Abstract
The breeding systems of many organisms are cryptic and difficult to investigate with observational data, yet they have profound effects on a species’ ecology, evolution, and genome organization. Genomic approaches offer a novel, indirect way to investigate breeding systems, specifically by studying the transmission of genetic information from parents to offspring. Here we exemplify this method through an assessment of self-fertilization vs. automictic parthenogenesis in Daphnia magna. Self-fertilization reduces heterozygosity by 50% compared to the parents, but under automixis, whereby two haploid products from a single meiosis fuse, the expected heterozygosity reduction depends on whether the two meiotic products are separated during meiosis I or II (i.e., central vs. terminal fusion). Reviewing the existing literature and incorporating recombination interference, we derive an interchromosomal and an intrachromosomal prediction of how to distinguish various forms of automixis from self-fertilization using offspring heterozygosity data. We then test these predictions using RAD-sequencing data on presumed automictic diapause offspring of so-called nonmale producing strains and compare them with “self-fertilized” offspring produced by within-clone mating. The results unequivocally show that these offspring were produced by automixis, mostly, but not exclusively, through terminal fusion. However, the results also show that this conclusion was only possible owing to genome-wide heterozygosity data, with phenotypic data as well as data from microsatellite markers yielding inconclusive or even misleading results. Our study thus demonstrates how to use the power of genomic approaches for elucidating breeding systems, and it provides the first demonstration of automictic parthenogenesis in Daphnia.
Keywords: genome-wide heterozygosity, breeding system, inbreeding, automixis, tychoparthenogenesis, Daphnia magna, nonmale producers
WHILE humans and most other mammals reproduce exclusively by sexual reproduction with sexes being determined by the well-known XY sex-chromosome system, the breeding systems of many other organisms, including many pests and parasites, remain unknown (Bell 1982; Normark 2003). The breeding system sensu lato, (including details of meiosis, e.g., recombination patterns and syngamy, e.g., levels of inbreeding, as well as their variants, e.g., modified meiosis in parthenogens) represents a key for understanding the biology of a species and has profound effects on its ecology, evolution, and genomics. Yet investigating breeding systems is often far from straightforward: Many species cannot easily be cultured and bred in the laboratory and observations of breeding behavior in nature are difficult. Even in species than can be bred in the laboratory, parts of the breeding system may be cryptic and not directly observable.
The advent of high-throughput genotyping methods opens an alternative possibility that can be used on a much larger array of species: indirect inference of the breeding system using genetic methods, which are based on differences among breeding systems in the transmission of genetic information from one generation to the next. In some cases, genome-wide information may not be needed. For instance, a few genetic markers such as microsatellites are sufficient to distinguish self-fertilization from outcrossing in hermaphrodites (e.g., David et al. 2007) or clonal from sexual reproduction in aphids (Delmotte et al. 2002). However, for a conclusive distinction between other breeding systems, a genome-wide approach may be essential. This is illustrated in the present paper for the distinction of self-fertilization vs. automictic parthenogenesis, comparing genomic data with microsatellite data and direct observations.
Self-fertilization and automictic parthenogenesis both reduce genome-wide heterozygosity among offspring compared to their parents, thereby increasing homozygosity due to identity by descent (Hartl and Clark 2007; Charlesworth and Willis 2009). Under self-fertilization, in which male and female gametes produced by the same, hermaphrodite individual fuse, the expected reduction in offspring heterozygosity for diploid, autosomal loci is 50% per generation. A similar heterozygosity reduction also occurs under some forms of automictic parthenogenesis (also called “automixis”). Automictic parthenogenesis is a common form of parthenogenetic (i.e., female-only) reproduction (Bell 1982; Mogie 1986; Suomalainen et al. 1987), in which offspring are produced by fusion of two products of a single meiosis. Examples are intratetrad mating in fungi or fusion of an egg cell with a polar body in animals (Suomalainen et al. 1987; Hood and Antonovics 2004; Stenberg and Saura 2009). A more detailed account of the different processes that are summarized under automixis is given below.
The distinction between automixis and self-fertilization is subtle both in terms of the expected heterozygosity reduction among offspring as well as with respect to the processes that lead to it. Both involve the fusion of two meiotic products produced by a single individual. Self-fertilization involves fusion of products of different, independent meioses and therefore parental alleles are sampled with replacement. In contrast, automixis involves fusion of the products of a single meiosis and therefore parental alleles are sampled without replacement. Sampling of parental alleles with replacement leads to the well-known Mendelian expectations of genotype frequencies (50% heterozygotes, 25% of each homozygote) among self-fertilized offspring. However, to understand the consequences of sampling of maternal alleles without replacement during automixis, we have to distinguish two cases: Under “central fusion” two products that have been separated during meiosis I (the first meiotic division) fuse, and under “terminal fusion” two products that have been separated during meiosis II fuse. Because homologous chromosomes (carrying different alleles at heterozygous loci) are separated during meiosis I, and sister chromatids (carrying identical alleles) are separated during meiosis II, central fusion tends to retains parental heterozygosity and terminal fusion tends to lead to fully homozygous genotypes. However, because recombination reshuffles alleles between homologous chromosomes, these expectations hold only for the centromere (at which sister chromatids are attached to each other). Expected offspring heterozygosity at loci far from the centromere attains 67% of parental heterozygosity for both central and terminal fusion. This is because, far from the centromere, alleles are distributed at random across sister and nonsister chromatids due to recombination, and therefore they are sampled randomly without replacement (once one meiotic product is chosen, two of the three remaining meiotic products contain the alternate allele) (Rizet and Engelmann 1949; Barratt et al. 1954; Suomalainen et al. 1987; Pearcy et al. 2006, 2011; Engelstädter et al. 2011).
Patterns of heterozygosity reduction between parents and offspring can thus be used to distinguish self-fertilization from automixis and/or central from terminal fusion. This approach has previously been used in a few organisms to address the question of whether automixis occurs via central or terminal fusion (Pearcy et al. 2006; Lampert et al. 2007; Oldroyd et al. 2008). However, differences in the realized levels of heterozygosity reduction among breeding systems depend on recombination rates and may be modulated by the degree of recombination interference and, if offspring heterozygosity is assessed at any later stage than the zygote, by differential survival of heterozygotes vs. homozygotes (i.e., viability selection, Wang and Hill 1999).
We therefore first derive two specific theoretical predictions of how to distinguish self-fertilized from automictic offspring and central from terminal fusion based on heterozygosity patterns. We then use the freshwater crustacean Daphnia magna to empirically assess and compare the consequences of self-fertilization and automixis for offspring heterozygosity. We use known, self-fertilized offspring as controls and compare them with offspring whose breeding system was initially unknown but could by the present study be identified as automictic. D. magna reproduces by cyclical parthenogenesis, in which clonal reproduction is intermitted by sexual reproduction. The clonal offspring may develop into males or females (environmental sex determination) and sexual reproduction always leads to the production of diapause stages (“ephippia”: structures formed by maternal tissue, usually encapsulating two diapausing embryos). Hence, “self-fertilized” offspring in diapause can easily be generated by growing clonal cultures to high population densities and letting males mate with their genetically identical sisters. We acknowledge that within-clone mating (mating of a female with a genetically identical male) may only genetically but not ecologically be equivalent to self-fertilization (fertilization between male and female organs of a single, hermaphrodite individual), but for simplicity, we do not distinguish between these terms in the present paper.
