Abstract
Escherichia coli messenger RNAs (mRNAs) are rapidly degraded immediately after bacteriophage T4 infection, and the host RNase E contributes to this process. Here, we found that a previously uncharacterized factor of T4 phage, Srd (Similarity with rpoD), was involved in T4-induced host mRNA degradation. The rapid decay of ompA and lpp mRNAs was partially alleviated and a decay intermediate of lpp mRNA rapidly accumulated in cells infected with T4 phage lacking srd. Exogenous expression of Srd in uninfected cells significantly accelerated the decay of these mRNAs. In addition, lpp(T) RNA, with a sequence identical to the decay intermediate of lpp mRNA and a triphosphate at 5′-end, was also destabilized by Srd. The destabilization of these RNAs by Srd was not observed in RNase E-defective cells. The initial cleavage of a primary transcript by RNase E can be either direct or dependent on the 5′-end of transcript. In the latter case, host RppH is required to convert the triphosphate at 5′-end to a monophosphate. lpp(T) RNA, but not lpp and ompA mRNAs, required RppH for Srd-stimulated degradation, indicating that Srd stimulates both 5′-end-dependent and -independent cleavage activities of RNase E. Furthermore, pull-down and immunoprecipitation analyses strongly suggested that Srd physically associates with the N-terminal half of RNase E containing the catalytic moiety and the membrane target sequence. Finally, the growth of T4 phage was significantly decreased by the disruption of srd. These results strongly suggest that the stimulation of RNase E activity by T4 Srd is required for efficient phage growth.
Keywords: Srd, RNase E, Escherichia coli, bacteriophage T4, mRNA decay
BACTERIOPHAGE T4 shuts off gene expression of the host, Escherichia coli, immediately after infection and quickly starts to express its own genes (Kutter et al. 1994). Multiple mechanisms, such as modifications of apparatuses for transcription and translation, are involved in this shift of gene expression from E. coli to T4 (Carlson et al. 1994). Our previous work revealed that the representative stable E. coli messenger RNAs (mRNAs), lpp and ompA, were rapidly degraded after T4 infection (T4-induced host mRNA degradation) and that RNase E, which is an essential endoribonuclease in E. coli (Marcaida et al. 2006), primarily functions in T4-induced host mRNA degradation (Ueno and Yonesaki 2004). This rapid degradation of host mRNAs may contribute to the quick shift from E. coli to T4 metabolism because it leads to immediate cessation of host gene expression and consequently generation of ribonucleotides and free ribosomes, each of which would stimulate transcription and translation of T4 genes. In fact, deficiency of RNase E resulted in a slow start of T4 gene transcription, reducing the level of transcription (Otsuka and Yonesaki 2005) and retarding the growth of T4 (Mudd et al. 1990a). In eukaryotic cells, the degradation of host mRNAs after viral infection, such as in alphaherpesvirus, gammaherpesvirus, or betacoronavirus, also contributes to the shutoff of host gene expression (Gaglia et al. 2012). In these cases, host mRNA degradation is initiated by a viral factor and requires host 5′–3′ exoribonuclease Xrn1. Therefore, the degradation of host mRNAs after viral infection may be a common mechanism in both prokaryotes and eukaryotes; infection with virus activates the host mRNA degradation machinery for the shift of gene expression from host to virus.
Similar to eukaryotic viruses described above, T4 gene product is required for T4-induced host mRNA degradation, and the T4 ∆tk2 mutant showed slower degradation of host mRNAs (Ueno and Yonesaki 2004). However, the effect of ∆tk2 on host mRNA degradation was partial, which suggests that multiple mechanisms are utilized for T4-induced host mRNA degradation. In this study, we found that host mRNAs were partially stabilized, that a decay intermediate rapidly accumulated in cells infected with a ∆(39-56)6 mutant lacking eight consecutive genes, and that the causal gene was srd (similarity with rpoD) encoding a 29-kDa protein. This gene was named srd because it partly shares sequence similarity with E. coli RNA polymerase sigma 70; Mosig et al. (1998) reported that exogenous expression of Srd in E. coli cells resulted in extremely slow growth and filamentation of cells. However, the function and the role of Srd in T4-infected cells remained unclear.
RNase E participates in maturation of transfer RNAs (tRNAs), processing of ribosomal RNAs (rRNAs), and turnover of bulk mRNAs in E. coli (Mudd et al. 1990b; Mackie 2013). It roughly consists of two domains, an N-terminal (1–529 aa) catalytic half (NTH) and a C-terminal (530–1036 aa) scaffold half (CTH); the former has a catalytic domain for RNase, and the latter is required for the targeting to the inner membrane, the interaction with RNA, and the binding of polynucleotide phosphorylase, enolase, and RhlB RNA helicase to form a multi-enzyme complex called the RNA degradosome (Callaghan et al. 2005; Khemici et al. 2008; Tsai et al. 2012; Mackie 2013). RNase E cleaves mRNAs at internal sites either directly or in a 5′-end-dependent manner (Bouvier and Carpousis 2011). The latter requires the removal of a pyrophosphate from a triphosphate at the 5′-end of the transcript by RNA pyrophosphohydrolase (RppH) prior to RNase E cleavages. Hundreds of mRNAs are the targets of the 5′-end-dependent mRNA degradation pathway because the disruption of rppH results in the increase of their amounts or stabilities (Deana et al. 2008). On the other hand, many other mRNAs appear to be degraded by RNase E using another mechanism, the direct cleavage, independently from RppH (Clarke et al. 2014).
Here, we strongly suggest that T4 Srd associates with the N-terminal half of RNase E and stimulates both the 5′-end-dependent and -independent mRNA degradation activities of RNase E. Because T4 phage lacking srd exhibited reduced growth, the degradation of host mRNAs by RNase E activity that Srd stimulates after infection is required for efficient phage growth.
Materials and Methods
Phages and bacterial strains
Wild-type bacteriophage T4 is T4D. A ∆(39-56)6 phage was kindly provided by H. Takahashi (University of Tokyo). ∆srd, ∆motB.2, ∆dda-dda.1, ∆dexA.1-dexA.2, ∆modA, and ∆dexA mutant phages were constructed using the insertion/substitution system (Selick et al. 1988). Briefly, a DNA fragment was amplified by polymerase chain reaction (PCR) using T4 DNA as the template and primers 1 and 2 (Supporting Information, Table S1). Next, the amplified fragment was used for a second PCR as the primer together with primer 3 using T4 DNA as the template. Consequently, the resulting fragment contained 5′ and 3′ flanking regions of target gene(s) that were deleted. This DNA fragment was inserted into pBSPL0+, and the mutant sequence was transferred into the T4 genome via homologous recombination between plasmid and T4 DNA.
