | |
| Female | 43 (53.1) | 19 (57.6) | 24 (50.0) | |
| LNs examined | 0.169 | |||
| ≥12 | 57 (70.4) | 26 (78.8) | 31 (64.6) | |
| <12 | 24 (29.6) | 7 (21.2) | 17 (35.4) | |
| pT4 | 1.000* | |||
| No | 75 (92.6) | 31 (93.9) | 44 (91.7) | |
| Yes | 6 (7.4) | 2 (6.1) | 4 (8.3) | |
| Lymphovascular invasion | 0.944 | |||
| No | 69 (85.2) | 28 (84.8) | 41 (85.4) | |
| Yes | 12 (14.8) | 5 (15.2) | 7 (14.6) | |
| Mucin production | 0.048 | |||
| No | 65 (80.2) | 23 (69.7) | 42 (87.5) | |
| Yes | 16 (19.8) | 10 (30.3) | 6 (12.5) | |
| Differentiation | 0.944 | |||
| Well and moderate | 69 (85.2) | 28 (84.8) | 41 (85.4) | |
| Poor | 12 (14.8) | 5 (15.2) | 7 (14.6) | |
| Location | 0.000 | |||
| Right colon | 43 (53.1) | 27 (81.8) | 16 (33.3) | |
| Left colon and rectum | 38 (46.9) | 6 (18.2) | 32 (66.7) | |
| KRAS mutations | 0.562 | |||
| No | 56 (69.1) | 24 (72.7) | 32 (66.7) | |
| Yes | 25 (30.9) | 9 (27.3) | 16 (33.3) | |
| MSI status | 0.893 | |||
| MSS | 40 (49.4) | 16 (48.5) | 24 (50.0) | |
| MSI | 41 (50.6) | 17 (51.5) | 24 (50.0) | |
| Recurrence | 0.029 | |||
| No | 53 (65.4) | 17 (51.5) | 36 (75.0) | |
| Yes | 28 (34.6) | 16 (48.5) | 12 (25.0) |
Fisher exact test
A Cox multiple regression model was used to assess the influence of all significant covariates on RFS. After controlling for age, sex, LNs examined, lymphovascular invasion, and differentiation, multivariate analysis confirmed that presence of IGFBP-3 methylation-high in PTs was significantly and independently associated with a worse prognosis (Table 4). The presence of IGFBP-3 methylation in PTs increased likelihood of developing a CRC recurrence by more than 6-fold (HR, 6.46; 95% CI, 1.51–27.70; P = 0.012) in the training set and doubled the likelihood (HR, 2.40; 95% CI, 1.10–5.25; P = 0.029) in the validation set.
Table 4. Multivariate Analysis for RFS in Training Set and Validation Set.
| Training Set (n = 34) | Validation Set (n = 81) | |||
|---|---|---|---|---|
|
|
|
|||
| Variable | HR (95% CI) | P | HR(95% CI) | P |
| Group | ||||
| High-risk vs low-risk | 6.46 (1.51–27.70) | 0.012 | 2.40 (1.10–5.25) | 0.029 |
| Age, yr | ||||
| ≥60 vs <60 | 0.44 (0.10–1.91) | 0.271 | 1.27 (0.51–3.13) | 0.612 |
| Sex | ||||
| Female vs male | 0.92 (0.20–4.32) | 0.916 | 0.72 (0.33–1.57) | 0.405 |
| LNs examined | ||||
| < 12 vs ≥12 | 1.90 (0.52–6.95) | 0.333 | 1.82 (0.77–4.30) | 0.172 |
| Lymphovascular invasion | ||||
| Yes vs no | 0.70 (0.13–3.84) | 0.676 | 3.29 (1.24–8.70) | 0.016 |
| Differentiation | ||||
| Poor vs well and moderate | 1.07 (0.28–4.18) | 0.920 | 0.63 (0.14–2.93) | 0.558 |
We next tested 462 LNs from the training set and 1179 LNs from the validation set for methylation of IGFBP-3 by QM-MSP to determine whether methylation of IGFBP-3 in LNs would add to the prognostic panel. IGFBP-3 was shown to be methylated (MI, ≥0.8) in at least one LN in 19.1% (22 of the 115) patients. One patient in the training set and 3 patients in the validation set had IGFBP-3 methylation in at least one LN and not in the PT. Methylation status was not significantly associated with RFS (log-rank P = 0.336). ROC analysis was used to identify cutoff values that had the highest sensitivity and specificity to predict recurrence, but no significant value was found.
