Abstract
The Drosophila Kelch protein is required to organize the ovarian ring canal cytoskeleton. Kelch binds and cross-links F-actin in vitro, and it also functions with Cullin 3 (Cul3) as a component of a ubiquitin E3 ligase. How these two activities contribute to cytoskeletal remodeling in vivo is not known. We used targeted mutagenesis to investigate the mechanism of Kelch function. We tested a model in which Cul3-dependent degradation of Kelch is required for its function, but we found no evidence to support this hypothesis. However, we found that mutant Kelch deficient in its ability to interact with Cul3 failed to rescue the kelch cytoskeletal defects, suggesting that ubiquitin ligase activity is the principal activity required in vivo. We also determined that the proteasome is required with Kelch to promote the ordered growth of the ring canal cytoskeleton. These results indicate that Kelch organizes the cytoskeleton in vivo by targeting a protein substrate for degradation by the proteasome.
Keywords: actin, oogenesis, cullin, Drosophila, proteasome
A crucial aspect of animal development is the ability of cells to change shape and remodel cellular structures in a precise, reproducible, and concerted manner. For example, the coordinated cellular movements of gastrulation require the assembly of actomyosin structures near the apical surfaces of epidermal cells, and the concerted contractility of these actomyosin assemblies provides the driving force for tissue morphogenesis (Solnica-Krezel and Sepich 2012). Developmental programs requiring changes in cell shape and structure ultimately act on cytoskeletal proteins and the factors that regulate them. Phosphorylation is one well-characterized mechanism that regulates cytoskeletal remodeling. Phosphorylation of membrane lipids is used to recruit cytoskeletal proteins to specific subcellular locations (Yin and Janmey 2003), while phosphorylation of cytoskeletal proteins can promote or inhibit their activity (Tan et al. 1992; Mizuno 2013). Protein ubiquitylation, in which the small protein ubiquitin is covalently conjugated to a target protein, can also regulate cytoskeletal proteins either by targeting them for destruction (Pintard et al. 2003; Razinia et al. 2011) or by inducing a change in protein activity (Hao et al. 2013; Yuan et al. 2014). In contrast to phosphorylation, however, relatively little is known about the mechanisms and scope of ubiquitylation in shaping the cytoskeleton.
Drosophila ovarian ring canals are remarkable cellular structures, serving to interconnect sibling germ cells that will produce a mature egg. Egg development takes place within a structure called an egg chamber, which consists of a 16-cell germline cyst surrounded by a layer of somatic follicle cells. Ring canals form during the four mitotic divisions that generate the 16-cell germline cyst; these divisions are different from most somatic cell divisions in that cytokinesis halts prior to abscission, resulting in persistent cleavage furrows that are transformed into ring canals through the recruitment of additional proteins. Of the 16 germ cells, one becomes the oocyte while the remaining 15 differentiate as nurse cells. The oocyte is transcriptionally silent and therefore depends on the nurse cells to synthesize and transport cellular components through the ring canals to the developing egg.
Since Drosophila oocytes grow to a large size—∼500 × 200 µm—the nurse cells and ring canals must also grow to support the growth of the oocyte. The growth of ring canals is dependent on a robust and dynamic F-actin cytoskeleton. Analysis of cytoskeletal genes demonstrated the importance of the F-actin cytoskeleton for ring canal growth and integrity: loss of ring canal F-actin in cheerio mutants results in destabilized ring canals (Robinson et al. 1997; Li et al. 1999; Sokol and Cooley 1999) and loss of F-actin polymerization activity when components of the Arp2/3 complex are mutated results in ring canals that fail to grow and also lack structural stability (Hudson and Cooley 2002).
The female-sterile gene kelch is required for organizing the F-actin cytoskeleton in ovarian ring canals (Xue and Cooley 1993). The kelch gene encodes a 76-kDa protein consisting of conserved BACK and BTB domains and a C-terminal domain consisting of six sequence repeats called Kelch repeats (KREP; Figure 2A). The BTB domain mediates dimerization (Stogios et al. 2005) and the KREP domain folds into a β-barrel structure (Supporting Information, Figure S1; Hudson and Cooley 2008). The kelch phenotype, along with homology between the Kelch KREP domain and the F-actin-binding protein scruin (Way et al. 1995), suggested that Kelch functioned as an F-actin binding protein, which is consistent with our previous finding that Kelch binds and cross-links F-actin in vitro (Kelso et al. 2002).
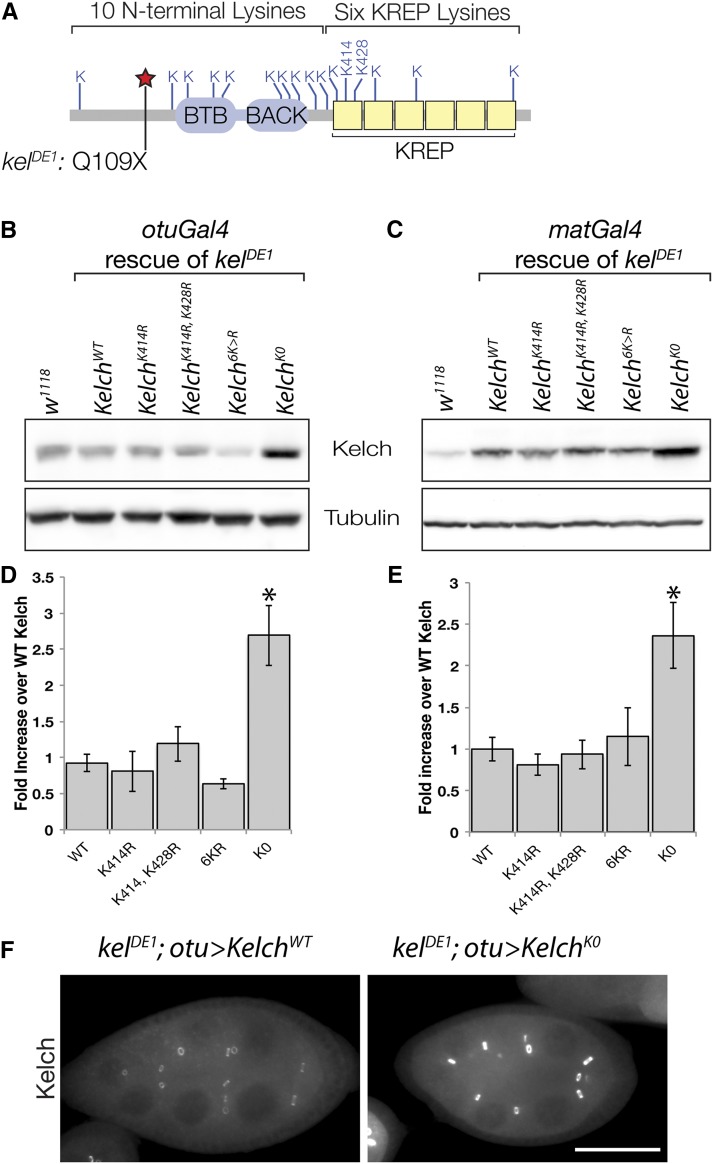
Figure 2.
Kelch is stabilized upon mutation of all lysine residues to arginine. (A) Schematic of Drosophila Kelch protein, with locations of lysine residues indicated. The location of the Q109X lesion in kelDE1 allele used in transgene rescue assays is also indicated. The allele truncates the protein before all functional domains. (B–E) Western analysis of protein levels from expression of the indicated mutant transgenes. cDNAs were expressed in the absence of endogenous Kelch using either weak (B, otuGal4) or strong (C, matGal4) germline drivers in a kelch null background. (D and E) Mean expression levels were measured relative to endogenous w1118 control, and the expression level of the WT cDNA in each experiment was set at 1. (*) Statistically significant difference compared to WT cDNA expression level (P < 0.01, one-way ANOVA, Dunnett multiple comparison test). Error bars represent SEM. (F) Egg chambers immunolabeled to reveal Kelch level. Comparably staged egg chambers were imaged with identical exposure times to reveal differences in Kelch abundance. Scale bar, 50 µm.
However, we found that Kelch is also a component of a cullin-RING E3 ubiquitin ligase (CRL) (Hudson and Cooley 2010). CRLs are multiprotein complexes that target specific substrate proteins for ubiquitylation, typically resulting in the degradation of the substrate by the proteasome (Petroski and Deshaies 2005). CRLs are assembled on a cullin scaffold protein. The cullin C terminus binds a small RING domain protein that recruits the ubiquitin E2 conjugating enzyme that catalyzes ubiquitin conjugation to a substrate. Cullins also associate with one or several proteins that function as substrate recognition subunits (SRS), and these components are responsible for binding specific substrate proteins. There are multiple cullins encoded in animal genomes, each of which associates with distinct classes of SRSs. CRLs assembled with the cullin 3 protein (Cul3) recruit BTB-domain proteins as SRS components through an interaction between the Cul3 N terminus and the BTB domain. We found that reducing Cul3 in germ cells resulted in a kelch-like ring canal phenotype and also provided evidence that Kelch and Cul3 function in a complex together at ring canals (Hudson and Cooley 2010). These results indicated that in addition to binding F-actin, Kelch functions as a substrate recognition subunit for a CRL assembled on a Cul3 scaffold (referred to as CRL3Kelch).
There are therefore two distinct mechanisms by which Kelch could promote cytoskeletal organization during ring canal growth: F-actin cross-linking and ubiquitin ligase activity. To determine how these mechanisms function in vivo to coordinate the growth and organization of the ring canal cytoskeleton, we undertook mutagenesis studies to test specific models of Kelch function. Like other SRS components of CRLs, Kelch itself is degraded through its association with Cul3; however, we found that this autocatalytic degradation of Kelch is not necessary for its function. In addition, we determined that when Kelch is specifically compromised for Cul3 binding, it is unable to rescue the kelch mutant phenotype, underscoring the importance of Kelch function as part of a CRL. Finally, we showed that reducing proteasome activity in the germline also results in a kelch-like phenotype, indicating that CRL3Kelch promotes ring canal growth and organization by targeting a substrate for ubiquitylation and degradation by the ubiquitin-proteasome system (UPS). Together, these results demonstrate that Kelch promotes ring canal cytoskeleton growth primarily through its activity as CRL component.