E. coli K-12 strains MH1 (sup0 araD139 hsdR ∆lacX74 rpsL), TY0807 (MH1 araD+) (Koga et al. 2011), and YT10 (MH1 zce726::Tn10) were used as wild types. YT10 or YT20 (YT10 ams1) was constructed by T4 GT7 phage transduction (Wilson et al. 1979) of the tetracycline-resistance marker from GW10 (Wachi et al. 1997) or by GT7 phage transduction of the tetracycline-resistance marker together with a temperature-sensitive mutation of ams1 from GW20 (Wachi et al. 1997). YT007 (MH1 ∆rppH::kan) was also constructed by GT7 phage transduction of the kanamycin-resistance marker from JW2798 (NIG). The kanamycin-resistance cassette of YT007 was removed by yeast Flp recombinase expressed from pCP20 to construct TY1005 (MH1 ∆rppH) (Cherepanov and Wackernagel 1995). ∆lpp::kan of JW1667 (NIG) was transferred into YT10, YT20, or TY1005 by GT7 phage transduction to construct TY1001 (MH1 zce726::Tn10 ∆lpp::kan), TY1002 (MH1 zce726::Tn10 ams1 ∆lpp::kan), or TY1006 (MH1 ∆rppH ∆lpp::kan), respectively. TY1007 (MH1 rne-FLAG-cat) and TY1008 (MH1 rne598-FLAG-cat) were also constructed by GT7 phage transduction of the chloramphenicol-resistance marker from TM338 (W3110 mlc rne-FLAG-cat) or TM529 (W3110 rne598-FLAG-cat), which were kindly provided by H. Aiba (Nagoya University).
Plasmids
To clone T4 srd, a 785-bp DNA fragment (11039–11803 of GenBank accession no. NC_000866) was amplified by PCR using T4 DNA as the template and primers 5′-ACGCGTCGACGTAAGATGTGAGAA and 5′-ACGCGTCGACTTATCCTCGGATAAG and digested with SalI. The resulting fragment was inserted into the SalI site of pBAD18 or pBAD33 (Guzman et al. 1995) to generate pBAD18-srd or pBAD33-srd, respectively. To construct pQE80L-srd-His, the DNA fragment encoding Srd with a His-tag at its C terminus was amplified by PCR with T4 DNA as the template and the primers 5′-GACTCCGGAATTCCGGGTAAGATGTGAGAA and 5′-GACCCAAGCTTGGGTTAGTGGTGATGGTGATGATGTCCTCGGATA, digested with EcoRI and HindIII and ligated into the corresponding sites of pQE80L (Qiagen). To generate pBSlpp(T), the primer 1 (5′-CTATTACGCCAGCTGGCGAAAAAAAATGGCGCACAATGTGC) and the primer 2 (5′-GGCCGATTCATTAATGCAGCTGGC) were first phosphorylated using T4 kinase with ATP. The phosphorylated primer 1 and the primer 3 (5′-CGCGCCCAGTACCAGTTTAGTAGCTTCCACACAACATACGAGCCGGAAGCAT) were used for PCR with E. coli MG1655 DNA as a template. The PCR-amplified fragment was used as the primer for the second PCR with the phosphorylated primer 2 and pBluescript II KS (+) as the template. Finally, the resulting fragment was ligated into the PvuII site of pBluescript II KS (+).
Determination of the deleted region in the ∆(39-56)6 phage
Two primers—5′-TCAACGCCATCTTCCAATCCAT located in motB.1 and 5′-ATCTTGCAGATGTTGAACAGT located in modB—were used for PCR. When ∆(39-56)6 phage DNA was used as a template, only a prominent band corresponding to 1.2 kb was produced, although 5.8-kb DNA was amplified with wild-type phage DNA as a template. The 1.2-kb DNA amplified with ∆(39-56)6 phage DNA was recovered from the gel and sequenced. The sequence context showed that the nucleotide at 7744 in the T4 genome was connected with that at 12,364, indicating that ∆(39-56)6 lacks a 4619-bp sequence.
RNA purification, Northern blot, and primer extension analyses
Isolation of total RNA and Northern blot analyses were carried out as described (Kai et al. 1996; Ueno and Yonesaki 2004). Cells were infected with T4 phage at a multiplicity of infection (m.o.i.) of 7 for examining the half-lives of mRNAs after infection. To measure half-lives of mRNAs in the absence of T4 infection, rifampicin was added to a final concentration of 500 µg/ml. Total RNAs extracted were electrophoresed through a 5% polyacrylamide gel containing 7 M urea, followed by Northern blotting with probes for lpp ompA or srd. Oligo probes for srd and lpp were 5′-32P-ACGCGTCGACTTATCCTCGGATAAG and 5′-32P-TTACTTGCGGTATTTAGTAG, respectively. Northern blot analyses were performed more than twice independently and a representative result is shown in Figure 1, Figure 2, Figure 3, Figure 4, and Figure 6. The relative amount of each RNA was normalized to 5S rRNA. Primer extension analysis for lpp RNA was performed using 5′-32P-GCATTGCGTTCACGTCGTTGCTCA as the primer.
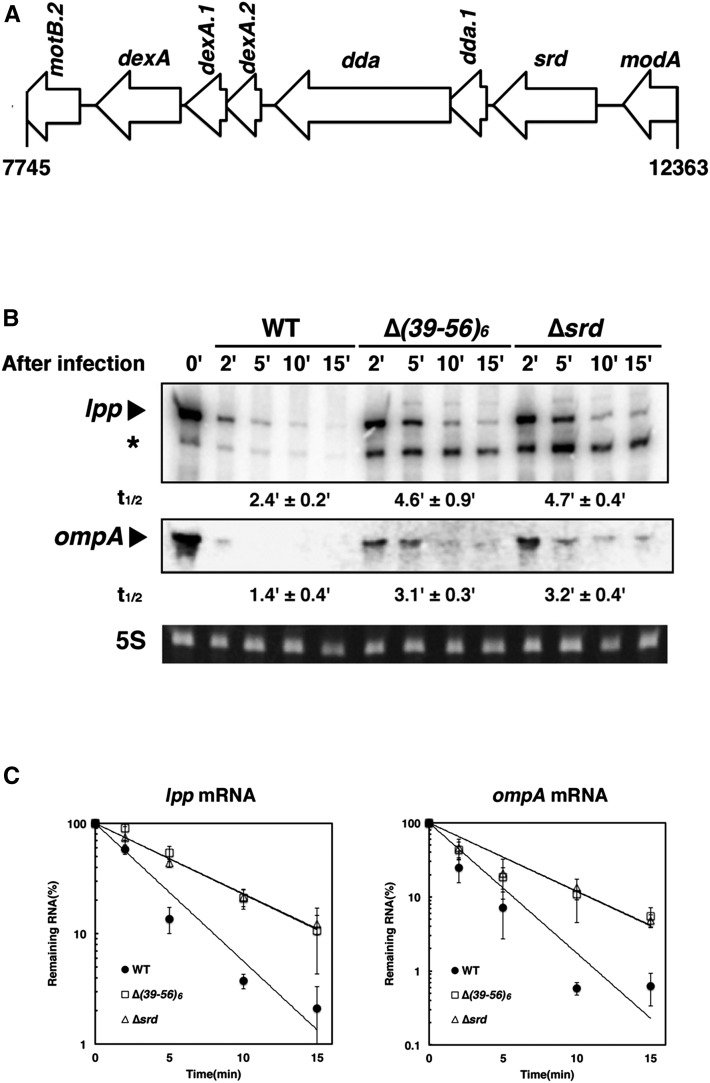
Figure 1.