While diapause stages can be produced clonally in some species of Daphnia (Hebert and Crease 1980), they were hitherto thought to be always produced by sexual reproduction in D. magna. However, we have previously found that some strains of D. magna do not produce males (“nonmale producing strains,” NMP), even when stimulated with a “male-inducing” hormone (Innes and Dunbrack 1993; Galimov et al. 2011). In natural populations, these strains still participate in sexual reproduction, but only via the female function, that is, by producing diapause eggs that have to be fertilized by males from other, male-producing (MP) strains (i.e., strains that produce both males and females with sex determined by the environment). When grown in isolation (i.e., in NMP-only cultures), females still produce the diapause capsules, but these are usually empty (i.e., do not contain viable embryos). Yet, very rarely, a few offspring hatch from these ephippia, indicating that a very low percentage of them do contain viable embryos (Galimov et al. 2011). The offspring are diploid and show segregation of maternal alleles, indicating that they are not produced clonally (Galimov et al. 2011). They may thus be produced either by within-clone mating through rare and undetected male production in the maternal NMP strain or by automictic parthenogenesis (Galimov et al. 2011). To evaluate these possibilities, we used (i) direct testing for the presence of males by phenotypic screening of large samples, (ii) crossing attempts between different NMP strains (if rare males are present they are expected to fertilize females of other NMP strains as well as their own), and (iii) an assessment of the heterozygosity patterns among offspring by microsatellite genotyping and restriction site-associated DNA (RAD) sequencing. Our results showed that only the genomic approach (RAD sequencing) could provide conclusive evidence for the mode of reproduction by which these offspring had been produced. More generally, our study thus serves to illustrate the observed and expected genome-wide patterns of heterozygosity reduction under automixis and self-fertilization and to provide evidence for the great potential of genomic approaches for elucidating cryptic breeding systems.
Expected Heterozygosity Reduction Under Automixis
The expected heterozygosity reduction under automixis has been described before (Rizet and Engelmann 1949; Barratt et al. 1954; Suomalainen et al. 1987; Pearcy et al. 2006, 2011; Engelstädter et al. 2011). However, different aspects are discussed in different papers, and the literature on breeding systems is rather disparate from the literature on genetic mapping in fungi or on mapping of centromeres either by natural or artificial automixis. Furthermore, in addition to central and terminal fusion, a further term “random fusion” is sometimes discussed, but its definition and effects on heterozygosity reduction require clarification. Finally, the effects of recombination interference on heterozygosity reduction have only rarely been considered in the breeding systems literature (e.g., Asher 1970; Nace et al. 1970). For these reasons, we briefly review here the literature on expected heterozygosity reduction under automixis with the focus on the comparison with self-fertilization. We identify two main predictions regarding expected heterozygosity patterns, an interchromosomal and an intrachromosomal one, which allow distinguishing automictic from self-fertilized offspring using genomic data. We also mathematically derive predictions on the intrachromosomal patterns of heterozygosity in offspring produced by terminal and central fusion, accounting for different degrees of recombination interference.
The terms central fusion and terminal fusion are derived from ordered tetrads (Tucker 1958; Suomalainen et al. 1987). In many fungi and algae, the four products of meiosis remain together in an envelope called “ascus,” with some of them retaining a specific order (Bos 1996): The four meiotic products of a diploid parent heterozygous A1A2 at a centromeric locus are ordered along a sequence A1_A1_A2_A2, with meiosis I explaining the central division and meiosis II the two terminal divisions (each division is indicated by an underscore). Hence, fusion of neighboring meiotic products during within-tetrad mating can either be terminal (leading to homozygous centromeric regions A1A1 or A2A2) or central (leading to heterozygous centromeric regions A1A2). However, because the effects on offspring heterozygosity are identical, the term central fusion is often used to describe the fusion of any two meiotic products that have been separated during meiosis I (or where meiosis I is suppressed, Asher 1970). Equivalently, the term terminal fusion is used to describe the fusion of any products that have been separated during meiosis II (or where meiosis II is suppressed, Asher 1970), not only in ordered tetrads.
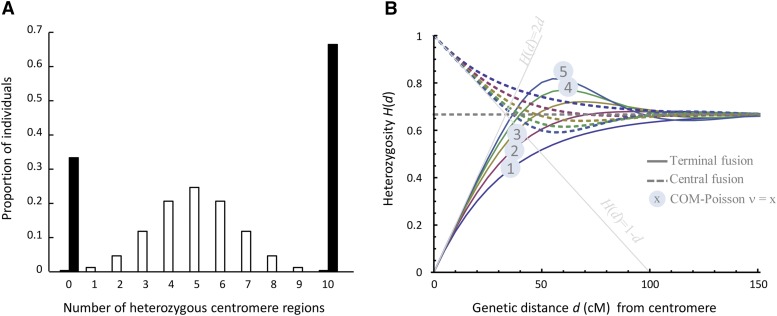
Random fusion can be defined as fusion of two randomly chosen products of a meiotic tetrad (Suomalainen et al. 1987; Pearcy et al. 2006; Lampert et al. 2007). Hence, with random fusion, 2/3 of the offspring are produced by central fusion and 1/3 by terminal fusion (once one meiotic product is chosen, only one of the three remaining products carries the same allele at the centromeric locus shown above, thus central fusion occurs with a probability of 2/3). Yet, in animals, meiosis typically leads to one oocyte and polar bodies, and automictic fusion usually (but not always, e.g., Seiler and Schäffer 1960) occurs between the oocyte and one of the polar bodies. However, the first polar body often decays rapidly or does not undergo meiosis II (e.g., in Daphnia, Zaffagnini and Sabelli 1972), and these details of the reproductive mode may change the proportion of offspring produced by central vs. terminal fusion even under random expectations (i.e., without specific mechanism favoring one over the other). It may therefore be more useful to distinguish cases in which both central fusion and terminal fusion occur, possibly in different proportions (we term this “mixed fusion”) from cases in which one of them is the exclusive mode of reproduction. With mixed fusion, any given offspring is produced by either central or terminal fusion (two specific meiotic products fuse or meiosis I or meiosis II is suppressed). This leads to a first general prediction, which should enable differentiating automixis from self-fertilization: Independently of whether automixis occurs by central or terminal fusion, the homozygosity of centromeric regions across different chromosomes should be 100% correlated within a given offspring (Figure 1). That is, either all centromeric regions should become homozygous (offspring produced by terminal fusion) or they should all retain parental heterozygosity (offspring produced by central fusion). In contrast, under self-fertilization, each centromeric region is expected to become homozygous or retain parental heterozygosity with an independent probability of 0.5 (i.e., independently of the heterozygosity of other centromeric regions in the same individual, Figure 1). A method to determine how the interchromosomal pattern can be assessed if the centromeric positions are unknown, is outlined in Supporting Information, File S1.
Figure 1.
Expected interchromosomal (A) and intrachromosomal patterns (B) of heterozygosity reduction in automictic offspring. (A) The proportion of individuals that retain parental heterozygosity at a given number (out of 10) centromeric regions. Solid bars represent automictic offspring, which should always have either 0 or 10 heterozygous centromeric regions (the relative proportion of individuals with heterozygous vs. homozygous regions depends on the proportion of offspring produced by central vs. terminal fusion; here 2/3 central fusion is assumed). The open bars represent self-fertilized controls. (B) Expected offspring heterozygosity as a function of the genetic distance from the centromere under central (dashed) and terminal (solid) fusion and different degrees of crossover interference (File S2). ν = 1 corresponds to no interference, and the two gray lines correspond to complete interference. The dashed gray line gives the expected heterozygosity for centromere–distal markers (2/3).
Second, within each chromosome, heterozygosity is expected to gradually increase from zero (terminal fusion) or decrease from 100% (central fusion) to 67% of parental heterozygosity with increasing genetic distance from the centromere (Figure 1) (Rizet and Engelmann 1949; Engelstädter et al. 2011; Pearcy et al. 2011). The leveling off at 67% under both terminal and central fusion occurs because, at large genetic distances from the centromere, recombination effectively distributes alleles at random across the sister and nonsister chromatids. Therefore, both terminal and central fusion result in random sampling without replacement of two alleles from four chromatids and thus to the expected heterozygosity of 67% (once one chromatid is chosen, two of the three remaining chromatids carry a different allele).
The transition from zero or 100% heterozygosity at the centromere to 67% heterozygosity in centromere–distant regions depends on the genetic map distance (i.e., the expected number of crossovers) and on the level of crossover interference (Figure 1, File S2) (Barratt et al. 1954; Nace et al. 1970; Zhao and Speed 1998). In File S2, we present an original derivation of this relationship, taking advantage of the flexibility of generalized Poisson distributions (Conway–Maxwell Poisson distribution, Sellers et al. 2012). With high degree of crossover interference, this relationship may be nonmonotonous (Figure 1). However, the initial slope of the change in heterozygosity close to the centromere is 2d (where d is the genetic distance in Morgan) under terminal fusion and –d under central fusion (Figure 1, File S2), irrespectively of the degree of interference. In contrast, under self-fertilization, expected heterozygosity is 50% of the parental heterozygosity and does not depend on the distance from the centromere nor on the level of crossover interference.