Materials and Methods
Genetics
To minimize transgene position effects on expression, wild-type and mutant Kelch cDNAs were cloned into a UASp vector modified for PhiC31 integration (gift of Mike Buszczak), and each was integrated into the attP2 PhiC31 integration site on chromosome 3 (Groth et al. 2004). For functional assays, we tested the ability of wild-type or mutant transgenes to rescue kelch null mutant ovaries using either the otuGal4 (P{otu-GAL4::VP16.R}1; Rorth 1998), or matGal4 (Bloomington no. 7063: P{matα4-GAL-VP16}V37; Kaltschmidt et al. 2000) germline drivers. Expression of Kelch using these Gal4 drivers results in Kelch levels that are either slightly lower or higher than endogenous Kelch, respectively, although both drivers fully rescue the fertility defect of the null kelDE1 allele when driving expression of wild-type cDNA. The UbG76V-GFP proteasome activity reporter construct was generated by cloning Ub-G76V-GFP (Dantuma et al. 2000; Addgene plasmid no. 11941) into pCasper3-Up2-RX polyA, a ubiquitin promoter P-element vector with a modified polylinker (gift of R. Fehon; Fehon et al. 1997), using EcoRI and NotI restriction sites. Transgenic flies were generated via P-element-mediated insertion. Injections of transgenes were performed at Rainbow Transgenics. The UASp-YFP::Cul3, UASp-FLAG::Cul3, and UASp-SBP-mCherry::Kelch lines were described previously (Hudson and Cooley 2010). For RNAi studies, we used the following shRNA lines from TRiP at Harvard Medical School (Ni et al. 2011): Prosβ5 (HMS00119); Rpn8 (GL00333); Prosα7 (HMS00068); Rpt2 (HMS00104); Rpn11 (HMS00071); Rpn12 (HMS01032); Cul3 (HMS01572); Nedd8 (HMS00818).
Fixation, immunofluorescence, and imaging
Ovaries were dissected in IMADS buffer (ionically matched Drosophila saline; Singleton and Woodruff 1994) and fixed for 10 min in 6% formaldehyde, 75 mM KCl, 25 mM NaCl, 3 mM MgCl2, and 17 mM potassium phosphate, pH 6.8 (Verheyen and Cooley 1994). Fixed tissue was washed in PBT (phosphate-buffered saline with 0.3% Triton X-100 and 0.5% BSA) and incubated with anti-Kelch Kel 1B (Xue and Cooley 1993) (1:5, Developmental Studies Hybridoma Bank). Secondary antibodies used were goat anti-mouse conjugated to Alexa-488, -568, or -647 (1:500, Invitrogen). F-actin was labeled with phalloidin conjugated to Alexa-488, -568, or -647 (∼30 nM incubation concentration, Invitrogen). Samples were washed in PBT and mounted on slides in ProLong Gold, or ProLong Diamond for samples relying on fluorescent proteins (both purchased from Invitrogen). Samples were imaged with a Leica SP5 or SP8 confocal microscope and a 40× 1.3 NA oil-immersion objective lens. Additional imaging was performed using Zeiss Axiovert 200 equipped with a CARV II spinning disc confocal imager, CoolSNAP HQ2 camera, and either a 20× 0.8 NA dry or 40× 1.2 NA water-immersion objective. For most images, acquisition settings and postacquisition processing were optimized to reveal structural features in egg chambers and ring canals. In figures where protein level comparisons are presented (Figure 1, A′–E′, and Figure 2F), images were acquired using fixed exposure times on a spinning disk confocal, and black/white levels of maximum-intensity projection images were set uniformly across all images using FIJI software.
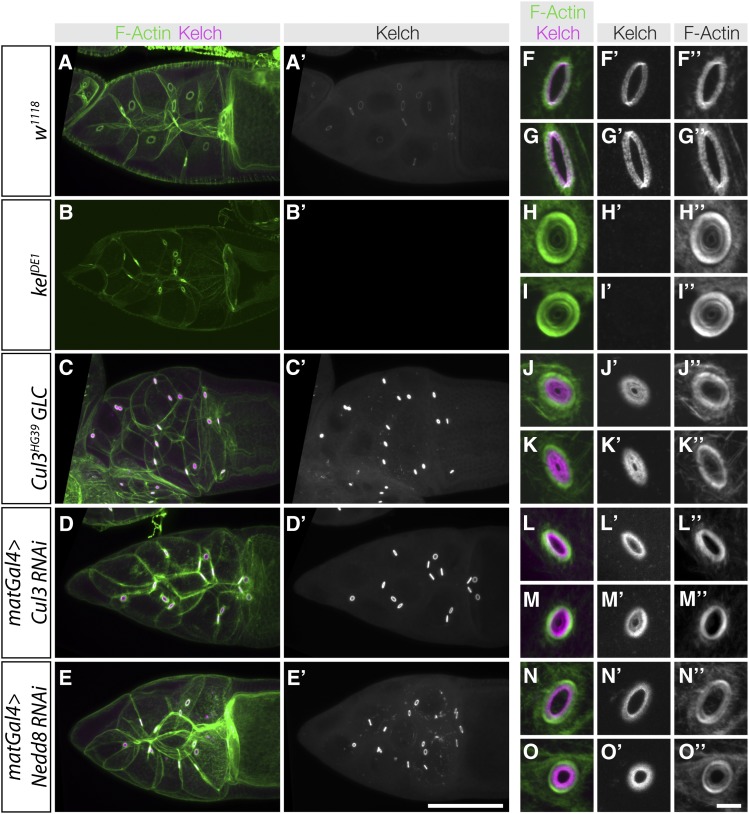
Figure 1.
Inhibition of Cul3 and Nedd8 in the germline results in kelch-like phenotypes. (A–E) Maximum intensity projection images showing ring canals from the genotypes listed on the left. In A′–E′, Kelch immunofluorescence images were acquired using identical imaging conditions; the images presented reflect the observed fluorescence intensities. Reduction of Cul3 or Nedd8 results in significant increases in Kelch accumulation at ring canals. (F–O) High-magnification laser confocal scanning images of ring canals from the genotypes listed on the left. Ring canals are representative images of each genotype and are not necessarily taken from the egg chambers shown in A–E. Displayed fluorescence intensity levels in F–O were optimized to show structural features of ring canals and do not reflect differences in abundance. (F–I) Wild-type and kelch mutant ring canals. In wild type, F-actin and Kelch are organized as a tight band at the ring canal, while kelch ring canals have a thick band of ring canal F-actin. (J–K) Cul3 germline clones have an F-actin disorganization phenotype that is similar to but not as severe as kelch, and abundant Kelch protein is found in the ring canal lumen. (L–M) Germline RNAi of Cul3 results in a phenotype similar to Cul3 germline clones. (N–O) RNAi of Nedd8 results in ring canals with disorganized F-actin and Kelch in the ring canal lumen, but these phenotypes are less severe than those in kelch and Cul3 mutants. Scale bars, 50 µm for egg chamber images and 10 µm for cropped ring canal images.
Western analysis and immunoprecipitation
Samples for Western analysis were prepared by homogenizing dissected ovaries in SDS sample buffer and loading approximately one ovary equivalent per lane on an 8.5% polyacrylamide gel. Gels were blotted to nitrocellulose, blocked in 5% nonfat dry milk in TBS-T (Tris-buffered saline with 0.1% Tween-20), and incubated with anti-Kelch Kel 1B (1:10) or anti-β-tubulin E7 (1:200, Developmental Studies Hybridoma Bank). Lysates for coprecipitation experiments were prepared by homogenizing samples with a Duall teflon homogenizer in a lysis buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 0.5% Triton X-100, 1 mM PMSF, and 5 µg/ml each of chymostatin, leupeptin, antipain, and pepstatin. Protein concentrations were determined by Bradford assay, and equal amounts of soluble lysate from each genotype (∼2 mg) were incubated with Streptavidin ultralink resin, washed in lysis buffer, and eluted with SDS sample buffer supplemented with 10 mM biotin. Samples were separated by SDS–PAGE, blotted to nitrocellulose, and probed with either Kel 1B or FLAG M2 (Sigma F1804, 1 µg/ml) antibodies. Western blots were quantified using the Gels tool in FIJI. Statistical analysis was performed using Prism 6 software.
Proteasome inhibition analysis
Images of control egg chambers or egg chambers experiencing proteasome inhibition (matGal4 > Prosβ5 RNAi or >Rpn8 RNAi) were analyzed in FIJI software. For UbG76V–GFP quantification analyses, maximum intensity projections were generated and the mean GFP fluorescence was measured for the whole area of the germline cells in individual egg chambers. GFP fluorescence was plotted as a function of egg chamber size (with five egg chamber area groupings—see Figure 7C), and the GFP fluorescence measurement for the control 3000 µm2 area grouping was normalized to 100 relative fluorescence units. To score the penetrance of the kelch-like ring canal phenotype, ring canals in mid-staged egg chambers (area grouping “3000 µm2”) were visually scored based on F-actin organization and were deemed kelch-like if they had a clearly thicker F-actin ring or the presence of an inner F-actin ring. For ring canal parameter analyses (measuring ring canal lumen span, diameter, and F-actin width and intensities—see Figure 7H and Figure S5), ring canals were analyzed from mid-staged egg chambers (area grouping “3000 µm2”) and Figure S5 describes how these key ring canal parameters were measured using F-actin intensity plots.