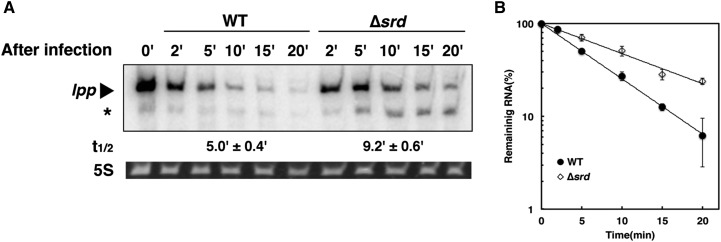
Involvement of srd in T4-induced host mRNA degradation. (A) Genetic organization of the chromosomal region deleted in the ∆(39-56)6 phage. (B) MH1 (wild-type) cells were grown in M9C medium (M9-glucose medium supplemented with 0.3% casamino acids, 1 µg/ml thiamine, and 20 µg/ml tryptophan) until the OD600 reached 0.5 at 37° and infected with T4 wild-type, ∆(39-56)6, or ∆srd mutant. Total RNAs extracted at the indicated times were analyzed by Northern blotting with probes for lpp or ompA. Ethidium bromide-stained 5S rRNA is shown as a loading control. Arrowheads and an asterisk indicate each full-length mRNA and a decay intermediate of lpp mRNA, respectively. (C) Quantification analyses of full-length mRNAs in B were performed. The signal intensity was quantified using the Image J program, the signal obtained at 0 min was set to 100%, and the percentage of full-length mRNA remaining at each time point was plotted. Data points represent the mean ± SD of triplicate measurements. The time required for a 50% reduction was taken as a half-life (t1/2) of each mRNA shown below B.
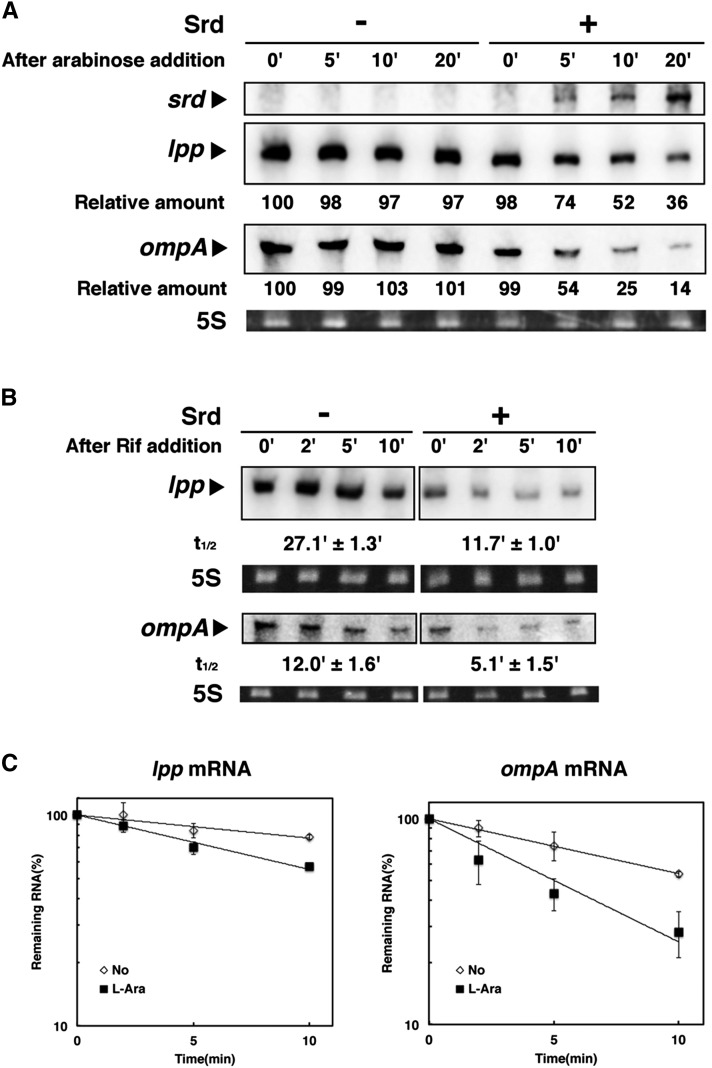
Figure 2.
Destabilization of lpp and ompA mRNAs by exogenous expression of Srd. (A) TY0807 (wild-type) cells harboring pBAD18 (–) or pBAD18-srd (+) were treated with 0.15% arabinose when the OD600 reached 0.5 at 30°. Total RNAs extracted at the indicated times were subjected to Northern blotting with probes for srd, lpp, or ompA mRNA. The intensity of the band at time 0 was set to 100%, and the relative intensity at each time is shown below. (B) TY0807 cells harboring pBAD18-srd were treated with (+) or without (–) 0.15% arabinose for 10 min when the OD600 reached 0.5 at 30°. Total RNAs extracted at the indicated times after addition of the transcription inhibitor rifampicin were subjected to Northern blotting with probes for lpp or ompA. (C) Quantification analyses of full-length mRNAs in B were performed. Data points represent the mean ± SD of triplicate measurements.
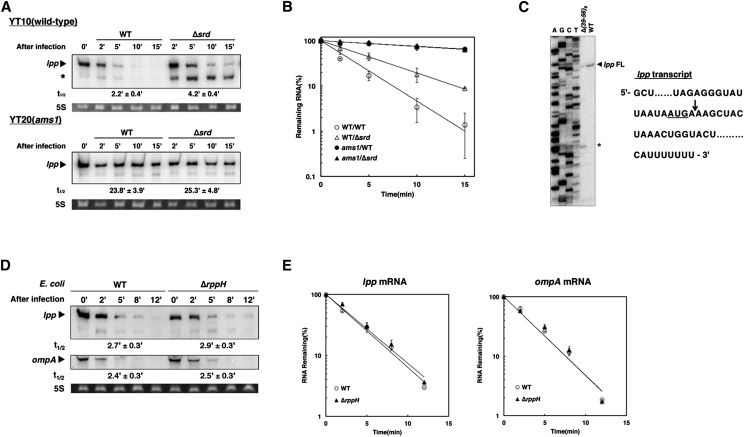
Figure 3.
Stimulation of 5′-end-independent cleavage activity of RNase E by Srd. (A) YT10 (wild-type) or its derivative, YT20 (ams1), cells were grown in M9C medium until the OD600 reached 0.3 at 30° and shifted to 44° for another 30 min and then infected with wild-type or ∆srd mutant. Total RNAs extracted at the indicated times were analyzed by Northern blotting with a probe for lpp. An arrowhead and an asterisk indicate the full-length lpp mRNA and its decay intermediate, respectively. (B) Quantification analysis of lpp mRNA in A was performed. Data points represent the mean ± SD of triplicate measurements. (C) Five micrograms of total RNAs extracted from MH1 cells infected with wild-type or ∆(39-56)6 mutant were used for primer extension analysis as described in Materials and Methods. “FL” or the asterisk denotes the full-length lpp mRNA or its decay intermediate. A set of sequence ladders for lpp obtained by dideoxy-sequencing with the same primer was run in parallel. The RNA sequence around the SD region and the start codon (underlined text) of lpp mRNA are shown at the right, and the arrow indicates the cleavage site generating the decay intermediate. (D) MH1 (WT) or TY1005 (∆rppH) cells were grown in M9C medium until the OD600 reached 0.5 at 37°, and total RNAs were extracted at the indicated times after infection with wild type and then analyzed by Northern blotting with probes for lpp or ompA. (E) Quantification analyses of full-length mRNAs in D were performed. Data points represent the mean ± SD of duplicate measurements.