Several other forms of automixis are defined and discussed elsewhere (Bell 1982; Mogie 1986; Suomalainen et al. 1987; Stenberg and Saura 2009; Archetti 2010; Lutes et al. 2010; Neiman et al. 2014; Nougué et al. 2015). Their effects on genome-wide heterozygosity reduction are often very different from self-fertilization (e.g., complete loss or complete retention of parental heterozygosity).
Materials and Methods
Origin of clones and outdoor experiments
We use the term “clone” to refer to a strain initiated by a single female and maintained by clonal reproduction. Clones used in this study originated from Russian populations known to contain NMP clones (Ast, BN, MZ, Vol; Galimov et al. 2011). They were classified as MP or NMP according to whether or not females of these clones produced male offspring during clonal reproduction when exposed to 400 nM methyl farnesoate, a juvenile hormone analog that has been shown to consistently induce male production in MP clones of D. magna (Olmstead and Leblanc 2002; see Galimov et al. 2011 for detailed methodology).
Outdoor mass cultures were carried out using two NMP treatments and two MP control treatments: (1) NMP single-clone cultures (“NMP_single”) each contained a single NMP clone. Because only one maternal clone was present, the ephippial offspring produced in these cultures were the result of within-clone mating (if rare males were present) or some form of parthenogenetic reproduction. This treatment was used (as in our earlier study, Galimov et al. 2011) to eliminate the possibility of clonal parthenogenesis by examining offspring for segregation of maternal alleles at microsatellite loci. Furthermore, the offspring of one culture (the culture that produced the largest number of offspring) were used to test for genome-wide patterns of heterozygosity using RAD sequencing. (2) NMP multiclone cultures (“NMP_mix”) contained two to four different NMP clones (distinguishable at microsatellite loci). They were used to test for the presence of rare males by testing for the occurrence of outcrossed offspring (i.e., crosses between different NMP clones). Outcrossing should occur in the presence of rare males, but not under automixis. Control treatments (3) “MP_single” and (4) “MP_mix,” containing single or two to four MP clones, respectively, were used to verify male production and outcrossing under the experimental conditions as well as to assess genome-wide heterozygosity in offspring produced by self-fertilization.
The outdoor cultures were set up under ambient conditions in the botanical garden of Fribourg, Switzerland (46°48′6.00′′N, 7°8′44.04′′E) by transferring ∼100 adult females of each clone into buckets containing 40 liters of artificial Daphnia medium (Klüttgen et al. 1994) as well as a 50-ml initial inoculum of natural microalgae and bacteria (50 μm filtered water from a local garden pond) as well as ∼100 g of fresh horse manure to provide nutrients. Some fresh unicellular green algae, Scenedesmus sp., were added intermittently throughout the experiment to keep densities high, and natural rain water gradually filled the buckets to ∼60 liters.
The experiment took place in two parts: A first batch of cultures was grown outside from March to November 2011, and a second batch from March/April 2013 to October 2013 (Table 1). In both batches, the clones reproduced mostly asexually during summer and fall, with intermittent production of males observed in the MP cultures and ephippia, both in MP and NMP cultures. Even though there was no systematic quantification of ephippia production in this experiment, we did not notice any obvious differences in numbers of ephippia produced between NMP and MP cultures. However, all opened ephippia from NMP cultures were empty (i.e., did not contain embryos), whereas almost all ephippia from MP cultures contained embryos (several dozens of ephippia from each of the two culture types were opened). The results of the first batch suggested the possibility of clonal selection leading to substantially unequal clone frequencies in multiclone cultures and thus reduced probabilities of outcrossing (assuming presence of males and random mating). We therefore intermittently (June, July 15, August 25, and September 17, 2013) restocked all multiclone cultures of the second batch by adding up to 100 nonephippial females of the less frequent clones, after estimating clone frequencies based on microsatellite genotypes of 25 individuals of each culture. The aim of this procedure was to equilibrate clone frequencies and thus to increase the likelihood of outcrossing if rare males were present. Finally, six NMP cultures of the first batch were used to phenotypically search for rare males using large samples (∼4000 individuals) taken at the end of the growing season (November 2011), with sex identified under a stereomicroscope. The same was also done for two MP control cultures.
Table 1. Origins of clones, sex rations, number of hatchlings, as well as numbers of within-clone and outcrossed offspring in each of the cultures.
| Bucket ID | Batch | Treatment | Origin of clones | N males | N females | N hatchlings | N genotyped | N within-clone offspring | N outcrossed offspring |
|---|---|---|---|---|---|---|---|---|---|
| V02 | 2011 | NMP_single | Vol | 7 | 2 | 2 | 0 | ||
| V03 | 2011 | NMP_single | MZ | 0 | |||||
| V04 | 2011 | NMP_single | Ast | 28 | 14 | 14 | 0 | ||
| V08 | 2011 | NMP_single | Vol | 0 | 4629 | 3 | 3 | 3 | 0 |
| V10 | 2011 | NMP_single | Ast | 0 | 5370 | 3 | 3 | 3 | 0 |
| V21 | 2011 | NMP_single | MZ | 8 | 5 | 5 | 0 | ||
| B11 | 2013 | NMP_single | Ast | 11 | |||||
| B12 | 2013 | NMP_single | Vol | 0 | |||||
| B13 | 2013 | NMP_single | MZ | 1 | |||||
| B14 | 2013 | NMP_single | Ast | 0 | |||||
| B15 | 2013 | NMP_single | Ast | 0 | |||||
| V01 | 2011 | NMP_mix | MZ, Vol | 13 | 3 | 3 (same parent) | 0 | ||
| V05 | 2011 | NMP_mix | BN, Vol | 11 | 7 | 7 (same parent) | 0 | ||
| V06 | 2011 | NMP_mix | MZ, Vol | 1 | 1 | 1 | 0 | ||
| V07 | 2011 | NMP_mix | BN, Vol | 0 | 5105 | 1 | |||
| V09 | 2011 | NMP_mix | MZ, Vol | 0 | 4256 | 10 | 3 | 3 (same parent) | 0 |
| V11 | 2011 | NMP_mix | BN (2x) | 0 | 5550 | 0 | |||
| V12 | 2011 | NMP_mix | Ast, BN, MZ, Vol | 2 | 2 | 1+1 (two different parents) | 0 | ||
| V15 | 2011 | NMP_mix | MZ (2x) | 0 | 1015 | 1 | |||
| V17 | 2011 | NMP_mix | MZ (2x) | 4 | 4 | 4 (same parent) | 0 | ||
| V19 | 2011 | NMP_mix | Ast, BN, MZ, Vol | 1 | |||||
| V20 | 2011 | NMP_mix | BN (2x) | 0 | |||||
| B20 | 2013 | NMP_mix | Ast, MZ, Vol | 1 | 1 | 1 | 0 | ||
| B21 | 2013 | NMP_mix | Ast (3x) | 0 | |||||
| B23 | 2013 | NMP_mix | Ast, MZ, Vol | 3 | 3 | 1+2 (two different parents) | 0 | ||
| B24 | 2013 | NMP_mix | Ast (3x) | 0 | |||||
| B26 | 2013 | NMP_mix | Ast, MZ, Vol | 1 | 1 | 1 | 0 | ||
| B27 | 2013 | NMP_mix | Ast (3x) | 0 | |||||
| B17 | 2013 | MP_single | MZ | 0 | |||||
| B18 | 2013 | MP_single | MZ | >30 | |||||
| B19 | 2013 | MP_single | MZ | >30 | |||||
| D069 | 2011 | MP_mix | BN, Vol | 53 | 440 | >30 | |||
| D096 | 2011 | MP_mix | BN, Vol | 82 | 232 | >30 | |||
| D141 | 2011 | MP_mix | BN, Vol | 142 | 224 | >30 | |||
| D202 | 2011 | MP_mix | BN, Vol | 109 | 220 | >30 | |||
| B22 | 2013 | MP_mix | MZ (4x) | >30 | 8 | 0 | 8 | ||
| B25 | 2013 | MP_mix | MZ (4x) | >30 | 8 | 1 | 7 | ||
| B28 | 2013 | MP_mix | MZ (4x) | >30 | 8 | 0 | 8 |
Empty cells indicate values that were not assessed in a given culture.