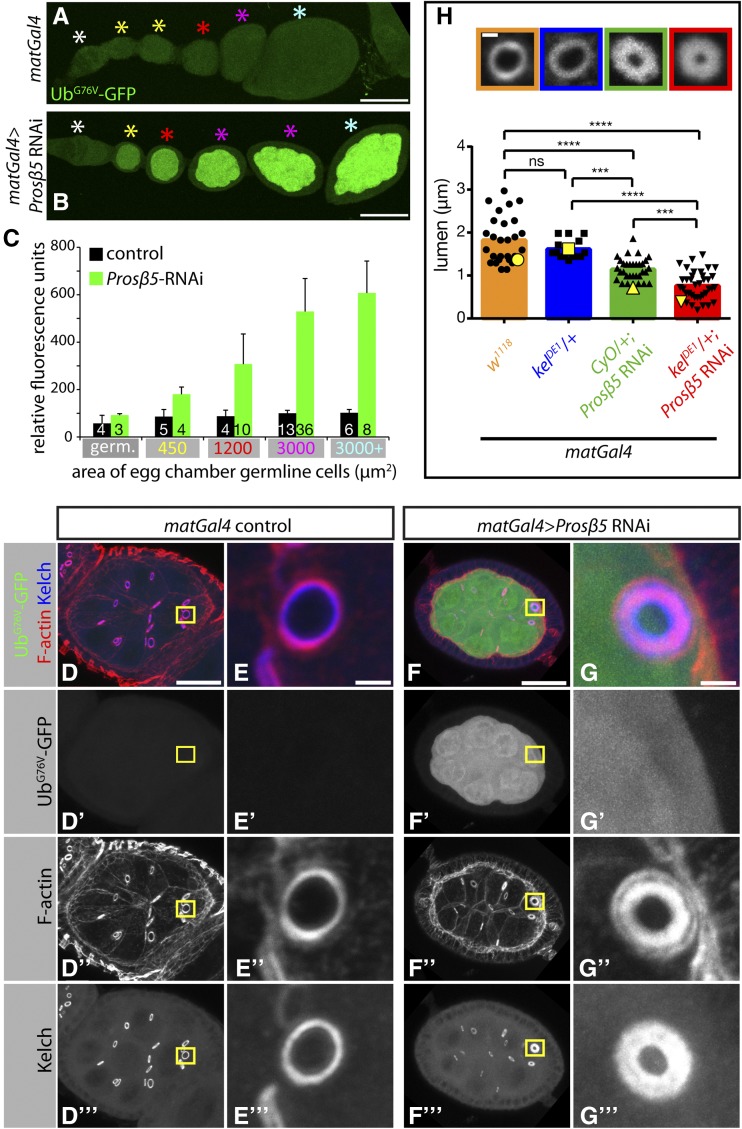
Figure 7.
Proteasome inhibition by RNAi leads to kelch-like ring canals. (A–C) Proteasome inhibition by RNAi is effective as evidenced by UbG76V–GFP reporter protein accumulation. Representative images showing UbG76V–GFP reporter protein levels in control egg chambers (A) or egg chambers expressing shRNAs targeting Prosβ5 proteasome subunit with matGal4 driver (B). (C) Quantification of UbG76V–GFP reporter protein levels. Mean GFP fluorescence of egg chamber germline cell area was measured for individual egg chambers. Egg chambers were grouped based on size (area, μm2) and the average GFP fluorescence for each area grouping is plotted. Area groupings on the x-axis indicate the maximum area cut-off for each particular group (e.g., 3000 μm2 group represents egg chambers 1201–3000 μm2 in size). Color-coded asterisks in A and B designate egg chambers that are representative of the area groupings plotted in C. The sample size is indicated at the base of each bar, and error bars represent standard deviation. (D–G) Representative images of egg chambers (D and F) or ring canals (E and G) expressing UbG76V–GFP reporter protein (D’–G’) and stained for F-actin (D′′–G′′) and Kelch (D′′′–G′′′). Note that the ring canal in G is kelch-like with a thicker F-actin ring (G”). (H) Reduction of one copy of kelch dominantly enhances the kelch-like ring canal phenotype observed upon proteasome inhibition. (Top) Representative images of ring canals stained for F-actin with color-coded boxes matching genotype displayed in graph below. (Graph) Quantification of ring canal lumen span (see Figure S5 for lumen span measurements). Bars represent mean of lumen span and black points represent all individual measurements. Yellow points correspond to individual ring canals shown in top panels. (***) P < 0.0005, (****) P < 0.0001, one-way ANOVA, Tukey’s multiple comparison test. E and G are insets of yellow boxes in D and F, respectively. Scale bars, 50 μm (A and B), 20 μm (D and F), 2 μm (E and G), 1 μm (H).
Data availability
Strains are available from the Bloomington Drosophila Stock Center or upon request. Plasmids are available upon request. The Supporting Information file contains five supplemental figures with legends.
Results
Ring canal phenotypes caused by CRL inhibition
We previously analyzed Cul3 function by creating germline clones (GLC) of Cul3 mutations (Hudson and Cooley 2010). We extended this analysis by carrying out RNAi against Cul3 using transgenic shRNA lines (Ni et al. 2011) and observed ring canal phenotypes similar to the GLC phenotype (Figure 1). A striking increase in Kelch protein accumulation at ring canals was apparent in egg chambers expressing the Cul3 shRNA compared to wild type, similar to the Cul3 GLC phenotype (Figure 1, A′, C′, and D′). Higher magnification imaging showed that wild-type ring canals contained Kelch and highly organized F-actin (Figure 1, A, F–G), while kelch mutant ring canals lacked Kelch and had disorganized F-actin and small lumens (Figure 1, B, H–I). Ring canals in Cul3 GLCs had kelch-like disorganized F-actin and also accumulation of Kelch protein in the lumen, both of which reduced lumen diameter (Figure 1, J–K). Germline-specific RNAi of Cul3 resulted in ring canals similar to the GLCs with Kelch enriched in the lumen, and a milder disruption of F-actin organization (Figure 1, L and M).
Nedd8 is a small, ubiquitin-like protein that is covalently attached to a specific lysine residue on cullin proteins, and this attachment is required for CRL activity (Wu et al. 2005; Lydeard et al. 2013). To determine whether Nedd8 was required at ring canals, we carried out RNAi against Nedd8 by expressing a Nedd8 shRNA construct in the germline. Nedd8 RNAi resulted in kelch- and Cul3-like ring canals, including a striking accumulation of Kelch protein at ring canals (Figure 1E′). Kelch protein was found in the ring canal lumens in Nedd8 RNAi egg chambers, and ring canal F-actin was disorganized (Figure 1, N–O). Kelch accumulation in the lumen of Nedd8 RNAi ring canals was less pronounced than Cul3 mutant or RNAi ring canals (Figure 1, J–O). This may be due to partial reduction of Nedd8 activity by RNAi. The finding that Nedd8 inhibition results in a phenotype similar to kelch and Cul3 suggests that CRL3Kelch activity as a ubiquitin ligase is required for the ordered growth of the ring canal cytoskeleton.
Functional analysis of Cul3-dependent Kelch degradation
A feature of CRLs is that the SRS is often itself ubiquitylated, leading to its destruction by the proteasome. SRS ubiquitylation occurs through an autocatalytic mechanism, and degradation of the SRS is dependent on the presence of its cognate cullin (Wirbelauer et al. 2000; Pintard et al. 2003; Li et al. 2004; de Bie and Ciechanover 2011). The physiological significance of SRS degradation is not clear, although it has been proposed to facilitate SRS exchange, allowing cells to assemble a diverse set of CRLs (Deshaies 1999). Kelch accumulates in ring canal lumens when Cul3 activity is reduced (Hudson and Cooley 2010; see also Figure 1, A′–D′), consistent with Kelch being autoubiquitylated and degraded by a similar mechanism.
Our results to this point demonstrate that Kelch is a multifunctional protein, with both ubiquitin ligase and F-actin cross-linking activities. We considered two basic models that could explain the mechanism of Kelch and Cul3 in organizing ring canal F-actin. First, F-actin cross-linking and ubiquitin ligase activity could be coupled in some way. This model is suggested by the Cul3 phenotype in which Kelch protein accumulates in a large aggregate in the ring canal lumens (Figure 1, J′–M′). Kelch could organize the ring canal cytoskeleton through its F-actin cross-linking activity and then require association with Cul3 to degrade it as a means to clear Kelch from the growing ring canal lumens. A second model would involve a more conventional role for Kelch as a component of a CRL3, in which the targeting of a distinct protein substrate is needed to organize the cytoskeleton during ring canal growth.
To test the first hypothesis that Cul3 association is primarily required to degrade Kelch and remove it from ring canal lumens, we mutated lysine residues in Kelch so that it would be resistant to autoubiquitylation and subsequent degradation. Our previous analysis of a construct lacking the Kelch-repeat domain suggested that the KREP domain was the target of ubiquitylation (Hudson and Cooley 2010), consistent with structural models showing that the KREP domain was likely to be positioned close to the E2 enzyme (Stogios et al. 2005; also see Figure S1A). Based on this result and structural modeling of the Kelch KREP domain we made an initial set of mutations targeting KREP lysine residues predicted to be surface exposed (Figure S1, B and C), as well as a construct in which all lysines in the Kelch-repeat domain were mutated to arginine (Figure 2A). We integrated both mutant and wild-type transgenes at attP2, a specific integration site on chromosome 3, and expressed the transgenes in kelchDE1, a null mutant background, using either a strong (matGal4) or weak (otuGal4) Gal4 driver (see Materials and Methods for details). Integration of the transgenes at the attP2 site was done to minimize position effects on transcription (Markstein et al. 2008), and we observed that independent lines of a given transgene produced consistent levels of protein expression. We previously showed that kelDE1 produces no detectable protein by Western blot (Xue and Cooley 1993), and sequence analysis revealed a stop codon at glutamine 109, upstream of the BTB domain (Figure 2A). This strategy allowed us to determine the steady-state levels and rescuing activities of transgenic proteins in the absence of endogenous Kelch.
Mutating one or both predicted surface-exposed lysines (K414 and K428) or all lysine residues (6K > R) in the KREP domain failed to stabilize the mutant Kelch protein relative to wild-type Kelch (Figure 2, B–E). All three of these transgenes produced proteins with an apparent stability similar to wild-type Kelch, and each rescued the kelch female-sterile phenotype, regardless of whether expression was driven by matGal4 or otuGal4 (data not shown). Since mutating lysines in the KREP domain did not block the autocatalytic degradation of Kelch, we reasoned that other lysines in Kelch could serve as sites of ubiquitylation. Therefore, we made a transgene expressing a Kelch protein in which all 16 lysines in Kelch were mutated to arginine; we refer to this mutant protein as KelchK0. Steady-state levels of KelchK0 protein were increased more than twofold relative to wild-type Kelch, suggesting that KelchK0 was resistant to Cul3-dependent degradation (Figure 2, B–E). In agreement with the Western analysis, egg chambers expressing only KelchK0 had elevated levels of Kelch in egg chambers (Figure 2F).