Discussion
We provide results of an analysis of 115 PTs and 1641 LNs from a cohort of 115 patients (40 of whom recurred) with stage II CRC, gathered from 2 independent tissue collections. The patients had not received any adjuvant chemotherapy, so the assessment of recurrence was not subject to potentially confounding contributions by predictive factors related to adjuvant treatment.
Our study indicates that the methylation of IGFBP-3 gene in PTs is associated with recurrence of stage II CRC and the IGFBP-3 methylation in LNs is not. This signature could be applied to all stage II patients independent of age, sex, LNs examined, lymphovascular invasion, and differentiation. In multivariate analysis for RFS, the methylation of IGFBP-3 was the only significant variable in the training set and 1 of the 2 significant variables in the validation set, superseding factors currently used to make decisions in the clinic. After 5 years, absolute differences in RFS between the patients with the low risk and the high risk were 76.4% and 58.3%, respectively As therapeutic options have broadened, more refined and accurate predictions of recurrence are needed to conduct treatment decision making. Although ensuring that 59.3% of patients with stage II CRC would not need the necessary treatment, our single gene signature would recommend additional medical intervention to 40.7% of the patients.
Numerous studies have demonstrated a more accurate prediction of the prognosis of patients by using a molecular analysis of the PT and various compartments such as the regional LNs and systemic circulation.15,34–41 Considering the high cost of these genome/epigenome-wide or multigene screenings, single biomarker analysis is not only a clinical necessity but also an economic requirement to keep the cost of cancer care in check. For example, the cost of a 12-gene assay (Oncotype DX Colon Cancer Assay, Genomic Health) is $3640 per sample,42 and the cost of Q-MSP is only about $20 per sample per gene. Previous attempts to correlate selected characteristics of PTs with recurrence have proven unsuccessful in CRC. The ability to use a single gene methylation pattern in predicting outcome could provide important information on both previously known and unknown biologic attributes in tumor characteristics. As an initial example, our study here focuses on stage II tumors in which the analysis defines IGFBP-3 gene methylation for prediction of recurrence. Our selection of multivariate patterns in IGFBP-3 methylation data from the PTs and examination of the value of such patterns in prediction of independent testing samples resulted in a relative high predictive accuracy. This represented an HR of 2.40 (95% CI, 1.10–5.25; P = 0.029) that is much higher than the known prognostic clinical covariates in CRC. This marker allows a relatively easy way of identifying patients at risk of recurrence, which would likely benefit most from adjuvant therapy. Moreover, the functional annotation for this gene provides insight into the underlying biologic mechanism that leads to early recurrence.
LN status is an important determinant of disease recurrence, survival, and treatment in CRC. The presence of disseminated tumor cells (DTCs) within regional LNs that are not detected on conventional histopathologic examination by using hematoxylin and eosin staining has been suspected to be markers of systemic tumor spread in these patients with CRCs.43 Therefore, detection of DTCs may help identify those patients with stage I or II CRC who are at high risk for recurrence and might benefit from adjuvant therapies. By using molecular detection techniques such as testing for mutations, methylation, or amplification of RNA, various studies have demonstrated DTCs in regional LNs in 20% to 50% of patients with LN-negative CRC on routine histopathologic analysis.14,19,44
Previous studies demonstrated that a considerable fraction of LNs did not resemble the PTs in CRCs in terms of various molecular analysis, such as the KRAS mutation status (up to 25% of discordance),45–47 MSI,48 and p16INK4a methylation.49 The similar discordance of molecular status between PT and LN deposits was also found in various malignancies (ie, lung cancer,50,51 breast cancer,52,53 gastric cancer,54 head and neck cancer,55 and thyroid cancer.56 On the basis of these studies, there is a debate in the field whether tumors from patients with cancer and matched LN metastases are different or not.