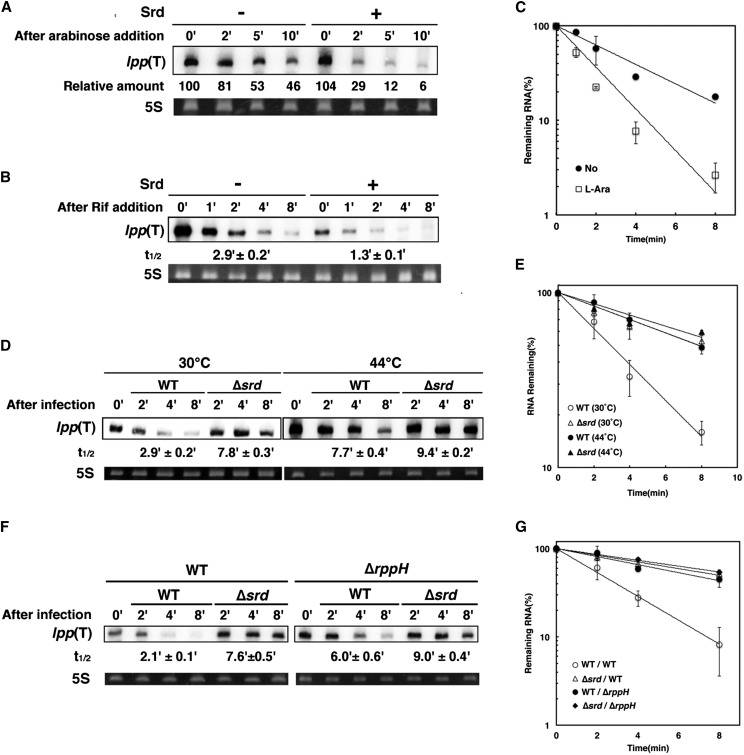
Figure 4.
Stimulation of 5′-end-dependent cleavage activity of RNase E by Srd. (A) TY1001 (∆lpp::kan) cells harboring pBSlpp(T) plus either pBAD33 (–) or pBAD33-srd (+) were grown at 30° until the OD600 reached 0.5 and supplemented with 1 mM IPTG. At 10 min after IPTG addition, cells were treated with 0.2% arabinose, and then total RNAs extracted at the indicated times were subjected to Northern blotting with a probe for lpp. The intensity of band at time 0 was set to 100%, and relative intensity at each time is shown below. (B) TY1001 cells harboring pBSlpp(T) and pBAD33-srd were grown at 30° until the OD600 reached 0.5 and treated with 1 mM IPTG. At 10 min after IPTG addition, cells were treated with (+) or without (–) 0.05% arabinose for another 10 min. Total RNAs extracted at the indicated times after addition of rifampicin were subjected to Northern blotting with a probe for lpp. (C) Quantification analysis of lpp(T) RNA in B was performed. Data points represent the mean ± SD of triplicate measurements. (D) TY1002 (∆lpp::kan ams1) cells harboring pBSlpp(T) were grown at 30° until the OD600 reached 0.3, divided into two aliquots, and incubated for 25 min at 30° or 44°, respectively. Then the cells were treated with 1 mM IPTG for 10 min and further divided into two aliquots followed by infection with wild-type or ∆srd mutant. Ten micrograms of total RNAs extracted at the indicated times after infection was subjected to Northern blotting with a probe for lpp. (E) Quantification analysis of lpp(T) RNA in D was performed. Data points represent the mean ± SD of five time measurements. (F) TY1001(∆lpp::kan) or TY1006 (∆lpp::kan ∆rppH) cells harboring pBSlpp(T) were grown at 30° until the OD600 reached 0.5. Then the cells were treated with 1 mM IPTG for 10 min followed by infection with wild-type or ∆srd mutant. Ten micrograms of total RNAs extracted at the indicated times after infection was subjected to Northern blotting with a probe for lpp. (G) Quantification analysis of full-length mRNAs in F was performed. Data points represent the mean ± SD of triplicate measurements.
Figure 6.
Effect of CTH of RNase E on T4-induced host mRNA degradation. (A) TY1008 (rne598-FLAG-cat) cells were grown at 30° in M9C medium until the OD600 reached 0.5, divided into two aliquots, and infected with wild-type or ∆srd mutant. Total RNAs extracted at the indicated times were subjected to Northern blotting with a probe for lpp. (B) Quantification analysis of lpp mRNA in A was performed. Data points represent the mean ± SD of duplicate measurements.
Immunoprecipitation, pull-down, and Western blot analyses
TY1007 or TY1008 cells harboring pQE80L-srd-His or pQ-orf2-95 encoding His-tagged IscR (Otsuka et al. 2010) were grown at 30° in 200 ml of LB medium containing 50 µg/ml ampicillin until the OD600 reached 0.6 and treated with 2 mM IPTG for 2 hr to induce His-tagged Srd or 20 µM IPTG for 1 hr to induce His-tagged IscR. Cells were harvested and suspended in 1.5 ml of lysis buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM DTT, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, and 1× protease inhibitor cocktail). One hundred microliters of the cell mixture was kept as a whole-cell extract. After sonication, the lysate was centrifuged at 20,400 × g for 20 min and separated into the supernatant as a soluble fraction and the pellet as an insoluble fraction. The pellet was suspended with 1.3 ml of TBS buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM DTT). The soluble fraction (1.3 ml) was treated with 20 µg/ml RNase A for 30 min at 25°, and then 0.6 ml was mixed with 20 µl of Ni-NTA Superflow agarose beads (Qiagen) or 20 µl of anti-FLAG M2 affinity agarose (Sigma-Aldrich) by end-over-end rotation overnight at 4°. The beads were washed four times with 0.6 ml of TBS buffer for anti-FLAG M2 agarose or TBS buffer containing 20 mM imidazole for Ni-NTA agarose, and then 45 µl of sample loading buffer was added to the beads. After boiling the samples, bound proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) [10% gel for full-length RNase E-FLAG, 12% gel for RNase E(1-598)-FLAG, or 15% gel for Srd-His and His-IscR] and electroblotted onto Immuno-Blot PVDF membrane (Bio-Rad). Western blot analysis was performed as previously described (Koga et al. 2011). Pull-down and immunoprecipitation analyses were performed at least three times, and a representative result is shown in Figure 5.
Figure 5.