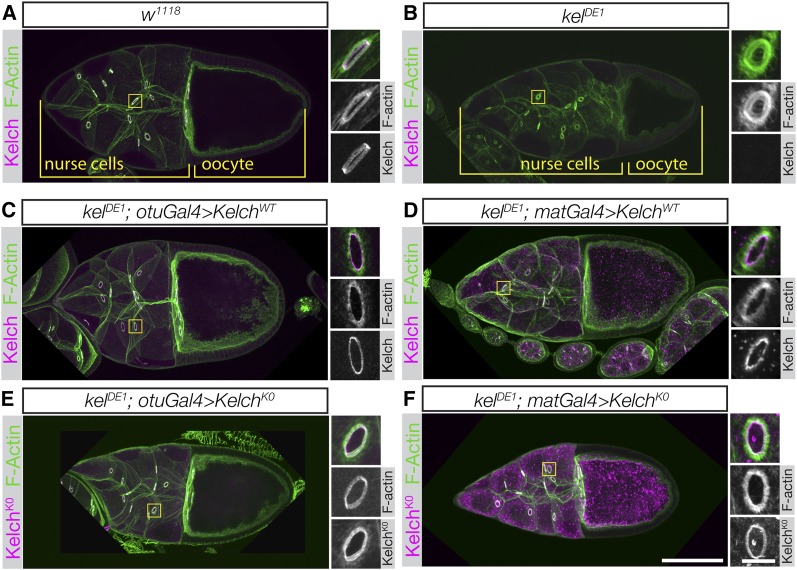
The ring canal phenotype in kelch mutant egg chambers results in the production of small oocytes (Figure 3B). We examined whether KelchK0 expressed at low (Figure 3E) or high (Figure 3F) levels could rescue kelch. The ring canal and small oocyte phenotypes are fully rescued by expression of wild-type Kelch expressed at either low or high levels of expression (Figure 3, C–D). At high levels of expression, wild-type Kelch accumulates in cytoplasmic aggregates (Figure 3D). The presence of these aggregates had no apparent effect on oogenesis. Aggregates formed when wild-type Kelch protein was overexpressed and often colocalized with Cul3 (see Figure 5, C–C′′′), but did not label with ubiquitin (A. M. Hudson and L. Cooley, unpublished data). Expression of KelchK0 restored oocyte size (Figure 3, E–F) and fertility. Examination of ring canals revealed that the KelchK0 protein did not accumulate in the ring canal lumens, as Kelch does in Cul3 mutant egg chambers. Cul3-dependent degradation of Kelch therefore does not appear to be necessary to remove Kelch from the ring canal lumen during growth. However, the accumulation of Kelch into cytoplasmic aggregates was more pronounced when KelchK0 was expressed at high levels compared to wild-type Kelch. The F-actin organization in these egg chambers was indistinguishable from wild type when KelchK0 was expressed at high levels using the matGal4 driver (compare Figures 3, D and F). Cul3-dependent degradation of Kelch therefore does not appear to be necessary to remove Kelch from the ring canal lumen during growth or to organize the ring canal actin cytoskeleton.
Figure 3.
Kelch protein resistant to autocatalytic degradation is fully functional. (A and B) Confocal maximum intensity projections of egg chambers labeled with Kelch antibody and fluorescent phalloidin. (A) Wild-type stage 10 egg chamber. Inset shows higher-resolution image of a ring canal, revealing the F-actin cytoskeleton and associated Kelch protein. (B) Stage 10 egg chamber from a kelDE1 female. The oocyte is considerably smaller than the comparably staged wild-type egg chamber in A, ring canals lack detectable Kelch protein, and the F-actin cytoskeleton largely occludes the lumens of the ring canals. (C and D) Ovaries from kelDE1 rescued with a wild-type cDNA driven at low (otuGal4, C) or high (matGal4, D) levels of expression. kelDE1 egg chambers rescued with otuGal4 resemble wild-type egg chambers. Ovaries rescued with high-level Kelch expression using matGal4 accumulate cytoplasmic aggregates of Kelch (D), but Kelch localizes to ring canals and rescues the cytoskeletal defects. (E and F) KelchK0 rescues the kelch ring canal cytoskeletal phenotype at both low (E, otuGal4) and high (F, matGal4) levels of expression. Accumulation of Kelch cytoplasmic aggregates was greatly enhanced with matGal4-mediated expression (F). Scale bars, 50 µm for egg chamber images and 10 µm for cropped ring canal images.
Figure 5.
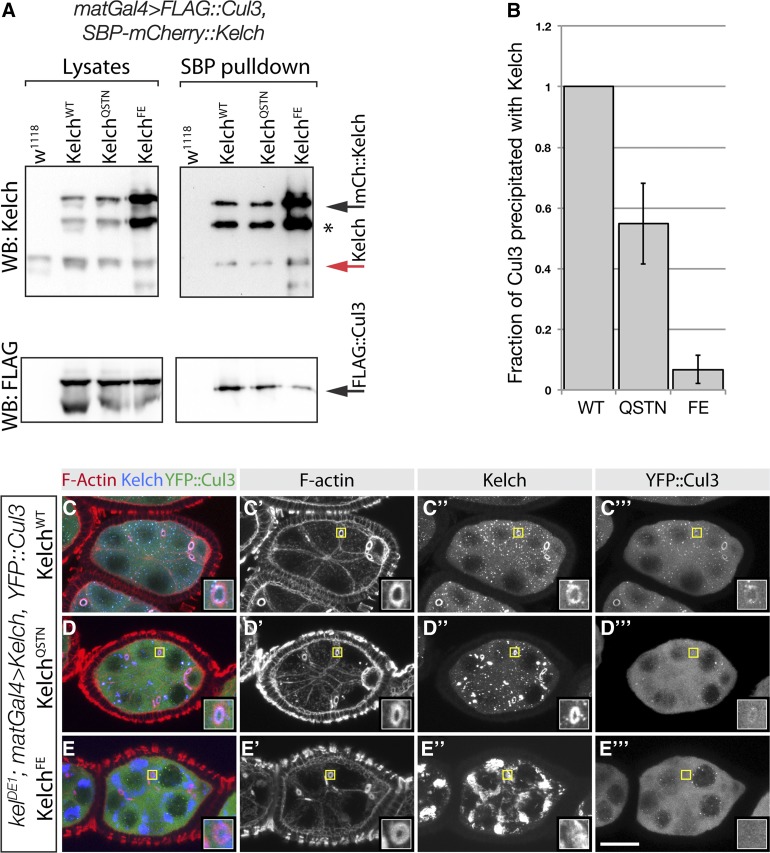
Mutations in the Kelch BTB domain disrupt the association of Kelch with Cul3 in vivo. (A) Western analysis of pulldown experiment to analyze complex formation between Cul3 and Kelch BTB mutants. Ovarian lysates were prepared from flies expressing SBP–mCherry-tagged Kelch proteins in a wild-type background and subjected to pulldowns with Streptavidin beads. Western blots of pulldowns and input lysates were probed with Kel 1B and FLAG M2 monoclonal antibodies. Red arrow indicates endogenous Kelch, the presence of which in the pulldown experiments is an indication of dimerization of endogenous Kelch with the tagged Kelch protein. Asterisk indicates a proteolytic product of the overexpressed SBP–mCherry::Kelch proteins. Black arrow indicates full-length SBP-mCherry::Kelch. (B) Analysis of Cul3 coprecipitation efficiency. Cul3 precipitation efficiency is plotted as a ratio of the amount of Cul3 precipitated to the amount of Kelch in the Streptavidin precipitates. The amount of Kelch and Cul3 precipitated from flies expressing tagged SBP–mCherry::KelchWT was set at 1. Error bars indicate the standard error of the mean from three independent experiments. The ratios obtained with each Kelch protein differed significantly from the other two (P < 0.05, one-way ANOVA, Tukey’s multiple comparison test). (C–E) Egg chambers from homozygous kelDE1 flies expressing the indicated Kelch protein and Venus::Cul3. Regions indicated by yellow boxes are shown as higher magnification insets. YFP::Cul3 localized to kelDE1 ring canals rescued by wild-type Kelch (C–C′′′) and KelchQSTN (D–D′′′), but not KelchFE (E–E′′′).
Functional dissection of Kelch molecular activities
To determine the importance of ubiquitin ligase activity, we carried out site-directed mutagenesis of Kelch. We focused on the BTB domain for this purpose, as structural analyses indicated that we could introduce mutations to specifically disrupt Cul3 binding without affecting dimerization, as these binding interfaces are distinct (Figure 4B). We expected such a mutant Kelch protein to retain function as a dimeric F-actin cross-linking protein, but to be unable to function as CRL3 substrate recognition subunit.
Figure 4.