At the end of each growing season (mid November 2011, end of October 2013 for the first and second batches, respectively), all ephippia that had accumulated at the bottom of the buckets were collected and overwintered (which is necessary for later hatching). Overwintering was done either outdoors in a small volume of water placed in the dark (first batch) or in a dark cold room at 0° (second batch). In the subsequent spring, hatching tests were carried out by transferring the ephippia to fresh Daphnia medium and keeping them under warm and high-light conditions (ambient Fribourg spring conditions in the first batch, ∼20° greenhouse conditions in the second batch). The containers were carefully checked for hatchlings at least every 3rd day, and hatchlings were removed and stored in ethanol at −20° for later genotyping or grown in isolation to establish cultures of offspring clones. Overall, the 2011 batch yielded more hatchlings than the 2013 batch, likely due to environmental effects during growth or hatching.
DNA extraction and microsatellite analysis
Genomic DNA was extracted using the HotSHOT protocol (Montero-Pau et al. 2008) and nine diagnostic microsatellite loci (Table S1) were used to distinguish outcrossed from nonoutcrossed offspring (the latter resulting from within-clone mating or parthenogenetic reproduction), as well as to check for segregation of markers that were heterozygous in the parent clones. We set up PCR reactions of 10 µl, using the Qiagen Multiplex PCR master mix (Qiagen, Venlo, The Netherlands). Cycling was performed following the recommendations of the manufacturer. Fragment lengths were analyzed using GeneMapper Software version 4.0 (Applied Biosystems, Foster City, CA) with GeneScan-500 LIZ as an internal size standard.
RAD sequencing
To obtain markers throughout the genome, at which heterozygosity could be assessed, we used RAD sequencing (Baird et al. 2008), using eight hatchlings from a single-clone NMP culture (clone AST-01-04, bucket V04), as well as 27 hatchlings from a single-clone MP culture (clone RM1-18 MP, bucket B19). Only eight offspring of an NMP clone were used because this was the highest number of offspring from a single-clone NMP culture that could successfully be grown in clonal culture in the laboratory before DNA extraction (several other hatchlings died before reproduction or were sterile). We used a RAD-sequencing protocol based on Etter et al. (2011) with a few modifications as specified below. Two libraries were prepared: one containing the offspring of the NMP single-clone culture, the other containing the offspring of the MP single-clone culture, with each offspring individually labeled. Each library also contained two independent replicates of the parental clone. The details of the RAD-sequencing protocol and the analysis pipeline including quality checks, alignment, SNP calling, and genotype calling, are explained in detail in File S3.
Inter- and intrachromosomal patterns of heterozygosity reduction
Putative centromere locations were inferred from the genetic map as corresponding to large, nonrecombining regions, of which each linkage group contains exactly one, except linkage group 3 which has two such regions (D. magna genetic map v4.0.1, File S6; M. Dukić et al., unpublished results). Centromeric regions were defined as consisting of all scaffolds (or parts of scaffolds) with the centimorgan position of these nonrecombining regions. Average heterozygosity as a function of the distance from the putative centromere was calculated for each chromosome arm separately by using a moving average, including markers within 5 cM on either side of the focal marker (but in all cases excluding markers at a distance of 0 cM from the centromeric regions). Subsequently, the averages and standard errors (SE) of these estimates were calculated across chromosome arms, and confidence limits were calculated as 1.96 SE.
To estimate the distance from the centromere of microsatellite loci, we first mapped each primer pair to the current D. magna assembly v2.4. Subsequently, we retrieved the position on the genetic map v4.0.1 of the closest marker on the same scaffold. In this way, we were able to obtain estimated map locations for six of the microsatellite loci (Table S2).
Probability of within-clone mating in the presence of rare males
The absence of outcrossed offspring in NMP multiclone cultures does not necessarily indicate the absence of rare males because a low number of offspring could, by chance be produced exclusively by within-clone mating. Hence, we calculated the probability of observing zero outcrossed offspring in the presence of rare males under the assumption of random mating among the clones present at the time of resting egg production in each NMP multiclone culture. Under random mating, the probability of within-clone mating of a given clone i in a given culture j is equal to its squared frequency, fi2, and the overall expected frequency of within-clone mated offspring is Σ(fi2), summed across all clones present in the culture. The probability of observing only offspring produced by within-clone mating among N offspring (i.e., the probability that despite the presence of males not a single outcrossed offspring was observed) then equals prj = [Σ(fi2)]N, and the combined probability across all cultures is the product Π(prj).
Because the frequencies of clones at the time of resting egg production were unknown, we assumed two contrasting scenarios: First, we assumed that all original parent clones were still present at equal frequency at the time of resting egg production. This scenario maximizes the probability of outcrossing. Therefore, we also used a second, more conservative scenario: We assumed that the frequency of each parent clone at the moment of resting egg production was equal to its proportional contribution to the offspring generation. For this second scenario, we only used buckets in which offspring from more than one parent clone were present (for the other buckets, the expected frequency of within-clone mated offspring under this scenario is 100%).
Data availability
All demultiplexed read data used for genotyping were submitted to the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA): BioProject ID PRJNA279333. The reference genome used for mapping and annotation is available at http://wfleabase.org/ (dmagna_v.2.4_20100422). The full raw and corrected SNP datasets, as well as the genetic map v4.0.1 are available as supporting information (File S4, File S5, and File S6).
Results
Sex ratios
We identified the sex of 25,925 NMP individuals of D. magna, sampled in late season from six outdoor cultures of NMP clones, but did not find a single male (Table 1). At the same time, cultures of MP clones contained between 10.8% and 38.8% males (mean across populations = 27.2%, SE = 6.1%, total N = 1502). Combined with data from our earlier study (Galimov et al. 2011), we have now identified the sex of 33,764 NMP individuals but did not find a single male. This yields an overall upper 95% confidence limit for the true proportion of males of ∼10−4 (Clopper–Pearson confidence interval 1 − (α/2)1/N). However, the experiment involved many more individuals than the ones that were checked (>105 individuals across the whole duration of the experiment and all NMP cultures combined). Thus the presence of rare males cannot entirely be excluded through these phenotypic observations alone.
Number of hatchlings
A total of 110 hatchlings were found in NMP cultures (between 0 and 28 per culture, Table 1). Of these, 61 were found in cultures containing just a single NMP clone and 49 in cultures containing multiple NMP clones. All MP control cultures except one yielded >30 offspring (Table 1), some of them even many more. Even though numbers of hatchlings >30 were not estimated systematically, this fits with our experience from similar experiments, where MP cultures usually yielded hundreds to thousands of hatchlings, though in rare cases only low numbers or even none (e.g., Haag and Ebert 2007).
Microsatellite genotypes of offspring from cultures containing single NMP clones
In total, we investigated microsatellite genotypes of 27 offspring from cultures containing single NMP clones. In all cases, these offspring showed segregation of maternal alleles (Table S1), thus excluding clonal parthenogenesis. Average heterozygosity across all cultures and loci was 0.61 (SE = 0.04), which is significantly different from 0.5 (N = 152, χ2 = 6.7, P = 0.0094), but not from 0.67 (χ2 = 2.9, P = 0.090). Nonetheless, two loci had heterozygosities that were significantly lower than 0.67 (locus B008: N = 5, heterozygosity = 0, binomial P = 0.005, locus B096: N = 21, heterozygosity = 0.36, SE = 0.10, χ2 = 9.2, P = 0.0022) and, in one case, even significantly lower than 0.5 (locus B008: binomial P = 0.031, locus B096: χ2 = 1.6, P = 0.20). Indeed heterogeneity among loci was significant (generalized linear model with binomial error distribution using Firth bias correction, likelihood ratio test, χ2 = 23.6, d.f. = 6, P = 0.0006). In contrast, offspring from different cultures or different individuals within cultures did not significantly vary in heterozygosity (tested in the same model as the loci effects, cultures: χ2 = 4.4, d.f. = 4, P = 0.35, individuals nested within cultures: χ2 = 20.9, d.f. = 22, P = 0.53). The heterogeneity among loci was at least partly explained by the distance from the centromere: The two loci with heterozygosities significantly lower than 0.67 (loci B008 and B096) were the two loci estimated to be most closely linked to a centromere (at 25.8 and 3.6 cM, respectively). All other loci had estimated distances from the centromere of >32 cM (Table S2).