Here, we reported the analysis of possible occult LN micrometastases from patients with CRC on the basis of the methylation of IGFBP-3 found in paired PTs. To our surprise, the experimental results show clearly that the LNs do not always carry the same methylation as the PT. The discordant cases for IGFBP-3 methylation status were all positive in the LN, but negative in the PT Only a few studies have described the presence of de novo methylation in LNs derived from CRCs. Our data indicate that LNs with or without metastases of a given tumor with methylated or unmethylated IGFBP-3 may differ in their IGFBP-3 methylation status. Many factors can be responsible for this incongruence: both related to the poorly understood biology and a well-recognized heterogeneity of the disease, clonal selection during the process of metastasis, factors related to the methodology. Because disseminated cells progress independently from the PT, a simple extrapolation from the PT methylation pattern to DTCs in LNs is impossible. Therefore, IGFBP-3 methylation does not seem suitable for determination of the occult metastases in LNs of CRC, and LN metastases not a reliable tool to determine the IGFBP-3 methylation status of CRC under routine diagnostic conditions as well. It is reasonable then, that our survival analyses show IGFBP-3 methylation in LNs did not correlate with tumor recurrence. Frequent methylation differences between PTs and LNs in CRC also question, at least to some extent, the role of PTs as a surrogate subject of the study for the systemic disease. All adjuvant therapies that just target genetic or epigenetic events in the PT are unlikely to eradicate DTCs because these cells may not uniformly share genetic or epigenetic changes that are acquired later on.
The data presented here support a model in which the propensity to recurrence reflects the predominant epigenetic state of a PT rattier than the emergence of rare cells in LNs with the metastatic phenotype. Furthermore, such findings are compatible with a newer model for parallel metastasis development.57
Limitations to our study include a retrospective analysis, a relative small sample size, and a 15-year duration of the specimen collection. However, these findings provide early insight regarding IGFBP-3 methylation and development of recurrence in patients with stage II CRC. The ability to identify patients with stage II CRC who have an increased risk of recurrence in the absence of chemotherapy offers the hope of broadening the currently arbitrary definition of patients who may benefit from intensive treatment or surveillance. If validated, the recommendation of adjuvant chemotherapy in resected CRC may be guided in the future by this prognostic marker for stage II cancer. The marker can also be tested with resected tumors other than stage II or with biopsy samples to potentially impact both adjuvant and neoadjuvant therapies. Finally, the IGFBP-3 gene that is methylated in tumors with a poor RFS is a potential target for the reasonable development of new cancer drugs, such as demethylating agent. Future researches are warranted to better understand the underlying biologic mechanisms that explain the association between IGFBP-3 methylation and tumor recurrence, and to evaluate the role of demethylating agent alone or in combination with traditional adjuvant systemic therapy in patients with high-risk stage II CRC. It should be noted that, although it yielded statistically significant results, our recurrence predictor was imperfect, suggesting that additional correlates of CRC behavior have yet to be identified or that clinical trials with higher statistical power will be required to generate more robust classifications.
Supplementary Material
Acknowledgments
We thank Kathy Bender and Joann Murphy for administrative support. We also thank Sharon Metzger-Gaud, Theresa Sanlorenzo-Caswell, and the Johns Hopkins Cancer Registry for assistance with the primary cancer databases.
Disclosure: Supported by the National Institutes of Health (Grant Nos. CA140599 and CA127141); the Society of University Surgeons Award, Jeannik M. Littlefield-American Association of Cancer Research grant in metastatic colon cancer (Grant No. K23 CA127141); and the National Natural Science Foundation of China (Grant Nos. 81000898 and 81472289).
Stephen B. Baylin has commercial grant funding and serves on the advisory board for MDx Health Inc and BioNumerik Pharmaceuticals Inc.
Footnotes
No potential conflicts of interest were disclosed by the other authors.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.annalsofsurgery.com).