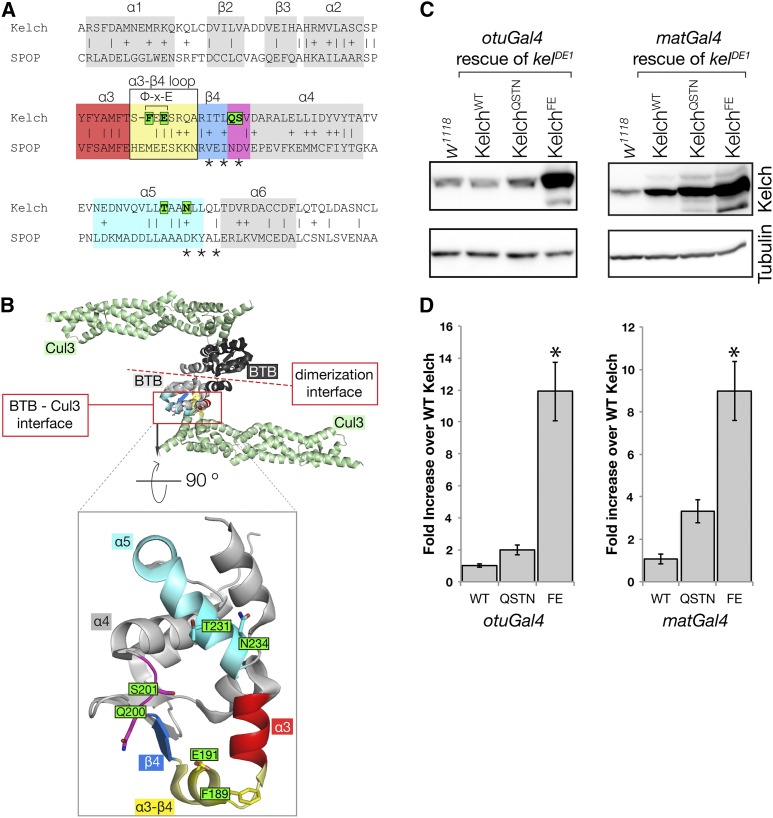
Mutagenesis of the Kelch BTB domain to disrupt Kelch–Cul3 binding. (A) Alignment of Kelch with the sequence SPOP, a BTB domain whose structure has been solved in complex with the Cul3 N-terminal domain. α-helix and β-sheet numbering are as in Errington et al. (2012). Residues mutated in this study are boxed in green; asterisks mark positions of residues mutated previously (Xu et al. 2003) to disrupt the BTB–Cul3 association. The Φ-X-E motif contributes significantly to BTB–Cul3 binding, as the hydrophobic residue inserts into a hydrophobic pocket in Cul3 and the glutamate residue forms a hydrogen bond with a Cul3 α-helix. Sequences highlighted by colored boxes correspond to similarly colored structural elements in B. (B) Structure of the SPOP BTB domain in complex with Cul3 (PDB 4EOZ; Errington et al. 2012). The structure is a heterotetramer consisting of two Cul3 N-terminal domains (green) and two BTB domains (black and gray). Two Cul3 α-helices make extensive contacts with one face of the BTB domain. Boxed region shows isolated view of Cul3 binding surface on the SPOP BTB domain. Kelch residues targeted in this study were generally not conserved in SPOP and were modeled into this representation using MacPyMol. The amino acid sequence positions targeted for mutagenesis are oriented toward Cul3, and each was found to contribute to the interaction surface in the solved structures (Errington et al. 2012; Canning et al. 2013; Ji and Prive 2013). (C) Western analysis of ovaries from kelDE1 females rescued with Kelch containing the indicated mutations in the BTB domain. (D) Mean protein expression levels measured relative to endogenous w1118 control. The expression level from the KelchWT cDNA rescue in each experiment was set at 1. Asterisk indicates statistically significant difference compared to expression from KelchWT cDNA (one-way ANOVA, Dunnett multiple comparison test, P < 0.001). Error bars indicate the standard error of the mean. QSTN, Q200A S201A T232A N234A; FE, F189E E191A.
BTB domains share a common structural fold with Skp1, which binds the Cul1 cullin and links it with an F-box SRS protein to form a CRL1 (Xu et al. 2003). Previous studies relied on this structural similarity to mutagenize the BTB domain in order to disrupt binding to Cul3 (Xu et al. 2003; Lee et al. 2010). We chose an initial set of residues to mutate based on these results in combination with residues predicted to be important for the BTB–Cul3 interaction based on further structural modeling (Stogios et al. 2005). Several structures of BTB–Cul3 complexes have subsequently been solved (Errington et al. 2012; Canning et al. 2013; Ji and Prive 2013), and these models confirmed that our mutagenesis approach targeted residues that contribute to the Cul3–BTB binding interface (Figure 4, A–B).
The BTB–Cul3 complex structure models highlighted three structural elements in the BTB domain that make significant contributions to the binding interaction: a loop between α-helix 3 and β-sheet 4, another turn between β-sheet 4 and α-helix 4, and residues in α-helix 5 (Figure 4, A–B). We initially targeted residues in the β4–α4 turn (Q200 and S201) as well as in α-helix 5 (T231 and N234). S201 and N234 were chosen based on correspondence with residues mutated in the CRL3 SRS proteins MEL-26 and KLHL20 that eliminated Cul3 binding (asterisks in Figure 4A; Xu et al. 2003; Lee et al. 2010); Q200 and T231 also contribute to the binding interface near these residues (Errington et al. 2012). We introduced constructs carrying either pair of residues mutated to alanine and expressed them in place of endogenous Kelch as in our lysine mutagenesis experiments. We expected that disrupting the interaction between Kelch and Cul3 would result in increased Kelch levels due to compromised Cul3-dependent degradation. We therefore examined Kelch protein levels as an initial test to determine whether Cul3 binding was compromised. Kelch proteins carrying either the Q200A/S201A pair (QS) or the T231A/N234A pair (TN) of mutated residues failed to accumulate relative to wild type (data not shown), but mutating all four residues to alanine (KelchQSTN) resulted in a Kelch protein that was stabilized relative to wild type, suggesting that this mutant was compromised for Cul3 binding (Figure 4, C–D). Later tests indicated that the KelchQSTN mutant protein retained some ability to bind Cul3 in vivo (see below), so we designed an additional BTB mutant targeting residues in a loop between α-helix 3 and β-sheet 4. Structural analysis identified a hydrophobic/acidic “Φ-X-E” amino acid triplet in this loop that is unstructured in crystals of BTB domains alone, but forms a short α-helix in crystals with BTB domains bound to Cul3 (Errington et al. 2012; Canning et al. 2013; Ji and Prive 2013). The hydrophobic Φ residue in this motif is buried in a nonpolar pocket on Cul3, and the highly conserved glutamic acid residue forms a hydrogen bond with Cul3. Changing the hydrophobic residue to a charged amino acid eliminated binding in vitro (Errington et al. 2012), and mutations in the glutamate residue of the closely related human Kelch protein KLHL3 have phenotypes consistent with reduced Cul3 binding (Boyden et al. 2012). Given these results we made a mutant in which the two analogous residues in Kelch were changed: F189 to glutamate and E191 to alanine, designated KelchFE (Figure 4A). As outlined below, this FE combination nearly eliminated the in vivo association between Cul3 and Kelch.
When we expressed KelchFE in a kelch null background, we found that it accumulated to significantly greater extent than wild type or KelchQSTN (Figure 4, C–D), consistent with the KelchFE mutation causing a more severe disruption of the Kelch–Cul3 interaction and associated autocatalytic destruction. Moreover, overexpression of KelchFE in the germline of wild-type flies driven by matGal4 resulted in a dominant-sterile phenotype in which the ring canals exhibited a kelch-like phenotype (Figure S2). This is consistent with the KelchFE mutant exerting a dominant-negative effect through its dimerization with wild-type Kelch considering that BTB dimerization is required for the activity of at least one other CRL3 (Zhang et al. 2005). In addition, dominant mutations in KLHL3, the human ortholog of Kelch, bear the analogous E191A mutation (Boyden et al. 2012). However, in our experiments KelchFE is expressed at levels significantly higher than those of endogenous Kelch, and so the mechanism may be more similar to overexpression of the Kelch KREP domain (Hudson and Cooley 2010). In the latter case, we hypothesized that the KREP domain remained bound to a ring canal substrate, trapping it in a complex protected from degradation mediated by the wild-type CRL3Kelch that was also present.
We next determined the extent to which the Cul3 interaction was disrupted in vivo with a coprecipitation assay using tagged forms of Cul3 and Kelch expressed in ovaries (Hudson and Cooley 2010). We made Strept-binding peptide (SBP)–mCherry-tagged forms of KelchQSTN and KelchFE and carried out pulldown experiments from ovaries overexpressing FLAG::Cul3 and either wild-type or mutant tagged Kelch protein. The higher molecular weight of the SBP-tagged form allowed us to distinguish it from endogenous Kelch that was also present in these experiments. Pulldown of tagged wild-type Kelch also precipitated endogenous Kelch, showing that the pulldown experiment served as an assay for the ability of tagged Kelch to dimerize (Figure 5A). Precipitates of wild-type Kelch also contained FLAG::Cul3. Pulldowns of tagged KelchQSTN and KelchFE contained endogenous Kelch, showing that the mutations did not disrupt the ability of the proteins to dimerize. Precipitates from flies expressing KelchQSTN contained about half as much FLAG::Cul3 as wild-type (Figure 5, A–B), indicating that the KelchQSTN protein retained some ability to bind Cul3. Precipitates from KelchFE contained elevated levels of KelchFE, consistent with the increased levels of KelchFE in lysates (Figure 5A). However, the amount of FLAG::Cul3 precipitated by KelchFE was reduced to <10% of wild-type Kelch (Figure 5B), indicating that the F189E/E191A mutant combination was more effective in disrupting binding between Kelch and Cul3.
Cul3 localizes to ring canals in a Kelch-dependent manner (Hudson and Cooley 2010); we used this dependence as a second in vivo test for BTB–Cul3 interaction. We examined localization of YFP::Cul3 in kelch mutant ovaries rescued with either wild-type or mutant kelch transgenes. YFP::Cul3 localized to ring canals in ovaries expressing wild-type Kelch (Figure 5C). In addition, YFP::Cul3 could also be observed in ring canals expressing only KelchQSTN, revealing that some Cul3 was able to associate with KelchQSTN at ring canals (Figure 5D). In contrast, we never observed YFP::Cul3 at ring canals in ovaries expressing only KelchFE (Figure 5E). Together, the protein stability results, coprecipitation experiments, and localization show that while KelchQSTN and KelchFE mutations were both impaired for Cul3 binding, the KelchFE mutation was more effective in disrupting the Kelch–Cul3 binding interaction.