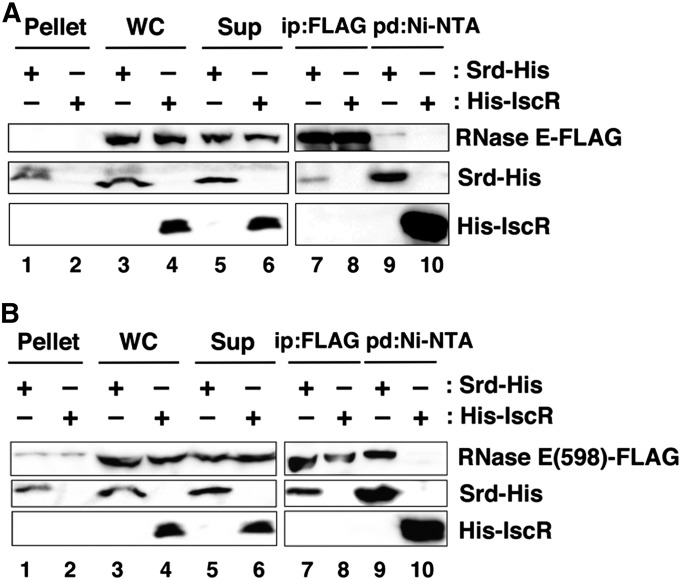
Physical association between RNase E and Srd in vivo. (A) TY1007 (rne-FLAG-cat) cells or (B) TY1008(rne598-FLAG-cat) cells harboring either pQE80L-srd-His or pQ-orf2-95 were grown at 30° until the OD600 reached 0.6, and IPTG was added to induce Srd-His or His-IscR. Preparation of cell extracts and fractions, immunoprecipitation with anti-FLAG M2 agarose beads, and pull-down with Ni-NTA beads were carried out as described in Materials and Methods. A total of 0.33% of whole-cell extract (WC, lanes 3 and 4), the insoluble fraction (Pellet, lanes 1 and 2), and the soluble fraction (Sup, lanes 5 and 6), and 33.3% of the bound fractions (lanes 7–10) was analyzed by Western blotting with antibodies against FLAG-tag (top) and His-tag (middle and bottom).
Data availability
File S1 contains Table S1 and Figures S1, S2, S3, S4, S5, and S6.
Results
Identification of a phage factor involved in T4-induced host mRNA degradation
To identify factor(s) involved in T4-induced host mRNA degradation, we examined several T4 deletion mutants that lack multiple genes in various genomic regions and found that the ∆(39-56)6 mutant affected the degradation of host mRNAs. MH1 (wild-type) cells were grown until the OD600 reached 0.5, and total RNAs were extracted from cells at appropriate times after infection and examined by Northern blot analysis. In cells infected with wild-type phage, the representative stable mRNAs lpp and ompA were rapidly degraded after infection with half-lives of 2.4 and 1.4 min, respectively (Figure 1, B and C). When infected with the ∆(39-56)6 mutant, the half-lives of lpp and ompA mRNAs were increased to 4.6 and 3.1 min, respectively. In addition, a decay intermediate of lpp mRNA was remarkably accumulated in ∆(39-56)6-infected cells. This result suggested that a gene lacking in ∆(39-56)6 was involved in T4-induced host mRNA degradation. The ∆(39-56)6 mutant was originally isolated by Homyk and Weil in 1974 (Homyk and Weil 1974), but the exact region deleted in the genome of this phage was not reported. We mapped the deletion by PCR and DNA sequencing and found it lacking a 4619-bp DNA sequence from nucleotides 7745 to 12,363 in the T4 genome, resulting in the lack of all or a part in eight ORFs (Figure 1A). To identify a gene responsible for T4-induced host mRNA degradation, we constructed six mutants that lacked one or a few of these ORFs and examined the stability of lpp mRNA and the accumulation of the decay intermediate after T4 infection (Figure 1B and Figure S1). The deletion mutants (except ∆srd) exhibited little effect on decay of lpp mRNA and no accumulation of the decay intermediate. In contrast, lpp and ompA mRNAs were stabilized in ∆srd-infected cells, and their half-lives were 4.7 and 3.2 min, similar to those of ∆(39-56)6-infected cells. In addition, the decay intermediate of lpp mRNA highly accumulated in ∆srd-infected cells. We also examined the effect of srd on stabilities of typical unstable mRNAs, rpsO, trxA, and rpsT, and observed that rpsO(P1-t1) and trxA mRNAs were degraded after infection with wild-type phage slightly faster than in uninfected cells or after infection with the ∆srd mutant, although the stabilities of rpsO(P1-RIII) and rpsT mRNAs had no significant differences among uninfected cells, cells infected with T4 wild type, and the ∆srd mutant (Figure S2). These results strongly suggested that srd is involved in T4-induced host mRNA degradation.
Effect of Srd on decay of lpp and ompA mRNAs
To examine the effect of Srd on decay of lpp and ompA mRNAs in uninfected cells, we constructed a plasmid, pBAD18-srd, expressing Srd under the control of an arabinose-inducible promoter. Total RNAs were extracted from cells harboring pBAD18-srd at 0, 5, 10, or 20 min after addition of arabinose and analyzed by Northern blotting. As seen in Figure 2A, the expression of srd mRNA was discernible at 5 min after addition of arabinose, and amounts of lpp and ompA mRNAs were reduced in contrast to the increase of srd mRNA. To further explore whether or not the reduction of these mRNAs is caused by the increase of decay rate, we measured stabilities of lpp and ompA mRNAs (Figure 2, B and C). At 10 min after Srd induction, a transcription inhibitor, rifampicin, was added to the culture, and total RNAs were extracted at appropriate times. In the absence of Srd, both lpp and ompA mRNAs were stable with half-lives of 27.1 and 12 min, respectively. In contrast, their half-lives were 11.7 min for lpp and 5.1 min for ompA in the presence of Srd, indicating that their decay rates were increased approximately twofold after Srd induction. Taken together with the result of Figure 1B, Srd destabilizes lpp and ompA mRNAs.
Effect of Srd on 5′-end-independent cleavage activity of RNase E
The decay intermediate of lpp mRNA accumulated in ∆srd-infected cells (Figure 1B) would be attributable to the cleavage by RNase E because full-length lpp mRNA was considerably stabilized in RNase E-defective cells (Ueno and Yonesaki 2004). To confirm this possibility, we first examined the effect of RNase E on decay of lpp mRNA in ∆srd-infected cells. Wild-type or temperature-sensitive RNase E mutant (ams1) cells were grown at a permissive temperature (30°) until the OD600 reached 0.3, and then the temperature was shifted to 44°, a nonpermissive temperature, for 30 min, followed by infection with wild-type or ∆srd mutant. RNAs extracted from cells at appropriate times after infection were analyzed by Northern blotting. Full-length lpp mRNA was stabilized, and the decay intermediate was hardly detected in RNase E-defective cells infected with ∆srd mutant as well as wild-type phage (Figure 3, A and B). The cleavage site to generate this decay intermediate was determined by primer extension analysis, and its 5′-terminus was two nucleotides downstream of the start codon of lpp mRNA (AUGA↓AA) (Figure 3C and Figure S3). RNase E has no canonical target sequence for cleavage, but preferentially cleaves at regions that are single-stranded with AU-rich sequences. These facts clearly support the idea that RNase E generates this decay intermediate. Finally, we examined the effect of RppH on decay of lpp and ompA mRNAs stimulated by Srd (Figure 3, D and E). After infection with wild-type phage, both mRNAs were degraded in ∆rppH cells as fast as in wild-type cells, indicating that RppH is not related to the degradation of their mRNAs. From these results, RNase E degrades these mRNAs through an RppH-independent manner in the presence of Srd.