References
- 1.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 4.Staib L, Link KH, Blatz A, et al. Surgery of colorectal cancer: surgical morbidity and five- and ten-year results in 2400 patients—monoinstitutional experience. World J Surg. 2002;26:59–66. doi: 10.1007/s00268-001-0182-5. [DOI] [PubMed] [Google Scholar]
- 5.Parc Y, Gueroult S, Mourra N, et al. Prognostic significance of microsatellite instability determined by immunohistochemical staining of MSH2 and MLH1 in sporadic T3N0M0 colon cancer. Gut. 2004;53:371–375. doi: 10.1136/gut.2003.019190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertagnolli MM, Redston M, Compton CC, et al. Microsatellite instability and loss of heterozygosity at chromosomal location 18q: prospective evaluation of biomarkers for stages II and III colon cancer—a study of CALGB 9581 and 89803. J Clin Oncol. 2011;29:3153–3162. doi: 10.1200/JCO.2010.33.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogunbiyi OA, Goodfellow PJ, Herfarth K, et al. Confirmation that chromosome 18q allelic loss in colon cancer is a prognostic indicator. J Clin Oncol. 1998;16:427–133. doi: 10.1200/JCO.1998.16.2.427. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Jatkoe T, Zhang Y, et al. Gene expression profiles and molecular markers to predict recurrence of Dukes' B colon cancer. J Clin Oncol. 2004;22:1564–1571. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 9.Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connell MJ, Lavery I, Yothers G, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28:3937–3944. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thebo JS, Senagore AJ, Reinhold DS, et al. Molecular staging of colorectal cancer: K-ras mutation analysis of lymph nodes upstages Dukes B patients. Dis Colon Rectum. 2000;43:155–159. doi: 10.1007/BF02236973. discussion 159–162. [DOI] [PubMed] [Google Scholar]
- 12.Tie J, Lipton L, Desai J, et al. KRAS mutation is associated with lung metastasis in patients with curatively resected colorectal cancer. Clin Cancer Res. 2011;17:1122–1130. doi: 10.1158/1078-0432.CCR-10-1720. [DOI] [PubMed] [Google Scholar]
- 13.Ogino S, Nosho K, Kirkner GJ, et al. PK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liefers GJ, Cleton-Jansen AM, van de Velde CJ, et al. Micrometastases and survival in stage II colorectal cancer. N Engl J Med. 1998;339:223–228. doi: 10.1056/NEJM199807233390403. [DOI] [PubMed] [Google Scholar]
- 15.Miyake Y, Yamamoto H, Fujiwara Y, et al. Extensive micrometastases to lymph nodes as a marker for rapid recurrence of colorectal cancer: a study of lymphatic mapping. Clin Cancer Res. 2001;7:1350–137. [PubMed] [Google Scholar]
- 16.Noura S, Yamamoto H, Ohnishi T, et al. Comparative detection of lymph node micrometastases of stage II colorectal cancer by reverse transcriptase polymerase chain reaction and immunohistochemistry. J Clin Oncol. 2002;20:4232–4241. doi: 10.1200/JCO.2002.10.023. [DOI] [PubMed] [Google Scholar]
- 17.Ho SB, Hyslop A, Albrecht R, et al. Quantification of colorectal cancer micrometastases in lymph nodes by nested and real-time reverse transcriptase-PCR analysis for carcinoembryonic antigen. Clin Cancer Res. 2004;10:5777–5784. doi: 10.1158/1078-0432.CCR-03-0507. [DOI] [PubMed] [Google Scholar]
- 18.Koyanagi K, Bilchik AJ, Saha S, et al. Prognostic relevance of occult nodal micrometastases and circulating tumor cells in colorectal cancer in a prospective multicenter trial. Clin Cancer Res. 2008;14:7391–7396. doi: 10.1158/1078-0432.CCR-08-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harder J, Engelstaedter V, Usadel H, et al. CpG-island methylation of the ER promoter in colorectal cancer: analysis of micrometastases in lymph nodes from UICC stage I and II patients. Br J Cancer. 2009;100:360–365. doi: 10.1038/sj.bjc.6604859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi JM, Dhir M, Guzzetta AA, et al. DNA methylation biomarker candidates for early detection of colon cancer. Tumour Biol. 2012;33:363–372. doi: 10.1007/s13277-011-0302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Culouscou JM, Remacle-Bonnet M, Garrouste F, et al. Simultaneous production of IGF-I and EGF competing growth factors by HT-29 human colon cancer line. Int J Cancer. 1987;40:646–652. doi: 10.1002/ijc.2910400513. [DOI] [PubMed] [Google Scholar]