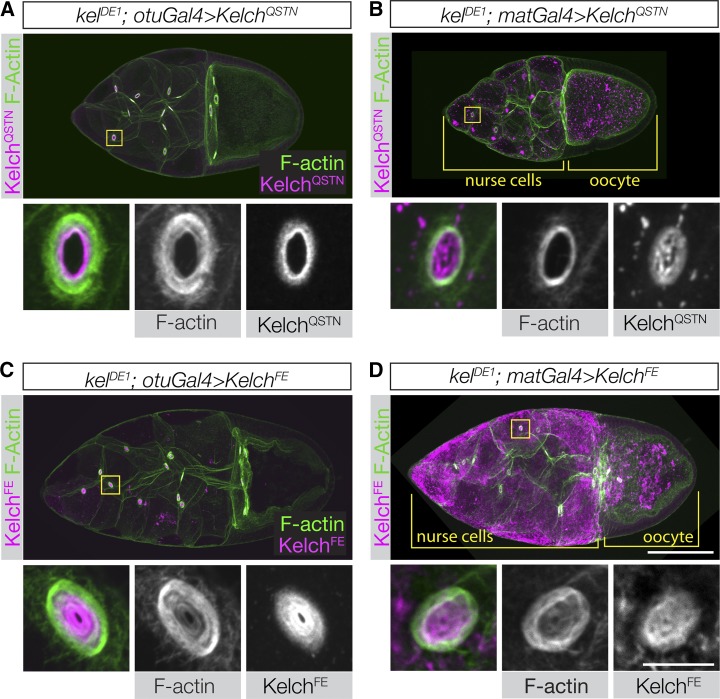
Having characterized the KelchQSTN and KelchFE mutant proteins with respect to Cul3 binding, we tested their ability to rescue the ring canal cytoskeletal defects. We examined rescue using expression levels close to the endogenous level (otuGal4) as well as at a higher level (matGal4). When KelchQSTN was used to rescue kelch using either driver, oocyte size appeared normal and flies were rescued to fertility. However, at the lower level of expression, ring canal F-actin thickness was intermediate between wild type and kelch (Figure 6A), indicating that KelchQSTN was not fully functional. When expressed at higher levels using matGal4, significant amounts of KelchQSTN mutant protein frequently accumulated in the ring canal lumens (Figure 6B), consistent with some reduction in Cul3 binding ability. These results indicate that the KelchQSTN mutant protein is partially compromised in its ability to interact with Cul3.
Figure 6.
Disrupting the Kelch–Cul3 interaction results in a failure to rescue the kelch mutant phenotype. (A–D) Confocal projections of egg chambers labeled with Kelch antibody and fluorescent phalloidin. (A and B) Egg chambers expressing KelchQSTN rescue the F-actin cytoskeletal defects of the kelDE1 mutant. (A) Expression at lower levels using otuGal4 occasionally resulted in an incomplete rescue, with some accumulation of KelchQSTN biased toward the ring canal lumen and an F-actin phenotype intermediate between wild type and kelch-like. (B) High-level expression of KelchQSTN driven by matGal4 fully rescued the ring canals with respect to F-actin organization. However, matGal4-driven KelchQSTN formed abundant cytoplasmic aggregates and tended to form aggregates of Kelch in the ring canal lumens (high magnification ring canal images in B). (C and D) Expression of KelchFE at either low (C) or high (D) levels failed to rescue the kelDE1 F-actin cytoskeletal defects (compare high-magnification ring canal images with those in Figure 3B). In addition, the KelchFE mutant protein accumulated in the ring canal lumen, colocalizing with the disorganized F-actin. When driven by matGal4, KelchFE also formed highly abundant aggregates throughout the germline cytoplasm. Scale bars, 50 µm for egg chamber images and 10 µm for cropped ring canal images.
In contrast, when we examined the ability of KelchFE to rescue, we found that at both low and high expression levels, KelchFE was unable to rescue the kelch null oocyte size and ring canal phenotype (Figure 6, C–D). Ring canal F-actin was indistinguishable from the kelDE1 mutant, and nurse cell-to-oocyte transport was severely compromised, resulting in small oocytes and sterility. The KelchFE protein localized to ring canals, but accumulated at highest levels in the ring canal lumen, similar to how endogenous Kelch localizes in Cul3 mutant clones (Hudson and Cooley 2010). Thus KelchFE, a protein that is specifically compromised in its ability to bind Cul3, cannot carry out its function in organizing the F-actin cytoskeleton. These results are consistent with a model in which the primary function of Kelch in ring canal morphogenesis is that of an SRS component of a CRL3.
Analysis of proteasome function at ring canals
In most cases, ubiquitylation by CRLs targets the substrate protein for destruction by the proteasome (Petroski and Deshaies 2005). There are exceptions to this outcome, however, including examples in which CRL-mediated ubiquitylation regulates cytoskeletal proteins through proteasome-independent pathways (Hao et al. 2013; Yuan et al. 2014). We hypothesized that if CRL3Kelch targets a ring canal substrate for degradation by the proteasome, then inhibition of the proteasome would cause a kelch-like ring canal phenotype. To test this, our approach was to inhibit the proteasome by expressing shRNAs (Ni et al. 2011) targeting proteasome subunit genes in postmitotic germline cells (with the matGal4 driver). To monitor the effectiveness of proteasome inhibition, we made transgenic flies expressing a proteasome activity reporter protein (UbG76V–GFP). The UbG76V–GFP fusion protein has an uncleavable ubiquitin moiety fused to GFP that becomes a target for polyubiquitylation and subsequent degradation through the ubiquitin fusion degradation pathway (Johnson et al. 1995; Dantuma et al. 2000). Under conditions of proteasome inhibition, the UbG76V–GFP protein will accumulate, so GFP fluorescence can be used as a readout of proteasome activity.
We found that expression of shRNAs targeting Prosβ5, a catalytic subunit of the proteasome (Belote and Zhong 2009; Finley 2009), led to an increase in UbG76V–GFP reporter protein, as shown by quantitation of GFP levels (Figure 7, A–C), indicating successful proteasome inhibition. Proteasome inhibition also led to arrest of egg chamber growth and eventual egg chamber degeneration (Figure 7, A and B, and Figure S3). Therefore, we compared ring canals of midstage egg chambers (Figure 7C, germline area group 3000 μm2) that had not begun degeneration (Figure 7, D–G). Proteasome inhibition resulted in kelch-like ring canals in which the actin cytoskeleton was dramatically thicker and/or often had the presence of an inner ring of F-actin (Figure 7G′′ and Figure S4A). We quantified the penetrance of the kelch-like ring canal phenotype and found that proteasome knockdown by Prosβ5 RNAi and Rpn8 RNAi (another subunit of the proteasome) resulted in 91 and 93% of ring canals being kelch-like, respectively (Figure S4B).
If Kelch and the proteasome function in a common pathway to ubiquitylate and degrade a ring canal substrate, we hypothesized that loss of one copy of kelch in addition to proteasome inhibition would enhance the kelch-like ring canal phenotype. Indeed, loss of one copy of kelch in addition to proteasome inhibition resulted in ring canals that were more kelch-like—marked by significantly smaller ring canal lumens, thicker F-actin rings, and increased F-actin staining in ring canal lumens—compared to ring canals experiencing only proteasome inhibition (Figure 7H and Figure S5). Taken together, these data indicate that Kelch and the proteasome function together to regulate the ring canal actin cytoskeleton.
Discussion
Drosophila Kelch is essential for the organization of the ring canal cytoskeleton
Mutations in kelch result in a highly disordered F-actin cytoskeleton in Drosophila ring canals. Our previous work documented biochemical and genetic evidence for distinct molecular mechanisms of Kelch that could explain this phenotype: F-actin cross-linking and ubiquitin ligase activity. In this article, we provide evidence that the ubiquitin ligase function of Kelch is critically important for the organization of the ring canal cytoskeleton. In addition, we present evidence that reducing proteasome activity in the germline results in a kelch-like ring canal phenotype, consistent with a model in which Kelch ubiquitylates a substrate for degradation by the proteasome. Together, these results establish the importance of the UPS in regulating the F-actin cytoskeleton in Drosophila.
Cul3-dependent proteolytic turnover is not essential for Kelch function
Kelch protein accumulates to high levels when Cul3 function is reduced (Hudson and Cooley 2010). Kelch levels are therefore likely to be controlled by autoubiquitylation and proteasome-mediated degradation. Similar results have been described for a number of other CRL SRS proteins (Zhou and Howley 1998; Galan and Peter 1999; Pintard et al. 2003; Li et al. 2004; Zhang et al. 2005). In addition, the extra Kelch is concentrated in the ring canal lumens, suggesting that Cul3-dependent turnover of Kelch could be required to remove it as the ring canals expand. This would be somewhat analogous to the regulation of the budding yeast F-Box protein Ctf13p. Ctf13p, in complex with the F-box-Cul1 linking protein Skp1p, performs an essential structural role in assembling the budding yeast kinetochore (Kaplan et al. 1997). Ctf13p is degraded through the action of the UPS, and degradation of Ctf13p is required for proper kinetochore assembly and function (Rodrigo-Brenni et al. 2004).
We tested whether Kelch ubiquitylation is required for its function by mutating lysine residues in Kelch to eliminate Cul3-dependent turnover. Mutating all lysine residues resulted in the stabilization of Kelch, consistent with a mechanism in which Kelch is targeted for destruction through Cul3-dependent ubiquitylation. However, we found that the lysine-less KelchK0 protein localized to ring canals and rescued the kelDE1 ring canal cytoskeletal defects, suggesting that mutation of all lysine residues did not have a deleterious effect on Kelch folding or function. Given that the ubiquitination-resistant KelchK0 was able to rescue kelch, we conclude that Cul3-dependent turnover of Kelch is not required to promote ring canal growth or F-actin organization.
Kelch functions as an E3 ubiquitin ligase in vivo
A number of SRS proteins have CRL-independent functions, with some notable examples involving cytoskeletal regulation. pVHL, the product of the VHL tumor-suppressor gene, functions as a SRS for a CRL2 ubiquitin ligase, targeting HIF1α, HIF2α, and other proteins for ubiquitylation and destruction (Frew and Krek 2008; Gossage et al. 2015). However, the pVHL protein also binds to and stabilizes microtubules (Hergovich et al. 2003; Thoma et al. 2009, 2010). The Caernorhabditis elegans MATH-BTB protein MEL-26, which functions as a CRL3 SRS targeting the microtubule-severing protein Katanin (Pintard et al. 2003), also has a Cul3-independent function in regulating the actin cytoskeleton during cytokinesis of early embryonic cell divisions (Luke-Glaser et al. 2005).