Microsatellite genotypes of offspring from cultures containing multiple clones
We obtained microsatellite genotypes of 25 offspring from cultures containing multiple NMP clones (Table 1). Among these, not a single offspring resulting from outcrossing between two of the parent clones was observed. Rather, all 25 offspring were produced by self-fertilization or automictic parthenogenesis: They showed segregation of maternal alleles, just as offspring of the single-clone cultures, but no sign of outcrossing between clones at diagnostic loci (Table 1, Table S1). In all but two of these cultures, all offspring found within the culture were produced by just one parent clone. Two cultures (V12 and B23) contained offspring from two different parent clones (Table 1), but nonetheless no outcrossed clone was observed (i.e., also these offspring were the result of self-fertilization or automictic parthenogenesis). In stark contrast, 23 of 24 genotyped offspring from cultures containing multiple MP clones were the result of outcrossing between two parent clones (Table 1, Table S1). The frequency of outcrossed offspring was even significantly higher (χ2 = 5.6, P = 0.018) than the 75% expected under random mating and equal frequencies of each of the four parent clones, likely due to inbreeding depression affecting hatching rates of inbred vs. outcrossed offspring.
Even though no outcrossed offspring was observed in the cultures containing multiple NMP clones, there is still a possibility that they were produced by the mating of a rare male with a female of the same clone. Assuming random mating of rare males with all females, the overall probability of observing zero outcrossed offspring among the 25 genotyped individuals from the cultures containing multiple NMP clones was calculated under two extreme scenarios (see Materials and Methods). Under the first scenario (assuming equal frequency of all introduced parent clones), this probability is very low (∼10−9). The second scenario (frequency of parent clones equal to their contribution to the offspring generation) could be assessed only for the five offspring from cultures V12 and B23, in which self-fertilized/automictic offspring from more than one parent were present. Under this scenario, the probability of observing zero outcrossed offspring among the five offspring in these cultures is 0.043.
Genome-wide patterns of heterozygosity assessed by RAD sequencing
RAD sequencing was carried out on eight offspring of the AST-01-04 NMP clone (the remaining offspring of this clone died or did not reproduce and therefore it was not possible to obtain sufficient amounts of DNA). Average genome-wide heterozygosity of these eight offspring was 0.54 (N = 2523 loci). It ranged from 0.24 to 0.67 among linkage groups and from 0.40 to 0.73 among individuals (Table S3). The relatively high variation among individuals and linkage groups was expected because only few recombination events occur per meiosis and chromosome (Routtu et al. 2010, 2014), so that many linked markers show identical inheritance patterns.
As a control, RAD sequencing was also carried out on 27 offspring of the RM1-18 MP clone. Average genome-wide heterozygosity among these offspring was 0.60 (N = 1610 loci), varying among linkage groups between 0.46 and 0.71 (Table S3). This was significantly higher than 0.5 (linkage groups as independent replicates, t = 3.7, d.f. = 9, P = 0.005), but also significantly lower than 2/3 (t = −2.7, d.f. = 9, P = 0.023, though not quite significantly so when linkage groups were weighed according to the number of loci: P = 0.067).
Interchromosomal patterns of heterozygosity at putative centromere regions
The analysis carried out here requires knowledge of centromere regions. We use putative centromere regions (large, nonrecombining regions as identified on each linkage group by the genetic map v4.0.1, which maps most scaffolds of the current D. magna assembly; M. Dukić et al., unpublished results). An equivalent analysis that does not require assumptions on putative centromere regions is presented in File S1.
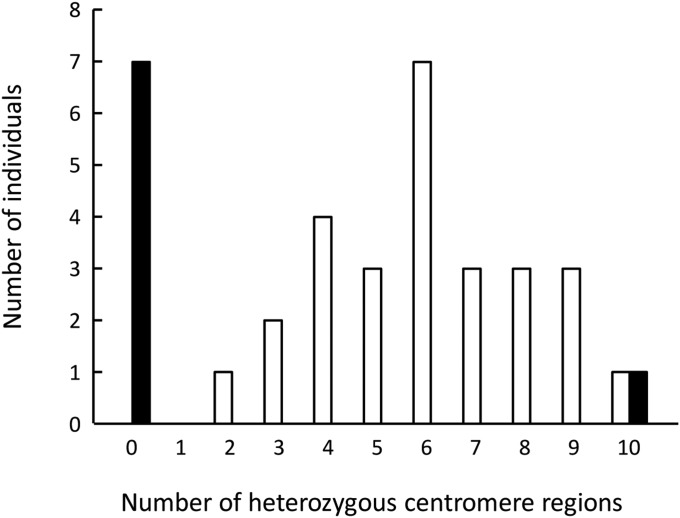
The putative centromere regions were either consistently homozygous (seven offspring) or heterozygous (one offspring: V04_04) across all 10 linkage groups (Figure 2). One of the linkage groups (LG3) contained two such regions, but only the region at 90.8 cM showed the same heterozygosity as the putative centromere regions of the remaining linkage groups. Hence, this, rather than the region at 62.5 cM, is the likely centromere region of LG3 (and only this region was considered for all other analyses). The interchromosomal pattern thus strongly suggests that V04_04 was produced by central fusion and the other seven offspring by terminal fusion.
Figure 2.
Observed number of individuals that retained parental heterozygosity at a given number (out of 10) of centromeric regions. Solid bars represent offspring of the AST-01-04 NMP clone, open bars, offspring of the RM1-18 MP clone. For LG3 only the region at 90.8 cM was considered.
In contrast, the putative centromere regions were not consistently homozygous or heterozygous across all linkage groups within individual offspring of the RM1-18 MP clone, except for one individual, in which all 10 centromere regions were heterozygous (Figure 2). Using the observed average heterozygosity of 0.60, the probability of observing this at least one time by chance among 27 offspring is ∼0.15 (p = 0.610 is the probability that an individual is heterozygous for the 10 centromeric regions, (1 − p)27 that none of the 27 offspring is heterozygous for the 10 centromeric regions, and thus 1− (1 − p)27 is the probability that at least 1 is heterozygous for the 10 centromeric regions).
Intrachromosomal patterns of heterozygosity
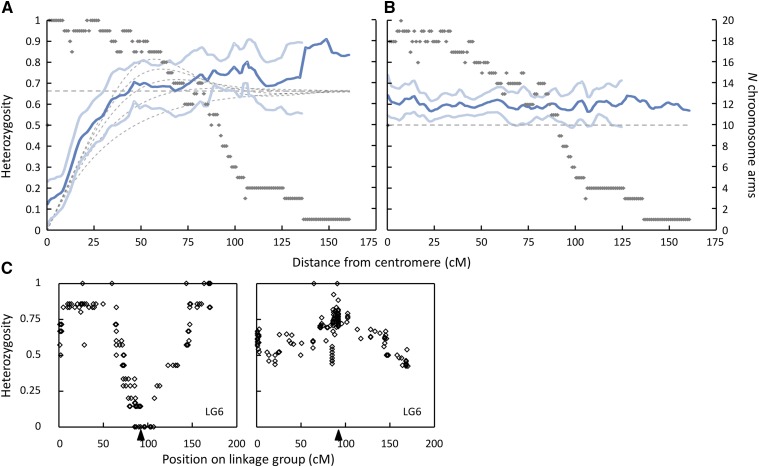
Among the seven offspring of the NMP clone that were presumably produced by terminal fusion, heterozygosity gradually increased with distance from the centromere (Figure 3) and reached an average heterozygosity of clearly >0.5 already at a distance of 50 cM. Heterozygosity in distal regions was even somewhat higher than the expected 0.67, though this was only marginally significant (the lower 95% confidence interval calculated by using chromosome arms as independent replicates mostly included 0.67). Heterozygosity of the individual produced by central fusion (offspring V04_04) averaged across all markers and all chromosome arms was 0.73, and 0.67 if only markers located >50 cM from the centromere were considered. In contrast, heterozygosity among the 27 offspring of the RM1-18 MP clone did not vary in any systematic way along the chromosomes nor according to the distance from the centromere (Figure 3).