- 22.Durrant LG, Watson SA, Hall A, et al. Co-stimulation of gastrointestinal tumour cell growth by gastrin, transforming growth factor alpha and insulin like growth factor-I. Br J Cancer. 1991;63:67–70. doi: 10.1038/bjc.1991.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo YS, Narayan S, Yallampalli C, et al. Characterization of insulin-like growth factor I receptors in human colon cancer. Gastroenterology. 1992;102:1101–1108. [PubMed] [Google Scholar]
- 24.Wu Y, Yakar S, Zhao L, et al. Circulating insulin-like growth factor-I levels regulate colon cancer growth and metastasis. Cancer Res. 2002;62:1030–1035. [PubMed] [Google Scholar]
- 25.Collard TJ, Guy M, Butt AJ, et al. Transcriptional upregulation of the insulin-like growth factor binding protein IGFBP-3 by sodium butyrate increases IGF-independent apoptosis in human colonic adenoma-derived epithelial cells. Carcinogenesis. 2003;24:393–401. doi: 10.1093/carcin/24.3.393. [DOI] [PubMed] [Google Scholar]
- 26.Ma J, Pollak MN, Giovannucci E, et al. Prospective study of colorectal cancer risk in men and plasma levels of insulin-like growth factor (IGF)-I and IGF-binding protein-3. J Natl Cancer Inst. 1999;91:620–625. doi: 10.1093/jnci/91.7.620. [DOI] [PubMed] [Google Scholar]
- 27.Giovannucci E, Pollak MN, Platz EA, et al. A prospective study of plasma insulin-like growth factor-1 and binding protein-3 and risk of colorectal neoplasia in women. Cancer Epidemiol Biomarkers Prev. 2000;9:345–349. [PubMed] [Google Scholar]
- 28.Haydon AM, Macinnis RJ, English DR, et al. Physical activity, insulin-like growth factor 1, insulin-like growth factor binding protein 3, and survival from colorectal cancer. Gut. 2006;55:689–694. doi: 10.1136/gut.2005.081547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Portier G, Elias D, Bouche O, et al. Multicenter randomized trial of adjuvant fluorouracil and folinic acid compared with surgery alone after resection of colorectal liver metastases: FFCD ACHBTH AURC 9002 trial. J Clin Oncol. 2006;24:4976–4982. doi: 10.1200/JCO.2006.06.8353. [DOI] [PubMed] [Google Scholar]
- 30.Yachida S, Mudali S, Martin SA, et al. Beta-catenin nuclear labeling is a common feature of sessile serrated adenomas and correlates with early neoplastic progression after BRAF activation. Am J Surg Pathol. 2009;33:1823–1832. doi: 10.1097/PAS.0b013e3181b6da19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietmaier W, Wallinger S, Bocker T, et al. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 32.Fackler MJ, McVeigh M, Mehrotra J, et al. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res. 2004;64:4442–4452. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 33.Brandes JC, Carraway H, Herman JG. Optimal primer design using the novel primer design program: MSPprimer provides accurate methylation analysis of the ATM promoter. Oncogene. 2007;26:6229–6237. doi: 10.1038/sj.onc.1210433. [DOI] [PubMed] [Google Scholar]
- 34.Venook AP, Niedzwiecki D, Lopatin M, et al. Biologic determinants of tumor recurrence in stage II colon cancer: validation study of the 12-gene recurrence score in cancer and leukemia group B (CALGB) 9581. J Clin Oncol. 2013;31:1775–1781. doi: 10.1200/JCO.2012.45.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maak M, Simon I, Nitsche U, et al. Independent validation of a prognostic genomic signature (ColoPrint) for patients with stage II colon cancer. Ann Surg. 2013;257:1053–1058. doi: 10.1097/SLA.0b013e31827c1180. [DOI] [PubMed] [Google Scholar]
- 36.Agesen TH, Sveen A, Merok MA, et al. ColoGuideEx: a robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 2012;61:1560–1567. doi: 10.1136/gutjnl-2011-301179. [DOI] [PubMed] [Google Scholar]
- 37.Bae S, Tie J, Desai J, et al. Microsatellite instability status is critical to analysis of survival in stage II colon cancer. J Clin Oncol. 2012;30:675–676. doi: 10.1200/JCO.2011.39.7000. author reply 676–677. [DOI] [PubMed] [Google Scholar]
- 38.Roth AD, Delorenzi M, Tejpar S, et al. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J Natl Cancer Inst. 2012;104:1635–1646. doi: 10.1093/jnci/djs427. [DOI] [PubMed] [Google Scholar]
- 39.Nitsche U, Rosenberg R, Balmert A, et al. Integrative marker analysis allows risk assessment for metastasis in stage II colon cancer. Ann Surg. 2012;256:763–771. doi: 10.1097/SLA.0b013e318272de87. discussion 771. [DOI] [PubMed] [Google Scholar]