For Kelch, phenotypic analysis of kelch and Cul3 mutants alone was not sufficient to determine the in vivo contributions of F-actin cross-linking and SRS activity. To examine these functions, we generated two mutant proteins, KelchFE and KelchQSTN, designed to specifically disrupt the Kelch–Cul3 binding interaction without affecting its ability to dimerize, bind F-actin, or bind substrates.
The mutations in KelchQSTN appear to result in partial loss of function with respect to Cul3 binding and CRL3 activity. This is somewhat surprising, since these mutations were chosen based on changes that were reported to completely disrupt the BTB–Cul3 interaction in the BTB proteins MEL-26 and KLHL20 (Xu et al. 2003; Lee et al. 2010). A set of six residues was mutated in those studies (Figure 4A, asterisks), although recent structural work showed that four of these are buried residues involved in maintaining the BTB structural fold (Errington et al. 2012). Two of the six residues make substantial contacts between the BTB domain and Cul3 (Errington et al. 2012), and these correspond to S201 and N234 in KelchQSTN. Q200 and T231 are nearby residues shown to also contribute to Cul3–BTB binding (Errington et al. 2012; Canning et al. 2013; Ji and Prive 2013). The rescue of the F-actin cytoskeletal phenotypes by KelchQSTN suggests that it is still capable of targeting its substrate for ubiquitylation.
We found that KelchFE did not associate with Cul3 at ring canals and was unable to rescue the kelch ring canal defects. Independent assays of Kelch function indicated that the KelchFE mutation affected only Cul3 association. Tagged KelchFE was able to dimerize with endogenous Kelch based on pulldown experiments, demonstrating the mutations did not disrupt BTB dimerization. KelchFE was also able to localize to ring canals (Figure 6 C, D), which is mediated by the Kelch-repeat domain (Robinson and Cooley 1997). Since these results indicate that KelchFE is specifically compromised for ubiquitin ligase activity, we conclude that the principal function of Kelch required for organizing the ring canal cytoskeleton is that of an E3 ubiquitin ligase.
We note that the ability of KelchFE to dimerize and localize to ring canals indicates that the domains important for F-actin cross-linking—the BTB dimerization interface and the Kelch repeat domain (which binds F-actin in vitro)—are functional. KelchFE should therefore retain F-actin cross-linking activity, yet it does not rescue F-actin organization defects in kelch mutants. These results suggest the possibility that F-actin cross-linking by Kelch is not functionally significant at ring canals and that the F-actin cross-linking activity of Kelch detected in vitro does not reflect its in vivo function.
We were not able to design mutations in Kelch to specifically target its F-actin cross-linking activity. We previously mapped the F-actin binding domain to Kelch-repeat 5 (Kelso et al. 2002); however, mutagenesis of repeat 5 would be difficult to interpret, as mutations in this region could also disrupt CRL3Kelch–substrate interactions. Structures of two Kelch-repeat domains in complex with substrate peptides show that substrate binding involves contacts with residues in each structural repeat (Lo et al. 2006; Padmanabhan et al. 2006; Schumacher et al. 2014). Moreover, attempts to specifically disrupt F-actin cross-linking by creating a dimerization-defective BTB domain would be similarly difficult to interpret, as dimer-defective mutations in the BTB domain of SPOP eliminated the ligase activity of CRL3SPOP (Zhuang et al. 2009).
The proteasome is required for ring canal cytoskeletal organization
The attachment of ubiquitin to a protein can lead to a number of different outcomes. The attachment of a lysine 48 (K48)-linked polyubiquitin chain, in which each added ubiquitin is attached to K48 of the preceding ubiquitin, marks the protein for degradation by the proteasome (Pickart and Fushman 2004; Ikeda et al. 2010). CRLs typically direct the attachment of K48-linked polyubiquitin chains (Petroski and Deshaies 2005), although there are some notable exceptions. Two mammalian CRL3s, CRL3KLHL9/KLHL13 and CRL3KLHL21, ubiquitylate the Aurora B kinase, a protein required for chromosome segregation and cytokinesis. CRL3KLHL21 attaches a single ubiquitin to Aurora B; monoubiquitylation is a nondestructive signal that, in this case, alters the subcellular localization of Aurora B instead of targeting it to the proteasome (Maerki et al. 2009). The CRL3KLHL9/KLHL13 ligase attaches a polyubiquitin chain of unknown linkage type to Aurora B, and this modification also results in a change in localization rather than degradation (Sumara et al. 2007). In yeast, ubiquitylation by a CRL results in reversible inhibition of its target without degradation: The transcription factor Met4 is ubiquitylated by the SCFMet30, and this results in its repression of Met4’s activity. Upon an environmental cue, Met4 is deubiquitylated and repression is relieved (Ouni et al. 2011).
To determine whether substrate ubiquitylation by CRL3Kelch results in proteasome-mediated degradation or an alternative outcome, we examined the requirements for the proteasome in ring canal cytoskeletal organization. Initial experiments using dominant temperature-sensitive mutations in genes encoding proteasome subunits (Saville and Belote 1993; Smyth and Belote 1999) or culturing egg chambers with proteasome inhibitors failed to induce kelch-like ring canal phenotypes (A. M. Hudson and L. Cooley, unpublished data), suggesting that ubiquitylation by CRL3Kelch might also function to regulate a substrate in a proteasome-independent manner. To investigate this possibility more rigorously, we developed a transgenic proteasome activity reporter, allowing us to determine the extent of proteasome inhibition in the ovary. In addition, we made use of recently developed transgenic shRNA lines that allow for potent, tissue-specific gene silencing (Ni et al. 2011) to target genes encoding proteasome subunits. Using these reagents together, we were able to document that inhibition of the proteasome results in a kelch-like phenotype. This result provides strong evidence that a substrate of CRL3Kelch is degraded by the proteasome to allow the ordered growth of the ring canal cytoskeleton. We note that the ring canal phenotype seen in kelch mutants is remarkably specific: the loss-of-function ovarian germline phenotypes of thousands of genes—perhaps a third of the genome—have been examined (see Hudson and Cooley 2014 for review of systematic genetic screens performed in the ovary), and yet only four genes are known to result in a kelch-like phenotype. Two of these are kelch and Cul3, while in the other two, Impα2 and orbit, the phenotypes appear to result from a failure to localize Kelch to ring canals (Gorjanacz et al. 2002; Mathe et al. 2003). Thus to our knowledge, all characterized mutations that result in a kelch-like phenotype are required for Kelch function in some manner. In addition, we found that the ring canal phenotype associated with Prosβ5 knockdown was dominantly enhanced by removal of one copy of kelch. Taken together, these results provide compelling evidence that the proteasome functions together with Kelch to organize the ring canal cytoskeleton, most likely by degrading a ubiquitylated substrate(s) of CRL3Kelch. The substrate may directly affect F-actin, for example, by enhancing F-actin polymerization or blocking F-actin depolymerzation. In either case, accumulation of substrate in a kelch mutant would lead to excess ring canal F-actin.
Shaping the cytoskeleton by the ubiquitin proteasome system
A number of examples of ubiquitin-mediated regulation of the cytoskeleton in cultured cells highlight the diverse mechanisms possible for UPS-mediated cytoskeletal regulation. Degradation of key regulatory proteins is one mechanism, and UPS-mediated destruction of Rho-family GTPases has been described (Wang et al. 2003; Chen et al. 2009). In addition, cytoskeletal structural proteins, such as the F-actin binding protein Filamin, have been shown to be targets of UPS-mediated destruction (Razinia et al. 2011). Finally, several actin-associated proteins are regulated by ubiquitin conjugation that alters the activity of the target proteins (Hao et al. 2013; Yuan et al. 2014). A future challenge will be to understand how ubiquitin-mediated regulation of the cytoskeleton is implemented during development. Our work defining the central role of the UPS in shaping the ring canal cytoskeleton presents an opportunity to investigate how UPS-mediated regulation of cytoskeleton can be integrated into a metazoan developmental program.
Supplementary Material
Acknowledgments
We thank Vladimir Polejaev at the Yale West Campus imaging center and Yves Chabu and Tian Xu for providing access to confocal microscopes. pCasper3-Up2-RX polyA was a gift from Rick Fehon, and pPW-attB was a gift from Mike Buszczak. Stocks obtained from the Bloomington Drosophila Stock Center (National Institutes of Health, NIH, P40OD018537) were used in this study. Antibodies were obtained from the Developmental Studies Hybridoma Bank at The University of Iowa, Iowa City. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks used in this study. This work was funded by NIH R01 GM043301 grant to L.C. K.M.M. was supported in part by the National Institute of General Medical Sciences NIH training grant T32 GM007223.
Footnotes
Communicating editor: M. F. Wolfner
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.181289/-/DC1.