Effect of Srd on 5′-end-dependent cleavage activity of RNase E
The decay intermediate of lpp mRNA should be processively degraded by RNase E in the presence of Srd because it is not observed in cells infected with wild-type phage. To investigate the effect of Srd on the decay intermediate of lpp mRNA, we constructed a plasmid, pBSlpp(T), expressing the truncated lpp RNA with a sequence identical to the decay intermediate of lpp mRNA from the lac promoter of pBluescript II KS (+) (Figure S3). First, we characterized the decay of lpp(T) RNA in the absence of Srd. At 44°, lpp(T) RNA had a half-life as short as 1.4 min in ∆lpp::kan cells expressing wild-type RNase E, while it was more stable with a half-life of 9.5 min in ∆lpp::kan ams1 cells defective in RNase E activity (Figure S4, A and B). lpp(T) RNA should have a triphosphate at the 5′-end, which differs from a decay intermediate of lpp mRNA with a 5′-monophosphate end. Therefore, the degradation of lpp(T) RNA may be dependent on RppH. To address this issue, we examined the effect of RppH on stability of lpp(T) RNA (Figure S4, C and D). The half-life of lpp(T) RNA in wild-type or ∆rppH cells was 2.8 or 7.1 min, respectively. Taken together, these results show that lpp(T) RNA is degraded by RNase E through an RppH-dependent pathway under normal conditions.
The effect of Srd on decay of lpp(T) RNA was examined by measuring the amount of lpp(T) RNA with or without induction of Srd (Figure 4A), and we found that the amount was considerably reduced after induction of Srd for 10 min. Furthermore, we checked the stability of lpp(T) RNA (Figure 4, B and C). The half-life with or without induction of Srd was 1.3 or 2.9 min, respectively, clearly demonstrating that Srd stimulates the degradation of lpp(T) RNA. Next, we confirmed that Srd stimulates the RNase E-dependent decay of lpp(T) after T4 infection (Figure 4, D and E). The half-life of lpp(T) RNA in the presence or absence of srd was 2.9 or 7.8 min, respectively, at 30°, while it was 7.7 or 9.4 min at 44°. Finally, we examined the effect of RppH on decay of lpp(T) RNA after T4 infection. When wild-type phage infected, lpp(T) RNA was significantly stabilized in ∆rppH cells (6.0 min), compared with wild-type cells (2.1 min) (Figure 4, F and G). These results indicate that T4 Srd, RNase E, and RppH are required for the rapid degradation of lpp(T) RNA after T4 infection. Taken together with all the results, Srd should stimulate both 5′-end-independent and 5′-end-dependent cleavage activities of RNase E either directly or indirectly.
Association of Srd and RNase E in vivo
The above results implied that Srd physically associated with RNase E. Hence, we carried out immunoprecipitation and pull-down analyses to examine their interaction in vivo. For these experiments, we used TY1007 cells in which the chromosomal rne was replaced with rne-FLAG encoding FLAG-tagged RNase E. Extracts prepared from TY1007 cells expressing His-tagged Srd or His-tagged IscR as a control were used for immunoprecipitation with FLAG antibody or pull-down with Ni-NTA. Before immunoprecipitation and pull-down analyses, we checked how much RNase E was solubilized because RNase E is membrane-associated. As seen in Figure 5A (lanes 1–6), the majority of RNase E was present in the soluble fraction under our experimental conditions. In the immunoprecipitation experiment, Srd-His was efficiently recovered together with RNase E-FLAG (lane 7), but His-IscR was not (lane 8). In the reciprocal pull-down experiment, RNase E-FLAG was precipitated together with Srd-His (lane 9), but not with His-IscR (lane 10). These results suggested the physical association of Srd and RNase E in vivo.
Dispensability of RNase E CTH for mRNA degradation stimulated by Srd
RNase E is composed of NTH (1–529) and CTH (530–1036) (Callaghan et al. 2005; Mackie 2013). We investigated whether or not CTH was indispensable for T4-induced host mRNA degradation. TY1008 cells expressing FLAG-tagged RNase E (1–598) containing the catalytic domain (1–529) and the membrane target sequence (segment A, 568–582) from the genome were infected with wild-type or ∆srd mutant, and total RNAs were extracted at appropriate times for Northern blot analysis. Consistent with the previous report (Ueno and Yonesaki 2004), RNase E lacking the C-terminal scaffold domain degraded full-length lpp mRNA more slowly than did normal RNase E, and its half-life was 5.0 min (compare to 2.4 min in Figure 1B) when infecting with wild-type phage (Figure 6, A and B). When infecting with the ∆srd mutant, full-length lpp mRNA was more stabilized with a half-life of 9.2 min, and the decay intermediate also accumulated. This result paralleled the result in Figure 1B, suggesting that the C-terminal scaffold domain (599–1036) is not necessary for E. coli mRNA degradation stimulated by Srd.
Prompted by the above result, we examined the activity of the N-terminal half (1–598) in association with Srd. As seen in Figure 5B, most of RNase E(598)-FLAG was present in the soluble fraction (lanes 1–6), like an RNase E-FLAG. Srd-His was precipitated by FLAG antibody together with RNase E(598)-FLAG (lane 7), but His-IscR was not (lane 8). In the pull-down experiments with Ni-NTA beads, RNase E(598)-FLAG was recovered in the bound fraction together with Srd-His (lane 9) but hardly with His-IscR (lane 10), indicating the association of Srd and the N-terminal half of RNase E containing the catalytic domain and the membrane target sequence in vivo.
Effect of srd on growth of T4 phage
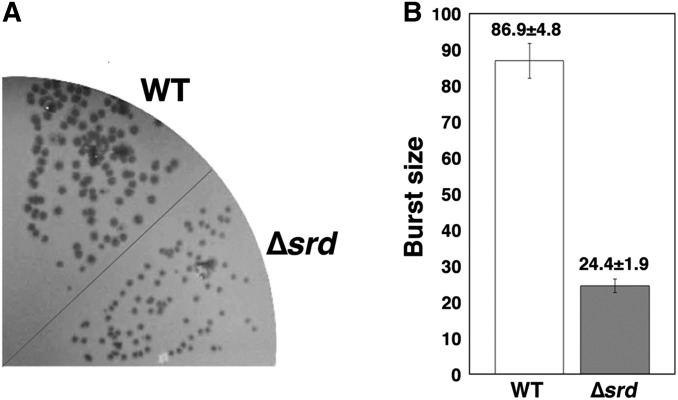
Finally, we investigated the effect of srd on T4 growth. Wild-type or ∆srd mutant was spread on plates seeded with MH1 (wild-type) cells as an indicator (Figure 7A). The ∆srd mutant formed smaller plaques than those of wild type. We also confirmed the stimulatory effect of srd on propagation of T4 phage by measuring burst size (Figure 7B). Consistently with the observation of plaque size, the burst size of the ∆srd mutant was 24.4 ± 1.9, while that of wild type was 86.9 ± 4.8. These results indicate that srd is required for efficient growth of T4 phage.