- 40.Rahbari NN, Bork U, Motschall E, et al. Molecular detection of tumor cells in regional lymph nodes is associated with disease recurrence and poor survival in node-negative colorectal cancer: a systematic review and meta-analysis. J Clin Oncol. 2012;30:60–70. doi: 10.1200/JCO.2011.36.9504. [DOI] [PubMed] [Google Scholar]
- 41.Iinuma H, Watanabe T, Mimori K, et al. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes' stage B and C colorectal cancer. J Clin Oncol. 2011;29:1547–1555. doi: 10.1200/JCO.2010.30.5151. [DOI] [PubMed] [Google Scholar]
- 42.Alberts SR, Yu TM, Behrens RJ, et al. Comparative economics of a 12-gene assay for predicting risk of recurrence in stage II colon cancer. Pharmacoeconomics. 2014;32:1231–1243. doi: 10.1007/s40273-014-0207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weitz J, Kienle P, Magener A, et al. Detection of disseminated colorectal cancer cells in lymph nodes, blood and bone marrow. Clin Cancer Res. 1999;5:1830–1836. [PubMed] [Google Scholar]
- 44.Sanchez-Cespedes M, Esteller M, Hibi K, et al. Molecular detection of neoplastic cells in lymph nodes of metastatic colorectal cancer patients predicts recurrence. Clin Cancer Res. 1999;5:2450–2454. [PubMed] [Google Scholar]
- 45.Al-Mulla F, Going JJ, Sowden ET, et al. Heterogeneity of mutant versus wild-type Ki-ras in primary and metastatic colorectal carcinomas, and association of codon-12 valine with early mortality. J Pathol. 1998;185:130–138. doi: 10.1002/(SICI)1096-9896(199806)185:2<130::AID-PATH85>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 46.Baldus SE, Schaefer KL, Engers R, et al. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16:790–799. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 47.Oliveira C, Velho S, Moutinho C, et al. KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene. 2007;26:158–163. doi: 10.1038/sj.onc.1209758. [DOI] [PubMed] [Google Scholar]
- 48.Messick CA, Church JM, Liu X, et al. Stage III colorectal cancer: molecular disparity between primary cancers and lymph node metastases. Ann Surg Oncol. 2010;17:425–131. doi: 10.1245/s10434-009-0783-z. [DOI] [PubMed] [Google Scholar]
- 49.Miranda E, Bianchi P, Destro A, et al. Genetic and epigenetic alterations in primary colorectal cancers and related lymph node and liver metastases. Cancer. 2013;119:266–276. doi: 10.1002/cncr.27722. [DOI] [PubMed] [Google Scholar]
- 50.Sasatomi E, Finkelstein SD, Woods JD, et al. Comparison of accumulated allele loss between primary tumor and lymph node metastasis in stage II non-small cell lung carcinoma: implications for the timing of lymph node metastasis and prognostic value. Cancer Res. 2002;62:2681–2689. [PubMed] [Google Scholar]
- 51.Park S, Holmes-Tisch AJ, Cho EY, et al. Discordance of molecular biomarkers associated with epidermal growth factor receptor pathway between primary tumors and lymph node metastasis in non-small cell lung cancer. J Thorac Oncol. 2009;4:809–815. doi: 10.1097/JTO.0b013e3181a94af4. [DOI] [PubMed] [Google Scholar]
- 52.Cardoso F, Di Leo A, Larsimont D, et al. Evaluation of HER2, p53, bcl-2, topoisomerase II-alpha, heat shock proteins 27 and 70 in primary breast cancer and metastatic ipsilateral axillary lymph nodes. Ann Oncol. 2001;12:615–620. doi: 10.1023/a:1011182524684. [DOI] [PubMed] [Google Scholar]
- 53.Zidan J, Dashkovsky I, Stayerman C, et al. Comparison of HER-2 overexpres-sion in primary breast cancer and metastatic sites and its effect on biological targeting therapy of metastatic disease. Br J Cancer. 2005;93:552–556. doi: 10.1038/sj.bjc.6602738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.An JY, Kim KM, Choi MG, et al. Prognostic role of p-mTOR expression in cancer tissues and metastatic lymph nodes in pT2b gastric cancer. Int J Cancer. 2010;126:2904–2913. doi: 10.1002/ijc.24872. [DOI] [PubMed] [Google Scholar]
- 55.Tabor MP, van Houten VM, Rummer JA, et al. Discordance of genetic alterations between primary head and neck tumors and corresponding metastases associated with mutational status of the TP53 gene. Genes Chromosomes Cancer. 2002;33:168–177. doi: 10.1002/gcc.10019. [DOI] [PubMed] [Google Scholar]
- 56.Kebebew E, Weng J, Bauer J, et al. The prevalence and prognostic value of BRAF mutation in thyroid cancer. Ann Surg. 2007;246:466–470. doi: 10.1097/SLA.0b013e318148563d. discussion 470–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