Literature Cited
- Belote J. M., Zhong L., 2009. Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity 103: 23–31. [DOI] [PubMed] [Google Scholar]
- Boyden L. M., Choi M., Choate K. A., Nelson-Williams C. J., Farhi A., et al. , 2012. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning P., Cooper C. D., Krojer T., Murray J. W., Pike A. C., et al. , 2013. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J. Biol. Chem. 288: 7803–7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yang Z., Meng M., Zhao Y., Dong N., et al. , 2009. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol. Cell 35: 841–855. [DOI] [PubMed] [Google Scholar]
- Dantuma N. P., Lindsten K., Glas R., Jellne M., Masucci M. G., 2000. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18: 538–543. [DOI] [PubMed] [Google Scholar]
- de Bie P., Ciechanover A., 2011. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 18: 1393–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15: 435–467. [DOI] [PubMed] [Google Scholar]
- Errington W. J., Khan M. Q., Bueler S. A., Rubinstein J. L., Chakrabartty A., et al. , 2012. Adaptor protein self-assembly drives the control of a cullin-RING ubiquitin ligase. Structure 20: 1141–1153. [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Oren T., LaJeunesse D. R., Melby T. E., McCartney B. M., 1997. Isolation of mutations in the Drosophila homologues of the human Neurofibromatosis 2 and yeast CDC42 genes using a simple and efficient reverse-genetic method. Genetics 146: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78: 477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frew I. J., Krek W., 2008. pVHL: a multipurpose adaptor protein. Sci. Signal. 1: pe30. [DOI] [PubMed] [Google Scholar]
- Galan J. M., Peter M., 1999. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96: 9124–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanacz M., Adam G., Torok I., Mechler B. M., Szlanka T., et al. , 2002. Importin-alpha 2 is critically required for the assembly of ring canals during Drosophila oogenesis. Dev. Biol. 251: 271–282. [DOI] [PubMed] [Google Scholar]
- Gossage L., Eisen T., Maher E. R., 2015. VHL, the story of a tumour suppressor gene. Nat. Rev. Cancer 15: 55–64. [DOI] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y. H., Doyle J. M., Ramanathan S., Gomez T. S., Jia D., et al. , 2013. Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell 152: 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Lisztwan J., Barry R., Ballschmieter P., Krek W., 2003. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat. Cell Biol. 5: 64–70. [DOI] [PubMed] [Google Scholar]
- Hudson A. M., Cooley L., 2002. A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J. Cell Biol. 156: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. M., Cooley L., 2008. Phylogenetic, structural and functional relationships between WD- and Kelch-repeat proteins. Subcell. Biochem. 48: 6–19. [DOI] [PubMed] [Google Scholar]
- Hudson A. M., Cooley L., 2010. Drosophila Kelch functions with Cullin-3 to organize the ring canal actin cytoskeleton. J. Cell Biol. 188: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. M., Cooley L., 2014. Methods for studying oogenesis. Methods 68: 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F., Crosetto N., Dikic I., 2010. What determines the specificity and outcomes of ubiquitin signaling? Cell 143: 677–681. [DOI] [PubMed] [Google Scholar]
- Ji A. X., Prive G. G., 2013. Crystal structure of KLHL3 in complex with Cullin3. PLoS One 8: e60445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. S., Ma P. C., Ota I. M., Varshavsky A., 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270: 17442–17456. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt J. A., Davidson C. M., Brown N. H., Brand A. H., 2000. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat. Cell Biol. 2: 7–12. [DOI] [PubMed] [Google Scholar]
- Kaplan K. B., Hyman A. A., Sorger P. K., 1997. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91: 491–500. [DOI] [PubMed] [Google Scholar]
- Kelso R. J., Hudson A. M., Cooley L., 2002. Drosophila Kelch regulates actin organization via Src64-dependent tyrosine phosphorylation. J. Cell Biol. 156: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. R., Yuan W. C., Ho H. C., Chen C. H., Shih H. M., et al. , 2010. The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 29: 1748–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. G., Serr M., Edwards K., Ludmann S., Yamamoto D., et al. , 1999. Filamin is required for ring canal assembly and actin organization during Drosophila oogenesis. J. Cell Biol. 146: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Gazdoiu S., Pan Z. Q., Fuchs S. Y., 2004. Stability of homologue of Slimb F-box protein is regulated by availability of its substrate. J. Biol. Chem. 279: 11074–11080. [DOI] [PubMed] [Google Scholar]
- Lo S. C., Li X., Henzl M. T., Beamer L. J., Hannink M., 2006. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 25: 3605–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S., Pintard L., Lu C., Mains P. E., Peter M., 2005. The BTB protein MEL-26 promotes cytokinesis in C. elegans by a CUL-3-independent mechanism. Curr. Biol. 15: 1605–1615. [DOI] [PubMed] [Google Scholar]
- Lydeard J. R., Schulman B. A., Harper J. W., 2013. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 14: 1050–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerki S., Olma M. H., Staubli T., Steigemann P., Gerlich D. W., et al. , 2009. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J. Cell Biol. 187: 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N., 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40: 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe E., Inoue Y. H., Palframan W., Brown G., Glover D. M., 2003. Orbit/Mast, the CLASP orthologue of Drosophila, is required for asymmetric stem cell and cystocyte divisions and development of the polarised microtubule network that interconnects oocyte and nurse cells during oogenesis. Development 130: 901–915. [DOI] [PubMed] [Google Scholar]
- Mizuno K., 2013. Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation. Cell. Signal. 25: 457–469. [DOI] [PubMed] [Google Scholar]
- Ni J. Q., Zhou R., Czech B., Liu L. P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouni I., Flick K., Kaiser P., 2011. Ubiquitin and transcription: the SCF/Met4 pathway, a (protein-) complex issue. Transcription 2: 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan B., Tong K. I., Ohta T., Nakamura Y., Scharlock M., et al. , 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21: 689–700. [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J., 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6: 9–20. [DOI] [PubMed] [Google Scholar]
- Pickart C. M., Fushman D., 2004. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8: 610–616. [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis J. H., Willems A., Johnson J. L., Srayko M., et al. , 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316. [DOI] [PubMed] [Google Scholar]
- Razinia Z., Baldassarre M., Bouaouina M., Lamsoul I., Lutz P. G., et al. , 2011. The E3 ubiquitin ligase specificity subunit ASB2alpha targets filamins for proteasomal degradation by interacting with the filamin actin-binding domain. J. Cell Sci. 124: 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. N., Cooley L., 1997. Drosophila kelch is an oligomeric ring canal actin organizer. J. Cell Biol. 138: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. N., Smith-Leiker T. A., Sokol N. S., Hudson A. M., Cooley L., 1997. Formation of the Drosophila ovarian ring canal inner rim depends on cheerio. Genetics 145: 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni M. C., Thomas S., Bouck D. C., Kaplan K. B., 2004. Sgt1p and Skp1p modulate the assembly and turnover of CBF3 complexes required for proper kinetochore function. Mol. Biol. Cell 15: 3366–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P., 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78: 113–118. [DOI] [PubMed] [Google Scholar]
- Saville K. J., Belote J. M., 1993. Identification of an essential gene, l(3)73Ai, with a dominant temperature-sensitive lethal allele, encoding a Drosophila proteasome subunit. Proc. Natl. Acad. Sci. USA 90: 8842–8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher F. R., Sorrell F. J., Alessi D. R., Bullock A. N., Kurz T., 2014. Structural and biochemical characterization of the KLHL3-WNK kinase interaction important in blood pressure regulation. Biochem. J. 460: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton K., Woodruff R. I., 1994. The osmolarity of adult Drosophila hemolymph and its effect on oocyte-nurse cell electrical polarity. Dev. Biol. 161: 154–167. [DOI] [PubMed] [Google Scholar]
- Smyth K. A., Belote J. M., 1999. The dominant temperature-sensitive lethal DTS7 of Drosophila melanogaster encodes an altered 20S proteasome beta-type subunit. Genetics 151: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol N. S., Cooley L., 1999. Drosophila filamin encoded by the cheerio locus is a component of ovarian ring canals. Curr. Biol. 9: 1221–1230. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L., Sepich D. S., 2012. Gastrulation: making and shaping germ layers. Annu. Rev. Cell Dev. Biol. 28: 687–717. [DOI] [PubMed] [Google Scholar]
- Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Prive G. G., 2005. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I., Quadroni M., Frei C., Olma M. H., Sumara G., et al. , 2007. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev. Cell 12: 887–900. [DOI] [PubMed] [Google Scholar]
- Tan J. L., Ravid S., Spudich J. A., 1992. Control of nonmuscle myosins by phosphorylation. Annu. Rev. Biochem. 61: 721–759. [DOI] [PubMed] [Google Scholar]
- Thoma C. R., Toso A., Gutbrodt K. L., Reggi S. P., Frew I. J., et al. , 2009. VHL loss causes spindle misorientation and chromosome instability. Nat. Cell Biol. 11: 994–1001. [DOI] [PubMed] [Google Scholar]
- Thoma C. R., Matov A., Gutbrodt K. L., Hoerner C. R., Smole Z., et al. , 2010. Quantitative image analysis identifies pVHL as a key regulator of microtubule dynamic instability. J. Cell Biol. 190: 991–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen E., Cooley L., 1994. Looking at oogenesis. Methods Cell Biol. 44: 545–561. [PubMed] [Google Scholar]
- Wang H. R., Zhang Y., Ozdamar B., Ogunjimi A. A., Alexandrova E., et al. , 2003. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302: 1775–1779. [DOI] [PubMed] [Google Scholar]
- Way M., Sanders M., Garcia C., Sakai J., Matsudaira P., 1995. Sequence and domain organization of scruin, an actin-cross-linking protein in the acrosomal process of Limulus sperm. J. Cell Biol. 128: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbelauer C., Sutterluty H., Blondel M., Gstaiger M., Peter M., et al. , 2000. The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. EMBO J. 19: 5362–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. T., Lin H. C., Hu Y. C., Chien C. T., 2005. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat. Cell Biol. 7: 1014–1020. [DOI] [PubMed] [Google Scholar]
- Xu L., Wei Y., Reboul J., Vaglio P., Shin T. H., et al. , 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425: 316–321. [DOI] [PubMed] [Google Scholar]
- Xue F., Cooley L., 1993. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72: 681–693. [DOI] [PubMed] [Google Scholar]
- Yin H. L., Janmey P. A., 2003. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65: 761–789. [DOI] [PubMed] [Google Scholar]
- Yuan W. C., Lee Y. R., Lin S. Y., Chang L. Y., Tan Y. P., et al. , 2014. K33-Linked polyubiquitination of coronin 7 by Cul3-KLHL20 ubiquitin E3 ligase regulates protein trafficking. Mol. Cell 54: 586–600. [DOI] [PubMed] [Google Scholar]
- Zhang D. D., Lo S. C., Sun Z., Habib G. M., Lieberman M. W., et al. , 2005. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem. 280: 30091–30099. [DOI] [PubMed] [Google Scholar]
- Zhou P., Howley P. M., 1998. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2: 571–580. [DOI] [PubMed] [Google Scholar]
- Zhuang M., Calabrese M. F., Liu J., Waddell M. B., Nourse A., et al. , 2009. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell 36: 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available from the Bloomington Drosophila Stock Center or upon request. Plasmids are available upon request. The Supporting Information file contains five supplemental figures with legends.