Figure 3.
Heterozygosity as a function of the distance from the centromere under (A) automixis (terminal fusion only, N = 7 offspring) and (B) self-fertilization (N = 27 offspring). Dark blue lines represent averages across all chromosome arms with N chromosome arms (gray dots) according to the secondary y-axis. Light blue lines represent the 95% confidence limits, and the dashed lines, the expected heterozygosity and asymptotes under different degrees of recombination interference (see Figure 1). (C) Realized heterozygosity along linkage group 6 (automictic offspring, left; self-fertilized offspring, right) for illustration. The black triangle shows the presumed centromere position. The patterns of all linkage groups are shown in Figure S1. All heterozygosities are expressed in percentage of parental heterozygosity.
Discussion
Reliable distinction of automixis from self-fertilization requires genomic data
Our results demonstrate that the ephippial offspring of the D. magna NMP clone investigated by RAD sequencing were produced by automixis, mostly but not exclusively by terminal fusion. The microsatellite results on the offspring of the other clones strongly suggest that this was also the case for the offspring of the other clones (lower heterozygosity at centromere–proximal markers). However, an unequivocal distinction from self-fertilization was only possible owing to the use of genomic data, which also allowed confirmation of the centromere locations. While the phenotypic results were suggestive of automixis (no males detected in NMP cultures, no cross-breeding observed in cultures containing multiple NMP clones), they were nonetheless not entirely conclusive. Very low male frequencies could not be excluded, despite large sample sizes, and the expectation that the presence of males would lead to outcrossed offspring was based on specific assumptions that could not be verified. Moreover, the average microsatellite heterozygosity was similar to the average genome-wide heterozygosity of the self-fertilized controls and therefore also inconclusive. Also at RAD loci, automictic offspring retained 54% of parental heterozygosity, due to a predominance of terminal rather central fusion and due to inclusion of both centromere–distal and centromere–proximal markers. If only average heterozygosity had been assessed, this could easily have been mistaken as consistent with self-fertilization rather than automixis (average observed heterozygosity in self-fertilized offspring was 60%). This shows that average offspring heterozygosity is not necessarily a reliable indicator of the breeding system.
Unequivocal evidence for automixis was obtained only when the interchromosomal and intrachromosomal patterns of genome-wide heterozygosity were analyzed. These patterns are clearly inconsistent with self-fertilization, as shown by our parallel analysis of self-fertilized controls. In addition, the interchromosomal patterns provide a direct estimate of the proportion of offspring produced by terminal vs. central fusion: Offspring produced by terminal fusion are homozygous at the centromeric regions of all chromosomes; offspring produced by central fusion retain full parental heterozygosity at all these regions (see also Oldroyd et al. 2008). Even if the positions of the centromeres are unknown, the interchromosomal patterns can be analyzed by investigating if specific segregation patterns (among individuals) occur consistently on all chromosomes (File S1).
The intrachromosomal patterns of heterozygosity in automictic offspring can be used to map the centromeres, an approach that has been used both in natural automicts (Barratt et al. 1954, 2004) and in organisms in which automixis can be induced artificially (or meiotic tetrads or half-tetrads can be recovered by other means) (Lindsley et al. 1956; Eppig and Eicher 1983; Johnson et al. 1996; Zhu et al. 2013). Also in our study, the intrachromosome heterozygosity patterns among offspring produced by terminal fusion confirm the presumed locations of centromeres in D. magna and indicate that all D. magna chromosomes are metacentric. Furthermore, our results also indicate that the centromere on LG3 is located at 90.8 cM rather than 62.5 cM (two large, nonrecombining regions are found on this linkage group; Dukić et al., unpublished results).
Even though each centromeric region did contain markers that were homozygous in all individuals produced by terminal fusion, there were also, in each of these regions, some markers that were not fully homozygous (average heterozygosity across centromeric regions was 12%, Figure 3). These 12% of unexpected genotypes are likely due to a combination of genotyping error (false heterozygote calls), erroneous mapping of reads from paralogous loci to single loci (e.g., centromeric markers that were heterozygous in all individuals, as found on several linkage groups, Figure S1), errors in the genetic map and/or noncollinearity between chromosomes in our study population compared to the clones on which the D. magna map is based. Errors in the genetic map and noncollinearity would have the effect that loci that were mapped by us to the centromeric regions are in reality not in these regions, which could explain their nonzero heterozygosity.
Automixis and diversity of breeding systems in Daphnia
While most Daphnia species have so far been thought to produce diapause stages exclusively by sexual reproduction, a few species regularly produce parthenogenetic diapause stages (e.g., obligate parthenogenetic strains of D. pulex; Hebert and Crease 1980). Yet, in these cases, offspring do not show segregation of maternal alleles and are therefore believed to be clonal offspring, just as the offspring resulting from parthenogenetic production of subitaneous (directly developing) eggs during the regular asexual part of the life cycle in Daphnia (Hebert and Ward 1972, but see comments on clonality below). Hence our results constitute the first demonstration of classical automixis in Daphnia.
The finding of rare automixis in NMP clones of D. magna is important for our understanding of the NMP/MP polymorphism. Nonmale producing clones of D. magna participate in sexual reproduction only through their female function (Galimov et al. 2011) and were therefore speculated to be unable to colonize new populations on their own. However, rare automixis may allow these populations to persist through the first period of diapause and may therefore allow single NMP clones to colonize new populations. This may be especially important in environments with frequent extinction–colonization dynamics.
However, there is no reason to believe that rare automixis is limited to NMP clones. Production of empty ephippia (shells of resting stages not containing embryos) is frequently observed also in MP clones, when females are grown in the laboratory in isolation from males and even in nature (Ebert 2005). Yet, possible rare automictic reproduction in these cultures would be much more difficult to detect than in NMP clones because MP clones regularly produce males under the conditions needed to stimulate ephippia production. Indeed, rare automictic parthenogenesis occurs in a large number of organisms in the form of rare, spontaneous hatching of unfertilized eggs (“tychoparthenogenesis”) with diploidy restored via automixis (Bell 1982; Schwander et al. 2010; Neiman et al. 2014).
In other species, automixis is a regular form of reproduction, but often exclusively or almost exclusively with central fusion. Examples are many fungal species, including yeast, in which central fusion during within-tetrad mating (= automixis) is assured by a mating type locus (unless there is a mating type switch or recombination between the centromere and the mating type locus) (Antonovics and Abrams 2004). Another example comes from several social insects, which can reproduce parthenogenetically by central fusion, and, additionally, show very low rates of crossover within the chromosomes (Baudry et al. 2004; Oldroyd et al. 2008; Rey et al. 2011). With very low recombination rates, central fusion effectively approaches clonality because central fusion assures that parental heterozygosity is retained at the centromeres, and low rates of crossover result only in a slow decay of heterozygosity in the centromere–distal regions.
Automixis has been hypothesized to represent an intermediate step in the evolutionary pathway from sexual to clonal reproduction (Schwander et al. 2010). According to this hypothesis, rare automictic reproduction (tychoparthenogenesis) with mixed fusion may become more frequent, with subsequent selection for increased rates of central fusion and repression of recombination. Parthenogenetic reproduction in Daphnia has indeed been termed an intermediate between clonal and automictic reproduction because subitaneous, parthenogenetic eggs are produced by a modified meiosis rather than by mitosis (Hiruta et al. 2010; Hiruta and Tochinai 2012). Specifically, the homologs pair and start to separate, but meiosis I is not completed, and sister chromatids of a diploid set of chromosomes are separated during meiosis II (Hiruta et al. 2010). In other words, meiosis I is suppressed, which is identical to central fusion (Asher 1970), but it is also indistinguishable from purely clonal reproduction as long as no recombination occurs during the paring of homologs (a low degree of exchange occurs in D. pulex, Omilian et al. 2006; Tucker et al. 2013). More generally, this suggests that the mechanism of clonal parthenogenesis may, also in other diploid organisms, be an extreme form of automixis (central fusion with no or very low levels of recombination) and thus a meiosis-derived process rather than production of fertile eggs by mitosis. This distinction is important because it implies that the evolution of asexuality in these taxa may have involved strong heterozygosity-reducing processes (terminal fusion, central fusion with recombination). Hence the classical view that clonal diploids maintain their heterozygosity at least on short evolutionary timescales (thus for instance avoiding inbreeding, e.g., Haag and Ebert 2004) may not have been true during the initial evolution of asexuality. Hence it may be necessary to more explicitly account for the mechanism of this transition to fully understand the selection pressures acting during the evolution of parthenogenesis from sexuality. Furthermore, the same processes may also be important for understanding the maintenance of asexuality: If clonal parthenogenesis is indeed meiosis derived, there may be residual rates of recombination during homolog pairing (Hiruta et al. 2010), such that transitions to homozygosity and loss of complementation may occur at higher rates than under purely mitotic parthenogenesis (Archetti 2004, 2010; Nougué et al. 2015).