Figure 7.
Growth of ∆srd mutant phage. (A) Equal number of wild-type and ∆srd mutant phage particles were plated onto a plate inoculated with MH1 (wild-type) cells and incubated at 37° overnight. (B) MH1 cells were grown in M9C medium until the OD600 reached 0.5 and infected with wild-type or ∆srd mutant at an m.o.i. of 0.1 at 37°. At 8 min, the cells were diluted 104-fold with fresh M9C medium and further incubated for 70 min. After the cells were lysed with chloroform, the total number of progeny phage was determined by plating after an appropriate dilution with MH1 as an indicator. The burst size shown in the bar graph is the ratio of the number of progeny to the number of input phage particles. Each value indicates the mean and standard deviation obtained from three independent experiments.
Discussion
In this study, we demonstrated that T4 Srd is involved in phage-induced host mRNA degradation in which E. coli RNase E plays a central role. RNase E participates in the turnover of bulk mRNAs as well as the processing of tRNAs and rRNAs in E. coli and many other proteobacteria (Ono and Kuwano 1979; Jain et al. 2002; Carpousis 2007; Tsai et al. 2012). Cleavage of mRNA by RNase E can occur via two pathways: 5′-end monophosphate-dependent and 5′-end independent (so-called “direct entry”) (Anupama et al. 2011; Bouvier and Carpousis 2011; Garrey and Mackie 2011). RppH is necessary for the former case (Celesnik et al. 2007; Deana et al. 2008; Bouvier and Carpousis 2011). RppH removes a pyrophosphate from a 5′-end triphosphate of mRNA and then RNase E cleaves RNA due to the preference of RNase E for 5′-monophosphorylated RNA. Hundreds of mRNAs are reported to be the targets of RppH, such as rpsT, yeiP, and trxB (Deana et al. 2008; Richards et al. 2012). Our study showed that the plasmid-borne lpp(T) RNA was degraded by RNase E in an RppH-dependent fashion (Figure S4), and that Srd stimulated this activity (Figure 4). In the 5′-end-independent pathway, RNase E first triggers the degradation of mRNA by cleavage at an internal site to generate an RNA fragment with a monophosphate at 5′-end, and the sequential degradation by RNase E follows in a 5′-end-dependent fashion. E. coli stable mRNAs ompA and lpp are known as examples for this pathway (Emory et al. 1992; Ow et al. 2003; Deana et al. 2008). In fact, after T4 infection, ompA and lpp mRNAs were degraded in ∆rppH cells as fast as those in wild-type cells (Figure 3, D and E). Therefore, Srd could stimulate 5′-end-independent cleavage activity of RNase E. Taken together with all the analyses of lpp, ompA and lpp(T) mRNAs, we conclude that T4 Srd stimulates both 5′-end-dependent and independent cleavage activities of RNase E either directly or indirectly.
The NTH (1–529) of RNase E is necessary and sufficient for cleavage and forms a tetrameric assembly of RNase E (Mackie 2013). The CTH (530–1036) acts mainly as a scaffold for RNA degradosome assembly and recruits its core constituents: polynucleotide phosphorylase, enolase, and RhlB RNA helicase (Carpousis 2007; Górna et al. 2012). We demonstrated that T4-induced host mRNA degradation occurred in the absence of the C-terminal half (599–1036) (Figure 6). Furthermore, immunoprecipitation and pull-down experiments showed that Srd was physically associated with the N-terminal half (1–598) of RNase E. Considering that the amount of RNase E did not change after T4 infection (Figure S5), Srd is strongly suggested to accelerate host mRNA degradation by stimulation of catalytic activity of RNase E. The N-terminal half (1–598) consists of two RNase H-like domains, an S1 domain, a 5′ sensor domain, a DNase I-like domain, a Zn-link domain, a small domain, and the membrane target sequence (Mackie 2013). To identify which domain of RNase E interacts with Srd may help toward understanding the stimulatory mechanism by Srd. Furthermore, it would be also valuable to measure the kinetics of RNA cleavage by RNase E with or without Srd in vitro.
To date, three E. coli factors to regulate RNase E activity, RraA, RraB, and ribosomal protein L4, have been identified, but all of them regulate RNase E activity negatively (Lee et al. 2003; Gao et al. 2006; Yeom et al. 2008a,b; Singh et al. 2009). Apart from these factors in E. coli, only one viral factor has been reported to regulate the degradation of mRNAs by RNase E. A protein kinase of bacteriophage T7 phosphorylates RNase E at its CTH and stabilizes a subset of RNase E substrates (Marchand et al. 2001). In this case, T7 protein kinase would not play a role in the shutoff of host gene expression because it regulates RNase E activity negatively. Therefore, T4 Srd would be the first example of a factor that stimulates RNase E activity and induces the degradation of host mRNAs after infection.
Three different classes of promoters—early, middle, and late—initiate T4 transcription (Miller et al. 2003). srd is located immediately downstream of an early promoter, which is supposed to be expressed at a very early time after T4 infection (Mathews 1994; Miller et al. 2003). Although our attempt for detecting endogenous expression of Srd was not successful (data not shown), we could detect endogenous srd mRNA by semiquantitative RT-PCR (Figure S6). This result indicates that the level of srd mRNA during infection is almost the same as that derived from pBAD18-srd. The observations that the ∆srd mutant forms smaller plaques (Figure 7A) and exhibits a much lower burst size (Figure 7B) in comparison to wild-type phage indicate that Srd is required for efficient growth of T4 phage and suggest that host mRNA degradation by Srd-stimulated RNase E activity should contribute to the transition of gene expression from host to T4 immediately after infection. However, the effect of srd on host mRNA degradation is partial (Figure 1B). Previously, T4 ∆tk2 mutant showed slower degradation of host mRNAs (Ueno and Yonesaki 2004), and the effect of ∆tk2 on host mRNA degradation was also partial. Considered together, T4 phage may employ multiple factors and mechanisms to sustain rapid degradation of host mRNAs.
Supplementary Material
Acknowledgments
We thank John W. Drake at the U.S. National Institute of Environmental Health Sciences for invaluable help with the manuscript and the staff of the Radioisotope Research Center at Toyonaka, Osaka University (where all of our experiments using radioisotopes were carried out) for the facilitation of our research. This work was supported in part by a grant from the program Grant-in-Aid for Young Scientists (B) (to Y.O.) and in part by a grant from the program Grants-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.Y.). The authors have no conflict of interest to declare.
Footnotes
Communicating editor: A. Hochschild
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.180364/-/DC1.