Inbreeding depression in self-fertilized and automictic offspring
In the self-fertilized offspring, observed heterozygosities were higher than the expected 50% for the majority of the linkage groups. This suggests that the parent clone carried loci contributing to inbreeding depression, that is, loci with recessive or partly recessive deleterious alleles on these linkage groups (Fu and Ritland 1994b). Indeed, the realized heterozygosities can deviate from the expected ones in inbred individuals due to selection, and such deviations are a form of inbreeding depression (Fu and Ritland 1994a; Wang and Hill 1999). Also the higher than expected number of outcrossed offspring in the cultures containing multiple MP clones is evidence for inbreeding depression in the control cultures.
The automictic offspring also showed signs of inbreeding depression: Only few hatchlings survived to adulthood and were sufficiently fecund so that they could successfully be taken into clonal culture (Table 1). Furthermore, observed offspring heterozygosities also tended to be higher than the expected ones, even after accounting for high levels of crossover interference. A closer examination of the contribution of selection to the genome-wide patterns of observed heterozygosity is not possible due to the low number of automictic offspring investigated, and also due to complicating effects of possible genotyping errors and other possible errors (alignment, mapping, collinearity, see above) in our analysis. Due to these uncertainties, our prediction that the initial increase in heterozygosity at short distances from the centromere should be 2d (where d is the genetic distance in morgans), if it is not influenced by selection, could not be evaluated with the present data. Nonetheless, the strong initial increase in heterozygosity at distances up to 100 cM from the centromere is inconsistent with the absence of both recombination interference and selection, but rather indicates the action of one or both of these processes. If a larger number of offspring is analyzed and selection is estimated independently (e.g., by analyzing loci at >100 cM from the centromere) or can be excluded (e.g., by investigating zygotes), the analysis of heterozygosity patterns among automictic offspring may be used to investigate the degree of crossover interference.
Conclusions
Overall, our study shows that the mode of reproduction in automictic vs. self-fertilizing species can be inferred from the heterozygosity patterns among offspring. However, our study also illustrates that it was only due to the availability of genomic rather than sparse marker data that these inferences were robust to the complicating effects of recombination interference and selection. The same applies to the distinction between terminal and central fusion in species that use a mix of these two modes of reproduction (not necessarily in the ratios corresponding to random fusion). More generally, our findings support the idea that obtaining genome-wide heterozygosity data from mothers and a limited number of offspring may be a widely applicable and accessible approach to study breeding systems in species with cryptic or mixed modes of reproduction.
Supplementary Material
Acknowledgments
We thank the Moscow Zoo and N. I. Skuratov for sampling permits and support and the Botanical Garden of Fribourg, J. Schöpfer, G. Jacob, B. Walser, J. Lohr, and D. Frey for help with outdoor cultures. We thank the Department of Biosystem Science and Engineering of the Eidgenössische Technische Hochschule Zurich, in particular C. Beisel and I. Nissen for Illumina sequencing of the RADseq libraries, and we gratefully acknowledge support by T. Mathieu, D. Deguedre, and the platform Terrain d’Expériences of the Laboratoire d'excellence (LabEx) Centre Méditerranéen de l'Environnement et de la Biodiversité (CeMEB), as well as M.-P. Dubois and the platform Service des marqueurs génétiques en écologie (SMGE) at CEFE. We also thank S. Gonzalez-Martinez and two anonymous reviewers for their constructive comments on an earlier version of the paper. This work was supported by the Swiss National Science Foundation (grant no. 31003A_138203), the Russian Foundation of Basic Research, Agence nationale de la recherche (project SilentAdapt), and by the European Union (Marie Curie career integration grant PCIG13-GA-2013-618961, DamaNMP). The authors declare no competing financial interests.
Footnotes
Communicating editor: S. Gonzalez-Martinez
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179879/-/DC1.
Literature Cited
- Antonovics J., Abrams J. Y., 2004. Intratetrad mating and the evolution of linkage relationships. Evolution 58: 702–709. [DOI] [PubMed] [Google Scholar]
- Archetti M., 2004. Recombination and loss of complementation: a more than two-fold cost for parthenogenesis. J. Evol. Biol. 17: 1084–1097. [DOI] [PubMed] [Google Scholar]
- Archetti M., 2010. Complementation, genetic conflict, and the evolution of sex and recombination. J. Hered. 101: S21–S33. [DOI] [PubMed] [Google Scholar]
- Asher J. H., 1970. Parthenogenesis and genetic variability. II. One-locus models for various diploid populations. Genetics 66: 369–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt R. W., Newmeyer D., Perkins D. D., Garnjobst L., 1954. Map construction in Neurospora crassa. Adv. Genet. 6: 1–93. [DOI] [PubMed] [Google Scholar]
- Baudry E., Kryger P., Allsopp M., Koeniger N., Vautrin D., et al. , 2004. Whole-genome scan in thelytokous-laying workers of the Cape honeybee (Apis mellifera capensis): Central fusion, reduced recombination rates and centromere mapping using half-tetrad analysis. Genetics 167: 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G., 1982. The Masterpiece of Nature: The Evolution and Genetics of Sexuality. University of California Press, Berkeley. [Google Scholar]
- Bos C. J. E. (Editor), 1996. Fungal Genetics: Principles and Practice. Marcel Dekker, New York. [Google Scholar]
- Charlesworth D., Willis J. H., 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10: 783–796. [DOI] [PubMed] [Google Scholar]
- David P., Pujol B., Viard F., Castella V., Goudet J., 2007. Reliable selfing rate estimates from imperfect population genetic data. Mol. Ecol. 16: 2474–2487. [DOI] [PubMed] [Google Scholar]
- Delmotte F., Leterme N., Gauthier J. P., Rispe C., Simon J. C., 2002. Genetic architecture of sexual and asexual populations of the aphid Rhopalosiphum padi based on allozyme and microsatellite markers. Mol. Ecol. 11: 711–723. [DOI] [PubMed] [Google Scholar]
- Ebert, D., 2005 Ecology, Epidemiology, and Evolution of Parasitism in Daphnia. Bethesda: National Library of Medicine National Center for Biotechnology Information, Bethesda, MD. Available at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books. [Google Scholar]
- Engelstädter J., Sandrock C., Vorburger C., 2011. Contagious parthenogenesis, automixis, and a sex determination meltdown. Evolution 65: 501–511. [DOI] [PubMed] [Google Scholar]
- Eppig J. T., Eicher E. M., 1983. Application of the ovarian teratoma mapping method in the mouse. Genetics 103: 797–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter P. D., Preston J. L., Bassham S., Cresko W. A., Johnson E. A., 2011. Local de novo assembly of RAD paired-end contigs using short sequencing reads. PLoS One 6: e18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.-B., Ritland K., 1994a Evidence for the partial dominance of viability genes in Mimulus guttatus. Genetics 136: 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.-B., Ritland K., 1994b On estimating the linkage of marker genes to viability genes controlling inbreeding depression. Theor. Appl. Genet. 88: 925–932. [DOI] [PubMed] [Google Scholar]
- Galimov Y., Walser B., Haag C. R., 2011. Frequency and inheritance of non male producing clones in Daphnia magna: Evolution towards sex specialization in a cyclical parthenogen? J. Evol. Biol. 24: 1572–1583. [DOI] [PubMed] [Google Scholar]