Literature Cited
- Anupama K., Krishna Leela J., Gowrishankar J., 2011. Two pathways for RNase E action in Escherichia coli in vivo and bypass of its essentiality in mutants defective for Rho-dependent transcription termination. Mol. Microbiol. 82: 1330–1348. [DOI] [PubMed] [Google Scholar]
- Bouvier M., Carpousis A. J., 2011. A tale of two mRNA degradation pathways mediated by RNase E. Mol. Microbiol. 82: 1305–1310. [DOI] [PubMed] [Google Scholar]
- Callaghan A. J., Marcaida M. J., Stead J. A., McDowall K. J., Scott W. G., et al. , 2005. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437: 1187–1191. [DOI] [PubMed] [Google Scholar]
- Carlson K., Raleigh E. A., Hattman S., 1994. Restriction and Modification, pp. 369–381 in Molecular Biology of Bacteriophage T4, edited by Karam J. D., Drake J. W., Kreuzer K. N., Mosig G., Hall D. H. et al American Society for Microbiology Press, Washington, DC. [Google Scholar]
- Carpousis A. J., 2007. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61: 71–87. [DOI] [PubMed] [Google Scholar]
- Celesnik H., Deana A., Belasco J. G., 2007. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol. Cell 27: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. P., Wackernagel W., 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158: 9–14. [DOI] [PubMed] [Google Scholar]
- Clarke J. E., Kime L., A. D. Romero, and K. J. McDowall, 2014. Direct entry by RNase E is a major pathway for the degradation and processing of RNA in Escherichia coli. Nucleic Acids Res. 42: 11733–11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A., Celesnik H., Belasco J. G., 2008. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451: 355–358. [DOI] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G., 1992. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 6: 135–148. [DOI] [PubMed] [Google Scholar]
- Gaglia M. M., Covarrubias S., Wong W., Glaunsinger B. A., 2012. A common strategy for host RNA degradation by divergent viruses. J. Virol. 86: 9527–9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Lee K., Zhao M., Qiu J., Zhan X., et al. , 2006. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol. Microbiol. 61: 394–406. [DOI] [PubMed] [Google Scholar]
- Garrey S. M., Mackie G. A., 2011. Roles of the 5′-phosphate sensor domain in RNase E. Mol. Microbiol. 80: 1613–1624. [DOI] [PubMed] [Google Scholar]
- Górna M. W., Carpousis A. J., Luisi B. F., 2012. From conformational chaos to robust regulation: the structure and function of the multi-enzyme RNA degradosome. Q. Rev. Biophys. 45: 105–145. [DOI] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J., 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homyk T., Jr, Weil J., 1974. Deletion analysis of two nonessential regions of the T4 genome. Virology 61: 505–523. [DOI] [PubMed] [Google Scholar]
- Jain C., Deana A., Belasco J. G., 2002. Consequences of RNase E scarcity in Escherichia coli. Mol. Microbiol. 43: 1053–1064. [DOI] [PubMed] [Google Scholar]
- Kai T., Selick H. E., Yonesaki T., 1996. Destabilization of bacteriophage T4 mRNAs by a mutation of gene 61.5. Genetics 144: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemici V., Poijak L., Luisi B. F., Carpousis A. J., 2008. The RNase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 70: 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M., Otsuka Y., Lemire S., Yonesaki T., 2011. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics 187: 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter, E., T. White, M. Kashlev, M. Uzan, J. McKinney et al., 1994 Effects on host genome structure and expression, pp. 357–368 in Molecular Biology of Bacteriophage T4, edited by J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall et al., American Society for Microbiology Press, Washington, DC.
- Lee K., Zhan X., Gao J., Qiu J., Feng Y., et al. , 2003. RraA: a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell 114: 623–634. [PubMed] [Google Scholar]
- Mackie G. A., 2013. RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 11: 45–57. [DOI] [PubMed] [Google Scholar]
- Marcaida M. J., DePristo M. A., Chandran V., Carpousis A. J., Luisi B. F., 2006. The RNA degradosome: life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 31: 359–365. [DOI] [PubMed] [Google Scholar]
- Marchand I., Nicholson A. W., Dreyfus M., 2001. Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol. Microbiol. 42: 767–776. [DOI] [PubMed] [Google Scholar]
- Mathews, C. K., 1994 An overview of the T4 developmental program, pp. 1–8 in Molecular Biology of Bacteriophage T4, edited by J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall et al., American Society for Microbiology Press, Washington, DC.
- Miller E. S., Kutter E., Mosig G., Arisaka F., Kunisawa T., et al. , 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67: 86–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosig G., Colowick N. E., Pietz B. C., 1998. Several new bacteriophage T4 genes, mapped by sequencing deletion endpoints between genes 56 (dCTPase) and dda (a DNA-dependent ATPase-helicase) modulate transcription. Gene 223: 143–155. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Carpousis A. J., Krisch H. M., 1990a Escherichia coli RNase E has a role in the decay of bacteriophage T4 mRNA. Genes Dev. 4: 873–881. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Krisch H. M., Higgins C. F., 1990b RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol. Microbiol. 4: 2127–2135. [DOI] [PubMed] [Google Scholar]
- Ono M., Kuwano M., 1979. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J. Mol. Biol. 129: 343–357. [DOI] [PubMed] [Google Scholar]
- Otsuka Y., Yonesaki T., 2005. A novel endoribonuclease, RNase LS, in Escherichia coli. Genetics 169: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y., Miki K., Koga M., Katayama N., Morimoto W., et al. , 2010. IscR regulates RNase LS activity by repressing rnlA transcription. Genetics 185: 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M. C., Perwez T., Kushner S. R., 2003. RNase G of Escherichia coli exhibits only limited functional overlap with its essential homologue, RNase E. Mol. Microbiol. 49: 607–622. [DOI] [PubMed] [Google Scholar]
- Richards J., Luciano D. J., Belasco J. G., 2012. Influence of translation on RppH-dependent mRNA degradation in Escherichia coli. Mol. Microbiol. 86: 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selick H. E., Kreuzer K. N., Alberts B. M., 1988. The bacteriophage T4 insertion/substitution vector system. J. Biol. Chem. 263: 11336–11347. [PubMed] [Google Scholar]
- Singh D., Chang S. J., Lin P. H., Averina O. V., Kaberdin V. R., et al. , 2009. Regulation of ribonuclease E activity by L4 ribosomal protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 106: 864–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-C., Du D., Domínguez-Malfavón L., Dimastrogiovanni D., Cross J., et al. , 2012. Recognition of the 70S ribosome and polysome by the RNA degradosome in Escherichia coli. Nucleic Acids Res. 40: 10417–10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H., Yonesaki T., 2004. Phage-induced change in the stability of mRNAs. Virology 329: 134–141. [DOI] [PubMed] [Google Scholar]
- Wachi M., Umitsuki G., Nagai L., 1997. Functional relationship between Escherichia coli RNase E and the CafA protein. Mol. Gen. Genet. 253: 515–519. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Young K. K. Y., Edlin G. J., Konigsberg W., 1979. High-frequency generalized transduction by bacteriophage T4. Nature 280: 80–82. [DOI] [PubMed] [Google Scholar]
- Yeom J. H., Go H., Shin E., Kim H. L., Han S. H., et al. , 2008a Inhibitory effects of RraA and RraB on RNase E-related enzymes imply conserved functions in the regulated enzymatic cleavage of RNA. FEMS Microbiol. Lett. 285: 10–15. [DOI] [PubMed] [Google Scholar]
- Yeom J. H., Shin E., Go H., Sim S. H., Seong M. J., et al. , 2008b Functional implications of the conserved action of regulators of ribonuclease activity. J. Microbiol. Biotechnol. 18: 1353–1356. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S1 contains Table S1 and Figures S1, S2, S3, S4, S5, and S6.