- Haag C. R., Ebert D., 2004. A new hypothesis to explain geographic parthenogenesis. Ann. Zool. Fenn. 41: 539–544. [Google Scholar]
- Haag C. R., Ebert D., 2007. Genotypic selection in Daphnia populations consisting of inbred sibships. J. Evol. Biol. 20: 881–891. [DOI] [PubMed] [Google Scholar]
- Hartl D. L., Clark A. G., 2007. Principles of Population Genetics. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Hebert P. D. N., Ward R. D., 1972. Inheritance during parthenogenesis in Daphnia magna. Genetics 71: 639–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P. D. N., Crease T., 1980. Clonal coexistence in Daphnia pulex (Leydig): another planktonic paradox. Science 207: 1363–1365. [Google Scholar]
- Hiruta C., Tochinai S., 2012. Spindle assembly and spatial distribution of γ-tubulin during abortive meiosis and cleavage division in the parthenogenetic water flea Daphnia pulex. Zoolog. Sci. 29: 733–737. [DOI] [PubMed] [Google Scholar]
- Hiruta C., Nishida C., Tochinai S., 2010. Abortive meiosis in the oogenesis of parthenogenetic Daphnia pulex. Chromosome Res. 18: 833–840. [DOI] [PubMed] [Google Scholar]
- Hood M. E., Antonovics J., 2004. Mating within the meiotic tetrad and the maintenance of genomic heterozygosity. Genetics 166: 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes D. J., Dunbrack R. L., 1993. Sex allocation variation in Daphnia pulex. J. Evol. Biol. 6: 559–575. [Google Scholar]
- Johnson S. L., Gates M. A., Johnson M., Talbot W. S., Horne S., et al. , 1996. Centromere-linkage analysis and consolidation of the Zebrafish genetic map. Genetics 142: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klüttgen B., Dulmer U., Engels M., Ratte H. T., 1994. ADaM, an artificial freshwater for the culture of zooplankton. Water Res. 28: 743–746. [Google Scholar]
- Lampert K. P., Lamatsch D. K., Fischer P., Epplen J. T., Nanda I., et al. , 2007. Automictic reproduction in interspecific hybrids of poeciliid fish. Curr. Biol. 17: 1948–1953. [DOI] [PubMed] [Google Scholar]
- Lindsley D. L., Fankhauser G., Humphrey R. R., 1956. Mapping centromeres in the axolotl. Genetics 41: 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutes A. A., Neaves W. B., Baumann D. P., Wiegraebe W., Baumann P., 2010. Sister chromosome pairing maintains heterozygosity in parthenogenetic lizards. Nature 464: 283–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogie M., 1986. Automixis: its distribution and status. Biol. J. Linn. Soc. Lond. 28: 321–329. [Google Scholar]
- Montero-Pau J., Gómez A., Muñoz J., 2008. Application of an inexpensive and high-throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs. Limnol. Oceanogr. Methods 6: 218–222. [Google Scholar]
- Nace G. W., Richards C. M., Asher J. H., 1970. Parthenogenesis and genetic variability. I. Linkage and inbreeding estimations in the frog, Rana pipiens. Genetics 66: 349–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman M., Sharbel T. F., Schwander T., 2014. Genetic causes of transitions from sexual reproduction to asexuality in plants and animals. J. Evol. Biol. 6(27): 1346–1359. [DOI] [PubMed] [Google Scholar]
- Normark B. B., 2003. The evolution of alternative genetic systems in insects. Annu. Rev. Entomol. 48: 397–423. [DOI] [PubMed] [Google Scholar]
- Nougué, O., N. O. Rode, R. Zahab, A. Ségard, L.-M. Chevin et al., 2015 Automixis in Artemia solving a century-old controversy. J. Evol. Biol., in revision ( 10.1111/jeb.12757). [DOI] [PubMed]
- Oldroyd B. P., Allsopp M. H., Gloag R. S., Lim J., Jordan L. A., et al. , 2008. Thelytokous parthenogenesis in unmated queen honeybees (Apis mellifera capensis): central fusion and high recombination rates. Genetics 180: 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead A. W., Leblanc G. A., 2002. Juvenoid hormone methyl farnesoate is a sex determinant in the crustacean Daphnia magna. J. Exp. Zool. 293: 736–739. [DOI] [PubMed] [Google Scholar]
- Omilian A. R., Cristescu M. E. A., Dudycha J. L., Lynch M., 2006. Ameiotic recombination in asexual lineages of Daphnia. Proc. Natl. Acad. Sci. USA 103: 18638–18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearcy M., Hardy O., Aron S., 2006. Thelytokous parthenogenesis and its consequences on inbreeding in an ant. Heredity 96: 377–382. [DOI] [PubMed] [Google Scholar]
- Pearcy M., Hardy O. J., Aron S., 2011. Automictic parthenogenesis and rate of transition to homozygosity. Heredity 107: 187–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey O., Loiseau A., Facon B., Foucaud J., Orivel J., et al. , 2011. Meiotic recombination dramatically decreased in thelytokous queens of the little fire ant and their sexually produced workers. Mol. Biol. Evol. 28: 2591–2601. [DOI] [PubMed] [Google Scholar]
- Rizet G., Engelmann C., 1949. Contribution à l’étude génétique d’un Ascomycète tétrasporé: Podospora anserina (Ces.). Rehm. Rev. Cytol. Biol. Veg. 11: 201–304. [Google Scholar]
- Routtu J., Jansen B., Colson I., De Meester L., Ebert D., 2010. The first-generation Daphnia magna linkage map. BMC Genomics 11: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routtu J., Hall M., Albere B., Beisel C., Bergeron R., et al. , 2014. An SNP-based second-generation genetic map of Daphnia magna and its application to QTL analysis of phenotypic traits. BMC Genomics 15: 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander T., Vuilleumier S., Dubman J., Crespi B. J., 2010. Positive feedback in the transition from sexual reproduction to parthenogenesis. Proc. Biol. Sci. 277: 1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler J., Schäffer K., 1960. Untersuchungen über die Entstehung der Parthenogenese bei Solenobia triquetrella F. R. (Lepidoptera, Psychidae). Chromosoma 11: 29–102.14444635 [Google Scholar]
- Sellers K. F., Borle S., Shmueli G., 2012. The COM-Poisson model for count data: a survey of methods and applications. Appl. Stochastic Models Data Anal. 28: 104–116. [Google Scholar]
- Stenberg P., Saura A., 2009. Cytology of asexual animals, pp. 63–74 in Lost Sex. The Evolutionary Biology of Parthenogenesis, edited by Schön I., Martens K., van Dijk P. Springer Science, Dordrecht, The Netherlands. [Google Scholar]
- Suomalainen E., Saura A., Lokki J., 1987. Cytology and Evolution in Parthenogenesis, CRC Press, Boca Raton, FL. [Google Scholar]
- Tucker A. E., Ackerman M. S., Eads B. D., Xu S., Lynch M., 2013. Population-genomic insights into the evolutionary origin and fate of obligately asexual Daphnia pulex. Proc. Natl. Acad. Sci. USA 110: 15740–15745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K. W., 1958. Automictic parthenogenesis in the honey bee. Genetics 43: 299–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. L., Hill W. G., 1999. Effect of selection against deleterious mutations on the decline in heterozygosity at neutral loci in closely inbreeding populations. Genetics 153: 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini F., Sabelli B., 1972. Karyologic observations on the maturation of the summer and winter eggs of Daphnia pulex and Daphnia middendorffiana. Chromosoma 36: 193–203. [DOI] [PubMed] [Google Scholar]
- Zhao H., Speed T. P., 1998. Statistical analysis of ordered tetrads. Genetics 150: 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Sun Y., Yu X., Tong J., 2013. Centromere localization for bighead carp (Aristichthys nobilis) through half-tetrad analysis in diploid gynogenetic families. PLoS One 8: e82950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All demultiplexed read data used for genotyping were submitted to the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA): BioProject ID PRJNA279333. The reference genome used for mapping and annotation is available at http://wfleabase.org/ (dmagna_v.2.4_20100422). The full raw and corrected SNP datasets, as well as the genetic map v4.0.1 are available as supporting information (File S4, File S5, and File S6).